Abstract
Maintenance of optimal bone mass is controlled through the concerted functions of several cell types, including bone resorbing osteoclasts. Osteoclasts function to remove calcified tissue during developmental bone modeling, and degrade bone at sites of damage during bone remodeling. Changes to bone homeostasis can arise with alterations in osteoclastogenesis and/or catabolic activity that are not offset by anabolic activity; thus, factors that regulate osteoclastogenesis and bone resorption are of interest to further our understanding of basic bone biology, and as potential targets for therapeutic intervention. Several key cytokines, including Rankl and M-csf, as well as co-stimulatory factors elicit kinase signaling cascades that promote osteoclastogenesis. These kinase cascades are offset by the action of protein phosphatases, including members of the serine/threonine phosphatase family. Here we review the functions of serine/threonine phosphatases and their control of osteoclast differentiation and function, while highlighting deficiencies in our understanding of this understudied class of proteins within the field.
Keywords: Rankl, M-csf, phosphatase inhibitor, kinase, osteoclast, bone remodeling, myeloid lineage
Osteoclasts and Bone Remodeling
Osteoclasts play a major role in the bone remodeling cycle, being responsible for the destruction of old and damaged bone (1). Osteoclasts are multinucleated cells derived from monocyte/macrophage lineage precursors (1). Two cytokines are integral for the maturation of precursors to bone resorbing osteoclasts: receptor activator of nuclear factor-κB (NF-κB) ligand (RANKL, Tnfsf, Trance, Opgl) (1,2) and macrophage colony-stimulating factor (M-CSF or Csf1) (1,3). Produced mainly by mesenchymal-lineage cells such as osteoblasts and osteocytes (1,4), RANKL and M-CSF (1,2) contribute to the proliferation, survival and differentiation of osteoclast precursors and promote cytoskeletal rearrangement required for proper bone resorption (1,3). Differentiation occurs when RANKL binds to its cognate receptor RANK and M-CSF binds to colony-stimulating factor 1 receptor (c-fms), eliciting kinase-mediated signaling pathways that initiate the process (5). Osteoprotegerin (OPG) acts as an inhibitor of RANKL (1,2), which can be enhanced by pro-inflammatory cytokines and prostaglandins that simultaneously suppress OPG activity, resulting in a net gain of osteoclast activity (1). Maintenance of optimal bone mass requires tight regulation of kinase cascades, with dysregulation contributing to tissue dysfunction.
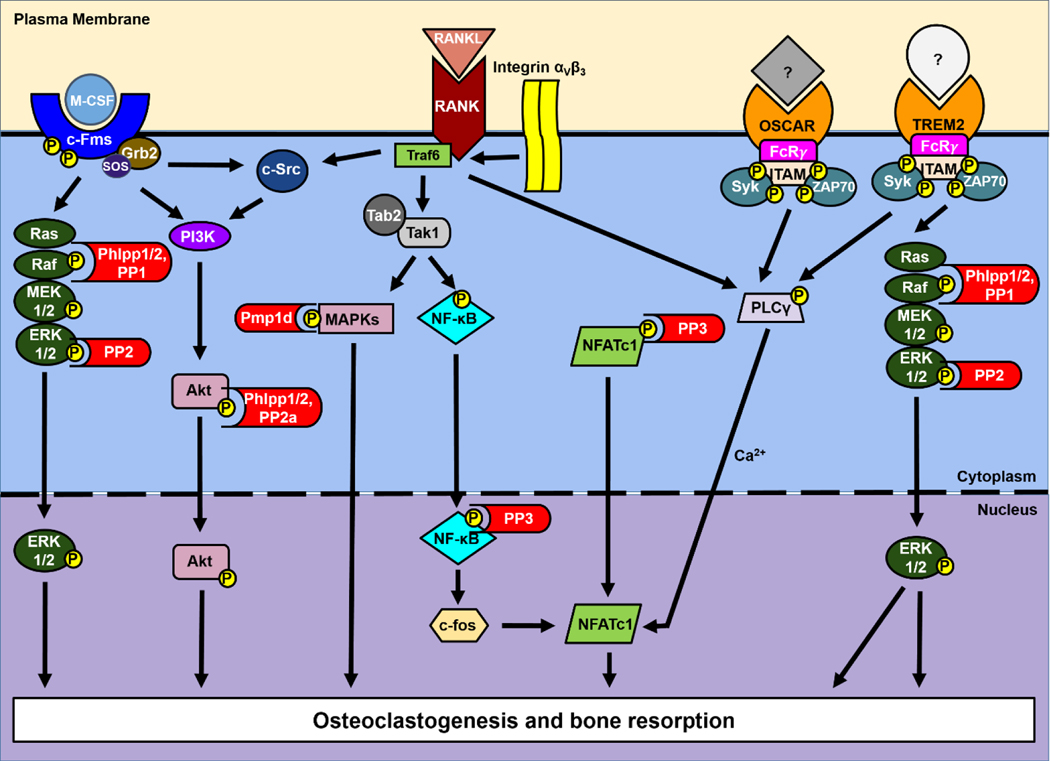
A wide array of kinase-dependent signaling pathways orchestrate osteoclast function (Figure 1), including those initiated following RANKL ligation to RANK. Signaling elicited by RANKL during osteoclastogenesis ultimately induces gene expression mediated by c-fos and nuclear factor-activated T cells c1 (NFATc1)-dependent transcription (6), but the impact of RANK activation first requires kinases activity. RANK signaling is facilitated via its association with tumor necrosis factor receptor (TNRF)-associated factor (TRAF) adaptor proteins, including TRAFs 1, 2, 3, 5 and 6. TRAF6 aids in activation of the kinase transforming growth factor β (Tgfβ)-activated kinase 1 (TAK1) via TAK binding protein 2 (TAB2) (7). This in turn activates downstream mitogen-activated protein kinases (MAPKs), such as p38 MAPK and JNK (Figure 1). Both canonical RelA/p50 and alternate RelB/p55 NF-κB signaling pathways are activated downstream of RANK via TRAF6 and TRAF2/5, respectively (4,6). TRAF6 activates the canonical pathway via TAK1-dependent phosphorylation of IκB kinase (IKK), leading to phosphorylation and degradation of inhibitory κB (IκB). Loss of IκB frees NF-κB and allows its translocation to the nucleus (8,9). In addition, TRAF6 recruits and induces activity of the receptor tyrosine kinase Proto-oncogene tyrosine-protein kinase (c-Src) via integrin αVβ3, leading to cytoskeletal reorganization, required for osteoclast-mediated bone resorption (4,10), as well as downstream activation of phosphoinositide 3-kinase (PI3K), and protein kinase B (Akt) (11). Activation of PI3K/Akt signaling downstream of RANKL induces expression of NFATc1 to facilitate osteoclast differentiation and promotes osteoclast migration (12,13). Thus, these kinase-mediated pathways are critical for RANKL-dependent osteoclast formation and function.
Figure 1. PSPs regulate signaling regulating osteoclastogenesis and bone resorption.
Multiple pathways downstream of M-CSF, RANKL and associated co-stimulator pathways are tempered by PSPs.
M-CSF-dependent signaling induced by ligand binding to c-fms is required for the definitive formation of osteoclasts and promotes their function and survival (Figure 1). This is highlighted by the op/op mouse phenotype, resulting from a point mutation within the Csf1 gene and production of non-functional M-CSF; these mice are characterized by lack of osteoclasts and severe osteopetrosis (14–16). PI3K, c-Src and growth factor receptor-bound protein 2/son of sevenless (Grb2/Sos) all associate with c-fms following receptor dimerization and autophosphorylation induced by M-CSF binding (17). Induction of PI3K activity plays a central role in osteoclast cytoskeletal organization and ruffled membrane formation (18), and c-Src activation plays a role in the regulation of resorptive organelles, such as the ruffled membrane (2). Akt, a downstream target of PI3K, mediates RANKL and/or M-CSF-stimulated proliferation and survival of osteoclast lineage cells (18). Association of the Grb2/Sos complex to c-fms results in a Ras-mediated MAPK signaling cascade ending in the phosphorylation and activation of extracellular signal-regulated kinase (ERK1/2), leading to osteoclast proliferation and survival (4,9).
While RANKL and M-CSF are sufficient to induce osteoclastogenesis, several co-stimulatory pathways help to promote osteoclast differentiation and bone remodeling. These include the immunoreceptor tyrosine-based activation motif (ITAM) costimulatory molecules Dap12, FcγR, and Oscar, which promote calcium-mediated activation of NFATc1 and greater osteoclast activity (Figure 1) (19). DNAX activating protein of 12 kDa (Dap12, also called Karap or Tyrobp) associates with triggering receptors expressed by myeloid cells (TREM)2 and contains a tyrosine-based motif that acts as a docking site for Syk and Zap70 tyrosine kinases. This promotes the recruitment and activation of P13K, phospholipase Cγ (PLCγ)1, and ERK1/2 pathways, resulting in the differentiation of osteoclast precursors, namely the fusion of macrophages into multinucleated giant cells (20,21). Fcγ receptor (FcγR) is constitutively expressed by monocytes, macrophages, and myeloid progenitor cells (22). Like Dap12, the FcγR chain contains ITAMs that serve as a docking site for Syk family kinases and Zap70 that recruit PLCγ1 to activate NFATc1 (22). The γ-chain of FcγR facilitates signaling and transport of IgG Fc receptors to the cell surface, though knowledge of the role of the latter in osteoclast activity is incomplete (19). Mice with severe osteopetrosis demonstrate the critical role of FcRγ and Dap12 during osteoclast activation when both factors are lacking (19). The phenotype is less pronounced in mice lacking only Dap12, whereas FcRγ-deficient mice show no disease phenotype, suggesting that FcRγ plays an important role in osteoclastogenesis in conjunction with Dap12 (19). Osteoclast-associated receptor (Oscar) is an IgG-like receptor upregulated in the presence of FcγR and acts as a costimulatory factor with a role in osteoclast differentiation (23). Association of Oscar with FcγR enhances activation of NFATc1 (23). While FcγR deficiency does not affect osteoclast development on its own, blocking Oscar activity inhibits osteoclastogenesis in vitro, suggesting that Oscar activated FcRγ-mediated signal transduction may not be essential for osteoclast development, but may be involved in an alternative signaling pathway for osteoclast differentiation (23). Taken together, the activation of Trem2 and Oscar by the binding of (as of yet) unknown ligands allows for the tyrosine phosphorylation of Dap12 and FcRγ, leading to the recruitment of kinases Syk and Zap70, which in turn activate PLCγ and calcium oscillations which activate NFATc1, with osteoclast differentiation as the concluding event (24).
Other cytokines can influence the differentiation and activity of osteoclasts. The T-cell cytokines interferon (Ifn) γ and interleukins (IL) 4 and 10 are known suppressors of osteoclast formation and all involve protein kinases within their signal cascades (1,18). Several studies identify many other factors involved in osteoclast differentiation and function including a variety of ILs, granulocyte macrophage colony-stimulating factor (GM-CSF), IFNβ, stromal cell-derived factor 1 (Sdf-1), monocyte chemoattractant protein 1 (Mcp-l), transforming growth factor β (Tgfβ), a range of Toll-like receptor ligands, Wnt ligands, and semaphorins (1,25,26). IL-1, like M-CSF and RANKL, has been shown to play a role in stimulating bone resorption (1,3). IL-1 activates NF-κB signaling and the JNK and p38 MAPK kinase pathways which induce the expression of canonical IL-1 target genes by transcriptional and posttranscriptional mechanisms (27). This is just a fraction of the complexity that can be found within the signaling pathways of bone remodeling and osteoclastogenesis, complexity that is deepened further by the involvement of protein serine/threonine phosphatases.
Serine/Threonine Protein Phosphatases
Activation of key kinase-activated signal transduction pathways facilitates osteoclast differentiation and their bone resorbing activity; in turn, these pathways must also be dampened by dephosphorylation of protein substrates. This action is performed by protein phosphatases. Whereas hundreds of protein kinases have been identified, only a handful of protein phosphatases are known (28). Though few in number, protein phosphatases are implicated in a plethora of cellular functions, including those involved in bone resorption. Protein phosphatases dephosphorylate amino acid residues via nucleophilic attack by a metal-activated water molecule, resulting in the hydrolysis of the substrate phosphate group (29). Water is split in the reaction, producing a phosphate ion and a protonated hydroxyl group on the amino acid residue (29). The activity of protein phosphatases are specified by regulatory subunits, interacting factors, and inhibitors. Protein phosphatases are classified by the kind of substrates to which they bind. These include protein serine/threonine phosphatases (PSPs), which are split into three major families: phosphoprotein phosphatases (PPPs), metal-dependent protein phosphatases (PPMs), and TFIIF-associating component of RNA polymerase II CTD phosphatase/ small CTD phosphatase (FCP/SCP) (Figure 2).
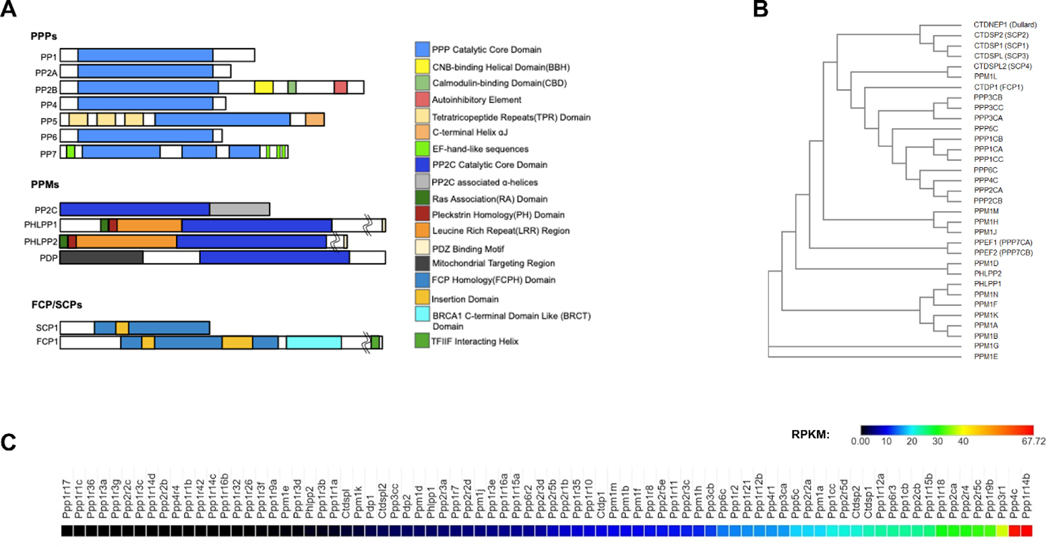
Figure 2. PSP Structure, relatedness and expression in murine osteoclasts.
(A) Conserved catalytic core domains and other functional domains of PSPs within each of three classes. (B) Phylogenetic tree comparing relatedness of PSPs was generated using Clustal Omega. See Supplemental Table 1 for sequences utilized. (C) High throughput RNA sequencing was performed using RNA generated from ex vivo generated murine osteoclasts from female C57Bl/6 mice. Shown is a heat map of all PSPs expressed.
Phosphoprotein Phosphatases
Phosphoprotein Phosphatases (PPPs) are the largest family of serine/threonine phosphatases. Within this family, each PPP is distinguished by highly conserved regulatory subunits that are unique to each type (Figure 2A). PPP binding is mediated by sequences called short linear motifs (SLiMs) that reside in regions devoid of secondary structure within regulatory subunits (30). SLiMs are unique to a single PPP, universal across species, degenerate in their amino acid sequence to allow for variations in affinities among regulatory subunits, and flexible such that use of different SLiMs would allow for more than one regulator to be accommodated on any one catalytic subunit (30). SLiMs for PPP members PP1 (KIQF and RVxF), PP2 (LSPIxE), and PP3 (LxVP) are known, but those for other PPPs are as of yet undiscovered or their method of recognizing other subunits is not understood (30). Notable members of the PPPs include PP1, PP2, Calcineurin, PP4, PP5, PP6, and PP7. The wide structural and functional variety of PPPs lends them well to an array of cellular activities and processes, including osteoclastogenesis and bone resorption.
Protein Phosphatase 1
Protein Phosphatase 1 (PP1) is a multi-protein complex composed of three catalytic subunits, PP1α, PP1β and PP1γ, and associated regulatory subunits (31). PP1 (PP1a, ppp1c) is universally expressed in eukaryotes and plays a role in numerous cellular processes such as meiosis, cell division, apoptosis, protein synthesis, metabolism, cytoskeletal reorganization and the regulation of membrane receptors and channels (32). The diverse functions of PP1 are made possible through use of varied regulatory subunits (32). PP1 enzymatic activity is inhibited by inhibitor-1, inhibitor-2, okadaic acid, microcystin, and tautomycetin (32,33). PP1 dephosphorylates a wide variety of substrates including Glycogen synthase (34), Glycogen phosphorylase (34), Calmodulin dependent protein kinase II (35), Per1/2 (36), Cenep (37), Atg16l1 (38) and FOXP3 (39). In osteoclasts, PP1 dephosphorylates the inhibitory Ser595 residue of Raf kinases, thereby promoting downstream MAPK signaling (Figure 1) (40,41).
While the role of PP1 isoforms within the osteoclast lineage is understudied, PP1 mutations resulting in human disease may provide a rationale for future study. Mutations of PPP1CB results in Noonan syndrome-like disorder with loose anagen hair 2 that is characterized by delayed bone age (Table 1) (42). Additionally, infiltrating T cells within the synovium of rheumatoid arthritis patients exhibit enhanced PP1 expression (39); thus, future assessment of functions PP1 within skeletal lineage cells may lead to impactful therapeutic treatments.
Table 1.
Human PSP gene alterations that have associated skeletal phenotypes. The Online Inheritance in Man (OM1M) database was used to identify human genetic alternations in serine/threonine phosphatase genes that have associated skeletal phenotypes.
| Ppase | NCBIGene No. | Mutation/Variant | Associated Skeletal Phenotype | Ref. No. |
|---|---|---|---|---|
| PPP1CB | 5500 | P49R or A56P substitution | Noonan syndrome-like disorder with loose anagen hair 2 (OMIM 617506) characterized by delayed bone age. short stature | (42) |
| PPP2R1A | 5513 | de novo R1S2W substitution | Skeletal overgrowth, craniofacial abnormalities | (64) |
| PPP2R5B | 5526 | de novo S161L deletion | Skeletal overgrowth, proximal interphalangeal joint swelling | (64) |
| PPP2R5C | 5527 | de novo T157 substitution | Skeletal overgrowth, craniofacial abnormalities | (64) |
| PPP2R5D | 5528 | de novo E19SK, W207R or P210R substitutions | Skeletal overgrowth, craniofacial abnormalities, syndactyly, polydactyly, scoliosis, hip dysplasia | (63.64) |
| PPP3CA | 5530 | F470L, A473T | arthrogryposis, cleft palate, craniosynostosis | (87) |
| PPM IB | 5495 | 2p21 deletion | Facial dysmorphism | (109) |
| PPM ID | 0493 | Truncations | Facial abnormalities, small hands -mil feet | (110) |
| CTDPI | 9150 | IVS6 + 389C-T | Facial dysmorphism | (131) |
Protein Phosphatase 2
Protein Phosphatase 2 (PP2, PP2a, ppp2ca, Nedlba, Rp-c) plays a role in development, cell mobility, proliferation, apoptosis, cytoskeletal dynamics, cell cycle control, and the regulation of several cell signaling pathways (28). PP2 is a heterotrimeric holoenzyme. It forms when a heterodimeric core enzyme, composed of a catalytic and scaffolding subunit, associate with a variable regulatory subunit (33). Regulatory subunits of PP2 are made up of 4 families: B through B’” all of which bind directly to the catalytic subunit (except B”’), each with two to five isoforms (28). This, in addition to the ɑ/β isoforms of the scaffolding and catalytic subunits, allows PP2 to act on substrates with precise specificity (28). Like PP1, PP2 is inhibited by okadaic acid and microcystin and is also inhibited by fostriecin (28). Notable substrates of PP2 include kinase suppressor of Ras (KSR), p21 activated kinase (PAK1) (43), Raf (44), mitogen-activated protein kinase kinase kinase 3 (MEKK3) (45), and RANKL pathway associated factors Calcium-calmodulin kinase IV (CaMKIV) (46), p38 MAPK(47), ERK1/2 (48), Pim-1(49), and Akt (50) among others.
PP2 also functions as a tumor suppressor (51–55) and its loss within hematopoietic stem cells (HSCs) of lymphoid cancers results in enhanced cell survival and proliferation (56–59), but little is known about how PP2 functions within non-transformed HSCs. One study demonstrated that PP2 dampens chemoattraction and migration of hematopoietic progenitor cells elicited by stromal cell derived factor 1 (Sdf1/Cxcl12) by limiting phosphorylation of Akt/PKB and Gsk3β (60).
Published studies document the role of PP2cα in bone homeostasis. Data obtained from high throughput sequencing of primary murine osteoclasts shows that the alpha catalytic subunit for PP2 is highly expressed as compared to other PSPs and their respective subunits (Figure 2C). PP2aα controls expression of RANKL and OPG by osteoblasts, and therefore indirectly modulates osteoclastogenesis (61). Wang et al. showed that PP2a inhibition via okadaic acid decreases osteoclastogenesis and bone destruction in a titanium particle-induced osteolysis model. Moreover, knockdown of PP2cα within RAW246.7 osteoclastogenesis assays suppresses activation of NF-κB and JNK and diminishes pit formation (62). These results suggest that PP2 is needed for optimal osteoclast activity, especially in the context of wear particle induced bone loss, but further study is needed to definitively assess the functions of PP2 within osteoclasts and bone homeostasis.
There are several clinical cases of PP2 regulatory subunit mutations in humans that result in an array of abnormal skeletal phenotypes, most notably skeletal overgrowth and craniofacial abnormalities (Table 1) (63,64). Several missense mutations of PPP2R5D result in various phenotypes, namely: hip dysplasia, scoliosis, finger syndactyly, and palate narrowing (63). Missense mutation of PPP2R1A resulted in similar phenotypes (63); thus, the skeletal manifestations imparted by mutations to PP2 regulatory subunits points to potential crucial roles for PP2 isoforms within skeletal lineage cells.
Calcineurin/Protein Phosphatase 3
Calcineurin (PP3, PP2b, Caln, Iecee) is the only PPP responsive to calcium ions in most tissues (65). It plays a role in calcium-dependent biological processes, including neurodevelopment, memory, immune response, cardiac hypertrophy, signal transduction, and muscle development (28,66). In terms of osteoclast activity, Calcineurin dephosphorylates NFATc1, the master osteoclastogenesis transcription factor, allowing its translocation to the nucleus (8,67) (Figure 1). Calcineurin consists of the catalytic subunit Calcineurin A (CNA) and the regulatory subunit Calcineurin B (CNB) (28). CNA contains an N-terminal phosphatase domain, CNB-binding helical domain (BBH), calcium-calmodulin (Ca2+-CaM)-binding motif, and an autoinhibitory element (Figure 2A) (28). CNA gains its activity when bound by Ca2+-CaM, displacing the autoinhibitory element (28,68), while CNB contains two Ca2+-binding domains for calcium mediated activity (28). Other than autoinhibition, small molecules exist that also inhibit Calcineurin. Cyclosporine A and FK506 (also known as tacrolimus) inhibit Calcineurin phosphatase activity by binding directly to CNA (69). Zinc similarly inhibits Calcineurin activity within the RANKL-induced NFATc1 pathway (8).
While the Calcineurin-dependent regulation of NFAT family transcription factors was originally described in T cells, emerging evidence demonstrates that this phosphatase also controls the fate of myeloid lineage cells. Calcineurin activity helps dictate the fate of HSCs from mesoderm progenitors during early embryonic development by facilitating Ca2+ signaling downstream of inositol 1,4,5-trisphosphate receptors (70). This is further refined by studies demonstrating that inhibition of Calcineurin diminished proliferation of CD34+ hematopoietic progenitor cells (71). Disruption of the NFAT/Calcineurin complex led to selective preferential expansion of myeloid progenitors (CD11B+ cells) and myeloid lineage cells (granulocyte, monocyte and dendritic cells) (72). Though Calcineurin-mediated NFAT activation directs CD33+ progenitor cells towards the dendritic cell lineage (73), inhibition of Calcineurin enhanced granulopoiesis (74), decreased lineage specification of progenitors into granulocyte subtypes (75) and negatively influenced the activation of mast cells through Stem cell factor (SCF)-mediated signaling cascade (76,77). Collectively, these studies demonstrate that NFAT/Calcineurin signaling represses myelopoiesis and may therefore limit the pool of osteoclast progenitors.
Numerous studies demonstrate the importance of Calcineurin as a positive regulator of osteoclast differentiation and activity. Several Calcineurin isoforms, including Aalpha, Abeta, Agamma and B1, are expressed by RAW246.7 throughout differentiation in vitro (78,79) and by primary mouse osteoclasts (Figure 2C). As stated previously, Calcineurin promotes osteoclastogenesis via its ability to dephosphorylate and promote the nuclear translocation of NFATc1 (67). To support this notion, overexpression of Calcineurin induces osteoclast differentiation and activity of ex vivo cultured primary osteoclast progenitors (79–81), which is blunted by co-expression of dominant negative NFATc1 (79). These data are further supported by observations that Calcineurin inhibitors cyclosporine and FK506, as well as expression of a Calcineurin inhibitory peptide, limit osteoclast differentiation (78,79). Moreover, expression of a constitutively active, Calcineurin-insensitive, NFATc1 construct induced osteoclast differentiation of RAW246.7 cells in the absence of RANKL (78). Calcineurin may also promote osteoclast differentiation and activity via its ability to control mTORC activity (82,83). Bone mass is similarly impacted by Calcineurin genetic deficiency, with CNAα germline null mice exhibiting decreased bone mass and elevated levels of TRAP+ multinucleated cells in vivo and enhanced ex vivo osteoclastogenesis (79). In addition, Calcineurin inhibitors cyclosporine A and FK506 induce bone loss in rodents (84) and osteoporosis in humans (85), but the cellular responses for this bone loss are unclear. Osteoclast-specific NFATc1 activity regulates bone mass (86), but while conditional deletion of Calcineurin in osteoblasts diminishes bone mass (86), the requirement of Calcineurin within osteoclasts has not been established. In humans, mutation of the Calcineurin auto-inhibitory domain results in arthrogryposis, cleft palate, craniosynostosis, short stature, and impaired intellectual development (Table 1) (87).
Protein Phosphatases 4, 5 and 6
Protein Phosphatase 4 (PP4, PP4c, ppp4c, Ppx, Pph3) and PP6 (ppp6c) are essential PPPs structurally similar to PP2 (Figure 2A, B) and are involved in similar processes, but function independently (28,88). The PP4 catalytic subunit associates with its own regulatory subunits, R1 and R2, to form distinct complexes (28,88), while PP6 forms a holoenzyme in a similar fashion to PP2 (28,89). Like PP2, PP4 and 6 are inhibited by okadaic acid, microcystin and fostriecin (33). PP6 downregulates TAK1 activation, an essential kinase for osteoclast differentiation (90,91). PP6 is expressed by murine osteoclasts (Figure 2C), but despite this, no studies have investigated the potential importance of PP6 within osteoclasts. Similarly, no studies seem to have focused on any potential effects of PP4 on osteoclast differentiation or activity despite its relatively high expression by ex vivo generated osteoclasts (Figure 2C).
Protein Phosphatase 5 (PP5, Ppp5c, Ppt) is unique in that it is encoded by a single gene and is not known to form complexes with regulatory subunits (28). While ubiquitously expressed in all tissues including osteoclasts (Figure 2C), it is highly expressed in the brain (28). It has roles in the regulation of cell proliferation, differentiation, migration, survival, death, DNA repair (28,92), and plays a significant role in hormone and stress-induced signaling (28,93). PP5 is auto-inhibited via interactions with tetratricopeptide repeats and a C-terminal helix alpha J region (Figure 2A) (28). These interactions are interrupted by heat shock protein (Hsp) 90 and fatty acids, releasing inhibition (28). Germline deletion of PP5 in mice results in reduced body weight and femur length (94). Bone mineral density, trabecular bone volume, and cortical thickness are negatively impacted as well (94). While much of this phenotype may be attributed to expression of PP5 by chondrocytes, the possible contributions of osteoclast function to this endochondral ossification defect have not been studied to date.
Protein Phosphatase 7
Protein Phosphatase 7 (PP7, At5g63870, Atpp7, Mgi19.12, Mgi19_12) has no known substrates, but is thought to play a role in sensory neuron function and/or development (95). Like Calcineurin, PP7 interacts with Ca2+-CaM via binding motif, but in an inhibitory fashion (28,96). PP7 also has EF-hand-like sequences that confer Ca2+ sensitivity (Figure 2A) (28,97). PP7 is autoinhibited by insertions within the core domain (Figure 2A), and excision of the insertions restores activity (28,96). The role PP7 plays within its associated tissues is not clearly understood, but it is thought that it regulates the dephosphorylation of G-protein-coupled receptors (33). PP7 is present only in the brain and retina (98), and RNA-seq data from primary murine osteoclasts would suggest that PP7 is not expressed by osteoclasts (Figure 2C). Because of the lack of expression, PP7 may not directly govern the generation and/or functions of osteoclasts.
Metal-dependent protein phosphatases
Metal-dependent protein phosphatases (PPMs) depend on manganese (Mn2+) or magnesium (Mg2+) ions for their activity, acting as the catalyst for dephosphorylation via activation of a water molecule (28). PPMs do not have associated regulatory subunits, but rather employ conserved domains and additional sequence motifs to act with specificity (28). Notable members are PP2c, pyruvate dehydrogenase phosphatase (Pdp), and the PH domain leucine rich repeat protein phosphatases (Phlpp). Like PPPs, some PPMs also play a part in osteoclastogenesis and bone resorption.
Protein Phosphatase 2C
Protein Phosphatase 2C (PP2C) is a very diverse class of PPM with over 20 known isoforms (28,97). Its primary role is in stress-signaling regulation, but it is also involved in several cellular functions including growth, differentiation, metabolism, survival, and apoptosis (28,99). Three additional α helices unique to PP2C associate with the core domain on the C-terminal side and are thought to contribute to substrate specificity or regulation (Figure 2A) (28). PP2Cs are resistant to all known PSP inhibitors, though initial studies identified putative candidates within the within National Cancer Institute Diversity Set (100). Detectable levels of all three PP2C isozymes, Ppm1a, Ppm1b and Ppm1g, are produced by primary mouse osteoclasts (Figure 2C), highlighting their potential role in bone resorption.
The PP2C family member Ppm1d (Iddgip, Jdvs, Pp2c-Delta, Wip1) is induced by p53 (56) and positively influences development of HSCs (56,101). This is supported by studies demonstrating that germline deletion of Ppm1d causes an HSC aging phenotype characterized by progressive declines in HSC numbers, reduced colony formation and self-renewal, reduced B and T lineage cells, expansion of myeloid progenitors, as well as a lack of bone marrow reconstitution capability (56). These effects may be due to the ability of Ppm1d to dephosphorylate and inactivate p38 MAPK and dampen mTOR activity (Figure 1) (102). Moreover, Ppm1d mutations occur with age and can lead to selective clonal expansion of HSCs conferring resistance to chemotherapy in hematological cancer patients (103). Similarly, germline deletion of mitochondrial localized PP2C phosphatase Ppm1k (Bdp, Msudmv, Pp2ckappa, Pp2cm, Ptmp, Ug0882e07) results in gradual decreases in HSC numbers concomitant with enhanced granulocyte-monocyte progenitors (GMPs) and modulates glycolysis, self-renewal and quiescence of HSCs (104); thus, Ppm1d and Ppm1k are critical regulators of HSC maintenance, but the impact of their deficiency on bone mass and osteoclastogenesis is unknown.
Ppm1a has a documented function within osteoclast lineage cells. Mice deficient in Ppm1a within myeloid lineage cells exhibit decreased bone mass and enhanced osteoclast numbers (105). Mechanistically, this was attributed to induction of RANK expression via elevated p38 MAPK activity (105). Ppm1a also dephosphorylates and inactivates Smad2/3 downstream of Tgfβ-induced signaling (106), implicating a function for Ppm1a in control of coupling bone resorption to bone formation (107). Ppm1a dampens bone morphogenic protein (BMP) signaling via dephosphorylation of Smad1 (108); thus, Ppm1a may also regulate osteoclastogenesis in this manner.
Mutation to several PPM genes induces skeletal phenotypes in humans. Deletion of the genomic region containing PPM1B results in facial dysmorphism (109), whereas truncating mutations to PPM1D result in Jansen de Vries syndrome characterized by facial abnormalities as well as small hands and feet (Table 1) (110); thus, further study of Ppm isoforms within skeletal lineage cells is warranted.
Pyruvate Dehydrogenase Phosphatase
Pyruvate Dehydrogenase Phosphatase (Pdp, Pdp1, Pdh, Pdpc, Ppm2c, Ppm2a) along with pyruvate dehydrogenase kinase (Pdk) and pyruvate dehydrogenase, form the pyruvate dehydrogenase complex (PDC) (Pelley 2006). The activity of Pdp is cyclical with Pdk; Pdk inhibits pyruvate dehydrogenase while PDC activates it (Pelley 2006). Pdp is stimulated by calcium ions and insulin and is inhibited by ATP, NADH and acetyl-CoA, which also activate Pdk (111). In addition to the conserved phosphatase domain, the first 70 N-terminal amino acids form a mitochondrial targeting region, where pyruvate dehydrogenase is localized (Figure 2A) (112). Although there are no described roles for Pdp in regulating osteoclast function, Pdp subunits are expressed by osteoclasts (Figure 2C) and by virtue of the ability to regulate cellular metabolism, PDP may likewise control osteoclast glycolytic activity.
PH Domain and Leucine Rich Protein Phosphatases
PH Domain and Leucine Rich Protein Phosphatases (PHLPP) is a relatively recent entry into the PPM family with two distinct isozymes: Phlpp1 (Plekhe1, Scop, Ppm3a) and Phlpp2 (Phlpp1, Ppm3b) (113–115). Both isozymes are composed of an N-terminal Ras association (RA) domain, a pleckstrin homology (PH) domain, a leucine rich repeat (LRR) region, a conserved PP2C phosphatase domain and a PDZ binding motif (113–115). The major difference between the two is a large N-terminal extension that is present in Phlpp1 but not in Phlpp2 (Figure 2A) (113–115). Phlpp is mainly involved in tumor suppression, apoptosis, histone regulation, and bone development (113–115). Substrates of Phlpp enzymes include: Akt, Raf1, Mst1, p70 S6K, as well as typical and atypical PKC isoforms (113). Amongst the of Akt isoforms, Phlpp1 primarily targets Ser474 Akt2 and Ser 472 Akt3, while Phlpp2 acts upon Ser473 Akt1 and Ser 472 Akt3 (113). Phlpp phosphatases also target threonine 387 of Mst1 (116). Phlpp gene expression is inhibited by histone deacetylase 3 (Hdac3) (117), translation is controlled by mTORC1, and the protein itself is inhibited via degradation by β-TrCP ubiquitination (114,115,118). Studies have shown that ubiquitination of Phlpp can be counteracted by ubiquitin specific peptidases 1, 12, and 46, preserving its stability (114,115).
Phlpp1 regulates a number of pathways that are involved with bone cell function. Ex vivo generated osteoclasts express both Phlpp1 and Phlpp2 transcripts (Figure 2C). Phlpp1 germline deficient mice display reduced snout-to-tail and tibia length as well as decreased bone mineral density and trabecular number (119). Conversely, conditional deletion of Phlpp1 within Ctskexpressing cells suppress osteoclast activity and enhances bone formation (120), but the utility of this model is questionable given the non-specific expression of Ctsk-Cre within osteoclasts and skeletal cells of mesenchymal origin (121–124). Future studies are required to definitively assess the functions of Phlpp1, as well as Phlpp2, within myeloid lineage cells and osteoclasts.
TFIIF-associating component of RNA polymerase II CTD Phosphatase/ small CTD phosphatase
The TFIIF-associating component of RNA polymerase II CTD Phosphatase/ small CTD phosphatase (FCP/SCP) class of PSPs use an aspartate based mechanism of catalysis, unlike the metal ion based mechanisms of PPPs and PPMs (33), and have only one major substrate: the carboxy-terminal domain (CTD) of RNA Polymerase II (28). There are eight known members, with Fcp1 (Fcp, Fcp1, Fcpx, Hbfqtl3, Ctdp1) and Scp1 (Sycp1, Ct8, Hom-tes-14, Ctsdp1) being major players (28). The conserved structural core of FCP/SCPs is the FCP homology (FCPH) domain (28,125). Within the domain are insertion domains thought to contribute to catalysis (28). In Fcp1, there is a BRCA1 C-terminal domain like (BRCT) domain that is C-terminal to the FCPH domain (126) and further down is a TFIIF interacting helix (Figure 2A) (125). Some members of FCP/SCP can be inhibited by BeF3- (33). There is evidence supporting the role of Scp1 (CTDSP1) and Scp2 (CTDSP2) in neurogenesis (127–130), but little is known about the role of FCP/SCP class of PSPs on myelogenesis and osteoclastogenesis. CTDP1 mutations result in facial dysmorphism accompanied by congenital cataracts and neuropathy (Table 1) (131).
Concluding Remarks
The intricate interplay between osteoclasts and osteoblasts modulates bone resorption and formation, thus regulating bone homeostasis. Disruption or loss of osteoclast activity tilts the balance of this equilibrium, leading to changes in bone mass. Despite the success of current therapeutics to enhance bone mass, the extremely rare yet adverse side effects of these therapies limit their clinical application; thus, new target for therapeutic intervention are needed.
Despite their ability to attenuate critical signaling pathways mediating osteoclast differentiation and function, little is known about the impact of PSPs on these processes. RNA-sequencing results suggest that, of the 80 PSPs identified, roughly half show significant expression by osteoclasts (e.g., RPKM>10). Of these, only Phlpp1, Calcinurin and PP2C, have been studied to a significant extent. Each of these PSPs has a unique function resulting in contrasting phenotypes when conditionally deleted within osteoclast lineage cells. This is likely due to differing substrates, with NFATc1 being the dominant Calcineurin substrate, and Phlpp1 primarily inactivating AGC kinases; thus, the functions of remaining individual PSPs within osteoclast lineage cells is of interest.
Inhibition of PSPs has been proposed as a therapeutic approach to treat neurodegenerative diseases (132,133) and cancer (134,135), and may likewise offer an avenue to regulate osteoclastogenesis and bone resorption. While a few small molecule inhibitor for specific PSPs have been identified, little is known about their effects within the osteoclast lineage. It would likely be beneficial to the study of PSPs and their functions within osteoclasts if inhibitors specific to each were developed, but this has historically been difficult. Within classes of PSPs, the phosphatase domain is highly conserved and known inhibitors are nonspecific. PP2C, for example, does not currently have any known inhibitors, lacks regulatory subunits, and has highly conserved phosphatase domains between its members. It may be possible to produce inhibitors that take advantage of differences within the different isoforms or specific subunits of PSPs in a given class, leading to a loss of function separate from the phosphatase domain itself.
In this review we have covered several PSPs that regulate the proliferation, differentiation, maintenance and function of osteoclasts. Modulation of these PSP-mediated signaling by reinvention of PSP inhibitors used for other diseases or against novel targets may therefore serve as a potential line of treatment for conditions where there is a dysregulation of bone homeostasis such osteoporosis and osteopetrosis.
Supplementary Material
Kinase signaling cascades that promote osteoclastogenesis are tempered by the action of protein phosphatases, including members of the serine/threonine phosphatase family.
Here we examine the impact of serine/threonine phosphatases on osteoclast differentiation and activity, while highlighting deficiencies in our understanding of this understudied class of proteins within the field.
Acknowledgments
The authors have no competing interests to declare. This work was made possible by a research grant from the National Institutes of Health (AR072634) and the University of Minnesota Board of Regents. These contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Abbreviations
- Rankl
receptor activator of nuclear factor-κB (NF-κB) ligand
- Rank
receptor activator of nuclear factor-κB (NF-κB)
- M-csf
macrophage colony-stimulating factor
- c-fms
colony-stimulating factor 1 receptor
- Opg
Osteoprotegerin
- Nfatc1
nuclear factor-activated T cells c1
- Traf
tumor necrosis factor receptor (Tnrf)-associated factor
- Tgfβ
kinase transforming growth factor β
- Tak1
transforming growth factor β (Tgfβ)-activated kinase 1
- MAPKs
mitogen-activated protein kinases
- Ikk
IκB kinase
- IκB
inhibitory κB
- c-Src
Proto-oncogene tyrosine-protein kinase
- PI3K
phosphoinositide 3-kinase
- Akt
protein kinase B
- Grb2/Sos
growth factor receptor-bound protein 2/son of sevenless
- Erk1/2
extracellular signal-regulated kinase
- ITAM
immunoreceptor tyrosine-based activation motif
- Dap12
DNAX activating protein of 12 kDa
- (TREM)2
triggering receptors expressed by myeloid cells
- (PLCγ)1
phospholipase Cγ
- FcγR
Fcγ receptor
- Oscar
Osteoclast-associated receptor
- Ifn
interferon
- Gm-csf
granulocyte macrophage colony-stimulating factor
- Sdf-1
stromal cell-derived factor 1
- Mcp-l
monocyte chemoattractant protein 1
- PSPs
protein serine/threonine phosphatases
- PPPs
phosphoprotein phosphatases
- PPMs
metal-dependent protein phosphatases
- FCP/SCP
TFIIF-associating component of RNA polymerase II CTD phosphatase/ small CTD phosphatase
- Pp
Protein Phosphatase 1
- Cna
Calcineurin A
- Cnb
Calcineurin B
- Ca2+-CaM
calcium-calmodulin
- Pdp
pyruvate dehydrogenase phosphatase
- Phlpp
PH domain leucine rich repeat protein phosphatases
- BMP
bone morphogenic protein
- PH
pleckstrin homology
- RA
Ras association
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Theill LE, Boyle WJ, and Penninger JM (2002) RANK-L and RANK: T cells, bone loss, and mammalian evolution. Annu Rev Immunol 20, 795–823 [DOI] [PubMed] [Google Scholar]
- 2.Ross FP, and Teitelbaum SL (2005) alphavbeta3 and macrophage colony-stimulating factor: partners in osteoclast biology. Immunol Rev 208, 88–105 [DOI] [PubMed] [Google Scholar]
- 3.Nakashima T, Hayashi M, Fukunaga T, Kurata K, Oh-Hora M, Feng JQ,Bonewald LF, Kodama T, Wutz A, Wagner EF, Penninger JM, and Takayanagi H. (2011) Evidence for osteocyte regulation of bone homeostasis through RANKL expression. Nat Med 17, 1231–1234 [DOI] [PubMed] [Google Scholar]
- 4.Kim JH, and Kim N. (2016) Signaling Pathways in Osteoclast Differentiation. Chonnam Med J 52, 12–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sobacchi C, Schulz A, Coxon FP, Villa A, and Helfrich MH (2013) Osteopetrosis: genetics, treatment and new insights into osteoclast function. Nat Rev Endocrinol 9, 522–536 [DOI] [PubMed] [Google Scholar]
- 6.Walsh MC, and Choi Y. (2014) Biology of the RANKL-RANK-OPG System in Immunity, Bone, and Beyond. Front Immunol 5, 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizukami J, Takaesu G, Akatsuka H, Sakurai H, Ninomiya-Tsuji J, Matsumoto K, and Sakurai N. (2002) Receptor activator of NF-kappaB ligand (RANKL) activates TAK1 mitogen-activated protein kinase kinase kinase through a signaling complex containing RANK, TAB2, and TRAF6. Mol Cell Biol 22, 992–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park KH, Park B, Yoon DS, Kwon SH, Shin DM, Lee JW, Lee HG, Shim JH, Park JH, and Lee JM (2013) Zinc inhibits osteoclast differentiation by suppression of Ca2+-Calcineurin-NFATc1 signaling pathway. Cell Commun Signal 11, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yavropoulou MP, and Yovos JG (2008) Osteoclastogenesis--current knowledge and future perspectives. J Musculoskelet Neuronal Interact 8, 204–216 [PubMed] [Google Scholar]
- 10.Izawa T, Zou W, Chappel JC, Ashley JW, Feng X, and Teitelbaum SL (2012) c-Src links a RANK/alphavbeta3 integrin complex to the osteoclast cytoskeleton. Mol Cell Biol 32, 2943–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King D, Yeomanson D, and Bryant HE (2015) PI3King the lock: targeting the PI3K/Akt/mTOR pathway as a novel therapeutic strategy in neuroblastoma. J Pediatr Hematol Oncol 37, 245–251 [DOI] [PubMed] [Google Scholar]
- 12.Moon JB, Kim JH, Kim K, Youn BU, Ko A, Lee SY, and Kim N. (2012) Akt induces osteoclast differentiation through regulating the GSK3beta/NFATc1 signaling cascade. J Immunol 188, 163–169 [DOI] [PubMed] [Google Scholar]
- 13.Sugatani T, Alvarez U, and Hruska KA (2003) PTEN regulates RANKL- and osteopontin-stimulated signal transduction during osteoclast differentiation and cell motility. J Biol Chem 278, 5001–5008 [DOI] [PubMed] [Google Scholar]
- 14.Marks SC Jr., and Lane PW (1976) Osteopetrosis, a new recessive skeletal mutation on chromosome 12 of the mouse. J Hered 67, 11–18 [DOI] [PubMed] [Google Scholar]
- 15.Wiktor-Jedrzejczak W, Bartocci A, Ferrante AW Jr., Ahmed-Ansari A, Sell KW, Pollard JW, and Stanley ER (1990) Total absence of colony-stimulating factor 1 in the macrophage-deficient osteopetrotic (op/op) mouse. Proc Natl Acad Sci U S A 87, 4828–4832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshida H, Hayashi S, Kunisada T, Ogawa M, Nishikawa S, Okamura H, Sudo T, Shultz LD, and Nishikawa S. (1990) The murine mutation osteopetrosis is in the coding region of the macrophage colony stimulating factor gene. Nature 345, 442–444 [DOI] [PubMed] [Google Scholar]
- 17.Feng X, and Teitelbaum SL (2013) Osteoclasts: New Insights. Bone Res 1, 11–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golden LH, and Insogna KL (2004) The expanding role of PI3-kinase in bone. Bone 34, 3–12 [DOI] [PubMed] [Google Scholar]
- 19.Karsten CM, and Kohl J. (2015) A bone to pick with Fc gamma receptors. Ann Transl Med 3, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colonna M. (2003) DAP12 signaling: from immune cells to bone modeling and brain myelination. J Clin Invest 111, 313–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helming L, Tomasello E, Kyriakides TR, Martinez FO, Takai T, Gordon S, and Vivier E. (2008) Essential role of DAP12 signaling in macrophage programming into a fusion-competent state. Sci Signal 1, ra11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bournazos S, Wang TT, and Ravetch JV (2016) The Role and Function of Fcgamma Receptors on Myeloid Cells. Microbiol Spectr 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nemeth K, Schoppet M, Al-Fakhri N, Helas S, Jessberger R, Hofbauer LC, and Goettsch C. (2011) The role of osteoclast-associated receptor in osteoimmunology. J Immunol 186, 13–18 [DOI] [PubMed] [Google Scholar]
- 24.Baron R. (2004) Arming the osteoclast. Nat Med 10, 458–460 [DOI] [PubMed] [Google Scholar]
- 25.Hayashi M, Nakashima T, Taniguchi M, Kodama T, Kumanogoh A, and Takayanagi H. (2012) Osteoprotection by semaphorin 3A. Nature 485, 69–74 [DOI] [PubMed] [Google Scholar]
- 26.Takayanagi H. (2009) Osteoimmunology and the effects of the immune system on bone. Nat Rev Rheumatol 5, 667–676 [DOI] [PubMed] [Google Scholar]
- 27.Weber A, Wasiliew P, and Kracht M. (2010) Interleukin-1 (IL-1) pathway. Sci Signal 3, cm1 [DOI] [PubMed] [Google Scholar]
- 28.Shi Y. (2009) Serine/threonine phosphatases: mechanism through structure. Cell 139, 468–484 [DOI] [PubMed] [Google Scholar]
- 29.Li X, Wilmanns M, Thornton J, and Kohn M. (2013) Elucidating human phosphatase-substrate networks. Sci Signal 6, rs10 [DOI] [PubMed] [Google Scholar]
- 30.Brautigan DL, and Shenolikar S. (2018) Protein Serine/Threonine Phosphatases: Keys to Unlocking Regulators and Substrates. Annu Rev Biochem 87, 921–964 [DOI] [PubMed] [Google Scholar]
- 31.Barker HM, Brewis ND, Street AJ, Spurr NK, and Cohen PT (1994) Three genes for protein phosphatase 1 map to different human chromosomes: sequence, expression and gene localisation of protein serine/threonine phosphatase 1 beta (PPP1CB). Biochim Biophys Acta 1220, 212–218 [DOI] [PubMed] [Google Scholar]
- 32.Ceulemans H, and Bollen M. (2004) Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol Rev 84, 1–39 [DOI] [PubMed] [Google Scholar]
- 33.Janssens V, and Goris J. (2001) Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J 353, 417–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ragolia L, and Begum N. (1998) Protein phosphatase-1 and insulin action. Mol Cell Biochem 182, 49–58 [PubMed] [Google Scholar]
- 35.Shioda N, and Fukunaga K. (2017) Physiological and Pathological Roles of CaMKIIPP1 Signaling in the Brain. Int J Mol Sci 19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee HM, Chen R, Kim H, Etchegaray JP, Weaver DR, and Lee C. (2011) The period of the circadian oscillator is primarily determined by the balance between casein kinase 1 and protein phosphatase 1. Proc Natl Acad Sci U S A 108, 16451–16456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu Z, Zhou X, Wang W, Deng W, Fang J, Hu H, Wang Z, Li S, Cui L, Shen J, Zhai L, Peng S, Wong J, Dong S, Yuan Z, Ou G, Zhang X, Xu P, Lou J, Yang N, Chen P, Xu RM, and Li G. (2015) Dynamic phosphorylation of CENP-A at Ser68 orchestrates its cell-cycle-dependent deposition at centromeres. Dev Cell 32, 68–81 [DOI] [PubMed] [Google Scholar]
- 38.Song H, Pu J, Wang L, Wu L, Xiao J, Liu Q, Chen J, Zhang M, Liu Y, Ni M, Mo J, Zheng Y, Wan D, Cai X, Cao Y, Xiao W, Ye L, Tu E, Lin Z, Wen J, Lu X, He J, Peng Y, Su J, Zhang H, Zhao Y, Lin M, and Zhang Z. (2015) ATG16L1 phosphorylation is oppositely regulated by CSNK2/casein kinase 2 and PPP1/protein phosphatase 1 which determines the fate of cardiomyocytes during hypoxia/reoxygenation. Autophagy 11, 1308–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nie H, Zheng Y, Li R, Guo TB, He D, Fang L, Liu X, Xiao L, Chen X, Wan B, Chin YE, and Zhang JZ (2013) Phosphorylation of FOXP3 controls regulatory T cell function and is inhibited by TNF-alpha in rheumatoid arthritis. Nat Med 19, 322–328 [DOI] [PubMed] [Google Scholar]
- 40.Umeki I, Niihori T, Abe T, Kanno SI, Okamoto N, Mizuno S, Kurosawa K, Nagasaki K, Yoshida M, Ohashi H, Inoue SI, Matsubara Y, Fujiwara I, Kure S, and Aoki Y. (2019) Delineation of LZTR1 mutation-positive patients with Noonan syndrome and identification of LZTR1 binding to RAF1-PPP1CB complexes. Hum Genet 138, 21–35 [DOI] [PubMed] [Google Scholar]
- 41.Young LC, Hartig N, Boned Del Rio I, Sari S, Ringham-Terry B, Wainwright JR, Jones GG, McCormick F, and Rodriguez-Viciana P. (2018) SHOC2-MRAS-PP1 complex positively regulates RAF activity and contributes to Noonan syndrome pathogenesis. Proc Natl Acad Sci U S A 115, E10576-E10585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gripp KW, Aldinger KA, Bennett JT, Baker L, Tusi J, Powell-Hamilton N, Stabley D, Sol-Church K, Timms AE, and Dobyns WB (2016) A novel rasopathy caused by recurrent de novo missense mutations in PPP1CB closely resembles Noonan syndrome with loose anagen hair. Am J Med Genet A 170, 2237–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bradley EW, Ruan MM, and Oursler MJ (2008) PAK1 is a novel MEK-independent raf target controlling expression of the IAP survivin in M-CSF-mediated osteoclast survival. J Cell Physiol 217, 752–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ory S, Zhou M, Conrads TP, Veenstra TD, and Morrison DK (2003) Protein phosphatase 2A positively regulates Ras signaling by dephosphorylating KSR1 and Raf-1 on critical 14–3-3 binding sites. Curr Biol 13, 1356–1364 [DOI] [PubMed] [Google Scholar]
- 45.Sun W, Wang H, Zhao X, Yu Y, Fan Y, Wang H, Wang X, Lu X, Zhang G, Fu S, and Yang J. (2010) Protein phosphatase 2A acts as a mitogen-activated protein kinase kinase kinase 3 (MEKK3) phosphatase to inhibit lysophosphatidic acid-induced IkappaB kinase beta/nuclear factor-kappaB activation. J Biol Chem 285, 21341–21348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Westphal RS, Anderson KA, Means AR, and Wadzinski BE (1998) A signaling complex of Ca2+-calmodulin-dependent protein kinase IV and protein phosphatase 2A. Science 280, 1258–1261 [DOI] [PubMed] [Google Scholar]
- 47.Lee T, Kim SJ, and Sumpio BE (2003) Role of PP2A in the regulation of p38 MAPK activation in bovine aortic endothelial cells exposed to cyclic strain. J Cell Physiol 194, 349–355 [DOI] [PubMed] [Google Scholar]
- 48.Roskoski R Jr. (2012) ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol Res 66, 105–143 [DOI] [PubMed] [Google Scholar]
- 49.Ma J, Arnold HK, Lilly MB, Sears RC, and Kraft AS (2007) Negative regulation of Pim-1 protein kinase levels by the B56beta subunit of PP2A. Oncogene 26, 5145–5153 [DOI] [PubMed] [Google Scholar]
- 50.Ivaska J, Nissinen L, Immonen N, Eriksson JE, Kahari VM, and Heino J. (2002) Integrin alpha 2 beta 1 promotes activation of protein phosphatase 2A and dephosphorylation of Akt and glycogen synthase kinase 3 beta. Mol Cell Biol 22, 1352–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arnold HK, and Sears RC (2008) A tumor suppressor role for PP2A-B56alpha through negative regulation of c-Myc and other key oncoproteins. Cancer Metastasis Rev 27, 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janssens V, Goris J, and Van Hoof C. (2005) PP2A: the expected tumor suppressor. Curr Opin Genet Dev 15, 34–41 [DOI] [PubMed] [Google Scholar]
- 53.Farrell AS, Allen-Petersen B, Daniel CJ, Wang X, Wang Z, Rodriguez S, Impey S, Oddo J, Vitek MP, Lopez C, Christensen DJ, Sheppard B, and Sears RC (2014) Targeting inhibitors of the tumor suppressor PP2A for the treatment of pancreatic cancer. Mol Cancer Res 12, 924–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perrotti D, and Neviani P. (2008) Protein phosphatase 2A (PP2A), a drugable tumor suppressor in Ph1(+) leukemias. Cancer Metastasis Rev 27, 159–168 [DOI] [PubMed] [Google Scholar]
- 55.Sangodkar J, Perl A, Tohme R, Kiselar J, Kastrinsky DB, Zaware N, Izadmehr S, Mazhar S, Wiredja DD, O’Connor CM, Hoon D, Dhawan NS, Schlatzer D, Yao S, Leonard D, Borczuk AC, Gokulrangan G, Wang L, Svenson E, Farrington CC, Yuan E, Avelar RA, Stachnik A, Smith B, Gidwani V, Giannini HM, McQuaid D, McClinch K, Wang Z, Levine AC, Sears RC, Chen EY, Duan Q, Datt M, Haider S, Ma’ayan A, DiFeo A, Sharma N, Galsky MD, Brautigan DL, Ioannou YA, Xu W, Chance MR, Ohlmeyer M, and Narla G. (2017) Activation of tumor suppressor protein PP2A inhibits KRAS-driven tumor growth. J Clin Invest 127, 2081–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Z, Yi W, Morita Y, Wang H, Cong Y, Liu JP, Xiao Z, Rudolph KL, Cheng T, and Ju Z. (2015) Wip1 deficiency impairs haematopoietic stem cell function via p53 and mTORC1 pathways. Nat Commun 6, 6808. [DOI] [PubMed] [Google Scholar]
- 57.Naughton R, Quiney C, Turner SD, and Cotter TG (2009) Bcr-Abl-mediated redox regulation of the PI3K/AKT pathway. Leukemia 23, 1432–1440 [DOI] [PubMed] [Google Scholar]
- 58.Neviani P, Harb JG, Oaks JJ, Santhanam R, Walker CJ, Ellis JJ, Ferenchak G, Dorrance AM, Paisie CA, Eiring AM, Ma Y, Mao HC, Zhang B, Wunderlich M, May PC, Sun C, Saddoughi SA, Bielawski J, Blum W, Klisovic RB, Solt JA, Byrd JC, Volinia S, Cortes J, Huettner CS, Koschmieder S, Holyoake TL, Devine S, Caligiuri MA, Croce CM, Garzon R, Ogretmen B, Arlinghaus RB, Chen CS, Bittman R, Hokland P, Roy DC, Milojkovic D, Apperley J, Goldman JM, Reid A, Mulloy JC, Bhatia R, Marcucci G, and Perrotti D. (2013) PP2A-activating drugs selectively eradicate TKI-resistant chronic myeloid leukemic stem cells. J Clin Invest 123, 4144–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salas A, Ponnusamy S, Senkal CE, Meyers-Needham M, Selvam SP, Saddoughi SA, Apohan E, Sentelle RD, Smith C, Gault CR, Obeid LM, El-Shewy HM, Oaks J, Santhanam R, Marcucci G, Baran Y, Mahajan S, Fernandes D, Stuart R, Perrotti D, and Ogretmen B. (2011) Sphingosine kinase-1 and sphingosine 1-phosphate receptor 2 mediate Bcr-Abl1 stability and drug resistance by modulation of protein phosphatase 2A. Blood 117, 5941–5952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Basu S, Ray NT, Atkinson SJ, and Broxmeyer HE (2007) Protein phosphatase 2A plays an important role in stromal cell-derived factor-1/CXC chemokine ligand 12-mediated migration and adhesion of CD34+ cells. J Immunol 179, 3075–3085 [DOI] [PubMed] [Google Scholar]
- 61.Okamura H, Yang D, Yoshida K, and Haneji T. (2013) Protein phosphatase 2A Calpha is involved in osteoclastogenesis by regulating RANKL and OPG expression in osteoblasts. FEBS Lett 587, 48–53 [DOI] [PubMed] [Google Scholar]
- 62.Wang L, Guo X, Zhou W, Ding Y, Shi J, Wu X, Liu Y, Xu Y, Yang H, and Geng D. (2018) Protein phosphatase 2A as a new target for downregulating osteoclastogenesis and alleviating titanium particle-induced bone resorption. Acta Biomater 73, 488–499 [DOI] [PubMed] [Google Scholar]
- 63.Houge G, Haesen D, Vissers LE, Mehta S, Parker MJ, Wright M, Vogt J, McKee S, Tolmie JL, Cordeiro N, Kleefstra T, Willemsen MH, Reijnders MR, Berland S, Hayman E, Lahat E, Brilstra EH, van Gassen KL, Zonneveld-Huijssoon E, de Bie CI, Hoischen A, Eichler EE, Holdhus R, Steen VM, Doskeland SO, Hurles ME, FitzPatrick DR, and Janssens V. (2015) B56delta-related protein phosphatase 2A dysfunction identified in patients with intellectual disability. J Clin Invest 125, 3051–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Loveday C, Tatton-Brown K, Clarke M, Westwood I, Renwick A, Ramsay E, Nemeth A, Campbell J, Joss S, Gardner M, Zachariou A, Elliott A, Ruark E, van Montfort R, Childhood Overgrowth C, and Rahman N. (2015) Mutations in the PP2A regulatory subunit B family genes PPP2R5B, PPP2R5C and PPP2R5D cause human overgrowth. Hum Mol Genet 24, 4775–4779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rusnak F, and Mertz P. (2000) Calcineurin: form and function. Physiol Rev 80, 1483–1521 [DOI] [PubMed] [Google Scholar]
- 66.Griffith JP, Kim JL, Kim EE, Sintchak MD, Thomson JA, Fitzgibbon MJ, Fleming MA, Caron PR, Hsiao K, and Navia MA (1995) X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12-FK506 complex. Cell 82, 507–522 [DOI] [PubMed] [Google Scholar]
- 67.Kim JH, and Kim N. (2014) Regulation of NFATc1 in Osteoclast Differentiation. J Bone Metab 21, 233–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cohen PT (2002) Protein phosphatase 1--targeted in many directions. J Cell Sci 115, 241–256 [DOI] [PubMed] [Google Scholar]
- 69.Azzi JR, Sayegh MH, and Mallat SG (2013) Calcineurin inhibitors: 40 years later, can’t live without. J Immunol 191, 5785–5791 [DOI] [PubMed] [Google Scholar]
- 70.Wang B, Liu J, Huang P, Xu K, Wang H, Wang X, Guo Z, and Xu L. (2017) Protein phosphatase 2A inhibition and subsequent cytoskeleton reorganization contributes to cell migration caused by microcystin-LR in human laryngeal epithelial cells (Hep-2). Environ Toxicol 32, 890–903 [DOI] [PubMed] [Google Scholar]
- 71.Kiani A, Habermann I, Haase M, Feldmann S, Boxberger S, Sanchez-Fernandez MA, Thiede C, Bornhauser M, and Ehninger G. (2004) Expression and regulation of NFAT (nuclear factors of activated T cells) in human CD34+ cells: down-regulation upon myeloid differentiation. J Leukoc Biol 76, 1057–1065 [DOI] [PubMed] [Google Scholar]
- 72.Fric J, Lim CX, Koh EG, Hofmann B, Chen J, Tay HS, Mohammad Isa SA, Mortellaro A, Ruedl C, and Ricciardi-Castagnoli P. (2012) Calcineurin/NFAT signalling inhibits myeloid haematopoiesis. EMBO Mol Med 4, 269–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Engels FH, Kreisel D, Faries MB, Bedrosian I, Koski GK, Cohen PA, and Czerniecki BJ (2000) Calcium ionophore activation of chronic myelogenous leukemia progenitor cells into dendritic cells is mediated by calcineurin phosphatase. Leuk Res 24, 795–804 [DOI] [PubMed] [Google Scholar]
- 74.Teng EC, Racioppi L, and Means AR (2011) A cell-intrinsic role for CaMKK2 in granulocyte lineage commitment and differentiation. J Leukoc Biol 90, 897–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fric J, Lim CX, Mertes A, Lee BT, Vigano E, Chen J, Zolezzi F, Poidinger M, Larbi A, Strobl H, Zelante T, and Ricciardi-Castagnoli P. (2014) Calcium and calcineurin-NFAT signaling regulate granulocyte-monocyte progenitor cell cycle via Flt3-L. Stem Cells 32, 3232–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ishizuka T, Chayama K, Takeda K, Hamelmann E, Terada N, Keller GM, Johnson GL, and Gelfand EW (1999) Mitogen-activated protein kinase activation through Fc epsilon receptor I and stem cell factor receptor is differentially regulated by phosphatidylinositol 3-kinase and calcineurin in mouse bone marrow-derived mast cells. J Immunol 162, 2087–2094 [PubMed] [Google Scholar]
- 77.Wu Z, Li Y, MacNeil AJ, Junkins RD, Berman JN, and Lin TJ (2013) Calcineurin-Rcan1 interaction contributes to stem cell factor-mediated mast cell activation. J Immunol 191, 5885–5894 [DOI] [PubMed] [Google Scholar]
- 78.Hirotani H, Tuohy NA, Woo JT, Stern PH, and Clipstone NA (2004) The calcineurin/nuclear factor of activated T cells signaling pathway regulates osteoclastogenesis in RAW264.7 cells. J Biol Chem 279, 13984–13992 [DOI] [PubMed] [Google Scholar]
- 79.Sun L, Peng Y, Zaidi N, Zhu LL, Iqbal J, Yamoah K, Wang X, Liu P, Abe E, Moonga BS, Epstein S, and Zaidi M. (2007) Evidence that calcineurin is required for the genesis of bone-resorbing osteoclasts. Am J Physiol Renal Physiol 292, F285–291 [DOI] [PubMed] [Google Scholar]
- 80.Sun L, Blair HC, Peng Y, Zaidi N, Adebanjo OA, Wu XB, Wu XY, Iqbal J, Epstein S, Abe E, Moonga BS, and Zaidi M. (2005) Calcineurin regulates bone formation by the osteoblast. Proc Natl Acad Sci U S A 102, 17130–17135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sun L, Moonga BS, Lu M, Zaidi N, Iqbal J, Blair HC, Epstein S, Abe E, Troen BR, Huang CL, and Zaidi M. (2003) Molecular cloning, expression, and function of osteoclastic calcineurin Aalpha. Am J Physiol Renal Physiol 284, F575–583 [DOI] [PubMed] [Google Scholar]
- 82.Huynh H, and Wan Y. (2018) mTORC1 impedes osteoclast differentiation via calcineurin and NFATc1. Commun Biol 1, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Y, Xu S, Li K, Tan K, Liang K, Wang J, Shen J, Zou W, Hu L, Cai D, Ding C, Li M, Xiao G, Liu B, Liu A, and Bai X. (2017) mTORC1 Inhibits NF-kappaB/NFATc1 Signaling and Prevents Osteoclast Precursor Differentiation, In Vitro and In Mice. J Bone Miner Res 32, 1829–1840 [DOI] [PubMed] [Google Scholar]
- 84.Cvetkovic M, Mann GN, Romero DF, Liang XG, Ma Y, Jee WS, and Epstein S. (1994) The deleterious effects of long-term cyclosporine A, cyclosporine G, and FK506 on bone mineral metabolism in vivo. Transplantation 57, 1231–1237 [DOI] [PubMed] [Google Scholar]
- 85.Epstein S, Inzerillo AM, Caminis J, and Zaidi M. (2003) Disorders associated with acute rapid and severe bone loss. J Bone Miner Res 18, 2083–2094 [DOI] [PubMed] [Google Scholar]
- 86.Winslow MM, Pan M, Starbuck M, Gallo EM, Deng L, Karsenty G, and Crabtree GR (2006) Calcineurin/NFAT signaling in osteoblasts regulates bone mass. Dev Cell 10, 771–782 [DOI] [PubMed] [Google Scholar]
- 87.Mizuguchi T, Nakashima M, Kato M, Okamoto N, Kurahashi H, Ekhilevitch N, Shiina M, Nishimura G, Shibata T, Matsuo M, Ikeda T, Ogata K, Tsuchida N, Mitsuhashi S, Miyatake S, Takata A, Miyake N, Hata K, Kaname T, Matsubara Y, Saitsu H, and Matsumoto N. (2018) Loss-of-function and gain-of-function mutations in PPP3CA cause two distinct disorders. Hum Mol Genet 27, 1421–1433 [DOI] [PubMed] [Google Scholar]
- 88.Stefansson B, Ohama T, Daugherty AE, and Brautigan DL (2008) Protein phosphatase 6 regulatory subunits composed of ankyrin repeat domains. Biochemistry 47, 1442–1451 [DOI] [PubMed] [Google Scholar]
- 89.Hinds TD Jr., and Sanchez ER (2008) Protein phosphatase 5. Int J Biochem Cell Biol 40, 2358–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kajino T, Ren H, Iemura S, Natsume T, Stefansson B, Brautigan DL, Matsumoto K, and Ninomiya-Tsuji J. (2006) Protein phosphatase 6 down-regulates TAK1 kinase activation in the IL-1 signaling pathway. J Biol Chem 281, 39891–39896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lamothe B, Lai Y, Xie M, Schneider MD, and Darnay BG (2013) TAK1 is essential for osteoclast differentiation and is an important modulator of cell death by apoptosis and necroptosis. Mol Cell Biol 33, 582–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Davies TH, Ning YM, and Sanchez ER (2005) Differential control of glucocorticoid receptor hormone-binding function by tetratricopeptide repeat (TPR) proteins and the immunosuppressive ligand FK506. Biochemistry 44, 2030–2038 [DOI] [PubMed] [Google Scholar]
- 93.Yang J, Roe SM, Cliff MJ, Williams MA, Ladbury JE, Cohen PT, and Barford D. (2005) Molecular basis for TPR domain-mediated regulation of protein phosphatase 5. EMBO J 24, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang J, Cao Y, Qiu B, Du J, Wang T, Wang C, Deng R, Shi X, Gao K, Xie Z, and Yong W. (2018) Ablation of protein phosphatase 5 (PP5) leads to enhanced both bone and cartilage development in mice. Cell Death Dis 9, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Herzig S, and Neumann J. (2000) Effects of serine/threonine protein phosphatases on ion channels in excitable membranes. Physiol Rev 80, 173–210 [DOI] [PubMed] [Google Scholar]
- 96.Andreeva AV, and Kutuzov MA (1999) RdgC/PP5-related phosphatases: novel components in signal transduction. Cell Signal 11, 555–562 [DOI] [PubMed] [Google Scholar]
- 97.Lammers T, and Lavi S. (2007) Role of type 2C protein phosphatases in growth regulation and in cellular stress signaling. Crit Rev Biochem Mol Biol 42, 437–461 [DOI] [PubMed] [Google Scholar]
- 98.Kutuzov MA, Evans DE, and Andreeva AV (1998) Expression and characterization of PP7, a novel plant protein Ser/Thr phosphatase distantly related to RdgC/PPEF and PP5. FEBS Lett 440, 147–152 [DOI] [PubMed] [Google Scholar]
- 99.Lu G, and Wang Y. (2008) Functional diversity of mammalian type 2C protein phosphatase isoforms: new tales from an old family. Clin Exp Pharmacol Physiol 35, 107–112 [DOI] [PubMed] [Google Scholar]
- 100.Rogers JP, Beuscher A. E. t., Flajolet M, McAvoy T, Nairn AC, Olson AJ, and Greengard P. (2006) Discovery of protein phosphatase 2C inhibitors by virtual screening. J Med Chem 49, 1658–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.He W, Wang X, Ni Y, Li Z, Liu W, Chang Z, Li H, Ju Z, and Li Z. (2020) Wip1 regulates hematopoietic stem cell development in the mouse embryo. Haematologica [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Uyanik B, Grigorash BB, Goloudina AR, and Demidov ON (2017) DNA damage-induced phosphatase Wip1 in regulation of hematopoiesis, immune system and inflammation. Cell Death Discov 3, 17018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McNerney ME, and Le Beau MM (2018) The Harmful Consequences of Increased Fitness in Hematopoietic Stem Cells. Cell Stem Cell 23, 634–635 [DOI] [PubMed] [Google Scholar]
- 104.Liu X, Zhang F, Zhang Y, Li X, Chen C, Zhou M, Yu Z, Liu Y, Zhao Y, Hao X, Tang Y, Zhu L, Liu L, Xie L, Gu H, Shao H, Xia F, Yin C, Tao M, Xie J, Zhang CC, Yang Y, Sun H, Chen GQ, and Zheng J. (2018) PPM1K Regulates Hematopoiesis and Leukemogenesis through CDC20-Mediated Ubiquitination of MEIS1 and p21. Cell Rep 23, 1461–1475 [DOI] [PubMed] [Google Scholar]
- 105.Kwon OC, Choi B, Lee EJ, Park JE, Lee EJ, Kim EY, Kim SM, Shin MK, Kim TH, Hong S, Lee CK, Yoo B, Robinson WH, Kim YG, and Chang E. (2020) Negative Regulation of Osteoclast Commitment by Intracellular Protein Phosphatase Magnesium-Dependent 1A. Arthritis Rheumatol 72, 750–760 [DOI] [PubMed] [Google Scholar]
- 106.Bourgeois B, Gilquin B, Tellier-Lebegue C, Ostlund C, Wu W, Perez J, El Hage P, Lallemand F, Worman HJ, and Zinn-Justin S. (2013) Inhibition of TGF-beta signaling at the nuclear envelope: characterization of interactions between MAN1, Smad2 and Smad3, and PPM1A. Sci Signal 6, ra49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Crane JL, Xian L, and Cao X. (2016) Role of TGF-beta Signaling in Coupling Bone Remodeling. Methods Mol Biol 1344, 287–300 [DOI] [PubMed] [Google Scholar]
- 108.Duan X, Liang YY, Feng XH, and Lin X. (2006) Protein serine/threonine phosphatase PPM1A dephosphorylates Smad1 in the bone morphogenetic protein signaling pathway. J Biol Chem 281, 36526–36532 [DOI] [PubMed] [Google Scholar]
- 109.Parvari R, Brodyansky I, Elpeleg O, Moses S, Landau D, and Hershkovitz E. (2001) A recessive contiguous gene deletion of chromosome 2p16 associated with cystinuria and a mitochondrial disease. Am J Hum Genet 69, 869–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jansen S, Geuer S, Pfundt R, Brough R, Ghongane P, Herkert JC, Marco EJ, Willemsen MH, Kleefstra T, Hannibal M, Shieh JT, Lynch SA, Flinter F, FitzPatrick DR, Gardham A, Bernhard B, Ragge N, Newbury-Ecob R, Bernier R, Kvarnung M, Magnusson EA, Wessels MW, van Slegtenhorst MA, Monaghan G, de Vries P, Veltman JA, Deciphering Developmental Disorders S, Lord CJ, Vissers LE, and de Vries BB (2017) De Novo Truncating Mutations in the Last and Penultimate Exons of PPM1D Cause an Intellectual Disability Syndrome. Am J Hum Genet 100, 650–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pelley JW (2006) Integrated Biochemistry, Elsevier [Google Scholar]
- 112.Huang B, Gudi R, Wu P, Harris RA, Hamilton J, and Popov KM (1998) Isoenzymes of pyruvate dehydrogenase phosphatase. DNA-derived amino acid sequences, expression, and regulation. J Biol Chem 273, 17680–17688 [DOI] [PubMed] [Google Scholar]
- 113.Brognard J, Sierecki E, Gao T, and Newton AC (2007) PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol Cell 25, 917–931 [DOI] [PubMed] [Google Scholar]
- 114.Gao T, Brognard J, and Newton AC (2008) The phosphatase PHLPP controls the cellular levels of protein kinase C. J Biol Chem 283, 6300–6311 [DOI] [PubMed] [Google Scholar]
- 115.Grzechnik AT, and Newton AC (2016) PHLPPing through history: a decade in the life of PHLPP phosphatases. Biochem Soc Trans 44, 1675–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Qiao M, Wang Y, Xu X, Lu J, Dong Y, Tao W, Stein J, Stein GS, Iglehart JD, Shi Q, and Pardee AB (2010) Mst1 is an interacting protein that mediates PHLPPs’ induced apoptosis. Mol Cell 38, 512–523 [DOI] [PubMed] [Google Scholar]
- 117.Bradley EW, Carpio LR, and Westendorf JJ (2013) Histone deacetylase 3 suppression increases PH domain and leucine-rich repeat phosphatase (Phlpp)1 expression in chondrocytes to suppress Akt signaling and matrix secretion. J Biol Chem 288, 9572–9582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Newton AC, and Trotman LC (2014) Turning off AKT: PHLPP as a drug target. Annu Rev Pharmacol Toxicol 54, 537–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bradley EW, Carpio LR, Newton AC, and Westendorf JJ (2015) Deletion of the PH-domain and Leucine-rich Repeat Protein Phosphatase 1 (Phlpp1) Increases Fibroblast Growth Factor (Fgf) 18 Expression and Promotes Chondrocyte Proliferation. J Biol Chem 290, 16272–16280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mattson AM, Begun DL, Molstad DHH, Meyer MA, Oursler MJ, Westendorf JJ, and Bradley EW (2019) Deficiency in the phosphatase PHLPP1 suppresses osteoclast-mediated bone resorption and enhances bone formation in mice. J Biol Chem 294, 11772–11784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chiu WS, McManus JF, Notini AJ, Cassady AI, Zajac JD, and Davey RA (2004) Transgenic mice that express Cre recombinase in osteoclasts. Genesis 39, 178–185 [DOI] [PubMed] [Google Scholar]
- 122.Debnath S, Yallowitz AR, McCormick J, Lalani S, Zhang T, Xu R, Li N, Liu Y, Yang YS, Eiseman M, Shim JH, Hameed M, Healey JH, Bostrom MP, Landau DA, and Greenblatt MB (2018) Discovery of a periosteal stem cell mediating intramembranous bone formation. Nature 562, 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nakamura T, Imai Y, Matsumoto T, Sato S, Takeuchi K, Igarashi K, Harada Y, Azuma Y, Krust A, Yamamoto Y, Nishina H, Takeda S, Takayanagi H, Metzger D, Kanno J, Takaoka K, Martin TJ, Chambon P, and Kato S. (2007) Estrogen prevents bone loss via estrogen receptor alpha and induction of Fas ligand in osteoclasts. Cell 130, 811–823 [DOI] [PubMed] [Google Scholar]
- 124.Ruiz P, Martin-Millan M, Gonzalez-Martin MC, Almeida M, Gonzalez-Macias J, and Ros MA (2016) CathepsinKCre mediated deletion of betacatenin results in dramatic loss of bone mass by targeting both osteoclasts and osteoblastic cells. Sci Rep 6, 36201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ghosh A, Shuman S, and Lima CD (2008) The structure of Fcp1, an essential RNA polymerase II CTD phosphatase. Mol Cell 32, 478–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang Y, Kim Y, Genoud N, Gao J, Kelly JW, Pfaff SL, Gill GN, Dixon JE, and Noel JP (2006) Determinants for dephosphorylation of the RNA polymerase II C-terminal domain by Scp1. Mol Cell 24, 759–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nesti E. (2015) Harnessing the master transcriptional repressor REST to reciprocally regulate neurogenesis. Neurogenesis (Austin) 2, e1055419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nesti E, Corson GM, McCleskey M, Oyer JA, and Mandel G. (2014) C-terminal domain small phosphatase 1 and MAP kinase reciprocally control REST stability and neuronal differentiation. Proc Natl Acad Sci U S A 111, E3929–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wohl SG, and Reh TA (2016) miR-124–9-9* potentiates Ascl1-induced reprogramming of cultured Muller glia. Glia 64, 743–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang H, Zhang L, and Sun T. (2018) Cohesive Regulation of Neural Progenitor Development by microRNA miR-26, Its Host Gene Ctdsp and Target Gene Emx2 in the Mouse Embryonic Cerebral Cortex. Front Mol Neurosci 11, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Varon R, Gooding R, Steglich C, Marns L, Tang H, Angelicheva D, Yong KK, Ambrugger P, Reinhold A, Morar B, Baas F, Kwa M, Tournev I, Guerguelcheva V, Kremensky I, Lochmuller H, Mullner-Eidenbock A, Merlini L, Neumann L, Burger J, Walter M, Swoboda K, Thomas PK, von Moers A, Risch N, and Kalaydjieva L. (2003) Partial deficiency of the C-terminal-domain phosphatase of RNA polymerase II is associated with congenital cataracts facial dysmorphism neuropathy syndrome. Nat Genet 35, 185–189 [DOI] [PubMed] [Google Scholar]
- 132.Clark AR, and Ohlmeyer M. (2019) Protein phosphatase 2A as a therapeutic target in inflammation and neurodegeneration. Pharmacol Ther 201, 181–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sundaram JR, Lee IC, and Shenolikar S. (2017) Translating protein phosphatase research into treatments for neurodegenerative diseases. Biochem Soc Trans 45, 101–112 [DOI] [PubMed] [Google Scholar]
- 134.Chen W, Wang Z, Jiang C, and Ding Y. (2013) PP2A-Mediated Anticancer Therapy. Gastroenterol Res Pract 2013, 675429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mazhar S, Taylor SE, Sangodkar J, and Narla G. (2019) Targeting PP2A in cancer: Combination therapies. Biochim Biophys Acta Mol Cell Res 1866, 51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.