Abstract

SARS-CoV-2 main protease (Mpro) is a cysteine protease that mediates the cleavage of viral polyproteins and is a validated antiviral drug target. Mpro is highly conserved among all seven human coronaviruses, with certain Mpro inhibitors having broad-spectrum antiviral activity. In this study, we designed two hybrid inhibitors UAWJ9-36-1 and UAWJ9-36-3 based on the superimposed X-ray crystal structures of SARS-CoV-2 Mpro with GC-376, telaprevir, and boceprevir. Both UAWJ9-36-1 and UAWJ9-36-3 showed potent binding and enzymatic inhibition against the Mpro’s from SARS-CoV-2, SARS-CoV, MERS-CoV, HCoV-OC43, HCoV-NL63, HCoV-229E, and HCoV-HKU1. Cell-based Flip-GFP Mpro assay results show that UAWJ9-36-1 and UAWJ9-36-3 inhibited the intracellular protease activity of SARS-CoV-2 Mpro. In addition, UAWJ9-36-1 and UAWJ9-36-3 had potent antiviral activity against SARS-CoV-2, HCoV-OC43, HCoV-NL63, and HCoV-229E, with UAWJ9-36-3 being more potent than GC-376 in inhibiting SARS-CoV-2. Selectivity profiling revealed that UAWJ9-36-1 and UAWJ9-36-3 had an improved selectivity index over that of GC-376 against host cysteine proteases calpain I and cathepsin L, but not cathepsin K. The X-ray crystal structures of SARS-CoV-2 Mpro with UAWJ9-36-1 and UAWJ9-36-3 were both solved at 1.9 Å, which validated our design hypothesis. Overall, hybrid inhibitors UAWJ9-36-1 and UAWJ9-36-3 are promising candidates to be further developed as broad-spectrum coronavirus antivirals.
Keywords: SARS-CoV-2, COVID-19, main protease, 3CL protease, antiviral
SARS-CoV-2 is the etiological agent of the COVID-19, and it is the third coronavirus that causes significant morbidity and mortality in humans. The other two highly pathogenic coronaviruses are SARS-CoV and MERS-CoV, with mortality rates of 9.7 and 34.3%,1 respectively. In addition, four common human coronaviruses including HCoV-OC43, HCoV-229E, HCoV-NL63, and HCoV-HKU1 also circulate among humans and cause common colds. SARS-CoV-2 is a single-stranded, positive-sense RNA virus that shares ∼80% sequence identity with SARS-CoV. Although the previous SARS and MERS outbreaks failed to fuel the development of coronavirus antivirals, the current COVID-19 pandemic is a reminder that broad-spectrum antivirals are needed to combat not only existing coronaviruses but also future emerging coronaviruses. In line with this, the viral polymerase and proteases are prominent targets for the development of broad-spectrum anti-coronavirus drugs.2 The viral polymerase inhibitor remdesivir was the first drug that received FDA approval for the treatment of COVID-19 infection, although the results from several clinical trials were not consistent.3−5 In addition, another viral polymerase inhibitor molnupiravir is currently being studied in a clinical trial.6,7 Molnupiravir was originally developed as an oral influenza drug.8
SARS-CoV-2 encodes two viral proteases, the main protease (Mpro) and the papain-like protease (PLpro), both of which are validated antiviral drug targets.9,10 Mpro and PLpro are cysteine proteases that cleave the viral polyproteins during viral replication. PLpro plays additional roles in antagonizing the host innate immune response through its deubiquitinating and deISG15ylating (interferon-induced gene 15) activities.11−13 The active site residues of Mpro across different coronaviruses are relatively conserved, and certain Mpro inhibitors have shown broad-spectrum antiviral activity. Among the Mpro inhibitors reported to date, the most advanced ones are GC-376,9,10,14,15 6j,16 PF-07304814,17 MI-09, MI-30,18 and the deuterated GC-376 (D2-GC-376)19 (Figure 1A). GC-376 showed in vivo antiviral efficacy in treating cats infected with lethal feline infectious peritonitis virus.14,15 A recent study found that GC-376 analog 6j protected mice from MERS-CoV infection.16 The same group also reported that a deuterated analog of GC-376 (D2-GC-376, compound 2 in the original publication) had in vivo antiviral efficacy in a SARS-CoV-2-infected mouse model.19 PF-07304814 is an α-hydroxyl ketone prodrug that was originally being developed by Pfizer as an antiviral drug for SARS-CoV.17 It has favorable pharmacokinetic properties and in vivo antiviral efficacy in the SARS-CoV-infected mouse model. PF-07304814 is currently in a phase I clinical trial for COVID-19.17 Two additional GC-376 analogs, MI-09 and MI-30, were recently reported to protect mice from lethal SARS-CoV-2 infection.18 These promising results highlight the translational potential of Mpro inhibitors as potent SARS-CoV-2 antivirals and validate Mpro as an antiviral drug target for coronaviruses.
Figure 1.

SARS-CoV-2 Mpro inhibitors. (A) Literature-reported SARS-CoV-2 Mpro inhibitors with in vivo antiviral efficacy. (B) Hybrid SARS-CoV-2 Mpro inhibitors UAWJ9-36-1 and UAWJ9-36-3 designed in this study.
Our previous high-throughput screening identified GC-376 and boceprevir as SARS-CoV-2 Mpro inhibitors with IC50 values of 0.03 and 4.13 μM, respectively.9 Telaprevir was less active and inhibited 31% of the Mpro enzymatic activity at 20 μM. We subsequently solved the X-ray crystal structure of SARS-CoV-2 Mpro with GC-376 and other hits including calpain inhibitors II and XII.9,10 Our results have been independently validated by others at about the same time. Fu et al. reported that GC-376 and boceprevir inhibited SARS-CoV-2 Mpro with IC50 values of 0.15 and 8.0 μM, respectively,20 and solved the X-ray crystal structure of SARS-CoV-2 Mpro with boceprevir. Vuong et al. showed that GC-376 and its active drug GC-373 inhibited SARS-CoV-2 Mpro with IC50 values of 0.40 and 0.19 μM, respectively.21 Although we reported telaprevir was a weak inhibitor of SARS-CoV-2 Mpro (IC50 > 20 μM), Kneller et al. showed that telaprevir inhibited SARS-CoV-2 Mpro with an IC50 of 18 μM and solved the X-ray crystal structure of SARS-CoV-2 Mpro with telaprevir.22 On the basis of the available X-ray cocrystal structures, we aim to further improve the enzymatic inhibition and cellular antiviral activity of SARS-CoV-2 Mpro inhibitors by structure-based drug design. Specifically, the design was guided by overlaying different Mpro inhibitors at the active site, and hybrid inhibitors were designed to integrate optimal substitutions at each binding pocket. UAWJ9-36-1 was designed as a hybrid of GC-376 and telaprevir, and UAWJ9-36-3 was designed as a hybrid of GC-376 and boceprevir (Figure 1B). Although UAWJ9-36-1 and UAWJ9-36-3 had enzymatic inhibition similar to that of GC-376 in the FRET assay, UAWJ9-36-3 had more potent enzymatic inhibition than GC-376 in the cell-based Flip-GFP Mpro assay. The cellular antiviral activity with infectious SARS-CoV-2 further confirmed the superior potency of UAWJ9-36-3 compared to those of UAWJ9-36-1 and GC-376. Hybrid inhibitors UAWJ9-36-1 and UAWJ9-36-3 also inhibited the Mpro from other known human coronaviruses including SARS-CoV, MERS-CoV, HCoV-OC43, HCoV-NL63, and HCoV-229E in the FRET-based enzymatic assay, and the binding was confirmed in the thermal shift binding assay. UAWJ9-36-1 and UAWJ9-36-3 also had potent antiviral activity against HCoV-OC43, HCoV-NL63, and HCoV-229E. Selectivity profiling revealed that UAWJ9-36-1 and UAWJ9-36-3 had improved selectivity compared to that of GC-376 against host cysteine proteases calpain I and cathepsin L. We solved the X-ray crystal structures of SARS-CoV-2 Mpro with UAWJ9-36-1 and UAWJ9-36-3, which validated our design hypothesis. Overall, designed hybrid inhibitors UAWJ9-36-1 and UAWJ9-36-3 are promising drug candidates for further development as broad-spectrum coronavirus antivirals.
Results and Discussion
Rational Design of SARS-CoV-2 Mpro Inhibitors
The superimposed cocrystal structures of GC-376 with telaprevir showed that the pyrrolidone from GC-376 and the norvaline from telaprevir fit in the S1 pocket (Figure 2A–C). Consistent with its substrate preference for glutamine at the P1 position, pyrrolidone is a preferred substitution where it forms two or three hydrogen bonds with the H163, E166, and F140 in the S1 pocket, while norvaline from telaprevir does not form any specific interaction. The leucine from GC-376 and the cyclopentylproline from telaprevir fit in the S2 hydrophobic pocket (Figure 2A–C). Since cyclopentylproline forms more hydrophobic interactions than leucine in the S2 pocket, we hypothesize that it might be a preferred substitution at the P2 position. The tert-leucine substitution at the P3 position of telaprevir was solvent-exposed. Because previous structure–activity relationship studies have shown that P3 substitution does not contribute significantly to the enzymatic inhibition,10 we decided to omit the P3 substitution. The carboxybenzyl (Cbz) group from GC-376 and the cyclohexane from telaprevir fit in the S4 pocket, and both are engaged in hydrophobic interactions. On the basis of the overlaying structures, we designed hybrid inhibitor UAWJ9-36-1, which integrates the favorable substitutions pyrrolidone at the P1, cyclopentylproline at P2, and benzyl at P4 position (Figure 2D). Using a similar strategy, UAWJ9-36-3 was designed as a hybrid of GC-376 and boceprevir, which contains dimethylcyclopropylproline at the P2 position (Figure 2A,E–G).
Figure 2.
Structure-guided design of SARS-CoV-2 Mpro hybrid inhibitors based on superimposed X-ray crystal structures. (A) X-ray crystal structure of SARS-CoV-2 Mpro with GC-376 (Protein Data Bank (PDB): 6WTT). (B) X-ray crystal structure of SARS-CoV-2 Mpro with telaprevir (PDB: 6XQS). (C) Overlaying X-ray crystal structures of Mpro with GC-376 and telaprevir. (D) Design of UAWJ9-36-1 as a hybrid of GC-376 and telaprevir. (E) X-ray crystal structure of SARS-CoV-2 Mpro with boceprevir (PDB: 6XQU). (F) Overlaying X-ray crystal structures of Mpro with GC-376 and boceprevir. (G) Design of UAWJ9-36-3 as a hybrid of GC-376 and boceprevir.
Synthesis of Hybrid Inhibitors UAWJ9-63-1 and UAWJ9-63-3
The synthesis of UAWJ9-63-1 and UAWJ9-63-3 started with commercially available amino esters 1 and 4 (Figure 3). Protecting the amine with the Cbz and subsequent hydrolysis of the ester gave carboxylic acid intermediates 2 and 5. Subsequent coupling with pyrrolidone intermediate 3, followed by reduction and oxidation, gave final products UAWJ9-36-1 and UAWJ9-36-3. It is noted that the first step Cbz protection of (1S,3aR,6aS)-ethyl octahydrocyclopenta[c]pyrrole-1-carboxylate hydrochloride (1) and methyl (1R,2S,5S)-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxylate hydrochloride (4) led to racemization of the α-chiral center, and H NMR showed a diastereomeric ratio (dr) of 1:1 (Figure S1). It is known from the synthesis of boceprevir, MI-09, and MI-30 that the α-chiral centers at compounds 1 and 4 are prone to racemization.18,23 The two diastereomers of final products UAWJ9-36-1 and UAWJ9-36-3 eluted as one broad peak in reverse-phase HPLC and could not be separated (Figure S2). As such, UAWJ9-36-1 and UAWJ9-36-3 were tested in the enzymatic assay and antiviral assay as a diastereomeric mixture (1:1).
Figure 3.
Synthesis of the hybrid inhibitors UAWJ9-36-1 and UAWJ9-36-3. *, Racemized chiral center.
Enzymatic Inhibition of UAWJ9-36-1 and UAWJ9-36-3 against the Mpro’s from Seven Human Coronaviruses
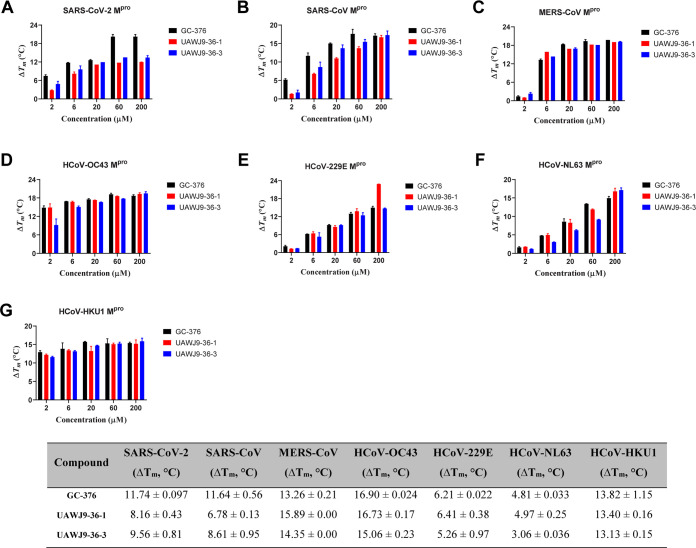
The enzymatic inhibition activities of UAWJ9-36-1 and UAWJ9-36-3 against the Mpro’s from all seven human coronaviruses, including SARS-CoV-2, SARS-CoV, MERS-CoV, HCoV-OC43, HCoV-229E, HCoV-NL63, and HCoV-HKU1, were tested in the FRET-based enzymatic assay (Figure 4). GC-376 was included as a control since it represents one of the most potent SARS-CoV-2 Mpro inhibitors reported so far. It was found that UAWJ9-36-1 and UAWJ9-36-3 were equally potent and had enzymatic inhibition activities comparable to that of GC-376 for all seven Mpro’s tested (Figure 4). UAWJ9-36-1 and UAWJ9-36-3 inhibited HCoV-NL63 Mpro with IC50 values of 0.36 and 0.45 μM, respectively, which were less potent compared to their inhibition of other Mpro’s. A thermal shift binding assay showed that UAWJ9-36-1 and UAWJ9-36-3 significantly increased the melting temperature shift (ΔTm) (Figure 5), indicating protein stabilization. Consistent with the enzymatic assay results, UAWJ9-36-1 and UAWJ9-36-3 were less potent in binding to HCoV-NL63 Mpro compared to the other Mpro’s. Overall, the enzymatic assay and the thermal shift binding assay found that UAWJ9-36-1 and UAWJ9-36-3 are potent inhibitors of the Mpro’s from all seven human coronaviruses.
Figure 4.
Enzymatic inhibition of GC-376, UAWJ9-36-1, and UAWJ9-36-3 against Mpro’s from all seven human coronaviruses. Data fittings of the proteolytic progression curves of the following: SARS-CoV-2 Mpro in the presence of GC-376 (A), UAWJ9-36-1 (B), and UAWJ9-36-3 (C); SARS-CoV Mpro in the presence of GC-376 (D), UAWJ9-36-1 (E), and UAWJ9-36-3 (F); MERS-CoV Mpro in the presence of GC-376 (G), UAWJ9-36-1 (H), and UAWJ9-36-3 (I). Dose–response curves of GC-376, UAWJ9-36-1, and UAWJ9-36-3 against Mpro from SARS-CoV-2 (J), SARS-CoV (K), MERS-CoV (L), HCoV-229E (M), HCoV-OC43 (N), HCoV-NL63 (O), and HCoV-HKU1 (P). Ratios of k2 (second rate constant) over KI (equilibrium dissociation constant) from kinetic studies and IC50 values from the dose–response curves are listed in the table at the bottom. Data are mean ± standard deviation of three replicates.
Figure 5.
Melting temperature shift (ΔTm) of Mpro’s from all seven human coronaviruses in the presence of the indicated concentrations of GC-376, UAWJ9-36-1, and UAWJ9-36-3: SARS-CoV-2 (A), SARS-CoV (B), MERS-CoV (C), HCoV-OC43 (D), HCoV-229E (E), HCoV-NL63 (F), and HCoV-HKU1 (G). ΔTm values of Mpro’s in the presence of 6 μM GC-376, UAWJ9-36-1, and UAWJ9-36-3 are listed in the table at the bottom. Data are mean ± standard deviation of two replicates.
Cellular Protease Inhibitory Activity of UAWJ9-36-1 and UAWJ9-36-3 in the Flip-GFP Mpro Assay
Although the FRET-based enzymatic assay is commonly used as a primary assay for the testing of SARS-CoV-2 Mpro inhibitors, the in vitro results from this assay might not have a direct correlation with cellular activity due to issues with drug efflux, cytotoxicity, membrane permeability, metabolism, off-target binding, and so on.24−26 As such, we developed the Flip-GFP assay to quantify the cellular protease inhibitory activity of UAWJ9-36-1 and UAWJ9-36-3 against SARS-CoV-2 Mpro (Figure 6). In the Flip-GFP assay, 293T cells were transfected with two plasmids, one expressing the SARS-CoV-2 Mpro and another expressing the Flip-GFP reporter construct with the Mpro cleavage site (Flip-GFP Mpro) (Figure 6A,B).27,28 Specifically, the Flip-GFP Mpro reporter construct expresses two GFP fragments, the β10–11 fragment and the β1–9 template. The β10–11 fragment contains an Mpro cleavage sequence (AVLQ↓SGFR). Upon cleavage by Mpro, the β11 strand will be able to assemble with the β1–9 template together with the β10 strand, leading to the restoration of green fluorescence signal (Figure 6A). The Flip-GFP Mpro plasmid also expresses the mCherry red fluorescence protein, which serves as an internal control to normalize the protein expression level (Figure 6B). As shown in Figure 6C, strong green fluorescence signals were only observed when there is a match between the protease and its corresponding substrate (second and eighth rows). No or minimal GFP signal was observed when there is a mismatch between the protease and its substrate (third, fourth, and fifth rows), no Mpro (sixth row), or the inactive Mpro (C145A) (seventh row). GC-376 showed dose-dependent inhibition activity in the Flip-GFP Mpro assay with an IC50 of 4.83 μM (Figure 6D,G). UAWJ9-36-1 was less active showing an IC50 of 11.10 μM (Figures 6E,6G), while UAWJ9-36-3 was more potent than GC-376 and had an IC50 value of 3.40 μM (Figures 6D,G). Overall, the Flip-GFP Mpro assay suggested that the UAWJ9-36-1 and UAWJ9-36-3 might have the cellular antiviral activity with a rank of potency in the order of UAWJ9-36-3 > GC-376 > UAWJ9-36-1.
Figure 6.
Cellular protease inhibitory activity of UAWJ9-36-1 and UAWJ9-36-3 in the Flip-GFP Mpro assay. (A) Principle of Flip-GFP assay. (B) Sequence of the flipped GFP β10–11 and construct of the Flip-GFP Mpro plasmid; the corresponding SARS-CoV-2 Mpro cleavage site between nsp4 and nsp5 was introduced into the plasmid. The arrow indicates the SARS-CoV-2 Mpro cleavage site. (C) FlipGFP-Mpro assay development. 293T cells were transfected with no plasmid (Ø) (first row); FlipGFP-PLpro and SARS-CoV-2 PLpro plasmids (second row); FlipGFP-PLpro and SARS-CoV-2 Mpro plasmids (third row); FlipGFP-Mpro and SARS-CoV-2 PLpro plasmids (fourth row); FlipGFP-TEV and SARS-CoV-2 Mpro plasmids (fifth row); FlipGFP-Mpro plasmid alone (sixth row); FlipGFP-Mpro and SARS-CoV-2 Mpro-C145A plasmids (seventh row); and FlipGFP-Mpro and SARS-CoV-2 Mpro plasmids (eighth row) (details are described in the “Cellular-Based FlipGFP Mpro Assay” section). (D–F) Representative images of FlipGFP-Mpro assay showed does-dependent decrease of GFP signal with the increasing concentration of compounds GC-376 (D), UAWJ9-36-1 (E), and UAWJ9-36-3 (F). (G) Dose–response curve of the inhibition of GFP signal over mCherry signal by compounds GC-376, UAWJ9-36-1, and UAWJ9-36-3; mCherry signal alone was used to calculate cytotoxicity.
Broad-Spectrum Antiviral Activity of UAWJ9-36-1 and UAWJ9-36-3 against SARS-CoV-2 and Human Coronaviruses HCoV-OC43, HCoV-229E, and HCoV-NL63
The antiviral activities of UAWJ9-36-1 and UAWJ9-36-3 against SARS-CoV-2 were tested using immunofluorescence assay in two cell lines, Vero E6 and Caco2-ACE2 (Figure 7A-D). Caco2-ACE2 expresses TMPRSS2 and is a physiologically relevant cell line for SARS-CoV-2 replication.29−31 It was found that UAWJ9-36-1 was less potent than GC-376 in inhibiting SARS-CoV-2 in both cell lines. Gratifyingly, UAWJ9-36-3 had improved antiviral activity than GC-376 and inhibited SARS-CoV-2 replication in Vero E6 cells and Caco2-ACE2 cells with EC50 values of 0.37 and 1.06 μM (Figure 7A–D). The relative antiviral activity of these three compounds was in agreement with the results from the cell-based Flip-GFP Mpro assay (Figure 6), suggesting that the Flip-GFP Mpro assay represents a viable assay to screen for Mpro inhibitors. The antiviral activities of UAWJ9-36-1 and UAWJ9-36-3 against HCoV-OC43 were tested in the plaque assay, and both were highly potent with EC50 values of 46 and 59 nM, respectively (Figure 7F,G). In comparison, GC-376 inhibited HCoV-OC43 with an EC50 value of 60 nM (Figure 7E). The antiviral activities against HCoV-229E and HCoV-NL63 were tested in the CPE assay (Figure 7H–P). UAWJ9-36-1 and UAWJ9-36-3 were equally potent in inhibiting HCoV-229E with EC50 values of 0.17 and 0.13 μM, respectively (Figure 7I,J). The antiviral activity of these two compounds against HCoV-NL63 was tested in two cell lines, Vero E6 and Huh-7 (Figure 7K–P). UAWJ9-36-1 and UAWJ9-36-3 were less potent in the Vero E6 cells than in the Huh-7 cells, which might be due to the drug efflux pump P-glycoprotein (P-gp) expressed on the Vero E6 cells.32 GC-376 and its analogs were previously shown to be the substrates of P-gp.17,24,33 To test this hypothesis, we repeated the antiviral assay in the presence of P-gp inhibitor CP-100356. It was found that the antiviral activity of UAWJ9-36-1 and UAWJ9-36-3 against HCoV-NL63 in Vero E6 cells increased in the presence of CP-100356 (Figure 7O–P).
Figure 7.
Antiviral activity of GC-376, UAWJ9-36-1, and UAWJ9-36-3 against SARS-CoV-2 and multiple HCoVs in cell culture. The analyses of antiviral activities of UAWJ9-36-1 (A, C) and UAWJ9-36-3 (B, D) against SARS-CoV-2 in immunofluorescence assay was carried out in Vero E6 or Caco2-ACE2 cells. The analyses of antiviral activities of GC-376 (E, H), UAWJ9-36-1 (F, I), and UAWJ9-36-3 (G, J) against HCoV-OC43 and HCoV-229E were carried out in plaque assay and in CPE assay, respectively. The antiviral activities of GC-376 (K, N), UAWJ9-36-1 (L, O), and UAWJ9-36-3 (M, P) against HCoV-NL63 in CPE assay was carried out in Huh-7 cells or Vero cells in the presence or absence of P-glycoprotein inhibitor CP-100356. EC50 values of GC-376, UAWJ9-36-1, and UAWJ9-36-3 against the coronaviruses tested in different types of cells and under different conditions are listed in the table at the bottom. EC50 curve fittings were obtained using log(concentration of inhibitors) vs percentage of positive control with variable slopes in prism 8. The cellular cytotoxicity test was carried out in each cell line used in the antiviral assays and the resulting curves were shown in blue. All data are mean ± standard deviation of three replicates.
Selectivity of UAWJ9-36-1 and UAWJ9-36-3 against human cysteine and serine proteases
Previous studies showed that GC-376 and its analogs also inhibit cathepsin L in addition to the SARS-CoV-2 Mpro.24,33 In addition, all three compounds GC-376, UAWJ9-36-1, and UAWJ9-36-3, contain aldehyde as a reactive warhead; therefore, there might be a potential concern with the off-target effect in inhibiting host cysteine proteases. To test this hypothesis, we profiled the selectivity of these two hybrid compounds against host cysteine proteases calpain I, cathepsin K, cathepsin L, and caspase-3, as well as the serine protease trypsin (Figure 8). GC-376 was included as a control. GC-376 was a potent inhibitor of calpain I with an IC50 of 0.074 μM, while UAWJ9-36-1 and UAWJ9-36-3 had drastically reduced inhibition activities with IC50 values of 16.56 and >20 μM, respectively (Figure 8A). Consistent with previous results, GC-376 was also a potent inhibitor of cathepsin L with an IC50 of 4.4 nM,24,33 while UAWJ9-36-1 and UAWJ9-36-3 were weak inhibitors of cathepsin L with IC50 values of 1.37 and 1.81 μM, respectively. GC-376 had potent inhibition activity against cathepsin K with an IC50 of 0.26 nM, and UAWJ9-36-1 and UAWJ9-36-3 had slightly improved selectivities but still had potent inhibition with IC50 values of 6.5 and 42 nM, respectively. None of the three compounds inhibited caspase-3 or trypsin (IC50 > 20 μM). Overall, UAWJ9-36-1 and UAWJ9-36-3 had a selectivity index improved over that of GC-376 in inhibiting calpain I and cathepsin L, but not cathepsin K.
Figure 8.
Selectivity of UAWJ9-36-1 and UAWJ9-36-3 against host cysteine and serine proteases. (A) IC50 values of UAWJ9-36-1 and UAWJ9-36-3 against host cysteine and serine proteases. Data are the mean ± standard error of two replications. aData from ref (34). bPan-caspase inhibitor Z-VAD-FMK was included as a positive control and IC50 was 0.10 ± 0.04 μM. (B) Selectivity heat map.
X-ray Crystal Structures of SARS-CoV-2 Mpro in Complex with UAWJ9-36-1 and UAWJ9-36-3
X-ray crystal structures of UAWJ9-36-1 and UAWJ9-36-3 with SARS-CoV-2 Mpro were both solved at 1.9 Å resolution (Figure 9, Table S1), revealing a binding pose that is consistent with our projections (Figure 2). In the P1 position, we found the pyrroldinone ring interacts with the S1 pocket, forming H-bonds with E166, H163, and the main chain of F140. The cyclopentylproline and dimethylcyclopropylproline moieties of UAWJ9-36-1 and UAWJ9-36-3 occupy the hydrophobic S2 sites almost identically to their telaprevir and boceprevir analogues (Figure 9E,F). Interestingly, the Cbz group adopts two different poses in UAWJ9-36-1 and UAWJ9-36-3. In UAWJ9-36-1, the Cbz group adheres to the substrate groove, covering the amide-binding segment of the S3 and S4 sites (Figure 9B). In UAWJ9-36-3, the Cbz flips up toward the weakly defined S3 site (Figure 9C). The variable binding conformation of the Cbz group has been observed in multiple structures of GC-376 and GC-376 analogues.10 It is not entirely clear what determines this pose, but given the nonspecific nature of the interactions at the S3 and S4 subsites, it is possible these populations exists in dynamic equilibrium, with individual poses promoted by the crystallization condition and nearby residues. Two different constructs of Mpro from our previous studies were used: the native Mpro for UAWJ9-36-1 and HM-Mpro (with two extra residues in the N-terminus) for UAWJ9-36-3. As the N-terminus of one protomer is in close proximity to the active site of the other protomer in the Mpro dimer, this resulted in differences in the conformation of E166 that interacts with the pyrroldinone ring of the inhibitor, which may in turn influence the conformation of the Cbz ring in a specific crystal structure. However, based on previous Mpro complex structures, both conformations of the Cbz may be relevant to the activity of the inhibitor, due to the flexible nature and the favorable protein interactions of both conformations.
Figure 9.
X-ray crystal structure of SARS-CoV-2 Mpro with hybrid inhibitors UAWJ9-36-1 and UAWJ9-36-3. (A) X-ray crystal structure of SARS-CoV-2 Mpro with GC-376 (PDB: 6WTT). (B) UAWJ9-36-1 (PDB: 7LYH) and (C) UAWJ9-36-3 (PDB ID: 7LYI) were both solved at 1.9 Å resolution. Unbiased Fo-Fc electron density map, shown in gray, is contoured at 2σ. (D) Design strategy for UAWJ9-36-1 and UAWJ9-36-3. (E) Superimposed binding pose of telaprevir (violet) in SARS-CoV-2 Mpro (PDB: 6XQS) with UAWJ9-36-1 (cyan). (F) Superimposed binding pose of boceprevir in SARS-CoV-2 Mpro (blue) (PDB: 6XQU) with UAWJ9-36-3 (magenta).
In parallel to our study, two compounds, MI-09 and MI-30 (Figure 1), with a similar design were reported to have both in vitro and in vivo antiviral activity against SARS-CoV-2 infection.18 The results from the hybrid molecules designed in this study UAWJ9-36-1 and UAWJ9-36-3 provided additional evidence to support the translational potential of this series of compounds. Highlights from this study include the following: (1) We solved the X-ray crystal structure of SARS-CoV-2 Mpro in complex with both cyclopentylproline-containg UAWJ9-36-1 and dimethylcyclopropylproline-containing UAWJ9-36-3, while the previous study only solved the X-ray crystal structure of the cyclopentylproline-containing analog MI-23.18 Since UAWJ9-36-3 demonstrated more potent antiviral activity than UAWJ9-36-1, the cocrystal structure with UAWJ9-36-3 is valuable in guiding the design of next generation of SARS-CoV-2 Mpro inhibitors. (2) We showed that the cell-based Flip-GFP Mpro assay is a viable assay that can be used to predict the cellular antiviral activity of Mpro inhibitors in a biosafety level 2 (BSL-2) facility. (3) UAWJ9-36-1 and UAWJ9-36-3 demonstrated broad-spectrum antiviral activity against not only SARS-CoV-2 but also common human coronaviruses HCoV-OC43, HCoV-NL63, and HCoV-229E. In addition, both UAWJ9-36-1 and UAWJ9-36-3 also had potent enzymatic inhibition against SARS-CoV and MERS-CoV Mpro’s, suggesting they might have antiviral activity against these two viruses. (4) Compared to GC-376 analogs reported to date,10,16,18,35,36UWAJ9-36-3 has superior cellular antiviral activity. (5) We profiled the selectivity of UAWJ9-36-1 and UAWJ9-36-3 against host cysteine proteases and showed that both compounds had a selectivity index improved over that of GC-376 against host proteases calpain I and cathepsin L, but not cathepsin K. Although GC-376 analogs such as MI-09, MI-23,18 and D2-GC-37619 were reported to have in vivo antiviral efficacy in a SARS-CoV-2 infection mouse model, their target selectivity and potential cytotoxicity have not been systematically studied, and their long-term side effects are unknown. As all these compounds contain an aldehyde as the reactive warhead, more attention should be given to profiling the selectivity against host cysteine proteases. Only one FDA-approved drug voxelotor contains an aldehyde, and many aldehyde-containing drug candidates were dropped out of clinical trials due to off-target effects.37
In summary, results from the hybrid inhibitors designed in this study, UAWJ9-36-1 and UAWJ9-36-3, coupled with the in vivo antiviral efficacy from analogs MI-09 and MI-30 reported recently,18 demonstrated that this series of compounds have great potential to be further developed as broad-spectrum coronavirus antivirals with an improved selectivity index.
Experimental Section
Materials and Methods
Chemistry
Chemicals were ordered from commercial sources and were used without further purification. Synthesis procedures for reactions described in Figure 3 are shown below. All final compounds were purified by flash column chromatography. 1H and 13C NMR spectra were recorded on a Bruker-400 spectrometer. Chemical shifts are reported in parts per million (ppm) referenced with respect to residual solvent CDCl3 (7.26 ppm) and internal standard tetramethylsilane (TMS, 0.00 ppm). The following abbreviations were used in reporting spectra: s, singlet; d, doublet; t, triplet; q, quartet; m, multiplet; dd, doublet of doublets. All reactions were carried out under N2 atmosphere unless otherwise stated. HPLC-grade solvents were used for all reactions. Flash column chromatography was carried out using silica gel (230–400 mesh, Merck). Low-resolution mass spectra were obtained using an ESI technique on a 3200 Q Trap LC/MS/MS system (Applied Biosystems). The purity was assessed by using Shimadzu LC-MS with Waters XTerra MS C-18 column (part no. 186000538, 50 × 2.1 mm2), at a flow rate of 0.3 mL/min (λ = 250 and 220 nm; mobile phase A, 0.1% formic acid in H2O, and mobile phase B, 0.1% formic in 60% isopropanol, 30% CH3CN, and 9.9% H2O). All compounds submitted for testing were confirmed to be >95.0% purity by LC-MS traces.
The solution of (1S,3aR,6aS)-ethyl octahydrocyclopenta[c]pyrrole-1-carboxylate hydrochloride (1) (5 mmol) and NaHCO3 (12 mmol) in THF/H2O (30 mL, THF/H2O = 2:1) was cooled with an ice bath and CbzCl (6 mmol) was added. The reaction was stirred until TLC shows complete consumption of the starting material. The mixture was extracted with CH2Cl2. The combined organic layer was separated, dried over anhydrous Na2SO4, filtered, and concentrated under reduced pressure. The crude product was used for the next step directly. NMRs showed a diastereomer (dr) mixture was obtained (dr = 1:1). 1H NMR (400 MHz, CDCl3) δ 7.41–7.30 (m, 5H), 5.22–5.01 (m, 2H), 4.26–4.00 (m, 3H), 3.82–3.74 (m, 1H), 3.43, 3.36 (dd, J = 10.8, 3.2 Hz, 1H), 2.80–2.62 (m, 2H), 2.05–1.95 (m, 1H), 1.90–1.74 (m, 2H), 1.67–1.45 (m, 3H), 1.29, 1.17 (t, J = 7.2 Hz, 3H). 13C NMR (100 MHz, CDCl3) δ 172.82, 172.67, 155.16, 154.57, 136.76, 136.60, 128.72, 128.58, 128.44, 128.38, 127.90, 127.48, 126.93, 66.96, 65.96, 65.69, 61.01, 60.96, 53.30, 52.78, 49.33, 48.16, 42.47, 41.51, 32.95, 32.84, 32.35, 32.26, 25.53, 14.18, 14.10. ESI-MS C18H24NO4: m/z (M + H+): 318.2 (calculated), 318.2 (found).
To the solution of the above crude product in THF/H2O (30 mL, THF/H2O = 2:1) at room temperature was added LiOH (7.5 mmol). The reaction was stirred until TLC shows complete consumption of the starting material. After removing THF, the aqueous layer was washed with hexane/ethyl acetate (hexane/ethyl acetate = 4:1), and the organic layer was discarded. Then, the aqueous layer was adjusted to slightly acidic pH with 1 N HCl and the mixture was extracted with CH2Cl2/MeOH (CH2Cl2/MeOH = 15:1). The combined organic layer was separated, dried over anhydrous Na2SO4, filtered, and concentrated under reduced pressure. The obtained acid, 2, was pure enough for later steps.
The solution of acid 2 (1.05 mmol) and amine salt 3 (1 mmol) in DMF was cooled to 0 °C with ice batch. DIPEA (4 mmol) was added, followed by HCTU (1.1 mmol). The reaction was warmed to room temperature and stirred overnight. The reaction was added brine and extracted with ethyl acetate. The combined organic layer was successively washed with 1 N HCl, saturated aqueous NaHCO3, and brine. Then, the organic layer was dried over anhydrous Na2SO4, filtered, and concentrated under reduced pressure. The crude product was used for the next step directly.
The solution of the above crude product in THF (20 mL) was cooled with an ice bath. LiBH4 (5 mmol) was added, followed by ethanol (5 mL). The reaction was warmed to room temperature and stirred overnight. After removing THF, the residue was dissolved in water and the pH was adjusted with 1 N HCl to be slightly acidic. The mixture was extracted with CH2Cl2/MeOH (CH2Cl2/MeOH = 15:1). The combined organic layer was separated, dried over anhydrous Na2SO4, filtered, and concentrated under reduced pressure. The crude product was used for the next step directly.
The solution of the above crude product in CH2Cl2 (20 mL) was cooled to 0 °C with an ice bath. NaHCO3 (1.5 mmol) was added, followed by Dess-Martin Periodinane (DMP) (1.5 mmol). The reaction was warmed to room temperature and stirred until TLC shows complete consumption of the starting material. The reaction was quenched with saturated aqueous Na2S2O3, followed by saturated aqueous NaHCO3. The mixture was extracted with CH2Cl2/MeOH (CH2Cl2/MeOH = 15:1). The combined organic layer was separated, dried over anhydrous Na2SO4, filtered, and concentrated under reduced pressure. The residue was purified by silica gel flash column chromatography (CH2Cl2 to CH2Cl2/MeOH = 15:1) to afford the target product UAWJ9-36-1.
UAWJ9-36-1
Yield: 58% from carboxylic acid 2. 1H NMR (400 MHz, CDCl3, isomers) δ 9.53, 9.19 (s, 1H), 8.52, 8.13 (s, 1H),7.35 (m, 5H), 6.24, 6.02 (s, 1H), 5.31–4.97 (m, 2H), 4.51–4.02 (m, 2H), 3.89–3.69 (m, 1H), 3.47–3.23 (m, 3H), 2.85–2.65 (m, 2H), 2.57–2.12 (m, 2H), 2.12–1.91 (m, 2H), 1.90–1.78 (m, 4H), 1.70–1.58 (m, 2H), 1.56–1.44 (m, 1H). 13C NMR (100 MHz, CDCl3, isomers) δ 200.11, 199.86, 180.06, 173.99, 173.50, 155.41, 154.73, 136.67, 128.54, 128.45, 127.96, 127.83, 127.79, 67.35–66.77 (m), 58.24, 57.81, 55.08, 53.29–52.96 (m), 50.11, 50.07, 48.23, 48.18, 42.70, 42.63, 41.74, 41.66, 40.60, 40.46, 38.51, 38.05, 32.59, 32.55, 31.84, 31.53, 29.70, 29.60, 28.98, 28.82, 25.41, 25.26. HRMS C23H30N3O5 calculated for m/z [M + H]+: 428.21855. Found: 428.21800.
Compound 5
The title compound was synthesized using the same procedure described above. The installation of Cbz also afforded a dr mixture (dr = 1:1). 1H NMR (400 MHz, CDCl3) δ 7.39–7.26 (m, 5H), 5.24–4.97 (m, 2H), 3.77, 3.62 (s, 3H), 3.76–3.72 (m, 1H), 3.55, 3.52 (d, J = 10.8 Hz, 1H), 1.90–1.81 (m, 1H), 1.46–1.39 (m, 2H), 1.05 (s, 3H), 0.98, 0.98 (s, 3H). 13C NMR (100 MHz, CDCl3) δ 172.67, 1172.52, 154.20, 153.63, 136.68, 136.58, 128.71, 128.56, 127.91, 127.63, 127.59, 66.98, 66.92, 59.88, 59.54, 52.31, 52.17, 46.89, 46.34, 32.04, 31.07, 27.32, 26.49, 26.26, 26.24, 19.41, 19.36, 12.55. ESI-MS C17H22NO4: m/z (M + H+): 304.2 (calculated), 304.2 (found).
UAWJ9-36-3
Yield: 52% from carboxylic acid 5. 1H NMR (400 MHz, CDCl3, isomers) δ 9.53, 9.14, (s, 1 H) 8.67, 8.20 (d, J = 4.0 Hz, 1H), 7.41–7.28 (m, 5H), 6.35–5.90 (m, 1H), 5.36–4.93 (m, 2H), 4.45–4.07 (m, 2H), 3.89–3.71 (m, 1H), 3.71–3.16 (m, 4H), 2.54–1.78 (m, 5H), 1.62–1.36 (m, 2H), 1.07 (s, 3H), 0.96, 0.94 (s, 3H). 13C NMR (100 MHz, CDCl3, isomers) δ 200.15, 199.84, 180.09, 180.07, 173.49, 172.88, 154.54, 153.93, 136.65, 136.58, 128.57, 12845, 128.44, 128.04, 127.99, 127.95, 127.58, 98.46 (hemiacetal), 67.47, 67.12, 67.04, 61.44, 61.40, 58.48, 57.92, 55.17, 53.07, 50.83, 47.25, 46.83, 40.72, 40.57, 38.67, 38.08, 32.96, 31.50, 29.79, 29.55, 29.07, 28.83, 27.44, 27.36, 26.31, 26.23, 26.15, 25.96, 19.32, 19.20, 12.62, 12.58. HRMS C23H30N3O5 calculated for m/z [M + H]+: 428.21855. Found: 428.21800.
Cell Lines and Viruses
Human rhabdomyosarcoma (RD, ATCC CCL-136), Vero C1008 (ATCC CRL-1586), Huh-7 (University of Pittsburgh), and HEK293T expressing ACE2 (293T-ACE2, BEI Resources, NR-52511) cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM). Human fibroblast Cell Line, MRC-5 (ATCC CCL-171) was maintained in Eagle’s minimum essential medium (EMEM, ATCC 30–2003). Both media were supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin antibiotics. Cells were kept at cell culture incubator (humidified, 5% CO2/95% air, 37 °C). The following reagents were obtained through BEI Resources, NIAID, NIH: human coronavirus, OC43, NR-52725; human coronavirus, NL63, NR-470. HCoV-OC43 was propagated in RD cells; HCoV-NL63 was initially propagated in 293T-ACE2 cells and accommodated in Vero E6 cells. HCoV-229E was obtained from Dr. Bart Tarbet (Utah State University) and amplified in Huh-7 or MRC-5 cells.
Protein Expression and Purification
The genes encoding SARS-CoV-2 main protease (accession no.: 7BUY_A), SARS-CoV main protease (accession no.: 6W79_A), MERS-CoV main protease (accession no.: 5C3N_B), HCoV-229E main protease (accession no.: P0C6X1), HCoV-OC43 main protease (accession no.: QDH43723), HCoV-NL63 main protease (accession no.: 5GWY_A), HCoV-HKU1 main protease (accession no.: 3D23_D) were purchased from GenScript (Piscataway, NJ) with Escherichia coli codon optimization and inserted into pET29a(+) plasmid. The Mpro genes were then subcloned into the pE-SUMO plasmid as previously described.10 The expression and purification of all Mpro’s followed the same procedures as previously described.33 Cathepsin K (catalog no. 219461) and cathepsin L (catalog no. 219402) were purchased from EMD Millipore. Calpain I (catalog no. C6108) and trypsin (catalog no. T6763) were purchased from Sigma-Aldrich, and caspas-3 (catalog no. 1083–25) was purchased from BioVision (Milpitas, CA).
Differential Scanning Fluorimetry (DSF)
Direct binding of GC-376, UAWJ9-36-1, and UAWJ9-36-3 with SARS-CoV-2, SARS-CoV, MERS-CoV, HCoV-OC43, HCoV-229E, HCoV-NL63, and HCoV-HKU1 Mpro’s was detected by differential scanning fluorimetry (DSF) using a Thermal Fisher QuantStudio 5 Real-Time PCR System as previously described33 with minor modifications. Mpro’s were diluted in a buffer containing 20 mM HEPES, pH 6.5, 120 mM NaCl, 0.4 mM EDTA, 4 mM DTT, and 20% glycerol to a final concentration of 4 μM and incubated with serial concentrations of compounds (0.06–200 μM) at 30 °C for 1 h. DMSO was included as a reference. SYPRO orange (1×, Thermal Fisher, catalog no.: S6650) was added, and the fluorescence signal was recorded under a temperature gradient ranging from 20 to 95 °C (incremental step of 0.05 °C s–1). The melting temperature (Tm) was calculated as the mid log of the transition phase from the native to the denatured protein using a Boltzmann model in Protein Thermal Shift Software v1.3. ΔTm was calculated by subtracting reference melting temperature of proteins in the presence of DMSO from the Tm in the presence of compounds.
Enzymatic Assays
To determine the IC50 values for GC-376, UAWJ9-36-1, and UAWJ9-36-3, 100 nM SARS-CoV-2, MERS-CoV, SARS-CoV, HCoV-OC43, HCoV-229E, HCoV-NL63, or HCoV-HKU1Mpro was incubated with serial concentrations of the compounds at 30 °C for 30 min in the reaction buffer containing 20 mM HEPES, pH 6.5, 120 mM NaCl, 0.4 mM EDTA, 4 mM DTT, and 20% glycerol. The proteolytic reactions were initiated by adding 10 μM of substrate peptide and recorded in Cytation 5 imaging reader (Thermo Fisher Scientific) with filters for excitation at 360/40 nm and emission at 460/40 nm for 1 h. The initial velocity of the proteolytic reaction was calculated by linear regression for the first 15 min of the kinetic progress curves. IC50 curve fittings were carried out using log(concentration of compounds) versus the initial velocity with variable slopes in Prism 8.
Kinetic studies of the proteolytic reaction progress curves with GC-376, UAWJ9-36-1, and UAWJ9-36-3 were carried out as follows: First, 5 nM SARS-CoV-2 Mpro, 60 nM MERS-CoV Mpro, or 5 nM SARS-CoV Mpro was added into 20 μM substrate peptide premixed with serial concentrations of the compounds in 200 μL of reaction buffer at 30 °C to initiate the proteolytic reaction. The reaction was monitored for 4 h. The progression curves were fitted as previously described.33 The first 90 min of the kinetic curves were used in the curve fittings as substrate depletion was observed when proteolytic reactions carried out longer than 90 min.
Trypsin assay reactions were carried out as previously described,34 with minor modifications; 100 μL of reaction solution containing 100 nM Trypsin (Millipore sigma, catalog no.: T6763), 50 mM HEPES (pH7.6), and serial concentrations of GC-376, UAWJ9-36-1, and UAWJ9-36-3 (0, 0.02, 0.06, 0.2, 0.6, 2, 6, and 20 μM) or Camostat (0, 0.002, 0.006, 0.02, 0.06, 0.2, 0.6, and 2 μM) were incubated at 30 °C for 30 min. The reactions were initiated by adding 100 μM Bz-Arg-AMC·HCl (BACHEM, Product No.: 4002540.0050). Fluorescence signal intensities were recorded for 20 min using a Biotek Cytation 3 plate reader (Thermo Fisher Scientific) with filters for excitation at 360/40 nm and emission at 460/40 nm, and the initial velocity was calculated for the first 10 min by linear regression. The IC50 values were determined by curve fittings using log(concentration of compounds) versus the initial velocity with variable slopes in Prism 8.
Calpain I, cathepsin L, and cathepsin K enzymatic assays were carried as previously described.34
The caspase-3 enzymatic assay was carried out as follows: 1 unit of caspase-3 protein was diluted into 1600 μL of reaction buffer (20 mM HEPES pH7.4, 2 mM EDTA, 0.1% CHAPS, and 5 mM DTT); 100 μL diluted protein was incubated with 1 μL various concentration of testing compounds for 30 min at 30 °C; the enzymatic reaction was initiated by adding 1 μL of 2 mM Ac-DEVD-AFC (Medchemexpress, catalog no. HY-P1005). The reaction was monitored a Molecular Devices SpectraMax iD3 plate reader with excitation at 400 nm and emission at 505 nm at 30 °C for 1 h. The IC50 values were calculated as described in the previous section.
Cellular-Based FlipGFP Mpro Assay
Plasmid pcDNA3-TEV-flipGFP-T2A-mCherry was purchased from Addgene (catalog no. 124429). SARS-CoV-2 Mpro cleavage site (AVLQSGFR) and SARS-CoV-2 PLpro cleavage site (LRGGAPTK) were introduced into pcDNA3-flipGFP-T2A-mCherry via overlapping PCRs to generate a fragment with SacI and HindIII sites at the ends. SARS-CoV-2 Mpro and PLPro expression plasmids pcDNA3.1 SARS-CoV-2 Mpro and pcDNA3.1 SARS-CoV-2 PLPro were ordered from Genscript (Piscataway NJ) with codon optimization. pcDNA3.1 SARS-CoV-2 Mpro-C145A was generated by site-directed Quikchange mutagenesis.
First, 293T cells were seeded in 96-well black, clear-bottomed Greiner plate (catalog no. 655090) and reached 70–90% confluency overnight. Next, 50 ng of pcDNA3-flipGFP-T2A-mCherry plasmid with TEV, PLpro, or Mpro cleavage site and 50 ng of protease expression plasmid pcDNA3.1 SARS-CoV-2 Mpro or SARS-CoV-2 PLpro were transfected into 293T cells with transfection reagent TransIT-293 (Mirus catalog no. MIR 2700) according to the manufacturer’s protocol. Three hours after transfection, 1 μL of testing compound was added to each well at 100-fold dilution. Two days after transfection, images were taken with Cytation 5 imaging reader (Biotek) using GFP and mCherry channels via 10× objective lens and were analyzed with Gen5 3.10 software (Biotek). SARS-CoV-2 Mpro protease activity was calculated as the ratio of GFP signal sum intensity over mCherry signal sum intensity. Testing compounds efficacy (IC50) in cells was calculated by plotting GFP/mCherry signal over the applied compound concentration with a 4-parameter dose–response function in prism 8. The mCherry signal alone in the presence of testing compounds was utilized to evaluate the compound cytotoxicity.
Antiviral Assays
The antiviral activities of GC-376, UAWJ9-36-1, and UAWJ9-36-3 against HCoV-229E and HCoV-NL63 were detected via the CPE assay as previously described.33,38 Briefly, near-confluent MRC-5 cells and Vero C1008 cells in 96-well plates were infected with 100 μL of HCoV-229E and HCoV-NL63 at desired dilutions and incubated at 33 or 37 °C for 1 h. Different concentrations of testing compounds (0, 0.015, 0.05, 0.15, 0.5, 1.5, 3, 5, and 15 μM) were added, and the infected cells were incubated for another 3–5 days until significant cytopathic effect was observed in the wells without compound (virus only). The growth medium was removed, and cells were stained with 0.1 mg/mL neutral red for 2 h. Excess dye was rinsed from the cells with PBS. The neutral red dye which was taken up was extracted from the cells with a buffer containing 50% ethanol and 1% glacial acetic acid. The absorbance of neutral red dye at 540 nm was measured on a spectrometer. The antiviral activity of GC-376, UAWJ9-36-1, and UAWJ9-36-3 was tested against HCoV-OC43 in plaque assay. RD cells were infected with HCoV-OC43 and incubated at 33 °C for 1 h to allow virus adsorption. The viral inoculum was removed, and an overlay containing 0.2% Avicel supplemented with 2% FBS in DMEM containing serial concentrations of testing compounds (0, 0.001, 0.003, 0.01, 0.03, 0.1, 0.3, and 1 μM) was added and incubated in the 33 °C incubator for 4–5 days. The plaque formation was detected by staining the cell monolayer with crystal violet, and the plaque areas were quantified using ImageJ. EC50 values were determined by plotting the percent CPE versus log10 compound concentrations from best-fit dose response curves with variable slope in Prism 8.
SARS-CoV-2 Mpro Crystallization and Structure Determination
SARS-CoV-2 Mpro and HM-Mpro protein was purified, and crystals were grown as previously described.9,10 X-ray diffraction data was collected on the Structural Biology Center 19-ID beamline at the Advanced Photon Source in Argonne, IL, and processed with the iMosflm. The CCP4 version of MOLREP was used to solve the structures of UAWJ9-36-1 + SARS-CoV-2 Mpro using 7KX5 as a reference model and UAWJ9-36-3 + SARS-CoV-2 HM-Mpro with 6XBI as a reference model. Structures were then refined with REFMAC5 and built with COOT.39,40 All protein structure figures were generated with PyMOL (Schrödinger, LLC).
Acknowledgments
This research was partially supported by the National Institutes of Health (NIH) (grants AI147325 and AI157046) and the Arizona Biomedical Research Centre Young Investigator grant (ADHS18-198859) to J.W. We thank Michael Kemp for assistance with crystallization and X-ray diffraction data collection. We also thank the staff members of the Advanced Photon Source of Argonne National Laboratory, particularly those at the Structural Biology Center (SBC), with X-ray diffraction data collection. SBC-CAT is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. The SARS-CoV-2 experiments were supported by a COVID-19 pilot grant from UTHSCSA and NIH grant AI151638 to Y.X. SARS-related Coronavirus 2, Isolate USA-WA1/2020 (NR-52281) was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsptsci.1c00099.
Crystallization statistics; H and C NMR; HPLC traces; HRMS (PDF)
Author Contributions
J.W. conceived and designed the study; Z.X. synthesized and characterized the compounds. M.S. carried out Mpro crystallization and structure determination, and analyzed the data with Y.C. Y.H. carried out the enzymatic assay, thermal shift binding assay, and antiviral assay against HCoV-OC43, HCoV-NL63, and HCoV-229E. C.M. expressed the SARS-CoV-2 Mpro and PLpro and carried out Flip-GFP assay and the counter-screening against host proteases. T.S. expressed the Mpro’s from MERS, NL63, 229E, and HKU1. X.M. and F.Z. carried out the SARS-CoV-2 immunofluorescence assay in Vero E6 and Caco2-ACE2 cells under the guidance of Y.X.; J.W. and Y.C. secured funding and supervised the study. J.W. wrote the manuscript with the input from others.
The authors declare the following competing financial interest(s): Z.X. and J.W. are inventors of a filled patent claiming the use of UAWJ9-36-1 and UAWJ9-36-3 and related compounds as potential COVID-19 antiviral drugs.
This article is made available via the ACS COVID-19 subset for unrestricted RESEARCH re-use and analyses in any form or by any means with acknowledgement of the original source. These permissions are granted for the duration of the World Health Organization (WHO) declaration of COVID-19 as a global pandemic.
Supplementary Material
References
- Petersen E.; Koopmans M.; Go U.; Hamer D. H.; Petrosillo N.; Castelli F.; Storgaard M.; Al Khalili S.; Simonsen L. (2020) Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect. Dis. 20, e238–e244. 10.1016/S1473-3099(20)30484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse J. S.; Lalonde T.; Xu S.; Liu W. R. (2020) Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. ChemBioChem 21, 730–738. 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J. H.; Tomashek K. M.; Dodd L. E.; Mehta A. K.; Zingman B. S.; Kalil A. C.; Hohmann E.; Chu H. Y.; Luetkemeyer A.; Kline S.; et al. (2020) Remdesivir for the treatment of Covid-19 - final report. N. Engl. J. Med. 383, 1813–1826. 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinner C. D.; Gottlieb R. L.; Criner G. J.; Arribas López J. R.; Cattelan A. M.; Soriano Viladomiu A.; Ogbuagu O.; Malhotra P.; Mullane K. M.; Castagna A.; et al. (2020) Effect of remdesivir vs standard care on clinical status at 11 days in patients with moderate COVID-19: a randomized clinical trial. JAMA 324, 1048–1057. 10.1001/jama.2020.16349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium W. H. O. S. T.; Pan H.; Peto R.; Henao-Restrepo A. M.; Preziosi M. P.; Sathiyamoorthy V.; Abdool Karim Q.; Alejandria M. M.; Hernandez Garcia C.; Kieny M. P.; et al. (2021) Repurposed antiviral drugs for Covid-19 - interim WHO solidarity trial results. N. Engl. J. Med. 384, 497–511. 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T. P.; Sims A. C.; Zhou S.; Graham R. L.; Pruijssers A. J.; Agostini M. L.; Leist S. R.; Schafer A.; Dinnon K. H. 3rd; Stevens L. J.; Chappell J. D.; Lu X.; Hughes T. M.; George A. S.; Hill C. S.; Montgomery S. A.; Brown A. J.; Bluemling G. R.; Natchus M. G.; Saindane M.; Kolykhalov A. A.; Painter G.; Harcourt J.; Tamin A.; Thornburg N. J.; Swanstrom R.; Denison M. R.; Baric R. S. (2020) An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 12, eabb5883. 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R. M.; Wolf J. D.; Plemper R. K. (2021) Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets. Nat. Microbiol. 6, 11–18. 10.1038/s41564-020-00835-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toots M.; Yoon J.-J.; Cox R. M.; Hart M.; Sticher Z. M.; Makhsous N.; Plesker R.; Barrena A. H.; Reddy P. G.; Mitchell D. G.; Shean R. C.; Bluemling G. R.; Kolykhalov A. A.; Greninger A. L.; Natchus M. G.; Painter G. R.; Plemper R. K. (2019) Characterization of orally efficacious influenza drug with high resistance barrier in ferrets and human airway epithelia. Sci. Transl. Med. 11, eaax5866. 10.1126/scitranslmed.aax5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C.; Sacco M. D.; Hurst B.; Townsend J. A.; Hu Y.; Szeto T.; Zhang X.; Tarbet B.; Marty M. T.; Chen Y.; Wang J. (2020) Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 30, 678–692. 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco M. D.; Ma C.; Lagarias P.; Gao A.; Townsend J. A.; Meng X.; Dube P.; Zhang X.; Hu Y.; Kitamura N.; Hurst B.; Tarbet B.; Marty M. T.; Kolocouris A.; Xiang Y.; Chen Y.; Wang J. (2020) Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M(pro) and cathepsin L. Sci. Adv. 6, eabe0751. 10.1126/sciadv.abe0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas B. T.; Durie I. A.; Murray J.; Longo J. E.; Miller H. C.; Crich D.; Hogan R. J.; Tripp R. A.; Pegan S. D. (2020) Characterization and noncovalent inhibition of the deubiquitinase and deISGylase activity of SARS-CoV-2 papain-like protease. ACS Infect. Dis. 6, 2099–2109. 10.1021/acsinfecdis.0c00168. [DOI] [PubMed] [Google Scholar]
- Shin D.; Mukherjee R.; Grewe D.; Bojkova D.; Baek K.; Bhattacharya A.; Schulz L.; Widera M.; Mehdipour A. R.; Tascher G.; Geurink P. P.; Wilhelm A.; van der Heden van Noort G. J.; Ovaa H.; Müller S.; Knobeloch K.-P.; Rajalingam K.; Schulman B. A.; Cinatl J.; Hummer G.; Ciesek S.; Dikic I. (2020) Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 587, 657–662. 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm T.; Ebert G.; Calleja D. J.; Allison C. C.; Richardson L. W.; Bernardini J. P.; Lu B. G.; Kuchel N. W.; Grohmann C.; Shibata Y.; et al. (2020) Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 39, e106275. 10.15252/embj.2020106275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N. C.; Kim Y.; Liu H.; Galasiti Kankanamalage A. C.; Eckstrand C.; Groutas W. C.; Bannasch M.; Meadows J. M.; Chang K. O. (2018) Efficacy of a 3C-like protease inhibitor in treating various forms of acquired feline infectious peritonitis. J. Feline Med. Surg. 20, 378–392. 10.1177/1098612X17729626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.; Liu H.; Galasiti Kankanamalage A. C.; Weerasekara S.; Hua D. H.; Groutas W. C.; Chang K. O.; Pedersen N. C. (2016) Reversal of the progression of fatal coronavirus infection in cats by a broad-spectrum coronavirus protease inhibitor. PLoS Pathog. 12, e1005531. 10.1371/journal.ppat.1005531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathnayake A. D.; Zheng J.; Kim Y.; Perera K. D.; Mackin S.; Meyerholz D. K.; Kashipathy M. M.; Battaile K. P.; Lovell S.; Perlman S.; Groutas W. C.; Chang K. O. (2020) 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice. Sci. Transl. Med. 12, eabc5332. 10.1126/scitranslmed.abc5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boras B., Jones R. M., Anson B. J., Arenson D., Aschenbrenner L., Bakowski M. A., Beutler N., Binder J., Chen E., and Eng H.. et al. (February 12, 2021) Discovery of a novel inhibitor of coronavirus 3CL protease as a clinical candidate for the potential treatment of COVID-19. bioRxiv (Pharmacology and Toxicology), 2020.2009.2012.293498, ver. 3. DOI: 10.1101/2020.09.12.293498.
- Qiao J.; Li Y. S.; Zeng R.; Liu F. L.; Luo R. H.; Huang C.; Wang Y. F.; Zhang J.; Quan B.; Shen C.; Mao X.; Liu X.; Sun W.; Yang W.; Ni X.; Wang K.; Xu L.; Duan Z. L.; Zou Q. C.; Zhang H. L.; Qu W.; Long Y. H.; Li M. H.; Yang R. C.; Liu X.; You J.; Zhou Y.; Yao R.; Li W. P.; Liu J. M.; Chen P.; Liu Y.; Lin G. F.; Yang X.; Zou J.; Li L.; Hu Y.; Lu G. W.; Li W. M.; Wei Y. Q.; Zheng Y. T.; Lei J.; Yang S. (2021) SARS-CoV-2 M(pro) inhibitors with antiviral activity in a transgenic mouse model. Science 371, 1374–1378. 10.1126/science.abf1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dampalla C. S., Zhang J., Perera K. D., Wong L.-Y. R., Meyerholz D. K., Nguyen H. N., Kashipathy M. M., Battaile K. P., Lovell S., and Kim Y.. et al. (February 5, 2021) Post-infection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection. bioRxiv (Microbiology), 2021.2002.2005.429937. DOI: 10.1101/2021.02.05.429937. [DOI] [PMC free article] [PubMed]
- Fu L.; Ye F.; Feng Y.; Yu F.; Wang Q.; Wu Y.; Zhao C.; Sun H.; Huang B.; Niu P.; Song H.; Shi Y.; Li X.; Tan W.; Qi J.; Gao G. F. (2020) Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 11, 4417. 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong W.; Khan M. B.; Fischer C.; Arutyunova E.; Lamer T.; Shields J.; Saffran H. A.; McKay R. T.; van Belkum M. J.; Joyce M. A.; Young H. S.; Tyrrell D. L.; Vederas J. C.; Lemieux M. J. (2020) Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 11, 4282. 10.1038/s41467-020-18096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneller D. W.; Galanie S.; Phillips G.; O’Neill H. M.; Coates L.; Kovalevsky A. (2020) Malleability of the SARS-CoV-2 3CL Mpro active-site cavity facilitates binding of clinical antivirals. Structure 28, 1313–1320. 10.1016/j.str.2020.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao D. S.; Arkala A. K. R.; Madhavi Y. V.; Nagaraju M.; Gade S. R.; Kumar U. K. S.; Bandichhor R.; Dahanukar V. H. (2015) Synthesis and process optimization of boceprevir: a protease inhibitor drug. Org. Process Res. Dev. 19, 1559–1567. 10.1021/op500065t. [DOI] [Google Scholar]
- Steuten K., Kim H., Widen J. C., Babin B. M., Onguka O., Lovell S., Bolgi O., Cerikan B., Neufeldt C. J., and Cortese M.. et al. (2021) Challenges for targeting SARS-CoV-2 proteases as a therapeutic strategy for COVID-19. ACS Infect. Dis. 10.1021/acsinfecdis.0c00815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C.; Hu Y.; Townsend J. A.; Lagarias P. I.; Marty M. T.; Kolocouris A.; Wang J. (2020) Ebselen, disulfiram, carmofur, PX-12, tideglusib, and shikonin are nonspecific promiscuous SARS-CoV-2 main protease inhibitors. ACS Pharmacol. Transl. Sci. 3, 1265–1277. 10.1021/acsptsci.0c00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C.; Wang J. (2021) Dipyridamole, chloroquine, montelukast sodium, candesartan, oxytetracycline, and atazanavir are not SARS-CoV-2 main protease inhibitors. Proc. Natl. Acad. Sci. U. S. A. 118, e2024420118. 10.1073/pnas.2024420118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Lidsky P., Xiao Y., Wu C.-T., Garcia-Knight M., Yang J., Nakayama T., Nayak J. V., Jackson P. K., Andino R., and Shu X. (March 16, 2020) Ethacridine inhibits SARS-CoV-2 by inactivating viral particles in cellular models. bioRxiv (Pharmacology and Toxicology), 2020.2010.2028.359042. DOI: 10.1101/2020.10.28.359042.
- Froggatt H. M.; Heaton B. E.; Heaton N. S. (2020) Development of a fluorescence-based, high-throughput SARS-CoV-2 3CLpro reporter assay. J. Virol. 94, e01265–01220. 10.1128/JVI.01265-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M.; Kleine-Weber H.; Schroeder S.; Kruger N.; Herrler T.; Erichsen S.; Schiergens T. S.; Herrler G.; Wu N. H.; Nitsche A.; et al. (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–278. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S.; Glowacka I.; Blazejewska P.; Soilleux E.; Allen P.; Danisch S.; Steffen I.; Choi S. Y.; Park Y.; Schneider H.; Schughart K.; Pohlmann S. (2010) TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. J. Virol. 84, 10016–10025. 10.1128/JVI.00239-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanifer M. L.; Kee C.; Cortese M.; Zumaran C. M.; Triana S.; Mukenhirn M.; Kraeusslich H. G.; Alexandrov T.; Bartenschlager R.; Boulant S. (2020) Critical role of type III interferon in controlling SARS-CoV-2 infection in human intestinal epithelial cells. Cell Rep. 32, 107863. 10.1016/j.celrep.2020.107863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa M. F.; Sillence D.; Ackerley C.; Lingwood C. (2004) Role of multiple drug resistance protein 1 in neutral but not acidic glycosphingolipid biosynthesis. J. Biol. Chem. 279, 7867–7876. 10.1074/jbc.M305645200. [DOI] [PubMed] [Google Scholar]
- Hu Y.; Ma C.; Szeto T.; Hurst B.; Tarbet B.; Wang J. (2021) Boceprevir, calpain inhibitors II and XII, and GC-376 have broad-spectrum antiviral activity against coronaviruses. ACS Infect. Dis. 7, 586–597. 10.1021/acsinfecdis.0c00761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura N., Sacco M. D., Ma C., Hu Y., Townsend J. A., Meng X., Zhang F., Zhang X., Ba M., and Szeto T.. et al. (2021) Expedited approach towards the rational design of non-covalent SARS-CoV-2 main protease inhibitors. J. Med. Chem. 10.1021/acs.jmedchem.1c00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatansever E. C.; Yang K. S.; Drelich A. K.; Kratch K. C.; Cho C.-C.; Kempaiah K. R.; Hsu J. C.; Mellott D. M.; Xu S.; Tseng C.-T. K.; Liu W. R. (2021) Bepridil is potent against SARS-CoV-2 in vitro. Proc. Natl. Acad. Sci. U. S. A. 118, e2012201118. 10.1073/pnas.2012201118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Lin D.; Kusov Y.; Nian Y.; Ma Q.; Wang J.; von Brunn A.; Leyssen P.; Lanko K.; Neyts J.; de Wilde A.; Snijder E. J.; Liu H.; Hilgenfeld R. (2020) Alpha-ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: structure-based design, synthesis, and activity assessment. J. Med. Chem. 63, 4562–4578. 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]
- Siklos M.; BenAissa M.; Thatcher G. R. J. (2015) Cysteine proteases as therapeutic targets: does selectivity matter? A systematic review of calpain and cathepsin inhibitors. Acta Pharm. Sin. B 5, 506–519. 10.1016/j.apsb.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.; Meng X.; Zhang F.; Xiang Y.; Wang J. (2021) The in vitro antiviral activity of lactoferrin against common human coronaviruses and SARS-CoV-2 is mediated by targeting the heparan sulfate co-receptor. Emerging Microbes Infect. 10, 317–330. 10.1080/22221751.2021.1888660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G. N.; Skubak P.; Lebedev A. A.; Pannu N. S.; Steiner R. A.; Nicholls R. A.; Winn M. D.; Long F.; Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr., Sect. D: Biol. Crystallogr. 67, 355–367. 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 60, 2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.