Abstract
Nicotine is the primary addictive agent in tobacco and P450 2A6 (gene name CYP2A6) is the primary catalyst of nicotine metabolism. It was proposed more than 20 years ago that individuals who metabolize nicotine poorly would smoke less, either fewer cigarettes per day or less intensely per cigarette, compared to smokers who metabolize nicotine more efficiently. These poor metabolizers would then be less likely to develop lung cancer due to their lower exposure to the many carcinogens delivered with nicotine in each puff of smoke. Numerous studies have reported that smokers who carry reduced activity or null CYP2A6 alleles do smoke less. Yet only in Asian populations, both Japanese and Chinese, which have a high prevalence of genetic variants has a link between CYP2A6, smoking dose, and lung cancer been established. In other ethnic groups, it has been challenging to confirm a direct link between P450 2A6–mediated nicotine metabolism and the risk of lung cancer. This challenge is due in part to the difficulty in accurately quantifying smoking dose and accurately predicting or measuring P450 2A6-mediated nicotine metabolism. Biomarkers of nicotine metabolism and smoking exposure, including the ratio of trans 3-hydroxycotine to cotinine, a measure of P450 2A6 activity and plasma cotinine or urinary total nicotine equivalents (TNE, the sum of nicotine and 6 metabolites) as measures of exposure are useful in addressing this challenge. However, to take full advantage of these biomarkers in the study of ethnic/racial differences in the risk of lung cancer requires the complete characterization of nicotine metabolism across ethnic/racial groups. Variation in metabolism pathways, other than those catalyzed by P450 2A6, can impact biomarkers of both nicotine metabolism and dose. This is clearly important for smokers with low levels of UGT2B10-catalyzed nicotine and cotinine glucuronidation, since UGT2B10 genotype influences plasma cotinine levels. Cotinine is not glucuronidated in 15% of African American smokers (compared to 1% of Whites) due to the prevalence of a UGT2B10 splice variant. This variant contributes significantly to the higher plasma cotinine levels per cigarette in this group and may also influence the accuracy of the 3HCOT to cotinine ratio as a measure of P4502A6 activity.
Table of Contents Figure
Introduction
Lung cancer deaths in the United States are estimated to be 158,080 in 2016 and the number worldwide will be more than 1.3 million.1, 2 Cigarette smoking is the cause of as much as 90% of this death toll.3 Yet despite a smoker’s overwhelming increased risk of lung cancer only 11–24% of smokers will develop the disease.3 In addition, for the same self-reported number of cigarettes per day (CPD), lung cancer risk differs across ethnic/racial groups.4 These data may lead one to conclude that susceptibility to the many lung carcinogens in each cigarette varies by ethnicity. But equally or likely more important may be differences in a smoker’s response to the nicotine present in each puff of smoke. Nicotine is essential to sustaining tobacco addiction5 and the activity of both the enzymes that metabolize it and the receptors that mediate its effects will vary across smokers.
It was proposed more than 20 years ago that individuals who metabolize nicotine poorly would smoke less, either fewer CPD or less intensely per cigarette, compared to those smokers who metabolize nicotine more efficiently.6, 7 These poor metabolizers would then be less likely to develop lung cancer due to their lower exposure to the many carcinogens delivered with nicotine in each puff of smoke. There are at least two challenges to establishing a relationship between nicotine metabolism, tobacco carcinogen exposure, and the risk of lung cancer. The first is the accurate quantitation of tobacco smoke exposure, which requires a better measure than CPD. The second is the accurate quantitation and characterization of nicotine metabolism.
This perspective summarizes nicotine metabolism, the effect of CYP2A6 genotype on smoking levels and lung cancer risk with a focus on ethnic/racial differences. The influence of ethnic/racial differences in nicotine metabolism on biomarkers of smoking exposure and on the biomarkers used to phenotype CYP2A6 is discussed. In addition, the association of CYP2A6 genotype with smoking dose is discussed with a specific emphasis on the effect of UGT2B10 genotype in African American smokers.
Urinary and plasma nicotine metabolites as biomarkers of smoking exposure
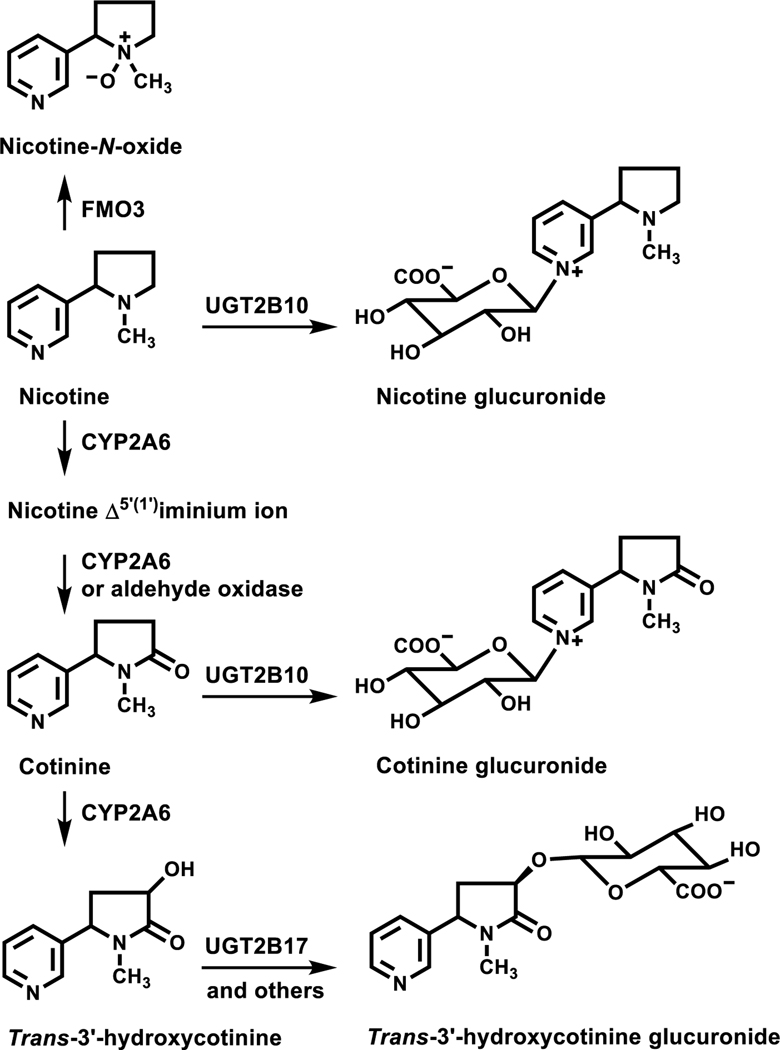
Nicotine is metabolized by three pathways; P450 2A6 catalyzed C-oxidation, UGT2B10-catalyzed N-glucuronidation, and FMO3-catalyzed N-oxidation (Figure 1).8–11 In the vast majority of smokers the primary pathway of metabolism is P450 2A6-catalyzed 5’-oxidation, which after a second oxidation generates cotinine, the predominant nicotine metabolite in smokers’ plasma. Cotinine is further metabolized to trans 3’-hydroxycotinine (3HCOT), the principal urinary metabolite of nicotine. Typically 10 to 30% of the 3HCOT excreted is an O-glucuronide conjugate.8, 9, 11 Cotinine and nicotine are excreted both free and as N-glucuronide conjugates8–10. The sum of the urinary concentrations of nicotine, cotinine, 3HCOT, nicotine N-glucuronide, cotinine N-glucuronide, 3HCOT-glucuronide, and nicotine N-oxide is referred to as total nicotine equivalents (TNE) and accounts for 85–90% of the nicotine dose.12, 13 The most abundant metabolite not included in TNE is 4-hydroxy-4-(3-pyridyl)-butanoic acid (7–9% of the nicotine dose).14 This metabolite may form by way of P450 2A6 catalyzed 2’-nicotine oxidation or by further metabolism of cotinine 15. Other minor metabolites in smokers urine include nornicotine (<1%), cotinine N-oxide (2–5%) and 5’-hydroxycotinine (~1.5%)8. Nornicotine is a product of P450 2A6-catalyzed nicotine metabolism but is also present in tobacco and tobacco smoke.16
Figure 1.
Nicotine metabolic pathways. Adapted from Murphy et al, Carcinogenesis (2014) 35
Cotinine and total cotinine (the sum of cotinine and its glucuronide conjugate) are used as biomarkers of smoking and nicotine exposure.17–19 However, variability in the urinary concentrations of cotinine and total cotinine across smokers will occur due to individual differences in metabolism of nicotine and cotinine.10, 17, 20 For many studies, TNE are a better biomarker of smoking exposure since the level will not be significantly affected by individual differences in metabolism.
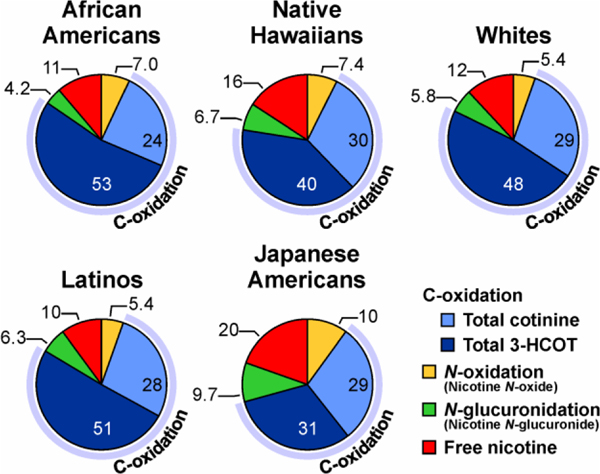
There are ethnic/racial differences in the activity of the enzymes that catalyze nicotine metabolism, and therefore the relative distribution of the metabolites excreted may vary across different populations.9 The unique pattern of urinary nicotine metabolism across five ethnic/racial groups is presented in Figure 2. These data are from a single study in which we quantified nicotine and the metabolites that make up TNE in over 2000 smokers in the Multiethnic Cohort study.9 The sections in the pie graphs represent the percentage of TNE excreted as unchanged nicotine and the products of the three nicotine metabolism pathways. In all groups, nicotine C-oxidation is by far the major pathway of metabolism, however the mean level for Japanese Americans and to a lesser extent Native Hawaiians is significantly lower. In addition, the amount of nicotine excreted unchanged is higher in these two groups compared to Whites, African Americans, and Latinos, among whom there was no significant difference in the proportion of nicotine metabolized by C-oxidation. Also of note is the lower level of nicotine N-glucuronidation in African Americans (4.2%). Characterizing and recognizing the ethnic specific difference in metabolism is critical to the correct interpretation of biomarkers of nicotine exposure and metabolism.
Figure 2.
Proportion of nicotine metabolized by C-oxidation, N-glucuronidation and N-oxidation in five ethnic/racial groups (n=2239 from a subset of the Multiethnic Cohort). The values are the molar percent of nicotine and six metabolites excreted in urine, and each slice of the pie is the mean percentage of the compound relative to TNE. From Murphy et al, Carcinogenesis (2014) 35 (11): 2526–2533. Used by permission of Oxford University Press.
The same compounds that make up the urinary TNE are present in smokers’ plasma. However, due to differences in their pharmacokinetic parameters the relative distribution is quite different. Nicotine has a relatively short half-life (100–150 min) and plasma levels are dependent on the time of the last cigarette smoked.8 Cotinine has a much longer half-life (770–1130 min) and in daily smokers plasma concentrations are relatively constant.8 Therefore, plasma cotinine is routinely used as a biomarker of tobacco exposure.17 The next most abundant metabolite is typically 3HCOT. Nicotine N-oxide, 3-HCOT glucuronide, nicotine N-glucuronide, and cotinine N-glucuronide are present in plasma, but at low levels (1–4% of the cotinine concentration).10, 21, 22 Despite the low levels of cotinine N-glucuronide in smokers’ plasma, the concentration of plasma cotinine is significantly influenced by the activity of UGT2B10, which is the sole catalyst of cotinine N-glucuronidation in smokers.10 Several years ago we reported that plasma cotinine levels were 20% higher in smokers who were heterozygous for a non-functional UGT2B10 variant, compared to those who did not carry this variant.10 In that study the majority of the smokers were White; low cotinine glucuronidation occurs much more frequently in African Americans.9, 23, 24 More recently, in a study of both African American and White smokers we observed that the mean serum cotinine concentration per TNE in smokers with no UGT2B10 activity was 50% higher than the mean level in smokers who have UGT2B10 activity. 25
P450 2A6 catalyzed nicotine metabolism
As noted above, P450 2A6 catalyzes the 5’-oxidation of nicotine. The iminium ion product of this reaction is further oxidized to cotinine, either by cytosolic aldehyde oxidase or P450 2A6.8, 26 Aldehyde oxidase is not required to form cotinine from nicotine, P450 2A6 alone may catalyze two oxidation of nicotine to generate cotinine.27 The oxidation of nicotine to the iminium ion and the subsequent oxidation of the iminium ion to cotinine by P450 2A6 may occur sequentially without the iminium ion leaving the active site of the enzyme.26 P450 2A6 also catalyzes the oxidation of cotinine to 3HCOT, but less efficiently than the 5’-oxidation of nicotine.16, 27, 28 Interestingly, in vitro the primary product of cotinine metabolism by either human liver microsomes or heterologously expressed P450 2A6 is N-(hydroxymethyl)norcotinine, which decomposes to norcotinine.28 In addition, 5’-hydroxycotinine is formed at 1.5 times the rate of 3HCOT. However, norcotinine and 5’-hydroxycotinine are minor nicotine metabolite in smokers.8 We have shown that P450 2A6 catalyzes the sequential oxidation of the nicotine Δ5’(‘1)iminium ion to 3HCOT, and that no 5’-hydroxycotinine and little norcotinine is formed by this pathway.26 Together these data have led us to suggest that a significant portion of the 3HCOT present in smokers may be generated by P450 2A6-catalyzed sequential oxidations of nicotine compared to direct metabolism of cotinine. Regardless of the pathway of 3HCOT formation, P450 2A6 is the most efficient catalyst of cotinine oxidation to 3HCOT and smokers null for the P450 2A6 gene (CYP2A6) excrete little 3HCOT.8, 29, 30 Based on these data and the relatively long half-life of cotinine compared to nicotine the plasma ratio of 3HCOT to cotinine has been used as a measure of P450 2A6.17 Likewise the ratio of total 3HCOT to total cotinine (“total” refers to the sum of the metabolite and its glucuronide conjugate), total 3HCOT/cotinine, or 3HCOT to cotinine in urine may be used to characterize P450 2A6 activity.9, 18, 31–33 Several studies have demonstrated the predicted relationship between these ratios and CYP2A6 genetic variants.31–35
UGT2B10 and nicotine and cotinine metabolism
In smokers, UGT2B10 is the catalyst of nicotine and cotinine N-glucuronidation. In vitro, heterologously expressed UGT1A4 also catalyzes the N-glucuronidation of both compounds.36, 37 However, UGT2B10 is a much more efficient catalyst of these reactions and UGT1A4 does not play a significant role in either nicotine or cotinine glucuronidation in smokers.9, 24, 38, 39 Essentially no nicotine or cotinine glucuronide conjugates are excreted by individuals who are homozygous for either of two UGT2B10 genetic variants (Table 1), the Asp67Tyr variant which has no catalytic activity and a splice variant which does not produce full length mRNA.9, 39, 40 When the genotypes for both variants are taken into account (risk score 2 in Table 1), heterozygous individuals excrete approximately 50% less cotinine and nicotine as glucuronide conjugates. The splice variant has an allele frequency of 37% in individuals of African descent, and therefore at least 14% of smokers with this ancestral background do not glucuronidate nicotine or cotinine.41 The exclusive role of UGT2B10 in the glucuronidation of cotinine and nicotine, allows one to phenotype a smoker’s UGT2B10 activity by quantifying either of these glucuronides relative to their aglycon in urine. The cotinine glucuronidation ratio is a more robust measure of UGT2B10 activity due to less variability in urinary cotinine concentrations compared to nicotine, which is more dependent on the time of last cigarette.
Table 1.
Geometric means of nicotine metabolite ratios stratified by UGT2B10 genotype9.
| UGT 2B10 Genotype | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Asp67Tyr Variant (rs6175900) | Splice variant (rs116294140) | Both variants | |||||||||||||
| ALL* | ALL* | African Americans | All | ||||||||||||
| N | means | (95% CI) | N | means | (95% CI) | N | means | (95% CI) | Genetic ScoreϮ | N | means | (95% CI) | |||
| Measures of Enzyme Activity | |||||||||||||||
| UGT2B10 | |||||||||||||||
| Nicotine glucuronide/ nicotine | GG | 2028 | 0.377 | (0.344–0.414) | AA | 1853 | 0.39 | (0.352–0.429) | 156 | 0.31 | (0.225–0.421) | 0 | 1664 | 0.45 | (0.402–0.496) |
| GT | 204 | 0.126 | (0.08–0.201) | CA | 333 | 0.23 | (0.128–0.411) | 166 | 0.19 | (0.143–0.264) | 1 | 503 | 0.19 | (0.155–0.224) | |
| TT§ | 2 | 0.00 | CC§ | 48 | 0.01 | (0.008–0.027) | 41 | 0.02 | (0.01–0.036) | 2§ | 67 | 0.00 | (0–0.001) | ||
| p-value** | <.0001 | <0.0001 | <.0001 | <0.0001 | |||||||||||
| Cotinine glucuronide/ cotinine | GG | 2031 | 1.12 | (1.07–1.17) | AA | 1857 | 1.28 | (1.22–1.38) | 157 | 1.10 | (0.97–1.24)† | 0 | 1668 | 1.38 | (1.33–1.44) |
| GT | 205 | 0.54 | (0.43–0.68) | CA | 333 | 0.63 | (0.5–0.80) | 166 | 0.48 | (0.42–0.54) | 1 | 504 | 0.68 | (0.63–0.73) | |
| TT§ | 1 | 0.01 | CC§ | 47 | 0.02 | (0.02–0.03) | 40 | 0.02 | (0.02–0.03) | 2§ | 65 | 0.01 | (0.005–0.013) | ||
| p-value** | <0.0001 | <0.0001 | <.0001 | <0.0001 | |||||||||||
| CYP2A6 | |||||||||||||||
| Total 3-HCOT/Total Cotinine | GG | 2032 | 1.38 | (1.34–1.43) | AA | 1858 | 1.33 | (1.29–1.38) | 157 | 1.65 | (1.469–1.853) | 0 | 1668 | 1.29 | (1.24–1.34) |
| GT | 205 | 1.84 | (1.56–2.18) | CA | 333 | 1.63 | (1.31–2.02) | 166 | 2.24 | (2.003–2.513) | 1 | 504 | 1.78 | (1.66–1.90) | |
| TT§ | 2 | 4.06 | (2.82–5.30) | CC§ | 48 | 3.76 | (3.03–4.67) | 41 | 4.75 | (3.794–5.952) | 2§ | 67 | 3.09 | (2.04–4.69) | |
| p-value** | 0.0011 | 0.0006 | <.0001 | <0.0001 | |||||||||||
| Total 3-HCOT/Cotinine | GG | 2031 | 3.21 | (3.10–3.32) | AA | 1857 | 3.23 | (3.11–3.35) | 157 | 3.78 | (3.35–4.26) | 0 | 1668 | 3.23 | (3.10–3.37) |
| GT | 205 | 3.35 | (2.82–3.98) | CA | 333 | 2.90 | (2.32–3.63) | 166 | 3.64 | (3.23–4.09) | 1 | 504 | 3.19 | (2.97–3.43) | |
| TT§ | 1 | 5.35 | CC§ | 47 | 3.72 | (2.97–4.66) | 40 | 4.52 | (3.57–5.72) | 2§ | 65 | 3.11 | (2.01–4.81) | ||
| p-value** | 0.631 | 0.6264 | 0.2706 | 0.9452 | |||||||||||
Incudes all 5 ethnic groups, values are adjusted for age, sex, creatinine (natural log), BMI (natural log), and race
Adjusted for age, sex, creatinine (natural log), BMI (natural log)
Genetic score 0, rs61750900 GG and rs116294140 AA; score 1, rs61750900 GT or rs116294140 CA; score 2, rs61750900 TT or rs116294140 CC, or both rs61750900 GT and rs116294140 CA
if n<10 the mean (minimum-maximum) are presented, not the geometric means and 95% Cis
FMO3 and nicotine metabolism
Nicotine N-oxidation is catalyzed by FMOs and the trans nicotine N-oxide excreted by smokers is the product of FMO3 metabolism.8 Only the trans isomer is found in smokers’ urine and human FMO3 is highly selective for the formation of this isomer. In a recent study of White smokers we reported a significant effect of FMO3 haplotype on plasma nicotine N-oxide concentration.22 Yet, by far the main contributor to nicotine N-oxide levels was CYP2A6 genotype due to its dominant effect on total nicotine metabolism in this population.22 Nicotine N-oxidation only accounts for a small percentage of hepatic nicotine metabolism but FMO3, unlike CYP2A6, is expressed in the brain and we recently measured nicotine N-oxidation activity in microsomal preparations from human brain tissue.22, 42, 43 In the same study, we reported that an FMO3 variant was associated with nicotine dependence, leading us to hypothesize that FMO3 activity in the brain affects tissue nicotine levels, which then influence a smoker’s level of addiction.22
Ethnic/racial differences in P450 2A6 and UGT2B10 activity, and nicotine metabolism and exposure biomarkers
The three primary enzymes that catalyze nicotine metabolism are polymorphic and, in the case of P450 2A6 and UGT2B10, the frequency of the variant alleles across ethnic/racial groups is quite different.31, 44 There are numerous genetic variants of CYP2A6 and several of these, including the deletion, CYP2A6*4, and CYP2A6*2, CYP2A6*7 and CYP2A6*12 produce little or no functional enzyme.44 Across ethnic/racial groups CYP2A6*4 is most prevalent in Japanese, with an allele frequency of 21%. In contrast, in people of European descent, the frequency of CYP2A6*4 is <2%, approximately the same frequency as CYP2A6*2. The CYP2A6 *7 variant is common in Asian (frequency 6–13%) but it is rare in individuals of European or African descent. 31, 44. The allele frequency of CYP2A6*12 ranges from 0.8 to 3.5% across racial/ethnic groups.31, 44. CYP2A6*9, a variant that results in decreased enzyme expression is present in all ethnic groups but its frequency varies from 6–8% in Africans and Europeans to 21% in Asian populations.31, 44. Whereas CYP2A6*17 is found in African Americans at a frequency of ~11%, but has not been found in individuals of African or Asian ancestry. 31, 44. There are fewer polymorphisms of UGT2B10, but two of these result in no functional enzyme; the Asp67Tyr variant and a splice variant, which is common in individuals of African descent.9, 39, 40 The splice variant is rare in individuals of European descent occurring at a frequency of about 0.3%. 25, 41 However, Latinos, Native Hawaiins and Japanese Americans have frequencies of 4.8 to 7.3%.9
It has been recognized for many years that smokers homozygous for CYP2A6 alleles that code for little or no functional enzyme excrete little cotinine and have lower levels of plasma cotinine for the same nicotine dose.44–48 Therefore, while plasma cotinine levels are clearly correlated with CPD,49 the plasma cotinine concentration of individual smokers will also depend on CYP2A6 genotype. This results in something of a catch 22 when trying to establish the relationship between P450 2A6 enzyme activity and smoking intensity using either plasma or urinary cotinine as a biomarker of tobacco exposure. The hypothesis is that reduced enzyme activity will result in reduced smoking and therefore lower cotinine levels, but a lack of P450 2A6 enzyme activity will also lead to lower cotinine concentrations (at the same level of nicotine exposure). The relationship between CYP2A6 genotype and cotinine is even more complicated since both the formation and the metabolism of cotinine are dependent on P450 2A6 activity. Smokers who have reduced activity CYP2A6 alleles or carry only a single null allele were found to have higher, not lower, plasma cotinine levels per TNE compared to smokers with no variant alleles.50 In contrast, individuals who were homozygous null for CYP2A6 (*4/*4) had much lower plasma cotinine per TNE.50 These data suggest that when some P450 2A6 activity is present then any variation in that activity has a greater effect on the metabolism of cotinine than on its formation. However, if a smoker has no active P450 2A6 enzyme, nicotine metabolism to cotinine will be reduced so significantly that cotinine can be easily cleared by enzymes other than P450 2A6. Therefore, among Japanese, a group with a high frequency of null or non-functional CYP2A6 alleles 31, 51, 52, plasma cotinine levels may on average be lower for the same level of smoking compared to other ethnic groups with much lower prevalence of these alleles. The use of TNE as a biomarker of nicotine dose and smoking exposure will minimize these ethnic specific difference inherent in the use of plasma cotinine.
In smokers, UGT2B10 genotype has not been shown to significantly affect plasma nicotine levels, nor is there any association with UGT2B10 genotype and TNE.20 However, as noted above plasma cotinine levels are influenced by UGT2B10 genotype. Recognizing the effect of UGT2B10 genotype on plasma cotinine levels is critical to establishing the correct relationship of smoking dose to cotinine levels. It has been long recognized that on average plasma cotinine concentrations are higher in African Americans compared to Whites with the same level of smoking or for similar environmental tobacco exposure exposure.53–55 After identifying lower levels of nicotine and cotinine glucuronidation in African Americans, Benowitz et al23 proposed that both lower cotinine glucuronidation and lower C-oxidation are likely to contribute to higher plasma cotinine in African Americans, but more recently it is often accepted that low P450 2A6 activity is responsible.50, 56 However, UGT2B10 genotype does affect plasma cotinine concentration10 and based on a 37% frequency of the UGT2B10 splice variant and 5% frequency of the Asp67Tyr variant, 16% of African Americans have no UGT2B10 activity and only 35% carry neither of these non-functional alleles.41 Therefore, it is likely that UGT2B10 genotype contributes significantly to the average higher plasma cotinine observed in African Americans relative to smokers of European descent.
The 3HCOT/cotinine ratio is a measure of CYP2A6 activity and there are some advantages to using this ratio over genotype (for example, the challenge of complete and accurate genotyping of this complex gene, and the inability of genotype to capture other influences on metabolism). The urinary total 3HCOT/cotinine or total 3HCOT/total cotinine ratios and the 3HCOT/cotinine plasma ratio do vary with CYP2A6 genotype across ethnic/racial groups as predicted.31–35 However, these ratios may also be affected by UGT2B10 genotype. Carriers of either the UGT2B10 splice variant or an Asp67Tyr variant were found to have higher total 3HCOT/total cotinine urine ratios due to decreased excretion of cotinine glucuronide by these smokers (Table 1).9 The total 3HCOT/cotinine ratio was not significantly affected by UGT2B10 genotype. In addition, smokers who do not glucuronidate cotinine (i.e., have no UGT2B10 activity) have significantly lower plasma 3HCOT/cotinine ratios due to the lower level of plasma cotinine in these individuals per TNE.25 Therefore, it is necessary that UGT2B10 genotype be taken into account when using the 3HCOT/cotinine ratio as a measure of CYP2A6 activity. This is particularly important for smokers of African descent due to the frequency of the UGT2B10 splice variant in this population.
The 3HCOT/cotinine ratio might also be influenced by genetic variants of the enzymes that glucuronidate 3HCOT, but unlike the glucuronidation of cotinine multiple UGTs catalyze this reaction.57 Among these is UGT2B17, which is polymorphic for a gene deletion. The frequency of the null UGT2B17 allele ranges from ~20% in some African populations to 90% in Japanese.58 However, despite the high frequency of the UGT2B17 null allele, the percentage of 3HCOT excreted as a glucuronide by Japanese Americans was only 30% lower than that by African American smokers.9 Therefore, the impact of the variant on the 3HCOT/cotinine ratio is expected to be minimal. In support of this conclusion, a study of over 500 African Americans smokers found that the UGT2B17 deletion did not significantly alter the plasma 3HCOT/cotinine ratio or the total 3HCOT/cotinine ratio in urine.59
Several recent studies in African Americans that investigate the effect of P450 2A6 activity (quantified by the 3HCOT/cotinine ratio) on smoking behavior have not included UGT2B10 genotype in their analysis. One pharmacokinetic study of nicotine metabolism grouped African Americans subjects into quartiles by plasma 3HCOT/cotinine ratio and found that the half-life of cotinine decreased from lowest to highest quartile. These data led the authors to conclude that higher plasma cotinine in African Americans compared to Whites is due to lower P450 2A6 activity.60 They stated that cotinine glucuronidation was not significantly correlated with the 3HCOT/cotinine ratio “suggesting no interaction” between the two. However, a significant positive correlation between the percentage of cotinine excreted as a glucuronide and the plasma 3HCOT/cotinine ratio was found for the data in the paper. That is, when the 3HCOT/cotinine ratio was low, the percentage of cotinine glucuronidated was low. Even more striking was the positive correlation observed between urinary cotinine glucuronide concentrations and the plasma 3HCOT/cotinine ratio. As the ratio decreased the amount of cotinine glucuronide excreted decreased. This is exactly the reverse of what one would expect if the ratio was only reflecting P450 2A6 activity, since as cotinine metabolism by C-oxidation decreases the amount of cotinine metabolized by N-glucuronidation would increase, not decrease. Taken together these data suggest that the 3HCOT/cotinine ratio is affected by the extent of cotinine glucuronidation. Low glucuronidation in African Americans will increase the plasma concentration of cotinine, leading to a lower 3HCOT/cotinine ratio, potentially resulting in misclassification of African American smokers for CYP2A6 activity. Some of the smokers in the lowest 3HCOT/cotinine quartile in this study may be UGT2B10 null. Data to support this conclusion is in a second publication by these authors56 and is discussed below.
In another publication looking at nicotine uptake and metabolism in African Americans the authors conclude from their data on plasma 3HCOT/cotinine ratios and the sum of cotinine and 3HCOT in plasma (as a measure of nicotine dose) that African Americans compared to Whites are “less likely to titrate their nicotine content” when they metabolize nicotine less efficiently.56 This conclusion conflicts with our data on CYP2A6 genotype and TNE in African Americans (Figure 3) and may be a misinterpretation of their data. Comparing plasma cotinine levels and the 3HCOT/cotinine ratio between African Americans and Whites and not considering UGT2B10 genotype is problematic. An unusual observation noted by the authors was that African Americans but not Whites in the lowest quartile of the 3HCOT/cotinine ratio had higher cotinine levels. The simplest explanation for this is that a significant number of the African American smokers in this quartile do not glucuronidate cotinine and therefore have higher plasma cotinine levels. The relatively higher cotinine would result in a lower 3HCOT/cotinine ratio, and some of these smokers may be misclassified as having low CYP2A6 activity. These data led us to question the conclusion of the paper, that African Americans do not alter their smoking as a function of CYP2A6 activity.
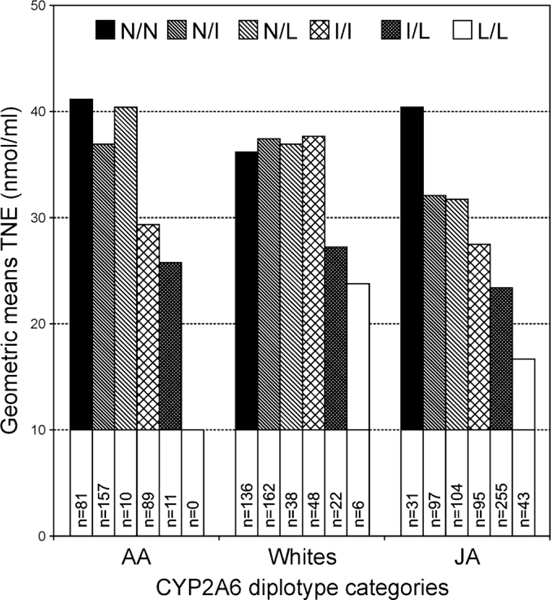
Figure 3.
TNE by CYP2A6 diplotype. Diplotypes categories are defined by the functional activity of each allele as follows: N (no variant allele or *1A +*14); I (intermediate activity), *1H, *1A, *9, *17, *23; L (little or no activity), *4, *1A+2, *1H+2, *12, *1H+*7, *7. Alleles as described http://www.cypalleles.ki.se/cyp2a6.htm. From Murphy et al, Carcinogenesis (2016) 37 (3): 269–279. Used by permission of Oxford University Press.
CYP2A6 genetic variants and smoking
In both Japanese and White populations, several studies have shown that self-reported CPD are lower for smokers who carry CYP2A6 variant alleles that code for little or no active P450 2A6 compared to smokers who carry none of these variant alleles.51, 61–65 Fujieda et al reported 2-fold lower CPD for Japanese smokers homozygous for CYP2A6*4 compared to smokers with no *4, *7, *9, *10 or *11 alleles.51 In addition, a GWAS in Japanese reported an association between the CYP2A6 deletion and CPD.65 Similarly, a GWAS of over 60,000 smokers of European descent found an association between CPD and a SNP in CYP2A6 that is linked to CYP2A6*2.64 However, in these as well as other populations, it has often not been possible to detect an effect of CYP2A6 genotype on CPD. In some populations this may be due to the low frequency of the CYP2A6 variants studied, or their modest effect on metabolism, but it is also due to the fact that CPD is a relatively crude measure of smoking dose. CYP2A6 genotype has been shown to influence smoking intensity (mean and total puff volume)66 and we have reported a significant association between TNE and CYP2A6 diplotype for African Americans, Latinos, Whites and Japanese Americans.31 That is, TNE values decreased with the predicted P450 2A6 activity for the diplotypes categories, across these ethnic/racial groups (Figure 3). Also, in a recent study of Shanghai Chinese we found a significant difference in mean TNE levels of smoker predicted by their genotype to be poor CYP2A6 metabolizers compared to predicted normal metabolizers.67 In the same study there was no significant difference in CPD across CYP2A6 predicted metabolizer groups. These data support TNE as a useful biomarker for the study of CYP2A6 and smoking dose. The use of TNE as a biomarker of smoking dose in future studies may allow a better assessment of the contribution of CYP2A6 genotype to smoking and lung cancer.
CYP2A6 genetic variants and lung cancer
An association of CYP2A6 genotype and lung cancer has been found consistently in Japanese smokers, a group with a relatively high frequency of CYP2A6 null alleles.51, 52, 68–70 However, a lack of statistical power in ethnic/racial groups with lower frequencies of CYP2A6 null alleles has made establishing an association in these groups more challenging. In addition, the robust association of genetic variation in the nicotinic receptor subunit CHRNA5 with lung cancer risk in is a potential confounding variable in some populations. The risk allele of CHRNA5, which is associated with lung cancer and smoking dose is common in smoker of European descent but rare in Asians and Africans.71, 72
The direct link between CYP2A6 and smoking dose and cancer has been established in a handful of studies in Asian populations. In Japanese smokers, studies have found a significant association between lung cancer, smoking and the CYP2A6 deletion variant, either alone or in combination with other reduced function variants, including CYP2A6 *7 and *9.51, 69. Also, the GWAS that found a significant association between the CYP2A6 deletion and CPD reported a modest association with lung cancer.65 Early studies in Chinese populations found no association between CYP2A6*4 and lung cancer, but several of these studies included non-smokers, and did not take into account other CYP2A6 variants and the lower frequency of CYP2A6*4 among Chinese compared to Japanese.73–75 Recently, we reported in a nested case control study in Shanghai Chinese that CYP2A6 genetic variants (*4,*7,*9,*1A) were associated with a reduced risk of lung cancer.67 In the same study the various diplotypes of these variants were confirmed by nicotine metabolism phenotype to confer reduced enzyme activity. The association between CYP2A6 genotype and lung cancer was no longer significant after adjusting for TNE, supporting the hypothesis that the association was the result of the relationship between nicotine metabolism and smoking dose.
In smokers of African and European ancestry studies that support a role of CYP2A6 in lung cancer risk are limited. A relatively large nested case control study in a European cohort reported a significant association between lung cancer and CYP2A6 *2, which codes for non-functional enzyme. An interaction with CPD was observed in that study.62 Another study that found an association of CYP2A6 with lung cancer in smokers of European ancestry was carried out with cases and controls selected from a GWAS that previously found an association with lung cancer and the CHRNA5-A3-B4 gene cluster.63, 76 An additive association of CYP2A6 genotype and a tag single nucleotide polymorphism in CHRNA5 with lung cancer risk and reported CPD was reported.63 An independent association with CYP2A6 genotype (*2,*4,*9,*12) was reported only in smokers of <20 CPD.63 Recently, in a study of two African American cohorts a reduced lung cancer risk was reported for carriers of reduced activity CYP2A6 alleles.77 Independently, we reported that CYP 2A6 genotype, including CYP*17 the most frequent variant in African Americans and most of the other alleles included in the lung cancer study are associated with a reduced level of TNE (Figure 3).31
Conclusions
As the primary catalyst of nicotine metabolism P450 2A6 clearly influences smoking dose and this in turn is likely the mechanism by which CYP2A6 genotype is associated with lung cancer. The use of quantitative measures of smoking dose, such as TNE, as well as improved measurement and interpretation of nicotine metabolism phenotypes should allow one to determine the contribution of CYP2A6 genotype to lung cancer risk across different ethnic/racial groups.
Acknowledgements
We thank Bob Carlson for editorial assistance.
Funding Sources
Funding for this research was provided by the National Cancer Institute (P01 CA138338).
Abbreviations
- 3HCOT
trans 3’-hydroxycotinine
- CPD
cigarettes per day
- TNE
total nicotine equivalents
Reference List
- 1.Siegel RL, Miller KD, and Jemal A. (2016) Cancer statistics, 2016. CA Cancer J Clin. 66, 7–30. [DOI] [PubMed] [Google Scholar]
- 2.Hecht SS (2012) Lung carcinogenesis by tobacco smoke. Int J Cancer. 131, 2724–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.International Agency for Research on Cancer (2004) Tobacco Smoke and Involuntary Smoking. In IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, vol. 83 pp 1179–1187, IARC, Lyon, FR. [PMC free article] [PubMed] [Google Scholar]
- 4.Haiman CA, Stram DO, Wilkens LR, Pike MC, Kolonel LN, Henderson BE, and Le Marchand L. (2006) Ethnic and racial differences in the smoking-related risk of lung cancer. N. Engl. J. Med. 354, 333–342. [DOI] [PubMed] [Google Scholar]
- 5.Benowitz NL (1999) Nicotine addiction. Prim. Care. 26, 611–631. [DOI] [PubMed] [Google Scholar]
- 6.Benowitz NL, Jacob P III, and Sachs DP (1995) Deficient C-oxidation of nicotine. Clin. Pharmacol. Ther. 57, 590–594. [DOI] [PubMed] [Google Scholar]
- 7.Tyndale RF and Sellers EM (2001) Variable CYP2A6-mediated nicotine metabolism alters smoking behavior and risk. Drug Metab Dispos. 29, 548–552. [PubMed] [Google Scholar]
- 8.Hukkanen J, Jacob P III, and Benowitz NL (2005) Metabolism and disposition kinetics of nicotine. Pharmacol. Rev. 57, 79–115. [DOI] [PubMed] [Google Scholar]
- 9.Murphy SE, Park SS, Thompson EF, Wilkens LR, Patel Y, Stram DO, and Le Marchand L. (2014) Nicotine N-glucuronidation relative to N-oxidation and C-oxidation and UGT2B10 genotype in five ethnic/racial groups. Carcinogenesis. 35, 2526–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berg JZ, von Weymarn LB, Thompson ET, Wickham KM, Weisensel NA, Hatsukami DK, and Murphy SE (2010) UGT2B10 genotype influences nicotine glucuronidation, oxidation and consumption. Cancer Epidemiology Biomarkers & Prevention. 19, 1423–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen G, Giambrone NE Jr., Dluzen DF, Muscat JE, Berg A, Gallagher CJ, and Lazarus P. (2010) Glucuronidation Genotypes and Nicotine Metabolic Phenotypes: Importance of Functional UGT2B10 and UGT2B17 Polymorphisms. Cancer Res. 70, 7543–7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scherer G, Engl J, Urban M, Gilch G, Janket D, and Riedel K. (2007) Relationship between machine-derived smoke yields and biomarkers in cigarette smokers in Germany. Regul. Toxicol. Pharmacol. 47, 171–183. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Liang Q, Mendes P, and Sarkar M. (2011) Is 24h nicotine equivalents a surrogate for smoke exposure based on its relationship with other biomarkers of exposure? Biomarkers. 16, 144–154. [DOI] [PubMed] [Google Scholar]
- 14.Hecht SS, Hatsukami DK, Bonilla LE, and Hochlater JB (1999) Quantitation of 4-oxo-4-(3-pyridyl)butanoic acid and enantiomers of 4-hydroxy-4-(3-pyridyl)butanoic acid in human urine: a substantial pathway of nicotine metabolism. Chem. Res. Toxicol. 12, 172–179. [DOI] [PubMed] [Google Scholar]
- 15.Hecht SS, Hochalter JB, Villalta PW, and Murphy SE (2000) 2’-Hydroxylation of nicotine by cytochrome P450 2A6 and human liver microsomes: Formation of a lung carcinogen precursor. Proc. Natl. Acad. Sci. 97, 12493–12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy SE, Raulinaitis V, and Brown KM (2005) Nicotine 5’-oxidation and methyl oxidation by P450 2A enzymes. Drug Metab Dispos. 33, 1166–1173. [DOI] [PubMed] [Google Scholar]
- 17.Benowitz NL, Hukkanen J, and Jacob P III (2009) Nicotine chemistry, metabolism, kinetics and biomarkers. Handb. Exp. Pharmacol. 192, 29–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Derby KS, Cuthrell K, Caberto C, Carmella SG, Franke AA, Hecht SS, Murphy SE, and Le Marchand L. (2008) Nicotine metabolism in three ethnic/racial groups with different risks of lung cancer. Cancer Epidemiology Biomarkers & Prevention. 17, 3526–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joseph AM, Hecht SS, Murphy SE, Carmella SG, Le CT, Zhang Y, Han S, and Hatsukami DK (2005) Relationships between cigarette consumption and biomarkers of tobacco toxin exposure. Cancer Epidemiology Biomarkers & Prevention. 14, 2963–2968. [DOI] [PubMed] [Google Scholar]
- 20.Patel YM, Stram DO, Wilkens LR, Park SS, Henderson BE, Le Marchand L, Haiman CA, and Murphy SE (2015) The contribution of common genetic variation to nicotine and cotinine glucuronidation in multiple ethnic/racial populations. Cancer Epidemiology Biomarkers & Prevention. 24, 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bloom AJ, von Weymarn LB, Martinez M, Bierut LJ, Goate A, and Murphy SE (2013) The contribution of common UGT2B10 and CYP2A6 alleles to variation in nicotine glucuronidation among European Americans. Pharmacogenet. Genomics. 23, 706–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teitelbaum AM, Murphy SE, Akk G, Baker TB, Germann A, von Weymarn LB, Bierut LJ, Goate A, Kharasch ED, and Bloom AJ (2016) Nicotine dependence is associated with functional variation in FMO3, an enzyme that metabolizes nicotine in the brain. Pharmacogenomics J. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benowitz NL, Perez-Stable EJ, Fong I, Modin G, Herrera B, and Jacob P III (1999) Ethnic differences in N-glucuronidation of nicotine and cotinine. Journal of Pharmacology and Experimental Therapeutics. 291, 1196–1203. [PubMed] [Google Scholar]
- 24.Berg JZ, Mason J, Boettcher AJ, Hatsukami DK, and Murphy SE (2010) Nicotine metabolism in African Americans and European Americans: variation in glucuronidation by ethnicity and UGT2B10 haplotype. Journal of Pharmacology and Experimental Therapeutics. 332, 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy SE, Sipe CK, Raddatz LM, Koopmeiners JS, Hatsukami DK, and Donny EC (2016) Low UGT2B10-catalyzed cotinine gluronidation results in higher serum cotinine in African American smokers. Cancer Epidemiol. Biomarkers & Prev. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Weymarn LB, Retzlaff C, and Murphy SE (2012) CYP2A6 and CYP2A13-catalyzed metabolism of the nicotine delta 1’(5’) iminium ion. Journal of Pharmacology and Experimental Therapeutics. 343, 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Weymarn LB, Brown KM, and Murphy SE (2006) Inactivation of CYP2A6 and CYP2A13 during nicotine metabolism. Journal of Pharmacology and Experimental Therapeutics. 316, 295–303. [DOI] [PubMed] [Google Scholar]
- 28.Brown KM, von Weymarn LB, and Murphy SE (2005) Identification of N-(hydroxymethyl)-norcotinine as a major product of cytochrome P450 2A6, but not cytochrome P450 2A13-catalyzed cotinine metabolism. Chem. Res. Toxicol. 18, 1792–1798. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X, Ameno K, Ameno S, Kinoshita H, Kubota T, Kumihashi M, Mostofa J, Iwahashi K, and Ijiri I. (2002) Effects of whole deletion of CYP2A6 on nicotine metabolism in humans. Drug Chem Toxicol. 25, 203–213. [DOI] [PubMed] [Google Scholar]
- 30.Yamanaka H, Nakajima M, Nishimura K, Yoshida R, Fukami T, Katoh M, and Yokoi T. (2004) Metabolic profile of nicotine in subjects whose CYP2A6 gene is deleted. Eur. J Pharm. Sci. 22, 419–425. [DOI] [PubMed] [Google Scholar]
- 31.Park SL, Tiirikainen MI, Patel YM, Wilkens LR, Stram DO, Le Marchand L, and Murphy SE (2016) Genetic determinants of CYP2A6 activity across racial/ethnic groups with different risks of lung cancer and effect on their smoking intensity. Carcinogenesis. 37, 269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swan GE, Lessov-Schlaggar CN, Bergen AW, He Y, Tyndale RF, and Benowitz NL (2009) Genetic and environmental influences on the ratio of 3’hydroxycotinine to cotinine in plasma and urine. Pharmacogenet. Genomics. 19, 388–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benowitz NL, Swan GE, Jacob P III, Lessov-Schlaggar CN, and Tyndale RF (2006) CYP2A6 genotype and the metabolism and disposition kinetics of nicotine. Clin. Pharmacol. Ther. 80, 457–467. [DOI] [PubMed] [Google Scholar]
- 34.Binnington MJ, Zhu AZ, Renner CC, Lanier AP, Hatsukami DK, Benowitz NL, and Tyndale RF (2012) CYP2A6 and CYP2B6 genetic variation and its association with nicotine metabolism in South Western Alaska Native people. Pharmacogenet. Genomics. 22, 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho M, Mwenifumbo JC, Al Koudsi N, Okuyemi K, Ahluwalia J, Benowitz NL, and Tyndale RF (2009) Association of nicotine metabolite ratio and CYP2A6 genotype with smoking cessation treatment in African-American light smokers. Clin. Pharmacol. Ther. 85, 635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuehl GE and Murphy SE (2003) N-Glucuronidation of Nicotine and Cotinine by human liver microsomes and Heterologously-Expressed UDP-glucuronosyltransferases. Drug Metab. Dispos. 31, 1361–1368. [DOI] [PubMed] [Google Scholar]
- 37.Nakajima M, Tanaka E, Kwon JT, and Yokoi T. (2002) Characterization of nicotine and cotinine N-glucuronidations in human liver microsomes. Drug Metab Dispos. 30, 1484–1490. [DOI] [PubMed] [Google Scholar]
- 38.Kaivosaari S, Toivonen P, Hesse LM, Koskinen M, Court MH, and Finel M. (2007) Nicotine Glucuronidation and the Human UDP-Glucuronosyltransferase UGT2B10. Mol. Pharmacol. 72, 761–768. [DOI] [PubMed] [Google Scholar]
- 39.Chen G, Blevins-Primeau AS, Dellinger RW, Muscat JE, and Lazarus P. (2007) Glucuronidation of nicotine and cotinine by UGT2B10: loss of function by the UGT2B10 Codon 67 (Asp>Tyr) polymorphism. Cancer Res. 67, 9024–9029. [DOI] [PubMed] [Google Scholar]
- 40.Fowler S, Kletzl H, Finel M, Manevski N, Schmid P, Tuerck D, Norcross RD, Hoener MC, Spleiss O, and Iglesias VA (2015) A UGT2B10 splicing polymorphism common in african populations may greatly increase drug exposure. Journal of Pharmacology and Experimental Therapeutics. 352, 358–367. [DOI] [PubMed] [Google Scholar]
- 41.NHLBI Exome Sequencing Project (ESP). (2016) Exome Variant Server. NHLBI Exome Sequencing Project (ESP) http://evs.gs.washington.edu/EVS/. [Google Scholar]
- 42.Zhang J and Cashman JR (2006) Quantitative analysis of FMO gene mRNA levels in human tissues. Drug Metab Dispos. 34, 19–26. [DOI] [PubMed] [Google Scholar]
- 43.Ferguson CS and Tyndale RF (2011) Cytochrome P450 enzymes in the brain: emerging evidence of biological significance. Trends Pharmacol. Sci. 32, 708–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mwenifumbo JC and Tyndale RF (2007) Genetic variability in CYP2A6 and the pharmacokinetics of nicotine. Pharmacogenomics. 8, 1385–1402. [DOI] [PubMed] [Google Scholar]
- 45.Nakajima M, Yamagishi S, Yamamoto H, Yamamoto T, Kuroiwa Y, and Yokoi T. (2000) Deficient cotinine formation from nicotine is attributed to the whole deletion of the CYP2A6 gene in humans. Clin Pharmacol. Ther. 67, 57–69. [DOI] [PubMed] [Google Scholar]
- 46.Benowitz NL, Griffin C, and Tyndale R. (2001) Deficient C-oxidation of nicotine continued. Clin. Pharmacol. Ther. 70, 567. [DOI] [PubMed] [Google Scholar]
- 47.Kitagawa K, Kunugita N, Katoh T, Yang MH, and Kawamoto T. (1999) The significance of the homozygous CYP2A6 deletion on nicotine metabolism: A new genotyping method of CYP2A6 using a single PCR-RFLP. Biochem. Biophys. Res. Commun. 262, 146–151. [DOI] [PubMed] [Google Scholar]
- 48.Bloom J, Hinrichs AL, Wang JC, von Weymarn LB, Bierut LJ, Goate A, and Murphy SE (2011) The Contribution of Common CYP2A6 Alleles to Variation in Nicotine Metabolism Among European Americans. Pharmacogenet. Genomics. 21, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benowitz NL (1994) Biomarkers of cigarette smoking. In Smoking and Tobacco Control Monograph No. 7. The FTC Cigarette Test Method for Determining, Tar, Nicotine and Carbon Monoxide Yeilds of U.S. Cigarettes. pp 93–111, U.S. Department of Health and Human Services, National Institutes of Health, National Cancer Institute, Bethesda, MD. [Google Scholar]
- 50.Zhu AZ, Renner CC, Hatsukami DK, Swan GE, Lerman C, Benowitz NL, and Tyndale RF (2013) The ability of plasma cotinine to predict nicotine and carcinogen exposure is altered by differences in CYP2A6: the influence of genetics, race, and sex. Cancer Epidemiology Biomarkers & Prevention. 22, 708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fujieda M, Yamazaki H, Saito T, Kiyotani K, Gyamfi MA, Sakurai M, Dosaka-Akita H, Sawamura Y, Yokota J, Kunitoh H, and Kamataki T. (2004) Evaluation of CYP2A6 genetic polymorphisms as determinants of smoking behavior and tobacco-related lung cancer risk in male Japanese smokers. Carcinogenesis. 25, 2451–2458. [DOI] [PubMed] [Google Scholar]
- 52.Hosono H, Kumondai M, Arai T, Sugimura H, Sasaki T, Hirasawa N, and Hiratsuka M. (2015) CYP2A6 genetic polymorphism is associated with decreased susceptibility to squamous cell lung cancer in Japanese smokers. Drug Metab Pharmacokinet. 30, 263–268. [DOI] [PubMed] [Google Scholar]
- 53.Perez-Stable EJ, Herrera B, Jacob P III, and Benowitz NL (1998) Nicotine metabolism and intake in black and white smokers. JAMA : the journal of the American Medical Association. 280, 152–156. [DOI] [PubMed] [Google Scholar]
- 54.Pirkle JL, Bernert JT, Caudill SP, Sosnoff CS, and Pechacek TF (2006) Trends in the exposure of nonsmokers in the U.S. population to secondhand smoke: 1988–2002. Environ. Health Perspect. 114, 853–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benowitz NL, Bernert JT, Caraballo RS, Holiday DB, and Wang J. (2009) Optimal serum cotinine levels for distinguishing cigarette smokers and nonsmokers within different racial/ethnic groups in the United States between 1999 and 2004. Am. J. Epidemiol. 169, 236–248. [DOI] [PubMed] [Google Scholar]
- 56.Ross KC, Gubner NR, Tyndale RF, Hawk LW Jr., Lerman C, George TP, Cinciripini P, Schnoll RA, and Benowitz NL (2016) Racial differences in the relationship between rate of nicotine metabolism and nicotine intake from cigarette smoking. Pharmacol. Biochem. Behav. 148, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen G, Giambrone NE, and Lazarus P. (2012) Glucuronidation of trans-3’-hydroxycotinine by UGT2B17 and UGT2B10. Pharmacogenet. Genomics. 22, 183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xue Y, Sun D, Daly A, Yang F, Zhou X, Zhao M, Huang N, Zerjal T, Lee C, Carter NP, Hurles ME, and Tyler-Smith C. (2008) Adaptive evolution of UGT2B17 copy-number variation. Am. J Hum. Genet. 83, 337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu AZ, Zhou Q, Cox LS, Ahluwalia JS, Benowitz NL, and Tyndale RF (2013) Variation in trans-3’-hydroxycotinine glucuronidation does not alter the nicotine metabolite ratio or nicotine intake. PLoS One. 8, e70938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benowitz NL, St Helen G, Dempsey DA, Jacob P III, and Tyndale RF (2016) Disposition kinetics and metabolism of nicotine and cotinine in African American smokers: impact of CYP2A6 genetic variation and enzymatic activity. Pharmacogenet. Genomics. 26, 340–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malaiyandi V, Sellers EM, and Tyndale RF (2005) Implications of CYP2A6 genetic variation for smoking behaviors and nicotine dependence. Clin. Pharmacol. Ther. 77, 145–158. [DOI] [PubMed] [Google Scholar]
- 62.Rotunno M, Yu K, Lubin JH, Consonni D, Pesatori AC, Goldstein AM, Goldin LR, Wacholder S, Welch R, Burdette L, Chanock SJ, Bertazzi PA, Tucker MA, Caporaso NE, Chatterjee N, Bergen AW, and Landi MT (2009) Phase I metabolic genes and risk of lung cancer: multiple polymorphisms and mRNA expression. PLoS One. 4, e5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wassenaar CA, Dong Q, Wei Q, Amos CI, Spitz MR, and Tyndale RF (2011) Relationship between CYP2A6 and CHRNA5-CHRNA3-CHRNB4 variation and smoking behaviors and lung cancer risk. J. Natl. Cancer Inst. 103, 1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, Sulem P, Rafnar T, Esko T, Walter S, Gieger C, Rawal R, Mangino M, Prokopenko I, Magi R, Keskitalo K, Gudjonsdottir IH, Gretarsdottir S, Stefansson H, Thompson JR, Aulchenko YS, Nelis M, Aben KK, den Heijer M, Dirksen A, Ashraf H, Soranzo N, Valdes AM, Steves C, Uitterlinden AG, Hofman A, Tonjes A, Kovacs P, Hottenga JJ, Willemsen G, Vogelzangs N, Doring A, Dahmen N, Nitz B, Pergadia ML, Saez B, De DV, Lezcano V, Garcia-Prats MD, Ripatti S, Perola M, Kettunen J, Hartikainen AL, Pouta A, Laitinen J, Isohanni M, Huei-Yi S, Allen M, Krestyaninova M, Hall AS, Jones GT, van Rij AM, Mueller T, Dieplinger B, Haltmayer M, Jonsson S, Matthiasson SE, Oskarsson H, Tyrfingsson T, Kiemeney LA, Mayordomo JI, Lindholt JS, Pedersen JH, Franklin WA, Wolf H, Montgomery GW, Heath AC, Martin NG, Madden PA, Giegling I, Rujescu D, Jarvelin MR, Salomaa V, Stumvoll M, Spector TD, Wichmann HE, Metspalu A, Samani NJ, Penninx BW, Oostra BA, Boomsma DI, Tiemeier H, van Duijn CM, Kaprio J, Gulcher JR, McCarthy MI, Peltonen L, Thorsteinsdottir U, and Stefansson K. (2010) Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat. Genet. 42, 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kumasaka N, Aoki M, Okada Y, Takahashi A, Ozaki K, Mushiroda T, Hirota T, Tamari M, Tanaka T, Nakamura Y, Kamatani N, and Kubo M. (2012) Haplotypes with copy number and single nucleotide polymorphisms in CYP2A6 locus are associated with smoking quantity in a Japanese population. PLoS One. 7, e44507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Strasser AA, Malaiyandi V, Hoffmann E, Tyndale RF, and Lerman C. (2007) An association of CYP2A6 genotype and smoking topography. Nicotine Tob Res. 9, 511–518. [DOI] [PubMed] [Google Scholar]
- 67.Yuan JM, Nelson HH, Butler LM, Carmella SG, Wang R, Kuriger-Laber JK, Adams-Haduch J, Hecht SS, Gao YT, and Murphy SE (2016) Genetic determinants of cytochrome P450 2A6 activity and biomarkers of tobacco smoke exposure in relation to risk of lung cancer development in the Shanghai cohort study. Int. J. Cancer. 138, 2161–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miyamoto M, Umetsu Y, Dosaka-Akita H, Sawamura Y, Yokota J, Kunitoh H, Nemoto N, Sato K, Ariyoshi N, and Kamataki T. (1999) CYP2A6 gene deletion reduces susceptibility to lung cancer. Biochem. Biophys. Res. Commun. 261, 658–660. [DOI] [PubMed] [Google Scholar]
- 69.Ariyoshi N, Miyamoto M, Umetsu Y, Kunitoh H, Dosaka-Akita H, Sawamura Y, Yokota J, Nemoto N, Sato K, and Kamataki T. (2002) Genetic polymorphism of CYP2A6 gene and tobacco-induced lung cancer risk in male smokers. Cancer Epidemiology Biomarkers & Prevention. 11, 890–894. [PubMed] [Google Scholar]
- 70.Liu YL, Xu Y, Li F, Chen H, and Guo SL (2013) CYP2A6 deletion polymorphism is associated with decreased susceptibility of lung cancer in Asian smokers: a meta-analysis. Tumour. Biol. 34, 2651–2657. [DOI] [PubMed] [Google Scholar]
- 71.Chen LS, Saccone NL, Culverhouse RC, Bracci PM, Chen CH, Dueker N, Han Y, Huang H, Jin G, Kohno T, Ma JZ, Przybeck TR, Sanders AR, Smith JA, Sung YJ, Wenzlaff AS, Wu C, Yoon D, Chen YT, Cheng YC, Cho YS, David SP, Duan J, Eaton CB, Furberg H, Goate AM, Gu D, Hansen HM, Hartz S, Hu Z, Kim YJ, Kittner SJ, Levinson DF, Mosley TH, Payne TJ, Rao DC, Rice JP, Rice TK, Schwantes-An TH, Shete SS, Shi J, Spitz MR, Sun YV, Tsai FJ, Wang JC, Wrensch MR, Xian H, Gejman PV, He J, Hunt SC, Kardia SL, Li MD, Lin D, Mitchell BD, Park T, Schwartz AG, Shen H, Wiencke JK, Wu JY, Yokota J, Amos CI, and Bierut LJ (2012) Smoking and genetic risk variation across populations of European, Asian, and African American ancestry--a meta-analysis of chromosome 15q25. Genet. Epidemiol. 36, 340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Le Marchand L, Derby KS, Murphy SE, Hecht SS, Hatsukami D, Carmella SG, Tiirikainen M, and Wang H. (2008) Smokers with the CHRNA lung cancer-associated variants are exposed to higher levels of nicotine equivalents and a carcinogenic tobacco-specific nitrosamine. Cancer Res. 68, 9137–9140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang H, Tan W, Hao B, Miao X, Zhou G, He F, and Lin D. (2003) Substantial reduction in risk of lung adenocarcinoma associated with genetic polymorphism in CYP2A13, the most active cytochrome P450 for the metabolic activation of tobacco-specific carcinogen NNK. Cancer Res. 63, 8057–8061. [PubMed] [Google Scholar]
- 74.Tan W, Chen GF, Xing DY, Song CY, Kadlubar FF, and Lin DX (2001) Frequency of CYP2A6 gene deletion and its relation to risk of lung and esophageal cancer in the Chinese population. Int. J. Cancer. 95, 96–101. [DOI] [PubMed] [Google Scholar]
- 75.Gu Y, Zhang S, Lai B, Zhan X, and Zhang Y. (2005) [Frequency of CYP2A6 gene deletion and its relation to risk of lung cancer]. Zhongguo Fei. Ai. Za Zhi. 8, 297–299. [DOI] [PubMed] [Google Scholar]
- 76.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, Sullivan K, Matakidou A, Wang Y, Mills G, Doheny K, Tsai YY, Chen WV, Shete S, Spitz MR, and Houlston RS (2008) Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet. 40, 616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wassenaar CA, Ye Y, Cai Q, Aldrich MC, Knight J, Spitz MR, Wu X, Blot WJ, and Tyndale RF (2015) CYP2A6 Reduced Activity Gene Variants Confer Reduction in Lung Cancer Risk in African American Smokers - Findings from Two Independent Populations. Carcinogenesis. 36, 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]