Abstract
Background
High-grade anal intraepithelial neoplasia (HGAIN; AIN2–3) is highly prevalent in HIV+ men, but only a minority of these lesions progress towards cancer. Currently, cancer progression risk cannot be established; therefore, no consensus exists on whether HGAIN should be treated. This study aimed to validate previously identified host cell DNA methylation markers for detection and cancer risk stratification of HGAIN.
Methods
A large independent cross-sectional series of 345 anal cancer, AIN3, AIN2, AIN1, and normal control biopsies of HIV+ men was tested for DNA methylation of 6 genes using quantitative methylation-specific PCR. We determined accuracy for detection of AIN3 and cancer (AIN3+) by univariable and multivariable logistic regression analysis, followed by leave-one-out cross-validation. Methylation levels were assessed in a series of 10 anal cancer cases with preceding HGAIN at similar anatomic locations, and compared with the cross-sectional series.
Results
Methylation levels of all genes increased with increasing severity of disease (P < .05). HGAIN revealed a heterogeneous methylation pattern, with a subset resembling cancer. ZNF582 showed highest accuracy (AUC = 0.88) for AIN3+ detection, slightly improved by addition of ASCL1 and SST (AUC = 0.89), forming a marker panel. In the longitudinal series, HGAIN preceding cancer displayed high methylation levels similar to cancers.
Conclusions
We validated the accuracy of 5 methylation markers for the detection of anal (pre-) cancer. High methylation levels in HGAIN were associated with progression to cancer. These markers provide a promising tool to identify HGAIN in need of treatment, preventing overtreatment of HGAIN with a low cancer progression risk.
Keywords: anal intraepithelial neoplasia, anal cancer, HIV, human papillomavirus, host cell DNA methylation markers
Independent validation of host cell DNA methylation markers associated with anal carcinogenesis demonstrates their significant prognostic value for treatment decision making of high-grade anal intraepithelial neoplasia. This is further emphasised by a longitudinal analysis-based association with progression to anal cancer.
(See the Editorial Commentary by Clifford and Alberts on pages 2164–6.)
Anal cancer is an increasing problem, especially in high-risk groups. Human immunodeficiency virus–positive (HIV+) men who have sex with men (MSM) have the highest incidence rates and a 78-fold higher risk of developing anal cancer compared with the general population [1, 2]. Anal cancer mainly comprises squamous cell carcinoma (SCC) and is caused by a persistent human papillomavirus (HPV) infection [3]. Similar to cervical carcinogenesis, anal SCC is preceded by precursor lesions: anal intraepithelial neoplasia (AIN; graded 1–3), which can develop perianally and in the anal canal [4]. High-grade AIN (HGAIN; AIN2–3), also called anal high-grade squamous intraepithelial lesions, are highly prevalent (23–35%) in HIV+ MSM [1, 5], for which screening using high-resolution anoscopy (HRA)–guided biopsies is the gold standard [6, 7]. However, only a minority of HGAIN eventually progresses to cancer [1, 8]. Since we are unable to determine the risk of progression, there are no consensus guidelines on screening or whether HGAIN should be treated. Current clinical practice in some countries of treating all HGAIN leads to considerable overtreatment of lesions that have a low risk of progression towards cancer. If we can better identify HGAIN with a high risk of progression towards cancer, we might be able to improve the effectiveness of anal cancer screening and treatment of HGAIN.
Host cell DNA methylation (ie, addition of methyl groups to cytosines in CpG-sites [hypermethylation]) is an epigenetic hallmark in HPV-induced carcinogenesis that can lead to inactivation of tumor suppressor genes [9]. Previously, we showed that host cell DNA methylation is associated with anal carcinogenesis and we identified several methylation markers for the detection of AIN3 and anal cancer [10]. These markers showed high methylation levels in anal SCC and a heterogeneous methylation pattern in HGAIN, displaying either high (cancer-like) or low methylation levels [10]. In analogy to our findings in the cervix, we hypothesized that precursor lesions with a cancer-like methylation pattern have a higher risk of progression towards cancer [11–13]. Providing further evidence of an association between high methylation levels and a high risk of progression would be an important step towards developing a risk-stratification tool.
The current study aimed to (1) validate the accuracy of previously identified methylation markers in detecting anal cancer and HGAIN in a large, independent cross-sectional series of tissue samples of HIV+ men and (2) to determine their association with progression to cancer in an international, longitudinal series of anal cancer cases with documented preceding HGAIN. In addition to the 5 best-performing markers identified in our previous study (ASCL1, SST, WDR17, ZIC1, and ZNF582) [10], we also tested LHX8 on anal samples, a recently discovered and well-performing marker in the cervix [14].
METHODS
Clinical Specimens and Ethics
This study involved a molecular analysis of a cross-sectional and a longitudinal series of formalin-fixed paraffin-embedded anal tissue samples.
The cross-sectional series, used for validation purposes, consisted of 345 samples, all from HIV+ men (Table 1). Anal SCC specimens (n = 30; including 2 verrucous carcinomas), obtained between 1999 and 2018, were retrieved from the pathology archives of the Amsterdam University Medical Centers (Amsterdam UMC) and 5 other hospitals in the Netherlands (see Acknowledgments). A total of 209 AIN biopsies (AIN1, n = 37; AIN2, n = 98; AIN3, n = 74), obtained between 2008 and 2017 in 205 men, were retrieved from the Amsterdam UMC pathology archive. For 4 patients, 2 biopsies from different lesions and histological AIN grades were included. Normal control samples (n = 106) consisted of biopsies with normal or reactive anal epithelium. These biopsies were taken from nonsuspected anal epithelium in 106 HIV+ men who also had biopsies taken from suspected lesions during screening for anal (pre-) cancer at the Amsterdam UMC between 2016 and 2018 as described before [10].
Table 1.
Tissue Samples, All From Human Immunodeficiency Virus-positive Men, Used in the Cross-sectional Series, Including Anatomic Location
| Histological Category | Total No. of Samples | Median Age, years | Anatomic Location, No. of Samples | ||
|---|---|---|---|---|---|
| Anal Canal | Perianal | Not Provided | |||
| Normal | 106 | 53 | 105 | 1 | 0 |
| AIN1 | 37 | 47 | 27 | 10 | 0 |
| AIN2 | 98 | 48 | 91 | 7 | 0 |
| AIN3 | 74 | 48 | 64 | 10 | 0 |
| SCC | 30 | 52 | 4 | 11 | 15 |
| Total | 345 | 50 | 291 | 39 | 15 |
Abbreviations: AIN1–AIN3, anal intraepithelial neoplasia (grades 1–3); Normal, normal control samples; SCC, anal squamous cell carcinoma.
The longitudinal series consisted of 40 biopsies of 10 patients (8 HIV+ men, 1 HIV-negative [HIV−] woman, 1 HIV− man) who developed (suspected) anal SCC over time. Each case comprised multiple consecutive biopsies taken from the same anatomic location, including 1 or more biopsies of histopathologically confirmed SCC or “highly suspicious for infiltrative growth” (endpoint) and all suitable and available biopsies preceding the endpoint diagnosis. To determine the corresponding anatomic location of the consecutive biopsies, medical records including HRA photo documentation and HPV genotyping results were reviewed. The biopsies, obtained between 2009 and 2019, were retrospectively identified and retrieved from the pathology archives of the Amsterdam UMC, and Onze Lieve Vrouwe Gasthuis, Amsterdam, the Netherlands; Homerton University Hospital, London, United Kingdom; and Helios St Elisabeth Hospital, Oberhausen, Germany.
This study followed the ethical guidelines of the Institutional Review Board of the Amsterdam UMC. Ethical approval was granted under reference number 05/031 (normal control samples) and 07/318 (AIN biopsies taken in the course of a triple-arm trial on AIN treatment) [15]. We adhered to the Code of Conduct for Responsible Use of Left-over Material of the Dutch Federation of Biomedical Scientific Societies and ethical approval was waived for use of archived biopsies (reference no. 17/151 [SCC], 18/341 [AIN biopsies], and 17/234 [longitudinal series]). For the longitudinal series, local ethical approval was granted by the NHS Health Research Authority, United Kingdom (IRAS ID 226196), and the Ethical Committee of the University Witten/Herdecke, Germany (reference no. 166/2017).
Histopathological Review, Human Papillomavirus Genotyping, and DNA Methylation Analysis
Histopathological review, HPV genotyping, and DNA methylation analysis were performed as described before (see Supplementary Methods) [10]. In short, a board-certified pathologist (C. J. M. v. N.) confirmed histopathological classification of all samples and p16INK4A immunohistochemistry was used if indicated. Bisulfite-converted sample DNA was analyzed for 6 methylation markers (ASCL1, LHX8, SST, WDR17, ZIC1, and ZNF582) [10, 14] using 2 multiplex quantitative methylation-specific polymerase chain reaction (qMSP) assays, each targeting 3 genes and the reference gene, β-actin (ACTB) [16, 17].
Statistical Analysis
Differences in methylation levels across the different histological categories (normal, AIN1–3, SCC) were visualized using boxplots and tested for statistical significance using the Kruskal-Wallis test, followed by pairwise Mann-Whitney U tests, with Bonferroni correction. Differences in methylation levels for anal canal versus perianal biopsies within histological categories were assessed using the Mann-Whitney U test and differences in distributions of HPV16 versus non-HPV16 samples between positive and negative methylation results were assessed using the Fisher’s exact test.
Evaluation of the diagnostic performance of the markers was performed as described before (see Supplementary Methods) [10]. In short, for the individual markers, we performed univariable (simple) logistic regression, while multivariable (multiple) logistic regression with forward and backward selection was used to obtain a marker panel. These analyses were performed on ≤AIN1 (normal control samples and AIN1) versus AIN3+ (AIN3 and anal SCC), which also allowed for comparison with our previous study. As additional confirmation, “normal” was compared with “anal SCC.” Performance of the models was visualized using receiver operating characteristic (ROC) curves and assessed through the area under the curve (AUC), as well as through the sensitivity and specificity at the Youden’s index (J) threshold (threshold that maximizes the sum of sensitivity and specificity). To evaluate the predictive performance of our models on samples outside the set, we performed leave-one-out cross-validation (LOOCV).
For the longitudinal series, methylation results were obtained by applying the multivariable logistic regression model fitted on the cross-sectional series and using the corresponding J-threshold.
In the AIN3+ analysis, the J-threshold is optimized for detecting SCC as well as all AIN3. For the purpose of trying to optimize identification of HGAINs that are likely to have a high risk of progression in the cross-sectional series, detection thresholds were adjusted to achieve maximum specificity. The adjustment was based on methylation levels of the (suspected) cancers and HGAIN in the longitudinal series, making sure none of these lesions were misclassified.
Statistical analyses were performed using R statistical software (version 3.5.1; R Foundation for Statistical Computing, Austria) with packages pROC and ggplot2 and IBM SPSS statistics software (version 24; IBM Corporation). Reported P values are 2-sided, with .05 as the significance threshold.
RESULTS
Human Papillomavirus Genotyping of Tissue Samples
All 209 AIN1–3 biopsies, 97% (29/30) of SCC, and 63% (67/106) of normal control samples were HPV positive (Supplementary Table 1). Of these HPV-positive biopsies, high-risk HPV (hrHPV) types [18, 19] were found in 30% of AIN1, 79% of AIN2, 89% of AIN3, and 90% of SCC. In HPV-positive normal control samples, 64% showed hrHPV positivity. Multiple HPV types were found in 37% of HPV-positive samples. HPV16 was the most predominant type, found in 39% of AIN2, 46% of AIN3, and 72% of HPV-positive anal SCC, and was the most frequent HPV type among the single infections.
Methylation Levels Across Histological Categories of Anal Disease
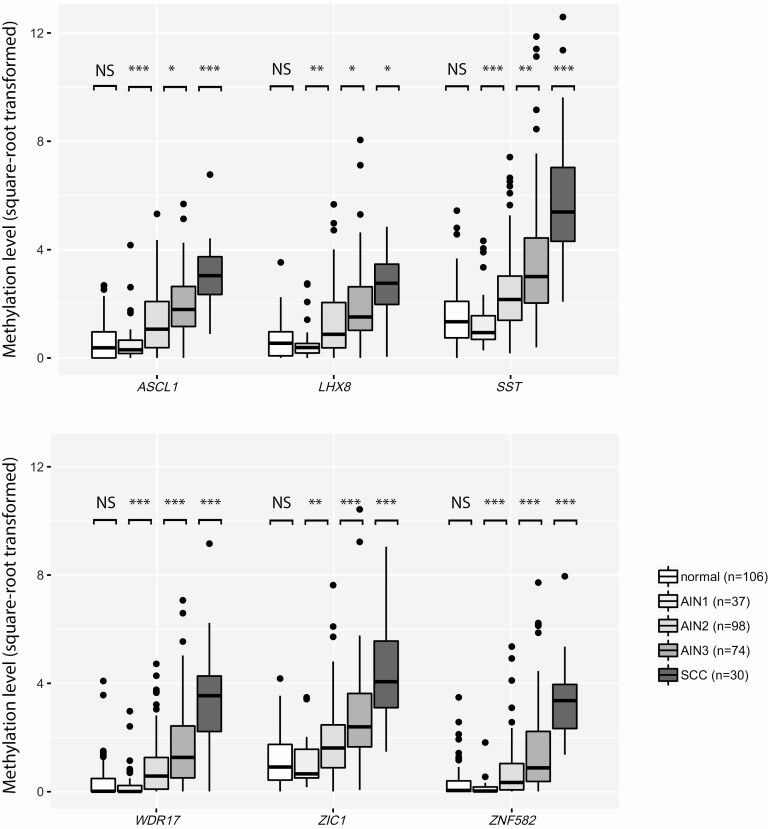
In the cross-sectional series, methylation levels of all markers differed significantly between histological categories (P < 8 × 10−19). For all consecutive AIN grades (AIN1–3) and SCC, methylation levels increased significantly with increasing severity of disease (P < .05) (Figure 1). Methylation levels did not significantly differ between anal canal and perianal biopsies in any histological category for any marker (P > .07) (Supplementary Table 2), although the proportion of perianal biopsies was generally low.
Figure 1.
Methylation levels increased with severity of anal disease. DNA methylation levels relative to a reference gene β-actin (square-root transformed ΔΔCq ratios; y axis) in the different histological categories of anal tissue samples of HIV-positive men (x axis) for 6 markers: ASCL1, LHX8, SST, WDR17, ZIC1, ZNF582. Differences between histological categories upon Kruskal-Wallis omnibus test, followed by post hoc testing using the Mann-Whitney U test and Bonferroni multiple testing correction: *P < .05, **P < .01, ***P < .001. ● outlier sample. Abbreviations: AIN1–AIN3, anal intraepithelial neoplasia (grades 1–3); HIV, human immunodeficiency virus; normal, normal control samples; NS, not significant; SCC, squamous cell carcinoma.
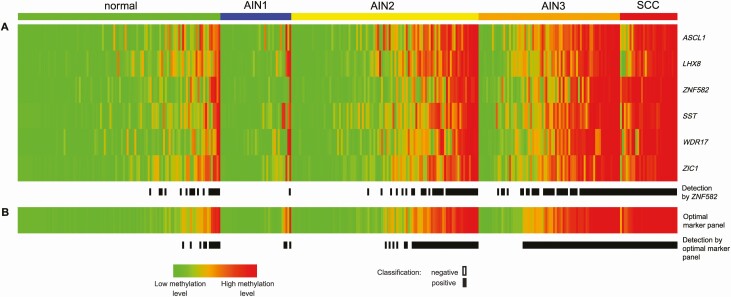
DNA methylation patterns of the 6 markers showed high methylation levels in all cancers and low methylation levels in the vast majority of normal control samples and AIN1 (Figure 2A). In AIN3, methylation levels varied greatly, exhibiting both low as well as high methylation levels similar to cancers. Whereas these biopsies were similar in histopathological grade, the methylation pattern proved to be heterogeneous. In AIN2, the same heterogeneous pattern was observed, although with a lower proportion of biopsies with cancer-like high methylation levels.
Figure 2.
A, DNA methylation pattern of the 6 individual methylation markers (rows: ASCL1, LHX8, SST, WDR17, ZIC1, ZNF582; based on LOOCV univariable regression for AIN3+ detection), per histological category in cross-sectional series. Methylation result per sample (column) in the different histological subgroups is displayed in color according to the PP from green (low methylation levels; PP of 0) to red (high methylation levels; PP of 1). In each group, samples are ordered based on their average predicted probability. Black boxes [detection by ZNF582] represent samples classified as methylation positive using ZNF582 alone (at the non-CV Youden index threshold ≥0.35). B, DNA methylation pattern of the optimal marker panel (ASCL1, SST, ZNF582; based on LOOCV multivariable regression for AIN3+ detection) and methylation positivity using the panel. Black boxes [detection by optimal marker panel] represent samples classified as methylation positive using the optimal marker panel (at the non-CV Youden index threshold ≥0.43). Endpoint: AIN3+ (AIN3 and anal SCC). Abbreviations: AIN1–AIN3, anal intraepithelial neoplasia (grades 1–3); LOOCV, leave-one-out cross-validated; non-CV, non–cross-validated; normal, normal control samples; PP, predicted probability; SCC, squamous cell carcinoma.
Diagnostic Performance of the Individual Methylation Markers
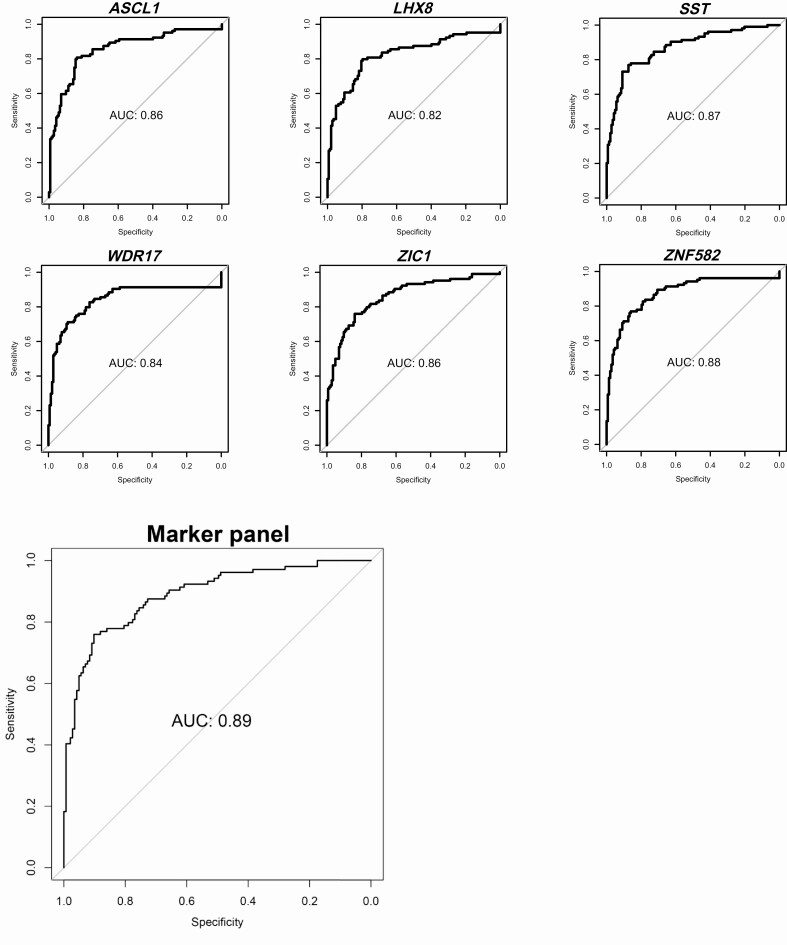
In the logistic regression analysis for AIN3+ detection (cases vs controls: AIN3+ [AIN3 and anal SCC; n = 104] vs ≤AIN1 [normal control samples and AIN1; n = 143]), all markers proved able to significantly distinguish cases from controls (P < .001). AUCs ranged from 0.84 to 0.89 (Table 2), with the highest AUC achieved by ZNF582 (AUC = 0.89), followed by SST (AUC = 0.88) and WDR17 (AUC = 0.87). ZNF582 was the only marker that classified all (30/30) cancers as methylation positive at the J-threshold (≥0.35), corresponding to an AIN3+ sensitivity and specificity of 76% and 87%, respectively. Using this threshold, ZNF582 classified 66% (49/74) of AIN3, 36% (35/98) of AIN2, 3% (1/37) of AIN1, and 16% (17/106) of normal control samples as methylation positive (Figure 2A: [Detection by ZNF582], black indicators). Upon leave-one-out cross-validation, we obtained AUC values from 0.82 to 0.88, with ZNF582 again achieving the highest AUC (AUC = 0.88) (Figure 3; Table 2) for AIN3+ detection. ZNF582 remained the only marker classifying all cancers as methylation positive at the J-threshold (≥0.32), corresponding to a sensitivity and specificity of 77% and 86%, respectively.
Table 2.
Logistic Regression Analysis on Diagnostic Performance for AIN3+ Detection: Univariable Regression of the 6 Individual Markers (ASCL1, LHX8, SST, WDR17, ZIC1, ZNF582)
| Methylation Marker | ASCL1 | LHX8 | SST | WDR17 | ZIC1 | ZNF582 |
|---|---|---|---|---|---|---|
| AUC (non-CV) (95% CI) | .87 (.82–.91) | .84 (.78–.89) | .88 (.83–.92) | .87 (.82–.91) | .87 (.82–.91) | .89 (.85–.93) |
| Sensitivity, % | 81 | 80 | 78 | 71 | 76 | 76 |
| Specificity, % | 85 | 80 | 87 | 90 | 84 | 87 |
| Missed SCC | 2 | 3 | 1 | 2 | 1 | 0 |
| AUC (LOOCV) | .86 | .82 | .87 | .84 | .86 | .88 |
| Sensitivity, % | 81 | 80 | 77 | 71 | 76 | 77 |
| Specificity, % | 84 | 80 | 87 | 89 | 84 | 86 |
| Missed SCC | 2 | 3 | 1 | 2 | 1 | 0 |
Non–CV, including 95% CI and LOOCV AUCs, are reported. Sensitivity and specificity are for the Youden index threshold. Endpoint: AIN3+ (AIN3 and anal SCC) in anal tissue samples of HIV-positive men.
Abbreviations: AIN3, anal intraepithelial neoplasia (grade 3); AUC, area under the receiver operating characteristic curve; CI, confidence interval; HIV, human immunodeficiency virus; LOOCV, leave-one-out cross-validated; nonCV, non–cross-validated; SCC, anal squamous cell carcinoma.
Figure 3.
Diagnostic performance visualized with ROC curves of the logistic regression analysis for AIN3+ detection: univariable regression (LOOCV) for the 6 individual methylation markers (ASCL1, LHX8, SST, WDR17, ZIC1, ZNF582) and multivariable regression (LOOCV) for the optimal marker panel (ASCL1, SST, ZNF582). Endpoint: AIN3+ (AIN3 and anal SCC). Abbreviations: AIN2–3, anal intraepithelial neoplasia (grade 2–3); AUC, area under the ROC curve; LOOCV, leave-one-out cross-validated; ROC, receiver operating characteristic.
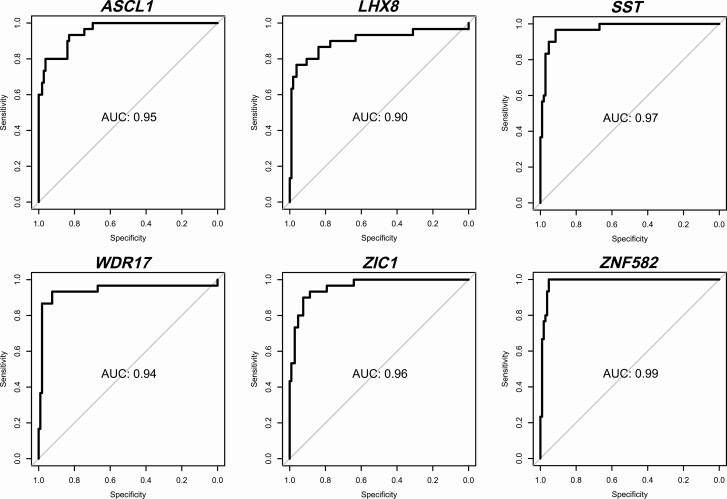
In the analysis for anal SCC detection (cases vs controls: anal SCC [n = 30] vs normal control samples [n = 106]), similar results were obtained (Figure 4; Table 3). ZNF582 attained the highest AUC (AUC = 0.99), with AUCs for the other markers ranging from 0.91 to 0.98. At the J-threshold (≥0.17) this resulted in a sensitivity of 100% and specificity of 95%. At this threshold, ZNF582 was the only marker able to classify all cancers as methylation positive, along with 36% (27/74) of AIN3, 19% (19/98) of AIN2, 3% (1/37) of AIN1, and 5% (5/106) of normal control samples. After cross-validation, AUCs ranged from 0.90 to 0.99 with ZNF582 performing best, providing a sensitivity of 100% and specificity of 95% at the J-threshold (≥0.16).
Figure 4.
Diagnostic performance visualized with ROC curves of the univariable regression analysis (LOOCV) for anal SCC detection (normal vs SCC) for the 6 individual methylation markers (ASCL1, LHX8, SST, WDR17, ZIC1, ZNF582). Endpoint: anal SCC. Abbreviations: AUC, area under the ROC curve; LOOCV, leave-one-out cross-validated; ROC, receiver operating characteristic; SCC, squamous cell carcinoma.
Table 3.
Univariable Logistic Regression Analysis on Diagnostic Performance for Anal Squamous Cell Carcinoma (SCC) Detection (Normal vs SCC) of the 6 Individual Markers (ASCL1, LHX8, SST, WDR17, ZIC1, ZNF582)
| Methylation Marker | ASCL1 | LHX8 | SST | WDR17 | ZIC1 | ZNF582 |
|---|---|---|---|---|---|---|
| AUC (non-CV) (95% CI) | .96 (.93–.99) | .91 (.85–.98) | .98 (.95–1.00) | .95 (.90–1.00) | .96 (.94–.99) | .99 (.97–1.00) |
| Sensitivity, % | 93 | 77 | 97 | 93 | 93 | 100 |
| Specificity, % | 83 | 96 | 92 | 93 | 90 | 95 |
| Missed SCC | 2 | 7 | 1 | 2 | 2 | 0 |
| AUC (LOOCV) | .95 | .90 | .97 | .94 | .96 | .99 |
| Sensitivity, % | 93 | 77 | 97 | 93 | 90 | 100 |
| Specificity, % | 83 | 96 | 92 | 92 | 92 | 95 |
| Missed SCC | 2 | 7 | 1 | 2 | 3 | 0 |
Non–CV, including 95% CI and LOOCV AUCs, are reported. Sensitivity and specificity are for the Youden index threshold. Endpoint: anal SCC in anal tissue samples of HIV-positive men.
Abbreviations: AUC, area under the receiver operating characteristic curve; CI, confidence interval; HIV, human immunodeficiency virus; LOOCV, leave-one-out cross-validated; non-CV, non–cross-validated.
Identification and Diagnostic Performance of a Marker Panel
In the multivariable logistic regression, both forward and backward selection for AIN3+ detection yielded the same model consisting of ASCL1, SST, and ZNF582 as the optimal marker panel, with an AUC of 0.90 (Figure 3; Table 4). Using the J-threshold (≥0.43), this panel provided a sensitivity of 78% and specificity of 90%. At this threshold, all cancers (30/30) were classified as methylation positive, 69% (51/74) of AIN3, 42% (41/98) of AIN2, 8% (3/37) of AIN1, and 10% (11/106) of normal control samples (Figure 2B: [Detection by optimal marker panel], black indicators). In the methylation-positive AIN3, significantly more cases were HPV16 positive than non–HPV16 positive as compared with methylation-negative AIN3 (P = .0001). Upon cross-validation at the J-threshold (≥0.44), we found an AUC of 0.89 with a sensitivity of 76% and specificity of 90% (Table 4), again with all cancers being classified as methylation positive. For anal SCC detection, multivariable logistic regression was not possible due to the relatively small number of SCC cases.
Table 4.
Logistic Regression Analysis on Diagnostic Performance for AIN3+ Detection: Multivariable Regression for Optimal Marker Panel (ZNF582, ASCL1, SST)
| Marker Panel | Non-CV | LOOCV |
|---|---|---|
| AUC (95% CI) | .90 (.86–.94) | .89 |
| Sensitivity, % | 78 | 76 |
| Specificity, % | 90 | 90 |
| Missed SCC | 0 | 0 |
Non–CV, including 95% CI and LOOCV AUCs, are reported. Sensitivity and specificity are for the Youden index threshold. Endpoint: AIN3+ (AIN3 and anal SCC) in anal tissue samples of HIV-positive men.
Abbreviations: AIN3, anal intraepithelial neoplasia (grade 3); AUC, area under the receiver operating characteristic curve; CI, confidence interval; HIV, human immunodeficiency virus; LOOCV, leave-one-out cross-validated; non-CV, non–cross-validated; SCC, anal squamous cell carcinoma.
Association of Methylation Positivity With Progression to Cancer
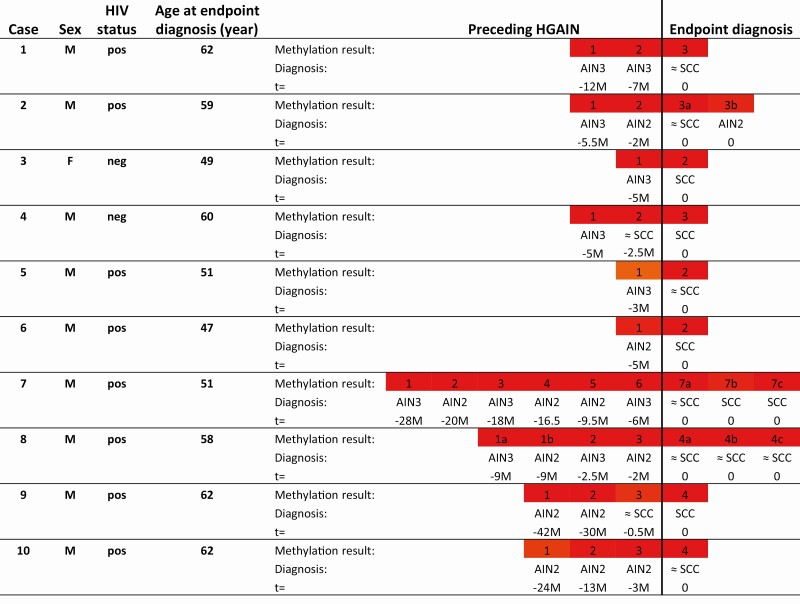
In the longitudinal series of 10 cases with (suspected) anal cancer and preceding HGAIN, all HPV types detected in (suspected) cancers were also found in the corresponding preceding HGAIN biopsies (Supplementary Table 3). In all cases, all HGAIN biopsies preceding the biopsies with endpoint diagnosis (suspected) SCC consistently showed high methylation levels, comparable to the highest levels in AIN3 and cancers in the cross-sectional series, even when taken several years before endpoint diagnosis (Figure 5). In addition, all of these biopsies were scored as methylation positive using the aforementioned 3-gene marker panel (ASCL1, SST, and ZNF582) at the J-threshold (Supplementary Table 3), as well as using ZNF582 alone (data not shown).
Figure 5.
DNA methylation pattern of longitudinal series per case (row). Numbers represent biopsy number corresponding to Supplementary Table 3 divided into Endpoint diagnosis and Preceding HGAIN. Endpoint diagnosis is SCC or suspected SCC: lesion with high suspicion for infiltrative growth (≈ SCC). In some cases, multiple biopsies were taken at the same time point, indicated with a letter (eg, 1a, 1b, etc). Methylation result per sample is displayed in color (similarly to the cross-sectional series; Figure 2) according to their PP from green (low methylation levels; PP of 0 [not applicable in longitudinal series]) to red (high methylation levels; PP of 1) at the non-CV marker panel Youden index threshold (≥0.43) for AIN3+ detection. All endpoint diagnosis samples and preceding HGAIN were classified as methylation positive. AIN1–AIN3, anal intraepithelial neoplasia (grades 1–3); F, female; HGAIN, high-grade anal intraepithelial neoplasia; HIV, human immunodeficiency virus; M, male; neg, negative; non-CV, non–cross-validated; pos, positive; PP, predicted probability; SCC, squamous cell carcinoma; t, time in months (M) before endpoint diagnosis.
Defining Detection Threshold Boundaries for Clinical Application
For the purpose of optimizing identification of HGAINs that are likely to have a high risk of progression to cancer, the detection threshold of the 3-gene marker panel could be adjusted to a maximum of ≥0.52, at which point all cancers and preceding HGAINs in the longitudinal series were scored as methylation positive, without missing any cancers in the cross-sectional series. At this threshold, 61% (45/74) of AIN3, 37% (36/98) of AIN2, 8% (3/37) of AIN1, and 8% (9/106) of normal control samples scored methylation positive.
DISCUSSION
The most important outcomes of this study were the validation of the accuracy of 5 host cell DNA methylation markers from our previous study [10] for the detection and cancer risk stratification of HGAIN in HIV+ men, and the finding that high methylation levels in precursor lesions are indeed associated with progression to cancer.
Using qMSP, we found that the diagnostic performance for AIN3+ detection was good and robust for all markers, as it not only held up after cross-validation (AUC = 0.82–0.88) but duplicated the performance of the same markers in our previous smaller study (AUC = 0.82–0.89), in which we analyzed 148 tissue samples [10]. Furthermore, we confirmed ZNF582 to be the most potent marker. The addition of genes ASCL1 and SST to ZNF582, forming a marker panel, slightly improved the performance for AIN3+ detection (AUC = 0.89) over ZNF582 alone. Importantly, both the marker panel as well as ZNF582 alone did not miss any cancers. LHX8, the newly tested marker, showed a similarly good performance as the markers in the previous study but did not have additive value. We acknowledge that omitting AIN2 in our analyses implicates an overestimation of specificity.
Consistent with our previous study, we established that histopathologically similar HGAIN samples display a heterogeneous methylation pattern, with both high (cancer-like) and low methylation levels. We hypothesized that the precursor lesions displaying high methylation levels were those having a higher risk of progression towards cancer. In this study, using a longitudinal series of biopsies from patients who developed (suspected) anal cancer over time, we were able to demonstrate that HGAIN preceding cancer consistently displayed methylation levels similar to cancers, even when biopsies were taken up to several years before cancer diagnosis. This supports our assumption that high methylation levels in HGAIN are associated with progression to cancer.
Hypermethylation of tumor suppressor genes appears to be an early event in HPV-induced carcinogenesis, with hypermethylation occurring up to at least 3.5 years before cancer diagnosis in one of our cases (Figure 5; Supplementary Table 3), making for an ideal screening tool [9, 20]. In comparable HPV-induced cervical (pre-) cancers, similarly high cancer-like methylation levels were found in a subset of precursor lesions of known high short-term cancer risk, comprising cervical intraepithelial neoplasia grade 2 or 3 with a persistent hrHPV infection (≥5 years) and/or a chromosome 3q gain [9, 11, 20–22].
For clinical application and resolving current overtreatment, the methylation test should ideally identify all HGAIN lesions with a high risk of progression towards cancer, while ignoring HGAIN lesions with a low risk of progression and not missing any cancers. Given the quantitative nature of the methylation test, thresholds can be adjusted to the clinical need. We provided a threshold for the methylation marker panel with maximum specificity, by which the panel identified all cancers in both the cross-sectional and in the longitudinal series, as well as all HGAIN lesions preceding cancer in the longitudinal series. With this threshold, 61% of AIN3 and 37% of AIN2 are identified as methylation positive. Currently, in some countries, treatment of all AIN2–3 lesions is recommended. Using this marker panel as a treatment decision-making tool and treating only the methylation-positive AIN2–3, treatment could be withheld in up to 39% of AIN3 and 63% of AIN2 cases. For future studies, determining the optimal detection threshold for clinical decision making with regard to treatment of HGAIN lesions is vital, providing long-term assurance that HGAIN lesions with negative methylation results will not progress to cancer and thus treatment for these lesions can be safely withheld.
We recognize that retrospective selection of HGAIN preceding anal cancer for the longitudinal series might render this series prone to error. Although anatomic locations of HGAIN preceding cancer were thoroughly reviewed and HPV genotypes matched, we cannot rule out that some of these lesions might not have been the exact same lesion, particularly in cases with long intervals between biopsies, such as in case 9 (Figure 5; Supplementary Table 3). Moreover, in cases with short intervals between HGAIN and endpoint diagnosis we cannot rule out that the preceding biopsies might already have been cancers that were misdiagnosed as HGAIN, rather than reflecting actual progression towards cancer. However, this mimics real clinical practice with regard to the difficulty in diagnosing anal cancer. In any case, our findings reinforce the idea that hypermethylation is an early event in anal carcinogenesis and emphasizes the need for clinical decision making based on more objective and reproducible biomarkers as compared with histopathology alone.
While several studies on DNA methylation in anal carcinogenesis have been published [23–29], we are the first to validate methylation markers in a large, independent cross-sectional series, as well as the first to provide evidence that high methylation levels are associated with progression to cancer in a longitudinal series.
In conclusion, we validated the accuracy of 5 host cell DNA methylation markers for the detection of anal (pre-) cancer in HIV+ men and established an optimal marker panel consisting of the genes ASCL1, SST, and ZNF582. High methylation levels in HGAIN were associated with progression to cancer. Therefore, these markers may provide a promising tool to identify which HGAIN lesions are in need of treatment, preventing overtreatment of HGAIN that has a low cancer progression risk. Prospective clinical studies are now warranted to assess the safety and efficacy of treatment decision making for HGAIN based on methylation analysis.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. R. D. M. S., J. M. P., and H. J. C. d. V. are principal investigators of the study. R. P. v. d. Z., O. R., H. J. C. d. V., J. M. P., and R. D. M. S. designed the study. R. P. v. d. Z., O. R., C. J. M. v. N., W. G. V. Q., H. J. C. d. V., J. M. P., and R. D. M. S. were involved in providing and collecting clinical material and data collection. M. N., T. C., M. S., and A. K. provided clinical data and samples for the longitudinal series. R. P. v. d. Z., A. P. v. S., and T. J. t. B. performed the laboratory work. C. J. L. M. M. and R. D. M. S. developed the qMSP assays used in this study. R. P. v. d. Z. and R. D. M. S. performed the data analysis. R. P. v. d. Z. and I. C.-T. performed the statistical analysis. R. P. v. d. Z. managed the database. R. P. v. d. Z., J. M. P., and R. D. M. S. drafted the manuscript. All authors critically reviewed the manuscript and approved the final version. All authors had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis and believe that the manuscript represents honest work. R. D. M. S. affirms that the manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; that any discrepancies from the study as planned have been explained; and had full responsibility for the decision to submit for publication.
Acknowledgments. The authors thank all participants for donating normal control biopsies; E. J. Kuyvenhoven, H. E. Nobel, K. C. M. Gosens, and M. L. Siegenbeek van Heukelom for collecting normal control biopsies; D. Gochez, W. Verlaat, and H. van den Munckhof for excellent technical advice and assistance; A. Leeman and M. E. van der Ende (Erasmus University Medical Center, Rotterdam), A. I. M. Hoepelman (University Medical Center Utrecht, Utrecht), L. B. S. Gelinck (Haaglanden Medical Center, Den Haag), W. F. W. Bierman (University Medical Center Groningen, Groningen), I. Cairo (Onze Lieve Vrouwe Gasthuis, Amsterdam), and S. M. E. Vrouenraets (Medical Center Slotervaart, Amsterdam) for providing archival tissue samples; and A. J. King (Dutch National Institute for Public Health and the Environment) for human papillomavirus (HPV) genotyping.
Disclaimer. The source of funding did not have any influence on the design of the study, collection, analysis, interpretation of the data, in writing the manuscript, or in the decision to submit the article for publication.
Financial support. This work was supported by the Aidsfonds (Dutch Aids Foundation) (High-Risk High-Gain call 2016; grant number P-26606) and the KWF Kankerbestrijding (Dutch Cancer Society) (grant number 2016-10781).
Potential conflicts of interest. R. D. M. S. and C. J. L. M. M. are minority stockholders of Self-screen B.V., a spin-off company of Vrije Universiteit University Medical Center, which owns patents on methylation markers and HPV detection. C. J. L. M. M. is part-time director of Self-screen B.V. since September 2017. He has received speakers’ fee from Glaxo Smith Kline (GSK), Qiagen, Sanofi Pasteur - Merck Sharp & Dohme (SPMSD)/Merck; served occasionally on the scientific advisory board (expert meeting) of GSK, Qiagen, SPMSD/Merck; has been coinvestigator on a SPMSD sponsored trial, of which his institute received research funding; has a very small number of Qiagen and MDxHealth shares; and was minority shareholder of Diassay B.V. until April 2016. H. J. C. d. V. received financial compensation or goods for research from Medigene, Gilead, and MSD; financial compensation for presentations from Abbott and Janssen; and financial compensation for advice to Medigene and Novartis. A. K. has received fees for lectures from MSD and InfectoPharm and has served as an advisory board member for Sanofi Pasteur, MSD, and AbbVie. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Machalek DA, Poynten M, Jin F, et al. Anal human papillomavirus infection and associated neoplastic lesions in men who have sex with men: a systematic review and meta-analysis. Lancet Oncol 2012; 13:487–500. [DOI] [PubMed] [Google Scholar]
- 2. Colón-López V, Shiels MS, Machin M, et al. Anal cancer risk among people with HIV infection in the United States. J Clin Oncol 2018; 36:68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hoots BE, Palefsky JM, Pimenta JM, Smith JS. Human papillomavirus type distribution in anal cancer and anal intraepithelial lesions. Int J Cancer 2009; 124:2375–83. [DOI] [PubMed] [Google Scholar]
- 4. Darragh TM, Winkler B. Anal cancer and cervical cancer screening: key differences. Cancer Cytopathol 2011; 119:5–19. [DOI] [PubMed] [Google Scholar]
- 5. Berry JM, Jay N, Cranston RD, et al. Progression of anal high-grade squamous intraepithelial lesions to invasive anal cancer among HIV-infected men who have sex with men. Int J Cancer 2014; 134:1147–55. [DOI] [PubMed] [Google Scholar]
- 6. Palefsky JM. Anal cancer prevention in HIV-positive men and women. Curr Opin Oncol 2009; 21:433–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schim van der Loeff MF, Mooij SH, Richel O, de Vries HJ, Prins JM. HPV and anal cancer in HIV-infected individuals: a review. Curr HIV/AIDS Rep 2014; 11:250–62. [DOI] [PubMed] [Google Scholar]
- 8. Lee GC, Kunitake H, Milch H, et al. What is the risk of anal carcinoma in patients with anal intraepithelial neoplasia III? Dis Colon Rectum 2018; 61:1350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Steenbergen RD, Snijders PJ, Heideman DA, Meijer CJ. Clinical implications of (epi)genetic changes in HPV-induced cervical precancerous lesions. Nat Rev Cancer 2014; 14:395–405. [DOI] [PubMed] [Google Scholar]
- 10. van der Zee RP, Richel O, van Noesel CJM, et al. Host cell deoxyribonucleic acid methylation markers for the detection of high-grade anal intraepithelial neoplasia and anal cancer. Clin Infect Dis 2019; 68:1110–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. De Strooper LM, Meijer CJ, Berkhof J, et al. Methylation analysis of the FAM19A4 gene in cervical scrapes is highly efficient in detecting cervical carcinomas and advanced CIN2/3 lesions. Cancer Prev Res (Phila) 2014; 7:1251–7. [DOI] [PubMed] [Google Scholar]
- 12. Verlaat W, Snijders PJF, Novianti PW, et al. Genome-wide DNA methylation profiling reveals methylation markers associated with 3q gain for detection of cervical precancer and cancer. Clin Cancer Res 2017; 23:3813–22. [DOI] [PubMed] [Google Scholar]
- 13. Bierkens M, Wilting SM, van Wieringen WN, et al. Chromosomal profiles of high-grade cervical intraepithelial neoplasia relate to duration of preceding high-risk human papillomavirus infection. Int J Cancer 2012; 131:E579–85. [DOI] [PubMed] [Google Scholar]
- 14. Verlaat W, Snoek BC, Heideman DAM, et al. Identification and validation of a 3-gene methylation classifier for HPV-based cervical screening on self-samples. Clin Cancer Res 2018; 24:3456–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Richel O, de Vries HJ, van Noesel CJ, Dijkgraaf MG, Prins JM. Comparison of imiquimod, topical fluorouracil, and electrocautery for the treatment of anal intraepithelial neoplasia in HIV-positive men who have sex with men: an open-label, randomised controlled trial. Lancet Oncol 2013; 14:346–53. [DOI] [PubMed] [Google Scholar]
- 16. Snellenberg S, De Strooper LM, Hesselink AT, et al. Development of a multiplex methylation-specific PCR as candidate triage test for women with an HPV-positive cervical scrape. BMC Cancer 2012; 12:551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008; 3:1101–8. [DOI] [PubMed] [Google Scholar]
- 18. Munoz N, Bosch FX, de Sanjose S, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med 2003; 348:518–27. [DOI] [PubMed] [Google Scholar]
- 19. Schiffman M, Clifford G, Buonaguro FM. Classification of weakly carcinogenic human papillomavirus types: addressing the limits of epidemiology at the borderline. Infect Agent Cancer 2009; 4:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Verlaat W, Van Leeuwen RW, Novianti PW, et al. Host-cell DNA methylation patterns during high-risk HPV-induced carcinogenesis reveal a heterogeneous nature of cervical pre-cancer. Epigenetics 2018; 13:769–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bierkens M, Hesselink AT, Meijer CJ, et al. CADM1 and MAL promoter methylation levels in hrHPV-positive cervical scrapes increase proportional to degree and duration of underlying cervical disease. Int J Cancer 2013; 133:1293–9. [DOI] [PubMed] [Google Scholar]
- 22. Wilting SM, Steenbergen RD, Tijssen M, et al. Chromosomal signatures of a subset of high-grade premalignant cervical lesions closely resemble invasive carcinomas. Cancer Res 2009; 69:647–55. [DOI] [PubMed] [Google Scholar]
- 23. Hernandez JM, Siegel EM, Riggs B, et al. DNA methylation profiling across the spectrum of HPV-associated anal squamous neoplasia. PLoS One 2012; 7:e50533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lorincz AT, Nathan M, Reuter C, et al. Methylation of HPV and a tumor suppressor gene reveals anal cancer and precursor lesions. Oncotarget 2017; 8:50510–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Siegel EM, Eschrich S, Winter K, et al. Epigenomic characterization of locally advanced anal cancer: a radiation therapy oncology group 98-11 specimen study. Dis Colon Rectum 2014; 57:941–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang J, Martins CR, Fansler ZB, et al. DNA methylation in anal intraepithelial lesions and anal squamous cell carcinoma. Clin Cancer Res 2005; 11: 6544–9. [DOI] [PubMed] [Google Scholar]
- 27. Molano M, Tabrizi SN, Garland SM, et al. ; SPANC Study Team . CpG methylation analysis of HPV16 in laser capture microdissected archival tissue and whole tissue sections from high grade anal squamous intraepithelial lesions: a potential disease biomarker. PLoS One 2016; 11:e0160673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wiley DJ, Huh J, Rao JY, et al. Methylation of human papillomavirus genomes in cells of anal epithelia of HIV-infected men. J Acquir Immune Defic Syndr 2005; 39:143–51. [PubMed] [Google Scholar]
- 29. Lahiri CD, Nguyen ML, Mehta CC, et al. Pilot study of markers for high-grade anal dysplasia in a southern cohort from the women’s interagency human immunodeficiency virus study. Clin Infect Dis 2020; 70:1121–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.