Abstract
The most frequent genetic alteration in acute myeloid leukemia (AML) is the mutation of nucleophosmin 1 (NPM1). Yet, its downstream oncogenic routes are not fully understood. Here, we report the identification of one long noncoding RNA (lncRNA) overexpressed in NPM1-mutated AML patients (named LONA) whose intracellular localization inversely reflects that of NPM1. While NPM1 is nuclear and LONA cytoplasmic in wild-type NPM1 AML cells, LONA becomes nuclear as mutant NPM1 moves toward the cytoplasm. Gain or loss of function combined with a genome-wide RNA-seq search identified a set of LONA mRNA targets encoding proteins involved in myeloid cell differentiation (including THSB1, MAFB, and ASB2) and interaction with its microenvironment. Consistently, LONA overexpression in mutant NPM1 established cell lines and primary AML cells exerts an anti-myeloid differentiation effect, whilst it exerts an opposite pro-myeloid differentiation effect in a wild type NPM1 setting. In vivo, LONA overexpression acts as an oncogenic lncRNA reducing the survival of mice transplanted with AML cells and rendering AML tumors more resistant to AraC chemotherapy.
These data indicate that mutation-dependent nuclear export of NPM1 leads to nuclear retention and consequent oncogenic functions of the overexpressed lncRNA LONA, thus uncovering a novel NPM1 mutation-dependent pathway in AML pathogenesis.
Subject terms: Acute myeloid leukaemia, Differentiation, Oncogenes
Introduction
The most frequent mutation observed in acute myeloid leukemia (AML) generates a truncated nucleophosmin (NPM1) protein in which the C-terminal nucleolar localization signal is mutated into a nuclear export signal [1–4]. As a consequence, NPM1 becomes cytoplasmic and has been named NPM1c + . Whether the mutation of NPM1 is oncogenic because of a nuclear loss of function [5, 6] or because of a cytoplasmic gain of function [7] is still a matter of intense debate [8]. It has been shown only recently that the cytoplasmic accumulation of NPM1 per se may play an active role in leukemogenesis through the regulation of genes encoding proteins required for myeloid differentiation [9]. Indeed, NPM1c + maintains a leukemic state through up-regulation of homeobox HOX genes which normally ensure myeloid differentiation [9]. However, the underlying molecular mechanism remains unknown. In normal cells, NPM1 is a ubiquitous protein possessing scaffolding properties modulating chromatin structure and regulating nuclear and nucleolar gene expression [10, 11]. Devoid of intrinsic transcriptional activity, NPM1 acts indirectly on nuclear gene expression through different partners including chromatin remodelers, genuine transcription factors, or non-coding (nc) RNAs [12].
Studies at the RNA level in cancer have been focused for a long time on messenger RNAs (mRNAs), the best known RNA family that represents only a small part of transcripts (2 to 5% of total RNAs) [13]. However, the human genome contains much more than just protein-coding genes. The majority of transcribed RNAs are ncRNAs, such as micro RNAs (miRNAs) and long noncoding RNAs (lncRNAs), playing key roles in cell fate [14]. While small ncRNAs, such as miRNAs, have well-established functions in cell and in oncogenesis, lncRNAs have only recently emerged as important players in physiological and pathological processes. LncRNAs were defined as RNA transcripts larger than 200 nt but devoid of protein-coding potential, and were initially considered as a transcriptional noise due to low evolutionary conservation and low expression levels [15, 16]. However, cumulative evidences now point toward lncRNA functional roles affecting almost all stages of gene regulation [17]. RNA quantification using high-throughput sequencing techniques has shown they have more specific spacio-temporal expression profiles than coding RNAs, with tissue-specific or development stage-dependent expression [18]. Functional studies, even if recent, has already demonstrated their involvement in major cell biology processes according to whether they are nuclear or cytoplasmic [19]. The majority of the nuclear lncRNAs described so far act through addressing chromatin remodeling complexes or transcription factors to their target genomic sequences [20]. One can distinguish lncRNAs acting in cis such as HOTTIP [21] regulating its neighboring genes, of those acting in trans such as HOTAIR controlling more distant loci [22]. Nuclear lncRNAs may also have an indirect role in gene regulation by sequestering or guiding transcription factors, or altering the allosteric conformation of proteins or nuclear domains [23–26]. Many lncRNAs also act in the cytoplasm where they regulate mRNA translation [27, 28] or stability [29, 30]. LncRNAs can also be precursors of small RNAs (including miRNAs). By their various mechanisms of actions, lncRNAs thus regulate numerous physiological processes: control of parental imprinting [31], HOX genes expression [22], cell pluripotency, or development of diverse organs or tissues [32, 33]. Their deregulation can also impact critical pathways involved in cancer development, progression, and resistance to treatment [14]. For example, we identified one lncRNA involved in acute myeloid leukemia (AML) t(2;11) translocation and leading to overexpression of the oncogenic miR-125b [34] whose upregulation induces leukemia in mouse [35]. More recently, by a genome-wide analysis (RNA-sequencing) of lncRNA expression profiles in a cohort of 40 AML patients, we described a signature composed of 12 lncRNAs differentially expressed between mutant and wild-type (wt) NPM1 patients [36]. However, whether the deregulation of these 12 lncRNAs is just a distant consequence of NPM1 mutation without functional roles, or whether they are directly regulated by NPM1 and play a causal role in AML remains unknown.
In the present study, we went further in the characterization of these lncRNAs and found that one of them plays critical role in AML pathogenesis through NPM1-dependent regulation of its subcellular location and consequent modulation of select genes.
Methods
Cell lines
The OCI-AML3, OCI-AML2, NB4, and HL-60 cell lines obtained from the Leibniz Institute DSMZ (DSMZ no. ACC 582, ACC 99, ACC 207, and ACC3, respectively) were grown in RPMI 1640 (IMDM for HL-60) medium supplemented with 10% (v/v) fetal bovine serum (FBS), 2mM L-glutamine, 100 U/mL penicillin and 100 U/mL streptomycin (Invitrogen). The engineered OCI-AML2 + NPM1c + cell line was grown in MEMα medium, GlutaMAX™ supplement, no nucleosides (32561094; Thermo Fisher Scientific) with 20% (v/v) FBS, 100 U/mL penicillin, and 100 U/mL streptomycin [37]. All cells were maintained at 37 °C in a humidified atmosphere containing 5% CO2.
AML samples
Eighteen AML samples were collected from patients registered at the HIMIP (Hémopathies malignes de l’Inserm Midi Pyrénées) (BB-0033-00060). Frozen samples were obtained after informed consent and the study was conducted in accordance with the Declaration of Helsinki. In accordance with French law, the HIMIP collection has been declared to the Ministry of Higher Education and Research (DC 2008-307 collection 1) and a transfer agreement has been obtained (AC 2008-129) after approbation by local ethical committees (the Comité de Protection des Personnes Sud-Ouest et Outremer II and the APHP (Assistance Publique - Hôpitaux de Paris) ethical committee). Clinical and biological annotations have also been declared to the CNIL (Comité National Informatique et Libertés). The cytogenetic and gene mutational profiles of all patients, whose samples were used in functional experiments, are provided in Supplemental Table 1.
AML primary cells were thawed in IMDM supplemented with 20% FBS and 10 µg/ml of DNAse I and cultured in StemSpan SFEM medium, supplemented with 10% BIT serum substitute (STEMCELL Technologies Inc.), 1% penyciline streptomycine (Gibco), 1% L-Glutamine (Gibco) and cytokines [rmScf (100 ng/ml), rhTPO (50 ng/ml), rhFLT3-ligand (40 ng/ml), rhIL-3 (10 ng/ml), and rhIL-6 (10 ng/ml)]. All cytokines were purchased from PeproTech. All cultures were performed at 37 °C and 5% CO2.
Subcellular fractionation
For cell lines, 12 million cells were washed with cold PBS, resuspended in Buffer A (10 mM HEPES-KOH pH 7.9; 1.5 mM MgCl2; 10 mM KCl, DTT and RNAse Inhibitor added freshly), and kept on ice for 10 min. Cells were transferred into a dounce homogenizer and dounced 15 times. For primary AML cells, 5 million cells were washed with cold PBS and resuspended in 500 µl of Buffer A with 50 µg/ml of digitonin.
Cells were centrifuged at 1000 g, 4 °C, for 10 min. The supernatant containing cytoplasmic RNAs was harvested for subsequent RNA extraction. The remaining pellet containing the nuclei was washed with Buffer A to further reduce contamination by cytoplasmic RNAs and then centrifuged again at 1000 g, 4 °C, for 10 min. The supernatant was discarded and the pellet containing nuclear RNAs was harvested for subsequent RNA extraction. For determining lncRNA localization following subcellular fractionation, equal volumes of cytoplasmic and nuclear RNAs were used. RNAs were reverse transcribed using Quantitect Reaction Kit (Qiagen). GAPDH or HPRT and SNORD44 were used as positive controls for cytoplasmic and nuclear RNA fractions, respectively (primer sequences are provided in Supplemental Table 2).
Apoptosis assay
Apoptosis was induced for 48 h with Cytarabine (Ara-C, Sigma) at a final concentration of 10 µM for OCI-AML3 cells or 500 nM for OCI-AML2 cells.
Cells were harvested and stained using Pacific Blue™ Annexin V Apoptosis Detection Kit with PI (Biolegend, #640928) according to the manufacturer’s instructions. Human AML blasts were treated with increasing doses of Ara-C for 72 h harvested and stained using APC™ Annexin V Apoptosis Detection Kit with PI (Biolegend #640930) according to the manufacturer’s instructions. AnnexinV stained samples were analyzed using a MACSQuant-10 flow cytometer (Miltenyi Biotec) and further analyses were performed using FlowJo Software.
Myeloid differentiation assays
For established cell lines, myeloid differentiation was induced for 48 hours by Vitamin D3 (1α,25-Dihydroxyvitamin D3, Sigma-Aldrich) at a final concentration of 100 nM. Cells were harvested and stained using CD11b-PeCy7 antibody (Biolegend #101215).
For patient samples, human blasts were mixed with methylcellulose-based medium (MethoCult H4034 Optimum, STEMCELL Technologies Inc), according to the manufacturer’s instructions, and seeded at a concentration of 100,000 cells/ml, in duplicate. At day 14, colonies were immunophenotyped by flow cytometry (MACS Quant10) using simultaneously APC™ Annexin V Apoptosis Detection Kit (Biolegend #640930) and FITC anti-human CD33 (Biolegend #366620), PE-Cy7 anti-human CD11b (Ozyme, BLE301322), BV421 anti-human CD14 (Biolegend #367144), PE anti-human CD15(Biolegend #301906) antibodies or their respective control isotypes.
Labeled samples were analyzed using a MACSQuant-10 flow cytometer (Miltenyi Biotec) and further analyses were performed using FlowJo Software.
Intravenous transplantation of LONA over- or downregulating cells in NSG mice
Animals were used in accordance to a protocol reviewed and approved by the Institutional Animal Care and Use Committee of Région Occitanie (France). NOD/LtSz-scid/IL-2Rγchainnull (NSG) mice were purchased at Charles River Laboratories. Mice (6 weeks old males) were sublethally treated with busulfan (20 mg/kg) 24 hr before injection of leukemic cells.
Two million of OCI-AML3 (PIG empty or PIG LONA), OCI-AML2 (PIG empty, PIG LONA, shRNA EMPTY or shRNA LONA 1), or OCI-AML2 + NPM1c + (PIG empty or PIG LONA) were intravenously injected in the tail vein.
Three million OCI-AML3 cells transfected with either GapmeR Neg A or GapmeR LONA 2 were injected 24 hours post-transfection in the tail vein.
At day 10 post-transplantation, OCI-AML2 (shRNA EMPTY or shRNA LONA 1) mice were treated by daily intraperitoneal (IP) injection of 30 mg/kg AraC for 5 days. One-day post-transplantation, OCI-AML3 (GapmeR Neg A or GapmeR LONA 2) mice were treated by daily intraperitoneal (IP) injection of 30 mg/kg AraC for 4 days.
Injected mice (7 per group) were daily monitored for disease symptoms (ruffled coat, hunched back, weakness, and reduced mobility) and were humanely sacrificed upon distress appearance according to European ethic protocols.
Results
Localization of the lncRNA LONA is dependent on NPM1 mutational status
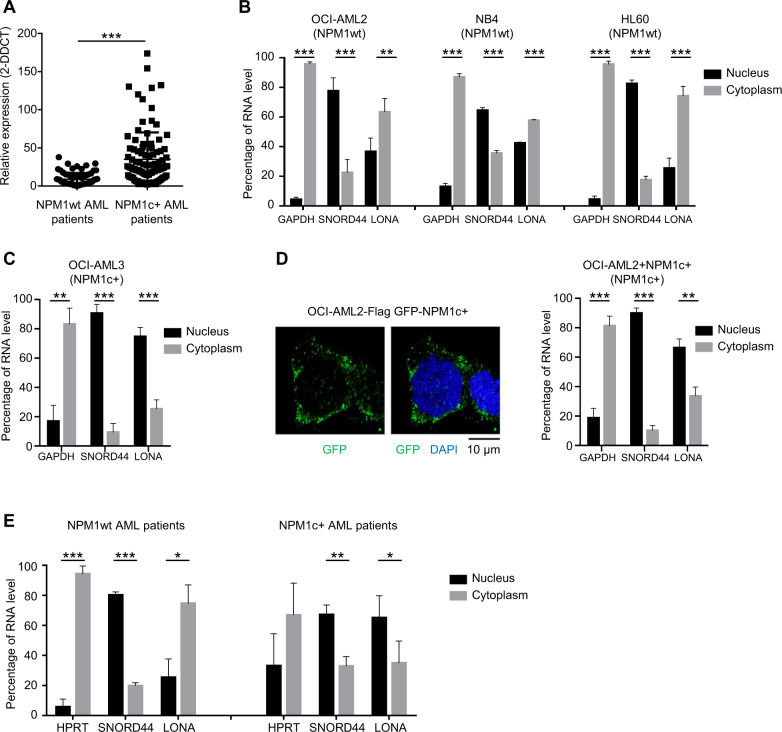
To guide us toward a lncRNA pertinent in NPM1-dependent leukemogenesis and because the pathologic behavior of mutant NPM1 is likely linked to its intracellular distribution, we first looked at the subcellular localization of lncRNAs in established AML cell lines harboring (NPM1c + ) or not (NPM1wt) a mutant NPM1 allele. Cell fractionation experiments showed that, among the 12 lncRNAs deregulated in NPM1c + patients [36], the subcellular distribution of one candidate (that we called LONA for lncRNA overexpressed in NPM1-mutated AML patients; Fig. 1A) exhibited a very intriguing pattern (Supplemental Fig. 1). LONA was mainly located in the cytoplasm of NPM1wt AML cell lines (OCI-AML2, NB4, HL-60) (Fig. 1B), but was almost exclusively detected in the nuclei of the NPM1c + cell line OCI-AML3 (Fig. 1C). To test whether LONA nuclear localization in NPM1c + cells is not just correlated to but is a direct consequence of NPM1 cytosolic translocation we used an OCI-AML2 cell line engineered to express a GFP-NPM1c + fusion protein (hereafter called OCI-AML2 + NPM1c + cells). Fluorescence visualization first confirmed that the GFP-NPM1c + fusion protein locates into the cytoplasm (Fig. 1D, left), and cell fractionation followed by RNA quantification then showed that LONA actually moves toward the nucleus as OCI-AML2 cells express the cytosolic GFP-NPM1c + fusion protein (right). Of note, the addition of NPM1c + in OCI-AML2 has no impact on LONA expression level (Supplemental Fig. 2).
Fig. 1. Subcellular localization of the LONA lncRNA is dependent on the NPM1 mutational status.
A LONA lncRNA is overexpressed in NPM1 mutated AML patients. Relative expression of the LONA lncRNA between NPM1 wild type (NPM1wt) and NPM1 mutated (NPM1c + ) AML patients. The LONA RNA level of 174 patients (80 AML patients NPM1wt and 94 AML patients NPM1c + ) were evaluated by large scale quantitative real-time PCR (Fluidigm 96.96 Dynamic Array) [36]. Results were normalized to the expression of RNAseP, 5 S rRNA, and MLN51 and presented as relative expression [2-ddCt] ± Standard Deviation (SD). ***P < 0.001. B LONA is mainly cytoplasmic in NPM1wt OCI-AML2, NB4, and HL60 cell lines, and C LONA is nuclear in NPM1c + OCI-AML3 cells. RNA level of LONA lncRNA in nuclear and cytoplasmic fractions were evaluated by RT-qPCR after cell fractionation. Results are presented as percentage of RNA level. GAPDH was used as a positive control for cytoplasmic RNA fraction and SNORD44 was used as a positive control for nuclear RNA fraction. Bar graph represents the average from four independent experiments ± Standard Deviation (SD). **P < 0.005, ***P < 0.001. Unpaired t-test. D Cytosolic localization of NPM1c + in the OCI-AML2 stably expressing Flag-GFP tagged NPM1c + (left). Cytospin cells were fixed and stained with DAPI. GFP fluorescence was examined with a confocal microscope. The mutant form of NPM1 leads to LONA nuclear delocalization (right). LONA RNA level in nuclear and cytoplasmic fractions were evaluated by RT-qPCR after cell fractionation of OCI-AML2 + NPM1c + cell line. Results are presented as percentage of RNA level. GAPDH was used as a positive control for cytoplasmic RNA fraction and SNORD44 was used as a positive control for nuclear RNA fraction. The bar graph represents the average from four independent experiments ± Standard Deviation (SD). **P < 0.005, ***P < 0.001. Unpaired t-test. E LONA is cytoplasmic in NPM1wt AML patients and mainly nuclear in NPM1c + AML patients. RNA level of LONA lncRNA in nuclear and cytoplasmic fractions were evaluated by RT-qPCR after cell fractionation. Results are presented as percentage of RNA level. HPRT was used as a positive control for cytoplasmic RNA fraction and SNORD44 was used as a positive control for nuclear RNA fraction. Bar graph represents the average from three independent experiments ± Standard Deviation (SD). **P < 0.005, ***P < 0.001. Unpaired t-test.
Next, to verify whether such nucleocytoplasmic cross-transport between mutant NPM1 and LONA exists also in patient samples, cell fractionations from either 3 NPM1wt (Fig. 1E, left) or 3 NPM1c + (right) primary AML cells were performed. The data revealed a NPM1c + -dependent nuclear localization of LONA in patient samples similar the one observed in established cell lines.
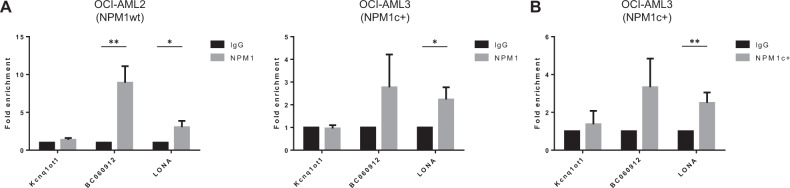
Both NPM1wt and NPM1c + bind LONA
To gain insight into the molecular mechanism that could explain the impact of NPM1 on LONA localization, we tested whether NPM1 (wt or c + ) could exist in complex with LONA. Through its carboxy-terminal nucleic-acid-binding domain, NPM1 is recognized as an RNA-binding protein interacting with diverse classes of RNAs, especially ribosomal RNAs [38] but also lncRNAs such as BC060912 [39]. As the mutation of NPM1 occurs into its carboxy-terminus, its capability of interacting with LONA (if so) could be affected. We, therefore, performed RNA immunoprecipitation (RIP) coupled to RT-qPCR experiments. The use of an NPM1 antibody which immunoprecipitates indifferently NPM1wt and NPM1c + first revealed that LONA can be co-precipitated with NPM1 in both NPM1wt OCI-AML2 (Fig. 2A, left) and NPM1c + OCI-AML3 (right) cell lines. However, these data could not inform as to whether NPM1c + has lost its capability of interacting with LONA since OCI-AML3 cells harbor only one mutated NPM1 allele and express NPM1wt from the other. We then used a validated antibody that recognizes specifically the mutant form of NPM1. LONA was equally co-precipitated with such NPM1c + -specific antibody, as is the case for the positive control BC060912 lncRNA, but not the negative control Kcnq1ot1 lncRNA. (Fig. 2B).
Fig. 2. Both NPM1wt and NPM1c + bind LONA.
A RNA Immunoprecipitation (RIP) experiments were performed in OCI-AML2 (left) and OCI-AML3 (right) cells using antibodies against NPM1wt and LONA levels were analyzed in co-precipitated RNA using RT-qPCR. The fold-enrichment of LONA in NPM1wt RIP is relative to its matching IgG control RIP. Kcnq1ot1 and BC060912 lncRNAs are used as negative and positive controls respectively. B RNA Immunoprecipitation (RIP) experiments were performed in OCI-AML3 using antibodies against NPM1c + and LONA levels were analyzed in co-precipitated RNA using RT-qPCR. The fold-enrichment of LONA in NPM1c + RIP is relative to its matching IgG control RIP. Kcnq1ot1 and BC060912 lncRNAs are used as negative and positive controls respectively. *P < 0.05, **P < 0.01. Unpaired t-test.
These data indicate that LONA exists in a complex with NPM1, independently of its mutational status.
Characterization of LONA full-length sequence
To go further into the description of LONA functional roles through molecular manipulation of its expression levels, we first performed rapid amplifications of its cDNA ends (5′ and 3′RACEs) to characterize its complete nucleotide sequence. LONA appears as an unspliced, 1992 nucleotides lncRNA located on chromosome 6 (Supplemental Fig. 3). Available online results from CAGE analysis on K562 AML cells are in agreement with the transcription start site we found by 5′ Race (Supplemental Fig. 3).
LONA downregulation enhances in vitro AML cells’ sensitivity to Ara-C-triggered apoptosis, independently of NPM1 mutational status
For gain of function approaches in established AML cells lines, the entire LONA sequence characterized above was inserted (PIG LONA) or not (PIG EMPTY) into the retroviral pMSCV-PIG vector containing a RNA polymerase II promoter driving LONA expression and a PGK promoter driving co-expression of puromycin resistance and GFP via a bicistronic mRNA [40]. Both OCI-AML3 and OCI-AML2 cells transduced with either PIG LONA or PIG EMPTY (negative control) viruses were GFP + sorted. Three independent transductions have been performed leading to a ~200-fold overexpression of LONA in OCI-AML3 cells and a ~15-fold in OCI-AML2 cells as compared to their PIG EMPTY negative control cells (Supplemental Fig. 4A, B). To check whether overexpressed LONA retains its differential subcellular localization in each cell line, a cell fractionation assay was performed. Overexpressed LONA remains mainly cytoplasmic in OCI-AML2 PIG LONA cells and predominantly nuclear in OCI-AML3 PIG LONA cells (Supplemental Fig. 4C, D).
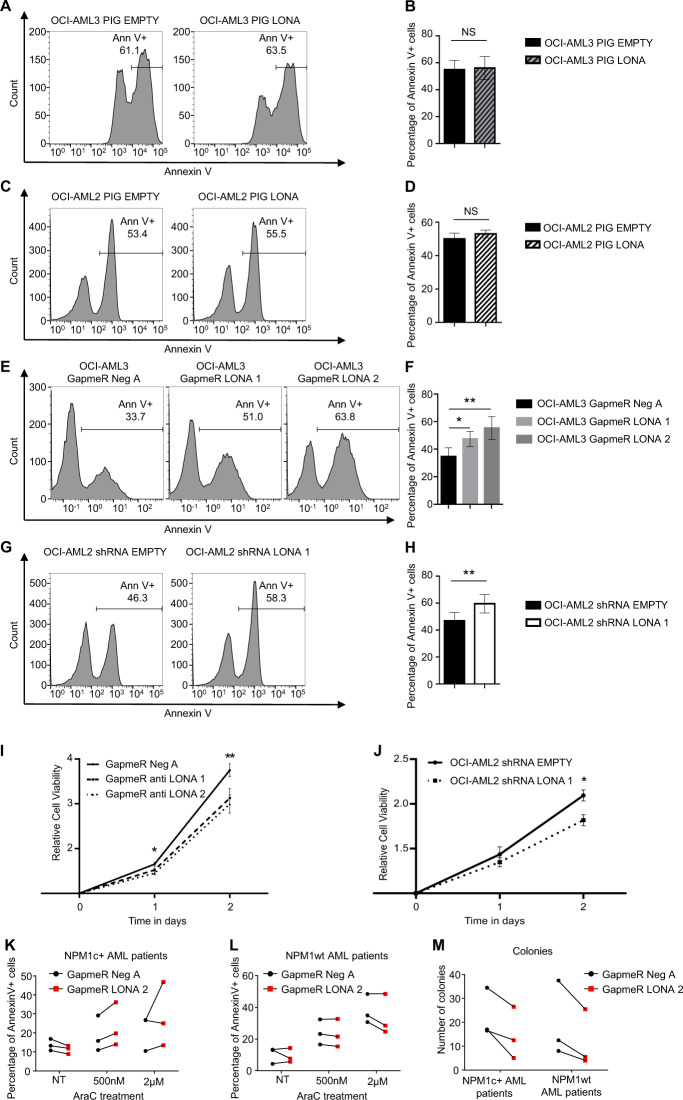
Using these models, cell death was triggered by Ara-C treatment for 48 h and the proportion of apoptotic cells was monitored by FACS analysis of annexinV stained cells. Gain of function approaches revealed no effect of LONA overexpression on both basal (not shown) and Ara-C-induced apoptosis (Fig. 3A–D), whatever the NPM1 mutational status.
Fig. 3. Downregulation of LONA decreases cell viability and enhances drug sensitivity, independently of the NPM1 mutational status.
A, B Upregulation of LONA does not influence drug sensitivity of OCI-AML3 (NPM1c + ) cell line. The OCI-AML3 (NPM1c + ) cell line was stably transduced with either the PIG EMPTY or the PIG LONA vectors and sorted for GFP + cells. GFP expression marks cells ectopically expressing LONA. Apoptosis was induced with Ara-C treatment (10 µM) and the percentage of apoptotic cells was assessed 2 days later by FACS using Annexin V staining. A One representative flow cytometry experiment is shown. B The histogram represents the average of apoptotic cells (annexin V + ) from four independent experiments ± Standard Deviation (SD). ns: no significant. Unpaired t-test. C, D Upregulation of LONA does not influence drug sensitivity of OCI-AML2 (NPM1wt) cell line. The OCI-AML2 (NPM1 wt) cell line was stably transduced with the PIG EMPTY vector or with the PIG LONA vector and sorted for GFP + cells. GFP expression marks cells ectopically expressing LONA. Apoptosis was induced with Ara-C treatment (500 nM), and the percentage of apoptotic cells was assessed 2 days later by FACS using Annexin V staining. C One representative flow cytometry experiment is shown. D The histogram represents the average of apoptotic cells (annexin V + ) from four independent experiments ± Standard Deviation (SD). Unpaired t-test. E, F Downregulation of LONA enhances drug sensitivity of OCI-AML3 (NPM1c + ) cell line. The OCI-AML3 (NPM1c + ) cell line transiently transfected with a negative control GapmeR with no target (Neg A) or with the two GapmeRs against LONA (LONA 1 or 2) were induced toward apoptosis by Ara-C treatment (10 µM) and the percentage of apoptotic cells was assessed after 2 days of treatment by annexin V staining in FACS. E One representative flow cytometry experiment is shown. F The histogram represents the average of apoptotic cells (annexin V + ) from four independent experiments ± Standard Deviation (SD). *P < 0.05, **P < 0.01. Unpaired t-test. G, H Downregulation of LONA enhances drug sensitivity of OCI-AML2 (NPM1wt) cell line. The OCI-AML2 (NPM1wt) stably transduced with shRNA EMPTY or shRNA LONA 1 were induced to apoptosis by Ara-C treatment (500 nM). Apoptosis was quantified 2 days later by flow cytometry with annexinV staining. G One representative flow cytometry experiment is shown. (H) The histogram represents the average of apoptotic cells (annexin V + ) from four independent experiments ± Standard Deviation (SD). **P < 0.01. Unpaired t-test. I, J In vitro cell viability of LONA downregulation in AML cells. I LONA downregulation decreases cell viability in OCI-AML3. The OCI-AML3 (NPM1c + ) transiently transfected with GapmeR Neg A or with GapmeRs LONA 1 or 2 were counted and seeded at 10,000 cells/well in 100 µL of complete medium. Cell viability was determined every 24 h during 3 days (J0, J1, J2) by bioluminescence using MTT reagent assay. Results are represented as relative cell viability from at least three independent experiments ± Standard error of the mean (SEM). *P < 0.05. **P < 0.01. Unpaired t-test. J LONA downregulation decreases cell viability in OCI-AML2. As above, cell viability of OCI-AML2 (NPM1wt) transduced with shRNA EMPTY or shRNA LONA 1 was determined during 3 days (J0, J1, J2) by bioluminescence using MTT reagent assay. Results are represented as relative cell viability, from three independent transductions ± Standard error of the mean (SEM). *P < 0.05. Unpaired t-test. K, L Downregulation of LONA does not influence drug sensitivity of primary AML cells. K Cells from three NPM1c + AML patients and L three NPM1wt AML patients were transiently transfected with GapmeR Neg A or with GapmeR LONA 2. One-day post transient transfection, cells were treated with increase amount of AraC. Three days later, apoptosis was assessed by FACS on fsc ssc gated live cells (see Supplemental Fig. 9) and the percentage of annexin V + cells was evaluated. Each line connects the two transfection conditions (GapmeR Neg A vs GapmeR LONA 2) for the same patient. NT, no treatment. M Downregulation of LONA reduces clonogenicity of primary AML cells. Primary cells from three NPM1c + AML patients and three NPM1wt AML patients were transiently transfected with GapmeR Neg A or GapmeR LONA 2. One day post-transfection, cells were seeded in methylcellulose media and 12 days later, the number of colonies was evaluated. Each line connects the two transfection conditions (GapmeR Neg A vs GapmeR LONA 2) for the same patient.
For loss of function approaches in established cell lines, we attempted to downregulate LONA expression by using two independent shRNAs (Supplemental Fig. 5A). Unfortunately, no effect on LONA expression could be visualized in OCI-AML3 cells. One probable explanation is that in these cells where LONA is mainly nuclear, the LONA-specific shRNAs cannot reach their target as the shRNA machinery resides in the cytoplasm (as previously shown [41]). Consistently, one of the two shRNAs did work well in OCI-AML2 cells where LONA is predominantly cytoplasmic, producing a 40% decrease in LONA expression as compared to shRNA EMPTY cells (Supplemental Fig. 6A, B). Therefore, transfections with two anti-LONA GapmeRs (1 and 2) or with a non-specific GapmeR (Neg A) whose mechanism of action involves the endogenous RNAse H and is able to target both cytoplasmic and nuclear RNAs were used as alternatives for extinguishing LONA in OCI-AML3 cells. An average of 40% and 50% decreases in LONA expression were observed following OCI-AML3 transient transfection with anti LONA GapmeRs 1 and 2, respectively, as compared to GapmeR Neg A which was used as a negative control (Supplemental Fig. 5B). However, although loss of functions approaches revealed no effect on apoptosis of unchallenged cells (not shown), an interesting Ara-C-sensitizing effect of LONA downregulation irrespective of NPM1 mutational status, was detected. Only 35% of GapmeR Neg A-transfected OCI-AML3 control cells were apoptotic (annexin V + ) following Ara-C treatment compared to 47% and 55% for anti-LONA GapmeRs 1 and 2-transfected cells (Fig. 3E, F). A similar enhancement of Ara-C-induced apoptosis was observed after shRNA-mediated downregulation of LONA in OCI-AML2 cells with only 45% of shRNA EMPTY control cells undergoing apoptosis compared to almost 60% for shRNA LONA 1 cells (Fig. 3G, H).
To test whether LONA has an impact on cell proliferation, we performed in vitro MTS assays and cell cycle analyses on both OCI-AML3 and OCI-AML2 cell lines upon LONA downregulation. Survival of both GapmeR-transfected OCI-AML3 cells and shRNA-transduced OCI-AML2 was decreased as compared to their respective control cells (Figs. 3I, J). This result is consistent with the fact that LONA downregulation enhances apoptosis of cells, independently of the NPM1 mutational status. As observed for apoptosis, no difference in survival were detected for both OCI-AML3 and OCI-AML2 cells overexpressing LONA, compared to their respective control cells (Supplemental Fig. 7A, B). Similarly, no difference in cell cycle distribution was observed in LONA overexpressing cells, although a slight but not significant increase in the proportion of G2/M cells overexpressing LONA could be detected when compared to control cells (Supplemental Fig. 7C, D).
For loss of function approaches in primary AML cells, LONA downregulation was efficiently achieved using GapmeRs transfections. An average of 80% decrease in LONA expression was observed following primary AML cells transient transfection with GapmeR LONA 2 as compared to GapmeR Neg A (Supplemental Fig. 8). Twenty-four hours after transient transfection, NPM1c + (n = 3) or NPM1wt (n = 3) primary AML cells were treated with increasing doses of AraC for 72 hours. FACS visualization (Annexin V + ) and quantification of apoptotic cells then revealed that, although an AraC cytotoxic dose-effect is seen, no significant difference is detected next to LONA downregulation in both NPM1wt and NPM1c + primary AML cells (Fig. 3K, L and Supplemental Fig. 9). We have then performed a colony formation assay to challenge the tumorigenic potential of LONA in primary AML cells. The results were much more convincing, with a significant decrease in colony formation next to LONA downregulation visible in all the 6 human primary samples (Fig. 3M).
LONA exerts opposite effects on myeloid differentiation depending on its subcellular location and NPM1 mutational status
One of the characteristics of AML cells is their blockade at an early stage of myeloid differentiation. The myeloblastic OCI-AML3 and OCI-AML2 cell lines are good models for studying myeloid differentiation because they are prone to Vitamin D3-induced monocytic differentiation. Their differentiation status can be easily appreciated by monitoring cell surface expression of the CD11b marker. We, therefore, explored the impacts of LONA gain or loss of function combined with NPM1 mutational status on myeloid differentiation by using our models.
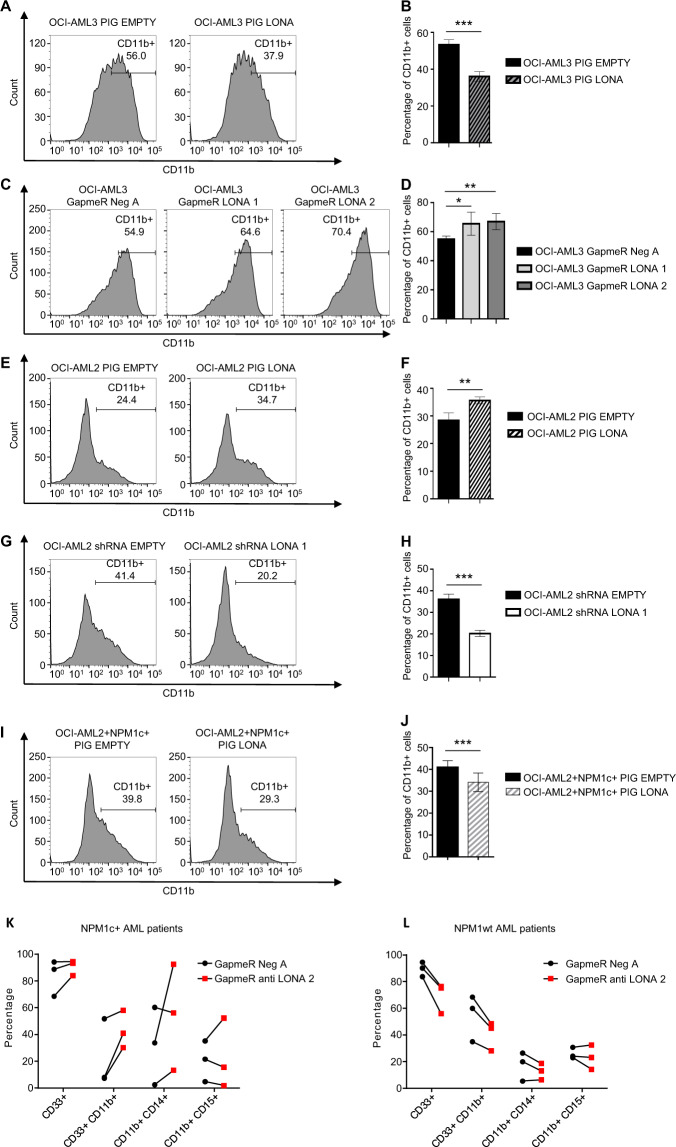
After 2 days in the presence of Vitamin D3, CD11b flow cytometry revealed a delay in myeloid differentiation of PIG-LONA OCI-AML3 cells, with less than 35% of CD11b positive cells compared to more than 55% for PIG-EMPTY control cells (Fig. 4A, B). Surprisingly, myeloid differentiation was in contrast pushed forward in PIG-LONA OCI-AML2 cells, with 36% of CD11b positive cells compared to 28% for PIG EMPTY control cells (Fig. 4E, F).
Fig. 4. Deregulation of LONA impairs myeloid differentiation, depending on the mutational status of NPM1.
A, B LONA overexpression delays myeloid differentiation of OCI-AML3 (NPM1c + ) cells. The OCI-AML3 PIG EMPTY or PIG LONA cells were induced towards myeloid differentiation by adding Vitamin D3 treatment (100 nM) and differentiation was assessed on day 2 post-induction by flow cytometry after staining with the myeloid marker CD11b. A One representative flow cytometry experiment is shown. B The bar graph represents the average of CD11b + myeloid cells from four independent experiments from three independent transductions ± Standard Deviation (SD). ***P < 0.001. Unpaired t-test. C, D LONA downregulation enhances myeloid differentiation of OCI-AML3 cells. The OCI-AML3 (NPM1c + ) cell line was transiently transfected with GapmeR Neg A or GapmeRs against LONA (LONA 1 or 2). One day after transient transfection, myeloid differentiation was induced by Vitamin D3 treatment (100 nM) and the percentage of differentiated myeloid cells was assessed 2 days later by CD11b staining by FACS. C One representative flow cytometry experiment is shown. D The bar graph represents the average of CD11b + myeloid cells from four independent experiments ± Standard Deviation (SD). *P < 0.05, **P < 0.01. Unpaired t-test. E, F LONA overexpression enhances myeloid differentiation of OCI-AML2 (NPM1wt) cells. The OCI-AML2 PIG EMPTY or PIG LONA cells were induced towards myeloid differentiation by adding Vitamin D3 treatment (100 nM) and differentiation was assessed on day 2 post-induction by flow cytometry after staining with the myeloid marker CD11b. (E) One representative flow cytometry experiment is shown. F The bar graph represents the average of CD11b + myeloid cells from four independent experiments from three independent transductions ± Standard Deviation (SD). **P < 0.01 Unpaired t-test. G, H LONA downregulation delays myeloid differentiation of OCI-AML2 cells. The OCI-AML2 (NPM1wt) cell line transduced with shRNA EMPTY or shRNA LONA 1 were induced toward myeloid differentiation upon Vitamin D3 treatment (100 nM) and the percentage of differentiated myeloid cells was assessed 2 days later by CD11b staining by FACS. G One representative flow cytometry experiment is shown. H The bar graph represents the average of CD11b + myeloid cells from four independent experiments from three independent transductions ±Standard Deviation (SD). ***P < 0.001. Unpaired t-test. I, J Addition of NPM1c + in OCI-AML2 cell line restores the phenotype observed in OCI-AML3 and leads to myeloid differentiation delays. The OCI-AML2 + NPM1c + cells were stably transduced with the PIG EMPTY or PIG LONA viruses and selected by puromycin treatment during 6 days (1 µg/ml). OCI-AML2 + NPM1c + PIG EMPTY and PIG LONA cells were induced toward myeloid differentiation by Vitamin D3 treatment (100 nM) and the percentage of differentiated myeloid cells was assessed 2 days later by CD11b staining by FACS. I One representative flow cytometry experiment is shown. J The bar graph represents the average of CD11b + myeloid cells from three independent experiments from two independent transductions ± Standard Deviation (SD). **P < 0.01 Unpaired t-test. K Primary cells from three NPM1c + AML patients and L three NPM1wt AML patients were transiently transfected with GapmeR Neg A or GapmeR LONA 2. One day post-transfection, cells were seeded in methylcellulose media with appropriate cytokines to induce myeloid differentiation. Fourteen days later, myeloid differentiation was assessed by FACS on live cells (Annexin V- cells) using CD33, CD11b, CD14, and CD15 myeloid markers.
In sharp contrast to the differentiation-arresting effect produced by LONA overexpression, LONA downregulation in OCI-AML3 cells provoked an opposite enhancement of Vitamin D3-driven myeloid differentiation, with up to ~67% of CD11b positive cells under LONA downregulation by GapmeRs LONA 1 or 2 but only ~55% for GapmeR Neg A control cells (Fig. 4C, D). The converse impact of LONA knockdown on OCI-AML2 differentiation was even more obvious. After 2 days under Vitamin D3, myeloid differentiation of OCI-AML2 cells transduced with shRNA LONA 1 was indeed severely affected with only ~19% of CD11b positive cells compared to ~36% for shRNA EMPTY control cells (Fig. 4G, H).
Thus, manipulation of LONA expression in two AML cell lines distinguishable by their endogenous NPM1 status suggests that cytosolic LONA favors myeloid differentiation in a wt NPM1 setting while nuclear LONA exerts an opposite inhibitory effect in a mutant NPM1 context. To further support a direct link between NPM1 and LONA, we then checked whether LONA-induced changes in differentiation could also be impinged by ectopic expression of a mutant NPM1 protein. To this end, OCI-AML2 + NPM1c + cells overexpressing (PIG LONA) or not (PIG EMPTY) LONA were engaged in a differentiation process. While the addition of NPM1c + to the OCI-AML2 cell line delays differentiation (Supplemental Fig. 10), this delay is even more pronounced following overexpression of LONA. Indeed, after two days under Vitamin D3, a differentiation delay similar to the one seen with PIG-LONA OCI-AML3 cells was observed (Fig. 4A, B), with less than 30% of CD11b positive PIG LONA OCI-AML2 + NPM1c + cells compared to 40% for PIG EMPTY control cells (Fig. 4I, J and supplemental Fig. 10). Thus, ectopic expression of a mutant and cytosolic NPM1 protein in wild-type cells phenocopies the response to LONA gain or loss of function observed in naturally occurring mutant NPM1 cells.
We next tested the consequences of LONA downregulation on the differentiation of NPM1c + or NPM1wt primary AML cells. Cells were transfected with either GapmeR LONA 2 or GapmeR Neg A and were seeded in methylcellulose media supplemented during 14 days with a cocktail of cytokines inducing myeloid differentiation. Cells were then FACS-monitored using the specific myeloid markers CD33, CD11b, CD14 (monocyte), and CD15 (granulocyte). NPM1c + primary AML cells transfected with GapmeR LONA 2, as compared to GapmeR Neg A, were globally more positives for myeloid markers (particularly for (CD11b) (Fig. 4K) suggesting that a LONA defect promotes myeloid differentiation. Conversely, LONA downregulation in NPM1wt primary AML cells more clearly delays myeloid differentiation, as the percentages of CD33 + and CD11b + cells become distinctly lower after transfection with GapmeR LONA 2 compared to GapmeR Neg A (Fig. 4L). Of note, LONA downregulation affected equally monocytic (CD11b + , CD14 + cells) and granulocytic (CD11b + , CD15 + ) differentiation in both NPM1wt and NPM1c + primary AML cells.
Together, these results demonstrate that cytosolic accumulation of mutant NPM1 provokes the nuclear import/sequestration of LONA and a consequent defect in myeloid differentiation.
Identification of LONA’s target genes
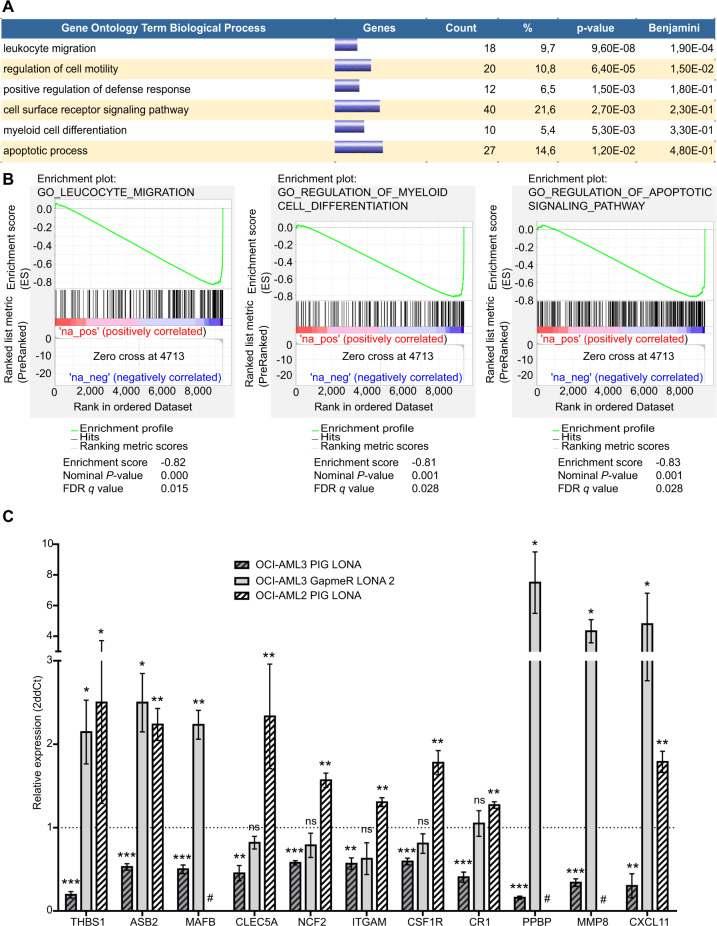
To identify genes potentially involved in the deregulations mediated by LONA, poly(A) RNA-sequencing was performed on RNA extracted from OCI-AML3 cells transduced with either PIG LONA or PIG EMPTY. Differential expression between the two cell lines was assessed using EdgeR and differentially expressed genes were selected according to the following criteria: fold change >1.5 and FDR < 0.05. Such analyses identified 34 upregulated and 172 downregulated genes in PIG LONA vs PIG EMPTY OCI-AML3 cells (supplemental Table 3). Gene ontology exploration of these differentially expressed genes using the DAVID software (Fig. 5A and supplemental Table 4) and confirmed for most of them by a GSEA analysis (Fig. 5B and supplemental Table 5) then revealed significant enrichments in pathways relevant for AML cells themselves and for interaction with their microenvironment such as “leucocyte migration”, “cell motility”, “apoptosis” or “defense response” and especially in “myeloid differentiation”. Of note, a number of deregulated genes enter also the “cell surface receptor signaling pathway” category including the well-known myeloid markers/receptors ITGAM (known as CD11b), CR1 (known as CD35), or CSF1R (known as M-CSF-R). Among the candidate genes found deregulated upon LONA overexpression we selected the most pertinent in myeloid differentiation and validated their deregulation by performing RT-qPCR on RNA extracted from OCI-AML3 and OCI-AML2 cells transduced with PIG LONA, from OCI-AML3 cells transfected with GapmeR LONA 2 and from their three respective control cells (Fig. 5C). Several of these selected genes could explain the differences observed on myeloid differentiation upon deregulation of LONA in the different models. For instance, THBS1 and ASB2 are known to promote myeloid differentiation and to inhibit growth of myeloid leukemia cells. THBS1 and ASB2 are downregulated in OCI-AML3 PIG-LONA cells (where LONA overexpression induces a blockade of myeloid differentiation in vitro) (Fig. 3A, B). At the opposite, these genes are upregulated after LONA downregulation in OCI-AML3 and in OCI-AML2 PIG-LONA (where LONA deregulation promotes myeloid differentiation (Fig. 3).
Fig. 5. LONA modulates the expression of key genes involved in myeloid differentiation.
A Gene ontology enrichment of LONA overexpression in OCI-AML3. RNA Sequencing was performed on two independent transductions of OCI-AML3 PIG EMPTY or OCI-AML3 PIG LONA. 206 genes were found differentially expressed between the 2 conditions and were analyzed using DAVID software for Gene Ontology enrichment. B GSEA plots for selected Gene Ontology gene sets, with black bars indicating gene sets represented among all genes ranked by (-log10*p-value)*ABS(logFC)*sign(LogFC) (PIG LONA versus PIG EMPTY cells). C Quantification by RT-qPCR of the candidate genes found deregulated by RNA-Seq. The mRNA expression levels of THBS1, ASB2, MAFB, CLEC5A, NCF2, ITGAM, CSF1R, CR1, PPBP, MMP8, and CXCL11 were measured in OCI-AML3 and OCI-AML2 cells infected with PIG EMPTY or PIG-LONA vectors, and in OCI-AML3 cells 2 days after transient transfection with Neg A or LONA 2 GapmeRs. The mRNA expression levels were evaluated by RT-qPCR, normalized to the expressions of TBP, MLN51 and HPRT and presented as 2-ΔΔCt ± SEM (n = 3) relative to control cells. # MAFB, PPBP, and MMP8 genes are not expressed in OCI-AML2 cell line. ns: no significant; *P < 0.05; **P < 0.01; ***P < 0.001. Unpaired t-test.
These results show that LONA is able to modulate the expression of genes involved in AML cell myeloid differentiation, survival, and interaction with its microenvironment.
LONA overexpression reduces the survival of transplanted NSG mice
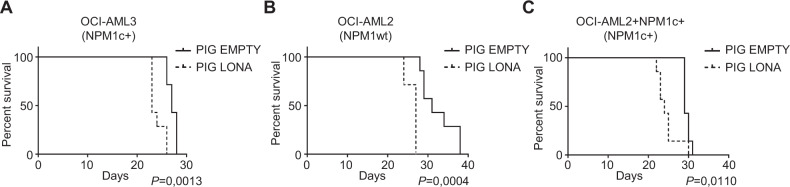
Because LONA overexpression in mutant NPM1 cells, but not in wild type cells, alters myeloid differentiation (a characteristic of AML tumor cells) and according to the RNAseq data showing an enrichment in LONA-deregulated genes involved in differentiation and in tumor microenvironment (a characteristic of AML tumors), we tested the impact of LONA overexpression on the aggressiveness of AML cells in vivo. NSG mice were first injected in the tail vein with either LONA-overexpressing (PIG LONA) or control (PIG EMPTY) NPM1c + OCI-AML3 cells. The survival of mice transplanted with LONA overexpressing cells was significantly reduced compared to that of mice transplanted with control cells (median survival 23 vs 27 days, P < 0.005) (Fig. 6A).
Fig. 6. LONA overexpression reduces survival in vivo.
Kaplan–Meier survival curves of NSG mice transplanted with (A) OCI-AML3, (B) OCI-AML2, or (C) OCI-AML2 + NPM1c + cells overexpressing LONA (PIG LONA) or control AML cells (PIG EMPTY). In total 2 × 106 cells were injected in the tail vein of NSG mice. Seven mice were injected per group. Mice were sacrificed when body weight was reduced by >20% or for signs of morbidity. Log rank test for 7 mice per group.
Next, to verify whether LONA is tumorigenic on its own or is solely dependent on the mutation of NPM1, two additional groups of NSG mice were similarly injected. One group with either LONA overexpressing or control NPM1wt OCI-AML2 cells, the other with LONA overexpressing or control OCI-AML2 + NPM1c + cells. In both groups, the survival of mice transplanted with LONA overexpressing cells was reduced compared to that of control mice with a median survival of 27 vs 31 days and 24 vs 29 days for NPM1wt OCI-AML2 and OCI-AML2 + NPM1c + cells, respectively (Fig. 6B, C).
Together, these data show that LONA is per se an oncogenic lncRNA implicated in leukemic tumorigenesis and aggressiveness.
LONA downregulation sensitizes to AraC treatment in transplanted NSG- mice
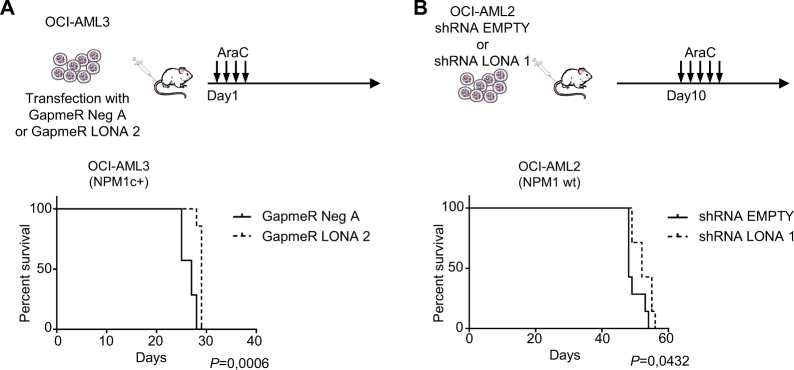
As LONA downregulation sensitizes cells to AraC treatment in vitro and is oncogenic when overexpressed, we evaluated the combination of its downregulation plus AraC treatment on the survival of NSG mice transplanted with AML cells. NSG mice were first injected in the tail vein with NPM1c + OCI-AML3 cells transfected with either GapmeR LONA 2 or GapmeR Neg A. The downregulation of LONA next to GapmeR transfection being transient, mice were treated one day after transplantation with 30 mg/kg of AraC for 4 days (Fig. 7A, top). Kaplan–Meir survival curves show that LONA downregulation in OCI-AML3 cells delays mice death (median survival 27 days for GapmeR Neg A vs 29 days for GapmeR LONA 2 mice) (Fig. 7A, bottom and Supplemental Fig. 11A). The impact of LONA downregulation was also tested in the NPM1wt context. NSG mice were similarly injected with either OCI-AML2 shRNA LONA 1 or shRNA EMPTY cells. As in these cells downregulation of LONA is persistent due to stable transduction with shRNAs, mice were left untreated for 10 days after injection and then treated with 30 mg/kg of AraC during 5 days (Fig. 7B, top). LONA downregulation in OCI-AML2 cells prolongs the survival of transplanted mice (median survival 48 days for OCI-AML2 shRNA EMPTY vs 52 days for OCI-AML2 shRNA LONA 1 mice, P < 0.05) (Fig. 7B, bottom and Supplemental Fig. 11B).
Fig. 7. LONA downregulation promotes survival in vivo upon AraC treatment.
A OCI-AML3 cells were transiently transfected with either GapmeR Neg A or GapmeR LONA 2. In total 2 × 106 cells were injected in the tail vein of NSG mice. Seven mice were injected per group. One-day post-transplantation, mice were treated with AraC (30 mg/kg) for 4 days. Mice were sacrificed when body weight was reduced by >20% or for signs of morbidity. Kaplan-Meier survival curve shows a better survival for mice transplanted with GapmeR LONA 2 OCI-AML3 cells compared to mice transplanted with control cells. Log-rank test for 7 mice per group. B 2 × 106 OCI-AML2 cells transduced with shRNA EMPTY or shRNA LONA 1 were injected in the tail vein of NSG mice. Seven mice were injected per group. Ten-day post-transplantation, mice were treated with AraC (30 mg/kg) for 5 days. Mice were sacrificed when body weight was reduced by >20% or for signs of morbidity. Kaplan–Meier survival curve shows a better survival for mice transplanted with shRNA LONA 1 OCI-AML2 cells compared to mice transplanted with control cells. Log-rank test for 7 mice per group.
Thus, despite the transient downregulation of LONA in OCI-AML3 mice and the incomplete efficacy of GapmeR and shRNA approaches, mice succumb with a slight delay after AraC treatment, suggesting that LONA downregulation may sensitize leukemic tumors to AraC chemotherapy.
Discussion
In this study, we report that both wild type and mutant NPM1 can exist in complex with the lncRNA LONA and that nuclear export of mutant NPM1 provokes nuclear import of LONA. Yet, at this stage, the possibility that NPM1 interacts with LONA through a third partner cannot be ruled out. Mechanistically, how could the NPM1 mutational status orchestrate the apparently counterintuitive cytonucleoplasmic cross-transport of LONA? One can postulate that the nucleocytoplasmic shuttling NPM1 protein is normally involved in LONA transport toward the cytosol, as is the case for ribosomal RNA [38]. One possibility then is that once synthetized into the cytoplasm, the mutant NPM1 protein devoid of its nucleolar localization signal (NoLS) to the benefit of an additional nuclear export signal (NES) does not shuttle between the cytoplasm and the nucleus as efficiently as wild type NPM1. As a consequence, mutant NPM1 can no longer go catch LONA into the nucleus thus compromising LONA nuclear export.
One important consequence of NPM1c + cytosolic relocation is that LONA switches from a cytosolic pro-myeloid differentiation to a nuclear anti-myeloid differentiation lncRNA. Our genome-wide data reveal that once into the nucleus, LONA controls the expression of a set of genes mastering myeloid differentiation. MAFB (MAF bZIP transcription factor B) is specifically expressed in the myeloid lineage and up-regulated across myeloid differentiation, from multipotent progenitors to macrophages [42]. THBS1 (Thrombospondin 1) is an adhesive matrix glycoprotein expressed by various cell types including macrophages and monocytes. THBS1 is known to inhibit cell growth and to induce myeloid differentiation of the promyelocytic AML cell line HL-60 [43]. It has also been shown that THBS1 is lowly expressed in AML patients [44]. MMP8 gene encodes the metalloproteinase 8. Its expression is significantly increased during monocyte differentiation into macrophages. MMP8 also modulates the polarization of macrophages in either M1 or M2 subtype. Monocyte-derived macrophages from MMP8-deficient mice express higher levels of M1 macrophages markers but lower levels of M2 macrophages markers than those from wild-type mice [45]. One of the most interesting candidate is ASB2 (Ankyrin repeat and SOCS Box Protein 2) which has been shown to inhibit growth and to promote myeloid differentiation of acute promyelocytic leukemia cells [46]. ASB2 is repressed by the oncogenic PML-RARα protein in acute promyelocytic leukemia cells but also in PLZF-RARα and AML1-ETO AML subtypes, suggesting that ASB2 repression in human AML may participate to the transformation process [46]. Moreover, forced expression of ASB2 in NB4 and MONMAC-6 AML cells reduces the proliferation and promotes myeloid differentiation upon ATRA treatment [47]. According to their known functions and expression patterns across myeloid differentiation, these genes are good candidates to explain LONA’s functions. However, whether they are direct targets of LONA remains to be explored. As LONA locates mostly in the nucleus of NPM1c + AML cells, one can hypothesize an implication in nuclear steps of gene expression id est epigenetic, transcriptional, or splicing events.
The overexpression of LONA does not seem to impact OCI-AML3 cell proliferation in vitro, as no differences in either cell survival or cell cycle completion were seen. However, it significantly reduces survival of transplanted NSG mice and appears involved in resistance to AraC chemotherapy in vivo. This suggests that LONA-overexpressing cells may need interactions with their microenvironment to enhance proliferation and to resist to AraC in vivo. Consistently, besides being involved in myeloid differentiation, many LONA’s target genes encode molecules critical for cell–matrix and cell–cell interactions. As a matter of fact, LONA regulates the expression of adhesion molecules such as the matrix glycoprotein THBS1, the metalloprotease MMP8 [48], or the CD11b integrin (encoded by ITGAM gene) [49]. LONA also regulates the expression of signaling molecules and chemokines such as CXCL11 [50] or PPBP that is also called CXCL7 [51].
What could be the function of LONA into the cytoplasm? Indeed, being mainly cytoplasmic in NPM1 wild-type cells, LONA could possess other functions in a physiological context. This idea is supported by the fact that some lncRNAs are multifunctional and harbor distinct roles depending on their subcellular localization. The most iconic example of a multifunctional lncRNA is HOTAIR. HOTAIR is mostly recognized for its nuclear function, regulating the recruitment of the repressive PRC2 complex on HOX genes promoters. But HOTAIR functions also in the cytoplasm where it acts as a scaffold for the recruitment of ubiquitin ligases [52]. Thus, investigating the interactome of cytoplasmic LONA could unveil new functions of this lncRNA.
Altogether, this study uncovers an original leukemic path involving a nucleocytoplasmic cross-transport between mutant NPM1 and the newly described and oncogenic long noncoding RNA LONA.
Supplementary information
SUPPLEMENTAL MATERIAL, FIGURES AND TABLES
Acknowledgements
We thank the flow cytometry, vectorology, and microscopy core facilities of U1037-CRCT for technical assistance and Anne-Marie Benot, Muriel Serthelon and Stéphanie Nevouet for their daily help about the administrative and financial management of the team. We thank Dr. V. De Mas for the management of the Biobank HIMIP. We thank all the members of the CRCT-CREFRE (Centre Régional d’Exploration Fonctionnelle et de Ressources Expérimentales) animal facility for technical assistance. We thank V. Fregona for his help with GSEA analysis. This work was supported by the Fondation ARC pour la Recherche sur le Cancer, the Association Laurette Fugain, the Ligue Régionale Contre le Cancer (Comités de la Haute-Garonne, du Gers et de l’Aude) and the LABEX TOUCAN. E.D.C. and M.G. were supported by a fellowship from LABEX TOUCAN. M.G. was supported by a fellowship from the SFH (Société Française d’Hématologie). We are grateful to our healthcare professionals for their boundless investment during the COVID-19 crisis.
Author contributions
M.B., M.G., and P.B. designed the study. E.D.C. performed PIG LONA cloning construction. W.H.C. engineered the OCI-AML2 + NPM1c + cell line and performed microscopy analysis. M.G. and M.B. performed and analyzed in vitro and in vivo experiments. M.B. analyzed the RNA sequencing data. M.B., M.G., and S.P. wrote the manuscript. M.B., P.B., S.P., H.T., C.T., and F.M. revised the content and approved the manuscript for publication. M.B., P.B., and S.P. were involved in obtaining funding.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-021-01307-0.
References
- 1.Falini B, Nicoletti I, Martelli MF, Mecucci C. Acute myeloid leukemia carrying cytoplasmic/mutated nucleophosmin (NPMc+ AML): Biologic and clinical features. Blood. 2007;109:874–85. doi: 10.1182/blood-2006-07-012252. [DOI] [PubMed] [Google Scholar]
- 2.Thiede C, Koch S, Creutzig E, Steudel C, Illmer T, Schaich M, et al. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML) Blood. 2006;107:4011–20. doi: 10.1182/blood-2005-08-3167. [DOI] [PubMed] [Google Scholar]
- 3.Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N. Engl J Med. 2005;352:254–66. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 4.Nishimura Y, Ohkubo T, Furuichi Y, Umekawa H. Tryptophans 286 and 288 in the C-terminal region of protein B23.1 are important for its nucleolar localization. Biosci Biotechnol Biochem. 2002;66:2239–42. doi: 10.1271/bbb.66.2239. [DOI] [PubMed] [Google Scholar]
- 5.Colombo E, Martinelli P, Zamponi R, Shing DC, Bonetti P, Luzi L, et al. Delocalization and destabilization of the Arf tumor suppressor by the leukemia-associated NPM mutant. Cancer Res. 2006;66:3044–50. doi: 10.1158/0008-5472.CAN-05-2378. [DOI] [PubMed] [Google Scholar]
- 6.Bonetti P, Davoli T, Sironi C, Amati B, Pelicci PG, Colombo E. Nucleophosmin and its AML-associated mutant regulate c-Myc turnover through Fbw7γ. J Cell Biol. 2008;182:19–26. doi: 10.1083/jcb.200711040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leong SM, Tan BX, Ahmad BB, Yan T, Chee LY, Ang ST, et al. Mutant nucleophosmin deregulates cell death and myeloid differentiation through excessive caspase-6 and -8 inhibition. Blood. 2010;116:3286–96. doi: 10.1182/blood-2009-12-256149. [DOI] [PubMed] [Google Scholar]
- 8.Sportoletti P. How does the NPM1 mutant induce leukemia? Pediatr Rep. 2011;3:11–13. doi: 10.4081/pr.2011.s2.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunetti L, Gundry MC, Sorcini D, Guzman AG, Huang YH, Ramabadran R, et al. Mutant NPM1 maintains the leukemic state through HOX expression. Cancer Cell. 2018;34:499–512. doi: 10.1016/j.ccell.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- 11.Lindström MS. NPM1/B23: a multifunctional chaperone in ribosome biogenesis and chromatin remodeling. Biochem Res Int. 2011;2011:195209. doi: 10.1155/2011/195209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Becker H, Marcucci G, Maharry K, Radmacher MD, Mrózek K, Margeson D, et al. Favorable prognostic impact of NPM1 mutations in older patients with cytogenetically normal de novo acute myeloid leukemia and associated gene- and microRNA-expression signatures: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:596–604. doi: 10.1200/JCO.2009.25.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibb EA, Vucic EA, Enfield KSS, Stewart GL, Lonergan KM, Kennett JY, et al. Human cancer long non-coding RNA transcriptomes. PLoS One. 2011;6:e25915. doi: 10.1371/journal.pone.0025915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–65. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kapranov P, St. Laurent G. Dark matter RNA: Existence, function, and controversy. Front Genet. 2012;3:60. doi: 10.3389/fgene.2012.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73:2491–509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cabili M, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, et al. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015;16:20. doi: 10.1186/s13059-015-0586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun S, Del Rosario BC, Szanto A, Ogawa Y, Jeon Y, Lee JT. Jpx RNA activates Xist by evicting CTCF. Cell. 2013;153:1537–51. doi: 10.1016/j.cell.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–84. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–30. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–88. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–7. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 28.Yoon J-H, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–55. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–30. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 31.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–46. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 32.Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012;13:971–83. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 34.Bousquet M, Quelen C, Rosati R, Mansat-De Mas V, La Starza R, Bastard C, et al. Myeloid cell differentiation arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. J Exp Med. 2008;205:2499–506. doi: 10.1084/jem.20080285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bousquet M, Harris MH, Zhou B, Lodish HF. MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci USA. 2010;107:21558–63. doi: 10.1073/pnas.1016611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Clara E, Gourvest M, Ma H, Vergez F, Tosolini M, Dejean S, et al. Long non-coding RNA expression profile in cytogenetically normal acute myeloid leukemia identifies a distinct signature and a new biomarker in NPM1-mutated patients. Haematologica. 2017;102:1718–26. doi: 10.3324/haematol.2017.171645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nabbouh AI, Hleihel RS, Saliba JL, Karam MM, Hamie MH, Wu HCJM, et al. Imidazoquinoxaline derivative EAPB0503: A promising drug targeting mutant nucleophosmin 1 in acute myeloid leukemia. Cancer. 2017;123:1662–73. doi: 10.1002/cncr.30515. [DOI] [PubMed] [Google Scholar]
- 38.Maggi LB, Kuchenruether M, Dadey DYA, Schwope RM, Grisendi S, Townsend RR, et al. Nucleophosmin serves as a rate-limiting nuclear export chaperone for the mammalian ribosome. Mol Cell Biol. 2008;28:7050–65. doi: 10.1128/MCB.01548-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo H, Sun Y, Wei G, Luo J, Yang X, Liu W, et al. Functional characterization of long noncoding RNA Lnc_bc060912 in human lung carcinoma cells. Biochemistry. 2015;54:2895–902. doi: 10.1021/acs.biochem.5b00259. [DOI] [PubMed] [Google Scholar]
- 40.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–84. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lennox KA, Behlke MA. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016;44:863–77. doi: 10.1093/nar/gkv1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelly LM. MafB is an inducer of monocytic differentiation. EMBO J. 2000;19:1987–97. doi: 10.1093/emboj/19.9.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bruel A, Touhami-Carrier M, Thomaidis A, Legrand C. Thrombospondin-1 (TSP-1) and TSP-1-derived heparin-binding peptides induce promyelocytic leukemia cell differentiation and apoptosis. Anticancer Res. 2005;25:757–64. [PubMed] [Google Scholar]
- 44.Zhu L, Li Q, Wang X, Liao J, Zhang W, Gao L, et al. THBS1 is a novel serum prognostic factors of acute myeloid leukemia. Front Oncol. 2020;9:1567. doi: 10.3389/fonc.2019.01567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wen G, Zhang C, Chen Q, Luong LA, Mustafa A, Ye S, et al. A novel role of matrix metalloproteinase-8 in macrophage differentiation and polarization. J Biol Chem. 2015;290:19158–72. doi: 10.1074/jbc.M114.634022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guibal FC, Moog-Lutz C, Smolewski P, Gioia YD, Darzynkiewicz Z, Lutz PG, et al. ASB-2 inhibits growth and promotes commitment in myeloid leukemia cells. J Biol Chem. 2002;277:218–24. doi: 10.1074/jbc.M108476200. [DOI] [PubMed] [Google Scholar]
- 47.Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–41. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Decock J, Hendrickx W, Thirkettle S, Gutiérrez-Fernández A, Robinson SD, Edwards DR. Pleiotropic functions of the tumor- and metastasis-suppressing matrix metalloproteinase-8 in mammary cancer in MMTV-PyMT transgenic mice. Breast Cancer Res. 2015;17:38. doi: 10.1186/s13058-015-0545-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang S, Zhang B, Fan W, Zhao Q, Yang L, Xin W, et al. Identification of prognostic genes in the acute myeloid leukemia microenvironment. Aging (Albany NY) 2019;11:10557–80. doi: 10.18632/aging.102477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hatfield K, Oyan AM, Ersvaer E, Kalland KH, Lassalle P, Gjertsen BT, et al. Primary human acute myeloid leukaemia cells increase the proliferation of microvascular endothelial cells through the release of soluble mediators. Br J Haematol. 2009;144:53–68. doi: 10.1111/j.1365-2141.2008.07411.x. [DOI] [PubMed] [Google Scholar]
- 51.Keeley EC, Mehrad B, Strieter RM. CXC chemokines in cancer angiogenesis and metastases. In: Advances in Cancer Research. Academic Press Inc., 2010; 106: 91–111. [DOI] [PMC free article] [PubMed]
- 52.Yoon J-H, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4:2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTAL MATERIAL, FIGURES AND TABLES