Abstract
The WNT signaling system governs critical processes during embryonic development and tissue homeostasis, and its dysfunction can lead to cancer. Details concerning selectivity and differences in relative binding affinities of 19 mammalian WNTs to the cysteine-rich domain (CRD) of their receptors—the ten mammalian Frizzleds (FZDs)—remain unclear. Here, we used eGFP-tagged mouse WNT-3A for a systematic analysis of WNT interaction with every human FZD paralogue in HEK293A cells. Employing HiBiT-tagged full-length FZDs, we studied eGFP-WNT-3A binding kinetics, saturation binding, and competition binding with commercially available WNTs in live HEK293A cells using a NanoBiT/BRET-based assay. Further, we generated receptor chimeras to dissect the contribution of the transmembrane core to WNT-CRD binding. Our data pinpoint distinct WNT-FZD selectivity and shed light on the complex WNT-FZD binding mechanism. The methodological development described herein reveals yet unappreciated details of the complexity of WNT signaling and WNT-FZD interactions, providing further details with respect to WNT-FZD selectivity.
Keywords: WNT, Frizzled, bioluminescence resonance energy transfer (BRET), NanoBiT/BRET, ligand binding, G protein-coupled receptor (GPCR)
The ten mammalian Frizzleds (FZD1–10) are G protein-coupled receptors (GPCRs) and form—together with Smoothened (SMO)—the class F of GPCRs.1,2 The 19 different WNT lipoglycoproteins are the main macromolecular ligands of FZDs, interacting with the extracellular cysteine-rich domain (CRD) of the receptor. WNT-FZD signaling orchestrates multiple processes during embryonic development, stem cell regulation, and adult tissue homeostasis.2 Additionally, aberrant WNT signaling is implicated in tumorigenesis and other pathologies.3,4 Whereas recent advances have resulted in a better understanding of the underlying mechanisms controlling WNT-induced FZD activation and signal initiation, the relative binding affinities and ligand–receptor selectivity remain largely unknown.5−13 The quantitative assessment of WNT binding has been limited by the strong lipophilicity of WNTs, which makes their purification challenging and necessitates detergents and serum for solubilization and stabilization of WNTs, respectively.14,15 Nevertheless, WNT-FZD interactions were studied using biochemical and biophysical assays as well as through the use of in silico calculations.16−22 These studies generally reported WNT-FZD binding affinities in the range of 1–100 nM, which is reasonable when considering the known affinities of proteinaceous ligands to other GPCRs.23,24 Nevertheless, it remains unclear how these values translate into the physiological reality, since the local concentration of WNTs at the receptors in vivo remains unknown and is likely to be highly context-dependent.25 Until recently, the assessment of ligand binding was based on WNT binding to the CRD rather than the full-length FZD, or the reported assays were not performed in live cells. However, progress has been made with the use of FRET- and cpGFP-based biosensors to demonstrate WNT-induced FZD conformational dynamics and receptor activation,7,12,26 where WNT binding to a full-length receptor was reflected by an outward movement of the transmembrane domain TM6. Furthermore, the generation of a functional eGFP-tagged WNT-3A provided for the first time a biologically relevant FZD ligand that could be used as a probe in quantitative binding assays in real-time using living cells.27,28
Here, using live cell analysis of transiently transfected HEK293A cells overexpressing HiBiT-tagged FZDs, we provide a comparative assessment of binding affinities of eGFP-WNT-3A to all human FZD paralogues using kinetic and saturation binding formats. Furthermore, using a competition binding assay, we have assessed binding affinities of unlabeled, commercially available WNT proteins to FZD4. Finally, we have also explored the contribution of the FZD transmembrane core for the binding of WNTs to the primary FZD-CRD binding site.29 Compared with the previously described BRET-based assay for Nluc-FZD4 and Nluc-FZD6,28 we have used a nanoluciferase complementation-based BRET binding approach here, termed NanoBiT/BRET. In this BRET assay format, the fluorescent WNT-3A binds FZDs that are N-terminally tagged with the 11-amino-acid HiBiT peptide. The addition of the complementary LgBiT to the system allows rapid and high-affinity association to the HiBiT peptide, forming a stable NanoBiT moiety with a luciferase activity. This setup allows targeted analysis of cell surface receptors due to the cell impermeability of LgBiT, thereby providing a system with less intracellular background luminescence. This method is a modification of a well-established NanoBRET binding assay to study ligand–receptor association,30 and has been lately employed to study ligand binding to Class A GPCRs and receptor tyrosine kinases.31−34 Our results demonstrate that eGFP-WNT-3A interacts with full-length human FZDs transiently overexpressed in live HEK293A cells in a paralogue selective manner. This concept was expanded to unlabeled WNTs in competition binding experiments suggesting a complex WNT-FZD selectivity profile. The binding data based on full-length FZDs, FZD-CD86, and FZD-FZD chimeras underline the complexity of the WNT-FZD interaction and suggest that the core regions of FZDs may contribute to receptor selectivity.
Results and Discussion
eGFP-WNT-3A/FZD Binding Kinetics
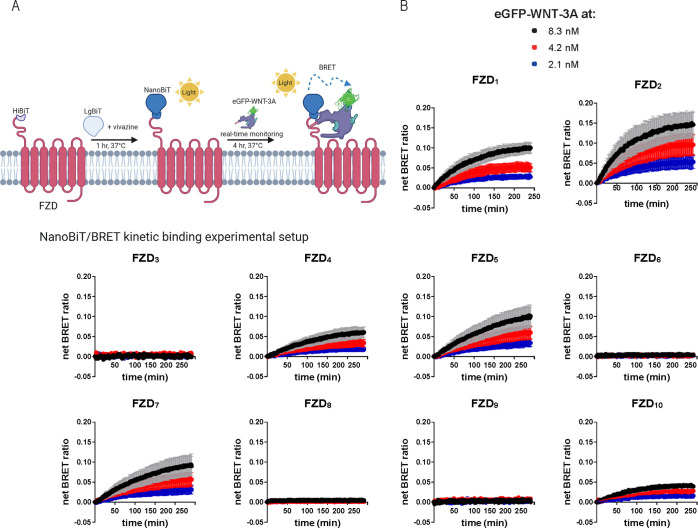
In order to establish a nanoluciferase complementation-dependent NanoBiT/BRET binding assay format to study all human FZD paralogues, we generated constructs for all 10 receptors carrying an N-terminal HiBiT tag (Figure S1A,B). Upon transient overexpression in HEK293A cells, all receptor constructs were detected at the cell surface, albeit with varying cell surface expression levels (Figure S1C). Additionally, HiBiT-tagged FZD1, FZD2, FZD4, FZD5, FZD7, FZD8, and FZD10 mediated WNT-3A-induced β-catenin-dependent signals as assessed by the TOPFlash reporter assay performed in HEK293T cells devoid of endogenous FZDs (ΔFZD1–10 HEK293T cells, Figure S1D(25)). In contrast, FZD3, FZD6, and FZD9 could not transduce WNT-3A-induced activation of this pathway, similar to what has been reported previously, yet with differing results for FZD9.28,35−38 Having verified the sequence and functionality of the HiBiT-tagged FZD constructs, HEK293A cells transiently overexpressing these FZDs were used in kinetic binding experiments. For these experiments, cells were first incubated with the complementary LgBiT protein and the luciferase substrate vivazine, and after 1 h, eGFP-WNT-3A was added to final concentrations of 2.1, 4.2, or 8.3 nM, and BRET readings were taken over a 4 h period at 37 °C (Figure 1A). The concentrations used for eGFP-WNT-3A were dictated by the maximal concentration that could be obtained for the eGFP-WNT-3A preparations and the assay format. We detected a saturable net BRET ratio indicative of eGFP-WNT-3A specific binding to HiBiT-tagged FZD1, FZD2, FZD4, FZD5, FZD7, and FZD10, with Kd values varying from 2.3 to 29.9 nM (Figure 1B, Table 1). In line with the TOPFlash data, no concentration-dependent increase in receptor–ligand BRET was detected for FZD3 and FZD9. Interestingly, FZD8, which maintained a strong WNT-3A-induced TOPFlash activity, and to a lesser extent FZD6, displayed very low but detectable binding that could be fitted to association curves over time (Figure S2A).
Figure 1.
eGFP-WNT-3A binding kinetics. A. The scheme depicts the experimental setup of the NanoBiT/BRET analysis of association kinetics between the HiBiT-tagged FZD and the eGFP-WNT-3A. Created with BioRender.com. B. Association kinetics of the eGFP-WNT-3A to human HiBiT-FZDs were determined by the detection of NanoBiT/BRET in transiently overexpressing living HEK293A cells over time. BRET was sampled once per 90 s for 240 min. Data points are presented as means ± SEM from n = 3 individual experiments, fitting a two or more hot ligand concentrations kinetics model. Experiments were performed with eGFP-WNT-3A batch 1.
Table 1. Kinetic and Saturation Binding Parameters of eGFP-WNT-3A Binding to All 10 Human HiBiT-Tagged FZD Paraloguesa.
| FZD1 | FZD2 | FZD3 | FZD4 | FZD5 | FZD6 | FZD7 | FZD8 | FZD9 | FZD10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Kinetic binding Kd (nM) ± SEM | 29.9 ± 1.5 | 5.4 ± 0.1 | n.d. | 9.4 ± 0.5 | 2.3 ± 0.2 | 10.2 ± 3.7 | 2.8 ± 0.2 | 17.8 ± 4.4 | n.d. | 4.3 ± 0.3 |
| Saturation binding Kd (nM) ± SEM | 36.7 ± 12.7 | 48.6 ± 8.2 | n.d. | 17.7 ± 7.2 | 14.9 ± 7.6 | 6.5 ± 5.7 | 24.9 ± 9.9 | 4.9 ± 3.1 | n.d. | 21.3 ± 9.0 |
eGFP-WNT-3A/FZD Binding Affinity at Equilibrium
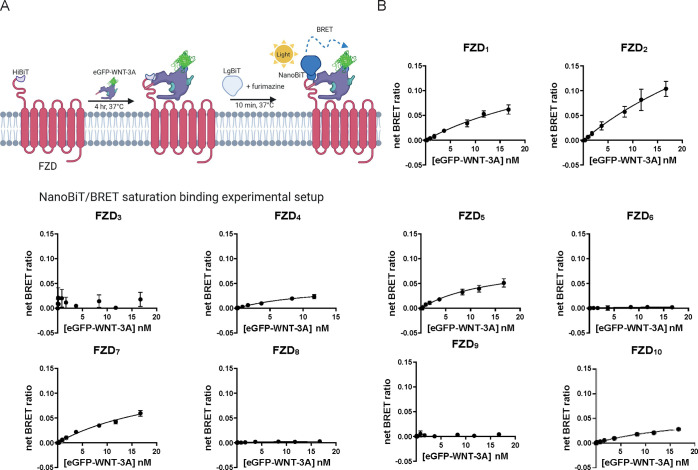
To define saturation binding affinity of eGFP-WNT-3A, we incubated human HiBiT-FZDs with a full concentration range (16.7 pM to 16.7 nM) of eGFP-WNT-3A for 240 min at 37 °C (Figure 2A). The net BRET ratio representing ligand–receptor binding increased in a clear, concentration-dependent manner for FZD1, FZD2, FZD4, FZD5, FZD7, and FZD10. Unfortunately, using transient overexpression of HiBiT-FZDs in HEK293A cells and the eGFP-WNT-3A with a limited maximal concentration in the conditioned medium, binding curves did not reach maximal asymptotic values but only came to near-saturable levels (Figure 2B). Again, detection of binding of eGFP-Wnt-3A to FZD6 and FZD8 was only marginally above background levels, as the net BRET values were low (Figure S2B). Similar to the kinetic binding assays, no quantifiable eGFP-WNT-3A binding was detected for FZD3 or FZD9. The affinities of eGFP-WNT-3A/FZD interactions were determined from linear regression curves showing near-saturable binding,39 and the Kd values are shown in Table 1. The reported saturation binding affinity values range from 4.9 to 48.6 nM, and they are in good agreement with the Kd values determined with kinetic binding for FZD1, FZD4, and FZD6. The degree of agreement is, however, only fair for FZD5, FZD8, and FZD10 and relatively poor for FZD2 and FZD7. Taken together, these kinetic and saturation binding data are in line with our previous results using fluorescence microscopy analysis, where no eGFP-WNT-3A association could be observed with C-terminally mCherry-tagged FZD6, and only a very weak association with FZD8 and FZD9 (FZD3 was not used).28 Although this fluorescence imaging-based method could provide an estimate of the relative ability of eGFP-WNT-3A to associate with different FZDs, accurate quantification of the binding affinities was not possible. Also, in that study, ΔFZD1–10 HEK293GFP-free cells overexpressing FZD8-mCherry (but not FZD6-mCherry), showed very faint binding of eGFP-WNT-3A, and this is also in agreement with the HiBiT-tagged system used here, which can detect very low level, but specific, binding to FZD8 (Figure S2). This is in agreement with a recent report claiming that ligand–receptor interaction using the HiBiT-tagged system allows detection of very weak interactions.32 Furthermore, in the case of FZD6, there are differences in reports of its ability to bind or respond to WNT-3A. Biochemical experiments failed to detect any association between WNT-3A and FZD6-CRD-IgG,16 and no eGFP-WNT-3A/Nluc-FZD6 interaction was detected in our previous NanoBRET study.28 However, it should be noted that, compared with the HiBiT-tagged systems, binding analyses with Nluc-tagged receptors can display reduced sensitivity for detection of weak interactions, as recently discussed.32 On the other hand, recombinant human WNT-3A induced a conformational change in FZD612 and affected the mobility of the receptor in the cell membrane as assessed by fluorescence recovery after photobleaching assay.40 Finally, the findings with respect to FZD8 are particularly intriguing, given the existing structural information on the WNT-3A-FZD8 CRD complex.41 It is currently unclear why such discrepancies exist for these two FZD paralogues.
Figure 2.
eGFP-WNT-3A saturation binding. A. The scheme depicts the experimental setup of NanoBiT/BRET analysis of equilibrium binding between the HiBiT-tagged FZD and the eGFP-WNT-3A. Created with BioRender.com. B. Saturation binding of the eGFP-WNT-3A to human HiBiT-FZDs was determined by the detection of NanoBiT/BRET in transiently overexpressing living HEK293A cells following 240 min incubation. Data points are presented as means ± SEM from n = 4 individual experiments, fitting a one-site specific model models. Experiments were performed with eGFP-WNT-3A batch 1.
In order to directly compare NanoBiT/BRET and NanoBRET binding assay formats, we used Nluc-FZD4 and HiBiT-FZD4 constructs for saturation binding (Figure S3A–C). NanoBiT/BRET experiments were performed in two different experimental paradigms, where LgBiT protein was added either directly after (setup 1, used throughout this study Figure S3B and Figure 2A) or for 10 min before (setup 2, Figure S3C) the 4 h incubation with eGFP-WNT-3A. This allowed us to test for any potential steric hindrance of WNT binding to FZD caused by the presence or complementation of nanoluciferase. In these comparisons, although the differences in Kd values were apparent, they did not reach statistical significance (Nluc-FZD4Kd (nM) ± SEM = 7.6 ± 3.6; HiBiT-FZD4 setup 1 Kd (nM) ± SEM = 11.9 ± 3.6; HiBiT-FZD4 setup 2 Kd ± SEM (nM) = 20.4 ± 7.6). Furthermore, eGFP-WNT-3A binding to Nluc-FZD4 (Figure S3A) resulted in lower maximal BRET (BRETmax) compared to either of the two HiBiT-FZD4 binding setups (Figure S3B,C) at similar luminescence levels (Nluc-FZD4 BRETmax ± SEM = 0.026 ± 0.006 vs HiBiT-FZD4 setup 1 BRETmax ± SEM = 0.049 ± 0.005, P = 0.0484; vs HiBiT-FZD4 setup 2 BRETmax ± SEM = 0.049 ± 0.004, P = 0.0349). This suggests that intracellular luminescence originating from receptors that are not accessible for the ligand reduces the assay’s dynamic range (see Figure S3D for expression analysis). In support of our choice to change from NanoBRET to the NanoBiT/BRET experimental setup, a recent study has shown that affinity measurements obtained from NanoBRET binding assays were generally less consistent in comparison with NanoBiT/BRET analyses.32
eGFP-WNT-3A Competition Binding with Untagged WNTs at FZD4
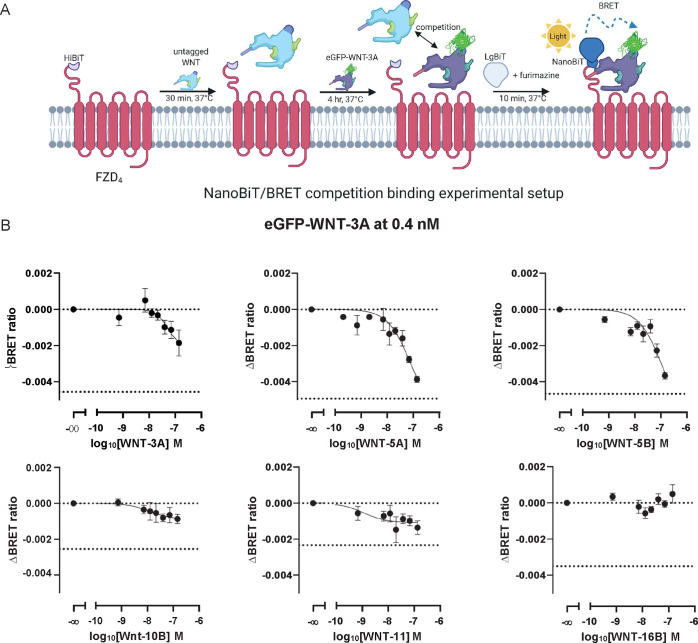
With the aim to understand the competitive nature of eGFP-WNT-3A binding to FZD, we combined eGFP-WNT-3A with increasing concentrations of several commercially available and purified untagged WNT proteins: WNT-3A, WNT-5A, WNT-5B, WNT-10B, WNT-11, and WNT-16B. Again, we have used HiBiT-FZD4 as our model receptor. In this assay setup, HEK293A cells transiently overexpressing HiBiT-FZD4 were preincubated with the untagged WNTs for 30 min before addition of eGPF-WNT-3A to a final concentration of 0.4 nM. Cells were then incubated for a further 4 h to allow a competitive equilibrium to be reached before addition of LgBiT and subsequent BRET measurement (Figure 3A). The results of these experiments are shown in Figure 3B and summarized in the Table 2, and show that the untagged WNTs competitively displaced eGFP-WNT-3A from FZD4 with different affinities and capacities. Interestingly, WNT-10B and WNT-11 had the highest affinities, but they showed moderate BRET signal decrease as indicated by remaining residual BRET. Intriguingly, WNT-3A presented higher binding affinity (lower Kd) but caused a lower reduction of BRET than WNT-5A and WNT-5B. WNT-16B did not compete with eGFP-WNT-3A in this FZD4-based assay. These results are in fair agreement with the recently published potencies and efficacies of WNTs in eliciting conformational changes in FZD4-cpGFP biosensor except for WNT-5B.12 Nevertheless, a similar rank order of affinities was obtained for WNT-3A, WNT-5A, and WNT-5B in binding to the isolated FZD4 CRD.17 Obviously, the insights into the mechanism of WNT–WNT competition for the primary binding site at FZDs remain obscure. We can only speculate that the process of functional ligand binding is more complex than for small-molecule ligands and other GPCRs. Along these lines, FZD oligomers associate or dissociate upon ligand addition,42−44 adding to the complexity of ligand binding analysis.45 Additionally, FZD coreceptors and various regulators could alter WNT interactions with FZD in HEK293 cells.25
Figure 3.
Competition binding between eGFP-WNT-3A and untagged WNTs at FZD4. A. The scheme depicts the experimental setup of NanoBiT/BRET analysis of competition binding between the eGFP-WNT-3A and commercially available untagged WNT-3A, WNT-5A, WNT-5B, WNT-10B, WNT-11, and WNT-16B. Created with BioRender.com. B. FZD4 binding of eGFP-WNT-3A at 0.4 nM in the presence of increasing concentrations of the untagged WNTs was determined by the detection of NanoBiT/BRET in transiently overexpressing living HEK293A cells following 240 min incubation. Data points are presented as means ± SEM from n = 3–6 individual experiments, fitting a three- or four-parameter model. Upper dashed line indicates the BRET ratio of eGFP-WNT-3A-only treated cells; lower dashed line indicates the BRET ratio of ligand-untreated cells (BRET donor only). Experiments were performed with eGFP-WNT-3A batch 2.
Table 2. Binding Properties of Various FZD Ligands in Competition with eGFP-WNT-3A Binding (0.4 nM) to HiBiT-FZD4a.
| WNT-3A | WNT-5A | WNT-5B | WNT-10B | WNT-11 | WNT-16B | |
|---|---|---|---|---|---|---|
| Competition binding pKi ± SEM | 7.26 ± 0.35 | 7.08 ± 0.21 | 6.93 ± 0.25 | 7.87 ± 0.55 | 8.90 ± 0.66 | n.d. |
| ΔBRET ± SEM | –0.002 ± 0.001 | –0.004 ± 0.0002 | –0.004 ± 0.0002 | –0.001 ± 0.0003 | –0.001 ± 0.0004 | n.d. |
| 0.4 nM eGFP- WNT-3A binding displaced (%) | 40.8 | 78.4 | 78.3 | 34.5 | 58.4 | n.d. |
Binding of eGFP-WNT-3A to FZD Chimeras
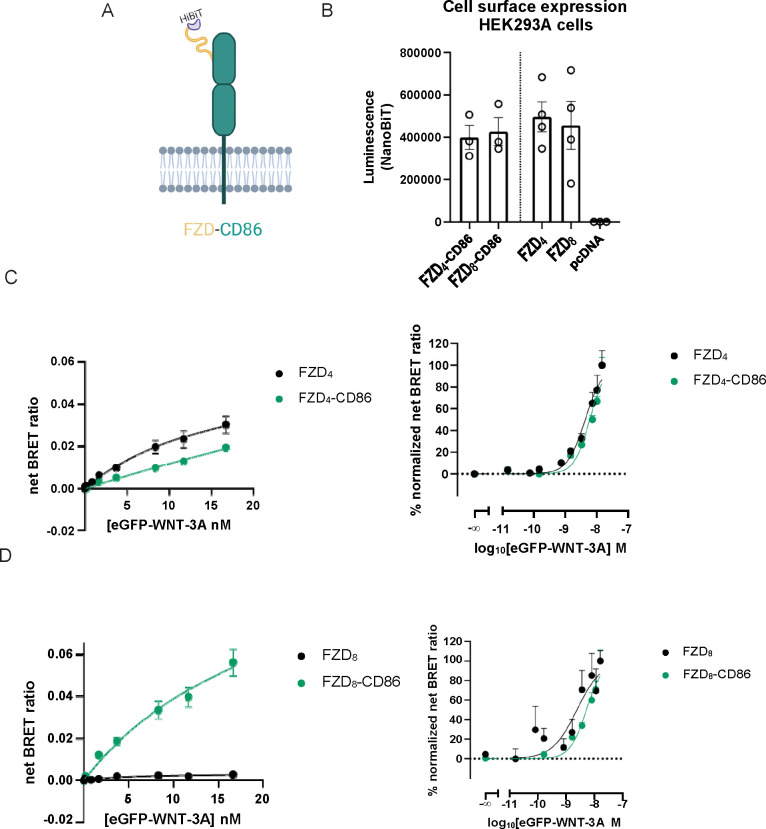
WNTs directly engage the CRD through protein–protein and protein–lipid interactions.41,46 However, it remains unclear whether non-CRD domains of FZDs contribute to WNT-FZD interaction.20 It has been hypothesized that the CRD simply serves the purpose of binding WNTs in order to bring them in close proximity with the receptor for additional binding/activation mechanisms.47 Indeed, the long and flexible nature of the linker connecting the CRD of FZDs to TM1 would tend to support such a hypothesis.48 The role of the FZD transmembrane domains in ligand binding, complex formation, receptor conformational changes, and signal transduction has been a subject of debate.5,8,9,49−52 Here, we seek to obtain a more mechanistic insight into the contribution of the transmembrane core to eGFP-WNT-3A binding. To this end, we generated two chimeric proteins fusing the N-terminal domain (NTD; CRD + linker) of one FZD with an unrelated CD86 single transmembrane domain spanning protein (Figure 4A). Specifically, we generated FZD4-CD86 and FZD8-CD86 chimeric proteins (Figure S4A). In this manner, we aimed to study the effect of a FZD core on eGFP-WNT-3A binding to the CRD. We validated the chimeras with regard to proper membrane trafficking upon transient overexpression in HEK293A cells and detected no difference in surface expression levels between FZD-CD86 and wild-type (WT) FZDs (Figure 4B).
Figure 4.
eGFP-WNT-3A binding to FZD-CD86 chimeras. A. The cartoon representations of the FZD-CD86 fusion proteins used in this study. eGFP-WNT-3A binding at equilibrium was assessed as depicted in Figure 2A. Created with BioRender.com. B. Cell surface expression of HiBiT-tagged FZD-CD86 chimeras as measured by NanoBiT luminescence (from the experiments summarized in Figure 2B and parts C–D). Data are presented as means ± SEM from n = 3–4 individual experiments. Expression data of FZD4, FZD8, and pcDNA are also depicted in SI Figure 1C. C. Saturation binding of eGFP-WNT-3A at FZD4-CD86 and FZD4 (data also present in Figure 2B) was determined by the detection of NanoBiT/BRET in transiently overexpressing living HEK293A cells following 240 min incubation. Data points are presented as means ± SEM from n = 3–4 individual experiments. eGFP-WNT-3A batch 1 was used in these experiments. D. Saturation binding of eGFP-WNT-3A at FZD8-CD86 and FZD8 (data also present in Figure 2B) was determined by the detection of NanoBiT/BRET in transiently overexpressing living HEK293A cells following 240 min incubation. Data points are presented as means ± SEM from n = 3–4 individual experiments. eGFP-WNT-3A batch 1 was used in the experiments. Linear scale data are fitted to a one-site specific binding model. Logarithmic-scale data are fitted to a normalized three- or four-parameter model. The right plot in every panel shows data normalized between 0% (BRETmin) and 100% (BRETmax) for each studied construct.
In the NanoBiT/BRET binding experiments with the FZD4-CD86 chimera, we could detect a concentration-dependent increase in the BRET signal indicative of eGFP-WNT-3A binding. Interestingly, eGFP-WNT-3A interacted with FZD4-CD86 with a visibly lower affinity (higher Kd) and a visibly lower maximal BRET (at the fixed concentrations used) than for an intact FZD4 protein (FZD4-CD86 Kd ± SEM (nM) = 141.9 ± 181.5, P = 0.0991; BRETmax ± SEM = 0.019 ± 0.001 vs FZD4 BRETmax ± SEM = 0.030 ± 0.004, P = 0.0622; Figure 4C). In order to emphasize differences in Kd for the tested receptors, the data were also normalized and are presented in a semilogarithmic presentation in Figure 4C (FZD4-CD86 pKd ± SEM = 8.19 ± 0.04 vs FZD4 pKd ± SEM = 8.31 ± 0.05, P = 0.0331). Importantly, differences in BRETmax for FZD4-CD86 and FZD4 cannot arise from differences in surface expression levels, as both studied receptors are similarly expressed (P = 0.3324).
Next, we performed a similar analysis for a FZD8-CD86 chimera. eGFP-WNT-3A binding to the chimera FZD8-CD86 compared to FZD8 at similar levels of receptor surface expressions (P = 0.8340; Figure 4B) did not differ in affinity in the analysis of non-normalized data (FZD8-CD86 Kd ± SEM (nM) = 22.9 ± 10.6, P = 0.6668; Figure 4D), but the difference reached statistical significance when comparing the normalized values (P = 0.0390; Figure 4D). Furthermore, the NanoBiT/BRET signal increased significantly when the FZD8 core was replaced by CD86. The NanoBiT/BRET signal (BRETmax) was in fact comparable to other WNT-3A-binding competent FZDs (FZD8-CD86 BRETmax ± SEM = 0.056 ± 0.006 vs FZD8 BRETmax ± SEM = 0.003 ± 0.0003, P = 0.0002; Figure 4D and Figure 2). Intriguingly, these findings are the opposite of what we observe for FZD4, where replacing the receptor core with CD86 visibly reduced maximal BRET (Figure 4C). The efficiency of resonance energy transfer depends on both orientation and distance between BRET donor and BRET acceptor.53 Thus, in the NanoBiT/BRET binding setup, the differences in BRETmax can be interpreted as distinct ligand–receptor conformations. This further suggests that the cores of FZD4 and FZD8 can differently contribute to WNT-FZD binding.
In addition to the FZD-CD86 chimeras detailed above, we also generated FZD-FZD chimeras. Specifically, we constructed FZD4-FZD6, FZD4-FZD8, FZD5-FZD6, FZD5-FZD7, and FZD6-FZD4 chimeric proteins (Figure S4B). In this manner, we have used at least one FZD paralogue from every FZD homology cluster. The transmembrane cores of FZD6 and FZD8 were selected to test whether they negatively affect eGFP-WNT-3A binding to the CRD of FZD4 and FZD5. On the other hand, the FZD7 core was chosen to assess if it positively modulates ligand binding to the CRD of FZD5. The rationale for this selection was based on the very weak NanoBiT/BRET signal seen for eGFP-WNT-3A binding to FZD6 and FZD8, and strong NanoBiT/BRET signal of eGFP-WNT- 3A/FZD7 association in the two assay paradigms described in this study (Figure 1B and Figure 2B). Additionally, the FZD6 core was replaced with the FZD4 core to assess whether eGFP-WNT-3A binding to FZD6 CRD would increase upon insertion of a core from an eGFP-WNT-3A binding-competent FZD paralogue (Figure 1B and Figure 2B). We validated the chimeras with regard to their proper membrane trafficking upon transient overexpression in HEK293A cells and detected that the FZD-FZD chimera proteins are relatively poorly expressed on the cell surface compared to WT receptors (Figure S4C). Binding affinities of fluorescent propranolol to HiBiT-tagged β2-adrenergic receptors vary depending on the protein expression levels, with higher expression levels generally resulting in slightly elevated Kd values (lower affinity).32 Furthermore, binding affinities of DKK1-eGFP proteins to the WNT coreceptor LRP6-mCherry measured by dual-color axial line-scanning FCS (axial lsFCS) were higher for lower, more physiologically relevant receptor expression levels.54 Our data for transient overexpression of FZD4 in HEK293A cells mostly support these notions, with some exceptions (Figure S4D). Overall, it needs to be emphasized that although the Kd is a thermodynamic parameter that should be constant for various expression levels, differences in the cellular context may lead to different functionalities of overexpressed receptors present on the cell surface. This in turn can affect the comparative analysis and interpretation of real-time ligand binding data.54 However, provided the availability of cell surface expression data in the experimental paradigm of the NanoBiT/BRET binding assay, we shed light on a potential role of the receptor core for WNT-CRD binding. Focusing on the FZD4 chimeras, eGFP-WNT-3A bound with a significantly higher affinity (lower Kd) to the FZD4-FZD6 and the FZD4-FZD8 chimeras compared to full-length FZD4 (FZD4-FZD6Kd ± SEM (nM) = 3.5 ± 1.8, P = 0.0255; FZD4-FZD8Kd ± SEM (nM) = 2.9 ± 1.3, P = 0.0371; Figure S4E). Interestingly, BRETmax values were visibly or significantly lower for both weakly expressed FZD4-FZD chimeras than for the intact FZD4 (FZD4-FZD6 BRETmax ± SEM = 0.021 ± 0.003, P = 0.0828; FZD4-FZD8 BRETmax ± SEM = 0.010 ± 0.001, P = 0.0192).
Additionally, our data showed that in comparison to WT FZD5, eGFP-WNT-3A binding to FZD5-FZD6 occurred with only visibly higher affinity, but this difference did not reach statistical significance (FZD5-FZD6Kd ± SEM (nM) = 4.4 ± 1.5, P = 0.0638; Figure S4F). Next, the affinity of eGFP-WNT-3A binding to FZD5-FZD7 was also not significantly different compared with WT FZD5 (FZD5-FZD7Kd ± SEM (nM) = 5.0 ± 2.4, P = 0.2622; Figure S4F). Moreover, the differences in BRETmax did not reach statistical significance (FZD5-FZD6 BRETmax ± SEM = 0.080 ± 0.014, P = 0.1715; FZD5-FZD7 BRETmax ± SEM = 0.098 ± 0.026, P = 0.2046; FZD5 BRETmax ± SEM = 0.051 ± 0.009).
In addition, our data indicated that eGFP-WNT-3A bound to FZD6-FZD4 with the same affinity (FZD6-FZD4Kd ± SEM (nM) = 5.8 ± 3.4, P = 0.9695; Figure S4G) as to FZD6 but with a significant, over 10-fold increase in the maximal BRET signal (FZD6-FZD4 BRETmax ± SEM = 0.030 ± 0.006 vs FZD6 BRETmax ± SEM = 0.002 ± 0.001, P = 0.056) arguing that the FZD6CRD can efficiently bind eGFP-WNT-3A in the context of a different receptor core. As mentioned before, it needs to be emphasized that receptor expression levels can affect ligand–receptor interaction. Along these lines, in classical BRET titration experiments, increasing BRET donor amounts (by increasing plasmid DNA amounts) with constant BRET acceptor levels (unchanged plasmid DNA amounts) leads to a decrease in BRETmax signal for specific donor–acceptor interactions.55 In contrast, no such relationship was found in our FZD-CD86 and FZD-FZD NanoBiT/BRET binding experiments, further supporting the notion that the FZD core has a differential role in WNT binding depending on the FZD paralogue.
Here, we have used the NanoBiT/BRET system to analyze the contribution of the FZD seven-transmembrane-spanning core to eGFP-WNT-3A binding to the CRD. We show that swapping the receptor core can have a substantial effect on the affinity or maximal BRET of eGFP-WNT-3A binding in the NanoBiT/BRET read-out. Our data, particularly from the FZD-CD86 experiments, argue that the seven-transmembrane-spanning core contributes to ligand binding for the tested FZDs, even though the details on the molecular level remain obscure.
This study adds substantial methodological advance to the pharmacological toolbox suitable for the study of the class F GPCRs and their coreceptors.28,54,56 We have demonstrated the vast potential of employing fluorescent WNTs and the NanoBiT/BRET binding technique for the pharmacological quantification of WNT-FZD interactions in live HEK293A cells despite the limitations that come with the low concentration of the tracer WNT in the conditioned medium preparation. The broader analysis of the selectivity of ligand–receptor interactions, WNT binding in the presence or absence of either FZD coreceptors, FZD-binding intracellular transducer proteins and at different FZD expression levels, can now be further investigated to understand the pluridimensionality of WNT-FZD system in a more physiologically relevant cell system.
Experimental Section
Cell Culture and Ligands
HEK293A cells (ATCC), HEK293F (Thermo Fisher Scientific, Waltham, MA, USA), HEK293T (DSMZ ACC-635), and ΔFZD1–10 HEK293T cells25 were cultured in DMEM supplemented with 10% FBS, 1% penicillin/streptomycin, and 1% l-glutamine (all from Thermo Fisher Scientific, Waltham, MA, USA) in a humidified CO2 incubator at 37 °C. All cell culture plastics were from Sarstedt (Nümbrecht, Germany), unless otherwise specified. The absence of mycoplasma contamination was routinely confirmed by PCR using 5′-GGCGAATGGGTGAGTAACACG-3′ and 5′-CGGATAACGCTTGCGACTATG-3′ primers detecting 16 S rRNA of mycoplasma in the media after 2–3 days of cell exposure. Untagged human WNT-3A, human/mouse WNT-5A, human WNT-5B, human WNT-10B, human WNT-11, and human WNT-16B were all from RnD Systems/Biotechne (#5036-WN, #645-WN, #7347-WN, #7196-WN, #6179-WN, and #7790-WN, Minneapolis, MI, USA). WNTs were dissolved at 100 μg/mL in filter-sterilized 0.1% BSA/PBS and stored at 4 °C. Molecular weights of the WNTs were as per supplier’s datasheets. Porcupine inhibitor C59 was from Abcam (#ab142216, Cambridge, UK).57 C59 was dissolved in DMSO at 5 mM and stored at −20 °C. The serial dilutions of WNTs were prepared in the protein-low binding tubes (Eppendorf, Hamburg, Germany).
Preparation of eGFP-WNT-3A CM
HEK293F suspension cells growing in serum-free Expi293 expression medium (60 mL, 2.5 × 106 cells/mL) were cotransfected with 10 μg of either pCS2+-WNT-3A or pCS2+-eGFP-WNT- 3A together with 50 μg of pCMV-His-Afamin plasmid using ScreenFect UP-293 (ScreenFect GmbH, Eggenstein-Leopoldshafen, Germany) according to the manufacturer’s instructions. The corresponding control CM was generated from cells transfected with pCS2+ plasmid.
Cells were first cleared from the HEK293F CM by centrifugation at 260 g (1200 rpm) for 10 min and then at 2800 g (4000 rpm) for 30 min to remove any remaining cellular debris and insoluble material. This “raw” CM then was concentrated 5-fold using Vivaspin turbo 15 centrifugal concentrators (30,000-molecular-weight-cutoff, Satorius AG, Göttingen, Germany) and exchanged to the desired cell culture medium using Sephadex G-25 PD10 desalting columns (GE Healthcare Bio-Science, Freiburg, Germany). The final concentration and integrity of eGFP-WNT-3A in the CM samples were determined using ELISA (GFP ELISA kit, #ab171581, Abcam) and SDS-PAGE/Western Blot analysis, respectively. Two eGFP-WNT-3A batches (eGFP-WNT-3A batch 1 final concentration: 16.7 nM; eGFP-WNT-3A batch 2 final concentration: 16.2 nM) were used in this study. Current WNT purification methods allow only limited WNT concentration to be obtained from CM.58 For validation of the eGFP-WNT-3A batches, please see Figure S5.
Plasmids
Generation of HiBiT-FZD4, HiBiT-FZD6, and Nluc-FZD4 has been described previously.28 Gibson cloning was used to generate other HiBiT-tagged receptor constructs using HiBiT-tagged backbone from HiBiT-FZD4 containing a 5-HT3A signal sequence. To generate chimeras, the N-terminal domains (NTD; CRD with a linker region) and the transmembrane cores were defined according to Frizzled structures predicted on GPCRdb (http://www.gpcrdb.org), and the constructs were generated with Gibson cloning. Nluc-CD86 used to generate FZD-CD86 chimeras was from Martin J. Lohse (Max-Delbrueck Center for Molecular Medicine, Berlin, Germany). Frizzled and CD86 signal peptides were defined with SignalIP-5.0 Server (http://www.cbs.dtu.dk/services/SignalP/). The constructs were validated by sequencing (Eurofins GATC, Konstanz, Germany). The details of the constructs used in this study are presented in Figure S1A,B and Figure S4A.
NanoBiT/BRET Binding
HEK293A cells were transiently transfected in suspension using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA). A total of 4 × 105 cells were transfected in 1 mL with 1000 ng of HiBiT-tagged FZDs or 10 ng of Nluc-FZD4 plasmid DNA. The cells (50 μL) were seeded onto a poly(d-lysine)-coated black 96-well cell culture plate with a solid flat bottom (Greiner BioOne). Next, 50 μL of complete DMEM medium was added to each well. Forty-eight hours post-transfection, the cells were washed once with 200 μL of Hanks’ balanced salt solution (HBSS; HyClone). In the kinetic binding experiments, the cells were preincubated with 50 μL of a mix of Nluc substrate vivazine (1:50 dilution; #N2581, Promega, Fitchburg, WI, USA) and LgBiT (1:100 dilution; #N2421, Promega, Fitchburg, WI, USA) in a complete, nonphenol red DMEM (HyClone) supplemented with 10 mm HEPES for 1 h at 37 °C without CO2. Subsequently, 50 μL of eGFP-WNT-3A conditioned medium or control medium supplemented with 5% FBS and 10 mm HEPES was added, and the BRET signal was measured every 90 s for 240 min at 37 °C (161 measurements, no CO2). In the saturation-binding experiments, the cells were incubated with different concentrations of eGFP-WNT-3A conditioned medium (90 μL) supplemented with 5% FBS and 10 mm HEPES for 240 min at 37 °C with no CO2. In the competition binding experiments, the cells were preincubated for 30 min at 37 °C with 80 μL of unlabeled WNT proteins at 37 °C with no CO2. Subsequently, 10 μL of eGFP-WNT-3A at a concentration of 3.6 nM (final concentration of 0.4 nM) were added, and the cells were incubated for further 240 min at 37 °C with no CO2. Next, for saturation and competition binding experiments, 10 μL of a mix of furimazine (1:10 dilution; #N2421, Promega, Fitchburg, WI, USA) and LgBiT (1:20 dilution; #N2421, Promega, Fitchburg, WI, USA) was added. For saturation binding experiments with Nluc-FZD4 furimazine was used at 1:1000 final dilution (#N1572, Promega, Fitchburg, WI, USA) and no LgBiT was added. The cells were incubated for another 10 min at 37 °C with no CO2 before the BRET measurements. The BRET ratio was determined as the ratio of light emitted by eGFP (energy acceptor) and light emitted by HiBiT-FZD1–10 or Nluc-FZD4 (energy donors). The net BRET ratio was calculated as the difference in BRET ratio between cells treated with eGFP-WNT-3A, and cells treated with vehicle. ΔBRET ratio in the competition binding experiment was calculated as the difference in BRET ratio of cells treated with vehicle (eGFP-WNT-3A only wells, no ΔBRET) and cells treated with WNTs. The BRET acceptor (bandpass filter, 535–30 nm) and BRET donor (bandpass filter, 475–30 nm) emission signals were measured using a CLARIOstar microplate reader (BMG, Ortenberg, Germany). Cell surface expression of HiBiT-tagged FZDs and total expression of Nluc-FZD4 was assessed by measuring luminescence of vehicle-treated wells (no BRET acceptor) in the NanoBiT/BRET or NanoBRET binding assays, respectively. eGFP fluorescence was measured prior to reading BRET (excitation, 470–15 nm; emission, 515–20 nm).
TOPFlash Reporter Gene Assay
ΔFZD1–10 HEK 293T cells were transfected in suspension (4 × 105 cells were transfected in 1 mL) with 700 ng of HiBiT-tagged receptor, 250 ng M50 Super 8× TOPFlash (#12456; Addgene, Watertown, MA, USA), and 50 ng pRL-TK Luc (#E2241, Promega, Fitchurg, WI, USA) and seeded (50 μL) onto a poly(d-lysine)-coated white 96-well cell culture plate with a solid flat bottom (Greiner BioOne). Next, 50 μL of complete DMEM medium was added to each well. Twenty-four hours after transfection, the medium was changed to starvation medium (DMEM without FBS) containing either 8.0 nM (300 ng/mL) WNT-3A or vehicle, and 10 nM C59. Twenty-four hours after stimulation, cells were lysed gently shaking with 20 μL 1× Passive Lysis Buffer (#E1910; Promega, Fitchurg, WI, USA) for 15 min. Subsequently, 20 μL of LAR II (Promega, E1910) was added to all wells after which luminescence (580–80 nm) was read, and then 20 μL of Stop & Glo (Promega, E1910) was added to all wells after which luminescence (480–80 nm) was read again with a CLARIOstar microplate reader (BMG, Ortenberg, Germany).
Data Analysis and Statistics
All data were analyzed in GraphPad Prism 8 (San Diego, CA, USA) using built-in equations. All data presented in this study come from n individual experiments (at least three biological replicates) with each individual experiment performed typically in duplicates (technical replicates) for each tested concentration/condition. Data points on the binding curves represent mean ± SEM. Saturation binding curves were fit using one-site-specific or total and nonspecific saturation nonlinear regression models (linear scale for eGFP-WNT-3A concentrations) or normalized three-parameter or normalized four-parameter nonlinear regression models (logarithmic scale for eGFP-WNT-3A concentrations with normalized net BRET ratio). The fitting models were selected based on an extra-sum-of-squares F-test (P < 0.05). Kinetic binding data were analyzed using the association model with two or more hot ligand concentrations. Binding affinity values (Kd) are presented as a best-fit Kd with SEM. Kd values were compared using an extra-sum-of-squares F-test (P < 0.05). Competition binding curves were analyzed using a three- or four-parameter nonlinear regression model to obtain equilibrium dissociation constant values pKi with SEM of unlabeled ligands as per the Cheng–Prusoff equation.59 Minimal BRET (BRETmin) and maximal BRET (BRETmax) were defined as the lowest and highest measured net BRET ratios, respectively. BRETmax values were compared using unpaired t-test. TOPFlash and cell surface expression data are presented as mean ± SEM. TOPFlash and cell surface expression data were analyzed for differences with Brown–Forsythe and Welch one-way analysis of variance (ANOVA); ** P ≤ 0.01, * P ≤ 0.05.
Acknowledgments
We thank Anna Krook for access to the CLARIOstar plate reader and Benoit Vanhollebeke, who kindly provided the ΔFZD1–10 cells used in this work.
Glossary
Abbreviations
- CM
conditioned medium
- CRD
cysteine-rich domain
- eGFP
enhanced green fluorescent protein
- FRET
Förster resonance energy transfer
- FZD
Frizzled
- GPCR
G protein-coupled receptor
- HBSS
Hank’s balanced salt solution
- NanoBiT/BRET
nanoluciferase binary technology/bioluminescence resonance energy transfer
- Nluc
nanoluciferase
- NTD
N-terminal domain
- TCF/LEF
T-cell factor/lymphoid enhancer-binding factor
- WNT
WNT/Int-1 family of proteins
- WT
wild-type
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsptsci.1c00084.
Detailed plasmid information, cell surface expression data, TOPFlash data, NanoBRET vs NanoBiT/BRET binding comparison data, detailed information about FZD-CD86 and FZD-FZD chimeras, eGFP-WNT-3A binding to FZD-FZD chimeras and eGFP-WNT-3A validation PDF)
Author Contributions
P.K. and G.S. and G.D. conceived the study, and P.K. and G.S. designed the study. S.M. and J.W. generated and validated the eGFP-WNT-3A conditioned media. P.K., R.S., and C.-F.B. performed experiments. P.K., S.M., and J.W. designed and prepared the figures. P.K., G.D., and G.S. wrote the manuscript. S.M., R.S., C.-F.B., and J.W. commented and contributed to the manuscript writing. G.S. supervised and coordinated the project with input from G.D. All authors have approved the final version of the manuscript.
This study was supported by Karolinska Institutet, the Swedish Cancer Society (20 0264P, CAN2017/561), the Swedish Research Council (2019–01190), Novo Nordisk Foundation (NNF20OC0063168, NNF17OC0026940, NNF19OC0056122), The Lars Hierta Memorial Foundation (FO2019–0086, FO2020–0304), The Alex and Eva Wallström Foundation for Scientific Research and Education (2020–00228), The Swedish Society of Medical Research (P19-0055), and the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation), project number 331351713–SFB 1324 (project A06 to G.D.). Funding for the open access publication charges has been covered by National Library of Sweden (BIBSAM).
The authors declare no competing financial interest.
Supplementary Material
References
- Foord S. M.; Bonner T. I.; Neubig R. R.; Rosser E. M.; Pin J. P.; Davenport A. P.; Spedding M.; Harmar A. J. (2005) International Union of Pharmacology. XLVI. G protein-coupled receptor list. Pharmacol. Rev. 57, 279–288. 10.1124/pr.57.2.5. [DOI] [PubMed] [Google Scholar]
- Schulte G. (2010) International Union of Basic and Clinical Pharmacology. LXXX. The class Frizzled receptors. Pharmacol. Rev. 62, 632–667. 10.1124/pr.110.002931. [DOI] [PubMed] [Google Scholar]
- Jung Y. S.; Park J. I. (2020) Wnt signaling in cancer: therapeutic targeting of Wnt signaling beyond beta-catenin and the destruction complex. Exp. Mol. Med. 52, 183–191. 10.1038/s12276-020-0380-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgy O.; Konigshoff M. (2018) The WNT signaling pathways in wound healing and fibrosis. Matrix Biol. 68–69, 67–80. 10.1016/j.matbio.2018.03.017. [DOI] [PubMed] [Google Scholar]
- Schulte G.; Kozielewicz P. (2020) Structural insight into Class F receptors - What have we learnt regarding agonist-induced activation?. Basic Clin. Pharmacol. Toxicol. 126, 17. 10.1111/bcpt.13235. [DOI] [PubMed] [Google Scholar]
- Schulte G.; Wright S. C. (2018) Frizzleds as GPCRs - More Conventional Than We Thought!. Trends Pharmacol. Sci. 39, 828–842. 10.1016/j.tips.2018.07.001. [DOI] [PubMed] [Google Scholar]
- Kozielewicz P.; Turku A.; Bowin C. F.; Petersen J.; Valnohova J.; Canizal M. C. A.; Ono Y.; Inoue A.; Hoffmann C.; Schulte G. (2020) Structural insight into small molecule action on Frizzleds. Nat. Commun. 11, 414. 10.1038/s41467-019-14149-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. C.; Kozielewicz P.; Kowalski-Jahn M.; Petersen J.; Bowin C. F.; Slodkowicz G.; Marti-Solano M.; Rodriguez D.; Hot B.; Okashah N.; et al. (2019) A conserved molecular switch in Class F receptors regulates receptor activation and pathway selection. Nat. Commun. 10, 667. 10.1038/s41467-019-08630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi N.; Mukherjee S.; Waghray D.; Janda C. Y.; Jude K. M.; Miao Y.; Burg J. S.; Aduri N. G.; Kossiakoff A. A.; Gati C.; et al. (2020) Structure of human Frizzled5 by fiducial-assisted cryo- EM supports a heterodimeric mechanism of canonical Wnt signaling. eLife 9, 9. 10.7554/eLife.58464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygaard R.; Yu J.; Kim J.; Ross D. R.; Parisi G.; Clarke O. B.; Virshup D. M.; Mancia F. (2021) Structural Basis of WLS/Evi-Mediated Wnt Transport and Secretion. Cell 184, 194–206. 10.1016/j.cell.2020.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowin C. F.; Inoue A.; Schulte G. (2019) WNT-3A-induced beta-catenin signaling does not require signaling through heterotrimeric G proteins. J. Biol. Chem. 294, 11677–11684. 10.1074/jbc.AC119.009412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schihada H.; Kowalski-Jahn M.; Turku A.; Schulte G. (2021) Deconvolution of WNT-induced Frizzled conformational dynamics with fluorescent biosensors. Biosens. Bioelectron. 177, 112948. 10.1016/j.bios.2020.112948. [DOI] [PubMed] [Google Scholar]
- Ma W.; Chen M.; Kang H.; Steinhart Z.; Angers S.; He X.; Kirschner M. W. (2020) Single-molecule dynamics of Dishevelled at the plasma membrane and Wnt pathway activation. Proc. Natl. Acad. Sci. U. S. A. 117, 16690–16701. 10.1073/pnas.1910547117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuysuz N.; van Bloois L.; van den Brink S.; Begthel H.; Verstegen M. M.; Cruz L. J.; Hui L.; van der Laan L. J.; de Jonge J.; Vries R.; et al. (2017) Lipid-mediated Wnt protein stabilization enables serum-free culture of human organ stem cells. Nat. Commun. 8, 14578. 10.1038/ncomms14578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara E.; Hirai H.; Yamamoto H.; Tamura-Kawakami K.; Matano M.; Kikuchi A.; Sato T.; Takagi J. (2016) Active and water-soluble form of lipidated Wnt protein is maintained by a serum glycoprotein afamin/alpha-albumin. eLife 5, 5. 10.7554/eLife.11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A.; Yamamoto H.; Sakane H.; Koyama H.; Kikuchi A. (2010) Wnt5a regulates distinct signalling pathways by binding to Frizzled2. EMBO J. 29, 41–54. 10.1038/emboj.2009.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijksterhuis J. P.; Baljinnyam B.; Stanger K.; Sercan H. O.; Ji Y.; Andres O.; Rubin J. S.; Hannoush R. N.; Schulte G. (2015) Systematic mapping of WNT-FZD protein interactions reveals functional selectivity by distinct WNT-FZD pairs. J. Biol. Chem. 290, 6789–6798. 10.1074/jbc.M114.612648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. H.; Nusse R. (2002) Ligand receptor interactions in the Wnt signaling pathway in Drosophila. J. Biol. Chem. 277, 41762–41769. 10.1074/jbc.M207850200. [DOI] [PubMed] [Google Scholar]
- Agostino M.; Pohl S. O.; Dharmarajan A. (2017) Structure-based prediction of Wnt binding affinities for Frizzled-type cysteine-rich domains. J. Biol. Chem. 292, 11218–11229. 10.1074/jbc.M117.786269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J. C.; Rattner A.; Smallwood P. M.; Nathans J. (1999) Biochemical characterization of Wnt- Frizzled interactions using a soluble, biologically active vertebrate Wnt protein. Proc. Natl. Acad. Sci. U. S. A. 96, 3546–3551. 10.1073/pnas.96.7.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain R. K.; Katoh M.; Medina A.; Steinbeisser H. (2005) Xenopus frizzled-4S, a splicing variant of Xfz4 is a context-dependent activator and inhibitor of Wnt/beta-catenin signaling. Cell Commun. Signaling 3, 12. 10.1186/1478-811X-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostino M.; Pohl S. O. (2019) Wnt Binding Affinity Prediction for Putative Frizzled-Type Cysteine-Rich Domains. Int. J. Mol. Sci. 20, 4168. 10.3390/ijms20174168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport A. P.; Scully C. C. G.; de Graaf C.; Brown A. J. H.; Maguire J. J. (2020) Advances in therapeutic peptides targeting G protein-coupled receptors. Nat. Rev. Drug Discovery 19, 389–413. 10.1038/s41573-020-0062-z. [DOI] [PubMed] [Google Scholar]
- Kaiser A.; Coin I. (2020) Capturing Peptide-GPCR Interactions and Their Dynamics. Molecules 25, 4724. 10.3390/molecules25204724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubelen M.; Bostaille N.; Cabochette P.; Gauquier A.; Tebabi P.; Dumitru A. C.; Koehler M.; Gut P.; Alsteens D.; Stainier D. Y. R.; et al. (2018) A molecular mechanism for Wnt ligand-specific signaling. Science 361, eaat1178. 10.1126/science.aat1178. [DOI] [PubMed] [Google Scholar]
- Wright S. C.; Canizal M. C. A.; Benkel T.; Simon K.; Le Gouill C.; Matricon P.; Namkung Y.; Lukasheva V.; Konig G. M.; Laporte S. A.; et al. (2018) FZD5 is a Galphaq-coupled receptor that exhibits the functional hallmarks of prototypical GPCRs. Sci. Signaling 11, eaar5536. 10.1126/scisignal.aar5536. [DOI] [PubMed] [Google Scholar]
- Takada R.; Mii Y.; Krayukhina E.; Maruyama Y.; Mio K.; Sasaki Y.; Shinkawa T.; Pack C. G.; Sako Y.; Sato C.; et al. (2018) Assembly of protein complexes restricts diffusion of Wnt3a proteins. Commun. Biol. 1, 165. 10.1038/s42003-018-0172-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesslowski J.; Kozielewicz P.; Wang X.; Cui H.; Schihada H.; Kranz D.; Karuna M. P.; Levkin P.; Gross J. C.; Boutros M.; et al. (2020) eGFP-tagged Wnt-3a enables functional analysis of Wnt trafficking and signaling and kinetic assessment of Wnt binding to full-length Frizzled. J. Biol. Chem. 295, 8759–8774. 10.1074/jbc.RA120.012892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan J. F.; Janda C. Y.; Garcia K. C. (2012) Structural architecture and functional evolution of Wnts. Dev. Cell 23, 227–232. 10.1016/j.devcel.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddart L. A.; Johnstone E. K.; Wheal A. J.; Goulding J.; Robers M. B.; Machleidt T.; Wood K. V.; Hill S. J.; Pfleger K. D. (2015) Application of BRET to monitor ligand binding to GPCRs. Nat. Methods 12, 661–663. 10.1038/nmeth.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soave M.; Heukers R.; Kellam B.; Woolard J.; Smit M. J.; Briddon S. J.; Hill S. J. (2020) Monitoring Allosteric Interactions with CXCR4 Using NanoBiT Conjugated Nanobodies. Cell Chem. Biol. 27, 1250–1261. 10.1016/j.chembiol.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursier M. E.; Levin S.; Zimmerman K.; Machleidt T.; Hurst R.; Butler B. L.; Eggers C. T.; Kirkland T. A.; Wood K. V.; Friedman Ohana R. (2020) The luminescent HiBiT peptide enables selective quantitation of G protein-coupled receptor ligand engagement and internalization in living cells. J. Biol. Chem. 295, 5124–5135. 10.1074/jbc.RA119.011952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soave M.; Kellam B.; Woolard J.; Briddon S. J.; Hill S. J. (2020) NanoBiT Complementation to Monitor Agonist-Induced Adenosine A1 Receptor Internalization. SLAS Discov 25, 186–194. 10.1177/2472555219880475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peach C. J.; Kilpatrick L. E.; Woolard J.; Hill S. J. (2021) Use of NanoBiT and NanoBRET to monitor fluorescent VEGF-A binding kinetics to VEGFR2/NRP1 heteromeric complexes in living cells. Br. J. Pharmacol. 1. 10.1111/bph.15426. [DOI] [PubMed] [Google Scholar]
- Ring L.; Neth P.; Weber C.; Steffens S.; Faussner A. (2014) beta-Catenin-dependent pathway activation by both promiscuous “canonical” WNT3a-, and specific “noncanonical” WNT4- and WNT5a-FZD receptor combinations with strong differences in LRP5 and LRP6 dependency. Cell. Signalling 26, 260–267. 10.1016/j.cellsig.2013.11.021. [DOI] [PubMed] [Google Scholar]
- Yu H.; Ye X.; Guo N.; Nathans J. (2012) Frizzled 2 and frizzled 7 function redundantly in convergent extension and closure of the ventricular septum and palate: evidence for a network of interacting genes. Development 139, 4383–4394. 10.1242/dev.083352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voloshanenko O.; Gmach P.; Winter J.; Kranz D.; Boutros M. (2017) Mapping of Wnt-Frizzled interactions by multiplex CRISPR targeting of receptor gene families. FASEB J. 31, 4832–4844. 10.1096/fj.201700144R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasawa T.; Yokokura H.; Kitajewski J.; Lombroso P. J. (2002) Frizzled-9 is activated by Wnt-2 and functions in Wnt/beta -catenin signaling. J. Biol. Chem. 277, 37479–37486. 10.1074/jbc.M205658200. [DOI] [PubMed] [Google Scholar]
- Kenakin T.A Pharmacology Primer - Techniques for More Effective and Strategic Drug Discovery, 5th ed., Fedor J., Ed.; Academic Press, 2018; pp 85. [Google Scholar]
- Kilander M. B.; Dahlstrom J.; Schulte G. (2014) Assessment of Frizzled 6 membrane mobility by FRAP supports G protein coupling and reveals WNT-Frizzled selectivity. Cell. Signalling 26, 1943–1949. 10.1016/j.cellsig.2014.05.012. [DOI] [PubMed] [Google Scholar]
- Hirai H.; Matoba K.; Mihara E.; Arimori T.; Takagi J. (2019) Crystal structure of a mammalian Wnt-frizzled complex. Nat. Struct. Mol. Biol. 26, 372–379. 10.1038/s41594-019-0216-z. [DOI] [PubMed] [Google Scholar]
- Petersen J.; Wright S. C.; Rodriguez D.; Matricon P.; Lahav N.; Vromen A.; Friedler A.; Stromqvist J.; Wennmalm S.; Carlsson J.; et al. (2017) Agonist-induced dimer dissociation as a macromolecular step in G protein-coupled receptor signaling. Nat. Commun. 8, 226. 10.1038/s41467-017-00253-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBruine Z. J.; Ke J.; Harikumar K. G.; Gu X.; Borowsky P.; Williams B. O.; Xu W.; Miller L. J.; Xu H. E.; Melcher K. (2017) Wnt5a promotes Frizzled-4 signalosome assembly by stabilizing cysteine-rich domain dimerization. Genes Dev. 31, 916–926. 10.1101/gad.298331.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang I.; Kim H. R.; Beaven A. H.; Kim J.; Ko S. B.; Lee G. R.; Lee H.; Im W.; Seok C.; Chung K. Y.; et al. (2018) Biophysical and functional characterization of Norrin signaling through Frizzled4. Proc. Natl. Acad. Sci. U. S. A. 115, 8787–8792. 10.1073/pnas.1805901115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovira X.; Vivo M.; Serra J.; Roche D.; Strange P. G.; Giraldo J. (2009) Modelling the interdependence between the stoichiometry of receptor oligomerization and ligand binding for a coexisting dimer/tetramer receptor system. Br. J. Pharmacol. 156, 28–35. 10.1111/j.1476-5381.2008.00031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda C. Y.; Waghray D.; Levin A. M.; Thomas C.; Garcia K. C. (2012) Structural basis of Wnt recognition by Frizzled. Science 337, 59–64. 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povelones M.; Nusse R. (2005) The role of the cysteine-rich domain of Frizzled in Wingless- Armadillo signaling. EMBO J. 24, 3493–3503. 10.1038/sj.emboj.7600817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Dong S.; Xu F. (2018) Structural and Druggability Landscape of Frizzled G Protein- Coupled Receptors. Trends Biochem. Sci. 43, 1033–1046. 10.1016/j.tibs.2018.09.002. [DOI] [PubMed] [Google Scholar]
- DeBruine Z. J.; Xu H. E.; Melcher K. (2017) Assembly and architecture of the Wnt/beta-catenin signalosome at the membrane. Br. J. Pharmacol. 174, 4564–4574. 10.1111/bph.14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozielewicz P.; Turku A.; Schulte G. (2020) Molecular Pharmacology of Class F Receptor Activation. Mol. Pharmacol. 97, 62–71. 10.1124/mol.119.117986. [DOI] [PubMed] [Google Scholar]
- Miao Y.; Ha A.; de Lau W.; Yuki K.; Santos A. J. M.; You C.; Geurts M. H.; Puschhof J.; Pleguezuelos-Manzano C.; Peng W. C.; et al. (2020) Next-Generation Surrogate Wnts Support Organoid Growth and Deconvolute Frizzled Pleiotropy In Vivo. Cell Stem Cell 27, 840–851. 10.1016/j.stem.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valnohova J.; Kowalski-Jahn M.; Sunahara R. K.; Schulte G. (2018) Functional dissection of the N- terminal extracellular domains of Frizzled 6 reveals their roles for receptor localization and Dishevelled recruitment. J. Biol. Chem. 293, 17875–17887. 10.1074/jbc.RA118.004763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihs F.; Wang J.; Pfleger K. D. G.; Dacres H. (2020) Experimental determination of the bioluminescence resonance energy transfer (BRET) Forster distances of NanoBRET and red-shifted BRET pairs. Anal Chim Acta X 6, 100059. 10.1016/j.acax.2020.100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert A. F.; Gao P.; Wesslowski J.; Wang X.; Rath J.; Nienhaus K.; Davidson G.; Nienhaus G. U. (2020) Measuring ligand-cell surface receptor affinities with axial line-scanning fluorescence correlation spectroscopy. eLife 9, 9. 10.7554/eLife.55286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan T. H.; Liu Q.; Li C.; Wu G.; Steyaert J.; Lambert N. A. (2015) BRET evidence that beta2 adrenergic receptors do not oligomerize in cells. Sci. Rep. 5, 10166. 10.1038/srep10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozielewicz P.; Bowin C. F.; Turku A.; Schulte G. (2020) A NanoBRET-Based Binding Assay for Smoothened Allows Real-time Analysis of Ligand Binding and Distinction of Two Binding Sites for BODIPY-cyclopamine. Mol. Pharmacol. 97, 23–34. 10.1124/mol.119.118158. [DOI] [PubMed] [Google Scholar]
- Proffitt K. D.; Madan B.; Ke Z.; Pendharkar V.; Ding L.; Lee M. A.; Hannoush R. N.; Virshup D. M. (2013) Pharmacological inhibition of the Wnt acyltransferase PORCN prevents growth of WNT-driven mammary cancer. Cancer Res. 73, 502–507. 10.1158/0008-5472.CAN-12-2258. [DOI] [PubMed] [Google Scholar]
- Willert K. H. (2008) Isolation and application of bioactive Wnt proteins. Methods Mol. Biol. 468, 17–29. 10.1007/978-1-59745-249-6_2. [DOI] [PubMed] [Google Scholar]
- Cheng Y.; Prusoff W. H. (1973) Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50% inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22, 3099–3108. 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.