Abstract
The emergence of SARS-CoV-2 and its rapid spread globally emphasizes the ever-present threat of emerging and re-emerging infectious diseases. In this review, the pathogen pyramid framework was utilized to identify the “unknown unknowns” associated with the emergence and rapid transmission of novel infectious disease agents. Given that the evolutionary origin of most of the emerging infectious disease agents can be traced to an animal source, we argue the need to integrate the “One Health” approach as a part of surveillance activities. The need for focusing on undertaking global and regional mapping activities to identify novel pathogens is discussed, given that there are an estimated 1.67 million unknown viruses, of which around 631,000 to 827,000 unknown viruses have the capacity to infect human beings. The emerging risks due to the ever-expanding interface between human, animals, both domestic and wildlife, and the environment are highlighted, these are largely driven by the need for safe habitation, growing food, developing infrastructure to support the increasing human population and desire for economic growth. The One Health approach provides a holistic way to address these cross-sectoral issues, by bridging institutional gaps, enumerating priority risk areas and pathogens, and highlighting putative risk factors for subsequent spillover events involving emerging and re-emerging infectious disease pathogens at the human-animal-environment interface.
Keywords: COVID-19, emerging infectious diseases, One Health, pandemics, pathogen pyramid, surveillance
The default response to infectious disease threats has been the development of preventive or curative options post facto, often in the face of rising disease burden, human suffering, and costs. Though the value of timely identification of emerging infectious diseases has been well-established in the context of transmission containment, even with the most sensitive of surveillance systems, it is difficult to identify a novel pathogen1. The evolutionary advantage held by microbes enables them to adapt rapidly to host species, and eventually spill over. In the 25 families of viruses that can potentially infect human beings, there are an estimated 1.67 million unknown viruses, of which, an estimated 631,000 to 827,000 unknown viruses have the capacity to infect human beings2,3. Given this wide array of potential spillover threats, and the absence of a risk stratified global virus atlas, it becomes important to improve disease surveillance, especially at the interfaces with the highest risk of novel pathogen emergence.
The most recent cross-species spillover of a novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has assumed pandemic proportions, resulting in almost 2.5 million deaths from the novel coronavirus disease (COVID-19) globally by the end of February 20214. This is the third instance of the emergence of a novel coronavirus, after the severe acute respiratory syndrome (SARS) in 2003 and Middle East respiratory coronavirus (MERS CoV) in 20125,6,7. The consistency with which these novel pathogens have transcended species and international borders, while a cause of grave concern, is also an indicator of a complex interplay of various factors at the human-animal-environment interface, through which the pathogens continue to expand their host-tropism. While some efforts at using computational approaches have shown encouraging signs, accurate prediction models, which can predict the potential spillover threat, as well as its impact on human health, are yet to be developed8,9,10.Although SARS-CoV-1, MERS-CoV and SARS-CoV-2 all share similar phylogenetic roots, yet the considerable heterogeneity between the pathogen transmission patterns, disease severity and case fatality rates gives rise to varying magnitudes of global health threats. What makes SARS-CoV-2 a truly unique threat is its high transmissibility, and ability to cause adverse health outcomes (severe disease requiring hospitalization, and deaths), especially in vulnerable population groups, such as the elderly or those with co-morbidities, while simultaneously keeping overall case fatality rates low enough to enable further spread11,12,13,14,15,16,17.
Using the pathogen pyramid to explore emerging infectious diseases
These novel microbes are new entrants in a long emerging pattern. Between 1940 and 2004, 335 pathogens have emerged, with 60 per cent having a zoonotic source, of which 71 per cent originate from wildlife18. Prediction models have identified emerging infectious disease (EID) hotspots in Africa, Latin America and South Asia19. Most of these models take into account the current understanding of disease dynamics, and assumptions with respect to some of the “known unknown” determinants. The “unknown unknowns”, which form a large part of the problem at hand, remain in the blind spot. As the rapidly expanding pandemic of COVID-19 has adequately proved, this is an oversight one cannot afford.
One of the approaches used to identify the origins of zoonotic diseases utilizes the ecologic “pathogen pyramid”. This approach identifies the pathogens’ transitions from being an environment- or animal-specific agent into a human-infecting one20,21. This approach identifies intermediate levels of adaptation, through which zoonotic microbes evolve into efficient human pathogens over time (summarized in the Figure).
Figure.
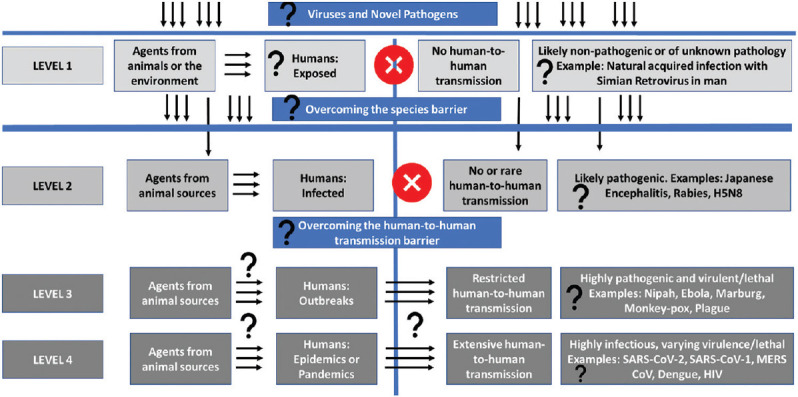
Using the “pathogen pyramid” schematic to identify the potential “unknown unknowns” in the emergence of novel infectious disease agents; Levels of “unknown unknowns” indicated by question marks.
At level 1 exposure, humans are exposed to a wide range of pathogens of environmental or animal origin, which may leave an immunologic trace, that are either unlikely, or not known to be disease causing pathogens. Naturally acquired infection with non-human simian retroviruses is a typical example22. Given the number of unknown viruses which exist at the human-animal-environment interface without crossing the species barrier effectively, or without adapting to a pathogenic role in human hosts, it is difficult to estimate the number and types of viruses which may be included in this level22.
Level 2 reflects the stage of infection, where zoonotic viruses can overcome the species barrier, invade human cells, and cause disease. However, the risk of human-to-human transmission, while theoretically possible, remains very low, as is seen in the case of diseases like Japanese encephalitis or rabies23,24,25,26. Another example is the influenza A(H5N8) virus, which is not known to exhibit human-to-human transmission, and does not show efficient transmission between ferrets, the animal model of choice for human influenza infections in human beings27,28,29,30,31.
At level 3, in addition to causing human infections, the zoonotic viral pathogens can cross the human-to-human transmission barrier and can establish limited transmission chains, resulting in outbreaks, which flare up and then eventually die down. This is the case for diseases like the plague, Nipah, Ebola or Marburg virus diseases, which are rapidly fatal.
At level 4, the pathogen can establish efficient human-to-human transmission chains, which can result in disease burden of epidemic or pandemic proportions, as has been observed for the ongoing COVID-19 pandemic, its predecessors MERS-CoV, SARS, or influenza. HIV can also be considered to be a pathogen at this level of the pathogen pyramid32,33,34. Levels 3 and 4 provide opportunities for interventions to disrupt transmission chains of emerging infectious diseases. This structure is referred to as a pyramid owing to the tapering number of pathogens from level 1 to level 4, with fewer pathogen members at each incremental level.
Emerging infectious diseases, zoonoses and “Disease X”
Tracing this ecological chain of uncertainties almost always leads to an animal source. The diseases identified by the WHO's priority research and development (R&D) blueprint in all zoonotic in nature and have been responsible for outbreaks in the recent past35. COVID-19 is the latest entrant in the list, having graduated from the category of Disease “X”. This ominously named category represents the currently unknown pathogens, which may emerge from unidentified spillover events, and result in outbreaks that may pose international threats. The risk of the emergence of these yet to be identified threats grow larger with increasing human population, loss of biodiversity, changing climates, aggressive land use for human habitation and agriculture, all of which contribute to the expanding interfaces between humans, animals and the environment36. The epidemiology of the H1N1 influenza virus outbreak in 2009, which contained genetic material from human, avian and swine origin, involved wildlife, pig farming, animal movement and farm workers37,38.
The specific animal-to-human spillover event for SARS-CoV-2 infection in man remains poorly understood. Hypothesized to have originated from horseshoe bats, the role and identity of any intermediate hosts in the transmission chain remains unclear39. Genetic studies propose two pathways of transmission: viral mutations in human after zoonotic transmission; or mutations in animal reservoirs before zoonotic transmission to human40. While the current evidence base is not substantive enough to distinguish which of these two trajectories was taken by SARS-CoV-2, the latter represents a more efficient transmission pathway, which is reflective of the threat posed by the “unknown unknowns”.
As human residence, agriculture, and consumption patterns continue to invade wildlife habitat, the pathophysiological aspects of these changing equations are often overshadowed by the economic imperative or cost-benefit rationalizations. Human encroachments into animal well-being and territory are usually considered under the framework of biodiversity and conservation, and the disease risk perspective often remains unexplored and undervalued. The role and risk of densification of animals through factory farming, animal markets, and wildlife trade should be evaluated, and disease surveillance conducted in these high-risk contexts. Densification further enhances the opportunities for viral replication and mutation41.
One Health approach and emerging infectious disease threats
Despite the emergence of some structured efforts at undertaking cross-sectoral approaches to build capacity for One Health response to deal with such disease X threats, these have remained scattered, with little governmental buy-in and horizontal integration42. Event-specific inter-sectoral platforms have been developed to coordinate responses in the aftermath of epidemic or pandemic threats. The National Task Force, Empowered Groups and Joint Monitoring Group to guide the government's response to COVID-19 in India, or the Inter-Ministerial Task Force and Joint Monitoring Group set up to coordinate the H5N1 influenza response represent such efforts43. These opportunities need to be capitalized on for strengthening systems responses across disciplines and departments. Often, linkages established during a crisis response tend to be neglected in the inter-epidemic period, when other competing priorities take precedence. It is important to institutionalize these One Health linkages to ensure continuity and enable initiation of preparedness and response activities in a timely fashion when the next disease threat inevitably emerges. This need for improved communications across the multiple sectors of human, animal, environmental and economic well-being, has been highlighted as a major lacuna by multiple priority-setting exercises undertaken globally44,45.
In addition to improving systems collaboration and coordination, there needs to be a concerted effort to invest in capacity building for developing cadres of scientists with One Health core competencies. A previous landscape analysis has shown that there are only a few research and capacity building programmes in place in South Asian countries, which can satisfy the core competencies expected of One Health efforts42. Although there has been some move to institutionalize the policy responses to infectious disease threats using the One Health approach, efforts at developing trans-sectoral training programmes or research and practice capacity building programmes have been limited. Without a competent corps of One Health scientist leaders, policymaking and response efforts will continue to remain constrained.
Dubbed as the once-in-a-century pandemic, COVID-19 shows a relentless march across countries, and is likely to affect the fragile health systems of low- and middle-income countries (LMICs) in more ways than just as an infectious contagion46. Modelling estimates have shown that as a fallout of the COVID-19 pandemic, in high burden settings, even under optimistic assumptions of transmission and mitigation of SARS-CoV-2, mortality from HIV, TB and malaria could increase by 10, 20 and 36 per cent, respectively over a five-year period47. Other estimates computed early in the COVID-19 pandemic revealed that even modest levels of reduction in coverage of maternal and child health services in LMICs were likely to result in over 250,000 additional child deaths and over 12,000 maternal deaths48. The Global Financing Facility estimated that childhood vaccinations were likely to drop by 50 per cent or more in many LMICs facing COVID-19 related lockdowns or service restrictions49. Another modelling effort showed that in a high-impact setting in Africa, for every COVID-19 death that could be attributed to SARS-CoV-2 infection through person-to-person transmission during routine childhood vaccinations, 84 under-five deaths due to vaccine preventable diseases, such as measles, could be avoided50.
The evolution of the COVID-19 threat has given us a unique opportunity to introspect on the narrative of reactive response driven epidemic management vis-à-vis the need to develop proactive prediction systems which can pre-empt novel spillovers and emergence or re-emergence of dangerous pathogens51. Further, the trajectory at which the pandemic has progressed in different countries has been slightly different, which also drives the issue of identifying and contextualizing evidence to base policy on. As identified by WHO, a one-size-fits-all approach to manage COVID-19 is likely to fail, given the wide variance in the epidemiology of COVID-19 globally52.
Global learnings from the pandemic response
There have been several positive takeaways from the global experiences of dealing with the COVID-19 threat. Global ability to devise technical solutions at a short notice has improved. The whole genome sequence of the virus was examined and rapidly shared on public domains53,54,55. Publications have come out in pre-print servers, cutting down the time to put evidence to use. This is one of the first instances where the evolving epidemiology in various countries is being studied in real time by a diverse group of researchers from across the world. Access to libraries of pre-tested compounds has accelerated drug development efforts, while computational approaches have helped identify in silico, which drugs would be a best fit to repurpose. Innovations in risk-mitigating technologies have allowed businesses, education and healthcare, to transition to the new normal56. The development of COVID-19 vaccines has also taken place at an unprecedented pace, with global and regional collaborations being developed to produce and disseminate the vaccines globally57,58. However, with the inevitable imbalance between demand and supply, we have also been witness to the emergence of the phenomenon of vaccine nationalism, especially in high-income countries, which has threatened the notions of global solidarity59,60,61.
Given the global implications of the spread of COVID-19, these technical solutions may fall short. Diplomatic and international cooperative solutions such as data sharing, using common protocols for rapid product (diagnostic, preventive, therapeutic) testing, and accelerated or common approval systems for deployment of medical products for diagnosis, prevention or management of COVID-19 or other novel spillover events need to be explored and encouraged. Cross-border and regional cooperation remain central to the discussion around developing solutions that work for all. Further, transparent systems, undergoing iterative improvement over time, based on accurate data, collected, and reported through sensitive surveillance systems, would also provide the means to ensure global solidarity in response to emergent infectious disease threats. In our global village, and infectious scourge at any corner of the world is a potential threat to health security for all62.
Using One Health to move from reactive to proactive response
The holistic view of health encouraged by a One Health perspective supports the inter-sectoral collaboration on human, animal, and environmental health. Novel and emerging infectious diseases is a perfect platform for incorporating this perspective63. It is also important to consider the potential for climate change to influence future pandemics64. With a change in climate, animal (including vectors) and plant species will shift as the suitability of different areas for specific biomes changes. This will likely change the interaction dynamics at the evolving human-animal-environment interfaces, as different species of animals will come into contact, under a changing ecosystem. Erratic weather patterns, droughts, heat waves and desynchronizations of life cycles of animals and plants may also alter the mix of animals in specific locations. New mixing patterns of different plant and animal species, and pathogens specific to each, has the potential for new cross-over infections between animals, with subsequent human spillovers65. Moreover, changes in the distribution of animals, environmental degradation itself, crop failure or livestock death, for example, may alter human interactions with wildlife, bringing them into closer contact or increasing hunting and eating of bushmeat, for instance66.
In February 2018, in its blueprint for research on priority diseases, the WHO prophetically added a new potential threat: Disease X35. The X stood for something unexpected, a microbe that had the potential to cause a future pandemic. This was based on advocacy from multiple health experts, who pointed out that it was a question of when and not if the next disease with epidemic potential would emerge from a hitherto unknown source, the most likely being a zoonotic one67. However, the common consensus was that it would be a pandemic influenza and some nations developed plans to deal with such outbreaks. A regional outbreak that progressed to a public health emergency of international concern and finally to a pandemic in a couple of months, killing millions in less than a year took the world by surprise4. Hence, given the continued threat of emerging infectious diseases, the global health community must review the successes, lessons learnt, and mistakes made over the past year to ensure that history does not repeat itself.
COVID-19 is a wakeup call which provides us an opportunity to learn and adapt, so that the impact of future pandemics can be mitigated68. It is obvious, that we need to focus on developing more precise early warning systems, which pre-empt outbreaks or spread of epidemics, and enable us to mount an effective response before sustaining losses in human and animal life, and economic well-being69. In addition, we must look at investing in strengthening health systems and interdepartmental collaborations; improving the sensitivity, breadth and fidelity of disease surveillance; and empowering communities to react and respond to outbreak threats in a more efficient manner70,71. Whilst the immediate focus should be directed towards containing the COVID-19 threat and minimizing the losses, both in terms of human lives and economic security, we must build up the bulwarks before the next novel dangerous pathogen comes calling at our doors. Just as Joshua Lederberg, the Nobel-laureate had stated, “the future of humanity and microbes likely will unfold as episodes of a suspense thriller that could be titled ‘Our Wits Versus Their Genes’”72.
Footnotes
Financial support & sponsorship: None.
Conflicts of Interest: None.
References
- 1.Morens DM, Folkers GK, Fauci AS. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430:242–9. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll D, Daszak P, Wolfe ND, Gao GF, Morel CM, Morzaria S, et al. The Global Virome Project. Science. 2018;359:872–4. doi: 10.1126/science.aap7463. [DOI] [PubMed] [Google Scholar]
- 3.Anthony SJ, Epstein JH, Murray KA, Navarrete-Macias I, Zambrana-Torrelio CM, Solovyov A, et al. A Strategy to estimate unknown viral diversity in mammals. mBio. 2013;4:e00598–13. doi: 10.1128/mBio.00598-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johns Hopkins, Coronavirus Resource Center. Data in motion. [accessed on March 1, 2021]. Available from: https://coronavirus.jhu.edu/
- 5.Zhong NS, Zheng BJ, Li YM, Poon LLM, Xie ZH, Chan KH, et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. The Lancet. 2003;362:1353–8. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alsahafi AJ, Cheng AC. The epidemiology of Middle East respiratory syndrome coronavirus in the Kingdom of Saudi Arabia, 2012–2015. Int J Infect Dis. 2016;45:1–4. doi: 10.1016/j.ijid.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New Engl J Med. 2012;367:1814–20. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 8.Olival KJ, Hosseini PR, Zambrana-Torrelio C, Ross N, Bogich TL, Daszak P. Host and viral traits predict zoonotic spillover from mammals. Nature. 2017;546:646–50. doi: 10.1038/nature22975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iacono GL, Cunningham AA, Fichet-Calvet E, Garry RF, Grant DS, Leach M, et al. A Unified Framework for the infection dynamics of zoonotic spillover and spread. PLOS Negl Trop Dis. 2016;10:e0004957. doi: 10.1371/journal.pntd.0004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eng CLP, Tong JC, Tan TW. Predicting host tropism of influenza A virus proteins using random forest. BMC Med Genomics. 2014;7(Suppl 3):S1. doi: 10.1186/1755-8794-7-S3-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imai N, Cori A, Dorigatti I, Baguelin M, Donnelly CA, Riley S, et al. Report 3: Transmissibility of 2019-nCoV. [accessed on February 15, 2020]. Available from: https://www.imperial.ac.uk/media/imperialcollege/medicine/sph/ide/gida-fellowships/Imperial-College-COVID19-transmissibility-25-01-2020.pdf .
- 12.Majumder MS, Mandl KD. Early transmissibility assessment of a novel coronavirus in Wuhan, China. SSRN. 2020 doi: 10.2139/ssrn.3524675. [Google Scholar]
- 13.The Japan Times. 44 more on Diamond Princess cruise ship test positive for COVID-19. 2020. [accessed on February 15, 2020]. Available from: https://www. japantimes.co.jp/news/2020/02/13/national/coronavirusdiamond-princess/
- 14.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–13. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ki M Task Force for 2019-nCoV. Epidemiologic characteristics of early cases with 2019 novel coronavirus (2019-nCoV) disease in Korea. Epidemiol Health. 2020;43:e2020007. doi: 10.4178/epih.e2020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsang KW, Ho PL, Ooi GC, Yee WK, Wang T, Chan-Yeung M, et al. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1977–85. doi: 10.1056/NEJMoa030666. [DOI] [PubMed] [Google Scholar]
- 17.Ryu S, Chun BC Korean Society of Epidemiology 2019-nCoV Task Force Team. An interim review of the epidemiological characteristics of 2019 novel coronavirus. Epidemiol Health. 2020;42:e2020006. doi: 10.4178/epih.e2020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, et al. Global trends in emerging infectious diseases. Nature. 2008;451:990–3. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woolhouse MEJ. Emerging diseases go global. Nature. 2008;451:898–9. doi: 10.1038/451898a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woolhouse M, Scott F, Hudson Z, Howey R, Chase-Topping M. Human viruses: discovery and emergence. Philoso Trans R Soc Lond B Biol Sci. 2012;367:2864–71. doi: 10.1098/rstb.2011.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolfe ND, Dunavan CP, Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–83. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woolhouse MEJ, Adair K, Brierley L. RNA viruses: a case study of the biology of emerging infectious diseases. Microbiol Spectr. 2013;1 doi: 10.1128/microbiolspec.OH-0001-2012. doi: 10.1128/microbiolspec.OH-0001-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaplan MM. Epidemiology of rabies. Nature; 1969;221:421–5. doi: 10.1038/221421a0. [DOI] [PubMed] [Google Scholar]
- 24.Sudarshan MK, Madhusudana SN, Mahendra BJ, Rao NSN, Ashwath Narayana DH, Abdul Rahman S, et al. Assessing the burden of human rabies in India: results of a national multi-center epidemiological survey. Int J Infec Dis. 2007;11:29–35. doi: 10.1016/j.ijid.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 25.Dhillon GP, Raina VK. Epidemiology of Japanese encephalitis in context with Indian scenario. J Indian Med Assoc. 2008;106:660–3. [PubMed] [Google Scholar]
- 26.Kakkar M, Chaturvedi S, Saxena VK, Dhole TN, Kumar A, Rogawski ET, et al. Identifying sources, pathways and risk drivers in ecosystems of Japanese Encephalitis in an epidemic-prone north Indian district. PLoS One. 2017;12:e0175745. doi: 10.1371/journal.pone.0175745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.World Health Organization. Assessment of risk associated with influenza A(H5N8) virus. [accessed on February 27, 2021]. Available from: http://www.who.int/influenza/human_animal_interface/avian_influenza/riskassessment_AH5N8_201611/en/
- 28.Richard M, Herfst S, van den Brand JMA, Lexmond P, Bestebroer TM, Rimmelzwaan GF, et al. Low virulence and lack of airborne transmission of the Dutch Highly Pathogenic Avian Influenza Virus H5N8 in Ferrets. PLoS One. 2015;10:e0129827. doi: 10.1371/journal.pone.0129827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaplan BS, Russier M, Jeevan T, Marathe B, Govorkova EA, Russell CJ, et al. Novel highly pathogenic avian A(H5N2) and A(H5N8) influenza viruses of Clade 2.3.4.4 from North America have limited capacity for replication and transmission in mammals. mSphere. 2016;1:e00003–16. doi: 10.1128/mSphere.00003-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pulit-Penaloza JA, Sun X, Creager HM, Zeng H, Belser JA, Maines TR, et al. Pathogenesis and Transmission of Novel Highly Pathogenic Avian Influenza H5N2 and H5N8 Viruses in Ferrets and Mice. J Virol. 2015;89:10286–93. doi: 10.1128/JVI.01438-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.European Centre for Disease Prevention and Control. First identification of human cases of avian influenza A(H5N8) infection. [accessed on February 27, 2021]. Available from: https://www.ecdc.europa.eu/sites/default/files/documents/First-identification-human-casesavian-influenza-A-H5N8-infection.pdf .
- 32.Fehervari Z. Origin story. [accessed on March 7, 2021]. Available from: https://www.nature.com/articles/d42859-018-00008-6 .
- 33.Peeters M, Honoré C, Huet T, Bedjabaga L, Ossari S, Bussi P, et al. Isolation and partial characterization of an HIV-related virus occurring naturally in chimpanzees in Gabon. AIDS. 1989;3:625–30. doi: 10.1097/00002030-198910000-00001. [DOI] [PubMed] [Google Scholar]
- 34.Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, et al. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–41. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 35.World Health Organization. Prioritizing diseases for research and development in emergency contexts. [accessed on April 14, 2020]. Available from: https://www.who.int/activities/prioritizing-diseases-forresearch-and-development-in-emergency-contexts .
- 36.Muehlenbein MP. Disease and Human/Animal Interactions. Annual Review of Anthropology. 2016;45:395–416. [Google Scholar]
- 37.World Health Organization. Influenza-like illness in the United States and Mexico. [accessed on March 1, 2021]. Available from: https://www.who.int/csr/don/2009_04_24/en/
- 38.New Scientist. Deadly new flu virus in US and Mexico may go pandemic. [accessed on March 1, 2021]. Available from: https://www.newscientist.com/article/dn17025-deadly-new-flu-virus-in-us-and-mexico-maygo-pandemic/
- 39.Chatterjee P, Nagi N, Agarwal A, Das B, Banerjee S, Sarkar S, et al. The 2019 novel coronavirus disease (COVID-19) pandemic: A review of the current evidence. Indian J Med Res. 2020;151:147–59. doi: 10.4103/ijmr.IJMR_519_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020;26:450–2. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones BA, Grace D, Kock R, Alonso S, Rushton J, Said MY, et al. Zoonosis emergence linked to agricultural intensification and environmental change. Proc Natl Acad Sci U S A. 2013;110:8399–404. doi: 10.1073/pnas.1208059110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKenzie JS, Dahal R, Kakkar M, Debnath N, Rahman M, Dorjee S, et al. One health research and training and government support for one health in South Asia. Infect Ecol Epidemiol. 2016;6:33842. doi: 10.3402/iee.v6.33842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chatterjee P, Kakkar M, Chaturvedi S. Integrating one health in national health policies of developing countries: India's lost opportunities. Infect Dis Poverty. 2016;5:87. doi: 10.1186/s40249-016-0181-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Standley CJ, Carlin EP, Sorrell EM, Barry AM, Bile E, Diakite AS, et al. Assessing health systems in Guinea for prevention and control of priority zoonotic diseases: A one health approach. One Health. 2019;7:100093. doi: 10.1016/j.onehlt.2019.100093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sorrell EM, El Azhari M, Maswdeh N, Kornblet S, Standley CJ, Katz RL, et al. Mapping of networks to detect priority zoonoses in Jordan. Front Public Health. 2015;3:219. doi: 10.3389/fpubh.2015.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gates B. Responding to Covid-19 - A once-in-a-century pandemic? N Eng J Med. 2020;382:1677–9. doi: 10.1056/NEJMp2003762. [DOI] [PubMed] [Google Scholar]
- 47.Hogan AB, Jewell BL, Sherrard-Smith E, Vesga JF, Watson OJ, Whittaker C, et al. Potential impact of the COVID-19 pandemic on HIV, tuberculosis, and malaria in low-income and middle-income countries: a modelling study. Lancet Glob Health. 2020;8:e1132–41. doi: 10.1016/S2214-109X(20)30288-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roberton T, Carter ED, Chou VB, Stegmuller AR, Jackson BD, Tam Y, et al. Early estimates of the indirect effects of the COVID-19 pandemic on maternal and child mortality in low-income and middle-income countries: a modelling study. Lancet Glob Health. 2020;8:e901–8. doi: 10.1016/S2214-109X(20)30229-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Global Financing Facility. Country briefs: Preserve essential health services during the Covid-19 pandemic. [accessed on February 28, 2021]. Available from: https://www.globalfinancingfacility.org/countrybriefs-preserve-essential-health-services-during-covid-19-pandemic .
- 50.Abbas K, Procter SR, Zandvoort K van, Clark A, Funk S, Mengistu T, et al. Routine childhood immunisation during the COVID-19 pandemic in Africa: a benefit–risk analysis of health benefits versus excess risk of SARS-CoV-2 infection. Lancet Glob Health. 2020;8:e1264–72. doi: 10.1016/S2214-109X(20)30308-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jonas O, Seifman R. Do we need a global virome project? Lancet Glob Health. 2019;7:e1314–6. doi: 10.1016/S2214-109X(19)30335-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.World Health Organization. Coronavirus disease 2019 (COVID-19) Situation Report – 42. [accessed on March 3, 2020]. Available from: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200302-sitrep-42-covid-19.pdf?sfvrsn=edd4f123_2 .
- 53.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–74. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu D, Zhu C, Ai L, He T, Wang Y, Ye F, et al. Genomic characterization and infectivity of a novel SARS-like coronavirus in Chinese bats. Emerg Microbes Infect. 2018;7:154. doi: 10.1038/s41426-018-0155-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.NCBI SARS-CoV-2 Resources. SARS-CoV-2 data. [accessed on March 2, 2020]. Available from: https://www.ncbi.nlm.nih.gov/genbank/sars-cov-2-seqs/
- 56.Harvard Business School, Working Knowledge. The one good thing caused by COVID-19: Innovation. 2020. [accessed on February 27, 2021]. Available from: http://hbswk.hbs.edu/item/the-one-good-thing-causedby-covid-19-innovation .
- 57.So AD, Woo J. Reserving coronavirus disease 2019 vaccines for global access: cross sectional analysis. BMJ. 2020;371:m4750. doi: 10.1136/bmj.m4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwartz JL. Equitable global access to coronavirus disease 2019 vaccines. BMJ. 2020;371:m4735. doi: 10.1136/bmj.m4735. [DOI] [PubMed] [Google Scholar]
- 59.Eaton L. Covid-19: WHO warns against “vaccine nationalism” or face further virus mutations. BMJ. 2021;372:n292. doi: 10.1136/bmj.n292. [DOI] [PubMed] [Google Scholar]
- 60.Fidler DP. Vaccine nationalism's politics. Science. 2020;369:749. doi: 10.1126/science.abe2275. [DOI] [PubMed] [Google Scholar]
- 61.Science. ‘Vaccine nationalism’ threatens global plan to distribute COVID-19 shots fairly. [accessed on March 7, 2021]. Available from: https://www.sciencemag.org/news/2020/07/vaccine-nationalismthreatens-global-plan-distribute-covid-19-shots-fairly .
- 62.Peckham R. COVID-19 and the anti-lessons of history. Lancet. 2020;395:850–2. doi: 10.1016/S0140-6736(20)30468-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mackenzie JS, Jeggo M. The one health approach-Why is it so important? Trop Med Infect Dis. 2019;4:88. doi: 10.3390/tropicalmed4020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zinsstag J, Crump L, Schelling E, Hattendorf J, Maidane YO, Ali KO, et al. Climate change and One Health. FEMS Microbiol Lett. 2018;365:fny085. doi: 10.1093/femsle/fny085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heffernan C. Climate change and multiple emerging infectious diseases. Vet J. 2018;234:43–7. doi: 10.1016/j.tvjl.2017.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wolfe ND, Daszak P, Kilpatrick AM, Burke DS. Bushmeat hunting, deforestation, and prediction of zoonotic disease. Emerg Infect Dis. 2005;11:1822–7. doi: 10.3201/eid1112.040789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simpson S, Kaufmann MC, Glozman V, Chakrabarti A. Disease X: accelerating the development of medical countermeasures for the next pandemic. Lancet Infect Dis. 2020;20:e108–15. doi: 10.1016/S1473-3099(20)30123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chatterjee P, Seth B, Biswas T. Hotspots of H1N1 influenza in India: analysis of reported cases and deaths (2010-2017) Trop Doct. 2020;50:166–9. doi: 10.1177/0049475519879357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McCall B. COVID-19 and artificial intelligence: protecting health-care workers and curbing the spread. Lancet Digit Health. 2020;2:e166–7. doi: 10.1016/S2589-7500(20)30054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Van den Broucke S. Why health promotion matters to the COVID-19 pandemic, and vice versa. Health Promot Int. 2020;35:181–6. doi: 10.1093/heapro/daaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gates B. The next epidemic — Lessons from Ebola. N Eng J Med. 2015;372:1381–4. doi: 10.1056/NEJMp1502918. [DOI] [PubMed] [Google Scholar]
- 72.Lederberg J. Infectious History. Science. 2000;288:287–93. doi: 10.1126/science.288.5464.287. [DOI] [PubMed] [Google Scholar]


