ABSTRACT
Plant growth and development is dependent on the regulation of classes of microRNAs (miRNAs) that have emerged as important gene regulators. These miRNAs can regulate plant gene expression to function. They play an important roles in biological homeostasis and environmental response controls. A wide range of plant biological and metabolic processes, including developmental timing, tissues specific development, and differentiation, depends on miRNAs. They perpetually regulate secondary metabolite functions in different plant family lines. Mapping of molecular phylogenies shows the distribution of secondary metabolism in the plant territory. More importantly, a lot of information related to miRNA regulatory processes in plants is revealed, but the role of miRNAs in secondary metabolism regulation and functions of the metabolites are still unclear. In this review, we pinnacle some potential miRNAs regulating the secondary metabolite biosynthesis activities in plants. This will provide an alternative knowledge for functional studies of secondary metabolism.
KEYWORDS: MicroRNAs, molecular phylogenies, primary metabolism and, Secondary metabolism
1. Introduction
Plants have been utilized as a source of nutrition, drug, and livelihood improvement over the year 1. Human population development is believed to directly relate to increasing demand for plants and plant activities, which give merits to expand the yield of plants for different purposes. 2 Advanced progressive logical strategies have also predicted an increase or decrease in plant functions and capacities. With this, it is revealed that these advanced strategies can help solve many plant-related issues to help bridge the gap between humans and plants. The advance strategic inputs have also shown the influence of miRNAs in managing primary biological processes, including metabolic procedures at the post-transcriptional level. The miRNAs are non-coding small RNAs composed of 20–23 nucleotides. 3 About thousands of these miRNAs have been identified in plants. 4 In plants, they are divided into two groups: conserved miRNAs that occur in different families and species-specific miRNAs characterized by low-level expression. 5 Different works have shown that miRNAs influence plant developmental activities such as blooming of leaves, seed emergence, and root morphology 1, not forgetting the hormonal reactions in the plants. 6 They also play a critical part in a wide range of biological processes, 7 such as developmental timing, 8 cell and tissue differentiation, 9 proliferation, 10 apoptosis, 11 and metabolism. 12 Their function at the plant’s biological level improves growth, development, and biological responses. At the gene expression level, they effectively obstruct the translation of their target gene (mRNAs). 13 More importantly, the miRNAs can control the expression of genes that encode transcription factors 14 and strain reaction proteins, which impact the biological processes in the plant. 15 They similarly control organic procedures in plants, such as upkeep of genome honesty, primary and secondary metabolisms, 16 development of signal transductions, 17 signaling pathways, 18 and adaptive responses to biotic and abiotic stresses. 19 Where these metabolites contribute to plants’ growth and development, the nutrient and beneficial properties of plants are also owed to the presence of an abundance of these metabolites. 20 The metabolites are a group of two types, primary and secondary. 21 The secondary metabolites regulated plant phytochemicals processes related to their interactions with the environment. 22 The secondary metabolites consist of phytochemical compounds, including terpenoids, alkaloids, phenolics, glycosides, tannins, and saponins. The metabolites are predicted to function in the interactions of plants and their environment. The metabolites are predicted to protect plants from a series of environmental conditions. 23 Specifically, flavonoids, terpenoids, and carotenoids are recognized for their dynamic functional roles in plant reproduction system, growth, development, and defense mechanisms. 24 Over the years, miRNAs significant role in regulating biosynthetic activities and secondary metabolites in plants has gained much attention. 16 This review presents updated knowledge of miRNA’s functions in regulating phenolypropanoid biosynthesis, terpenoid biosynthesis, alkaloid biosynthesis, chlorophyll biosynthesis, carotenoids biosynthesis, and genes targeted by miRNAs.
2. Molecular phylogenies distribution of secondary metabolites in plants
Over the years, botanists have reconstructed a tree of existence for the plant kingdom usage of nucleotide sequences, plastid, and nuclear marker genes. 25 Ensuing this, molecular phylogenies can map the distribution of secondary metabolism in the plant territory. It is suggested that plants accumulate a positive secondary metabolism type, making it probable that their synthesis and functions are genetic. This would advocate that the corresponding pathways and miRNAs have been advanced all through early growth. 26 In Figure 1, we show the molecular phylogeny of some selected significant plant families. The lines show the position of synthesizing flavonoids and other phenolics deriving from phenylalanine. This suggests that the secondary plant metabolites existed from the early ages (Figure 1 A), linking the secondary metabolites gene families to the miRNAs’ possible functions in secondary metabolites (Figure 1B,C). Considering the most corresponding main enzymes, phenylalanine ammonia-lyase (PAL) and chalcone synthase (CHS) have been detected in plants. 27 Terpenoids, such as mono-, sesqui-, tri-, and tetraterpenes, and steroids are also existing from lower plants to more higher plants, suggesting that the generate pathways of the main energetic substance isoprene and the cyclization of C-5 units had been present already in plants ancestry.28,29 Alkaloids have a more excellent patchy distribution, and distinct alkaloidal sorts are usually precise for certain taxon groups; therefore, they had been regularly used as chemotaxonomic markers. Alkaloids can be detected in plants but are predominant in angiosperms. 30 Because many alkaloids are neurotoxins, which interfere with pursuit of neuronal sign transduction, we speculate that the diversification of alkaloids in plants coevolved with the rapid diversification of tertiary periods. The distribution of alkaloids provides a clear example of the perception of secondary metabolism dynamics with limited occurrences.
Figure 1.
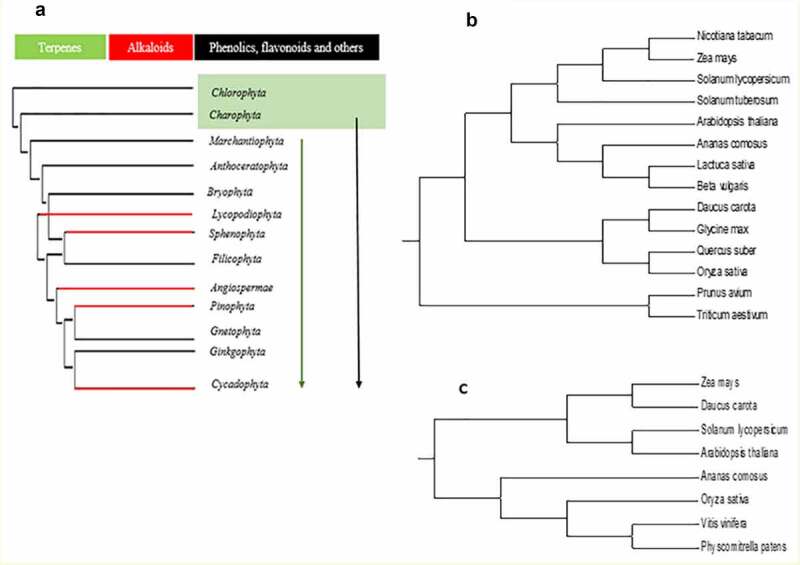
Molecular phylogeny of terrestrial plants. Groups of biological species that produce terpenes are marked in green, terpenoids in red and phenolics, flavonoids, and others in black (a). Molecular phylogeny of PAL (b) and CHS (c) secondary from their corresponding genes’ nucleotides sequences
3. Regulatory function of miRNAs in secondary metabolism activities
Many scientific questions are asked: under what condition are secondary metabolites produced and can the production be regulated by miRNAs? A large number of miRNAs control genes (translation factors and structure genes) and biological activities including environmental change reactions that affect natural procedures in plants. 31 They function to regulate plant’s biological processes such as maintenance of genome integrity, primary and secondary metabolism, development, signal transduction and pathways, homeostasis, innate and immunity, and adaptive responses to environmental change. 32 The secondary metabolites are groups of phytochemicals that regulate various processes related to the plants’ interaction and their environment. 33 They are mixtures assimilated with terpenoids, alkaloids, phenolics, glycosides, tannins, saponins and shield plants from biotic and abiotic stressors. 34 Plants integrate secondary metabolism categories to help in self-protection. 35 The generation of metabolites can be overseen by the miRNAs since the miRNAs can positively or negatively regulate the production of the anticipated metabolites while limiting the production of toxic metabolites and new metabolites that can be produced. 36 Stimulated plants initiate change in quality articulation and generation of guarded metabolites, and miRNAs control these activities. For example, Solanum tuberosum L. under light improvement revealed that light-responsive miRNAs could significantly control alkaloid digestion, lipid biosynthesis, and cellulose catabolism. 37 In addition, transcriptomes analysis of Swertia chirayita shows recognizable proof that miRNAs regulate metabolite biosynthesis. 38 The miRNAs consisted of miR-156a, miR-166a, miR-166b, miR-168, miR-11071, and miR-11320, that regulates metabolites biosynthesis. 38 Moreover, in Swertia chirayita, it is predicted that homeobox-leucine zipper encoded a protein with a potential relationship with metabolites biosynthesis process. 34 Similarly, the funtion of miR-774 and miR-1126 in numerous plant biological processes includes the development of secondary metabolic pathways. 39 In summary, these illustrations indicate that specific miRNAs are associated with the secondary metabolism activities in response to the influence of plant growth and differentiation.
3.1. miRNAs/phenylpropanoid biosynthesis
Phenylpropanoids are the largest class of plant secondary metabolites. They are derived from the aromatic amino acid phenylalanine. 40 They mainly include flavonoids, monolignols, phenolic acids, stilbenes, and coumarins. 41 It is broadly disseminated in most plant realms revealing its fundamental functions in the secondary metabolites. They improve plants through cell walls, protection against high light and UV radiation, phytoalexins against pathogens, and pollinator interactions. 42 Its pathway serves as a rich source of metabolites activities in plants, which is required for the biosynthesis of lignin, flavonoids, coumarins, and hydroxycinnamic acids. 43
Moreover, the phenylpropanoid pathway serves as a brilliant structure for developing through genetically manipulating complex natural product pathways and responds to environmental changes in plants. 44,45 An example is the overexpression of miR-7695 to boost disease resistance to a fungal pathogen that causes blast disease in Magnaporthe oryzae plant. 46 Interestingly, the blocking of miR-858a activity can result in the upregulation of flavonoid-specific target genes like AtMYB12 and AtCHS1 which is involved in the phenylpropanoid pathway process. 47 Besides, miR-159, miR-172, and miR-530 obtained from the roots and leaves also regulate secondary metabolite associated with mRNAs of Chlorophytum borivilianum, Oryza sativa, and Arabidopsis thaliana. 48
Interestingly, the target genes of miR-9662, miR-894, miR-172, and miR-166 are revealed to be involved in regulating the plant phenylpropanoid saponin metabolic biosynthetic process, but an activated miR-8154 and miR-5298b also increase the phenylpropanoid content. 48 In summary, this phenylpropanoid is being broadly disseminated in the plant realm. It assumes a tremendous fundamental functional work in a plant, such as improving growth and cell differentiation of cell walls. The miRNAs discussed in phenylpropanoid functional activities regulate plant growth and development. With this, much attention should be given to phenylpropanoid potential miRNAs since most agronomic crops and model plants are exposed to light and other inhibitory conditions in practice.
However, specifically in flavonols, it is exceptionally unpredictable. In Arabidopsis thaliana, different MYB transcription factors control flavonols biosynthesis process, including MYB11, and MYB12. 49 These MYBs influence three explicit flavonols in Arabidopsis under which miR-858a is expressed extensively in all the tissues, which means that the expression of miR-858a may directly control the MYBs and the flavonol production in the Arabidopsis leaf tissue. 6 Similarly, miR-156 regulates flavonoid biosynthesis in Arabidopsis. miR-156-SPL9 directly influenced anthocyanin production through targeting genes encoding AP-1-like transcription factor (PAP1) and dihydroflavonol 4-reductase. 50 During the flavonoid expansion, miR-160 upregulate Auxin response factor 17 (ARF17) and downregulate Auxin response factor 18 (ARF18), to response to different factors affecting root development. 51 MiR-828 act as a negative regulator of anthocyanin and polyphenolics biosynthesis, regulated the conspicuous red fruit trait of Ananas comosus var. Bracteatus. It directs post-transcriptional gene silencing of MYB family members associated with UV light- and sugar-signaling. 52 Two flavonoid responsive genes, Cryptochrome1 (CRY 1) and Cryptochrome2 (CRY 2), is upregulated by miR-172, to which they mediate the expression and stimulation of CONSTANS 67 independently during the photoperiodic flowering time. 53 In grapevine berry fruit, it is revealed that the up-regulation of miR-3627 and miR-4376 facilitates anthocyanin accumulation. 54 Several miRNAs including, miR-171, miR-166i, miR-159e, miR-845, and miR-396e, were identified to control carotenoids and flavonoids biosynthesis. 55
3.2. miRNAs/terpenoid biosynthesis
The largest group of natural products are the terpenoids, used as aroma compounds and natural flavor enhancers. 56 The terpenoids are synthesized through two independent pathways: cytosolic mevalonic acid (MVA) and plastidial methylerythritol phosphate (MEP) pathway.57 Terpenoid compounds are produced in high abundance under biotic stress as compared to abiotic stress. 58 In Ananas comosus var. Bracteatus leaves, it is revealed through transcription analysis that about 43 identified unigenes encoded enzymes are involved in the terpenoid backbone biosynthesis. 57 Moreover, miR-2919, miR-5251, miR-838, miR-5021, and miR-5658 from Ferula gummosis control for terpene biosynthesis. However, the transcription factors SPL7, SPL11, and ATHB13 from Arabidopsis predicted to function in terpenoid biosynthesis are putatively regulated by miR-5021 and miR-5658. 59 It is predicted that miR-5021 and miR-414 function to regulate terpenoid backbone biosynthesis and sesquiterpenoid and triterpenoid biosynthesis, respectively, whereas miR-7540, miR-5183, miR-6449, miR-5255, miR-5491, and miR-6435 target downstream of catalysts in mono, sesqui-, di-, and tri-terpenoids of triterpenoid biosynthesis. 60 In the same vein, Persicaria minor has a high level of secondary metabolite compounds, particularly terpenoids, where pmi-miR-6173 and pmi-miR-6300 are downregulated, but miR-396a and miR-398 f/g are upregulated. 61
Moreover, in Arabidopsis thaliana and Pogostemon Cablin, transcription factor, SPL9 and SPL10 targeted by miR-156 are predicted to be an activator for terpene synthesis. 62 Overexpression of miR-156 results in the downregulation of the sesquiterpenes level in Arabidopsis thaliana and Pogostemon Cablin. 62 MiRNAs are predicted to be involved in sesquiterpenes biosynthesis pathways through a computational prediction approach in Xanthium strumarium. 62 Similarly, the overexpression of miR-393 in Arabidopsis thaliana changes the glucosinolate and camalexin levels through perturbation of auxin signal pathway. 63 Table 1 presents some miRNAs predicted to regulate terpenoid biosynthesis in some plant species. These miRNAs will be very useful in terpenoid biosynthesis, regulating these plant species’ metabolic and biological processes.
Table 1.
Probable miRNAs regulation in terpenoid biosynthesis with target gene and their functions
| MicroRNAs | Targets | Functions | Ref. |
|---|---|---|---|
|
miR-164a-d miR-528 miR-319e miR-159 |
4-hydroxy-3-methylbut-2-enyl-diphosphate synthase. | It is essential for chloroplast development and required for the salicylic acid (SA)-mediated disease resistance to biotrophic pathogen. | 64 |
|
miR-9662a-p miR-894 |
Heptaprenyl diphosphate synthase | Supplies heptaprenyl diphosphate, the precursor for the side chain of the isoprenoid quinone menaquinone | 48,58,65 |
|
miR-319a miR-166 J miR-156 g miR156m miR-156e |
Diphosphomevalonate decarboxylase and SPL transcription factor |
Accumulation of anthocyanins, whereas reduced miR156 activity results in high levels of flavonols | 48,55,66 |
| miR-171a-3p | Phosphomevalonate kinase | Transfers a chemical group, e.g., a methyl group, to another compound (acceptor) and catalyzes phosphate transfer to a second substrate. | 48,58 |
| miR-172d-3p | 1-deoxy-D-xylulose-5-phosphate synthase | A limiting enzyme for plastidic isoprenoid biosynthesis and essential for chloroplast development. | 48,58,67 |
| miR-172 c | Farnesyl diphosphate synthase and geranylgeranyl diphosphate synthase | Geranylgeranyl diphosphate (GGPP) synthase activity. The enzyme catalyzes the synthesis of GGPP from farnesyl diphosphate and isopentenyl diphosphate. | 48,55,68 |
|
miR-167 g-5p miR-167 c miR-167 f-5p |
Isopentenyl-diphosphate Delta-isomerase | It is involved in the pathway chlorophyll biosynthesis, which is part of Porphyrin-containing metabolism compounds. | 48,55,69 |
|
miR-167 c miR-159 c |
Hydroxymethylglutaryl-CoA synthase | Condenses acetyl-CoA with acetoacetyl-CoA to form HMG-CoA, which is the substrate for HMG-CoA reductase | 61,70 |
|
miR-396-3p miR-894 miR-9662a miR-159 miR-172 c |
Farnesyl-diphosphate synthase and geranylgeranyl diphosphate synthase | Flavonoid biosynthesis | 55 |
|
miR396-3p miR-894 |
Trans-nonaprenyl-diphosphate synthase | Flavonoid biosynthesis | 55 |
3.3. miRNAs/alkaloids biosynthesis
Alkaloids are nitrogen-containing low molecular-weight compounds. They are mostly derived from amino acids. 71 Alkaloids are recognized to play a significant role in plant defense against pathogens. 72 Over the years, about 12,000 alkaloids have been characterized in these highly diverse and heterogeneous secondary metabolites. 73 The alkaloids consist of indole, piperidine, tropane, purine, pyrrolizidine, imidazole, quinolizidine, isoquinoline, and pyrrolidine compounds. 72 These compounds are created through different metabolic pathways in the alkaloids. 74 However, genome-based technological advancement has added to our current study's understanding of alkaloid biosynthetic pathways and functions. 34 Consistent information on miRNAs’ functional roles regulating alkaloid biosynthesis and its accumulation in plant’s monarchy is on the rise. 34 Research conducted in 2014 revealed different miRNAs regulating alkaloid biosynthesis pathway in Opium poppy plant (species of flowering plant in the family of Papaveraceae), 75 including pso-miR-13, pso-miR-2161, and pso-miR-408. The pso-miRNA-2161 acts as an intermediate molecule in benzylisoquinoline alkaloids biosynthesis targeted with mRNA encoding S-adenosyl-L-methionine, 30-hydroxy-N-methylcoclaurine, and 40-O-methyltransferase enzymes which convert S-norcoclaurine into S-reticuline. 76 Correspondingly, pso-miR-13 is evaluated to target mRNA 7-O-methyltransferase, which is predicted to convert S-reticuline to morphinan alkaloids. 34
Interestingly, most of the endogenous targets of miRNAs obstruct the function of corresponding miRNAs by inhibiting miRNAs’ binding with their authentic target genes. 77 For example, in poisonous Taxus baccata, 78 two predicted paclitaxel biosynthetic genes, Taxane 13α hydroxylase and Taxane 2α-O-benzoyltransferase, cleavage target of miR-164 and miR-171 to obstructs the poisonous content. 79 Mentha is revealed to have miR-156, miR-414, and miR-5021 responsible for regulating essential oil biosynthesis. However, miR-156 is specially revealed to participate in flavone and alkaloids biosynthesis. 80 A better understanding of miRNAs regulating alkaloid and molecular regulation of these predicted miRNAs can improve plant growth, such as integrated by plants, to protect self-protection.
3.4. miRNAs/chlorophyll biosynthesis
The chlorophyll content is critical for plant pigments involved in light engagement and energy transfer during photosynthesis. 81 They are essential for generating reactive oxygen for plant species, essential for plant growth and development. 82 The precursors of chlorophyll biosynthesis involve oxygen, which enables the activity of secondary metabolite in a specific plant. 82 It has been well documented that multiple pathways regulate chlorophyll biosynthesis at transcriptional and post-transcriptional levels, which means that chlorophyll biosynthesis is involved in the secondary and primary metabolic activities. 83 Most studies have revealed that diploid and autotetraploid plant leaves can decrease chlorophyll content to activate the secondary and primary metabolic activities.84
Meanwhile, some miRNAs that are predicted to target chlorophyll biosynthesis can increase the superoxide dismutase activity, solubility protein content, relative conductivity, sugar content, proline contents, and malondialdehyde contents.85 In Ananas comosus var. bracteatus, it is revealed that Ab-miR-124 is predicted to target the trihelix transcription factor, which acts as a molecular switch in response to light signals and related to chlorophyll biosynthesis.86 More importantly, miR-124 is revealed to be significantly down-regulated in the white leaves of Ananas comosus var. bracteatus leaves. Furthermore, miR-127, miR-160, miR-161, and miR-163 are predicted to target divinyl chlorophyllide a 8-vinyl-reductase (DVR) which is down-regulated in the complete white leaves in var. Bracteatus.86 miR-156 is revealed to participate in the vegetative changing phase by down-regulating several chlorophyll Squamosa promoter binding protein-like (SPL) transcription factors.87 MiR-159 targets Proto-oncogene like 1 transcription factor, which is required for further development in light. Phytochrome interacting factor 3 (PIF3) is a negative regulator in chloroplast development and decreases the expression level of chlorophyll biosynthetic and photosynthetic gene activities. Moreover, Phytochrome interacting factor 1 (PIF1) also represses chlorophyll biosynthesis. 88 Another study has revealed that the chlorophyll biosynthetic and photosynthetic genes could be directly targeted by PIF3. 89 These findings further support the theory that PIF3 acts as a negative regulator in chloroplast development by directly repressing chlorophyll biosynthetic and photosynthetic genes.89 The transcriptional level of SCL genes is regulated by miRNAs.90 In Arabidopsis, miR-171 is predicted to regulate chlorophyll biosynthesis activities positively in the light but down-regulate chlorophyll biosynthesis when targeting Scarecrow-Like 6 (SCLs6).91 However, the overexpression of bol-miR171b in transgenic broccoli exhibited dark green leaves with a high chlorophyll content.92 miR-171 upregulates Protochlorophyllide oxidoreductase (POR), a key enzyme in chlorophyll biosynthesis, as shown in Table 2, but the SCLs gene in different plant species functions differently, notwithstanding its general functions in plants. 94 Similarly, overexpression of miR-171 c targeting scl6, scl22, and scl27 showed higher chlorophyll content and POR expression levels.94 In addition, miR-171, targeting rSCL27, expresses to reduce the chlorophyll content and POR expression levels. On the other hand, the downregulation of POR expression reduces the chlorophyll content.95 MiR-171-SCL module is also predicted to mediate gibberellin-dependent effects on chlorophyll biosynthesis due to its regulation of DELLA proteins and POR expression in the light, but not in the dark. Finally, SCLs induce the expression of miR-171 genes, revealing a regulatory feedback loop.96 This demonstrates that the overexpression of miR-171 could affect the chlorophyll content in transgenic plants. In the table, we put together miRNAs that regulate chlorophyll biosynthetic activities in some plant species and their target gene function.
Table 2.
The roles of chlorophyll biosynthetic miRNAs in plants and their target genes
| MicroRNAs | Regulation | Species | Target | Phenotypes | Target Function | Ref. |
|---|---|---|---|---|---|---|
| miR-171b | Up | Broccoli | SCL | Dark green | Plant information transmission and signal transduction | 92 |
| miR-171b | Up | Arabidopsis thaliana | SCL | Silique | Signal transduction | 93 |
|
miR-127 miR-160 miR-161 miR-163 |
Down | Ananas comosus var. bracteatus | DVR add function | White | Catalyzes the conversion of divinyl chlorophyllide to monovinyl chlorophyllide | 86 |
| miR-171 | Down | Arabidopsis thaliana | POR | Pale Green | Catalyzes the photoreduction of protochlorophyllide (Pchlide) to chlorophyllide. |
94 91 |
3.5. miRNAs/carotenoids biosynthesis
Plants are natural chemical factories that synthesize health-promoting micronutrients like carotenoids.97 They are lipophilic compounds that play a critical role in pigmentation,98 photosynthesis,99 and development.100 Carotenoids promote health, sexual behavior in plant reproduction, forming colors and flavors.22 In plants, carotenoids help capture light, offer photoprotection, and signal control over gene expression.101 Carotenoid biosynthetic nature is required throughout the plant’s life cycle. The structures and levels of carotenoids are exceptionally tuned to the environment and the stages of development.22 Carotenoids are combined and sequestered in plastid organelles, for example, dim developed etioplasts, light-developed chloroplasts, leucoplasts from roots, amyloplasts from seeds, and chromoplasts from natural products.102 It has been proven by research that most plants have high carotenoid content. 103 The carotenoid content in complete green leaves of Ananas comosus var. bracteatus is significantly higher than in the complete white leaves in three leaf developmental stages.86
Moreover, SET Domain Group 8 (SDG8) varies in histone-3lysine-4 trimethylation surrounded by Carotenoid isomerase (CRTISO) gene and lutein biosynthesis. 104 The sedge and CRTISO reduced lutcin in leaves but tend to accumulate cis-carotenoids in etiolated seedlings. Overexpression of miRNA-156 enhanced lutein and β carotene in Brassica napus seeds and shoots reproduction. 105 In Arabidopsis, miR-156 regulates carotenoids SPL families, which further regulates other genes through complex gene regulation. Microarray analysis revealed that Histone lysine methyltransferase could control the expression of SPL-15. 106 miRNA-156 is upregulated by 24-epibrassinolide, a high-lightening link between SDG8 regulation, shot branching, and carotenoids.32,105 Another analysis of the lycopene β-cyclase gene targeted by miR-1857 is involved in carotenoid biosynthesis. 107
5. MicroRNAs FORWARD FOR SECONDARY AND PRIMARY METABOLITES ACTIVITIES
It is meaningful to look into the intertwining functions of the primary metabolites and secondary metabolites in the plant to give a clear distinction of their activities through cell differentiation, growth, and development. Primary metabolites differed from secondary metabolites because they are required at every stage of the plant’s growth process.108 The precursor molecules for secondary metabolites biosynthesis are a channel from primary metabolites. 109 The primary metabolite biosynthetic regulation pathway is pressing advantage over the secondary metabolite for that the primary metabolite biosynthetic pathway is well explored at the transcriptional, post-transcriptional, and DNA levels. Still, the secondary metabolic pathway is narrowed at the transcriptional level. 110 The secondary metabolic pathways’ activities are recently on the rise at the post-transcriptional level, which is the miRNAs interaction. As shown in Figure 2, we display the explored direction of the two metabolic pathways from the plants to their specific metabolites. Consequently, most of the research has focused on miRNAs’ role during the primary metabolism of growth and development. Moreover, these miRNAs of primary metabolism and some other miRNAs have been reported for their essential role during secondary metabolism, for instance, the SPL-miRNA-156 activities in secondary metabolites. 34
Figure 2.
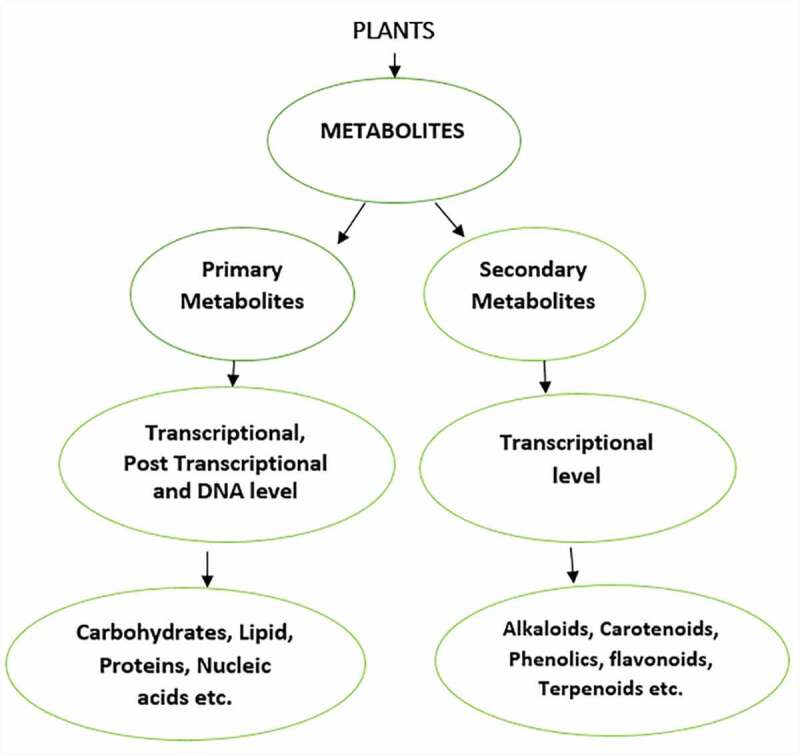
Systematic level occurrences of primary and secondary metabolites. The secondary metabolites through transcriptional level, give rise to different metabolites. The primary metabolites consist of transcriptional and post-transcriptional level through which proteins, lipids and carbohydrates are observed
Similarly, miR-4995 targets 3-deoxy-7-phosphoheptulonate synthase in the initial step of the phenylpropanoid pathway for picrosides-I biosynthesis. 34 The 3-deoxy-7-phosphoheptulonate enzyme also initiates the phenylpropanoid pathway process. This enzyme holds the key to the pathway’s progress as its regulation of cinnamic acid production, which affects picrosides-I content in plants. 34 Regarding these regulatory roles, the modification in the miRNAs’ expression level would help control the biosynthesis activities of secondary metabolites in plants. For example, SPL9 and TCP3 transcription factors play a significant role in secondary metabolism regulation, 16 miRNAs targeting these genes would be ideal candidates for such an approach. 34 However, identifying and understanding the three-dimensional and temporal expression of other miRNAs might also regulate the effort at the branch point of primary and secondary metabolic pathways.
6. Conclusion
Owing to the wide range of the biosynthetic pathways of the secondary metabolism, biological significance in plants is essential. Secondary metabolites as written and their regulatory miRNAs functions unveiled are a helpful biological research approach. However, on the other hand, computational deep sequencing technology methods for miRNAs predictions have huge statistics, which can also be considered. With all these, the knowledge of miRNAs regulation of secondary metabolism in the plant will increase plants’ physical and biological functional knowledge. These studies will help the entire biosynthetic pathway’s metabolic studies to give a clear understanding of biosynthetic phytochemicals such as secondary metabolites.
Acknowledgments
We are thankful to Sichuan Agricultural University, China, for providing support to the present study.
Funding Statement
This work was supported by the National Natural Science Foundation of China, 31770743; 31971704 (http://www.nsfc.gov.cn) National Natural Science Foundation of China [31770743; 31971704].
Disclosure statement
The authors declare no conflict of interest in this research.
Author contributions
MOA conceived the idea and designed the manuscript. ZX, MM, and FR collected literatures and contributed to the writing of the manuscript. MJ critically advised and evaluated the manuscript for further correction.
Data Availability
Biochemistry of Plant Secondary Metabolism and Functions of Plant Secondary Metabolites and their Exploitation in Biotechnology, DOI: https://doi.org/10.1016/S0167-7799(00)01454-2
References
- 1.Piironen V, Lindsay DG, Miettinen TA, Toivo J, Lampi AM.. Plant sterols: biosynthesis, biological function and their importance to human nutrition. J Sci Food Agric. 2000;80:1–9. [Google Scholar]
- 2.Schippmann U, Leaman DJ, Cunningham A. 2002. Impact of cultivation and gathering of medicinal plants on biodiversity: global trends and issues. Biodiversity and the ecosystem approach in agriculture, forestry and fisheries. [Google Scholar]
- 3.Katahira J, Yoneda Y. Nucleocytoplasmic transport of microRNAs and related small RNAs. Traffic. 2011;12(11):1468–1474. doi: 10.1111/j.1600-0854.2011.01211.x. [DOI] [PubMed] [Google Scholar]
- 4.Zhang B, Wang QJ. MicroRNA‐based biotechnology for plant improvement. J Cell Physiol. 2015;230:1–15. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma D, Tiwari M, Pandey A, Bhatia C, Sharma A, Trivedi PK. MicroRNA858 is a potential regulator of phenylpropanoid pathway and plant development. Plant Physiol. 2016;171(2):944–959. doi: 10.1104/pp.15.01831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149(3):515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ambros V. MicroRNAs and developmental timing. Curr Opin Genet Dev. 2011;21(4):511–517. doi: 10.1016/j.gde.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bentwich I. A postulated role for microRNA in cellular differentiation. The FASEB Journal. 2005;19(8):875–879. doi: 10.1096/fj.04-3609hyp. [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez RE, Schommer C, Palatnik JF. Control of cell proliferation by microRNAs in plants. Curr Opin Plant Biol. 2016;34:68–76. doi: 10.1016/j.pbi.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Liu L, Zhang G, Liang Z, Liu X, Li T, Fan J, Bia J, Wang Y. MicroRNA-15b enhances hypoxia/reoxygenation-induced apoptosis of cardiomyocytes via a mitochondrial apoptotic pathway. Apoptosis. 2014;19(1):19–29. doi: 10.1007/s10495-013-0899-2. [DOI] [PubMed] [Google Scholar]
- 12.Boehm M, Slack FJ. MicroRNA control of lifespan and metabolism. Cell Cycle. 2006;5(8):837–840. doi: 10.4161/cc.5.8.2688. [DOI] [PubMed] [Google Scholar]
- 13.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proceedings of the National Academy of Sciences;104:9667–9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dugas DV, Bartel BJ. MicroRNA regulation of gene expression in plants. Curr Opin Plant Biol. 2004;7(5):512–520. doi: 10.1016/j.pbi.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 15.Budak H, Akpinar BA. Plant miRNAs: biogenesis, organization and origins. Funct Integr Genomics. 2015;15(5):523–531. doi: 10.1007/s10142-015-0451-2. [DOI] [PubMed] [Google Scholar]
- 16.Bulgakov VP, Avramenko TV. New opportunities for the regulation of secondary metabolism in plants: focus on microRNAs. Biotechnol Lett. 2015;37(9):1719–1727. doi: 10.1007/s10529-015-1863-8. [DOI] [PubMed] [Google Scholar]
- 17.Bolle C. The role of GRAS proteins in plant signal transduction and development. Plants. 2004;218(5):683–692. doi: 10.1007/s00425-004-1203-z. [DOI] [PubMed] [Google Scholar]
- 18.Meng Y, Ma X, Chen D, Wu P, Chen M. MicroRNA-mediated signaling involved in plant root development. Biochem Biophys Res Commun. 2010;393(3):345–349. doi: 10.1016/j.bbrc.2010.01.129. [DOI] [PubMed] [Google Scholar]
- 19.Shukla LI, Chinnusamy V, Sunkar RJ. The role of microRNAs and other endogenous small RNAs in plant stress responses. Biochim Biophys Acta (BBA) Gene Regul Mech. 2008;1779(11):743–748. doi: 10.1016/j.bbagrm.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Kennedy DO, Wightman EL. Herbal extracts and phytochemicals: plant secondary metabolites and the enhancement of human brain function. Advances in Nutrition. 2011;2:32–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanada K, Sawada Y, Kuromori T, Klausnitzer R, Saito K, Toyoda T, Hirai MY. Functional compensation of primary and secondary metabolites by duplicate genes in Arabidopsis thaliana. Mol Biol Evol. 2011;28(1):377–382. doi: 10.1093/molbev/msq204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartwal A, Mall R, Lohani P, Guru S, Arora SJ. Role of secondary metabolites and brassinosteroids in plant defense against environmental stresses. Plant Growth Rregulation. 2013;32:216–232. [Google Scholar]
- 23.Kabera JN, Semana E, Mussa AR, He XJ. Plant secondary metabolites: biosynthesis, classification, function and pharmacological properties. Pharm Pharmacol. 2014;2:377–392. [Google Scholar]
- 24.Pagare S, Bhatia M, Tripathi N, Pagare S, Bansal YJ. Secondary metabolites of plants and their role: overview. Curr Trends Biotechnol Pharm. 2015;9:293–304. [Google Scholar]
- 25.Agarwal M, Shrivastava N, Padh H. Advances in molecular marker techniques and their applications in plant sciences. Plant Critical Reviews. 2008;27:617–631. [DOI] [PubMed] [Google Scholar]
- 26.Samanta S, Thakur JK. Importance of mediator complex in the regulation and integration of diverse signaling pathways in plants. Front Plant Sci. 2015;6:757. doi: 10.3389/fpls.2015.00757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alokam S, Li Y, Li W, Chinnappa C, Reid DM. Photoregulation of phenylalanine ammonia-lyase (PAL) and chalcone synthase (CHS) in the accumulation of anthocyanin in alpine and prairie ecotypes of Stellaria longipes under varied R/FR. Physiol Plant. 2002;116(4):531–538. doi: 10.1034/j.1399-3054.2002.1160412.x. [DOI] [Google Scholar]
- 28.Dewick PM. The biosynthesis of C 5–C 25 terpenoid compounds. Nat Prod Rep. 2002;19(2):181–222. doi: 10.1039/b002685i. [DOI] [PubMed] [Google Scholar]
- 29.Wink M. 2015. Evolution of secondary metabolism in plants. Ecological Biochemistry: environment and Interspecies Interactions. [Google Scholar]
- 30.Liscombe DK, MacLeod BP, Loukanina N, Nandi OI, Facchini PJ. Evidence for the monophyletic evolution of benzylisoquinoline alkaloid biosynthesis in angiosperms. Phytochemistry. 2005;66(11):1374–1393. doi: 10.1016/j.phytochem.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 31.Mazzucotelli E, Mastrangelo AM, Crosatti C, Guerra D, Stanca AM, Cattivelli L. Abiotic stress response in plants: when post-transcriptional and post-translational regulations control transcription. Plant Science. 2008;174(4):420–431. doi: 10.1016/j.plantsci.2008.02.005. [DOI] [Google Scholar]
- 32.Djami-Tchatchou AT, Sanan-Mishra N, Ntushelo K, Dubery IA. Functional roles of microRNAs in agronomically important plants—potential as targets for crop improvement and protection. Front Plant Sci. 2017;8:378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahajan V, Mahajan A, Pagoch S, Bedi Y, Gandhi S. microRNA mediated regulation of plant secondary metabolism: an in silico analysis. Natural Science, Biology and Medicine. 2011;2. [Google Scholar]
- 34.Gupta OP, Karkute SG, Banerjee S, Meena NL, Dahuja A. Contemporary understanding of miRNA-based regulation of secondary metabolites biosynthesis in plants. Front Plant Sci. 2017;8:374. doi: 10.3389/fpls.2017.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gholipour M, Kohnehrouz BB. Molecular cloning and in silico analysis of rps7 gene from the Lactuca sativa. Jordan J Biol Sci. 2017;10:279. [Google Scholar]
- 36.Sabzehzari M, Naghavi M. Phyto-miRNAs-based regulation of metabolites biosynthesis in medicinal plants. Gene. 2019;682:13–24. doi: 10.1016/j.gene.2018.09.049. [DOI] [PubMed] [Google Scholar]
- 37.Qiao Y, Zhang J, Zhang J, Wang Z, Ran A, Guo H, Wang D, Zhang J. Integrated RNA-seq and sRNA-seq analysis reveals miRNA effects on secondary metabolism in Solanum tuberosum L. Molecular Genetics. 2017;292:37–52. [DOI] [PubMed] [Google Scholar]
- 38.Padhan JK, Kumar P, Sood H, Chauhan RS. Prospecting NGS-transcriptomes to assess regulation of miRNA-mediated secondary metabolites biosynthesis in Swertia chirayita, a medicinal herb of the North-Western Himalayas. Phytomedicines. 2016;8:219–228. [Google Scholar]
- 39.Han J, Kong M, Xie H, Sun Q, Nan Z, Zhang Q, Pan JB. Identification of miRNAs and their targets in wheat (Triticum aestivum L.) by EST analysis. Genet Mol Res. 2013;12(3):805. doi: 10.4238/2013.September.19.11. [DOI] [PubMed] [Google Scholar]
- 40.Croteau R, Kutchan TM, Lewis NG. Natural products (secondary metabolites). Biochemistry Molecular Biology of Plants. 2000;24:1250–1319. [Google Scholar]
- 41.Solecka DJ. Role of phenylpropanoid compounds in plant responses to different stress factors. Acta Physiol Plant. 1997;19(3):257–268. doi: 10.1007/s11738-997-0001-1. [DOI] [Google Scholar]
- 42.Zhu H, Zhou Y, Castillo-González C, Lu A, Ge C, Zhao Y-T, Zhang X. Bidirectional processing of pri-miRNAs with branched terminal loops by Arabidopsis Dicer-like1. Nat Struct Mol Biol. 2013;20(9):1106. doi: 10.1038/nsmb.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fraser CM, Chapple C. The phenylpropanoid pathway in Arabidopsis. The Arabidopsis Book/American Society of Plant Biologists. 2011. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K. Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol. 2011;11(1):163. doi: 10.1186/1471-2229-11-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ibdah M, Zhang X-H, Schmidt J, Vogt T. A novel Mg2+-dependent O-methyltransferase in the phenylpropanoid metabolism of Mesembryanthemum crystallinum. Biol Chem. 2003;278(45):43961–43972. doi: 10.1074/jbc.M304932200. [DOI] [PubMed] [Google Scholar]
- 46.Li Y, Lu Y-G, Shi Y, Wu L, Xu Y-J, Huang F, Fan J, Zhang Y, Guo XY, Zhao JQ, et al. Multiple rice MicroRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol. 2014;164(2):1077–1092. doi: 10.1104/pp.113.230052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma A, Badola PK, Bhatia C, Sharma D, Trivedi PK. miRNA-encoded peptide, miPEP858, regulates plant growth and development in Arabidopsis. Nature Plants. 2019;642561. [Google Scholar]
- 48.Marcela V-H, Gerardo V-M, Agustín A-RC, Antonio G-M-M, Oscar R, Diego C-P, Cruz-Hernández A. MicroRNAs associated with secondary metabolites production. Plant Physiological Aspects of Phenolic Compounds: intechOpen. 2019. [Google Scholar]
- 49.Kim J, Park JH, Lim CJ, Lim JY, Ryu J-Y, Lee B-W, Choi J-P, Kim W, Bom L, Ha Y, et al. Small RNA and transcriptome deep sequencing proffers insight into floral gene regulation in Rosa cultivars. BMC Genomics. 2012;613(1):657. doi: 10.1186/1471-2164-13-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gou J-Y, Felippes FF, Liu C-J, Weigel D, Wang J-W. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell. 2011;23(4):1512–1522. doi: 10.1105/tpc.111.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wójcik AM, Nodine MD, Gaj MD. miR160 and miR166/165 contribute to the LEC2-mediated auxin response involved in the somatic embryogenesis induction in Arabidopsis. Front Plant Sci. 2017;8:2024. doi: 10.3389/fpls.2017.02024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rock CD. A role for MIR828 in pineapple fruit development. FResearch. 2020;9:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Díaz-Manzano FE, Cabrera J, Ripoll -J-J, Del Olmo I, Andrés MF, Silva AC, Barcala M, Sánchez M, Ruíz-Ferrer V, De Almeida-engler J. A role for the gene regulatory module microRNA172/TARGET OF EARLY ACTIVATION TAGGED 1/FLOWERING LOCUST (miRNA172/TOE1/FT) in the feeding sites induced by Meloidogyne javanica in Arabidopsis thaliana. New Phytol. 2018;217(2):813. doi: 10.1111/nph.14839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sunitha S, Loyola R, Alcalde JA, Arce-Johnson P, Matus JT, Rock CD. The role of UV-B light on small RNA activity during grapevine berry development. Genes, Genomes, Genetics. 2019;9:769–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kajal M, Singh K. Small RNA profiling for identification of miRNAs involved in regulation of saponins biosynthesis in Chlorophytum borivilianum. BMC Plant Biol. 2017;17(1):265. doi: 10.1186/s12870-017-1214-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caputi L, Aprea E. Use of terpenoids as natural flavouring compounds in food industry. Recent Patents on Food, Nutrition Agriculture. 2011;3(1):9–16. doi: 10.2174/2212798411103010009. [DOI] [PubMed] [Google Scholar]
- 57.Ma J, Kanakala S, He Y, Zhang J, Zhong X. Transcriptome sequence analysis of an ornamental plant, Ananas comosus var. bracteatus, revealed the potential unigenes involved in terpenoid and phenylpropanoid biosynthesis. PloS One. 2015;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ee S-F, Mohamed-Hussein Z-A, Othman R, Shaharuddin NA, Ismail I, Zainal Z. Functional characterization of sesquiterpene synthase from Polygonum minus. The Scientific World Journal. 2014;2014:1–11. doi: 10.1155/2014/840592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Najafabadi AS, Naghavi MR. Mining Ferula gummosa transcriptome to identify miRNAs involved in the regulation and biosynthesis of terpenes. Gene. 2018;645:41–47. doi: 10.1016/j.gene.2017.12.035. [DOI] [PubMed] [Google Scholar]
- 60.Fan R, Li Y, Li C, Zhang Y. Differential microRNA analysis of glandular trichomes and young leaves in Xanthium strumarium L. reveals their putative roles in regulating terpenoid biosynthesis. PLoS One. 2015;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Samad AFA, Rahnamaie-Tajadod R, Sajad M, Jani J, Murad AMA, Noor NM, Ismail I. Regulation of terpenoid biosynthesis by miRNA in Persicaria minor induced by Fusarium oxysporum. BMC Genomics. 2019;20(1):586. doi: 10.1186/s12864-019-5954-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu Z-X, Wang L-J, Zhao B, Shan C-M, Zhang Y-H, Chen D-F, Chen X-Y. Progressive regulation of sesquiterpene biosynthesis in Arabidopsis and Patchouli (Pogostemon cablin) by the miR156-targeted SPL transcription factors. Mol Plant. 2015;8(1):98–110. doi: 10.1016/j.molp.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 63.Robert‐Seilaniantz A, MacLean D, Jikumaru Y, Hill L, Yamaguchi S, Kamiya Y, Jones J. The microRNA miR393 re‐directs secondary metabolite biosynthesis away from camalexin and towards glucosinolates. The Plant. 2011;67:218–231. [DOI] [PubMed] [Google Scholar]
- 64.Kidner CA, Martienssen RA. The developmental role of microRNA in plants. Curr Opin Plant Biol. 2005;8(1):38–44. doi: 10.1016/j.pbi.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 65.Desai J, Liu YL, Wei H, Liu W, Ko TP, Guo RT, Oldfield E. Back cover: structure, function, and inhibition of Staphylococcus aureus heptaprenyl diphosphate synthase (ChemMedChem 17/2016). ChemMedChem. 2016;11(17):1968. doi: 10.1002/cmdc.201600430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu M, Lu S. Plastoquinone and ubiquinone in plants: biosynthesis, physiological function and metabolic engineering. Front Plant Sci. 2016;7:1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mandel MA, Feldmann KA, Herrera‐Estrella L, Rocha‐Sosa M, León P. CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. The Plant Journal. 1996;9(5):649–658. doi: 10.1046/j.1365-313X.1996.9050649.x. [DOI] [PubMed] [Google Scholar]
- 68.Li C, Zhu Y, Guo X, Sun C, Luo H, Song J, Li Y, Wang L, Qian J, Chen S. Transcriptome analysis reveals ginsenosides biosynthetic genes, microRNAs and simple sequence repeats in Panax ginseng C. A. Meyer. BMC Genomics. 2013;14(1):245. doi: 10.1186/1471-2164-14-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramos-Valdivia AC, Van Der Heijden R, Verpoorte R. Isopentenyl diphosphate isomerase: a core enzyme in isoprenoid biosynthesis. A review of its biochemistry and function. Nat Prod Rep. 1997;14(6):591–603. doi: 10.1039/np9971400591. [DOI] [PubMed] [Google Scholar]
- 70.Akdogan G, Tufekci ED, Uranbey S, Unver T. miRNA-based drought regulation in wheat. Functional Integrative Genomics. 2016;16(3):221–233. doi: 10.1007/s10142-015-0452-1. [DOI] [PubMed] [Google Scholar]
- 71.De Luca V, St Pierre B. The cell and developmental biology of alkaloid biosynthesis. Trends Plant Sci. 2000;5(4):168–173. doi: 10.1016/S1360-1385(00)01575-2. [DOI] [PubMed] [Google Scholar]
- 72.Kaur R, Arora S. Alkaloids-important therapeutic secondary metabolites of plant origin. Critical Reviews. 2015;2:1–8. [Google Scholar]
- 73.Cazares P. Functional genomics of monoterpenoid indole alkaloid biosynthesis in Rauvolfia serpentina. 2016. [Google Scholar]
- 74.Yang L, Stöckigt J. Trends for diverse production strategies of plant medicinal alkaloids. Nat Prod Rep. 2010;27(10):1469–1479. doi: 10.1039/c005378c. [DOI] [PubMed] [Google Scholar]
- 75.Wheeler A, Bugs P. Heteroptera associated with developing and mature capsules (fruits) of Papaver somniferum L. (papaveraceae). Proceedings of the Entomological Society of Washington 2019; 121:511–524. [Google Scholar]
- 76.Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39(8):1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 77.Wu HJ, Wang ZM, Wang M, Wang XJ. Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant physiology. 2013, 161(4):1875–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anadón A, Martínez-Larrañaga MR, Ares I, Martínez MA. Poisonous Plants of the Europe. In: Veterinary Toxicology. Elsevier; 2018. p. 891–909. [Google Scholar]
- 79.Hao DC, Yang L, Xiao PG, Liu M. Identification of Taxus microRNAs and their targets with high-throughput sequencing and degradome analysis. Physiol Plant. 2012;146(4):388–403. doi: 10.1111/j.1399-3054.2012.01668.x. [DOI] [PubMed] [Google Scholar]
- 80.Singh N, Srivastava S, Shasany AK, Sharma A. Identification of miRNAs and their targets involved in the secondary metabolic pathways of Mentha spp. Comput Biol Chem. 2016;64:154–162. doi: 10.1016/j.compbiolchem.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 81.Papageorgiou GC. 2007. Chlorophyll a fluorescence: a signature of photosynthesis. Springer Science & Business Media. [Google Scholar]
- 82.Rejeb KB, Abdelly C, Savouré A. How reactive oxygen species and proline face stress together. Plant Physiol Biochem. 2014;80:278–284. doi: 10.1016/j.plaphy.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 83.Mochizuki N, Tanaka R, Grimm B, Masuda T, Moulin M, Smith AG, Tanka A, Terrry MJ. The cell biology of tetrapyrroles: a life and death struggle. Trends Plant Science. 2010;15(9):488–498. doi: 10.1016/j.tplants.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 84.Yang P-M, Huang Q-C, Qin G-Y, Zhao S-P, Zhou J-G. Different drought-stress responses in photosynthesis and reactive oxygen metabolism between autotetraploid and diploid rice. Plants. 2014;52:193–202. [Google Scholar]
- 85.Niu S, Wang Y, Zhao Z, Deng M, Cao L, Yang L, Fan G. Transcriptome and degradome of microRNAs and their targets in response to drought stress in the plants of a diploid and its autotetraploid Paulownia australis. PloS one. 2016. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xiong -Y-Y, Ma J, He Y-H, Lin Z, Li X, Yu S-M, Zhang Y. High-throughput sequencing analysis revealed the regulation patterns of small RNAs on the development of A. comosus var. bracteatus leaves. Sci Rep. 2018;8(1):1–11. doi: 10.1038/s41598-018-20261-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yu S, Galvão VC, Zhang Y-C, Horrer D, Zhang T-Q, Hao Y-H, Wang S, Schmid M, Wang J-W. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA PROMOTER BINDING–LIKE transcription factors. Plant Cell. 2012;24(8):3320–3332. doi: 10.1105/tpc.112.101014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu X, Chen C-Y, Wang K-C, Luo M, Tai R, Yuan L, Zhao M, Yang S, Tian G, Cui Y. PHYTOCHROME INTERACTING FACTOR3 associates with the histone deacetylase HDA15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings. Plant Cell. 2013;25(4):1258–1273. doi: 10.1105/tpc.113.109710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sentandreu Benavent M. Functional profiling of PIF3 regulated genes in the dark/Análisis funcional de los genes regulados por PIF3 en oscuridad. 2013. [Google Scholar]
- 90.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Journal of Science. 2002;297:2053–2056. [DOI] [PubMed] [Google Scholar]
- 91.Ma Z, Hu X, Cai W, Huang W, Zhou X, Luo Q, Yang H, Wang J, Huang J. Arabidopsis miR171-targeted scarecrow-like proteins bind to GT cis-elements and mediate gibberellin-regulated chlorophyll biosynthesis under light conditions. PLos One. 2014;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li H, Zhang Q, Li L, Yuan J, Wang Y, Wu M, et al. Ectopic overexpression of bol-miR171b increases chlorophyll content and results in sterility in broccoli (Brassica oleracea L var. Italica). 2018. Vol. 66: 9588–9597. [DOI] [PubMed] [Google Scholar]
- 93.Li L, Xue M, Yi H. Uncovering microRNA-mediated response to SO2 stress in Arabidopsis thaliana by deep sequencing. Hazardous Materials. 2016;316:178–185. doi: 10.1016/j.jhazmat.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 94.Kobayashi K, Masuda T. Transcriptional control for the chlorophyll metabolism. Metabolism, structure function of plant tetrapyrroles: control mechanisms of chlorophyll biosynthesis analysis of chlorophyll-binding proteins. 2019;91:133–161. [Google Scholar]
- 95.Patel P, Yadav K, Ganapathi RT. Small and hungry: microRNAs in micronutrient homeostasis of plants. MicroRNA. 2017;6(1):22–41. doi: 10.2174/2211536606666170117160338. [DOI] [PubMed] [Google Scholar]
- 96.Gabruk M, Mysliwa-Kurdziel B. Light-dependent protochlorophyllide oxidoreductase: phylogeny, regulation, and catalytic properties. Biochemistry. 2015;54(34):5255–5262. doi: 10.1021/acs.biochem.5b00704. [DOI] [PubMed] [Google Scholar]
- 97.Šramková Z, Gregová E, Šturdík E. Chemical composition and nutritional quality of wheat grain. Acta Chimica Slovaca. 2009;2:115–138. [Google Scholar]
- 98.Richter TKS. Discovery, biosynthesis and evolutionary history of sioxanthin, a novel glycosylated carotenoid from marine bacteria Salinispora. UC San Diego; 2014. [Google Scholar]
- 99.Saini RK, Keum Y-SJFC. Carotenoid extraction methods: a review of recent developments. Food Chemistry. 2018;240:90–103. doi: 10.1016/j.foodchem.2017.07.099. [DOI] [PubMed] [Google Scholar]
- 100.Schoefs B. Chlorophyll and carotenoid analysis in food products. Properties of the pigments and methods of analysis. Trends Food Sci Technol. 2002;13(11):361–371. doi: 10.1016/S0924-2244(02)00182-6. [DOI] [Google Scholar]
- 101.Esteban R, Moran JF, Becerril JM, García-Plazaola JI. Versatility of carotenoids: an integrated view on diversity, evolution, functional roles and environmental interactions. Environmental Experimental Botany. 2015;119:63–75. doi: 10.1016/j.envexpbot.2015.04.009. [DOI] [Google Scholar]
- 102.Solymosi K, Schoefs B. Prolamellar body: a unique plastid compartment, which does not only occur in dark-grown leaves. Plant cell organelles—selected topics. Trivandrum, India: Research Signpost; 2008. 151–202. [Google Scholar]
- 103.Pogson B, McDonald KA, Truong M, Britton G, DellaPenna D. Arabidopsis carotenoid mutants demonstrate that lutein is not essential for photosynthesis in higher plants. Plant Cell. 1996;8(9):1627–1639. doi: 10.1105/tpc.8.9.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wei S, Yu B, Gruber MY, Khachatourians GG, Hegedus DD, Hannoufa A. Enhanced seed carotenoid levels and branching in transgenic Brassica napus expressing the Arabidopsis miR156b gene. Agricultural Food Chemistry. 2010;58(17):9572–9578. doi: 10.1021/jf102635f. [DOI] [PubMed] [Google Scholar]
- 105.Wei S, Gruber MY, Yu B, Gao M-J, Khachatourians GG, Hegedus DD, Parkin IAP, Hannoufa A. Arabidopsis mutant sk156 reveals complex regulation of SPL15 in a miR156-controlled gene network. BMC Plant Biol. 2012;12(1):169. doi: 10.1186/1471-2229-12-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Baranski R, Cazzonelli C. Carotenoid biosynthesis and regulation in plants. Wiley Online Library; 2016. [Google Scholar]
- 107.Xu Q, Liu Y, Zhu A, Wu X, Ye J, Yu K, Guo W, Deng X. Discovery and comparative profiling of microRNAs in a sweet orange red-flesh mutant and its wild type. BMC Genomics. 2010;11(1):246. doi: 10.1186/1471-2164-11-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bourgaud F, Gravot A, Milesi S, Gontier E. Production of plant secondary metabolites: a historical perspective. Plant Science. 2001;161(5):839–851. doi: 10.1016/S0168-9452(01)00490-3. [DOI] [Google Scholar]
- 109.Zangerl AR, Berenbaum MR. Genetic variation in primary metabolites of Pastinaca sativa; can herbivores act as selective agents?. Chem Ecol. 2004;30(10):1985–2002. doi: 10.1023/B:JOEC.0000045590.28631.00. [DOI] [PubMed] [Google Scholar]
- 110.Kliebenstein D. Secondary metabolites and plant/environment interactions: a view through Arabidopsis thaliana tinged glasses. Plant, Cell and Environment. 2004;27(6):675–684. doi: 10.1111/j.1365-3040.2004.01180.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Biochemistry of Plant Secondary Metabolism and Functions of Plant Secondary Metabolites and their Exploitation in Biotechnology, DOI: https://doi.org/10.1016/S0167-7799(00)01454-2


