ABSTRACT
Understanding the pharmacokinetic (PK) properties of a drug, such as clearance, is a crucial step for evaluating efficacy. The PK of therapeutic antibodies can be complex and is influenced by interactions with the target, Fc-receptors, anti-drug antibodies, and antibody intrinsic factors. A growing body of literature has linked biophysical properties of antibodies, particularly nonspecific-binding propensity, hydrophobicity and charged regions to rapid clearance in preclinical species and selected human PK studies. A clear understanding of the connection between biophysical properties and their impact on PK would allow for early selection and optimization of antibodies and reduce costly attrition during clinical trials due to sub-optimal human clearance. Due to the difficulty in obtaining large and unbiased human PK data, previous studies have focused mostly on preclinical PK. For this study, we obtained and curated the most comprehensive clinical PK dataset to date and calculated accurate estimates of linear clearance for 64 monoclonal antibodies ranging from investigational candidates in Phase 2 trials to marketed products. This allows for the first time a deep analysis of the influence of biophysical and sequence-based in silico properties directly on human clearance. We use statistical analysis and a Random Forest classifier to identify properties that have the greatest influence in our dataset. Our findings indicate that in vitro poly-specificity assay and in silico estimated isoelectric point can discriminate fast and slow clearing antibodies, extending previous observations on preclinical clearance. This provides a simple yet powerful approach to select antibodies with desirable PK during early-stage screening.
KEYWORDS: Clearance, mAb, monoclonal antibody, pharmacokinetics, biophysical properties, clinical pharmacokinetics
Introduction
Monoclonal antibodies (mAbs) can bind with high affinity and specificity to a broad range of protein targets, making them desirable molecules for drug development. Due to their large molecular weight, they have poor oral bioavailability and are typically delivered via injection. The IgG family of antibodies, with the exception of IgG3, have a long endogenous in vivo half-life (~18-21 days) in humans as a result of pH-dependent binding to the neonatal Fc receptor (FcRn), which enables recycling from endocytic compartments.1–3 Their long half-lives allow less frequent dosing strategies, and their effector functions may offer a substantial benefit for patients across numerous indications. Pharmaceutical companies have, therefore, invested heavily in research and development of IgG-based therapies.4
In practice, the half-lives of engineered antibodies can vary greatly from as long as four weeks to as little as a few days. Half-lives can depend on dose, route of administration, patient population, and other aspects of the study design.3,5,6 Furthermore, many antibodies exhibit non-linear pharmacokinetic (PK) profiles, and therefore half-life cannot be unambiguously determined from a given experiment. A key factor that influences the half-life is antibody clearance, which quantifies the rate at which a drug is removed from circulation, providing a useful measure for how long the antibody remains in the body after dosing.
Previously published studies have associated changes in antibody biophysical properties, including mAbs Fv charge and hydrophobicity, with improved clearance,7–10 suggesting that mAbs can be engineered early in the discovery process to improve the likelihood of observing favorable PK at later stages. Similarly, correlations shown between early-stage developability assays, including affinity-capture self-interaction nanoparticle spectroscopy (ACSINS), poly-specificity reagent (PSR), and mAbs clearance, may offer a way to prioritize antibodies during early-stage screening.11–13 However, elucidating the relationship between antibody biophysical properties and favorable human clearance remains a challenge. In a recent study investigating the solution behavior and PK for several antibodies, no correlation was found between measured in vitro properties, calculated pI, or calculated charge and the human PK values obtained from package inserts.14 At present, the best preclinical proxy for human antibody clearance comes from studies of non-human primates, such as cynomolgus monkeys,15,16 which, however, are performed on only a few promising molecules due to strict ethical guidelines and associated long timelines and high costs.17 While progress has also been made in predicting human clearance from early-stage in vivo models in animals such as FcRn transgenic mice and minipigs,18,19 these studies can be influenced by a number of factors including target specificity, volume of distribution, low affinity of competing mouse antibodies in transgenic mice, and the range of sequence and structure variability in the antibodies tested.
Here, we use published human clearance data from clinical trials to identify key in silico and in vitro approaches that can be used to triage antibodies during early-stage screening. At the time this work was conducted, there were 75 approved mAbs and over 200 in Phases 2 and 3 undergoing clinical trials.20,21 Despite this substantial increase in clinical-stage antibodies in recent years, obtaining a large, unbiased dataset of publicly available antibody clearance from humans remains a difficult task because different studies and reports are published at different times during clinical development prior to or after submission in different populations at different dose levels. Therefore, a curation step was performed on the collected data, and statistical modeling was used to obtain high confidence and unbiased estimates of linear clearance for 64 mAbs. Using a set of 28 in silico properties calculated from antibody structure and 12 measurements from in vitro developability assays published by Jain et al.,22 we applied the random forest machine learning algorithm to identify biophysical properties that were best able to distinguish between normal clearing and fast clearing antibodies in our data, with the goal of identifying a set of property cutoffs that can be used for antibody prioritization.
Results
Obtaining linear clearance values for clinical-stage mAbs
Clinical-stage human PK data for therapeutic mAbs were obtained from two databases, the Clarivate Analytics Integrity database and the Elsevier Pharmapendium.23,24 To allow relevant comparison between distinct mAbs, the data was curated and converted to uniform units, resulting in a comprehensive set of paired clearance rates and dose concentrations for an initial collection of 87 therapeutic antibodies, with a median of 10 measurements per molecule. For 22 antibodies, we observed more than 100 times difference between the highest and lowest reported clearance, with extremes typically coming from a single reference source. An outlier analysis was performed, keeping values of clearance below 72.8 mL/day/kg and greater than 0.35 mL/day/kg. For eight antibodies where the median lay outside this range, we retained all data as we reasoned, based on the median values, that atypical clearance values for these antibodies were more likely due to intrinsically high clearance rather than spurious outliers (Supplementary Figure S1). We manually removed most of these high clearance biologics from further analysis, as they either could not be fit or were not typical monoclonal antibodies and could not be compared to the more typical monoclonals.
For mAbs with sufficient data, non-linear least-squares regression was used to model the relationship between dose and clearance in order to obtain an estimate of linear clearance at high-dose ranges, where all receptors are presumed to be saturated (Figure 1a, Supplementary Figure S2). Our final dataset consisted of 64 mAbs for which the clearance rates could be compared without the confounding effects of target-mediated drug disposition (TMDD) (Table 1). This was necessary in order to analyze the contribution of antibody “intrinsic” factors, such as biophysical properties, to clearance.
Figure 1.
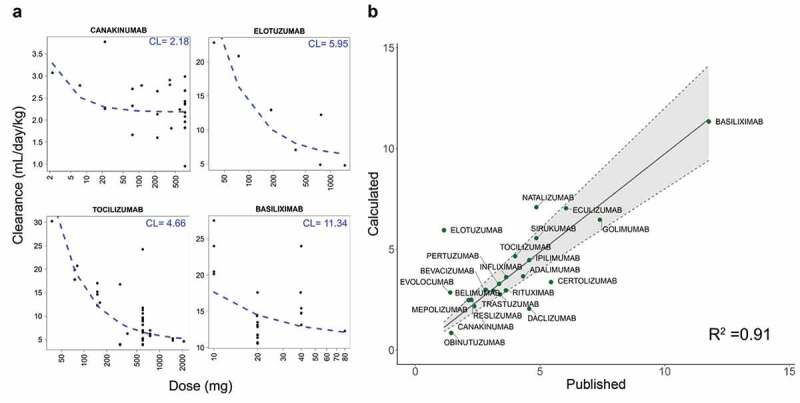
A. Clearance (CL) is plotted against dose for four antibodies. Regression line is shown in dashed blue. Estimated linear clearance is indicated in blue in the upper right. Dose values are shown on a log scale for clarity. B. Comparison of calculated clearance values with data from published literature. Gray area indicates 20% error from published values, with R2 shown in the lower right
Table 1.
Linear clearance rate estimates with residual standard error (RSE) reported for 64 therapeutic antibodies. XI, ZU & HU indicate chimeric, humanized or human origin. aAntibody drug conjugate, bpegylated Fab
| mAb | Clearance (mL/day/kg) | RSE | Isotype | Origin | mAb | Clearance (mL/day/kg) | RSE | Isotype | Origin | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Abituzumab | 3.52 | 2.63 | G2 | ZU | Guselkumab | 4.35 | 0.31 | G1 | HU | ||
| Adalimumab | 3.66 | 0.40 | G1 | HU | Infliximab | 3.62 | 0.48 | G1 | XI | ||
| Alirocumab | 3.16 | 0.28 | G1 | HU | Ipilimumab | 4.46 | 0.43 | G1 | HU | ||
| Atezolizumab | 2.70 | 1.20 | G1 | ZU | Matuzumab | 5.58 | 1.87 | G1 | ZU | ||
| Basiliximab | 11.34 | 1.00 | G1 | XI | Mavrilimumab | 4.33 | 0.32 | G4 | HU | ||
| Bavituximab | 31.22 | 0.74 | G1 | XI | Mepolizumab | 2.48 | 0.26 | G1 | ZU | ||
| Belimumab | 2.95 | 0.23 | G1 | HU | Natalizumab | 7.09 | 1.82 | G4 | ZU | ||
| Bevacizumab | 2.99 | 0.40 | G1 | ZU | Necitumumab | 2.35 | 1.11 | G1 | HU | ||
| Brentuximab vedotina | 22.19 | 0.40 | G1 | XI | Nimotuzumab | 4.64 | 1.44 | G1 | ZU | ||
| Brodalumab | 2.99 | 0.01 | G2 | HU | Nivolumab | 3.32 | 0.96 | G4 | HU | ||
| Canakinumab | 2.18 | 0.35 | G1 | HU | Obinutuzumab | 0.85 | 0.82 | G1 | ZU | ||
| Carlumab | 12.82 | 1.30 | G1 | HU | Ofatumumab | 2.90 | 2.51 | G1 | HU | ||
| Certolizumab pegolb | 3.37 | 0.14 | G1 | ZU | Onartuzumab | 6.96 | 0.03 | G1 | ZU | ||
| Cetuximab | 7.88 | 1.32 | G1 | XI | Pembrolizumab | 3.16 | 0.07 | G4 | ZU | ||
| Cixutumumab | 3.25 | 0.81 | G1 | HU | Pertuzumab | 3.29 | 0.46 | G1 | ZU | ||
| Daclizumab | 2.05 | 0.98 | G1 | ZU | Ponezumab | 2.12 | 0.23 | G2 | ZU | ||
| Dalotuzumab | 7.42 | 0.93 | G1 | ZU | Ramucirumab | 3.01 | 1.00 | G1 | HU | ||
| Daratumumab | 1.60 | 1.64 | G1 | HU | Reslizumab | 2.49 | 0.37 | G4 | ZU | ||
| Duligotuzumab | 6.38 | 0.44 | G1 | ZU | Rilotumumab | 2.76 | 0.29 | G2 | HU | ||
| Eculizumab | 7.04 | 1.26 | G2 | ZU | Rituximab | 2.96 | 1.70 | G1 | XI | ||
| Efalizumab | 0.50 | 176 | G1 | ZU | Secukinumab | 2.02 | 0.36 | G1 | HU | ||
| Eldelumab | 5.54 | 0.47 | G1 | HU | Sifalimumab | 2.55 | 0.18 | G1 | HU | ||
| Elotuzumab | 5.95 | 1.06 | G1 | ZU | Siltuximab | 4.00 | 0.79 | G1 | XI | ||
| Emibetuzumab | 4.02 | 0.19 | G4 | ZU | Sirukumab | 5.56 | 0.30 | G1 | HU | ||
| Etrolizumab | 3.56 | 0.16 | G1 | ZU | Tabalumab | 1.70 | 1.67 | G4 | HU | ||
| Evolocumab | 2.86 | 0.03 | G2 | HU | Tanezumab | 2.45 | 0.30 | G2 | ZU | ||
| Farletuzumab | 2.87 | 0.43 | G1 | ZU | Tigatuzumab | 4.90 | 0.50 | G1 | ZU | ||
| Ficlatuzumab | 5.07 | 0.47 | G1 | ZU | Tocilizumab | 4.66 | 0.85 | G1 | ZU | ||
| Figitumumab | 4.20 | 1.36 | G2 | HU | Trastuzumab | 2.76 | 1.49 | G1 | ZU | ||
| Ganitumab | 12.59 | 1.34 | G1 | HU | Tremelimumab | 3.45 | 0.27 | G2 | HU | ||
| Girentuximab | 5.23 | 0.75 | G1 | XI | Vedolizumab | 2.10 | 2.18 | G1 | ZU | ||
| Golimumab | 6.47 | 0.36 | G1 | HU | Veltuzumab | 1.00 | 0.54 | G1 | ZU | ||
As an additional confirmation that our method of obtaining linear clearance values is valid, the clearance values we obtained were compared to human clearance for 22 antibodies for which the published literature3,12,25–29 supplied reliable clearance estimates. We found strong agreement (within 20%) between our calculated clearance and the published values for 16 of the antibodies (Figure 1b). This indicates that the regression model can indeed be successfully used to obtain estimates of linear clearance for further analysis.
Influence of antibody origin and isotype on clearance
As we are primarily interested in understanding the correlates of antibody intrinsic linear clearance mechanisms, such as biophysical properties, we sought to analyze what role other, potentially confounding clearance mechanisms such as anti-drug antibodies (ADA) might play in our calculated clearance values. Antibody drugs, as “foreign” proteins, can induce ADA in humans, and ADA can affect the PK of protein drugs. We observed a statistically significant higher clearance among chimeric mAbs compared to either human or humanized ones, in our dataset (Figure 2a), and wondered whether this was due to differences in antibody intrinsic factors or to the increased “foreignness” of the chimeric antibodies. A comprehensive review of therapeutic mAbs30 found that while some murine or chimeric antibodies had faster clearance, overall no significant differences in the population PK of chimeric, humanized, and fully human antibodies were noted. In support of this, the observation that two mAbs with fast clearance, carlumab and ganitumab, (>10 mL/day/kg) were of human origin suggests the influence of a particular sequence, structural or biophysical motifs, rather than murine origin per se, on fast clearance. Furthermore, the clearance reported in well-designed clinical PK studies, as used in this dataset, filters out the effect of ADA, either by focusing on the first weeks after first dosing in drug naïve subjects, or by the design of the PK study and analysis itself, excluding ADA positive subjects or by investigating ADA positivity as a covariate in a population PK analysis.30,31 A high incidence of clearing ADA was not observed in clinical PK studies in drug-naïve subjects, for the fast clearing antibodies in our dataset.32–34 As consequently we can exclude ADAs, we attribute the difference we observe (Figure 2a) to different sequence motifs or biophysical properties of the chimeric antibodies. In further support of this, another fast clearing mAb in our dataset is brentuximab vedotin, a chimeric antibody conjugated to the cytotoxin MMAE, which could increase its hydrophobicity, leading to a higher clearance.35 This underscores the fact that molecule properties affect the PK of antibody-based therapeutics, independently of origin.
Figure 2.
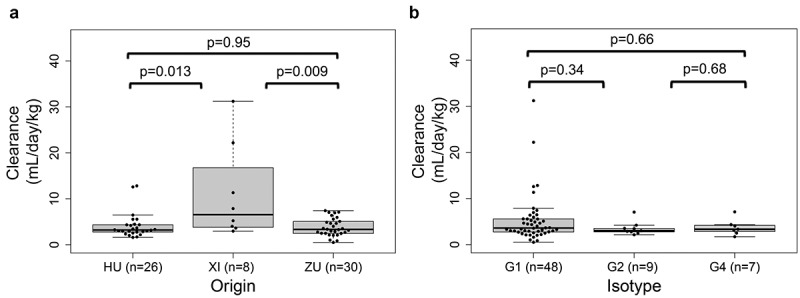
A. Box and whisker plots of calculated clearances separated by antibody origin (HU for human, XI for chimeric, ZU for humanized). A two-sided Wilcoxon rank sum test was used to determine the p-values shown. B. Box and whisker plots of calculated clearances separated by IgG isotype
We further compared clearance rates across different IgG isotypes. As Fc-receptor mediated elimination is thought to play a large role in linear clearance,30 differences in clearance across isotypes might be expected. No significant difference was observed between the isotypes, yet clearance of IgG1 mAbs had the most variability and represented all antibodies with fast clearance (Figure 2b). As the sample size for IgG2 and IgG4 is small, it cannot be unequivocally determined if the absence of high clearance rates in these isotypes is meaningful. However, it remains possible and likely that the lack of high clearance rates observed among IgG2 and IgG4 may be due to their biological properties, for example reduced Fc receptor binding in these isotypes.36,37 Larger clinical datasets for these isotypes are needed to serve as a point of future exploration. In this study, we focus on the IgG1 isotype when appropriate, to ensure properties affecting clearance are not simply correlated with isotype.
Impact of developability parameters on clearance
The study by Jain et al.22 introduced a set of developability “red flags”, whereby highly correlated assays were clustered together, and for each antibody a binary score was assigned to each cluster, based on whether a poor developability measure was present among the assays. Approved antibodies were shown to have the fewest total flags. In this study, we used the same clustering among in vitro assays and flag assignment and analyzed the relationship with clearance. Although all of our 64 study mAbs were represented in the set of 137 mAbs evaluated by Jain et al., they were expressed as IgG1 by the authors, regardless of their isotype in the clinic. We therefore restricted our analysis of in vitro properties on the 48 natively IgG1 antibodies in our full dataset, since measurements for the non-IgG1 antibodies may not be appropriate for assessing their impact on in vivo clearance. We observed that, for the 48 IgG1 mAbs, which included 31 approved drugs, 7 in Phase 3, and 10 in Phase 2, those with no flags had overall significantly slower clearance compared to those with one or more red flags (Figure 3). Considering that in order to progress to later stage clinical trials and eventually to market approval, mAbs must achieve sufficient human exposure to support a therapeutic effect, it is consistent with Jain et al.’s methodology that clearance rates are lower in mAbs with fewer flags. It is intriguing that the set of biophysical properties22 that they chose to examine might be causally related to some clearance mechanisms, and we explore this further below.
Figure 3.

Boxplot comparing clearance rates for antibodies with zero developability flags vs one or more developability red flags. Approved antibodies are shown in red, Phase 2 antibodies in green and Phase 3 in blue
Correlation of biophysical properties with mAb clearance
To study the relationship between mAb biophysical properties and clearance, we again leveraged the rich dataset published by Jain et al.22 using the results of 12 in vitro assay measurements. Assays performed in the study include measures of nonspecific binding, aggregation, hydrophobic interactions, expression, and thermal stability.
For each antibody, we additionally calculated in silico biophysical descriptors based on homology modeled structures of the variable fragment (Fv) domain obtained using the Molecular Operating Environment (MOE 2019.01).38 The 23 descriptors calculated include various measures of hydrophobicity and charge, including hydrophobic patch area, and structure-based isoelectric point. The same homology models were then used to calculate five in silico descriptors for antibody developability included in the Therapeutic Antibody Profiler (TAP) developed by Raybould et al.,39 which were implemented internally. All properties can be found in Supplementary Table 1. Among our 40 total in vitro and in silico properties, we found no strong correlation of any single property with linear clearance (Figure 4), although the strongest correlations that were observed were also statistically significant (Supplementary Figure S3). When assessing the properties on the 48 IgG1 antibodies, the highest Pearson correlation observed is 0.51 for the PSR assay. Of the in silico descriptors, the highest absolute Pearson correlation observed is 0.31 for pI_3D (described below). This is not unexpected, as the biophysical factors determining clearance are expected to be complex and likely interdependent.
Figure 4.
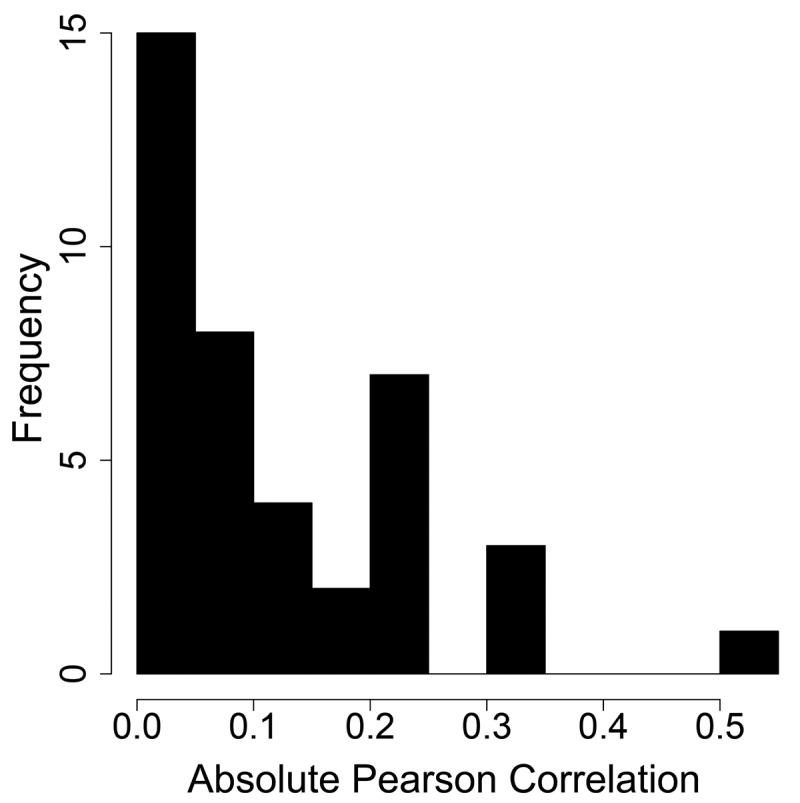
Histogram showing the absolute value of the correlations between clearance and all 40 biophysical properties. The highest correlation found is 0.51 for PSR
Next, we examined whether pairs of properties could be used to classify antibodies in a discrete fashion as having either fast or normal clearance. Determining a threshold for normal human clearance is a challenge because clinical-stage mAbs typically have been selected based on promising preclinical studies in non-human primates and are enriched for acceptable clearance rates. The lack of publicly shared preclinical datasets further limits the availability of additional information that could be used to probe the true range of observed mAb clearance during development. In this study, an appropriate linear clearance threshold was determined based on the distribution of the available clinical data. For the majority of antibodies, 59 of 64 mAbs, the clearance rate was below 8 mL/day/kg, in agreement with typical ranges that have been described for therapeutic antibodies,1,12,15 and followed a log-normal distribution (Figure 5). The remaining five mAbs had exceptionally high clearance rates and were excluded from the subsequent analysis. The mode of the log-normal distribution was at 2.53 mL/day/kg, matching closely with the 2.8 mL/day/kg clearance rate of an endogenous antibody reported by Ryman and Meibohm.1 Antibodies with clearance below 2.5 mL/day/kg were defined as having slow clearance, but many approved antibodies above this threshold have acceptable clearance properties in the clinic. The mean clearance was 3.7 mL/day/kg and the standard deviation was 1.7 mL/day/kg. We therefore selected as our clearance threshold a value of 5.4 mL/day/kg, corresponding to one standard deviation above the mean, which separated the data into 48 (75%) normal clearing antibodies and 16 (25%) fast clearing antibodies. A linear clearance threshold of 10 mL/day/kg in cynomolgus monkeys was previously introduced by Hotzel and colleagues for fast clearing antibodies in this species.11 Applying allometric scaling, we determined that this threshold corresponds to a human clearance of around 5.25 mL/day/kg, further supporting the value of 5.4 mL/day/kg used in this study.
Figure 5.
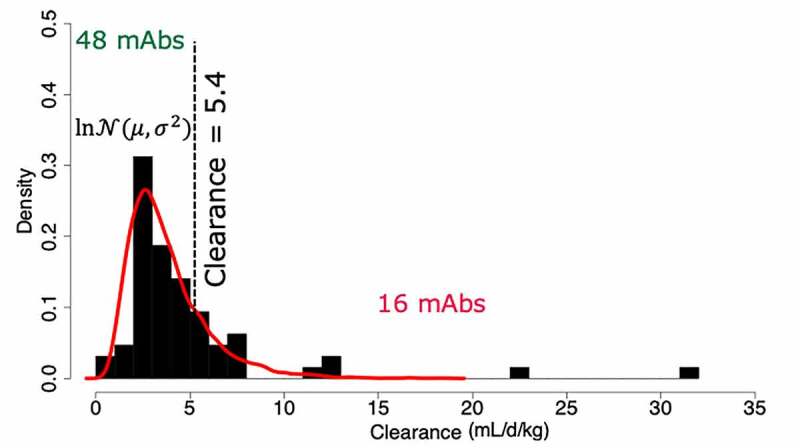
Histogram of calculated clearance rates with the log-normal fit to the data in red separating the data into 48 normal antibodies, and 16 fast clearing. A threshold of 5.4 mL/day/kg was selected, above which clearance is considered fast
In work performed by Sharma et al.,9 the authors proposed that either excess hydrophobicity or extreme charge values can potentially lead to nonspecific binding and cause faster clearance. They identified criteria that distinguished between slow and fast clearing mAbs in cynomolgus monkeys using the net charge of the antibody Fv region at pH 5.5 (fvcharge5.5) and the sum of the hydrophobic index along several CDRs (HI_sum). We performed a similar analysis here for human clearance with the classification determined as described above, using the features identified as relevant by Sharma and colleagues, with values calculated with a custom implementation using MOE 2019.01. The criteria used for cynomolgus clearance captured only 24 (50%) of our normal clearing antibodies (Figure 6a, solid-line red box), not indicating any significant advantage in prioritizing antibodies within the box. By adapting the thresholds of the original model for cynomolgus monkeys such that antibodies with normal human clearance were required only to have HI_sum below 10 (Figure 6a, dashed red box), we captured 43 (90%) of the normal clearing antibodies (including identifying all but one of the very slow clearing mAbs, blue dots in Figure 5a) while excluding 8 (50%) of fast clearing antibodies. Thus, we achieved similar or somewhat improved performance for identifying antibodies with normal human clearance as Sharma et al. achieved for cynomolgus monkey, but with slightly poorer specificity for identifying fast clearing antibodies. Furthermore, Figure 6a suggests that for this dataset, HI_sum does not help discriminate, as we had to adapt to a very large range (HI_sum <10) and this range still excluded slow clearing antibodies, including a very slow clearing mAb that would have been retained using only the fvcharge5.5 criteria.
Figure 6.
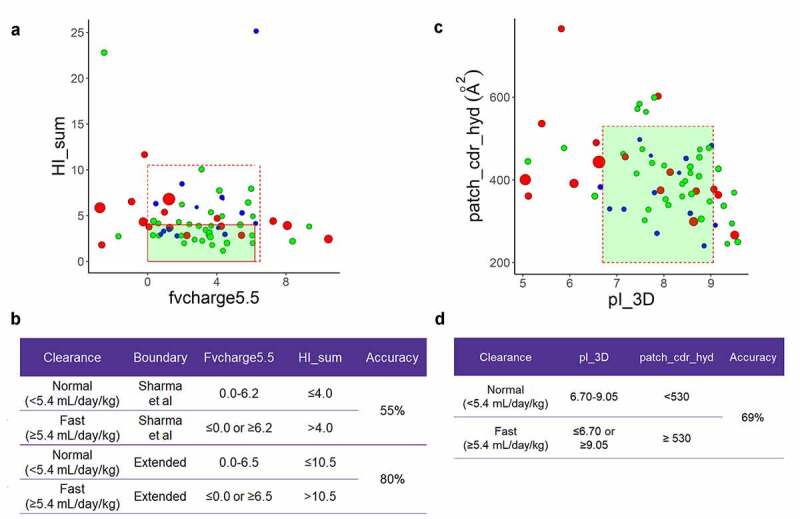
a. Fv charge at pH 5.5 and hydrophobic index sum (HI_sum) of L1, L3 and H3 as described in Sharma et al.9 The red solid box corresponds to the boundary for antibodies with normal clearance in cynomolgus monkeys as described by Sharma et al. The red dashed box indicates the extended area. Antibodies colored blue have clearance less than or equal 2.5 mL/day/kg, antibodies colored green have clearance between 2.5 and 5.4 mL/day/kg. Antibodies with fast clearance (>5.4 mL/day/kg) are colored in red. Marker size is proportional to the magnitude of the clearance. b. Cutoff criteria for Fv charge and HI_sum. The accuracy is presented, defined as the percentage of all mAbs that were correctly grouped using these criteria. c. Surface area of hydrophobic patches near CDRs (patch_cdr_hyd) and structure-based predicted isoelectric point (pI_3D). The red dashed box indicates boundaries for the enrichment box. Antibodies colored as in part A. d. Cutoff criteria and accuracy for patch_cdr_hyd and pI_3D
In an effort to apply the same biophysical theory but to improve the performance further, we evaluated whether the structure-based features calculated from homology models would improve discrimination compared to sequence-based features. For hydrophobicity, in place of HI_sum we selected patch_cdr_hyd, which is the surface area of the hydrophobic patches near the complementarity-determining regions (CDRs) averaged over a conformational ensemble, and has previously been shown to be correlated with developability.40 In place of the fvcharge5.5, we used the structure-based pI prediction where the residue pKa values are influenced by their 3D environment, and the values are averaged over the same conformational ensemble (pI_3D). Using these structure-based descriptors with the thresholds shown in Figure 6c and D, we improved discrimination of fast clearing antibodies, excluding 11 of 16 (69%) while retaining 86% of very slow clearing mAbs (blue dots, Figure 6c). However, correct identification of normal human clearance was not improved using these features and thresholds (67% correctly identified).
Application of machine learning algorithm to identify biophysical properties influencing human clearance
As neither single biophysical properties nor pairs of properties previously identified for preclinical clearance data appeared to optimally discriminate normal and fast human clearance, we turned to machine learning to identify more complex relationships between biophysical properties and human clearance. While the available data are insufficient to build a true predictive model, our goal was simply to identify properties that have significant influence and potentially meaningful, testable relationships with human clearance.
We implemented the random forest machine learning algorithm to identify biophysical properties that most strongly discriminate linear clearance. In brief, the random forest used as input features the in vitro and in silico biophysical properties to train a large number of decision trees, each of which tries to predict the linear clearance of mAbs. While a single tree produces high bias for the input data, a collection of many trees constructed from subsampled data and subsets of input features can be averaged to make a classification with minimal bias. The extent to which each individual tree is accurate for classifying the data in the training dataset is used to rank the features in order of importance (Supplementary Figure S4). Due to the relatively small size of our dataset, we used the random forest to perform a classification task, rather than regression, binning mAbs into two groups corresponding to normal clearance and fast clearance, with a threshold of 5.4 ml/day/kg described above.
Since the in vitro assay values22 are only appropriate to use for IgG1 antibodies, we trained our random forest model on only the IgG1 antibodies, which consisted of 34 normal clearing and 14 fast clearing antibodies. Multiple runs of the random forest produced variation among top properties, likely because of the small size of the training set. We therefore performed 10,000 repetitions of the random forest and tabulated the number of times a biophysical property was ranked among the top five most important. This yielded a consistent set of top-ranked properties for classification (Supplementary Figure S5). The top ranked biophysical property was the in silico calculated sequence-based isoelectric point (pI_seq). However, we noticed that many of the top properties were strongly correlated. In particular, among the properties occurring most frequently in the top 10, four were strongly correlated with pI_seq, resulting in other properties having lower rank. We therefore clustered all properties by Spearman correlation (Supplementary Figure S6), and iteratively selected the best property in each cluster. The final tabulated collection of random forest runs produced a set of rankings of uncorrelated properties, with isoelectric point (pI_seq) and poly-specificity reagent (PSR) appearing among the top five uncorrelated properties in every run (Figure 7a). Notably, both pI_seq and PSR have been identified as predictive of preclinical clearance in previous studies.10,13,41
Figure 7.
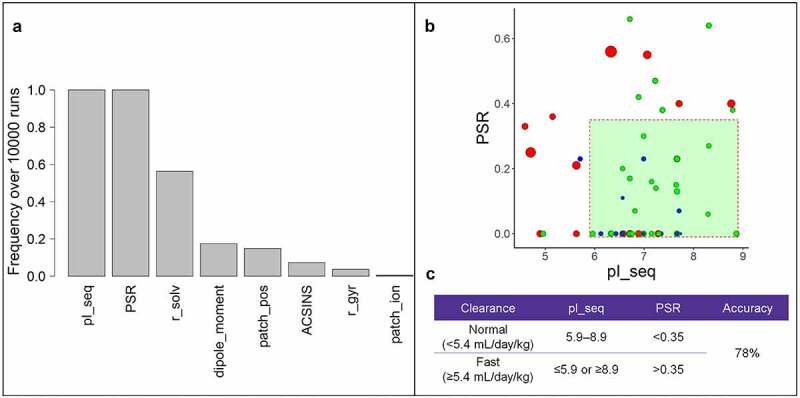
A. Properties with highest impact on clearance after 10000 runs of the random forest. B. Sequence based isoelectric point using the modeled Fv region (pI_seq) plotted against the poly specificity reagent (PSR). Antibodies colored blue have clearance less than or equal 2.5 mL/day/kg, antibodies colored green have clearance between 2.5 and 5.4 mL/day/kg. Antibodies with fast clearance >5.4 mL/day/kg are colored in red. Marker size is proportional to the magnitude of the clearance. Marker type indicates antibody isotype. C. Cutoff criteria and accuracy for separating normal clearing and fast clearing antibodies for pI_seq and PSR
While the random forest algorithm is able to determine important features, it does not provide a specific set of threshold values for mAb classification (Supplementary Figure S7), We therefore again plotted the clearance values as above, this time using the features identified by the random forest as the axes (Figure 7b). Using this visualization, we were able to identify threshold values for the PSR assay and pI_seq (Figure 7c) that captured 83% of antibodies with normal human clearance (similar to the performance of HI_sum and fvcharge5.5 for normal cynomolgus clearance9), including capturing all but one of the very slow clearing antibodies (blue dots, Figure 7b). Furthermore, 63% of fast clearing antibodies were excluded using these features and thresholds. It is notable that this performance applied to all isotypes in the dataset, although the algorithm was run on only the subset of 48 IgG1 antibodies.
Discussion
Antibody PK involves complex interactions between an antibody and its physiological environment. Publicly available datasets describing human PK of mAbs are limited in the number of antibodies described and lack expert curation to ensure that reported experimental conditions and values are directly comparable. Although a well-curated database of PK properties for therapeutic mAbs does not exist, we have shown that thoughtful selection and curation of a large dataset are able to provide comparable and reasonably accurate values of linear clearance that can be used for further analysis. Our approach relies on the observation that, for most therapeutic mAbs showing TMDD, the clearance decreases with dose, reaching a plateau at a higher dose when all receptors are saturated. While this work does not attempt to measure effects of target mediated clearance, an accurate model of clearance as a function of the dose could provide such insight as well, which has potential for improving clearance prediction in preclinical and clinical studies. We focused here on linear clearance, as a basis for understanding antibody intrinsic, non-target mediated effects on clearance. By compiling a large amount of human PK data, carefully standardizing the units, and applying a stringent statistical modeling approach, we were able to approximate linear clearance for 64 mAbs of varied isotype and clinical phase, the most comprehensive such dataset for human clearance values published as of now, to the best of our knowledge.
In this panel of 64 mAbs for which biophysical properties were characterized, we did not observe in human clearance values the strong correlations between clearance and the PSR and ACSINS assays that have previously been shown in C57BL/6 and Tg32 mice,12,13,42 nor any strong correlation with 40 measured in vitro and calculated in silico properties (Figure 2 and Supplementary Figure S3). Using criteria shown to be useful in discriminating normal and fast clearance in cynomolgus monkey,9 while adapting the threshold values, yielded reasonable discrimination of antibodies with normal clearance, and better than random but not very good discrimination of fast clearing antibodies (Figure 6a,b). Discrimination of fast clearing antibodies was somewhat improved by using structure-based properties calculated in silico using MOE (Figure 6c,d). Overall, it is challenging even with this well-curated dataset to identify properties that may discriminate fast and normal clearance, not least because the dataset is heavily skewed (75%) toward antibodies with normal and slow clearance, as expected for marketed and late-stage therapeutics.
Given the skewed dataset and the combinatorially large number of potentially useful properties and property combinations, we used a machine learning algorithm to systematically evaluate the influence of each property on clearance, while accounting for correlated properties. Using this approach, the algorithm identified PSR and pI_seq as properties that most strongly influence the rate of linear clearance in humans, in line with previous observations.10,13,41 We were able to determine threshold values that displayed reasonably good performance in identifying antibodies with normal clearance and especially at excluding antibodies with fast clearance (Figure 7). Even if not all normal clearing antibodies can be identified (existence of false negatives), it would be very useful to simply filter out fast clearing antibodies at an early discovery stage, where many diverse hits will meet the desired pharmacological profile but are generally uncharacterized in relevant PK models until much later. While we obtained similar performance as reported previously for cynomolgus PK data, the precise properties that give the best performance on each preclinical or clinical dataset vary, perhaps due to subtle species-specific differences. While preclinical models, especially non-human primates, remain a powerful tool to predict human clearance, these results highlight the importance of investigating the biophysical causes and correlates of human clearance rates directly on human datasets.
It is notable that in studies of both preclinical8–11,13,15 and clinical clearance,12 nonspecific binding, hydrophobicity and/or charge appear to be relevant to clearance. Sharma et al.9 did not perform poly-specificity assays, but commented that regions of high hydrophobicity and extreme charge can cause nonspecific binding leading to faster clearance, and found that HI_sum and fvcharge5.5 together can separate antibodies having fast clearance from those having normal clearance in cynomolgus monkeys. Kelly et al.13 performed nonspecificity assays and found that the higher the level of PSR binding, the faster the measured clearance for 16 antibodies in mice (Pearson correlation of 0.73). However, when they applied HI_sum and fvcharge5.5, the properties were not able to separate or enrich for slow, normal, or fast clearance. In the work by Igawa et al.,10 it was determined that unusually high pI values correspond to fast clearance in cynomolgus monkeys for human IgG4 antibodies. However, Kingsbury et al.14 observed no correlation between measured or calculated pI values to human clearance and half-life values obtained from the package inserts of about 40 antibodies, similar to our observation (Supplementary Figure S3). They included the TAP parameters39 in their analysis and found they were not useful for predicting either solution behavior or PK; similarly, we found that the TAP charge descriptors SFvCSP, PPC, PNC do not appear to be useful for PK predictions. For the current dataset of 64 clinical-stage antibodies, we found that PSR binding in combination with the calculated pI_seq was the most useful predictor for clearance in humans and can enrich for slow clearance and separate the fast clearing antibodies from those with normal clearance. The HI_sum and fvcharge5.5 descriptors together were able to enrich for normal clearance versus fast; however, HI_sum makes no contribution and the enrichment is entirely driven by fvcharge5.5. We found that the patch_cdr_hyd descriptor is able to filter some fast clearing antibodies while retaining all slow clearing antibodies, and, when used in combination with pI_3D, can enrich for slow clearance versus fast almost as well as PSR/pI_seq, but perhaps not as well for normal clearance as does fvcharge5.5.
Overall, when selecting therapeutic antibody candidates, they would ideally not have high measured values of PSR binding nor a high calculated hydrophobicity or an extreme charge. In the absence of PSR binding or other poly-specificity experiments, an in silico Fv charge descriptor such as pI_3D can likely enrich for slow clearance and filter out fast clearing antibodies, while an Fv hydrophobicity descriptor such as patch_cdr_hyd may filter additional fast clearing antibodies, as we have demonstrated with the current dataset. Furthermore, antibodies with high values for patch_cdr_hyd or other hydrophobicity descriptors are best avoided due to developability concerns such as aggregation40,43,44 and because human IgG1 antibodies with relatively low charge descriptor values for the variable region have been shown to have increased viscosity.9,45–47 Selected candidate mAbs may be further optimized for longer half-lives by determining variants that bind in pH-dependent manner to the FcRn7 and demonstrate relatively low nonspecific binding11,13 or by investigation using animal models.8,15
We expect that the true relationships between the biophysical properties of antibodies and their in vivo PK is driven by a complex combination of multiple factors; therefore, large and diverse datasets are necessary in order to improve the predictive power of machine learning approaches. It is unlikely that any single entity (e.g., biopharmaceutical company, academic group) currently possesses sufficient data to generate robust results to this challenging problem. The value of understanding the relationship between biophysics and PK of mAbs will enable the more efficient use of drug development resources, thereby facilitating the development of more efficacious therapies for patients in need. To achieve these insights, the biopharmaceutical industry would greatly benefit from having additional data available through precompetitive sharing, and by maintaining a single database of high-quality PK data for clinical-stage antibodies. Further investigation of preclinical and especially clinical mAbs clearance on larger datasets is required. We believe this analysis, based on curated clinical PK data for 64 advanced and approved mAbs, may serve as a useful resource of linear clearance data for different antibodies in humans and seed further cooperation and investigations in this important area.
Materials and methods
Selecting antibody PK data
All PK data for mAbs were collated from the Integrity database from Clarivate Analytics and PharmaPendium from Elsevier.23,24 These databases contain data on drug development from a variety of published primary sources, and provide multiple values for dose, clearance, and method of administration for antibody drugs. The units reported varied by study and were converted to standard units of mg for dose, and milliliters per day per kilogram for the clearance rate, with an assumed 75 kg as the average adult weight and 1.65 m2 for the body surface area. Our analysis focuses on clearance rather than half-life because it is a primary PK parameter, and because neither half-lives nor volumes of distribution were explicitly reported in these databases. Additionally, in order to avoid discrepancies in the relationship between the administered dose and the reported clearance due to method of administration, we focus only on data associated with intravenous injection of the drug.
Outlier removal of antibody clearance rates
Selected antibody clearance data were analyzed on a log scale. Measurements of clearance were required to be within two times the interquartile range (IQR) of the entire dataset. For 78 antibodies, clearance rates below 0.35 and above 72.8 mL/day/kg were excluded. The interquartile range is the difference between the 25th and 75th percentile of the data, and therefore includes the middle 50% of the dataset. Using a threshold based on IQR provides a measure of variability from the median and captures both the range and density of the data, without assuming a probability distribution. The other eight antibodies had a median that lay outside these bounds, likely due to intrinsically higher clearance, and no outlier removal was applied. Several of these were removed from further analysis, as they were antibody fragments or murine monoclonals rather than human Fc-containing monoclonals.
Determining linear clearance
Since mAb clearance often depends on dose, a statistical model was used to obtain the linear clearance from the data. Clearance () and dose () for individual antibodies were fitted by a non-linear least-squares regression using the stats package in the R language for statistical computing
The asymptote, a, estimates the clearance at dose ranges at which the receptor is presumably saturated and the PK disposition is apparently independent of receptor concentration. This approach captures the relationship between dose and clearance without assuming a specific PK model for each drug, and thus provides an unbiased estimate across multiple therapeutics. Antibodies were retained for further analysis if at least three data points were available and a positive value for the asymptote was returned.
Linear regression was used to compare with available published values, using the “lm” function in R to obtain the equation of the line and residual standard error.
Fitting a clearance probability distribution
A histogram of the clearance data was produced in R, using 23 bins based on the Freedman-Diaconis rule:48
where is the bin width as determined by the IQR
The parameters of the log-normal distribution were calculated from the logarithm transformed bulk data, using values up to the first gap among bins, which occurred at clearance of 8 mL/day/kg.
where and . A log-normal distribution was then generated in R using the “rlnorm” function in the stats package with 5000 data points: .
Allometric scaling for evaluating human clearance
The human clearance threshold assessed in this study was compared to data published by Hotzel et al.11 by simple allometric scaling between cynomolgus monkeys and humans using the following equation as described by Deng et al.15
where is the clearance rate for an antibody, is the body weight, and is an allometric scaling exponent. The average body weight used was 3 kg for cynomolgus and 75 kg for humans, with allometric exponent of 0.8 as previously described.19
Antibody homology model generation
Homology models of the variable region of the antibodies studied were created using the Antibody Modeler application in MOE 2019.01 with default settings.38 For each model, the Fv sequence was supplied and the framework structure from the Protein Data Bank (PDB)49 with highest identity to the query used as the template. For each CDR, the CDR template structure of the same type and length and highest identity to the query from the PDB was grafted onto the template. Any remaining mutated residues were modeled by sidechain packing rotamer exploration, and the final model was energy minimized with the Amber10 forcefield.50 Of the 64 antibodies modeled, 27 were 100% identical to their respective chimeric template and the average identity of the 37 remaining antibodies to their light- and heavy-chain templates ranged from 87% to 100%. Also, using these CDR-grafted chimeric templates ensures that there are no gaps in the alignments used for homology modeling. This automated method in combination with property calculations based on conformational sampling averages was shown to be effective by Jetha et al.40 for modeling antibodies and performing developability calculations.
Property calculations on 3D models
In silico biophysical properties of the antibodies were calculated on the Fv models using the Protein Properties application in MOE 2019.01. For each, the ensemble sampling method was used to produce 100 conformations with LowModeMD51 and Protonate3D,52 allowing flexible CDRs and sidechains as well as alternate protonation states. The descriptors calculated were averaged over this ensemble of 100 conformations for each antibody to provide values based on averaged models and reduce potential sensitivity from using a single homology model. In particular, the sequence based pI_seq descriptor is calculated using the algorithm of Sillero and Ribeiro,53 as is the pI_3D descriptor but with pKa values obtained by the PROPKA method54 on the ensemble of Fv models. The patch_cdr_hyd descriptor is calculated as the sum of the surface area of the hydrophobic patches involving any part of the CDR surface averaged over the 100 ensemble conformations.
Five additional properties from the Therapeutic Antibody Profiler39 (TAP) were calculated using an internal implementation written in Python. TAP properties are described in the Supplementary Table 1.
Random forest analysis
The random forest analysis was performed in R (randomForest package), using 1000 decision trees trained on the 48 IgG1 antibodies classified as normal or fast clearing. The number of properties selected for each tree was determined by the square root of the total number of biophysical properties in the analysis, the recommended approach for random forest classification.55 For the initial dataset, this yielded six properties, which decreased to three for the uncorrelated properties. Uncorrelated properties, determined by Pearson correlation, were selected by running the random forest 10000 times and selecting the most important property among correlated groups (Supplementary Figure S6). This step was done iteratively in order to allow for changes due to removal of properties in the previous runs. The top five most influential properties were considered for enrichment analysis.
Supplementary Material
Acknowledgments
The authors would like to thank the following individuals for their intellectual contributions and assistance with revision of the manuscript: Theresa Goletz, Anup Zutshi, Ariele Viacava Follis, and Shira Warszawski.
Notes
Boris Grinshpun is currently employed at Certara Strategic Consulting, Princeton, New Jersey, USA.
David Nannemann is currently employed at Rosetta Design Group LLC, Burlington VT, USA.
Abbreviations
| ACSINS | Affinity-capture self-interaction nanoparticle spectroscopy |
| ADA | Anti-drug antibodies |
| CDR | Complementarity-determining region |
| CL | Clearance |
| Fc | Crystalizable fragment |
| FcRn | Neonatal Fc receptor |
| Fv | Variable fragment |
| Fvcharge5.5 | Net antibody Fv charge at pH 5.5 |
| HI_sum | Hydrophobic index along three CDRs |
| IgG | Immunoglobulin G |
| mAb | Monoclonal antibody |
| MMAE | Monomethyl-Auristatin E |
| Patch_cdr_hyd | Surface area of the hydrophobic patches near the CDRs |
| pI | Isoelectric point |
| pI_3D | Structure-based pI prediction |
| pI_seq | Sequence-based pI prediction |
| PK | Pharmacokinetics |
| PSR | Poly-specificity reagent |
| TAP | Therapeutic antibody profiler |
| TMDD | Target-mediated drug disposition |
Disclosure statement
The authors report no conflict of interest.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
References
- 1.Ryman JT, Meibohm B.. Pharmacokinetics of monoclonal antibodies. CPT Pharmacometrics Syst Pharmacol. 2017;6(9):576–12. doi: 10.1002/psp4.12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Challa DK, Velmurugan R, Ober RJ, Sally Ward E.. FcRn: from molecular interactions to regulation of IgG pharmacokinetics and functions [Internet]. Daeron M, Nimmerjahn F, editors Fc Receptors. Cham: Springer International Publishing; 2014. 249–72. cited 2019 August5]. page . Available from: 10.1007/978-3-319-07911-0_12 [DOI] [PubMed] [Google Scholar]
- 3.Keizer RJ, Huitema ADR, Schellens JHM, Beijnen JH. Clinical pharmacokinetics of therapeutic monoclonal antibodies. Clin Pharmacokinet. 2010;49(8):493–507. doi: 10.2165/11531280-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 4.Strohl WR. Current progress in innovative engineered antibodies. Protein Cell. 2018;9(1):86–120. doi: 10.1007/s13238-017-0457-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, et al. DrugBank 5.0: a major update to the drugbank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–82. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lévêque D, Wisniewski SW, Jehl F. Pharmacokinetics of therapeutic monoclonal antibodies used in oncology. Anticancer Res. 2005;25:2327–43. [PubMed] [Google Scholar]
- 7.Schoch A, Kettenberger H, Mundigl O, Winter G, Engert J, Heinrich J, Emrich T. Charge-mediated influence of the antibody variable domain on FcRn-dependent pharmacokinetics. PNAS. 2015;112(19):5997–6002. doi: 10.1073/pnas.1408766112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yadav DB, Sharma VK, Boswell CA, Hotzel I, Tesar D, Shang Y, Ying Y, Fischer SK, Grogan JL, Chiang EY, et al. Evaluating the use of antibody variable region (Fv) charge as a risk assessment tool for predicting typical cynomolgus monkey pharmacokinetics. J Biol Chem. 2015;290(50):29732–41. doi: 10.1074/jbc.M115.692434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma VK, Patapoff TW, Kabakoff B, Pai S, Hilario E, Zhang B, Li C, Borisov O, Kelley RF, Chorny I, et al. In silico selection of therapeutic antibodies for development: viscosity, clearance, and chemical stability. Proc Natl Acad Sci U S A. 2014;111(52):18601–06. doi: 10.1073/pnas.1421779112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Igawa T, Tsunoda H, Tachibana T, Maeda A, Mimoto F, Moriyama C, Nanami M, Sekimori Y, Nabuchi Y, Aso Y, et al. Reduced elimination of IgG antibodies by engineering the variable region. Protein Eng Des Sel. 2010;23(5):385–92. doi: 10.1093/protein/gzq009. [DOI] [PubMed] [Google Scholar]
- 11.Hötzel I, Theil F-P, Bernstein LJ, Prabhu S, Deng R, Quintana L, Lutman J, Sibia R, Chan P, Bumbaca D, et al. A strategy for risk mitigation of antibodies with fast clearance. mAbs. 2012;4(6):753–60. doi: 10.4161/mabs.22189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avery LB, Wade J, Wang M, Tam A, King A, Piche-Nicholas N, Kavosi MS, Penn S, Cirelli D, Kurz JC, et al. Establishing in vitro in vivo correlations to screen monoclonal antibodies for physicochemical properties related to favorable human pharmacokinetics. MAbs. 2018;10(2):244–55. doi: 10.1080/19420862.2017.1417718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelly RL, Sun T, Jain T, Caffry I, Yu Y, Cao Y, Lynaugh H, Brown M, Vásquez M, Wittrup KD, et al. High throughput cross-interaction measures for human IgG1 antibodies correlate with clearance rates in mice. MAbs. 2015;7(4):770–77. doi: 10.1080/19420862.2015.1043503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kingsbury JS, Saini A, Auclair SM, Fu L, Lantz MM, Halloran KT, Calero-Rubio C, Schwenger W, Airiau CY, Zhang J, et al. A single molecular descriptor to predict solution behavior of therapeutic antibodies. Science Advances. 2020;6(32):eabb0372. doi: 10.1126/sciadv.abb0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng R, Iyer S, Theil F-P, Mortensen DL, Fielder PJ, Prabhu S. Projecting human pharmacokinetics of therapeutic antibodies from nonclinical data. MAbs. 2011;3(1):61–66. doi: 10.4161/mabs.3.1.13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bussiere JL. Species selection considerations for preclinical toxicology studies for biotherapeutics. Expert Opin Drug Metab Toxicol. 2008;4(7):871–77. doi: 10.1517/17425255.4.7.871. [DOI] [PubMed] [Google Scholar]
- 17.Final opinion on the need for non-human primates in biomedical research, production and testing of products and devices. SCHEER [Internet] 2017; Available from: https://ec.europa.eu/environment/chemicals/lab_animals/pdf/Scheer_may2017.pdf
- 18.Zheng Y, Tesar DB, Benincosa L, Birnböck H, Boswell CA, Bumbaca D, Cowan KJ, Danilenko DM, Daugherty AL, Fielder PJ, et al. Minipig as a potential translatable model for monoclonal antibody pharmacokinetics after intravenous and subcutaneous administration. MAbs. 2012;4(2):243–55. doi: 10.4161/mabs.4.2.19387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avery LB, Wang M, Kavosi MS, Joyce A, Kurz JC, Fan Y-Y, Dowty ME, Zhang M, Zhang Y, Cheng A, et al. Utility of a human FcRn transgenic mouse model in drug discovery for early assessment and prediction of human pharmacokinetics of monoclonal antibodies. MAbs. 2016;8(6):1064–78. doi: 10.1080/19420862.2016.1193660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raybould Matthew I J, Marks Claire, Lewis Alan P, Shi Jiye, Bujotzek Alexander, Taddese Bruck, Charlotte M Deane , Thera-SAbDab: the Therapeutic Structural Antibody Database, Nucleic Acids Research, Volume 48, Issue D1, 08 January 2020, Pages D383–D388, 10.1093/nar/gkz827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drugs@FDA : FDA approved drug products [Internet]. Silver Spring (MD): U.S. Food and Drug Administration, Center for Drug Evaluation and Research; [cited 2019 August5]. Available from: https://www.accessdata.fda.gov/scripts/cder/daf/ [Google Scholar]
- 22.Jain T, Sun T, Durand S, Hall A, Houston NR, Nett JH, Sharkey B, Bobrowicz B, Caffry I, Yu Y, et al. Biophysical properties of the clinical-stage antibody landscape. Proc Natl Acad Sci USA. 2017;114(5):944–49. doi: 10.1073/pnas.1616408114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Integrity, a cortellis solution [Internet]. Clarivate Analytics [cited 2019 August5]; Available from: https://clarivate.com/cortellis/solutions/pre-clinical-intelligence-analytics/
- 24.Pharmapendium [Internet]. Amsterdam (Netherlands): Elsevier; [cited 2019 March11]. Available from: https://www.pharmapendium.com [Google Scholar]
- 25.Clinical pharmacology and biopharmaceutics reviews - Elotuzumab [Internet]. Silver Spring (MD): U.S. Food and Drug Administration, Center for Drug Evaluation and Research; 2015. [cited 2019 August6]. Available from: https://www.accessdata.fda.gov/drugsatfda_docs/nda/2015/761035Orig1s000ClinPharmR.pdf [Google Scholar]
- 26.Xu Z, Bouman-Thio E, Comisar C, Frederick B, Van Hartingsveldt B, Marini JC, Davis HM, Zhou H. Pharmacokinetics, pharmacodynamics and safety of a human anti-IL-6 monoclonal antibody (sirukumab) in healthy subjects in a first-in-human study. Br J Clin Pharmacol. 2011;72(2):270–81. doi: 10.1111/j.1365-2125.2011.03964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuchimanchi M, Grover A, Emery MG, Somaratne R, Wasserman SM, Gibbs JP, Doshi S. Population pharmacokinetics and exposure-response modeling and simulation for evolocumab in healthy volunteers and patients with hypercholesterolemia. J Pharmacokinet Pharmacodyn. 2018;45(3):505–22. doi: 10.1007/s10928-018-9592-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matera MG, Rogliani P, Calzetta L, Cazzola M. Pharmacokinetic/pharmacodynamic profile of reslizumab in asthma. Expert Opin Drug Metab Toxicol. 2018;14(2):239–45. doi: 10.1080/17425255.2018.1421170. [DOI] [PubMed] [Google Scholar]
- 29.Smith DA, Minthorn EA, Beerahee M. Pharmacokinetics and pharmacodynamics of mepolizumab, an anti-interleukin-5 monoclonal antibody. Clin Pharmacokinet. 2011;50(4):215–27. doi: 10.2165/11584340-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 30.Dirks NL, Meibohm B. Population pharmacokinetics of therapeutic monoclonal antibodies. Clin Pharmacokinet. 2010;49(10):633–59. doi: 10.2165/11535960-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 31.Bajaj G, Suryawanshi S, Roy A, Gupta M. Evaluation of covariate effects on pharmacokinetics of monoclonal antibodies in oncology. Br J Clin Pharmacol. 2019;85(9):2045–58. doi: 10.1111/bcp.13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerber DE, Stopeck AT, Wong L, Rosen LS, Thorpe PE, Shan JS, Ibrahim NK. Phase I safety and pharmacokinetic study of bavituximab, a chimeric phosphatidylserine-targeting monoclonal antibody, in patients with advanced solid tumors. Clin Cancer Res. 2011;17(21):6888–96. doi: 10.1158/1078-0432.CCR-11-1074. [DOI] [PubMed] [Google Scholar]
- 33.Sandhu SK, Papadopoulos K, Fong PC, Patnaik A, Messiou C, Olmos D, Wang G, Tromp BJ, Puchalski TA, Balkwill F, et al. A first-in-human, first-in-class, phase I study of carlumab (CNTO 888), a human monoclonal antibody against CC-chemokine ligand 2 in patients with solid tumors. Cancer Chemother Pharmacol. 2013;71(4):1041–50. doi: 10.1007/s00280-013-2099-8. [DOI] [PubMed] [Google Scholar]
- 34.Tolcher AW, Sarantopoulos J, Patnaik A, Papadopoulos K, Lin C-C, Rodon J, Murphy B, Roth B, McCaffery I, Gorski KS, et al. Phase I, pharmacokinetic, and pharmacodynamic study of AMG 479, a fully human monoclonal antibody to insulin-like growth factor receptor 1. JCO. 2009;27(34):5800–07. doi: 10.1200/JCO.2009.23.6745. [DOI] [PubMed] [Google Scholar]
- 35.Kamath AV, Iyer S. Preclinical pharmacokinetic considerations for the development of antibody drug conjugates. Pharm Res. 2015;32(11):3470–79. doi: 10.1007/s11095-014-1584-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X, Song X, Li K, Zhang T. FcγR-binding is an important functional attribute for immune checkpoint antibodies in cancer immunotherapy. Front Immunol. Internet] 2019. [cited 2021 January4]; 10. Available from ;. https://www.frontiersin.org/articles/10.3389/fimmu.2019.00292/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warncke M, Calzascia T, Coulot M, Balke N, Touil R, Kolbinger F, Heusser C. Different adaptations of IgG effector function in human and nonhuman primates and implications for therapeutic antibody treatment. The Journal of Immunology. 2012;188(9):4405–11. doi: 10.4049/jimmunol.1200090. [DOI] [PubMed] [Google Scholar]
- 38.Molecular Operating Environment (MOE) . Montreal. QC (Canada): Chemical Computing Group ULC; 2019. [Google Scholar]
- 39.Raybould MIJ, Marks C, Krawczyk K, Taddese B, Nowak J, Lewis AP, Bujotzek A, Shi J, Deane CM. Five computational developability guidelines for therapeutic antibody profiling. Proceedings of the National Academy of Sciences 2019; 116:4025–4030. doi: 10.1073/pnas.1810576116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jetha A, Thorsteinson N, Jmeian Y, Jeganathan A, Giblin P, Fransson J. Homology modeling and structure-based design improve hydrophobic interaction chromatography behavior of integrin binding antibodies. mAbs. 2018;10(6):890–900. doi: 10.1080/19420862.2018.1475871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Datta-Mannan A, Lu J, Witcher DR, Leung D, Tang Y, Wroblewski VJ. The interplay of non-specific binding, target-mediated clearance and FcRn interactions on the pharmacokinetics of humanized antibodies. mAbs. 2015;7(6):1084–93. doi: 10.1080/19420862.2015.1075109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones HM, Zhang Z, Jasper P, Luo H, Avery LB, King LE, Neubert H, Barton HA, Betts AM, Webster R, et al. Model for the prediction of monoclonal antibody pharmacokinetics from in Vitro data. CPT: Pharmacometrics & Systems Pharmacology. 2019;8:738–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sankar K, Krystek SR, Carl SM, Day T, Maier JKX, Sankar K, SR K, Sm C, Day T, Maier JKX. AggScore: prediction of aggregation-prone regions in proteins based on the distribution of surface patches. Proteins: Structure, Function, and Bioinformatics. 2018;86(11):1147–56. doi: 10.1002/prot.25594. [DOI] [PubMed] [Google Scholar]
- 44.Chennamsetty N, Voynov V, Kayser V, Helk B, Trout BL. Design of therapeutic proteins with enhanced stability. PNAS. 2009;106(29):11937–42. doi: 10.1073/pnas.0904191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Apgar JR, Tam ASP, Sorm R, Moesta S, King AC, Yang H, Kelleher K, Murphy D, D’Antona AM, Yan G, et al. Modeling and mitigation of high-concentration antibody viscosity through structure-based computer-aided protein design. PLOS ONE. 2020;15(5):e0232713. doi: 10.1371/journal.pone.0232713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nichols P, Li L, Kumar S, Buck PM, Singh SK, Goswami S, Balthazor B, Conley TR, Sek D, Allen MJ. Rational design of viscosity reducing mutants of a monoclonal antibody: hydrophobic versus electrostatic inter-molecular interactions. MAbs. 2015;7(1):212–30. doi: 10.4161/19420862.2014.985504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agrawal NJ, Helk B, Kumar S, Mody N, Sathish HA, Samra HS, Buck PM, Li L, Trout BL. Computational tool for the early screening of monoclonal antibodies for their viscosities. MAbs. 2016;8(1):43–48. doi: 10.1080/19420862.2015.1099773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Freedman D, Diaconis P. On the histogram as a density estimator: l2theory. Z Wahrscheinlichkeitstheorie verw Gebiete. 1981;57(4):453–76. doi: 10.1007/BF01025868. [DOI] [Google Scholar]
- 49.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28(1):235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, et al. AMBER 10. University of California, San Francisco; 2008. [Google Scholar]
- 51.Labute P. LowModeMD—implicit low-mode velocity filtering applied to conformational search of macrocycles and protein loops. J Chem Inf Model. 2010;50(5):792–800. doi: 10.1021/ci900508k. [DOI] [PubMed] [Google Scholar]
- 52.Labute P. Protonate3D: assignment of ionization states and hydrogen coordinates to macromolecular structures. Proteins. 2009;75(1):187–205. doi: 10.1002/prot.22234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sillero A, Ribeiro JM. Isoelectric points of proteins: theoretical determination. Anal Biochem. 1989;179(2):319–25. doi: 10.1016/0003-2697(89)90136-X. [DOI] [PubMed] [Google Scholar]
- 54.Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pKa values. Proteins: Structure, Function, and Bioinformatics. 2005;61(4):704–21. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 55.Hastie T, Tibshirani R, Friedman J. The elements of statistical learning: data mining, inference, and prediction. second. New York: Springer; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


