ABSTRACT
Zika virus (ZIKV) is an emerging mosquito-borne flavivirus that poses significant threats to global public health. Macrophages and dendritic cells are both key sentinel cells in the host immune response and play critical roles in the pathogenesis of flavivirus infections. Recent studies showed that ZIKV could productively infect monocyte-derived dendritic cells (moDCs), but the role of macrophages in ZIKV infection remains incompletely understood. In this study, we first compared ZIKV infection in monocyte-derived macrophages (MDMs) and moDCs derived from the same donors. We demonstrated that while both MDMs and moDCs were susceptible to epidemic (Puerto Rico) and pre-epidemic (Uganda) strains of ZIKV, virus replication was largely restricted in MDMs but not in moDCs. ZIKV induced significant apoptosis in moDCs but not MDMs. The restricted virus replication in MDMs was not due to inefficient virus entry but was related to post-entry events in the viral replication cycle. In stark contrast with moDCs, ZIKV failed to inhibit STAT1 and STAT2 phosphorylation in MDMs. This resulted in the lack of efficient antagonism of the host type I interferon-mediated antiviral responses. Importantly, depletion of STAT2 but not STAT1 in MDMs significantly rescued the replication of ZIKV and the prototype flavivirus yellow fever virus. Overall, our findings revealed a differential interplay between macrophages and dendritic cells with ZIKV. While dendritic cells may be exploited by ZIKV to facilitate virus replication, macrophages restricted ZIKV infection.
KEYWORDS: Dendritic cells, flavivirus, macrophages, interferon signaling, STAT1, STAT2, Zika
Introduction
Zika virus (ZIKV) is an emerging mosquito-borne flavivirus that has raised global concerns in recent years [1]. In addition to the transmission of ZIKV by Aedes mosquito, it can also be sexually and vertically transmitted from person to person. While most ZIKV-infected patients are asymptomatic or mildly symptomatic, some adult patients develop severe neurological complications such as Guillain–Barré syndrome, and fetuses may develop congenital microcephaly and anomalies [1,2]. Since 2015, ZIKV has rapidly spread to over 87 countries and territories and remains a potential risk of future epidemics as new lineages continue to emerge [3,4].
Macrophages and dendritic cells are key antigen-presenting cells that play pivotal roles in the pathogenesis of flavivirus infections. They act as mediators of innate and adaptive immunity. Depletion of macrophages in types I and II interferon (IFN) receptor-deficient mice results in a significantly higher level of systemic dengue virus (DENV) titer [5]. Mice lacking type I IFN receptor expression on CD11+ dendritic cells and LysM+ macrophages develop fatal DENV infection [6]. Dendritic cells prime B and T cell responses against flavivirus infections via upregulation of major histocompatibility complex (MHC) and co-stimulatory molecules such as CD80, CD83, and CD86, as well as production of cytokines and chemokines [7–9]. Moreover, macrophages and dendritic cells are direct targets of DENV, West Nile virus, and Japanese encephalitis virus (JEV) that support virus replication and dissemination as Trojan horses [10–15]. In mosquito-borne transmission, DENV infects macrophages, resident CD103+ classical dendritic cells (cDCs), and Ly6C-/CD11b+ cDCs, and recruits monocytes to further differentiate into inflammatory dendritic cells and macrophages [16,17]. Importantly, certain tissue-resident macrophages derived from monocytes have been illustrated to be susceptible to ZIKV infection [18]. For example, ZIKV is able to efficiently infect and replicate in primary placental macrophages (Hofbauer cells), resulting in dissemination across the placental barrier and vertical transmission [19,20]. In the dermis, blood monocytes differentiate into monocyte-derived macrophages (MDMs) or monocyte-derived tissue-resident macrophages [16,21]. Mosquito-borne flaviviruses may therefore be able to target these cells to facilitate virus dissemination.
IFN signaling plays essential roles in protecting the host from viral infection. Flaviviruses possess IFN antagonism strategies to suppress the host IFN response and facilitate virus replication. ZIKV infection impairs type I and III IFN signaling and suppresses the induction of downstream interferon-stimulated genes (ISGs) through virus-mediated STAT2 degradation [22-24]. Moreover, ZIKV antagonizes type I IFN signaling through blockade of STAT1 and STAT2 phosphorylation and productively infects human monocyte-derived dendritic cells (moDCs) [24-26]. Despite extensive studies on flavivirus antagonism, the role of IFN signaling upon ZIKV infection in human MDMs remains incompletely defined.
Extending on these preliminary findings, in the present study, we utilized human primary MDMs as a model to investigate the role of macrophages in ZIKV infection. Our results demonstrated that ZIKV replication in MDMs was largely restricted compared to that in moDCs. We further found that ZIKV infection in MDMs did not efficiently suppress type I IFN-mediated antiviral responses and failed to inhibit IFN-α-induced STAT1 and STAT2 phosphorylation. Importantly, depletion of STAT2 but not STAT1 in MDMs significantly rescued ZIKV and yellow fever virus (YFV) replication. Collectively, our results indicated that human MDMs restrict flavivirus replication through a STAT2-dependent mechanism. These differential interplays between MDMs and moDCs with ZIKV provided novel insights into our understanding on the virus-host interaction in ZIKV infection.
Materials and methods
Cells and viruses
VeroE6 was purchased from the American Type Culture Collection (ATCC) and maintained in Dulcecco’s Modified Eagle’s Medium (DMEM), supplemented with 10% fetal bovine serum (FBS), 100 units/mL of penicillin and 100μg/mL of streptomycin (5%CO2 at 37°C). ZIKVPR (Puerto Rico strain PRVABC59) was obtained from a patient in the recent South American epidemic (kindly provided by Brandy Russell and Barbara Johnson, Centers for Disease Control and Prevention, USA) and ZIKVU (ZIKVAF-976 Uganda strain) was isolated from a nonhuman primate in Uganda in 1947 (kindly provided by Tatjana Avšič Županc, University of Ljubljana, Slovenia, the European Virus Archive) [27,28]. Yellow fever virus 17D (YFV) was available in our laboratory and cultured as previously described [28].
Preparation of human primary monocyte-derived macrophages (MDMs) and monocyte-derived dendritic cells (moDCs)
Healthy volunteer blood samples were collected from Hong Kong Red Cross Blood Transfusion Service according to a protocol approved by the Institutional Review Board of the University of Hong Kong. Human peripheral blood mononuclear cells (PBMCs) were isolated from the buffy coats as we previously described [29]. MDMs were differentiated from PBMCs by providing RPMI-1640 supplemented with 10% FBS, 100 units/mL of penicillin, 100μg/mL of streptomycin, 1% sodium pyruvate, 1% non-essential amino acids, 1U/mL GM-CSF (Cell Sciences, Canton, MA, USA) [30]. MoDCs were differentiated and maintained as the same condition as MDMs with IL-4 (R&D systems) [31].
Virus infections
MDMs and moDCs were differentiated in complete RPMI-1640 with specific growth factors for 6–7 days before infection. For immunofluorescence microscopy experiment, MDMs and moDCs were uninfected or infected with ZIKVPR or ZIKVU [multiplicity of infection (MOI) = 1.0] for 2 h at 37°C. After 2 h incubation, the virus inoculum was washed off and the cells were maintained with fresh RPMI-1640. For flow cytometry analysis, MDMs and moDCs were uninfected or infected with ZIKVPR or ZIKVU (MOI=0.1 or MOI=1.0) for 2 h at 37°C. For viral replication kinetics evaluation, cells were infected with ZIKVPR, ZIKVU, or YFV (MOI=0.01 or 1.0) for 2 h at 37°C. For extended period replication kinetics and cell viability evaluation, the cells were infected with ZIKVPR or ZIKVU (MOI=0.1) for 2 h at 37°C. For IL-13 treatment experiment, after 24 h treatment with IL-13, the cells were infected with ZIKVPR or ZIKVU (MOI=0.01) for 2 h at 37°C. For virus entry capability experiment, the cells were incubated with ZIKVPR at an MOI of 10.0 in an ice bath for 2 h, followed by incubation with 37°C for 30 min. After 30 min incubation, the virus inoculum was washed off and the cell lysates were collected for further detection. For ISGs evaluation experiment, the cells were uninfected or infected with ZIKVPR (MOI=10.0) for 6 h, followed by mock treatment or treatment with IFN-α for 6 h. For Western blots experiment, the cells were infected with ZIKVPR (MOI=1.0) for 1 h at 37°C. For small interfering RNA (siRNA) knockdown experiment, after transfection with siRNA, the cells were infected with ZIKVPR or YFV (MOI=1.0) for 1 h at 37°C.
Immunofluorescence staining and confocal microscopy
Immunostaining and confocal microscopy were performed as we previously described [32,33]. Briefly, MDMs and moDCs cultured in 24-well plate were digested by trypsin and re-seeded into chamber slide. After one day of culture in the RPMI-1640 with growth factors, the cells were uninfected or infected with ZIKVPR or ZIKVU (MOI=1.0) for 2 h at 37°C. At 48hpi, the infected MDMs and moDCs were washed with phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde. The cells were permeabilized for 10 min with 0.2% Triton X-100 and blocked with Dako blocking buffer for 30 min at room temperature. The cells were subsequently incubated with mouse 4G2 monoclonal antibody (MERCK) against envelope (E) protein of pan-flavivirus or mouse J2 monoclonal antibody (SCICONS) against double-stranded RNA (dsRNA) diluted with Dako antibody diluent overnight at 4°C, followed by incubation with goat anti-mouse Fluor 488 immunoglobulin G (IgG) (Abcam) secondary antibody diluted with Dako antibody diluent for 1 h. The nuclei of the cell were stained by DAPI nucleic acid stain (Thermo Fisher Scientific) for 10 min. The slides were imaged with confocal microscopy using a Carl Zeiss LSM 880 system (Zeiss, Oberkochen, Germany).
Flow cytometry analysis
Flow cytometry was performed according to standard protocols as we previously described [34,35]. In brief, MDMs and moDCs were infected with ZIKVPR or ZIKVU (MOI=0.1 or MOI=1.0) for 2 h at 37°C. The infected cells were detached with EDTA at 24hpi and 48hpi and fixed with 4% paraformaldehyde. The cells were then permeabilized for 10 min with 0.2% Triton X-100 and blocked with PBS supplemented with 2% FBS. The cells were incubated with primary antibodies (4G2 or J2) for 1 h at 4°C, followed by incubation with secondary antibody (goat anti-mouse Fluor 488 IgG) for 45 min at 4°C. Multi-color flow cytometry acquisition was performed using the FACS Canto II Analyzer (BD Biosciences), and the data were analyzed by FlowJo software.
RNA extraction and quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Cellular RNA extraction, reverse transcription, and quantitative PCR were performed as we described previously [36]. For evaluation of viral genome copy in supernatants and cell lysates, real-time qRT-PCR was used to quantify ZIKVPR or ZIKVU genome copy with LightCycler 96 Real-Time PCR System (Roche) as we described previously [37,38]. For host genes analysis, the cells were lysed using the RLT buffer provided from RNA extraction kit (TAKARA). The extracted RNA was reverse transcribed with Transcriptor First Strand cDNA Synthesis Kit (Roche). The levels of cellular gene expression were normalized to GAPDH and presented as per GAPDH in gene expressions.
Virus replication kinetics
MDMs, moDCs, and VeroE6 (2 × 105 cells per well of 24-well plate) were infected with ZIKVPR, ZIKVU, or YFV [MOI=0.01 (2 × 103 PFU) or MOI=1.0 (2 × 105 PFU)] for 2 h at 37°C and washed with PBS three times, followed by incubation with fresh medium. The cell lysates and supernatants were harvested for determining the genome copy and live infectious virus by qRT-PCR and plaque assays. For extended period viral replication kinetics, the cells were infected with ZIKVPR or ZIKVU (MOI=0.1) for 2 h at 37°C. The supernatants were collected at the indicated days post infection for determining virus titer by plaque assays.
Evaluation of cell viability and caspase-3/7 activity
Cell viability and Caspase-3/7 activity of mock- or virus-infected MDMs and moDCs were determined using CellTiterGlo assays (Promega, Madison, USA) and CaspaseGlo 3/7 assays (Promega, Madison, USA), respectively [39]. Briefly, the cells were infected with ZIKVPR or ZIKVU (MOI=0.1) for 2 h at 37°C. The cells were lysed together with culture supernatants at a 1:1 ratio with the CellTiterGlo reagent or CaspaseGlo 3/7 reagent at the indicated days post infection and placed on an orbital shaker for 10 min to induce cells lysis. The plates were read by measuring the luminescence signal with the VectorX3 multi-label plate reader (PerkinElmer, Waltham, MA, USA) as we previously described [40].
IL-13 treatment of MDMs
MDMs were untreated or treated with IL-13 of 20 and 50 ng/mL for 24 h and the cell lysates were harvested for testing expression of ZIKV entry-related host factors by qRT-PCR. MDMs treated with IL-13 of 50 ng/mL and moDCs treated with the same amount of DMSO were infected with ZIKVPR or ZIKVU (MOI=0.01). The cell lysates of the infected cells were collected at 24hpi for determining the viral genome copy by qRT-PCR.
Evaluation of viral entry capability
MDMs and moDCs were incubated with ZIKVPR (MOI=10.0) in an ice bath for 2 h, followed by incubation at 37°C. After 30 min incubation, the virus inoculum was discarded and the cells were washed three times with PBS. The cell lysates were collected for further detection.
Recombinant human IFN-α treatment
MDMs and moDCs were uninfected or infected with ZIKVPR (MOI=10.0) for 6 h at 37°C. After 6 h, the cells were subsequently washed with PBS and were untreated or treated with 1000U/mL of recombinant human IFN-α (PBL Assay Science). After 6 h of treatment, the cell lysates were harvested for detecting expression of interferon-stimulated genes (ISGs) by qRT-PCR.
Western blots and analysis
MDMs and moDCs were infected with ZIKVPR (MOI=1.0) at 37°C. At 48hpi, the cells were untreated or treated with 1000U/mL of recombinant human IFN-α for 30 min and then lysed with RIPA buffer supplemented with Protease Inhibitor Cocktail and Phosphatase Inhibitor Cocktail II (Thermo Fisher Scientific). The expression levels of STAT1, STAT2, STAT1 phosphotyrosine residue 701 (pSTAT1), STAT2 phosphotyrosine residue 689 (pSTAT2), and β-actin were detected using anti-STAT1 (BD Transduction Laboratories), anti-STAT2 (SANTA CRUZ BIOTECHNOLOGY), anti-pSTAT1 (Cell Signaling Technology), anti-pSTAT2 (Millipore), and anti-β-actin (Sigma-Aldrich) primary antibodies. The intensity of the bands was calculated using Image J software to generate the histogram. For evaluation of knockdown efficiency of siRNA transfection, MDMs and moDCs were transfected with scrambled siRNA or siRNA targeting STAT1 or STAT2. The expression of STAT1, STAT2, and β-actin were detected by Western blots.
Co-immunoprecipitation (co-IP) assays
The co-IP assays were performed using Pierce Crosslink Magnetic IP/co-IP kit according to manufacturer’s instruction (Thermo Fisher Scientific). Briefly, MDMs and moDCs were infected with ZIKVPR at an MOI of 5.0. After 24hpi, the whole cell lysates were incubated with protein A/G magnetic beads pre-crosslinked with anti-ZIKV NS5 antibody (GeneTex) or anti-STAT2 antibody (SANTA CRUZ BIOTECHNOLOGY). After incubation, the supernatants and beads were magnetically separated. The elution buffer from the kit was added into the beads and magnetically separated. The supernatants were harvested for detecting the expression of ZIKV NS5, STAT2, and β-actin by Western blots as we described earlier.
Transfection of siRNA
MDMs and moDCs were generated from two different healthy donors. The cells were transfected with siRNA targeting STAT1 or STAT2, as well scrambled siRNA as a negative control, followed by infection with ZIKVPR at an MOI of 1.0 as we previously described [41]. At 2hpi and 24hpi, the cell lysates were harvested for determining viral genome copy by qRT-PCR and the data was presented as fold change in viral genome copy of siRNA targeting STAT1 or STAT2 transfected cells relative to scrambled siRNA transfected cells. The supernatants were harvested for determining live infectious virus titer by plaque assays.
siRNA knockdown and Poly IC treatment
MDMs and moDCs were transfected with 70nM siRNA targeting STAT1 or STAT2 obtained from Dharmacon using RNAiMAX (Thermo Fisher Scientific), with scrambled siRNA as negative controls, followed by transfection with 5ug of Poly IC per well. After 12 h, the cell lysates were harvested for detecting host gene expressions by qRT-PCR, including IFIT1, MX1, ISG15, IRF1, and IP10.
Statistical analysis
The data are presented as mean ± standard deviation. Student’s t-test and one-way ANOVA were used for statistical analysis. Differences were considered significant when p < 0.05. * indicated p < 0.05, ** indicated p < 0.01, *** indicated p < 0.001, and **** represented p < 0.0001.
Results
ZIKV replication is supported by moDCs but restricted in MDMs
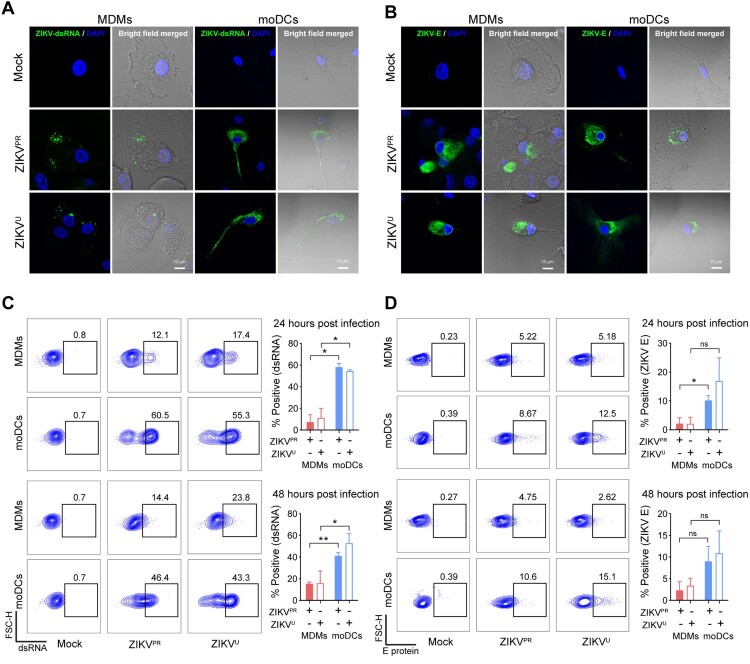
In this study, we used human primary MDMs and moDCs from healthy donors as models of peripheral macrophages and dendritic cells, respectively. First, we tested the susceptibility of MDMs to ZIKV infection using ZIKVPR or ZIKVU. moDCs from the same donors were included for comparison. Confocal microscopy revealed that viral double-stranded RNA (dsRNA) diffusely distributed as small puncta in the cytoplasm of ZIKVPR- or ZIKVU-infected MDMs (Figure 1A). The expression of ZIKV envelope (E) protein was detected to predominantly accumulate at the perinuclear regions in MDMs upon infection (Figure 1B). The distributions of viral dsRNA and E protein were similar in ZIKVPR-infected and ZIKVU-infected MDMs. The overall cellular distribution pattern of viral dsRNA and E protein in ZIKV-infected MDMs largely resembled that of ZIKV-infected moDCs (Figure 1A and 1B).
Figure 1.
MDMs are susceptible to ZIKV infection. Human monocyte-derived macrophages (MDMs) and dendritic cells (moDCs) were uninfected or infected with ZIKVPR or ZIKVU at an MOI of 1.0. At 48 h post-infection (hpi), the cells were fixed with 4% paraformaldehyde and then permeabilized with 0.2% Triton X-100. (A) Viral double-stranded RNA (dsRNA) and (B) envelope (E) protein were identified with J2 and 4G2 antibodies, respectively, and imaged with confocal microscopy. In parallel, MDMs and moDCs were infected with ZIKVPR or ZIKVU at an MOI of 0.1. At 24hpi and 48hpi, ZIKV-infected and mock-infected cells were fixed and stained with J2 or 4G2, and subsequently applied to flow cytometry to determine expression levels of (C) viral dsRNA and (D) E protein. The dot-plot showed data of one representative donor. Statistical analyses in all panels were performed with Student’s t-test and the differences were considered significant when p < 0.05. *p < 0.05, **p < 0.01. ns, not significant. The histogram showed the mean values and standard deviations from three donors. Bars represented 10 μm.
We further quantitatively examined ZIKV infection in MDMs with flow cytometry. Upon ZIKVPR or ZIKVU infection at 0.1 MOI, the mean percentage of dsRNA-positive MDMs was 7.37% (ZIKVPR-infected MDMs vs moDCs at 24hpi: P=0.0108) and 11.27% (ZIKVU-infected MDMs vs moDCs at 24hpi: P=0.0198) at 24hpi, which increased to 15.45% (ZIKVPR-infected MDMs vs moDCs at 48hpi: P=0.0089) and 15.88% (ZIKVU-infected MDMs vs moDCs at 48hpi: P=0.0312) at 48hpi, respectively (Figure 1C). Compared with viral dsRNA, the viral E protein was expressed at a lower percentage in ZIKVPR-infected and ZIKVU-infected MDMs at both 24hpi and 48hpi (Figure 1D). At 1.0 MOI, the expression of viral dsRNA and E protein in the infected MDMs and moDCs modestly increased in comparison to that infected at 0.1 MOI (Figure S1). Interestingly, the expression of viral dsRNA and E protein in ZIKV-infected MDMs were in general lower than those of ZIKV-infected moDCs (Figure 1 and Figure S1).
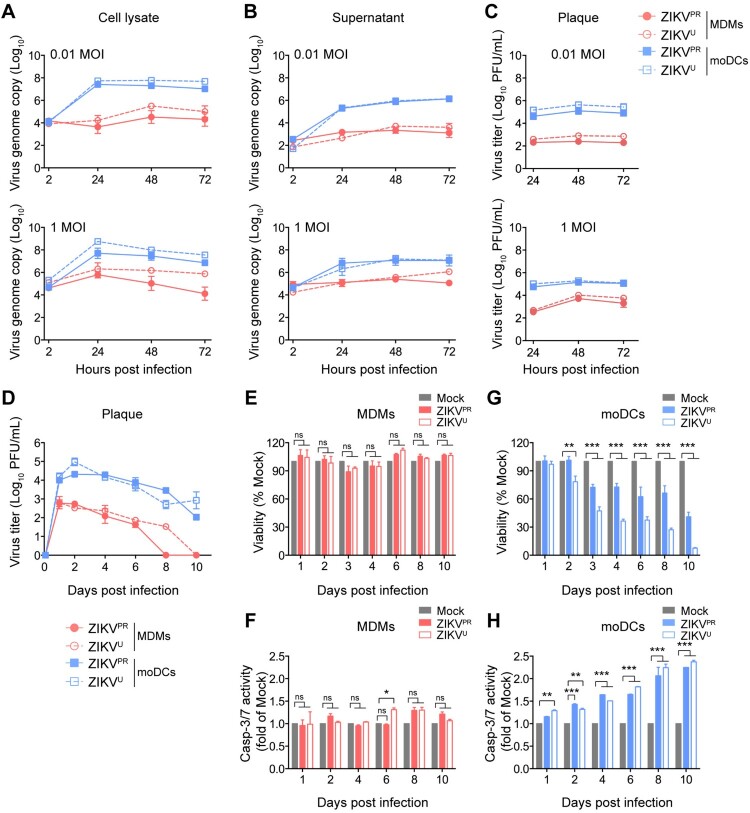
Next, we examined the replication kinetics of ZIKV in MDMs and moDCs by determining the viral genome copy and infectious virus titer at different time points. In agreement with the flow cytometry results, moDCs supported robust ZIKV replication with a 4–5 log10 increase in viral genome copy (Figures 2A and 2B) over the 72 h incubation period. In contrast, ZIKV replicated modestly in MDMs with only 1–2 log10 increase in viral genome copy over the same period (Figures 2A and 2B). Similarly, the peak infectious virus titer reached approximately 105–106 PFU/mL in the infected moDCs, but only 102–104 PFU/mL in the infected MDMs, depending on the virus inoculum (Figure 2C). Notably, although the replication efficiency of ZIKV in moDCs was higher than that in MDMs, ZIKV replication in both cell types was less efficient than that in VeroE6 cells (Figure S2). In addition, the replication pattern of yellow fever virus (YFV) in MDMs, moDCs, and VeroE6 was largely comparable to that of ZIKV. Intriguingly, comparable levels of viral RNA were detected in the cell lysates of ZIKV-infected MDMs and moDCs at 2hpi, suggesting that the discrepancies in ZIKV replication were not due to virus entry (Figure 2A). Since the infectious virus titer remained detectable at 72hpi without significant reduction in both ZIKV-infected MDMs and moDCs, we further explored ZIKV infection in these cell types at later time points. As shown in Figure 2D, ZIKV-infected moDCs continued to release infectious virus particles that were largely sustained in the supernatants even at day 10 post infection, whereas the production of infectious virus particles was rapidly halted in ZIKV-infected MDMs and fell below the detection limit at day 8–10 post infection (Figure 2D). The decrease in virus titer in the infected MDMs was not due to ZIKV-induced cytotoxicity since the viability and caspase-3/7 activity of ZIKV-infected MDMs remained similar compared to the mock-infected cells (Figures 2E and 2F). Meanwhile, the viability of ZIKV-infected moDCs dropped to 40.93% and 7.57% of mock for ZIKVPR- and ZIKVU-infected moDCs at day 10 post infection, respectively (Figure 2G). Moreover, moDCs were susceptible to ZIKV-induced apoptosis as indicated by the significantly elevated caspase-3/7 activity (Figure 2H). Taken together, our data suggested that while moDCs supported ZIKV replication, ZIKV replication in MDMs was largely restricted.
Figure 2.
MDMs but not moDCs restrict ZIKV replication. MDMs and moDCs were infected with ZIKVPR or ZIKVU at an MOI of 0.01 or 1.0. Viral genome copy in the (A) cell lysates and (B) supernatants were determined by qRT-PCR. (C) The live infectious virus titer in the supernatants was measured by plaque assays. (D) MDMs and moDCs were infected with ZIKVPR or ZIKVU at an MOI of 0.1. The supernatants were harvested for determining the infectious virus titer by plaque assays at the indicated days post-infection. The (E) cell viability and (F) caspase-3/7 activities of uninfected and ZIKV-infected MDMs; and (G) cell viability and (H) caspase-3/7 activities of uninfected and ZIKV-infected moDCs were determined at the indicated time points by CellTiterGlo and CaspaseGlo 3/7 assays, respectively. Data represented mean and standard deviations from 3–6 donors. Statistical analyses in all panels were performed with two way-ANOVA and the differences were considered significant when p < 0.05. *p < 0.05,**p < 0.01, and ***p < 0.001.
Restricted ZIKV replication in MDMs is not due to inefficient virus entry
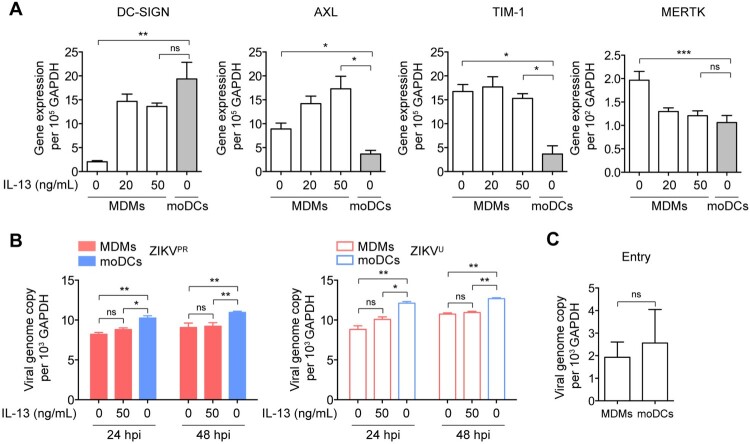
To further evaluate whether the restricted virus replication in MDMs is due to inefficient virus entry, we measured the endogenous expression levels of ZIKV-related entry factors in MDMs, including dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN) [42], AXL receptor tyrosine kinase (AXL) [43-45], T-cell immunoglobulin and mucin domain 1 (TIM-1) [42], and MERTK [46]. Our data demonstrated that MDMs expressed significantly higher levels of AXL, TIM-1, and MERTK, but less DC-SIGN than moDCs (Figure 3A). We next tested whether the restricted ZIKV infection in MDMs was due to the lower expression level of DC-SIGN by treating MDMs with IL-13, which is known to upregulate DC-SIGN expression [47,48], followed by infecting the cells with ZIKV. As shown in Figure 3A, IL-13 treatment substantially upregulated the expression of DC-SIGN in MDMs to levels comparable with that in moDCs. Interestingly, despite a comparable expression level of DC-SIGN and higher expression levels of AXL and TIM-1, ZIKV replication remained significantly less efficient in MDMs than that in moDCs (Figure 3B). In addition, the difference in replication was not a result of differential entry efficiency as ZIKV entry in MDMs and moDCs at 1hpi was not significantly different (Figure 3C). Together, our data indicate that MDMs restrict ZIKV replication at post-entry events in the viral replication cycle.
Figure 3.
Restricted ZIKV replication in MDMs is not due to inefficient virus entry. (A) MDMs were untreated or treated with IL-13 (20 and 50 ng/mL) for 24 h and then subjected to determine the expression levels of ZIKV-related entry factors, including DC-SIGN, AXL, TIM-1, and MERTK by qRT-PCR. moDCs were included as a control. (B) MDMs were untreated or treated with IL-13 (50 ng/mL) for 24 h prior to infection with ZIKVPR or ZIKVU at an MOI of 0.01. The viral genome copy in the cell lysates was determined by qRT-PCR at 24hpi and 48hpi. (C) Viral entry capability in MDMs and moDCs were determined by qRT-PCR and normalized with GAPDH. Data represented mean and standard deviations from 3 donors. Statistical analyses in all panels were performed with one way-ANOVA and the differences were considered significant when p < 0.05. *p < 0.05, **p < 0.01, and ***p < 0.001. ns, not significant.
ZIKV infection does not antagonize type I IFN-mediated responses in MDMs
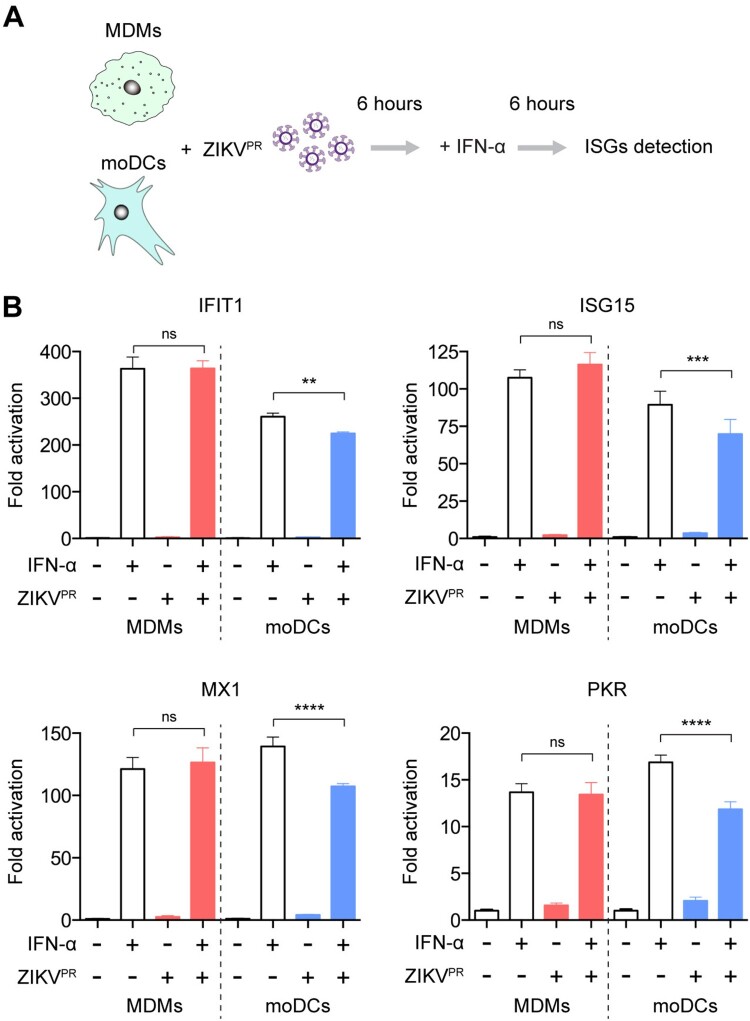
ZIKV has been demonstrated to antagonize the host IFN response to achieve optimal virus replication [22,23,26,49]. We next asked whether the restricted ZIKV replication could be attributed to inefficient viral antagonism of the IFN response in MDMs. MDMs or moDCs were infected with ZIKVPR for 6 h at a multiplicity of infection (MOI) of 10.0 and subsequently treated with IFN-α for an additional 6 h (Figure 4A). The cell lysates were harvested to evaluate the expression of interferon-stimulated genes (ISGs), including IFIT1, ISG15, MX1, and PKR. IFN-α treatment upregulated the expressions of all four ISGs in MDMs and moDCs (Figure 4B). In ZIKV-infected moDCs, the upregulated expressions of ISGs through IFN-α treatment were significantly attenuated (IFIT1: from 260-fold to 224-fold; ISG15: from 89-fold to 69-fold; MX1: from 139-fold to 107-fold; PKR: from 17-fold to 12-fold). In stark contrast, ZIKV infection did not attenuate the expressions of any of the evaluated ISGs in MDMs (Figure 4B). These results suggest that ZIKV infection insufficiently antagonizes type I IFN-mediated antiviral responses in MDMs, which results in restricted viral replication in these cells.
Figure 4.
ZIKV infection does not antagonize type I IFN-mediated responses in MDMs. (A) Schematic illustration of human recombinant IFN-α treatment and ZIKV infection. (B) MDMs and moDCs were uninfected or infected with ZIKVPR at an MOI of 10.0 for 6 h, followed by mock treatment or treatment with 1000U/mL of recombinant human IFN-α for 6 h. Cell lysates were collected for detection of ISGs induction using qRT-PCR, including IFIT1, ISG15, MX1, and PKR; the fold activations were calculated as compared to mock groups. Data represented mean and standard deviations from 3 donors. Statistical analyses in all panels were performed with one way-ANOVA and the differences were considered significant when p < 0.05. **p < 0.01, ***p < 0.001, and ****p < 0.0001. ns, not significant.
ZIKV infection does not inhibit the phosphorylation of STAT1 and STAT2 in MDMs
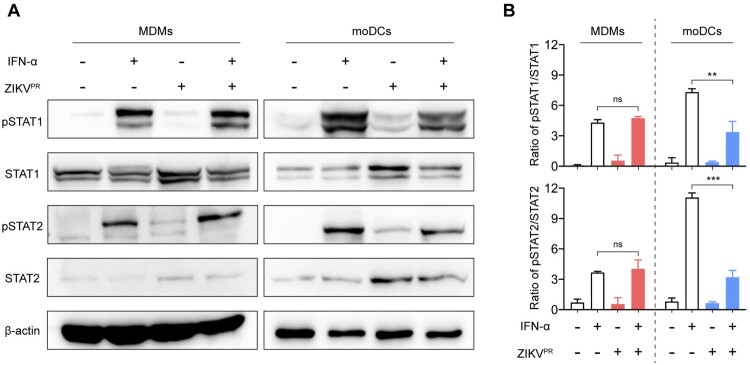
It has been recently reported that ZIKV subverts type I IFN signaling through (i) virus-induced degradation of STAT2 [22-24] or (ii) blockade of STAT1 or STAT2 phosphorylation [24,26]. However, it remains unknown whether these IFN antagonism mechanisms are applicable to MDMs. To this end, we treated mock- or ZIKVPR-infected MDMs or moDCs with IFN-α for 30 min at 48hpi. The expression levels of STAT1, STAT2, and their phosphorylation were examined by Western blots. In mock-infected MDMs and moDCs, IFN-α treatment substantially promoted STAT1 and STAT2 phosphorylation. Upon ZIKV infection, the upregulated amount of pSTAT1 and pSTAT2 were significantly reduced in moDCs from 7.3 folds to 3.4 folds and 11.1 folds to 3.2 folds, respectively (Figure 5A and 5B). In contrast, ZIKV infection in MDMs did not counteract the phosphorylation of STAT1 or STAT2 stimulated by IFN-α treatment (Figure 5A and 5B). Previous studies reported that ZIKV NS5 played critical roles in viral antagonism of type I IFN signaling through degradation of STAT2. Thus, we examined the interaction of endogenous STAT2 with ZIKV NS5 in the infected MDMs and moDCs. As illustrated in Figure S3, we observed a much higher level of NS5 protein expression in the infected moDCs comparing to the infected MDMs. However, we did not detect any appreciable level of interaction between endogenous STAT2 and NS5 in both the infected MDMs and moDCs with the co-IP assays (Figure S3). Collectively, these results demonstrate that ZIKV does not efficiently antagonize the phosphorylation of STAT1 or STAT2 in MDMs, which may contribute to the attenuated type I IFN-mediated antiviral responses in these cells.
Figure 5.
ZIKV infection fails to inhibit the phosphorylation of STAT1 and STAT2 in MDMs. (A) MDMs and moDCs were uninfected or infected with ZIKVPR at an MOI of 1.0. At 48hpi, the cells were untreated or treated with 1000U/mL of recombinant human IFN-α for 40 min. The cell lysates were harvested for detecting expressions of pSTAT1, pSTAT2, STAT1, STAT2, and β-actin by Western blots. Representative blots represented data from three independent experiments. (B) Quantitation was calculated as the ratio of pSTAT/STAT protein from three independent experiments. Statistical analyses in all panels were performed with one way-ANOVA and the differences were considered significant when p < 0.05. **p < 0.01, and ***p < 0.001. ns, not significant.
Depletion of STAT2 but not STAT1 rescues ZIKV and YFV replication in MDMs
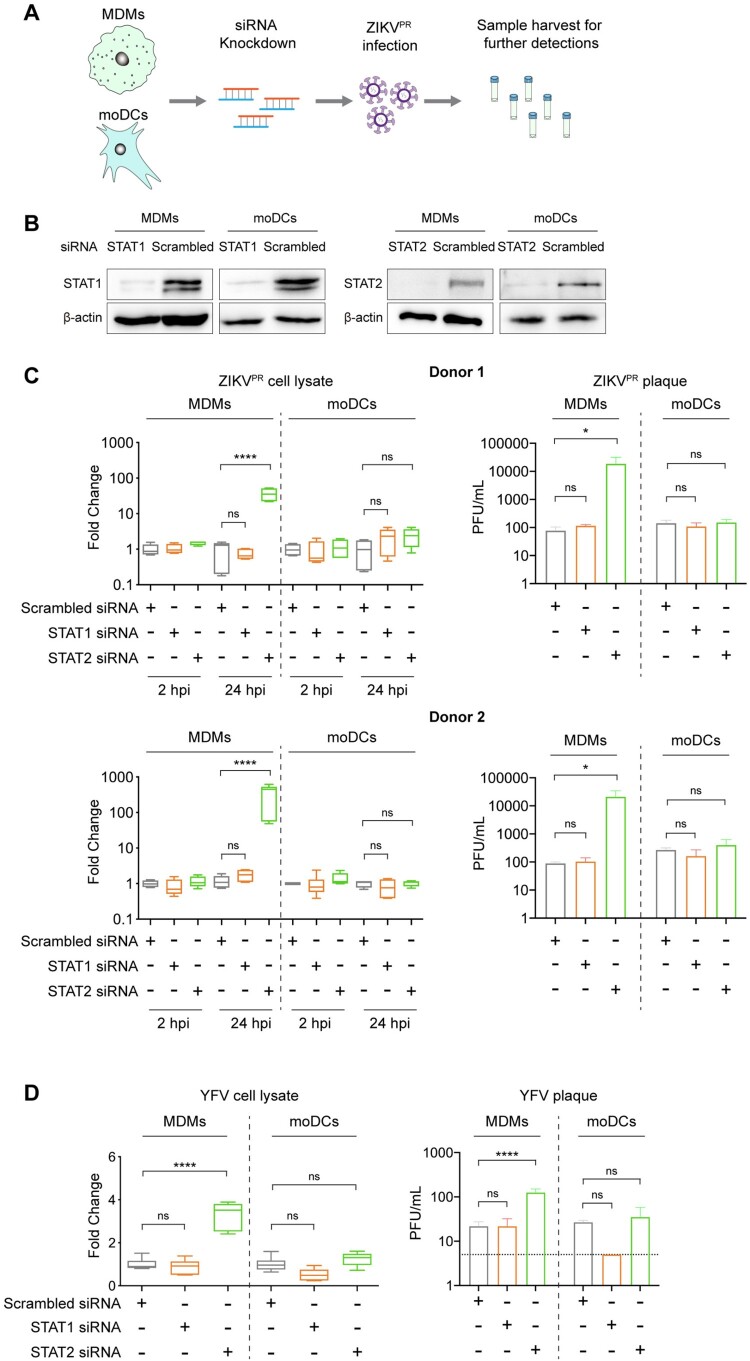
Next, we performed loss-of-function studies to further investigate the role of STAT1 and STAT2 on ZIKV replication in MDMs. We transfected MDMs and moDCs with scrambled siRNA or siRNA targeting STAT1 or STAT2 (Figure 6A). At 24 h post-transfection, the siRNA-treated cells were infected with ZIKVPR. The cell lysates and supernatants were harvested for determining virus replication at 2hpi and 24hpi. Western blots showed that the knockdown efficiencies of STAT1 and STAT2 in MDMs were comparable to those in moDCs (Figure 6B). We further showed that the siRNA knockdown of STAT1 and STAT2 was specific, and was able to interfere with IFN signaling in MDMs and moDCs (Figure S4 and S5). Interestingly, the depletion of STAT1 did not modulate ZIKV replication in either MDMs or moDCs from both donors, indicating a dispensable role of STAT1 on regulating ZIKV replication in these two cell types (Figure 6C). Importantly, depletion of STAT2 in MDMs resulted in a 37-fold to 354-fold increase in viral genome copy and 2-log increase in infectious virus titer compared to scrambled siRNA-treated MDMs. In contrast, STAT2 knockdown in moDCs showed an insignificant impact on ZIKV replication when compared with the scrambled siRNA-treated moDCs (Figure 6C).
Figure 6.
Depletion of STAT2 but not STAT1 rescues ZIKV and YFV replication in MDMs. (A) Schematic illustration of siRNA transfection and ZIKV infection. (B) MDMs and moDCs were transfected with scrambled siRNA or siRNA targeting STAT1 or STAT2. The knockdown efficiency of siRNA transfection was determined by Western blots. (C) MDMs and moDCs were generated from 2 donors. The cells were transfected with scrambled siRNA or siRNA targeting STAT1 or STAT2, followed by infection with ZIKVPR at an MOI of 1.0. At 2hpi and 24hpi, the cell lysates and supernatants were harvested for detection of viral genome copy and live infectious virus titer by qRT-PCR and plaque assays, respectively. (D) MDMs and moDCs were transfected with the same procedure as mentioned above and then infected with YFV at an MOI of 1.0. The cell lysates and supernatants were collected for further detection at 24hpi. Statistical analyses in all panels were performed with one way-ANOVA and the differences were considered significant when p < 0.05. *p < 0.05, and ****p < 0.0001. ns, not significant.
We further examined whether this observation could be applicable to other flaviviruses. In line with our results obtained using ZIKV, qRT-PCR and plaque assays demonstrated that the replication of YFV significantly increased in MDMs upon genetic depletion of STAT2 but not STAT1, whereas viral replication in moDCs was not significantly rescued after either STAT1 or STAT2 knockdown (Figure 6D). Overall, these results suggest that MDMs but not moDCs restrict ZIKV and YFV in a STAT2-dependent manner, implicating potential discrepancies of regulating viral replication in MDMs and moDCs.
Discussion
Macrophages and dendritic cells are key sentinel cell types, playing critical roles in the host immune response against virus infection. However, the detailed interplay between human primary macrophages and ZIKV remains elusive. In this study, we demonstrated that ZIKV replication was largely restricted in MDMs but not in moDCs. The underlying mechanism of the restricted ZIKV replication in MDMs was not due to inadequate virus entry. Our data further illustrated that ZIKV infection in MDMs did not efficiently suppress type I IFN-mediated antiviral response and failed to antagonize STAT1 and STAT2 phosphorylation induced by IFN-α treatment. Moreover, depletion of STAT2 but not STAT1 substantially rescued ZIKV and YFV replication in MDMs. These findings suggest that MDMs restrict flavivirus replication in a STAT2-dependent manner that is not antagonized during virus infection. Overall, our study provided novel insights into the role of human macrophages in the restriction of flavivirus infection.
Macrophages and dendritic cells are reported to be susceptible to various flaviviruses. However, although mouse bone marrow-derived dendritic cells (bmDCs) and macrophages (bmMs) are permissive to JEV infection, the susceptibility of bmMs is much higher than that of bmDCs [12]. Interestingly, in comparison with human moDCs, human MDMs and mature dendritic cells (mDCs) are less susceptible to DENV infection [50]. Notably, the mechanism of the discrepant JEV and DENV replication between macrophages and dendritic cells was not explored in those studies. Here, we demonstrated that ZIKV replication was largely restricted in human primary MDMs but not in moDCs. In addition, we illustrated that the restricted ZIKV replication in MDMs occurred at post-entry events of the viral replication cycle. Since Ly6C+ blood monocytes enter the dermis and become MDMs or monocyte-derived tissue-resident macrophages, which are targeted by flaviviruses when the infected mosquitos bite [18,21], our findings suggested that MDMs may play indispensable roles in restricting the initial wave of ZIKV infection to limit viral dissemination in the infected host.
Flaviviruses have evolved diverse IFN antagonism mechanisms targeting multiple stages of the IFN pathway to facilitate virus replication and spread. The NS5 of many flaviviruses, such as DENV and YFV, interacts and promotes STAT2 degradation through UBR4 or binding to other E3 ligase TRIM23, thereby suppressing the antiviral responses [51,52]. Recent studies reported that ZIKV infection antagonizes type I IFN signaling by targeting STAT2 for proteasomal degradation [22,23]. At the same time, viral antagonism of type I IFN has been demonstrated to be cell type-dependent, which could determine the susceptibility of viral infection and in turn affect tissue tropism [53,54]. In the present study, ZIKV infection was shown to suppress type I IFN-mediated response and inhibited phosphorylation of STAT1 and STAT2 in moDCs, which is in line with previous findings [26]. However, our data illustrated that the ZIKV-mediated inhibitory effect on IFN response was surprisingly not observed in MDMs, which contributed to inefficient virus replication in these cells. Previous studies reported that ZIKV NS5 played critical roles in antagonizing type I IFN signaling through degradation of STAT2. However, NS5 expression in the infected MDMs was inefficient and we did not detect any interaction between endogenous STAT2 and NS5 in the infected MDMs. The low level of NS5 in the infected MDMs may be due to the lack of virus replication in these cells, which should be recognized as a potential problem in interpreting the MDM results. Alternatively, the restricted viral replication may have resulted in the low expression level of NS5 in ZIKV-infected MDMs. Further studies should be conducted to investigate the detailed mechanisms of the cell type-dependent IFN-antagonism against ZIKV infection.
STAT1 and STAT2 are critical transcription factors required for the transcriptional activation of IFN-stimulated antiviral genes in type I, II, and III IFN signaling [55]. However, STAT2 is relatively diverse across different species [56,57]. In addition, although STAT1 and STAT2 interact with each other and lead to the activation of downstream antiviral response, STAT2 is likely a key mediator in the host defense mechanism. STAT2-mediated antiviral responses attenuate DENV infection in mice even in the absence of STAT1 [58]. Further, STAT2 can overcome mouse cytomegalovirus (MCMV) pM27-mediated degradation to limit the virus spread in mice with STAT2-dependent immune responses [59]. Here, we provided the first evidence that demonstrated the essential role of STAT2 against ZIKV and YFV infection in MDMs. The exact mechanism of how STAT2 overcomes ZIKV antagonism in MDMs should be further explored.
Overall, our findings provided novel insights into how ZIKV differentially interacts with the two sentinel cell types in the host immune system and underscoring the restriction role of MDMs against ZIKV infection.
Supplementary Material
Acknowledgments
We thank the staff at the Core Facility of The University of Hong Kong for facilitating this study.
Funding Statement
This study was partly supported by funding from the Research Grants Council General Research Fund (17123920 and 17123319), the University Grants Committee, Hong Kong Special Administrative Region, China; the Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Diseases and Research Capability on Antimicrobial Resistance for Department of Health of the Hong Kong Special Administrative Region Government; Sanming Project of Medicine in Shenzhen, China (SZSM201911014); the High Level-Hospital Program, Health Commission of Guangdong Province, China; and the Major Science and Technology Program of Hainan Province (ZDKJ202003); and the donations of Lee Wan Keung Charity Foundation Ltd, the Shaw Foundation Hong Kong, Michael Seak-Kan Tong, Marina Man-Wai Lee, the Hong Kong Hainan Commercial Association South China Microbiology Research Fund, and Lo Ying Shek Chi Wai Foundation. The funding sources had no role in the study design, data collection, analysis, interpretation, or writing of the report.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Chan JF, Choi GK, Yip CC, et al. Zika fever and congenital Zika syndrome: An unexpected emerging arboviral disease. J Infect. 2016 May;72(5):507–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duffy MR, Chen TH, Hancock WT, et al. Zika virus outbreak on Yap Island, federated states of Micronesia. N Engl J Med. 2009 Jun 11;360(24):2536–2543. [DOI] [PubMed] [Google Scholar]
- 3.Kasprzykowski JI, Fukutani KF, Fabio H, et al. A recursive sub-typing screening surveillance system detects the appearance of the ZIKV African lineage in Brazil: Is there a risk of a new epidemic? Int J Infect Dis. 2020 Jul;96:579–581. [DOI] [PubMed] [Google Scholar]
- 4.WHO . ZIKA epidemiology update July 2019. https://wwwwhoint/emergencies/diseases/zika/zika-epidemiology-update-july-2019pdf?ua=1. 2019.
- 5.Fink K, Ng C, Nkenfou C, et al. Depletion of macrophages in mice results in higher dengue virus titers and highlights the role of macrophages for virus control. Eur J Immunol. 2009 Oct;39(10):2809–2821. [DOI] [PubMed] [Google Scholar]
- 6.Zust R, Toh YX, Valdes I, et al. Type I interferon signals in macrophages and dendritic cells control dengue virus infection: implications for a new mouse model to test dengue vaccines. J Virol. 2014 Jul;88(13):7276–7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Libraty DH, Pichyangkul S, Ajariyakhajorn C, et al. Human dendritic cells are activated by dengue virus infection: enhancement by gamma interferon and implications for disease pathogenesis. J Virol. 2001 Apr;75(8):3501–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dejnirattisai W, Duangchinda T, Lin CL, et al. A complex interplay among virus, dendritic cells, T cells, and cytokines in dengue virus infections. J Immunol. 2008 Nov 1;181(9):5865–5874. [DOI] [PubMed] [Google Scholar]
- 9.Querec T, Bennouna S, Alkan S, et al. Yellow fever vaccine YF-17D activates multiple dendritic cell subsets via TLR2, 7, 8, and 9 to stimulate polyvalent immunity. J Exp Med. 2006 Feb 20;203(2):413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samuel MA, Diamond MS.. Pathogenesis of West Nile Virus infection: a balance between virulence, innate and adaptive immunity, and viral evasion. J Virol. 2006 Oct;80(19):9349–9360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dutta K, Mishra MK, Nazmi A, et al. Minocycline differentially modulates macrophage mediated peripheral immune response following Japanese encephalitis virus infection. Immunobiology. 2010 Nov;215(11):884–893. [DOI] [PubMed] [Google Scholar]
- 12.Aleyas AG, George JA, Han YW, et al. Functional modulation of dendritic cells and macrophages by Japanese encephalitis virus through MyD88 adaptor molecule-dependent and -independent pathways. J Immunol. 2009 Aug 15;183(4):2462–2474. [DOI] [PubMed] [Google Scholar]
- 13.Colpitts TM, Conway MJ, Montgomery RR, et al. West Nile Virus: biology, transmission, and human infection. Clin Microbiol Rev. 2012 Oct;25(4):635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kyle JL, Beatty PR, Harris E.. Dengue virus infects macrophages and dendritic cells in a mouse model of infection. J Infect Dis. 2007 Jun 15;195(12):1808–1817. [DOI] [PubMed] [Google Scholar]
- 15.Pham AM, Langlois RA, TenOever BR.. Replication in cells of hematopoietic origin is necessary for dengue virus dissemination. PLoS Pathog. 2012 Jan;8(1):e1002465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmid MA, Harris E.. Monocyte recruitment to the dermis and differentiation to dendritic cells increases the targets for dengue virus replication. PLoS Pathog. 2014 Dec;10(12):e1004541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murray PJ, Wynn TA.. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011 Oct 14;11(11):723–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Italiani P, Boraschi D.. From monocytes to M1/M2 macrophages: phenotypical vs. functional differentiation. Front Immunol. 2014;5(514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jurado KA, Simoni MK, Tang Z, et al. Zika virus productively infects primary human placenta-specific macrophages. JCI Insight. 2016 Aug 18;1(13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quicke KM, Bowen JR, Johnson EL, et al. Zika virus infects human placental macrophages. Cell Host Microbe. 2016 Jul 13;20(1):83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pingen M, Schmid MA, Harris E, et al. Mosquito biting modulates skin response to virus infection. Trends Parasitol. 2017 Aug;33(8):645–657. [DOI] [PubMed] [Google Scholar]
- 22.Grant A, Ponia SS, Tripathi S, et al. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe. 2016 Jun 8;19(6):882–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar A, Hou S, Airo AM, et al. Zika virus inhibits type-I interferon production and downstream signaling. EMBO Rep. 2016 Dec;17(12):1766–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hertzog J, Dias Junior AG, Rigby RE, et al. Infection with a Brazilian isolate of Zika virus generates RIG-I stimulatory RNA and the viral NS5 protein blocks type I IFN induction and signaling. Eur J Immunol. 2018 Jul;48(7):1120–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vielle NJ, Zumkehr B, Garcia-Nicolas O, et al. Silent infection of human dendritic cells by African and asian strains of Zika virus. Sci Rep. 2018 Apr 3;8(1):5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowen JR, Quicke KM, Maddur MS, et al. Zika virus antagonizes type I interferon responses during infection of human dendritic cells. PLoS Pathog. 2017 Feb;13(2):e1006164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu H, Yuen TTT, Chik KKH, et al. Targeting the inositol-requiring enzyme-1 pathway efficiently reverts Zika virus-induced neurogenesis and spermatogenesis marker perturbations. ACS Infect Dis. 2020 Jul 10;6(7):1745–1758. [DOI] [PubMed] [Google Scholar]
- 28.Zhu Z, Chu H, Wen L, et al. Targeting SUMO modification of the Non-structural protein 5 of Zika virus as a host-targeting antiviral strategy. Int J Mol Sci. 2019 Jan 17;20(2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu H, Wang JJ, Qi M, et al. Tetherin/BST-2 is essential for the formation of the intracellular virus-containing compartment in HIV-infected macrophages. Cell Host Microbe. 2012 Sep 13;12(3):360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou J, Chu H, Li C, et al. Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J Infect Dis. 2014 May 1;209(9):1331–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chu H, Zhou J, Wong BH, et al. Productive replication of Middle East respiratory syndrome coronavirus in monocyte-derived dendritic cells modulates innate immune response. Virology. 2014 Apr;454-455:197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu H, Chan JF, Yuen TT, et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020 May;1(1):e14–e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chu H, Chan JF, Wang Y, et al. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: An Ex vivo study With implications for the pathogenesis of COVID-19. Clin Infect Dis. 2020 Sep 12;71(6):1400–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chu H, Zhou J, Wong BH, et al. Middle east respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J Infect Dis. 2016 Mar 15;213(6):904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu H, Hu B, Huang X, et al. Host and viral determinants for efficient SARS-CoV-2 infection of the human lung. Nat Commun. 2021 Jan 8;12(1):134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan JF, Zhu Z, Chu H, et al. The celecoxib derivative kinase inhibitor AR-12 (OSU-03012) inhibits Zika virus via down-regulation of the PI3K/Akt pathway and protects Zika virus-infected A129 mice: A host-targeting treatment strategy. Antiviral Res. 2018 Dec;160:38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan JF, Yip CC, Tee KM, et al. Improved detection of Zika virus RNA in human and animal specimens by a novel, highly sensitive and specific real-time RT-PCR assay targeting the 5'-untranslated region of Zika virus. Trop Med Int Health. 2017 May;22(5):594–603. [DOI] [PubMed] [Google Scholar]
- 38.Yuan S, Chan JF, den-Haan H, et al. Structure-based discovery of clinically approved drugs as Zika virus NS2B-NS3 protease inhibitors that potently inhibit Zika virus infection in vitro and in vivo. Antiviral Res. 2017 Sep;145:33–43. [DOI] [PubMed] [Google Scholar]
- 39.Chan JF, Chik KK, Yuan S, et al. Novel antiviral activity and mechanism of bromocriptine as a Zika virus NS2B-NS3 protease inhibitor. Antiviral Res. 2017 May;141:29–37. [DOI] [PubMed] [Google Scholar]
- 40.Yang D, Chu H, Hou Y, et al. Attenuated interferon and pro-inflammatory response in SARS-CoV-2-infected human dendritic cells is associated with viral antagonism of STAT1 phosphorylation. J Infect Dis. 2020 Jun 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chu H, Chan CM, Zhang X, et al. Middle East respiratory syndrome coronavirus and bat coronavirus HKU9 both can utilize GRP78 for attachment onto host cells. J Biol Chem. 2018 Jul 27;293(30):11709–11726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamel R, Dejarnac O, Wichit S, et al. Biology of Zika virus infection in human skin cells. J Virol. 2015 Sep;89(17):8880–8896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meertens L, Labeau A, Dejarnac O, et al. Axl mediates ZIKA virus entry in human glial cells and modulates innate immune responses. Cell Rep. 2017 Jan 10;18(2):324–333. [DOI] [PubMed] [Google Scholar]
- 44.Richard AS, Shim BS, Kwon YC, et al. AXL-dependent infection of human fetal endothelial cells distinguishes Zika virus from other pathogenic flaviviruses. Proc Natl Acad Sci U S A. 2017 Feb 21;114(8):2024–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strange DP, Jiyarom B, Pourhabibi Zarandi N, et al. Axl promotes Zika virus entry and Modulates the antiviral State of human sertoli cells. mBio. 2019 Jul 16;10(4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watanabe M, Buth JE, Vishlaghi N, et al. Self-organized cerebral organoids with human-specific features predict effective drugs to combat Zika virus infection. Cell Rep. 2017 Oct 10;21(2):517–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rappocciolo G, Jenkins FJ, Hensler HR, et al. DC-SIGN is a receptor for human herpesvirus 8 on dendritic cells and macrophages. J Immunol. 2006 Feb 1;176(3):1741–1749. [DOI] [PubMed] [Google Scholar]
- 48.Soilleux EJ, Morris LS, Leslie G, et al. Constitutive and induced expression of DC-SIGN on dendritic cell and macrophage subpopulations in situ and in vitro. J Leukoc Biol. 2002 Mar;71(3):445–457. [PubMed] [Google Scholar]
- 49.Xia H, Luo H, Shan C, et al. An evolutionary NS1 mutation enhances Zika virus evasion of host interferon induction. Nat Commun. 2018 Jan 29;9(1):414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flipse J, Torres S, Diosa-Toro M, et al. Dengue tropism for macrophages and dendritic cells: the host cell effect. J Gen Virol. 2016 Jul;97(7):1531–1536. [DOI] [PubMed] [Google Scholar]
- 51.Morrison J, Laurent-Rolle M, Maestre AM, et al. Dengue virus co-opts UBR4 to degrade STAT2 and antagonize type I interferon signaling. PLoS Pathog. 2013 Mar;9(3):e1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laurent-Rolle M, Morrison J, Rajsbaum R, et al. The interferon signaling antagonist function of yellow fever virus NS5 protein is activated by type I interferon. Cell Host Microbe. 2014 Sep 10;16(3):314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Boxel-Dezaire AH, Rani MR, Stark GR.. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006 Sep;25(3):361–372. [DOI] [PubMed] [Google Scholar]
- 54.Zhao L, Rose KM, Elliott R, et al. Cell-type-specific type I interferon antagonism influences organ tropism of murine coronavirus. J Virol. 2011 Oct;85(19):10058–10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoffmann HH, Schneider WM, Rice CM.. Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol. 2015 Mar;36(3):124–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paulson M, Pisharody S, Pan L, et al. Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J Biol Chem. 1999 Sep 3;274(36):25343–9. [DOI] [PubMed] [Google Scholar]
- 57.Parisien JP, Lau JF, Horvath CM.. STAT2 acts as a host range determinant for species-specific paramyxovirus interferon antagonism and simian virus 5 replication. J Virol. 2002 Jul;76(13):6435–6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perry ST, Buck MD, Lada SM, et al. STAT2 mediates innate immunity to dengue virus in the absence of STAT1 via the type I interferon receptor. PLoS Pathog. 2011 Feb;7(2):e1001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Le-Trilling VTK, Wohlgemuth K, Ruckborn MU, et al. STAT2-Dependent immune responses ensure host survival despite the presence of a potent viral antagonist. J Virol. 2018 Jul 15;92(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.