Abstract
The application of small amounts of natural plant growth hormones, such as gibberellins (GAs), can increase the productivity and quality of many vegetable and fruit crops. However, gibberellin growth hormones usage is limited by the high cost of their production, which is currently based on fermentation of a natural fungal producer Fusarium fujikuroi that produces a mix of several GAs. We explored the potential of the oleaginous yeast Yarrowia lipolytica to produce specific profiles of GAs. Firstly, the production of the GA-precursor ent-kaurenoic acid (KA) at 3.75 mg/L was achieved by expression of biosynthetic enzymes from the plant Arabidopsis thaliana and upregulation of the mevalonate (MVA) pathway.
We then built a GA4-producing strain by extending the GA-biosynthetic pathway and upregulating the MVA-pathway further, resulting in 17.29 mg/L GA4. Additional expression of the F. fujikoroi GA-biosynthetic enzymes resulted in the production of GA7 (trace amounts) and GA3 (2.93 mg/L). Lastly, through protein engineering and the expression of additional KA-biosynthetic genes, we increased the GA3-production 4.4-fold resulting in 12.81 mg/L. The developed system presents a promising resource for the recombinant production of specific gibberellins, identifying bottlenecks in GA biosynthesis, and discovering new GA biosynthetic genes.
Classification
Biological Sciences, Applied Biological Sciences.
Keywords: Gibberellins, Plant growth hormones, Isoprenoids, Oleaginous yeast, Yarrowia lipolytica
Highlights
-
•
A complete biosynthetic pathway towards gibberellins was reconstructed in a microbial host
-
•
The pathway towards ent-kaurenoic acid consisted of Arabidopsis thaliana enzymes
-
•
The pathway from ent-kaurenoic acid to gibberellins GA3, GA4 and GA7 consisted of Fusarium fujikuroi enzymes
-
•
Y. lipolytica expressed 14 heterologous genes for gibberellins biosynthesis and had 5 genome edits for improved mevalonate flux
-
•
The strains produced up to 12 mg/L of GA3 and up to 17 mg/L GA4 in small-scale cultivations
1. Introduction
Gibberellins (GAs) are diterpene phytohormones involved in plant developmental processes like seed germination, pollen maturation, stem elongation, flower formation, and fruit development (Kato, 1955; Lang, 1956; Harrington et al., 1957; Kahn et al., 1957; Wittwer et al., 1957). Of the at least 126 known gibberellins, some are bioactive while others are inert side-products or biosynthetic intermediates (MacMillan, 2001). Bioactive gibberellins include GA3 (gibberellic acid), GA4, and GA7; they are used in commercial agricultural products, either pure or as mixtures (Camara et al., 2018). Spraying with gibberellins increases the size of seedless grapes, blueberries, and pears (Casanova et al., 2009; Ito et al., 2016; Zang et al., 2016), as well as the salt stress tolerance of soybeans, maize, and sugarcane (Tuna et al., 2008; Hamayun et al., 2010; Shomeili et al., 2011). Valent Biosciences, a commercial provider of GA-products, lists more than 40 plant species that can be enhanced by gibberellins (Rademacher, 2015). Gibberellins are also applied in beer brewing to enhance the malting process (MacLeod and Millar, 1962). According to some sources, the estimated global market size for gibberellins is USD 548.9 million as of 2016 (Gibberellins Market | Industry Analysis Report, 2016).
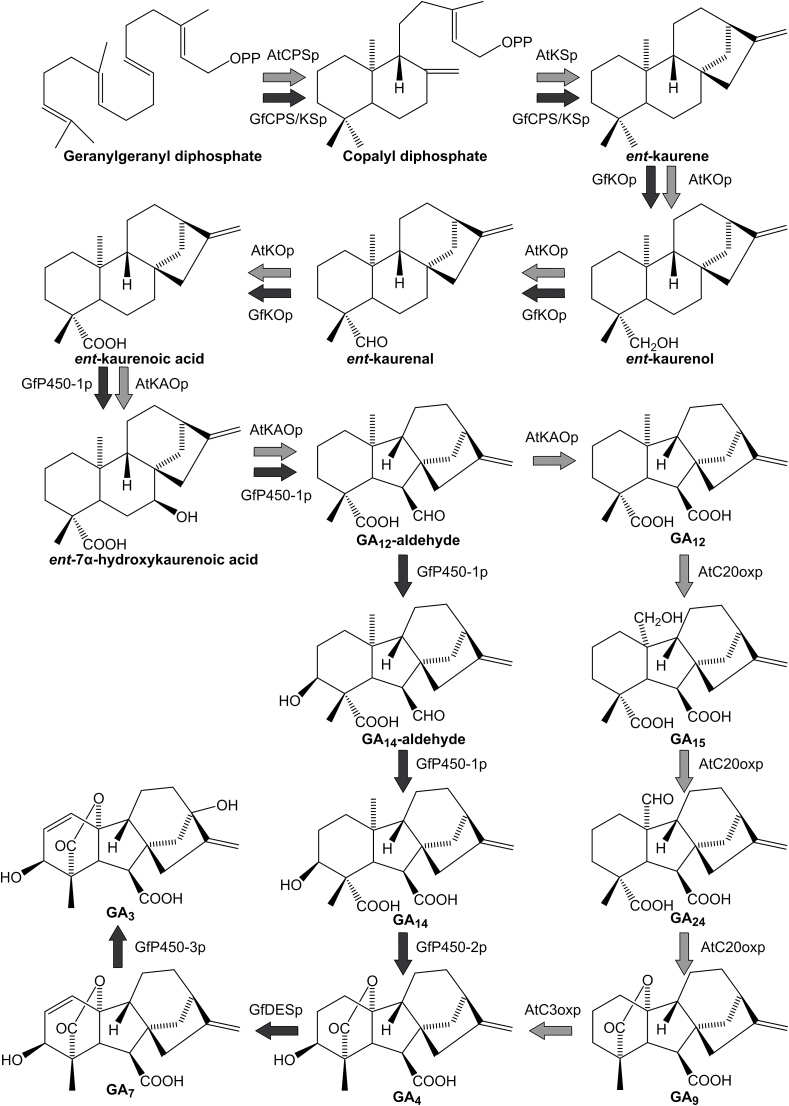
In plants, gibberellins are mainly produced through the plastidial methylerythritol pathway (MEP), while in fungi, they are produced through the mevalonate pathway (MVA) (Kasahara et al., 2002; Albermann et al., 2013b). The initial steps of gibberellin biosynthesis from geranylgeranyl diphosphate (GGPP) to GA12-aldehyde are similar in fungi and plants. Cyclization of GGPP into ent-copalyl diphosphate and then into ent-kaurene is catalyzed by the copalyl diphosphate synthase (CPSp) and the ent-kaurene synthase (KSp) in plants. In fungi, one bi-functional synthase (CPS/KSp) forms ent-kaurene (Fig. 1) (Sun and Kamiya, 1994; Tudzynski et al., 1998, 2001; Yamaguchi et al., 1998). Ent-kaurene is hydroxylated to ent-kaurenol, deprotonated to ent-kaurenal, and then modified to ent-kaurenoic acid (KA) by the ent-kaurene oxidase (KOp) (Helliwell et al., 1999; Tudzynski et al., 2001). Ent-kaurenoic acid oxidase (KAOp) then catalyzes the formation of GA12-aldehyde via the intermediate ent-7α-hydroxykarenoic acid (Helliwell et al., 2001a). In plants, KAOp also forms GA12 and is located in the ER-membrane, while CPSp, KSp, and KOp are located in plastids (Helliwell et al., 2001b). In fungi, KAOp (P450-1p) converts GA12-aldehyde into GA14-aldehyde and then into GA14 (Rojas et al., 2001). In plants, the GA 20-oxidase can convert GA12 into GA9 via GA15 and GA24, while GA 3-oxidase converts GA9 into the bioactive GA4 (Lange et al., 1994; Williams et al., 1998). In fungi, GA14 is converted to GA4 by a cytochrome P450 monooxygenase (P450-2p) (Tudzynski et al., 2002). Subsequently, GA4 is converted to GA7 by the GA desaturase (DESp) and then to GA3 by another monooxygenase (P450-3p) (Tudzynski et al., 2003).
Fig. 1.
The biosynthetic pathways for bioactive GA4, GA7, and GA3. Black arrows indicate GA-biosynthetic enzymes from F. fujikuroi (Gf), and grey arrows indicate GA-biosynthetic enzymes from A. thaliana (At). CPSp, copalyl disphosphate synthase. KSp, ent-kaurene synthase. CPS/KSp, bifunctional copalyl diphosphate synthase and ent-kaurene synthase. KOp, ent-kaurene oxidase. KAOp, ent-kaurenoic acid oxidase. C20oxp, GA carbon 20 oxidase. C3oxp, GA carbon 3 oxidase. P450-1p, P450-2p, P450-3p, cytochromes P450. DES, GA desaturase.
While several fungi species like Sphaceloma manihoticola or Phaeosphaeria sp. and bacteria species like Rhizobium phaseoli or Azospirillum lipoferum can produce gibberellins, the industrially used microbe for gibberellin production is the fungus Fusarium fujikuroi (formerly Gibberella fujikuroi) (Rademacher and Graebe, 1979; Atzorn et al., 1988; Bottini et al., 1989; Sassa et al., 1994; Shi et al., 2017). Submerged fermentation is most common as solid-state fermentation is complicated to scale-up (Camara et al., 2018). Titers of 3.9 g/L GA3 for an eight-day submerged fermentation with fungal mycelium immobilized on calcium-polygalacturonate beads have been reported in the literature (Escamilla S et al., 2000). While GA3 from submerged fermentation can be recovered with column-based adsorption or liquid-liquid extraction, it is more challenging to separate GA4 from GA7 due to their high structural similarity (Rademacher, 2015; Camara et al., 2018). Since GA4 degrades faster than GA3 and GA7, it can be more useful for some applications, yet most GA4 products contain at least some GA7 (Rademacher, 2015).
While literature describes mutated strains of F. fujikuroi that are able to produce predominately GA7 at 234 mg/L or solely GA4 at 420 mg/L during ten days of fermentation, it is unclear whether these strains can be used industrially (Lale and Gadre, 2010; Albermann et al., 2013a). Although genome data and transformation procedures are available for F. fujikuroi, the molecular toolkit for genome modification is still limited compared to more conventional host organisms (Wiemann et al., 2013; Hwang and Ahn, 2016; Bashyal et al., 2017; Shi et al., 2019). Furthermore, commercial efforts to produce pure GA4 with S. manihoticola have failed (Rademacher, 2015). Moving the production into a heterologous host with more molecular tools available and established procedures for industrial-scale production could enable improved control of relative levels of bioactive GAs, better titers, and reduced production costs.
The oleaginous yeast species Yarrowia lipolytica is a promising host for the production of lipids and other compounds derived from acetyl- or acyl-CoAs, due to their high endogenous levels (Darvishi et al., 2018; Marella et al., 2018). Furthermore, Y. lipolytica offers other benefits due to GRAS status being granted to several production strains, amenability to genetic engineering, sequenced genomes, ability to express functional P450 enzymes, and the presence of lipid bodies for terpene sequestration (Dujon et al., 2004; Christen and Sauer, 2011; Groenewald et al., 2014; Gao et al., 2017; Holkenbrink et al., 2018). Indeed, Y. lipolytica has already been engineered to produce a variety of terpenoids, such as d-limonene, α-farnesene, betulinic acid, lycopene, and astaxanthin (Matthäus et al., 2014; Cao et al., 2016; Yang et al., 2016; Gao et al., 2017; Kildegaard et al., 2017; Sun et al., 2019). Y. lipolytica was used industrially to produce omega 3-fatty acids by DuPont (Zhu and Jackson, 2015).
This study demonstrates the production of bioactive gibberellins in Y. lipolytica by optimizing ent-kaurenoic acid production and establishing heterologous pathways combining enzymes originating from both fungi and plants for the complete biosynthesis of bioactive gibberellins GA3, GA4, and GA7.
2. Materials and methods
2.1. Strains and media
Y. lipolytica strain GB20/ST3683 (mus51Δ (ku70Δ), nugm-Htg2, ndh2i, lys11−, leu2−, ura3−, MatB) was a gift from Volker Zickermann (Goethe University Medical School, Institute of Biochemistry II, Germany) (Angerer et al., 2014). Y. lipolytica strains were grown on Yeast extract Peptone Dextrose (YPD) or synthetic drop-out media (SC) medium at 30°C. YPD medium contained 20 g/L peptone, 10 g/L yeast extract, 20 g/L glucose. For growth on solid media, YPD and SC medium were supplemented with 20 g/L agar. When necessary, the YPD medium was supplemented with antibiotics hygromycin (50 mg/L) or nourseothricin (250 mg/L). Cultivation of the recombinant strains for isoprenoid production was performed in Yeast extract Peptone medium containing 80 g/L glucose instead of 20 g/L glucose (YP+8%D).
Escherichia coli strain DH5α was used for DNA manipulations. Cells were cultivated at 37°C on Lysogeny Broth (LB) medium supplemented with 100 mg/L ampicillin for plasmid selection.
The chemicals were obtained, if not indicated otherwise, from Sigma-Aldrich. Nourseothricin was purchased from Jena BioScience GmbH (Germany).
2.2. Plasmid construction
The primers, biobricks and plasmids used in this study are listed in Supplementary Tables S4, S5, and S6. The biobricks were amplified with PCR using Phusion U polymerase (Thermo Scientific) and assembled into the EasyClone vectors with USER cloning (Holkenbrink et al., 2018). The USER reactions were transformed into E. coli and correct assemblies were verified by sequencing. Genes encoding Arabidopsis thaliana AtCPS (GenBank: AAA53632.1), AtKS (GenBank: AAC39443.1), AtKO (GenBank: AAC39507.1), AtKAO2 (GenBank: ABJ17103.1), AtC3ox1 (GenBank: AAO64762.1), AtC20ox1 (GenBank: AEE36106.1), AtATR2 (GenBank: AEE85737.1), Fusarium (Gibberella) fujikuroi GfCPS/KS (GenBank: BAA84917.1), GfP450-1 (GenBank: CAA75565.1), GfP450-2 (GenBank: CAA75566.1), GfP450-3 (GenBank: CAA75567.1), GfP450-4 (GfKO; GenBank: CCT69175.1), GfDES (GenBank: CAD10289.1), GfCPR (GenBank: CAE09055.1), GfCyb5 (GenBank: KLO84088.1), GfCybRed (GenBank: KLO80170.1), and Synechococcus sp. SsGGPPs7 (NCBI Reference Sequence: WP_011429285.1) were codon-optimized for Y. lipolytica and synthesized as GeneArt String DNA fragments or Synthetic Genes by Thermo Fisher Scientific (see Supplementary sequences for full DNA sequences). The software ChloroP v1.1 was used to predict chloroplast targeting protein sequences (Emanuelsson et al., 1999). The software InterProScan was used to predict the presence of protein transmembrane regions (Jones et al., 2014; Blum et al., 2021).
A Hygromycin B resistance cassette included in BioBrick BB2080 was cloned into plasmids pCfB6409, pCfB6410, pCfB6411, pCfB6412, pCfB6413, pCfB6414, and pCfB6415 between SacI and HpaI restriction sites in order to build plasmids from pCfB6601 to pCfB6607 (Supplementary Table S6).
Vectors pCfB6607 and pCfB6408 were constructed via point mutation of parental plasmids using primers indicated in Supplementary Table S4 and following indications of QuickChange II Site-Directed Mutagenesis Kit (Agilent Technologies).
2.3. Yeast strain construction
All strains used in this study are listed in Supplementary Table S7. The plasmids for targeted integration were NotI-linearized and transformed into Y. lipolytica cells using an adapted protocol based on the published lithium acetate transformation protocol (Holkenbrink et al., 2018). In detail, the parental strain was spread over a YPD agar and grown for 24 h at 30°C prior to transformation. Cells were scraped from the plate and washed with water. Subsequently, a certain amount of cells (volume corresponding to OD600 9.2 per transformation) was resuspended in 100 μL of a transformation mix containing 43.8% poly-ethylene glycol (PEG 3350), 100 mM dithiothreitol, 0.25 g/L single-stranded DNA and 0.1 M LiAc. DNA was then added (a minimum of 500 ng). After a heat-shock at 39°C for 1 h, cells were pelleted for 5 min at 3000 rpm. The transformation mix was removed, and cell pellets were resuspended in 1 mL YPD and allowed to recover for 2 h at 30°C, 250 rpm agitation. Finally, cells were pelleted for 5 min at 3000 rpm, resuspended in 100 μL sterile water, and plated on selective medium. Plates were incubated at 30°C for 2–5 days until colonies appeared.
All strains were verified by PCR using insert-specific oligos in combination with oligos specific to the regions outside the different recombination sites. The CreA-loxP-mediated selection marker loop-out was performed as described previously (Holkenbrink et al., 2018). The elimination of the selection marker was verified by PCR in addition to phenotypic tests.
2.4. Cultivation
For KA-quantification, single colonies were inoculated from fresh plates into 3 mL YPD in 24-well plates with air-penetrable lids (EnzyScreen, NL) and grown for 24 h at 30°C and 300 rpm agitation as pre-cultures. For cultivation, the required volume of the pre-culture was transferred to 50 ml YP+8%D in 250 mL baffled shake flasks for an initial optical density at 600 nm (OD600) of 1.0. The shake flask cultures were cultivated for 72 h at 30°C with 300 rpm agitation.
For GA-quantification, single colonies were inoculated from fresh plates into 2.5 or 3 mL YPD in 24-well plates with air-penetrable lids and grown for 16–24 h at 30°C and 300 rpm agitation as pre-cultures. For cultivation, the required volume of the pre-culture was transferred to 2.5 or 3 mL YP+8%D in 24-well plates for an initial optical density at 600 nm (OD600) of 0.1. The plates were cultivated for 72 h at 30°C with 300 rpm agitation. By the end of the cultivation, either cell dry weight, OD600, or both were measured. OD600 was measured using a NanoPhotometer (Implen GmbH, Germany). All cultivations were done in biological triplicates.
For the feeding experiments with GA9, GA4 or GA7, pre-cultures and cultivation were done under the same conditions as for GA-quantification. During the cultivation step, 100 mg/L of either GA9, GA4 or GA7 was added to the media (3 or 2.5 ml of YP+8% glucose). The stock solutions of GA9, GA4, and GA7 were 5 mg/mL in absolute ethanol, which yielded a final ethanol concentration of 2% in the media.
For growth profiling of selected strains, pre-culture preparations from single colonies were inoculated from fresh plates into 2.5 YPD in 24-well plates with air-penetrable lids and grown for ~16 h at 30°C and 300 rpm agitation. Then the required volume of pre-culture was transferred to 1 mL of YP+8%D in 24-roundwell plates (Enzyscreen, CR1424f) with air-penetrable lids. The plates were cultivated, and their growth continuously measured in a Growth Profiler 960 (Enzyscreen) at 30°C with 225 rpm agitation for 72 h. All growth profiling experiments were conducted in biological triplicates.
2.5. Analytical methods
For the KA-quantification, GA12-detection, GA-analysis of ST6513 and ST6514, and feeding experiments, the supernatant was sampled directly in the case of cell-free media or extracted by the following protocol. 300 μL of cultivation broth was transferred into a 2 ml microtube (Sarstedt). 500 μL of 0.5–0.75 mm acid-washed glass beads and 1.2 mL of acetonitrile were added to each tube. Thereafter, the cells were disrupted with a Precellys®24 homogenizer (Bertin Corp.) four times at 5500 rpm for 10 s, with samples being kept on ice in between rounds of breaking. The samples were then shaken for 10 min at room temperature. Lastly, the samples were centrifuged, and the supernatant fractions were sampled for analysis.
For quantification of GA9, GA4, GA7, and GA3 for all other strains, 1 mL of broth were transferred to a 2 mL microtube. The samples were centrifuged, and the supernatant was extracted for analysis. Samples were diluted in water prior to analysis.
Furthermore, to investigate the presence of GA9, GA4, GA7, and GA3 in ST8580 cell pellets, 1 mL of broth was transferred 2 mL microtube. The cells were pelleted by centrifugation, and the supernatant was removed. The cell pellets were washed twice by resuspension in 1 mL of water, centrifugation, and removal of the liquid phase. Thereafter, 500 μl of 0.5–0.75 mm acid-washed glass beads and 1 mL of acetonitrile were added to each tube. Subsequently, the cells were disrupted with a Precellys®24 homogenizer four times at 5500 rpm for 10 s, with samples being kept on ice in between rounds of breaking. The samples were then shaken for 10 min in a DVX-2500 Multi-tube vortexer at room temperature. Lastly, the tubes were centrifuged, and the acetonitrile fractions were sampled for analysis.
The quantitative LC-MS data for the feeding experiments, KA-quantification, GA-quantification for ST6513 and ST6514, and GA12 mass confirmation can be seen in the supplementary methods.
The LC-MS/MS analysis of GAs for all other strains was performed on a Vanquish Duo UHPLC binary system (Thermo Fisher Scientific, USA) coupled to IDX-Orbitrap Mass Spectrometer (Thermo Fisher Scientific, USA). The chromatographic separation was achieved using a Waters ACQUITY BEH C18 (10 cm × 2.1 mm, 1.7 μm) equipped with an ACQUITY BEH C18 guard column kept at 40 C and using a flow rate of 0.35 mL/min. The mobile phases consisted of MilliQ© water + O.1% formic acid (A) and acetonitrile + 0.1% formic acid (B). The initial composition was 2%B held for 0.8 min, followed by a linear gradient till 5% in 3.3 min, and afterward, 100%B was reached in 10 min and held for 1 min before going back to initial conditions. Re-equilibration time was 2.7 min. The MS measurement was done in negative-heated electrospray ionization (HESI) mode with a voltage of 2500 V acquiring in full MS/MS spectra (Data dependent Acquisition-driven MS/MS) in the mass range of 70–1000 Da. The authentic GA3 (012 2503), GA4 (012 2532), GA7 (012 2543), and GA9 (012 2663) standards were acquired from OlChemIm s.r.o. (Czech Republic) and the detection limits of all GAs were 0.05 mg/L.
3. Results
3.1. Establishing the biosynthetic pathway to ent-kaurenoic acid
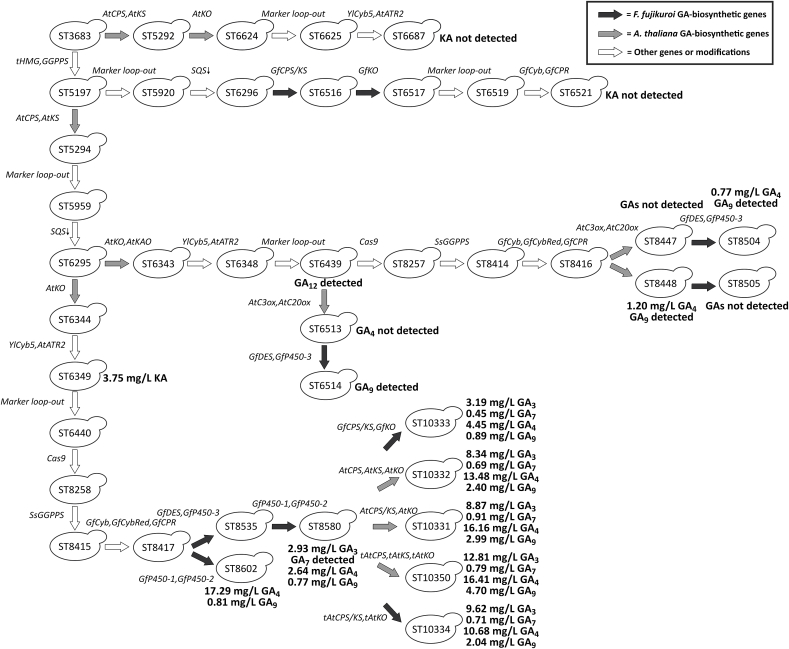
We began with evaluating plant and fungal biosynthetic pathways for ent-kaurenoic acid production in Y. lipolytica. All genes that were not native to Y. lipolytica were codon-optimized. To produce the GA-precursor ent-kaurenoic acid via the plant pathway, we expressed the genes encoding copalyl diphosphate synthase (AtCPS), ent-kaurene synthase (AtKS), ent-kaurene oxidase (AtKO), NADPH cytochrome P450 reductase (AtATR2) from A. thaliana, and the native Y. lipolytica cytochrome b5 (YlCyb5) to generate strain ST6687 (Fig. 2). The genes encoding AtCPS, AtKS, and AtKO were full-length with the native plastidial signal peptides intact. Furthermore, an MVA-pathway optimized strain ST6349 was constructed that expressed the same genes as ST6687, a truncated version of the native 3-hydroxy-3-methyl-glutaryl-coenzyme A reductase (tHMG), and the native geranylgeranyl diphosphate synthase (GGPPS), while the endogenous squalene synthase gene was downregulated by truncating its promoter down to 50 base pairs (SQS↓) (Kildegaard et al., 2017). The HMG-reductase (HMGp) that reduces HMG-CoA to mevalonate was previously reported as one of the flux-controlling enzymes in the MVA-pathway (Cao et al., 2016, 2017; Yang et al., 2016). Furthermore, in Saccharomyces cerevisiae, removing the N-terminal transmembrane domain of HMGp resulted in a non-regulated protein retaining its catalytic activity (Donald et al., 1997).
Fig. 2.
Overview of the strain engineering process of Y. lipolytica for GA-production. Italicized genes were expressed or overexpressed. SQS↓, downregulation of the squalene synthase gene by promoter truncation. ‘Marker loop-out’, removal of selection markers by Cre/loxP-recombination.
Overexpression of GGPPSp has been shown to increase the production of β-carotene in Y. lipolytica and gibberellins in F. fujikuroi (Albermann et al., 2013b; Kildegaard et al., 2017). The final modification in the MVA-optimized strains is the truncation of the native squalene promoter, which was previously shown to increase β-carotene production in Y. lipolytica (Kildegaard et al., 2017).
To produce ent-kaurenoic acid via the fungal pathway, we expressed the genes encoding the bifunctional F. fujikuroi copalyl disphosphate synthase/ent-kaurene synthase (GfCPS/KS), F. fujikuroi ent-kaurene oxidase (GfKO), F. fujikuroi NADPH cytochrome P450 reductase (GfCPR) and F. fujikuroi cytochrome b5 (GfCyb5), while tHMG and GGPPS were overexpressed, and the squalene promoter truncated (SQS↓), which generated ST6521.
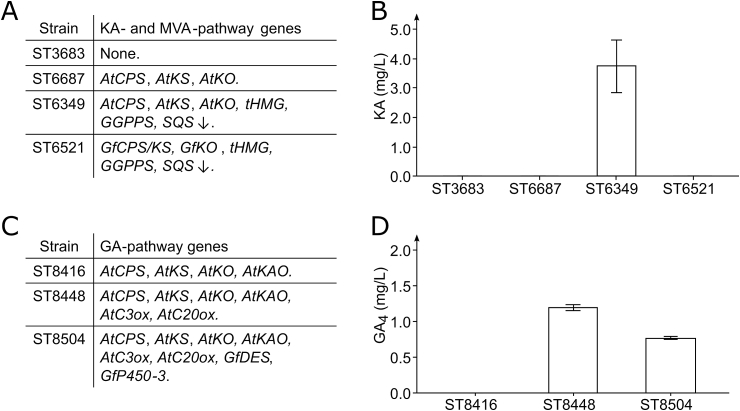
The MVA-optimized strain ST6349 expressing the plant KA-biosynthetic pathway produced 3.75 mg/L KA, while no KA was detected for the non-optimized strain ST6687 (Fig. 3A and B). Surprisingly, the strain ST6521 with optimized MVA-pathway and expressing fungal enzymes did not produce measurable amounts of KA.
Fig. 3.
KA-production by engineered Y. lipolytica strains. A. Overview of KA-producing strains showing which MVA-pathway genes were overexpressed or downregulated and which KA-biosynthetic genes were expressed. B. KA-production for ST3683, ST6687, ST6349, and ST6521. C. Overview of GA-producing strains showing which GA-biosynthetic genes were expressed. D. GA4-production for ST8416, ST8448, and ST8504. Three biological replicates were used to calculate titer averages and standard deviations for all strains.
3.2. Production of the early gibberellin intermediates
Since KA production was achieved with the plant pathway enzymes, we sought to reconstruct the pathway towards bioactive GAs with mainly plant-derived enzymes. The strain ST6439 for production of the inert intermediate GA12 was constructed based on the ent-kaurene producer ST6295 and further expression of AtKO, A. thaliana ent-kaurenoic acid oxidase (AtKAO), YlCyb5, and AtATR2. After cultivation, a compound identified as GA12 could be detected for ST6439 based on targeted ion mass and retention time (Fig. S1). Since no standard was available, the amount of putative GA12 could not be quantified.
3.3. Introduction of the biosynthetic pathway for bioactive gibberellin production
As the next step, we wanted to extend the pathway from GA12 to the biologically active gibberellin GA4. For this, we expressed codon-optimized versions of the A. thaliana GA C20-oxidase (AtC20ox) and GA C3-oxidase (AtC3ox) in the GA12-producing strain ST6439 to generate ST6513 that should be able to produce GA4 from GA12 via the intermediates GA15, GA24, and GA9. However, the cultivation of ST6513 did not result in detectable amounts of GA4. However, we still attempted to further extend the pathway from GA4 to GA7 and to GA3 by expressing fungal enzymes GfP450-3p and GfDESp generating ST6514.
While GA9 was detected for ST6514, neither GA4, GA7, nor GA3 were detected (Table S2). This indicated that AtC20oxp was active in ST6514 and catalyzed the formation of GA9 from GA12. To discern whether the three biosynthetic enzymes, AtC3oxp, GfDESp, and GfP450-3p, were active, we conducted several precursor feeding experiments. When strain ST6514 was cultivated in a medium supplemented with GA9, it produced detectable amounts of GA4, which was not the case for the control strain ST3683 (Table S2). This confirmed that AtC3oxp was active in ST6514 and that the formation of GA4 was limited by GA9-supply. When the ST6514 was supplemented with GA4, the presence of GA7 was detected; However, GA7 was also detected when ST3683 and cell-free media were supplemented with GA4 (Table S2 and S3). This suggests that GA4 is either spontaneously converted to GA7 or that the commercial GA4 product contains small amounts of GA7. The manufactures (OlChemIm s.r.o.) note that the GA4 standard we used is ≥90% pure, which indicates the presence of impurities (OlchemIm s.r.o.|Products, 2021). Furthermore, other providers note that their GA4 standards contain at least some GA7 (Sigma-Aldrich|Gibberellin A4, 2021). Indeed, GA4 and GA7 are difficult to separate during purification, which may explain such impurities (Rademacher, 2015; Camara et al., 2018). Supplementing the cultivation medium of ST6514 with GA7 did not lead to a detectable amount of GA3 (Table S2). In summary, the results indicate that AtC20oxP and AtC3oxp were active, while GfP450-3p was likely inactive when expressed in ST6514.
3.4. Production of GA4 by further engineering
We decided to perform additional metabolic engineering of the yeast strain ST6349 to increase the flux towards the GA-biosynthesis. To simplify the genome editing, we integrated a codon-optimized cas9 gene from Streptococcus pyogenes into GA12-producing strain ST6439, resulting in strain ST8257 (Fig. 2). This enabled marker-free integration of DNA constructs (Holkenbrink et al., 2018). Firstly, to further boost the production of GGPP, a codon-optimized version of the Synechococcus sp. GGPP synthase (SsGGPPS7) was expressed in ST8257, giving strain ST8414. Expression of SsGGPPS7 was previously shown to substantially increase carotenoid production in Y. lipolytica (Tramontin et al., 2019). Secondly, three cytochromes P450 partners: cytochrome b5, cytochrome b5 reductase, and cytochrome P450 reductase (GfCyb5, GfCyb5Red, GfCPR) from F. fujikuroi were expressed to promote the electron transfer from NADPH to F. fujikuroi cytochromes P450 for the last steps towards GA3, resulting in strain ST8416. Finally, AtC20ox and AtC3ox were expressed under stronger promoters to overcome the potential rate-limitation of these steps. The genes AtC20ox and AtC3ox were expressed under the control of the promoters PrExp and PrTefintron, respectively, to generate strain ST8447 and under promoters PrTefintron and PrGpd, respectively, to generate ST8448. Both strains should be capable of producing GA4, albeit potentially at different levels due to the different promoter pairs used. Furthermore, the PrTefintron promoter is currently the strongest known constitutive promoter for Y. lipolytica (Tai and Stephanopoulos, 2013; Holkenbrink et al., 2018).
Cultivation of ST8448 resulted in the production of 1.20 mg/L GA4 and trace amounts of GA9 (Fig. 3C and D, and S2A), whereas no GA4 or GA9 could be detected for ST8447. The difference between the strains may be explained by the low expression level of AtC20ox from the PrExp promoter in ST8447 (Holkenbrink et al., 2018). After establishing de novo production of GA4 in yeast, we proceeded to complete the biosynthetic pathway towards GA7 and GA3. For this, GfDES and GfP450-3 were expressed in both strains ST8447 and ST8448, resulting in strains ST8504 and ST8505, respectively. Cultivation of ST8504 resulted in the production of 0.77 mg/L GA4 and trace amounts of GA9, while neither GA7 nor GA3 were detected (Fig. 3C and D, and S2A). None of these four compounds were detected in strain ST8505. This contradicts the previous results, where only the parental strain of ST8505 produced GA4.
Feeding of ST8504 with GA7 did not result in detectable amounts of GA3, which suggested that GfP450-3p was inactive in this strain (Table S3). This strain also grew very poorly when supplemented with GA7 and 2% ethanol (3 g dry cell weight/L) compared to without (15 g dry cell weight/L). Furthermore, GA4 was also detected when both ST3683 and cell-free media were supplemented with GA7, possibly due to product impurities (Table S3).
3.5. Production of GA3, GA4, and GA7 by the introduction of fungal cytochromes P450
Since no GA3 could be detected by expressing the plant genes AtC20ox, AtC3ox, and fungal genes GfDES and GfP450-3, we sought to reconstruct the biosynthetic pathway KA to GA3, GA7, and GA4 with fungal enzymes.
We built on top of the KA-producing strain ST6440, into which, through several rounds of transformation, we integrated the genes encoding for Cas9, SsGGPPS, and P450 auxiliary proteins GfCyb5, GfCybRed, GfCPR, generating ST8417. Since Cas9 expression may be toxic to yeast, we investigated the growth profiles of the Cas9 expressing strain ST8258 and its parental strain ST6440, however, no reductions in the growth of ST8258 compared to ST6440 were found (Fig. S3A and S3B).
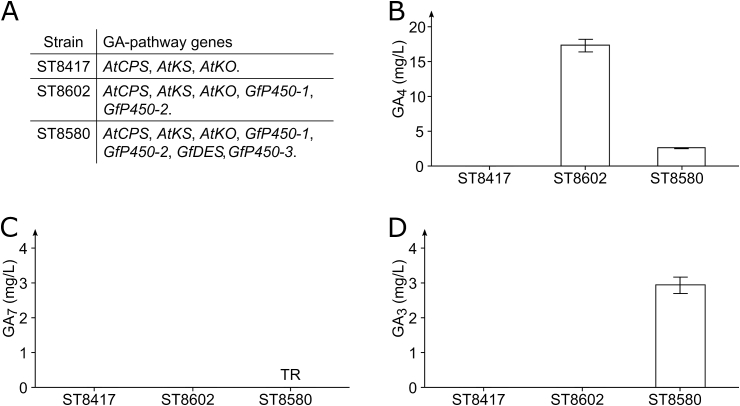
Furthermore, we expressed two fungal genes GfP450-1 and GfP450-2, encoding cytochromes P450 that should convert KA into GA4. The resulting strain ST8602 produced 17.29 mg/L GA4 and 0.81 mg/L GA9 (Fig. 4A and B, and S2B). The presence of GA9 is likely due to the oxygenation of GA12-aldehyde by GfP450-1p resulting in GA12, which is then processed into GA9 by the action of GfP450-2p (Tudzynski et al., 2002). The titer of GA4 in strain ST8602 expressing GfP450-1 and GfP450-2 was 14.4-fold greater than what was achieved for the analogous strain ST8448 expressing AtC20ox and AtC3ox. As the next step, the strain ST8580 was built that combined all the modifications as in ST8602 and additionally expressed GfP450-3 and GfDES. Cultivation of ST8580 resulted in the production of 2.93 mg/L GA3, trace amounts of GA7, 2.64 mg/L GA4, and 0.77 mg/L GA9 (Fig. 4A–D, and S2B). The identities of the compounds were confirmed by retention time and high-resolution mass-spectrometry compared to authentic standards (Fig. S4). Furthermore, analysis of the cell fraction of ST8580 did not show detectable amounts of GA7 and only trace amounts of GA3, GA4, and GA9, indicating that all GAs were secreted. Lastly, we investigated the growth profiles of the GA-producing strains ST8580 and ST8602 compared to their parental strain ST8417. There were no significant differences in growth (Fig. S3A and S3C).
Fig. 4.
GA-production by engineered Y. lipolytica strains. A. Overview of GA-producing strains showing which GA-biosynthetic genes were expressed. B. GA4-production for ST8417, ST8602, and ST8580. C. GA7-production for ST8417, ST8602, and ST8580. D. GA3-production for ST8417, ST8602, and ST8580. TR, trace amounts. Three biological replicates were used to calculate titer averages and standard deviations for all strains.
3.6. Improved GA-production by increasing early GA-pathway flux
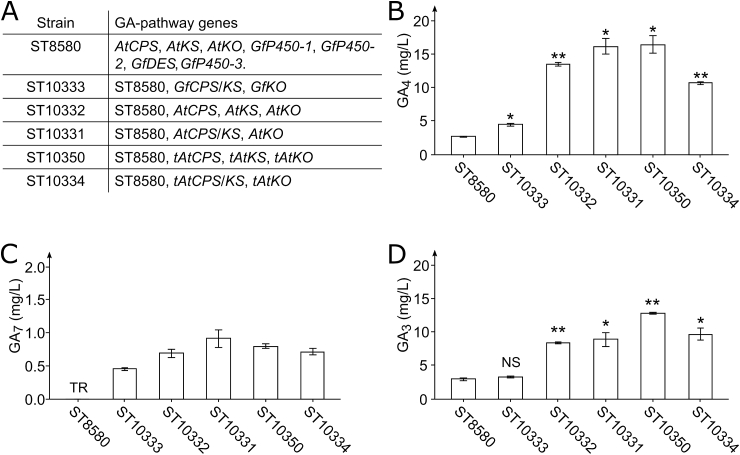
To increase GA-production, we expressed either GfCPS/KS and GfKS or AtCPS, AtKS, and AtKO in ST8580, generating strains ST10333 and ST10332, respectively. A fused enzyme consisting of AtCPS C-terminally linked to the N-terminal of AtKS (AtCPS/KS) was expressed together with AtKO in ST8580, generating ST10331. Furthermore, truncated variants of the A. thaliana KA-biosynthetic enzymes with N-terminal plastidial targeting sequences removed (tAtCPS, tAtKS, tAtKO) were expressed under individual promoters or with tAtCPS fused to tAtKS (tAtCPS/KS) alongside the individual expression of tAtKO in ST8580 generating ST10350 and ST10334, respectively. The ChloroP software and earlier reports were used to predict the plastidial targeting sequences of AtCPSp and AtKOp based on which the 2–60 and 2–28 amino acids were removed to make tAtCPSp and tAtKOp, respectively (Emanuelsson et al., 1999; Helliwell et al., 2001b). Of note, a transmembrane region at amino acid positions 6–24 was predicted for AtKOp by the InterProScan software, whereas no transmembrane regions were predicted for pAtCPS and pAtKS (Jones et al., 2014; Blum et al., 2021). No plastidial targeting sequence for AtKSp was predicted by ChloroP; however, earlier reports confirm the presence of a plastidial targeting sequence in AtKSp and suggest that the 44 first amino acids of AtKSp are involved in the translocation (Yamaguchi et al., 1998; Helliwell et al., 2001b). ST10350 exhibited the most enhanced GA-production with 12.81 mg/L GA3, 16.41 mg/L GA4, and 4.70 mg/L GA9, a 4.4-, 6.2- and 6.1- fold increase over ST8580, respectively (Fig. 5). Interestingly, all strains constructed from ST8580 demonstrated improved growth rates compared to ST8580 on YP+8%D media, likely due to the restoration of uracil prototrophy (Fig. S3A and S3D).
Fig. 5.
Y. lipolytica strains engineered for improved GA-production. A. Overview of GA-producing strains showing which GA-biosynthetic genes were expressed. B. GA4-production for ST8580, ST10333, ST10332, ST10331, ST10350, and ST10334. C. GA7-production for ST8580, ST10333, ST10332, ST10331, ST10350, and ST10334. D. GA3-production for ST8580, ST10333, ST10332, ST10331, ST10350, and ST10334. TR, trace amounts. Three biological replicates were used to calculate titer averages and standard deviations for all strains. Statistical significance (two-tailed student’s t-test) compared to ST8580 is represented by asterisks (*, p < 0.05. **, p < 0.001. NS, not significant).
4. Discussion
The gibberellins GA4, GA7, and GA3 are plant hormones with several applications in agriculture. The current production methods for these GAs are based on fermentation of the fungus F. fujikuroi (Shi et al., 2017). We have demonstrated the production of GA3, GA4, and GA7 in recombinant yeast Y. lipolytica by expression of biosynthetic pathway enzymes from F. fujikuroi and A. thaliana.
The production of KA was established by improving the flux towards GGPP and expression of AtCPS, AtKS, and AtKO. Interestingly, AtKOp has previously been functionally characterized by expression in S. cerevisiae, demonstrating its functionality in another yeast species (Helliwell et al., 1999). Yet, the presence of cofactors, substrate, and pH-levels may differ in Y. lipolytica compared to the native host environment, which may affect the activity of heterologous enzymes. Indeed, in vitro kinetic assays of N-terminally truncated AtCPS demonstrate optimal catalytic rates at 0.1 mM of the cofactor Mg2+ with decreasing enzymatic activity at both greater and lesser Mg2+-concentrations. A similar inhibitory effect is exerted by the substrate GGPP, while the optimal pH for truncated AtCPS was 7.5–8 (Prisic and Peters, 2007). Differences in such factors could also explain why the expression of GfCPS/KS and GfKO did not result in KA-production. Furthermore, there exists different algorithms for codon-optimization and it has been shown that codon-optimization can sometimes result in decreased expression or impaired folding of heterologous proteins (Fath et al., 2011; Komar A. A., 2019; Saito et al., 2019; Fu et al., 2020).
It would be interesting for future research to investigate the activity of heterologous KA-biosynthetic genes from various sources and with DNA sequences based on different codon-optimization algorithms in Y. lipolytica. Notably, the first two enzymes of KA-biosynthesis, AtCPSp, and AtKSp, are located in plastids in plants, while AtKOp is located on the outer plastidial membrane, and the rest of the GA-biosynthetic enzymes are located in the ER and cytosol (Helliwell et al., 2001b; Hedden and Sponsel, 2015). In the case of AtCPSp, the protein is processed into the mature enzyme during transport into the chloroplast, likely by removal of the N-terminal plastidial targeting peptide, which may yield a better performing enzyme (Sun and Kamiya, 1994; Helliwell et al., 2001b). Indeed, the highest GA3 production was achieved by expression of tAtCPS, tAtKS, and tAtKO, which suggests that N-terminal truncation may be a viable strategy for increasing enzyme activity.
Furthermore, the removal of plastidial targeting peptides from heterologous diterpene synthases has also been successfully used to construct yeast cell factories for the production of 13R-manoyl oxide (Zhang et al., 2019). In fact, targeting diterpene synthases to the plant cytosol by removal of targeting peptides alongside MVA-pathway upregulation resulted in increased production of both taxadiene, 13R-manoyl oxide, and forskolin in Nicotiana benthamiana compared to the expression of nontruncated diterpene synthases and MEP-pathway upregulation (De La Peña and Sattely, 2020). Interestingly, a transmembrane domain within the truncated region was predicted for tAtKOp, which likely renders the protein cytosolic upon removal.
Additionally, expression of fused AtCPS/KS or tAtCPS/KS together with AtKO or tAtKO, respectively, also led to improved GA-production. The fusion of the CPSp and KSp from Salvia miltiorrhiza led to enhanced production of ent-kaurene in S. cerevisiae (Su et al., 2016). Interestingly, certain mosses and liverworts have naturally occurring bifunctional CPS/KSp enzymes similar to F. fujikuroi (Hayashi et al., 2006; Kawaide et al., 2011). Therefore, enzymatic fusion could be further explored to improve GA-production in Y. lipolytica.
The further expression of AtKAOp resulted in the production of a compound putatively identified as GA12. This enzyme has previously been characterized and co-expressed with either AtATR1 or AtATR2 in S. cerevisiae, where co-expression with AtATR2 led to increased relative amounts of GA12 (Helliwell et al., 2001a). This indicates that optimal redox partner pairing with cytochromes P450 like AtKAOp can lead to improved activity in heterologous systems. Although GA12 is considered to be biologically inert, it is implicated in long-range GA signaling in A. thaliana, since GA12 may be transported via plant vascular tissue, converted to bioactive GAs, and thereby activate GA signaling cascades in receiving tissues, resulting in decreased accumulation of growth inhibitory DELLA proteins (Regnault et al., 2015). Therefore, agricultural GA12-application could potentially be useful to enhance plant growth. Furthermore, GA12 can be converted by the “Gain of Function in ABA-modulated Seed Germination 2”-enzyme (AtGas2p) to GA12 16, 17-dihydro-16α-ol (DHGA12), which can enhance seed germination and seedling development in A. thaliana (Liu et al., 2019). Cell factories for the production of GA12 and bioactive GA12-derivatives may therefore become useful in the future.
While we attempted to complete the biosynthetic pathway towards GA12 to GA3 by expression of AtC20ox, AtC3ox, GfDES, and GfP450-3 to generate ST6514, neither GA4, GA7, nor GA3 were produced by this strain. The feeding studies indicated that GfP450-3p was not active in ST6514. However, GA3 was produced when GfP450-3p was expressed in ST8580. Interestingly, ST6514 expressed only AtATR2 and YlCyb5, while ST8580 expressed the fungal redox partners GfCyb5, GfCybRed, and GfCPR in addition to the prior. Therefore, the presence of specific redox partners may be necessary to support the activity of F. fujikuroi cytochromes P450. Indeed, previous research has demonstrated that the activity of heterologous cytochromes P450 in yeast depends heavily on their co-expressed redox partners. The cytochrome P450 FoCYP53A19p from Fusarium oxysporum could convert 3-hydroxybenzoic acid to 3,4-dihydroxybenzoic acid when paired with an F. oxysporum CPR but not when paired with an S. cerevisiae or Candida albicans CPR (Durairaj et al., 2015). Furthermore, the activity of heterologous cytochromes P450 could be affected by codon-optimization and promoter choice, since the available constitutive promoters for Y. lipolytica vary greatly in strength (Holkenbrink et al., 2018).
The GA3-titer of 12.81 mg/L reported herein is still very low compared to the highest reported titer of 3.9 g/L for the natural producer F. fujikuroi (Escamilla S et al., 2000). However, we demonstrate that GA-production of our strains can be improved considerably by engineering the KA-biosynthetic pathway, and many existing strategies for terpenoid overproduction could be utilized for further improvements.
Indeed, Y. lipolytica is a relatively new host for recombinant terpene production, but it has already outperformed S. cerevisiae for the production of β-carotene (Larroude et al., 2018; Moser and Pichler, 2019). This is likely due to the higher acetyl-CoA flux and the lipophilic environment in Y. lipolytica, which may serve to sequester and store lipophilic terpenes (Christen and Sauer, 2011). Therefore, Y. lipolytica holds great potential as a microbial cell factory for terpenoid production.
5. Conclusions
Production of bioactive gibberellins GA3, GA4, and GA7 in Y. lipolytica was achieved by expressing a GA-biosynthetic pathway with both plant and fungal enzymes. The complete biosynthesis of GA3, GA4, and GA7 was done by heterologous expression of A. thaliana enzymes for KA-production and F. fujikuroi enzymes for the subsequent biosynthetic steps. We demonstrate that the GA-production could be enhanced by further engineering of the yeast chassis. This yeast-based platform holds the potential to improve the future production of bioactive GAs.
Author statements
Kanchana R. Kildegaard: Conceptualization, methodology, investigation, formal analysis, supervision, writing – original draft, writing – review & editing, funding acquisition. Jonathan A. Arnesen: methodology, investigation, validation, formal analysis, visualization, writing – original draft, writing – review & editing. Belén Adiego-Pérez: Investigation, writing – review & editing. Daniela Rago: methodology, investigation, data curation, visualization. Mette Kristensen: methodology, investigation, data curation, writing – review & editing. Andreas K. Klitgaard: methodology, investigation, data curation, visualization, writing – review & editing. Esben H. Hansen: resources, methodology, writing – review & editing. Jørgen Hansen: resources, writing – review & editing. Irina Borodina: Conceptualization, methodology, supervision, writing – original draft, writing – review & editing, supervision, funding acquisition.
Declaration of competing interest
No conflicts of interest.
Acknowledgments
The research was funded by the Novo Nordisk Foundation (Grant agreement NNF15OC0016592, NNF20CC0035580, and NNF20OC0060809). IB and KRK acknowledge the financial support from the European Union’s Horizon 2020 research and innovation programs (European Research Council, YEAST-TRANS project No 757384 and OLEFINE project No 760798). We are grateful to Larissa Ribeiro Ramos Tramontin for the kind gift of pCfB8092 and to Dr. Mahsa Babaei for the kind gift of PR-27422 and PR-27423.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ymben.2021.03.010.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Albermann S., Elter T., Teubner A., Krischke W., Hirth T., Tudzynski B. Characterization of novel mutants with an altered gibberellin spectrum in comparison to different wild-type strains of Fusarium fujikuroi. Appl. Microbiol. Biotechnol. 2013;97:7779–7790. doi: 10.1007/s00253-013-4917-7. [DOI] [PubMed] [Google Scholar]
- Albermann S., Linnemannstöns P., Tudzynski B. Strategies for strain improvement in Fusarium fujikuroi: overexpression and localization of key enzymes of the isoprenoid pathway and their impact on gibberellin biosynthesis. Appl. Microbiol. Biotechnol. 2013;97:2979–2995. doi: 10.1007/s00253-012-4377-5. [DOI] [PubMed] [Google Scholar]
- Angerer H., Radermacher M., Mankowska M., Steger M., Zwicker K., Heide H. The LYR protein subunit NB4M/NDUFA6 of mitochondrial complex I anchors an acyl carrier protein and is essential for catalytic activity. Proc. Natl. Acad. Sci. 2014;111:5207–5212. doi: 10.1073/pnas.1322438111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atzorn R., Crozier A., Wheeler C.T., Sandberg G. Production of gibberellins and indole-3-acetic acid by Rhizobium phaseoli in relation to nodulation of Phaseolus vulgaris roots. Planta. 1988;175:532–538. doi: 10.1007/BF00393076. [DOI] [PubMed] [Google Scholar]
- Bashyal B.M., Rawat K., Sharma S., Kulshreshtha D., Gopala Krishnan S., Singh A.K. Whole genome sequencing of Fusarium fujikuroi provides insight into the role of secretory proteins and cell wall degrading enzymes in causing bakanae disease of rice. Front. Plant Sci. 2017;8:1–12. doi: 10.3389/fpls.2017.02013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum M., Chang H.-Y., Chuguransky S., Grego T., Kandasaamy S., Mitchell A. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021;49:D344–D354. doi: 10.1093/nar/gkaa977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottini R., Fulchieri M., Pearce D., Pharis R.P. Identification of gibberellins A1, A3, and iso-A3 in cultures of Azospirillum lipoferum. Plant Physiol. 1989;90:45–47. doi: 10.1104/pp.90.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camara M.C., Vandenberghe L.P.S., Rodrigues C., de Oliveira J., Faulds C., Bertrand E. Current advances in gibberellic acid (GA3) production, patented technologies and potential applications. Planta. 2018;248:1049–1062. doi: 10.1007/s00425-018-2959-x. [DOI] [PubMed] [Google Scholar]
- Cao X., Lv Y.B., Chen J., Imanaka T., Wei L.J., Hua Q. Metabolic engineering of oleaginous yeast Yarrowia lipolytica for limonene overproduction. Biotechnol. Biofuels. 2016;9:1–11. doi: 10.1186/s13068-016-0626-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Wei L.J., Lin J.Y., Hua Q. Enhancing linalool production by engineering oleaginous yeast Yarrowia lipolytica. Bioresour. Technol. 2017;245:1641–1644. doi: 10.1016/j.biortech.2017.06.105. [DOI] [PubMed] [Google Scholar]
- Casanova L., Casanova R., Moret A., Agustí M. The application of gibberellic acid increases berry size of “Emperatriz” seedless grape. Spanish J. Agric. Res. 2009;7:919. doi: 10.5424/sjar/2009074-1105. [DOI] [Google Scholar]
- Christen S., Sauer U. Intracellular characterization of aerobic glucose metabolism in seven yeast species by 13C flux analysis and metabolomics. FEMS Yeast Res. 2011;11:263–272. doi: 10.1111/j.1567-1364.2010.00713.x. [DOI] [PubMed] [Google Scholar]
- Darvishi F., Ariana M., Marella E.R., Borodina I. Advances in synthetic biology of oleaginous yeast Yarrowia lipolytica for producing non-native chemicals. Appl. Microbiol. Biotechnol. 2018;102:5925–5938. doi: 10.1007/s00253-018-9099-x. [DOI] [PubMed] [Google Scholar]
- De La Peña R., Sattely E.S. Rerouting plant terpene biosynthesis enables momilactone pathway elucidation. Nat. Chem. Biol. 2020;17:205–212. doi: 10.1038/s41589-020-00669-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald K.A.G., Hampton R.Y., Fritz I.B. Effects of overproduction of the catalytic domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase on squalene synthesis in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1997;63:3341–3344. doi: 10.1128/aem.63.9.3341-3344.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B., Sherman D., Fischer G., Durrens P., Casaregola S., Lafontaine I. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- Durairaj P., Jung E., Park H.H., Kim B.G., Yun H. Comparative functional characterization of a novel benzoate hydroxylase cytochrome P450 of Fusarium oxysporum. Enzym. Microb. Technol. 2015;70:58–65. doi: 10.1016/j.enzmictec.2014.12.013. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Heijne G. Von. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escamilla S E.M., Dendooven L., Magaña I.P., Parra S R., De la Torre M. Optimization of gibberellic acid production by immobilized Gibberella fujikuroi mycelium in fluidized bioreactors. J. Biotechnol. 2000;76:147–155. doi: 10.1016/S0168-1656(99)00182-0. [DOI] [PubMed] [Google Scholar]
- Fath S., Bauer A.P., Liss M., Spriestersbach A., Maertens B., Hahn P. Multiparameter RNA and codon optimization: a standardized tool to assess and enhance autologous mammalian gene expression. PloS One. 2011;6:1–14. doi: 10.1371/journal.pone.0017596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Liang Y., Zhong X., Pan Z.L., Huang L., Zhang H.L. Codon optimization with deep learning to enhance protein expression. Sci. Rep. 2020;10:1–9. doi: 10.1038/s41598-020-74091-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S., Tong Y., Zhu L., Ge M., Zhang Y., Chen D. Iterative integration of multiple-copy pathway genes in Yarrowia lipolytica for heterologous β-carotene production. Metab. Eng. 2017;41:192–201. doi: 10.1016/j.ymben.2017.04.004. [DOI] [PubMed] [Google Scholar]
- Gibberellins Market Industry analysis report. 2016. https://www.grandviewresearch.com/industry-analysis/gibberellins-market Available at:
- Groenewald M., Boekhout T., Neuvéglise C., Gaillardin C., Van Dijck P.W.M., Wyss M. Yarrowia lipolytica: safety assessment of an oleaginous yeast with a great industrial potential. Crit. Rev. Microbiol. 2014;40:187–206. doi: 10.3109/1040841X.2013.770386. [DOI] [PubMed] [Google Scholar]
- Hamayun M., Khan S.A., Khan A.L., Shin J.-H., Ahmad B., Shin D.-H. Exogenous gibberellic acid reprograms soybean to higher growth and salt stress tolerance. J. Agric. Food Chem. 2010;58:7226–7232. doi: 10.1021/jf101221t. [DOI] [PubMed] [Google Scholar]
- Harrington J.F., Rappaport L., Hood K.J. Influence of gibberellins on stem elongation and flowering of endive. Science. 1957;125:601–602. doi: 10.1126/science.125.3248.601-a. [DOI] [Google Scholar]
- Hayashi K., Kawaide H., Notomi M., Sakigi Y., Matsuo A., Nozaki H. Identification and functional analysis of bifunctional ent-kaurene synthase from the moss Physcomitrella patens. FEBS Lett. 2006;580:6175–6181. doi: 10.1016/j.febslet.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Hedden P., Sponsel V. A century of gibberellin research. J. Plant Growth Regul. 2015;34:740–760. doi: 10.1007/s00344-015-9546-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell C.A., Chandler P.M., Poole A., Dennis E.S., Peacock W.J. The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc. Natl. Acad. Sci. 2001;98:2065–2070. doi: 10.1073/pnas.98.4.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell C.A., Poole A., James Peacock W., Dennis E.S. Arabidopsis ent-kaurene oxidase catalyzes three steps of gibberellin biosynthesis. Plant Physiol. 1999;119:507–510. doi: 10.1104/pp.119.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell C.A., Sullivan J.A., Mould R.M., Gray J.C., James Peacock W., Dennis E.S. A plastid envelope location of Arabidopsis ent-kaurene oxidase links the plastid and endoplasmic reticulum steps of the gibberellin biosynthesis pathway. Plant J. 2001;28:201–208. doi: 10.1046/j.1365-313X.2001.01150.x. [DOI] [PubMed] [Google Scholar]
- Holkenbrink C., Dam M.I., Kildegaard K.R., Beder J., Dahlin J., Doménech Belda D. EasyCloneYALI: CRISPR/Cas9-Based synthetic toolbox for engineering of the yeast Yarrowia lipolytica. Biotechnol. J. 2018;13:1–8. doi: 10.1002/biot.201700543. [DOI] [PubMed] [Google Scholar]
- Hwang I.S., Ahn I.-P. Multi-homologous recombination-based gene manipulation in the rice pathogen Fusarium fujikuroi. Plant Pathol. J. 2016;32:173–181. doi: 10.5423/PPJ.OA.12.2015.0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito A., Sakamoto D., Itai A., Nishijima T., Oyama-Okubo N., Nakamura Y. Effects of GA3+4 and GA4+7 application either alone or combined with prohexadione-Ca on fruit development of Japanese pear ‘kosui’. Horticulture J. 2016;85:201–208. doi: 10.2503/hortj.MI-107. [DOI] [Google Scholar]
- Jones P., Binns D., Chang H.-Y., Fraser M., Li W., McAnulla C. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn A., Goss J.A., Smith D.E. Effect of gibberellin on germination of lettuce seed. Science. 1957;125:645–646. doi: 10.1126/science.125.3249.645. [DOI] [PubMed] [Google Scholar]
- Kasahara H., Hanada A., Kuzuyama T., Takagi M., Kamiya Y., Yamaguchi S. Contribution of the mevalonate and methylerythritol phosphate pathways to the biosynthesis of gibberellins in Arabidopsis. J. Biol. Chem. 2002;277:45188–45194. doi: 10.1074/jbc.M208659200. [DOI] [PubMed] [Google Scholar]
- Kato Y. Responses of plant cells to gibberellin. Bot. Gaz. 1955;117:16–24. doi: 10.1086/335884. [DOI] [Google Scholar]
- Kawaide H., Hayashi K.I., Kawanabe R., Sakigi Y., Matsuo A., Natsume M. Identification of the single amino acid involved in quenching the ent-kauranyl cation by a water molecule in ent-kaurene synthase of Physcomitrella patens. FEBS J. 2011;278:123–133. doi: 10.1111/j.1742-4658.2010.07938.x. [DOI] [PubMed] [Google Scholar]
- Kildegaard K.R., Adiego-Pérez B., Doménech Belda D., Khangura J.K., Holkenbrink C., Borodina I. Engineering of Yarrowia lipolytica for production of astaxanthin. Synth. Syst. Biotechnol. 2017;2:287–294. doi: 10.1016/j.synbio.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar A.A. Synonymous codon usage—a guide for Co-translational protein folding in the cell. Mol. Biol. 2019;53:777–790. doi: 10.1134/S0026893319060098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lale G., Gadre R. Enhanced production of gibberellin A4 (GA4) by a mutant of Gibberella fujikuroi in wheat gluten medium. J. Ind. Microbiol. Biotechnol. 2010;37:297–306. doi: 10.1007/s10295-009-0673-1. [DOI] [PubMed] [Google Scholar]
- Lang A. Induction of flower formation in biennial hyoscyamus by treatment with gibberellin. Naturwissenschaften. 1956;43:284–285. doi: 10.1007/BF00622495. [DOI] [Google Scholar]
- Lange T., Hedden P., Graebe J.E. Expression cloning of a gibberellin 20-oxidase, a multifunctional enzyme involved in gibberellin biosynthesis. Proc. Natl. Acad. Sci. 1994;91:8552–8556. doi: 10.1073/pnas.91.18.8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larroude M., Celinska E., Back A., Thomas S., Nicaud J.M., Ledesma-Amaro R. A synthetic biology approach to transform Yarrowia lipolytica into a competitive biotechnological producer of β-carotene. Biotechnol. Bioeng. 2018;115:464–472. doi: 10.1002/bit.26473. [DOI] [PubMed] [Google Scholar]
- Liu H., Guo S., Lu M., Zhang Y., Li J., Wang W. Biosynthesis of DHGA12 and its roles in Arabidopsis seedling establishment. Nat. Commun. 2019;10:1–14. doi: 10.1038/s41467-019-09467-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod A.M., Millar A.S. Effects of gibberellic acid on barley endosperm. J. Inst. Brew. 1962;68:322–332. doi: 10.1002/j.2050-0416.1962.tb01873.x. [DOI] [Google Scholar]
- MacMillan J. Occurrence of gibberellins in vascular plants, fungi, and bacteria. J. Plant Growth Regul. 2001;20:387–442. doi: 10.1007/s003440010038. [DOI] [PubMed] [Google Scholar]
- Marella E.R., Holkenbrink C., Siewers V., Borodina I. Engineering microbial fatty acid metabolism for biofuels and biochemicals. Curr. Opin. Biotechnol. 2018;50:39–46. doi: 10.1016/j.copbio.2017.10.002. [DOI] [PubMed] [Google Scholar]
- Matthäus F., Ketelhot M., Gatter M., Barth G. Production of lycopene in the non-carotenoid-producing yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 2014;80:1660–1669. doi: 10.1128/AEM.03167-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser S., Pichler H. Identifying and engineering the ideal microbial terpenoid production host. Appl. Microbiol. Biotechnol. 2019;103:5501–5516. doi: 10.1007/s00253-019-09892-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OlchemIm s.r.o. Products. 2021. https://www.olchemim.cz/Products.aspx?idc=1&idp=4 Available at:
- Prisic S., Peters R.J. Synergistic substrate inhibition of ent-copalyl diphosphate synthase: a potential feed-forward inhibition mechanism limiting gibberellin metabolism. Plant Physiol. 2007;144:445–454. doi: 10.1104/pp.106.095208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher W. Plant growth regulators: backgrounds and uses in plant production. J. Plant Growth Regul. 2015;34:845–872. doi: 10.1007/s00344-015-9541-6. [DOI] [Google Scholar]
- Rademacher W., Graebe J.E. Gibberellin A4 produced by Sphaceloma manihoticola, the cause of the superelongation disease of cassava (Manihotesculenta) Biochem. Biophys. Res. Commun. 1979;91:35–40. doi: 10.1016/0006-291X(79)90579-5. [DOI] [PubMed] [Google Scholar]
- Regnault T., Davière J.-M., Wild M., Sakvarelidze-Achard L., Heintz D., Carrera Bergua E. The gibberellin precursor GA12 acts as a long-distance growth signal in Arabidopsis. Nature Plants. 2015;1:1–6. doi: 10.1038/nplants.2015.73. [DOI] [PubMed] [Google Scholar]
- Rojas M.C., Hedden P., Gaskin P., Tudzynski B. The P450-1 gene of Gibberella fujikuroi encodes a multifunctional enzyme in gibberellin biosynthesis. Proc. Natl. Acad. Sci. 2001;98:5838–5843. doi: 10.1073/pnas.091096298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y., Kitagawa W., Kumagai T., Tajima N., Nishimiya Y., Tamano K. Developing a codon optimization method for improved expression of recombinant proteins in actinobacteria. Sci. Rep. 2019;9:1–10. doi: 10.1038/s41598-019-44500-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassa T., Kawaide H., Takarada T. Identification of gibberellins A4 , A9 , and A24 from Phaeosphaeria sp. L487 cultured in a chemically defined medium. Biosci. Biotechnol. Biochem. 1994;58:438–439. doi: 10.1271/bbb.58.438. [DOI] [Google Scholar]
- Shi T.Q., Gao J., Wang W.J., Wang K.F., Xu G.Q., Huang H. CRISPR/Cas9-Based genome editing in the filamentous fungus Fusarium fujikuroi and its application in strain engineering for gibberellic acid production. ACS Synth. Biol. 2019;8:445–454. doi: 10.1021/acssynbio.8b00478. [DOI] [PubMed] [Google Scholar]
- Shi T.Q., Peng H., Zeng S.Y., Ji R.Y., Shi K., Huang H. Microbial production of plant hormones: opportunities and challenges. Bioengineered. 2017;8:124–128. doi: 10.1080/21655979.2016.1212138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomeili M., Nabipour M., Meskarbashee M., Memari H.R. Effects of gibberellic acid on sugarcane plants exposed to salinity under a hydroponic system. Afr. J. Plant Sci. 2011;5:609–616. doi: 10.5897/AJPS.9000093. [DOI] [Google Scholar]
- Sigma-Aldrich Gibberellin A4. 2021. https://www.sigmaaldrich.com/catalog/product/sigma/g7276?lang=en®ion=DK Available at:
- Su P., Tong Y., Cheng Q., Hu Y., Zhang M., Yang J. Functional characterization of ent-copalyl diphosphate synthase, kaurene synthase and kaurene oxidase in the Salvia miltiorrhiza gibberellin biosynthetic pathway. Sci. Rep. 2016;6:23057. doi: 10.1038/srep23057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Zhang C., Nan W., Li D., Ke D., Lu W. Glycerol improves heterologous biosynthesis of betulinic acid in engineered Yarrowia lipolytica. Chem. Eng. Sci. 2019;196:82–90. doi: 10.1016/j.ces.2018.10.052. [DOI] [Google Scholar]
- Sun T.P., Kamiya Y. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell. 1994;6:1509–1518. doi: 10.1105/tpc.6.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai M., Stephanopoulos G. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab. Eng. 2013;15:1–9. doi: 10.1016/j.ymben.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Tramontin L.R.R., Kildegaard K.R., Sudarsan S., Borodina I. Enhancement of astaxanthin biosynthesis in oleaginous yeast Yarrowia lipolytica via microalgal pathway. Microorganisms. 2019;7:472. doi: 10.3390/microorganisms7100472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudzynski B., Hedden P., Carrera E., Gaskin P. The P450-4 gene of Gibberella fujikuroi encodes ent-kaurene oxidase in the gibberellin biosynthesis pathway. Appl. Environ. Microbiol. 2001;67:3514–3522. doi: 10.1128/AEM.67.8.3514-3522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudzynski B., Kawaide H., Kamiya Y. Gibberellin biosynthesis in Gibberella fujikuroi: cloning and characterization of the copalyl diphosphate synthase gene. Curr. Genet. 1998;34:234–240. doi: 10.1007/s002940050392. [DOI] [PubMed] [Google Scholar]
- Tudzynski B., Mihlan M., Rojas M.C., Linnemannstöns P., Gaskin P., Hedden P. Characterization of the final two genes of the gibberellin biosynthesis gene cluster of Gibberella fujikuroi. J. Biol. Chem. 2003;278:28635–28643. doi: 10.1074/jbc.M301927200. [DOI] [PubMed] [Google Scholar]
- Tudzynski B., Rojas M.C., Gaskin P., Hedden P. The gibberellin 20-oxidase of Gibberella fujikuroi is a multifunctional monooxygenase. J. Biol. Chem. 2002;277:21246–21253. doi: 10.1074/jbc.M201651200. [DOI] [PubMed] [Google Scholar]
- Tuna A.L., Kaya C., Dikilitas M., Higgs D. The combined effects of gibberellic acid and salinity on some antioxidant enzyme activities, plant growth parameters and nutritional status in maize plants. Environ. Exp. Bot. 2008;62:1–9. doi: 10.1016/j.envexpbot.2007.06.007. [DOI] [Google Scholar]
- Wiemann P., Sieber C.M.K., von Bargen K.W., Studt L., Niehaus E.M., Espino J.J. Deciphering the cryptic genome: genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J., Phillips A.L., Gaskin P., Hedden P. Function and substrate specificity of the gibberellin 3β-hydroxylase encoded by the Arabidopsis GA4 gene. Plant Physiol. 1998;117:559–563. doi: 10.1104/pp.117.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittwer S.H., Bukovac M.J., Sell H.M., Weller L.E. Some effects of gibberellin on flowering and fruit setting. Plant Physiol. 1957;32:39–41. doi: 10.1104/pp.32.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Sun T., Kawaide H., Kamiya Y. The GA2 locus of Arabidopsis thaliana encodes ent-kaurene synthase of gibberellin biosynthesis. Plant Physiol. 1998;116:1271–1278. doi: 10.1104/pp.116.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Nambou K., Wei L., Hua Q. Heterologous production of α-farnesene in metabolically engineered strains of Yarrowia lipolytica. Bioresour. Technol. 2016;216:1040–1048. doi: 10.1016/j.biortech.2016.06.028. [DOI] [PubMed] [Google Scholar]
- Zang Y.X., Chun I.J., Zhang L.L., Hong S.B., Zheng W.W., Xu K. Effect of gibberellic acid application on plant growth attributes, return bloom, and fruit quality of rabbiteye blueberry. Sci. Hortic. 2016;200:13–18. doi: 10.1016/j.scienta.2015.12.057. [DOI] [Google Scholar]
- Zhang C., Ju H., Lu C.-Z., Zhao F., Liu J., Guo X. High-titer production of 13R-manoyl oxide in metabolically engineered Saccharomyces cerevisiae. Microb. Cell Factories. 2019;18:73. doi: 10.1186/s12934-019-1123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Jackson E.N. Metabolic engineering of Yarrowia lipolytica for industrial applications. Curr. Opin. Biotechnol. 2015;36:65–72. doi: 10.1016/j.copbio.2015.08.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.