ABSTRACT
Long non-coding RNAs (lncRNAs) in plants are emerging as new players in biotic stress responses. Pathogen-associated lncRNAs have been broadly identified and functionally characterized in multiple species. However, herbivore-responsive lncRNAs in plants are poorly investigated. Our recent study revealed that lncRNAs also play roles in plant defense against herbivores in wild tobacco. Here, we identified armyworm (AW)-elicited lncRNAs in monocot rice by employing a similar approach. A total of 238 lncRNAs were found to be differentially expressed (DE) in AW-treated plants relative to control plants. The cis effect of these DE lncRNAs was predicted. Interestingly, one DE lncRNA was identified from the antisense transcripts of the jasmonate ZIM-domain gene JAZ10.
KEYWORDS: Rice, herbivore, long non-coding RNA
Long non-coding RNAs (lncRNAs) are only beginning to be recognized in plants because of their low transcriptional abundance and previously considered “junk transcripts.” The rapid development of sequencing technology and functional analysis has uncovered the mystery of lncRNAs. LncRNAs have been documented to be involved in plant defense against pathogens. A large number of lncRNAs were identified in multiple plant species that are responsive to pathogen infections. Examples include the fungal pathogen-elicited lncRNAs in Arabidopsis, cotton, and rice;1–3 the bacterial pathogen-elicited lncRNAs in Arabidopsis and rice;4,5 and the virus-elicited lncRNAs in rice and tomato.6,7 Silencing or the overexpression of some pathogen-elicited lncRNAs alters plant resistance to these pathogens.2,4–6 Mechanism study revealed that these lncRNAs could regulate transcription machineries or hormone-mediated defense pathways. The long intergenic noncoding RNA (lincRNA) ELENA1 could directly interact with Mediator subunit 19a and promote the expression of pathogenesis-related genes.4 In rice, the lincRNA ALEX1 regulates Xanthomonas oryzae pv. Oryzae resistance by activating jasmonate (JA) signaling.5 In addition to pathogens, plants are also frequently suffered by herbivores in nature. However, herbivore-induced lncRNAs have yet to be explored. In cotton, 1,331 lncRNAs were found to be differentially expressed (DE) in aphid-infested leaves relative to control leaves.8 Our recent work showed that lncRNAs in wild tobacco (Nicotiana attenuata) are also responsive to herbivore attacks.9 JA signaling plays a central role in plant defense against herbivores.10 Silencing of two early-responsive lincRNAs, namely, JAL1 and JAL3, attenuates JA-mediated plant resistance to Manduca sexta larvae.
In the current work, we explored herbivore-elicited lncRNAs in monocot rice. A generalist herbivore Mythimna separata (armyworm, AW) was used in this study. The feeding of AW was mimicked by mechanical wounding following AW larval oral secretion (OS) treatment.11 The first fully expanded leaf was treated, and samples were collected 0.25, 0.5, 1, 3, 8, and 24 h after treatment. Leaves from nontreated plants at each timepoint were used as controls. Five replicates were performed for each timepoint, and combined as one sample for strand-specific RNA library construction. After RNA sequencing, the bioinformatics pipeline described previously was used to identify the transcription levels of genes and lncRNAs (Supplemental Material; raw data have been deposited in BIG Data Center under accession number CRA004044).9 All sequenced reads of each sample were aligned to the rice reference genome. The assembled transcript isoforms were compared with all protein coding gene models. Transcripts shorter than 200 bp or those with an open reading frame longer than 67 aa were discarded. The remaining transcripts were defined as lncRNAs. The fold change (FC) of genes and lncRNAs in the AW-treated leaves relative to the control leaves was calculated. If FC > 2 in at least two timepoints and FC > 3 in at least one timepoint, then the gene or lncRNA was defined as DE. A total of 559 DE genes were down-regulated by AW attack while 6,261 DE genes were up-regulated (Figure 1a). Gene ontology (GO) enrichment analysis showed that down-regulated genes were enriched in growth-related terms and that up-regulated genes were enriched in JA signaling pathway and defense response terms (Figure 1b and c).12 These results suggested that herbivore elicitation may activate JA-mediated plant defenses but suppress plant growth. A total of 372 DE lncRNAs were identified, and 78% of them were up-regulated. The natural antisense transcripts (NATs) are the most abundant (Figure 1d). According to the time course expression profile, some of the DE lncRNAs are early responders during herbivory while some of them respond to herbivore attacks late (Figure 1e). Hence, DE lncRNAs may play a different role in plant–herbivore interactions.
Figure 1.
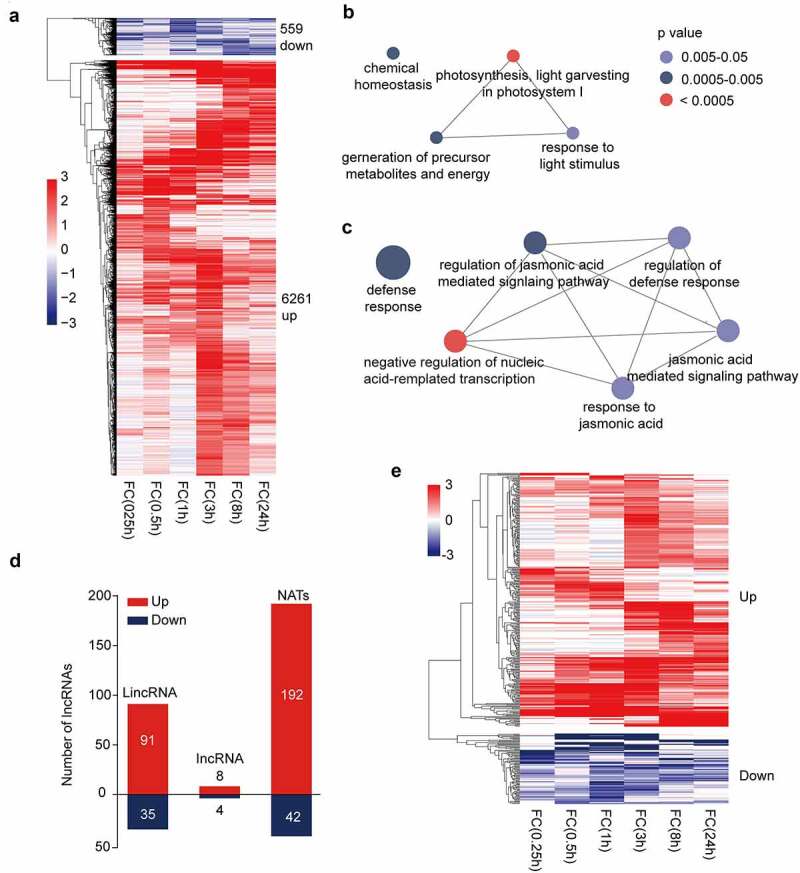
Differentially expressed (DE) genes and lncRNAs in armyworm (AW)-treated plants relative to control plants and gene ontology (GO) analysis of DE genes. (a) Heatmap representing the transcript abundance fold change (FC) of DE genes in AW-treated plants relative to control plants. Leaves were harvested 0.25, 0.5, 1, 3, 8, and 24 h after wounding and then treated with AW oral secretion. Leaves from untreated plants were used as control. Up or down, treated leaves relative to control leaves. (b) GO analysis of down-regulated genes in A. GO analysis was performed by ClueGO. The size of the bubble corresponds to the number of mapped genes. (c) GO analysis of up-regulated genes in A. (d) Numbers of up- and down-regulated lncRNAs in AW-treated plants and control plants. (e) Heatmap representing the transcript abundance FC of DE lncRNAs in AW-treated plants relative to control plants
LncRNAs are known to act in cis to regulate the expression of nearby genes.13 To predict the potential function of herbivore-elicited lncRNAs, this study investigated the neighboring genes of DE lncRNAs. The host genes of DE NATs and DE intronic RNAs (incRNAs) were first identified. Interestingly, the expression pattern of these host genes is similar to their associated lncRNAs (Figure 2a and b). For instance, the jasmonate ZIM-domain protein (JAZ) is one of the JA co-receptors that regulates JA signaling by directly interacting with JA-responsive transcription factors.14 The antisense transcript of JAZ10 gene (NAT-JAZ10) was identified as a DE lncRNA in the AW-treated plants (Figure 2c). The expression of JAZ10 gene was significantly increased after the AW treatment (Figure 2d). Consistently, the transcription levels of NAT-JAZ10 were extremely low in normal conditions, but they dramatically increased after the AW treatment (Figure 2e). Given the key role of JA in herbivore resistance, JAZ10 and its NAT pair may be involved in rice and AW interactions. The genes located 100 kb upstream and downstream of DE lincRNAs were subsequently identified, and the DE genes from them were used for the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. The results revealed that these DE genes neighboring DE lincRNAs were enriched in many primary and secondary metabolisms that are likely to be involved in rice–herbivore interactions, such as the biosynthesis of amino acids, carbon metabolism, and alpha-linolenic acid metabolism (Figure 2f).
Figure 2.
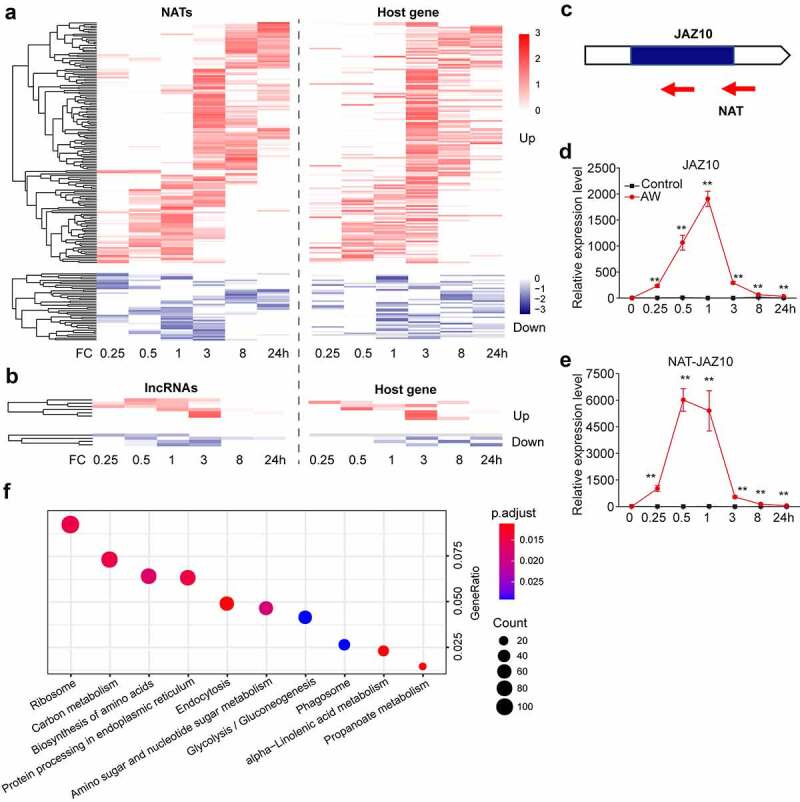
Prediction of cis effect of DE lncRNAs and the expression of JAZ10 and its associated lncRNA after AW treatment (a) Expression pattern of DE-NATs and their host genes after AW treatment. FC, fold change. (b) Expression pattern of DE incRNAs and their host genes after AW treatment. (c) Schematic diagram of JAZ10 and its natural antisense transcripts. Mean transcript abundance (± SE, n = 5) of JAZ10 (d) and NAT-JAZ10 (e) in AW-treated and control leaves. Transcript levels were analyzed by reverse transcription-quantitative PCR (RT-qPCR). The primers used for RT-qPCR are listed in the Supplemental Material. Asterisks indicate significant differences in the treated plants relative to the control plants (**, p < .01; Student’s t-test). (f) KEGG enrichment analysis of DE genes neighboring DE lincRNAs. GeneRatio, ratio of gene numbers in a particular term to all gene numbers. The size of each circle represents the protein numbers of the pathways. The color density indicates the significance of the pathway enrichment (p < .05)
In summary, we identified lncRNAs that are responsive to herbivores in monocot rice by RNA sequencing. The potential function of these lncRNAs was predicted. One of the lncRNAs, NAT-JAZ10, was highlighted and found to be potentially interesting to be studied further. This identification will enrich our understanding of plant–herbivore interactions and provide new candidates for dissecting herbivore-induced defense networks.
Supplementary Material
Funding Statement
This work was supported by the National Natural Science Foundation of China [31901955]; Max Planck Partner Group Program (to R.L.); the Hundred-Talent Program of Zhejiang University (to R.L.).
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
References
- 1.Zhu QH, Stephen S, Taylor J, Helliwell CA, Wang MB.. Long noncoding RNAs responsive to Fusarium oxysporum infection in Arabidopsis thaliana. New Phytol. 2014;201:1–4. doi: 10.1111/nph.12537. [DOI] [PubMed] [Google Scholar]
- 2.Zhang L, Wang M, Li N, Wang H, Qiu P, Pei L, Xu Z, Wang T, Gao E, Liu J, et al. Long noncoding RNAs involve in resistance to Verticillium dahliae, a fungal disease in cotton. Plant Biotechnol J. 2018;16(6):1172–1185. doi: 10.1111/pbi.12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang LL, Jin JJ, Li LH, Qu SH.. Long non-coding RNAs responsive to blast fungus infection in rice. Rice. 2020;13:77. doi: 10.1186/s12284-020-00437-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seo JS, Sun HX, Park BS, Huang CH, Yeh SD, Jung C, Chua N-H. ELF18-INDUCED LONG-NONCODING RNA associates with mediator to enhance expression of innate immune response genes in Arabidopsis. Plant Cell. 2017;29:1024–1038. doi: 10.1105/tpc.16.00886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu Y, Zhou YF, Feng YZ, He H, Lian JP, Yang YW, Lei M-Q, Zhang Y-C, Chen Y-Q. Transcriptional landscape of pathogen-responsive lncRNAs in rice unveils the role of ALEX1 in jasmonate pathway and disease resistance. Plant Biotechnol J. 2020;18:679–690. doi: 10.1111/pbi.13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J, Yu W, Yang Y, Li X, Chen T, Liu T, Ma N, Yang X, Liu R, Zhang B, et al. Genome-wide analysis of tomato long non-coding RNAs and identification as endogenous target mimic for microRNA in response to TYLCV infection. Sci Rep. 2015;5(1):16946. doi: 10.1038/srep16946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang T, Liang Q, Li C, Fu S, Kundu JK, Zhou X, Wu J. Transcriptome analysis of rice reveals the lncRNA-mRNA regulatory network in response to rice black-streaked dwarf virus infection. Viruses-Basel. 2020;12:951. doi: 10.3390/v12090951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, Yang Z, Feng P, Zhong X, Ma Q, Su Q, Wang X, Li C, Yang Y. Identification and the potential roles of long non-coding RNAs in cotton leaves damaged by Aphis gossypii. Plant Growth Regul. 2019;88(3):215–225. doi: 10.1007/s10725-019-00500-7. [DOI] [Google Scholar]
- 9.Li R, Jin J, Xu J, Wang L, Li J, Lou Y, Baldwin IT. Long non-coding RNAs associate with jasmonate-mediated plant defence against herbivores. Plant Cell Environ. 2021;44(3):982–994. doi: 10.1111/pce.13952. [DOI] [PubMed] [Google Scholar]
- 10.Erb M, Reymond P. Molecular interactions between plants and insect herbivores. Annu Rev Plant Biol. 2019;70:527–557. doi: 10.1146/annurev-arplant-050718-095910. [DOI] [PubMed] [Google Scholar]
- 11.Xu J, Wang X, Zu H, Zeng X, Baldwin IT, Lou Y, Li R. Molecular dissection of rice phytohormone signaling involved in resistance to a piercing-sucking herbivore. New Phytol. 2021;230:1639–1652. doi: 10.1111/nph.17251. [DOI] [PubMed] [Google Scholar]
- 12.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman W-H, Pagès F, Trajanoski Z, Galon J, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howe GA, Major IT, Koo AJ. Modularity in jasmonate signaling for multistress resilience. Annu Rev Plant Biol. 2018;69:387–415. doi: 10.1146/annurev-arplant-042817-040047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


