Abstract
Many SARS-CoV-2 variants with naturally acquired mutations have emerged. These mutations can affect viral properties such as infectivity and immune resistance. Although the sensitivity of naturally occurring SARS-CoV-2 variants to humoral immunity has been investigated, sensitivity to human leukocyte antigen (HLA)-restricted cellular immunity remains largely unexplored. Here, we demonstrate that two recently emerging mutations in the receptor-binding domain of the SARS-CoV-2 spike protein, L452R (in B.1.427/429 and B.1.617) and Y453F (in B.1.1.298), confer escape from HLA-A24-restricted cellular immunity. These mutations reinforce affinity toward the host entry receptor ACE2. Notably, the L452R mutation increases spike stability, viral infectivity, viral fusogenicity, and thereby promotes viral replication. These data suggest that HLA-restricted cellular immunity potentially affects the evolution of viral phenotypes and that a further threat of the SARS-CoV-2 pandemic is escape from cellular immunity.
Keywords: SARS-CoV-2, COVID-19, cellular immunity, spike protein, receptor-binding motif, naturally occurring variants, B.1.427/429, B.1.1.298, L452R, Y453F
Graphical abstract
Motozono and G2P-Japan Consortium et al. show that the emerging mutations L452R and Y453F in the SARS-CoV-2 spike receptor-binding motif evade HLA-A24-restricted cellular immunity. L452R increases spike stability, viral infectivity, and viral fusogenicity, thereby promoting viral replication. These data suggest that HLA-restricted cellular immunity potentially affects evolution of viral phenotypes.
Introduction
Coronavirus disease 2019 (COVID-19) is an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Since an unusual outbreak in Wuhan, Hubei Province, China, in December 2019 (Wu et al., 2020; Zhou et al., 2020), SARS-CoV-2 has rapidly spread worldwide, and as of May 2021, COVID-19 remains an ongoing pandemic: more than one-hundred million cases of SARS-CoV-2 infection have been reported worldwide, with more than three million people dying of COVID-19 (WHO, 2020a).
A variety of SARS-CoV-2 mutants have emerged during the pandemic and some have dominantly spread (reviewed in Plante et al., 2021b). A well-studied SARS-CoV-2 mutant harbors a D614G substitution in the spike (S) protein. Recent studies have revealed that the D614G mutation increases the binding affinity of SARS-CoV-2 to ACE2, the SARS-CoV-2 receptor (Ozono et al., 2021; Yurkovetskiy et al., 2020; Zhou et al., 2021); infectivity (Ozono et al., 2021; Yurkovetskiy et al., 2020; Zhou et al., 2021), fitness (Hou et al., 2020; Plante et al., 2021a; Zhou et al., 2021), and transmissibility in the human population (Volz et al., 2021) are also enhanced. However, there is no evidence suggesting that the D614G variant is associated with viral pathogenicity or lethality (Hou et al., 2020; Korber et al., 2020; Plante et al., 2021a). Additionally, since fall of 2020, new SARS-CoV-2 variants such as B.1.1.7 (also known as a variant of concern 202012/01 or 20I/501Y.V1), B.1.351 (also known as 20H/501Y.V2), and P.1 (also known as 501Y.V3) lineages have emerged in the UK, South Africa, and Brazil, respectively, and have rapidly spread worldwide (CDC, 2020). At the end of 2020, another lineage, B.1.427/429 (also known as CAL.20C), has been predominant, particularly in California state, USA (Deng et al., 2021; Zhang et al., 2021). Moreover, cross-species viral infection can accelerate the emergence of diverse viruses ([reviewed in Banerjee et al., 2021 and Parrish et al., 2008). In the case of SARS-CoV-2, various mammals, such as nonhuman primates (Chandrashekar et al., 2020; Munster et al., 2020; Yu et al., 2020) and carnivores (Halfmann et al., 2020; Kim et al., 2020; Shi et al., 2020), can become infected (Damas et al., 2020; Martínez-Hernández et al., 2020; OIE, 2021). Strikingly, the emergence of a SARS-CoV-2 variant, B.1.1.298, is likely associated with the outbreak in farmed minks in Denmark (Koopmans, 2021; WHO, 2020b), and phylogenetic analysis has provided evidence of mink-to-human transmission of SARS-CoV-2 within Danish mink farms (Oude Munnink et al., 2021). Because newly emerging variants can potentially change viral infectivity, transmissibility, and pathogenicity, deep monitoring of SARS-CoV-2 strains circulating globally and locally and evaluation of the effects of detected mutations on virological characteristics are urgent and crucial.
The emergence of mutated viruses is mainly due to error-prone viral replication, and the spread of emerged variants can be attributed to the escape from immune selective pressures (reviewed in Duffy et al., 2008). In fact, several SARS-CoV-2 mutants may be resistant to the neutralization mediated by antibodies from COVID-19 patients (Baum et al., 2020; Chen et al., 2021; Liu et al., 2021c; McCarthy et al., 2021; Weisblum et al., 2020) as well as those from vaccinated individuals (Liu et al., 2021b). Although the B1.1.7 variant is sensitive to convalescent and vaccinated sera (Collier et al., 2021; Garcia-Beltran et al., 2021; Shen et al., 2021; Supasa et al., 2021; Wang et al., 2021b), the B.1.351 and P.1 variants are relatively resistant to anti-SARS-CoV-2 humoral immunity (Garcia-Beltran et al., 2021; Hoffmann et al., 2021a; Wang et al., 2021b).
In addition to the humoral immunity mediated by neutralizing antibodies, another protection system against pathogens is the cellular immunity due to cytotoxic T lymphocytes (CTLs) (reviewed in Fryer et al., 2012 and Leslie et al., 2004). CTLs recognize nonself epitopes present on virus-infected cells via human leukocyte antigen (HLA) class I molecules; thus, CTL-mediated antiviral immunity is HLA restricted (reviewed in La Gruta et al., 2018). Recent studies have reported HLA-restricted SARS-CoV-2-derived epitopes that can be recognized by human CTLs (Kared et al., 2021; Kiyotani et al., 2020; Nelde et al., 2021; Schulien et al., 2021; Wilson et al., 2021). Furthermore, Le Bert et al. found that the functionality of virus-specific cellular immunity correlates inversely with COVID-19 severity (Le Bert et al., 2021). Therefore, it is conceivable that HLA-restricted CTLs play crucial roles in controlling SARS-CoV-2 infection and COVID-19 disorders, though compared to humoral immune responses, it remains unclear whether SARS-CoV-2 variants can escape cellular immunity.
In this study, we investigated the possibility of the emergence of SARS-CoV-2 mutants that are able to escape HLA-restricted cellular immunity. We demonstrate that at least two naturally occurring substitutions in the receptor-binding motif (RBM; residues 438–506) of the SARS-CoV-2 S protein, L452R and Y453F, which were identified in the two major variants, B.1.427/429 (L452R) and B1.1.298 (Y453F), can be resistant to cellular immunity in the context of HLA-A24, an allele of HLA-I. More intriguingly, the L452R and Y453F mutants enhance binding affinity for ACE2, and experiments using pseudoviruses show that the L452R substitution increases viral infectivity. Furthermore, we artificially generated SARS-CoV-2 harboring these point mutations by reverse genetics and show that the L452R mutant enhances viral replication capacity.
Results
Evasion from HLA-A24-restricted CTL responses by acquisition of mutations in the RBM of the SARS-CoV-2 S protein
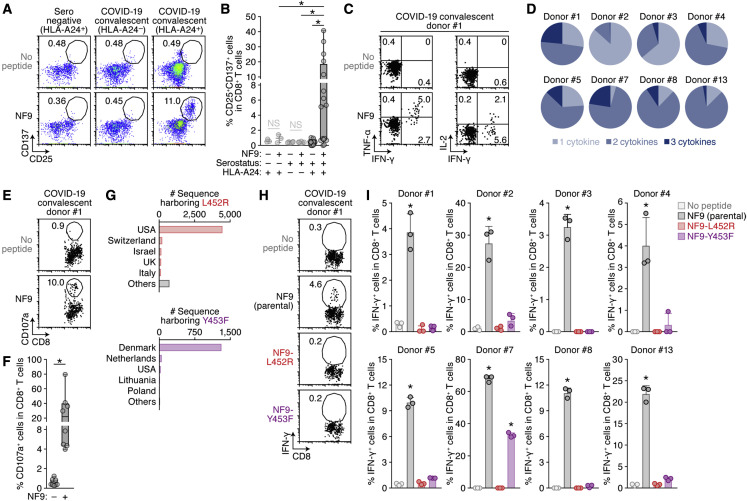
We set out to address the possibility of the emergence of naturally occurring mutants that can confer resistance to antigen recognition by HLA-restricted cellular immunity. A bioinformatic study has suggested that the 9-mer peptide in the RBM, NYNYLYRLF (we designate this peptide “NF9”), which spans 448–456 of the S protein, is the potential epitope presented by HLA-A24 (Kiyotani et al., 2020), an HLA-I allele widely distributed globally and particularly predominant in East and Southeast Asian areas (Table S1). Additionally, three immunological analyses using COVID-19 convalescent samples have shown that the NF9 peptide is an immunodominant epitope presented by HLA-A∗24:02 (Gao et al., 2021; Hu et al., 2020; Kared et al., 2021). To verify these observations, we obtained peripheral blood mononuclear cells (PBMCs) from 16 COVID-19 convalescent donors with HLA-A∗24:02 and stimulated these cells with the NF9 peptide (Table S2). As shown in Figure 1 A, two activation markers, CD25 and CD137, were upregulated in a fraction of CD8+ T cells in response to stimulation with NF9. In the 16 samples of the CD8+ T cells from COVID-19 convalescent patients with HLA-A∗24:02, the percentage of CD25+CD137+ cells in the presence of the NF9 peptide (median 5.2%) was significantly higher than that in its absence (median 0.44%) (Figure 1B; p = 0.0003 by Wilcoxon signed-rank test). In contrast, stimulation with the NF9 peptide did not upregulate CD25 and CD137 on the CD8+ T cells of three seronegative samples with HLA-A∗24:02, and the percentage of CD25+CD137+ cells in seronegative samples (median 0.93%) was significantly lower than that in COVID-19 convalescent samples (Figure 1B; p = 0.0003 by Wilcoxon signed-rank test). We also verified that the NF9 peptide did not activate the CD8+ T cells in six COVID-19 convalescent samples with non-HLA-A24 (median 0.43%) (Figure 1B; p = 0.031 by Wilcoxon signed-rank test versus HLA-A24-positive). Consistent with previous reports (Gao et al., 2021; Hu et al., 2020; Kared et al., 2021; Kiyotani et al., 2020), our data suggest that the NF9 peptide is an immunodominant HLA-A∗24:02-restricted epitope recognized by the CD8+ T cells of COVID-19 convalescents in our cohort. In particular, a convalescent sample from donor #13 exhibited a prominent CD8+ T cell-mediated response triggered by the NF9 peptide (Figure 1B; Table S2). Notably, the PBMCs of donor #13 were collected 68 days after PCR positivity (Table S2), suggesting that NF9-mediated CD8+ T cell immunity can be maintained for a relatively long period and that NF9 is an important epitope.
Figure 1.
Escape of two naturally occurring SARS-CoV-2 mutations from S RBM-specific CD8+ T cells
(A and B) Detection of HLA-A24-restricted NF9-specific CTLs. HLA-A24-positive CTL lines of 3 seronegative donors and 16 COVID-19 convalescent donors as well as 6 HLA-A24-negative COVID-19 convalescent donors (listed in Table S2) were stimulated with or without 1 μM NF9 peptide (NYNYLYRLF, residues 448–456 of the SARS-CoV-2 S protein). (A) Representative FACS plots showing surface expression of CD25 and CD137 on the CD8+ T cell subset (i.e., CD3+CD8+ cells) of an HLA-A24-positive seronegative donor (left), an HLA-A24-negative COVID-19 convalescent donor (middle), and an HLA-A24-positive COVID-19 convalescent donor #1 (right). (B) The median of the percentage of CD25+CD137+ cells among CD8+ T cells.
(C and D) Multifunctionality of HLA-A24-restricted NF9-specific CTLs. HLA-A24-positive CTL lines of eight COVID-19 convalescent patients were stimulated with or without 10 nM NF9 peptide. Representative FACS plots showing intracellular expression of IFN-γ, TNF-α, and IL-2 in the CD8+ T cell subset of COVID-19 convalescent donor #1 (C) and pie charts showing the proportion of cytokine-positive cells in each convalescent sample (D) are shown.
(E and F) Potential killing activity of HLA-A24-restricted NF9-specific CTLs. HLA-A24-positive CTL lines of eight COVID-19 convalescent donors were stimulated with C1R-A2402 cells pulsed with or without 10 nM NF9 peptide. Representative FACS plots showing surface expression of CD107a in the CD8+ T cell subset of a COVID-19 convalescent donor #1 (E) and the median of the percentage of CD107a+ cells in CD8+ T cells (F) are shown.
(G) Distribution of the L452R and Y453F mutants during the pandemic. Shown are the top five countries where variants harboring the L452R (top) and Y453F (bottom) mutations are found. The raw data are summarized in Table S3.
(H and I) Mutations conferring escape from HLA-A24-restricted NF9-specific CTLs. HLA-A24-positive CTL lines of eight COVID-19 convalescent donors were stimulated with 1 nM NF9 peptide or its derivatives: NF9-L452R (NYNYRYRLF) and NF9-Y453F (NYNYLFRLF). Representative FACS plots showing intracellular expression of IFN-γ in the CD8+ T cell subset of a COVID-19 convalescent donor #1 (H) and the mean percentage of IFN-γ+ cells in CD8+ T cells (I) are shown.
In (A, E, and H), the numbers in the FACS plot represent the percentage of gated cells among CD8+ T cells. In (C), the number represents the percentage of cells in each quadrant.
In (B), a statistically significant difference (∗p < 0.05) is determined by the Wilcoxon signed-rank test. NS, no statistical significance.
In (F), each symbol of the COVID-19 convalescent data represents the mean of technical triplicates. Statistically significant differences (∗p < 0.05) between COVID-19 convalescent samples with and without the NF9 peptide are determined by the Wilcoxon signed-rank test.
In (I), the assay was performed in triplicate, and the means are shown with the SD. Statistically significant differences (∗p < 0.05) versus “no peptide” are determined by ANOVA, with multiple comparisons by Bonferroni correction.
We then assessed the profile of cytokine production by NF9 stimulation. As depicted in Figure 1C, stimulation with the NF9 peptide induced the production of IFN-γ, TNF-α, and IL-2 by the CD8+ T cells from a COVID-19 convalescent patient. According to analysis using eight COVID-19 convalescent samples, the CD8+ T cells produced multiple cytokines in response to NF9 stimulation (Figure 1D), demonstrating the multifunctional nature of NF9-specific CD8+ T cells of COVID-19 convalescent patients. Moreover, the cytotoxic potential of NF9-specific CD8+ T cells was assessed by staining with surface CD107a, a degranulation marker (Figure 1E). As shown in Figure 1F, the percentage of CD107a+ cells among the CD8+ T cell population treated with the NF9 peptide (median 22.1%) was significantly higher than that among cells not treated with the NF9 peptide (median 0.88%) (p = 0.0078 by Wilcoxon signed-rank test), suggesting the cytotoxic potential of NF9-specific CD8+ T cells.
Next, we analyzed the diversity of SARS-CoV-2 during the current pandemic to assess the presence of naturally occurring variants harboring mutations in this region (residues 448–456 of the S protein). We downloaded 750,243 viral genome sequences from the Global Initiative on Sharing All Influenza Data (GISAID) database (https://www.gisaid.org; as of March 15, 2021). The L452R substitution was most frequent among the sequences analyzed (5,677 sequence), and 1,380 of the sequences reported contain the Y453F substitution (Table 1 ). Notably, lineages B.1.427/429 and B.1.1.298 (CDC, 2020) of the PANGO lineages (https://cov-lineages.org/index.html) mainly harbor the L452R and Y453F mutations, respectively (Figure 1G; Table S3). We then assessed the possibility that these two mutations affect the binding affinity of NF9 to HLA-A24 molecules. In silico analyses using five tools (Andreatta and Nielsen, 2016; Karosiene et al., 2012; Lundegaard et al., 2008; Nielsen et al., 2003; Reynisson et al., 2020; Zhang et al., 2009) predicted that the L452R mutation decreases the binding affinity of NF9 to HLA-A24, and four out of the five tools predicted that the Y453F mutation decreases the binding affinity (Figure S1A).
Table 1.
Naturally occurring mutations in residues 448–456 of the SARS-CoV-2 S protein
| Mutation | # sequence | Date first detected | Country first detected | Dominant PANGO lineage | Date first detected in the dominant lineage | Country first detected in the dominant lineage | Nonhuman hosts detecteda |
|---|---|---|---|---|---|---|---|
| L452R | 5,677 | March 17, 2020 | Denmark | B.1.427/429 | July 6, 2020 | Mexico | gorilla (1) |
| Y453F | 1,380 | April 20, 2020 | Denmark | B.1.1.298 | April 20, 2020 | Denmark | mink (339) and cat (3) |
| N450K | 181 | March 27, 2020 | UK | B.1.214.2 | December 5, 2020 | UK | - |
| L452M | 140 | April 13, 2020 | Japan | B.1.22/36 | July 2, 2020 | Netherlands | mink (32) |
| L452Q | 111 | March 6, 2020 | Spain | B.1.1.374 | October 23, 2020 | Belarus | - |
The number in parentheses indicates the number of sequences reported.
To experimentally address the possibility that these naturally occurring mutations, L452R and Y453F in the NF9 region, confer evasion ability from NF9-specific CD8+ T cells in HLA-A24-positive COVID-19 convalescents, two NF9 derivatives containing either substitution (NF9-L452R and NF9-Y453F) were prepared and used for stimulation experiments. As shown in Figure S1B, parental NF9 induced IFN-γ expression in a dose-dependent manner. In contrast, the level of IFN-γ expression induction by the NF9-Y453F derivative was significantly lower than that by parental NF9, and intriguingly, the NF9-L452R derivative did not induce IFN-γ expression, even at the highest concentration tested (10 nM) (Figure S1C). Among the eight HLA-A24-positive COVID-19 convalescent samples tested, parental NF9 peptide significantly induced IFN-γ expression. In contrast, the NF9-L452R derivative did not activate all CD8+ T cells tested, and seven out of the eight HLA-A24-positive COVID-19 convalescent samples failed to be activated by the NF9-Y453F derivative (Figures 1H and 1I). Altogether, these results suggest that the NF9 peptide, which is derived from the SARS-CoV-2 S protein RBM, is an immunodominant epitope of HLA-A24 and that two naturally occurring mutants, L452R and Y453F, are able to evade HLA-A24-restricted cellular immunity.
Augmentation of the binding affinity for ACE2 by the L452 and Y453 mutations
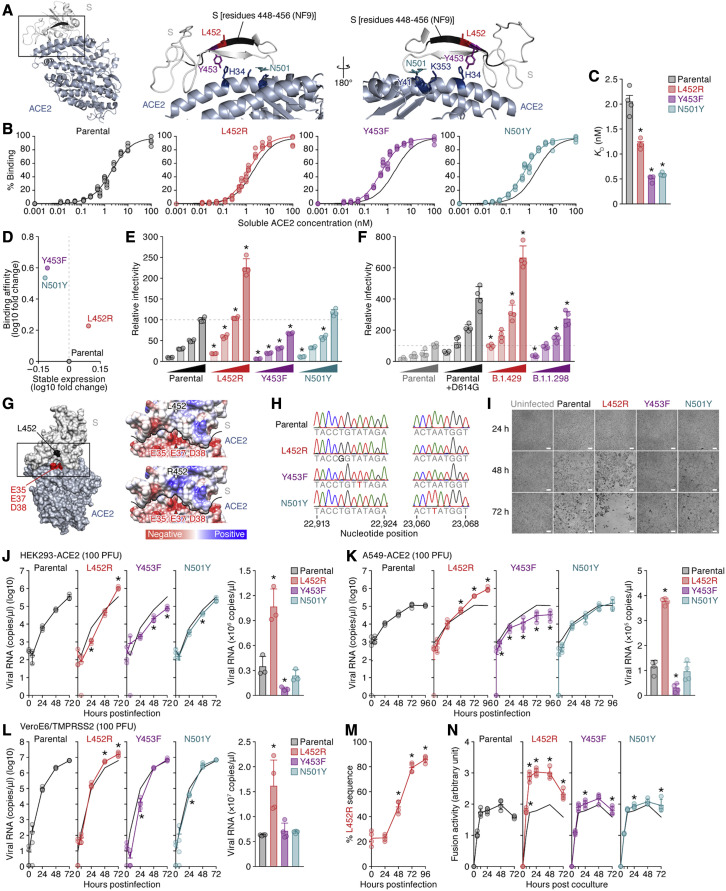
We next sought to determine whether the mutations of interest affect the efficacy of virus infection. Structural analyses have shown that the Y453 and N501 residues in the RBM are located at the interface between the SARS-CoV-2 RBM and human ACE2 and directly contribute to human ACE2 binding but that the L452 residue is not located at this interface (Lan et al., 2020; Wang et al., 2020; Zhao et al., 2020) (Figure 2 A). To directly assess the effect of these RBM mutations on binding to ACE2, we prepared yeasts expressing the parental SARS-CoV-2 receptor-binding domain (RBD) (residues 336–528) and derivatives (L452R, Y453F, and N501Y) and performed an in vitro binding assay using the yeast surface display of the RBD and soluble ACE2 protein. Consistent with recent studies, including ours (Supasa et al., 2021; Zahradník et al., 2021b), the N501Y mutation, which is a common mutation in B1.1.7, B1.351, and P.1 variants (reviewed in Plante et al., 2021b), as well as the Y453F mutation (Bayarri-Olmos et al., 2021; Zahradník et al., 2021b), significantly increased binding affinity for human ACE2 (Figures 2B and 2C; RBD parental K D = 2.05 ± 0.26 nM, RBD N501Y K D = 0.59 ± 0.03 nM, and RBD Y453F K D = 0.51 ± 0.06 nM). We also found that the L452R mutant significantly increased the binding affinity to human ACE2 (Figures 2B and 2C; RBD L452R K D = 1.20 ± 0.06 nM). Intriguingly, the L452R mutations increased surface expression, which reflects protein stability (Traxlmayr and Obinger, 2012), but the Y453F and N501Y mutations decreased it (Figure 2D).
Figure 2.
Increase in the binding affinity to ACE2, viral infectivity, viral fusogenicity, and viral replication capacity by the L452 mutation
(A) Location of the NF9 peptide (residues 448–456) in the cocrystal structure of the SARS-CoV-2 S and human ACE2 proteins (PDB: 6M17) (Yan et al., 2020). An overview (left), an enlarged view of the boxed area in the left panel (middle), and a view of the middle panel rotated 180° on the y axis (right) are shown. Residues 448–456 of SARS-CoV-2 S (corresponding to the NF9 peptide) are shown in black.
(B–D) Binding affinity of SARS-CoV-2 S RBD to ACE2 by yeast surface display. The percentage of the binding of the SARS-CoV-2 S RBD expressed on yeast to soluble ACE2 (B) and the KD values (C) are shown. Assays were performed in quadruplicate. (D) The level of stable expression of the SARS-CoV-2 RBD on yeast (x axis) and the binding affinity toward ACE2 (y axis) compared to the parental RBD. In (B), the fitting curve of parental RBD is shown as black lines in all panels.
(E and F) Pseudovirus assay. The HIV-1-based reporter virus pseudotyped with the parental SARS-CoV-2 S or its derivatives (E, L452R, Y453F, and N501Y; F, D614G, B.1.429 [S13I/W152C/L452R/D614G], and B.1.1.298 [HV69-70del/Y453F/D614G]) was inoculated into HEK293 cells transiently expressing human ACE2 and TMPRSS2 at four different doses (1, 3, 5, and 10 ng p24 antigens). Percentages of infectivity compared to the virus pseudotyped with parental S (10 ng p24 antigen) are shown.
(G) Gain of electrostatic complementarity by the L452R substitution. Left: the surface structure of SARS-CoV-2 S and ACE2 (PDB: 6M17) (Yan et al., 2020). Residue 452 of SARS-CoV-2 S and the negatively charged patch on ACE2 (residues E35, E37, and D38) are indicated in black and red. The boxed area is enlarged in the upper right panel. Right: Coulombic surface coloring of the structures of SARS-CoV-2 S and ACE2 (PDB: 6M17) (Yan et al., 2020) (top) and a model of the L452R substitution (bottom). The black line indicates the border between SARS-CoV-2 S and ACE2.
(H) Chromatograms of the mutated regions of SARS-CoV-2 viruses artificially generated by reverse genetics. Chromatograms of nucleotide positions 22,913–22,924 (left) and 23,060–23,068 (right) of parental SARS-CoV-2 (strain WK-521; GISAID ID: EPI_ISL_408667) and the L452R (T22917G in nucleotide), Y453F (A22920T in nucleotide), and N501Y (A23063T in nucleotide) mutants are shown.
(I–L) Growth kinetics of parental SARS-CoV-2 and SARS-CoV-2 mutants. Parental SARS-CoV-2 and the L452R, Y453F, and N501Y mutants (100 plaque-forming units [PFU]) were inoculated into HEK293-ACE2 cells (I and J), A549-ACE2 cells (K), and VeroE6/TMPRSS2 cells (L), and the copy number of viral RNA in the culture supernatant was quantified by real-time PCR. (I) Representative bright-field images of HEK293-ACE2 cells uninfected or infected with the viruses indicated at 24, 48, or 72 h post-infection are also shown. Bars, 200 μm. (J–L) Left: the growth curve of the viruses inoculated. The result for the parental virus is shown in all panels as a black line. Right: the amount of viral RNA in the culture supernatant at 72 h post-infection. Assays were performed in triplicate (J) or quadruplicate (K and L).
(M) Competition assay. Parental virus and the L452R mutant were mixed at a 1:1 ratio based on PFU, and the mixture was inoculated into HEK293-ACE2 cells. The percentage of L452R mutant at each time point was analyzed as described in STAR Methods. The data are shown as the average of four biological replicates.
(N) SARS-CoV-2 S-based fusion assay. Effector cells (S-expressing cells) and target cells (ACE2-expressing cells) were prepared, and the fusion activity was measured as described in STAR Methods. Assays were performed in quadruplicate, and fusion activity (arbitrary unit) is shown.
In (C), statistically significant differences (∗p < 0.05) versus parental S are determined by the Mann-Whitney U test.
In (E and F), statistically significant differences (∗p < 0.05) versus parental S (E) and the D614G mutant (F) at the same dose were determined by Student’s t test.
In (J–L and N), statistically significant differences (∗p < 0.05) versus parental virus (J–L) or parental S (N) were determined by Student’s t test.
In (M), values at respective time points were compared with those at the last time point using a two-tailed, paired Student’s t test, and an asterisk denotes familywise error rate <0.05 using the Holm test.
See also Figure S1.
Increase in pseudovirus infectivity by the L452R mutation
To directly analyze the effect of these mutations on viral infectivity, we prepared an HIV-1-based reporter virus pseudotyped with the SARS-CoV-2 S protein and its mutants as well as HEK293 cells transiently expressing human ACE2 and TMPRSS2. As shown in Figure 2E, although the N501Y mutation slightly affected viral infectivity in this assay, the L452R mutation significantly increased it compared to the parental S protein. In contrast to the yeast display assay (Figures 2B and 2C), the infectivity of the Y453F mutant was significantly lower than that of the parental S protein (Figure 2E). Similar to the observations using human ACE2 (Figure 2E), the L452R mutant, but not the other mutants including the Y453F mutant, significantly increased infection efficacy using mink ACE2 as the receptor (Figure S1D). Moreover, we demonstrated that the infectivity of the reporter virus pseudotyped with the S protein of the B.1.429 variant, which harbors the L452R mutation, was significantly higher than those of parental S as well as the D614G-harboring S, while that of the B.1.1.298 variant, which harbors the Y453F mutation, was not (Figure 2F). Altogether, these findings suggest that the L452R substitution increases the binding affinity of the SARS-CoV-2 RBD toward human ACE2, protein stability, and viral infectivity. Although the L452 residue is not directly located at the binding interface (Figure 2A), structural analysis and in silico mutagenesis suggest that the L452R substitution promotes electrostatic complementarity (Selzer et al., 2000) (Figure 2G). Because residue 452 is located in close proximity to a negatively charged patch of ACE2 residues (E35, E37, and D38), the increase in viral infectivity caused by the L452R substitution can be attributed to an increase in electrostatic interaction with ACE2.
Promotion of SARS-CoV-2 replication in cell cultures by the L452 mutation
To investigate the effect of RBM mutations on viral replication, we artificially generated recombinant SARS-CoV-2 viruses that harbor the mutations of interest as well as parental recombinant virus by a reverse genetics system (Torii et al., 2021). The nucleotide similarity of SARS-CoV-2 strain WK-521 (GISAID ID: EPI_ISL_408667) (Matsuyama et al., 2020), the backbone of the artificially generated recombinant SARS-CoV-2, to strain Wuhan-Hu-1 (GenBank: NC_045512.2) (Wu et al., 2020) is 99.91% (27 nucleotide difference); the sequences encoding the S protein between these two strains are identical, indicating that strain WK-521 is a SARS-CoV-2 prototype. We verified insertion of the targeted mutations in the viruses generated by direct sequencing (Figure 2H) and performed virus replication assays using these recombinant viruses. As shown in Figure 2I, the cytotoxicity of the L452R mutant was higher than that of the other mutants and the parental virus. We also found that the growth rates of the L452R mutant in VeroE6/TMPRSS2 cells (Figure 2J), HEK293 cells stably expressing human ACE2 (HEK293-ACE2; Figure 2K), and A549 cells stably expressing human ACE2 (A549-ACE2; Figure 2L) were significantly higher than those of the parental virus. The predominance of the L452R mutant was observed at different doses (Figure S1E). Moreover, a competition assay using the parental virus and the L452R mutant showed that the L452R mutant expanded more predominantly than the parental virus (Figure 2M). Together with the results of the binding assay (Figures 2B–2D) and the assay using pseudoviruses (Figures 2E, 2F, and S1E), our results suggest that the L452R mutation promotes viral replication.
Increase in viral fusogenicity by the L452R mutation
We next tested how the L452R mutation increases viral infectivity and cytotoxicity by performing a SARS-CoV-2 S-based fusion assay. As shown in Figure S1F, the surface expression levels of parental S and its derivatives on effector cells were comparable. Using the SARS-CoV-2 S-based fusion assay, we demonstrated that the L452R mutation significantly increased fusion efficacy compared to the parental S, as well as the other mutants (Figure 2N). Our data suggest that the L452R mutation promotes viral replication by increasing viral fusogenicity.
Dynamics of the spread of RBM mutants during the current pandemic
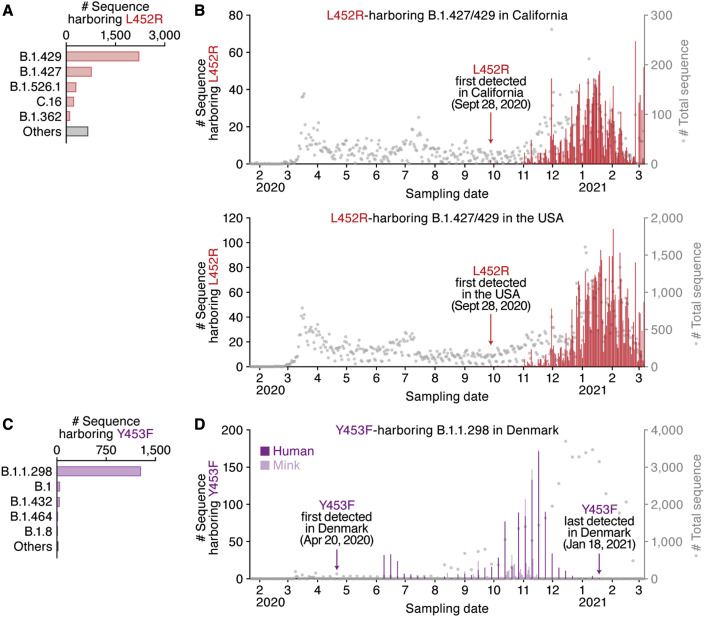
We finally assessed the epidemic dynamics of naturally occurring variants containing the L452 and Y453 substitutions. The L452R mutants were mainly found (3,967 sequences) in the B.1.427/B.1.429 lineage, which forms a single clade (Deng et al., 2021) (Figure 3 A; Table S4). Although the L452R mutant was first detected in the B.1.39 lineage in Denmark on March 17, 2020 (GISAID ID: EPI_ISL_429311) (Table 1), this variant did not spread. The oldest sequence that contains the L452R mutation in the B.1.427/B.1.429 lineage was isolated in Quintana Roo state, Mexico, on July 6, 2020 (GISAID ID: EPI_ISL_942929) (Table 1), and L452R-harboring mutants were first collected in California, USA, on September 28, 2020 (GISAID ID: EPI_ISL_730092 and EPI_ISL_730345) (Figure 3B). The B.1.427/B.1.429 lineage harboring the L452R mutation started expanding in California at the beginning of November 2020 (Figure 3B, top). In 2021, this lineage expanded throughout the USA, and it is currently one of the most predominant lineages in the country (Figure 3B, bottom; Table S5).
Figure 3.
Epidemic dynamics of the B.1.1.298 and B.1.427/429 lineages during the pandemic
The PANGO lineages harboring L452R (A and B) and Y453F (C and D) and their epidemic dynamics are summarized.
(A and C) Distribution of the L452R and Y453F mutants during the pandemic. The top five PANGO lineages (https://cov-lineages.org/index.html) that harbor the L452R (A) and Y453F (C) mutations are shown. The raw data are summarized in Table S3.
(B and D) Epidemic dynamics of the L452R-harboring B.1.427/429 lineage in California, USA (B, top) and the USA (B, bottom), and the Y453F-harboring B.1.1.298 lineage in Denmark (D). The numbers of sequences harboring mutations per day (left y axis, bars) and the numbers of total sequences per day (right y axis, dots) from January 22, 2020 to March 6, 2021 are summarized. Note that an L452R variant isolated from gorillas and three Y453F variants isolated from cats are not included.
Regarding the Y453F mutation, 1,274 of the 1,380 mutated sequences belong to the B.1.1.298 lineage, which has been exclusively detected in Denmark (Table S4). The oldest sequence in the B.1.1.298 lineage that contains the Y453F mutation was isolated from a human in Denmark on April 20, 2020 (GISAID ID: EPI_ISL_714253) (Figure 3C). Intriguingly, the B.1.1.298 variants containing either Y453 or F453 were detected not only in humans but also in minks (Figure 3D). Phylogenetic analysis of the entire genome sequences of the B.1.1.298 lineage suggested multiple SARS-CoV-2 transmissions between humans and minks (Figure S2). Additionally, the three sequences that contain the Y453F mutation were isolated from cats in Denmark: two identical sequences (GISAID ID: EPI_ISL_683164 and EPI_ISL_683166) and another sequence (GISAID ID: EPI_ISL_683165) of a distinct origin (Figure S2). These results suggest that this SARS-CoV-2 variant has transmitted from humans to cats multiple times and that some of them may have spread among the Danish cat population. Nevertheless, the epidemic of a fraction of the B.1.1.298 lineage containing the Y453F mutation in Denmark peaked from October to November 2020 and then gradually declined (Figure 3D). The variant containing the Y453F mutation was last collected in Denmark on January 18, 2021 (GISAID ID: EPI_ISL_925998), and it has not been reported worldwide since (Figure 3D).
Discussion
In the present study, we demonstrate that at least two naturally occurring mutations in the SARS-CoV-2 RBM, L452R and Y453F, escape HLA-restricted cellular immunity and further promote affinity toward the viral receptor ACE2. We also demonstrate that the L452R mutation increases the stability of the S protein and viral infectivity and thereby enhances viral replication. Our data suggest that the L452R mutant escapes HLA-A24-restricted cellular immunity and further strengthens its infectivity. Consistent with our findings, Deng et al. performed a pseudovirus assay and showed that the L452R mutation increases viral infectivity (Deng et al., 2021). However, the mechanism of action was not revealed. Here, we demonstrated that the L452R mutation increases viral fusogenicity.
Lines of recent evidence show the emergence of SARS-CoV-2 variants that evade anti-SARS-CoV-2 neutralizing humoral immunity (Baum et al., 2020; Chen et al., 2021; Garcia-Beltran et al., 2021; Hoffmann et al., 2021a; Liu et al., 2021b, 2021c; McCarthy et al., 2021; Wang et al., 2021a; Weisblum et al., 2020), forewarning the risk of the spread of immune escape variants. In addition to humoral immunity, Zuo et al. reported that functional SARS-CoV-2-specific cellular immune responses are retained for at least 6 months after infection (Zuo et al., 2021). Le Bert et al. also found that functional cellular immune responses can contribute to controlling the progression of COVID-19 (Le Bert et al., 2021). These observations suggest the importance of cellular immunity that elicits efficient antiviral effects. However, the possibility of the emergence of SARS-CoV-2 variants that are able to evade cellular immunity has not yet been addressed. Here, we demonstrate that the L452R and Y453F mutations contribute to escape from HLA-restricted cellular immunity. In addition to our findings, recent papers have documented that the L452R (Deng et al., 2021; Li et al., 2020) and Y453F (Baum et al., 2020; Hoffmann et al., 2021b) substitutions may confer resistance to neutralizing antibodies, suggesting that these mutants can evade both humoral and HLA-restricted cellular immunity. Furthermore, we demonstrate that the L452R mutation significantly improves viral replication capacity by increasing binding affinity to human ACE2 as well as S protein stability. Altogether, our findings suggest that the emergence of variants that can escape HLA-restricted cellular immunity and further enhance viral replication capacity is a potential risk for deterioration of the situation regarding the COVID-19 pandemic.
Previous papers have reported that the L452R (Garcia-Beltran et al., 2021; Liu et al., 2021c) and Y453F mutations (Baum et al., 2020; Garcia-Beltran et al., 2021) had less effect on the sensitivity to neutralizing antibodies (e.g., convalescent/vaccinated sera and monoclonal antibodies). On the other hand, a recent study has shown that the B.1.427/429 variant, which harbors the L452R mutation, is 2–6.7-fold more resistant to neutralizing antibodies than the non-L452R prototypic virus (Deng et al., 2021). Although the effects of L452R and Y453F mutations on sensitivity to neutralizing antibodies are controversial, our data demonstrate that these mutations evade HLA-A24-mediated cellular immunity. Therefore, the L452R/Y453F mutations would more crucially impact HLA-A24-mediated cellular immunity than sensitivity to neutralizing antibodies.
As suggested in previous reports (Koopmans, 2021; Oude Munnink et al., 2021; WHO, 2020b), our data show that the B.1.1.298 variant possessing the Y453F substitution is closely associated with the outbreak among minks in Denmark. Although it remains unclear whether the emergence of the Y453F mutant may be associated with evasion from acquired immunity in minks, we demonstrate here that this mutation does confer resistance to HLA-A24-restricted human cellular immunity. Because the Y453F mutation did not increase infection efficacy when using mink ACE2, our results suggest that the emergence of this mutation is not due to enhanced viral fitness in minks. Nevertheless, the host range of SARS-CoV-2 in terms of the use of ACE2 molecules as receptors mediating infection is broad in a variety of mammals (Liu et al., 2021a; OIE, 2021). More importantly, although murine ACE2 cannot be used for infection of prototype SARS-CoV-2, recent studies have revealed that some SARS-CoV-2 variants, including B.1.351 and P.1, have gained the ability to utilize murine ACE2 for infection, expanding their host range to mice (Li et al., 2021; Montagutelli et al., 2021). In addition to evasion from acquired immunity in humans, zoonotic and zooanthroponotic SARS-CoV-2 transmission can contribute to the accumulation of mutations in spreading viruses and further impact viral phenotypes, including infectivity, replication efficacy, pathogenicity, transmissibility, and even host range. Therefore, surveillance of novel variant emergence, even in nonhuman mammals, and assessment of the potential of these viruses to adapt to nonhuman ACE2 for infection receptors will be critical.
In contrast to the B.1.1.298 variant, the B.1.427/429 variant harboring the L452R substitution appears to have emerged during spread in the human population. Because the L452R mutation reinforces binding to human ACE2 and further enhances viral replication capacity, this variant might have emerged to improve viral fitness in humans. Another possibility is that the L452R mutant developed to evade HLA-A24-restricted cellular immunity: HLA-A24 is relatively predominant in East Asian individuals (Gonzalez-Galarza et al., 2020), and the proportion of Asian Americans in the USA is highest in California (CDC, 2019). For instance, the proportion of HLA-A24-positive individuals is ∼20% in San Diego (Moore et al., 2018), California, where approximately 280,000 SARS-CoV-2 infection cases have been reported thus far (https://coronavirus.jhu.edu; as of May 22, 2021). Because the B.1.427/429 variant harboring L452R mutation has been predominantly spreading in California, it may be assumed that the emergence of the L452R mutant (or the B.1.427/429 lineage) was driven by HLA-A24-mediated cellular immunity.
In addition to escape from antiviral acquired immunity, recent studies have shown that variants emerging during the current pandemic, particularly the B.1.1.7 variant, can even increase SARS-CoV-2 pathogenicity and the mortality of COVID-19 (Challen et al., 2021; Davies et al., 2021; Grint et al., 2021). Importantly, HLA-A24 individuals are relatively highly frequent in East and Southeast Asian countries such as Japan (Japanese, allele frequency = 0.364, n = 1,550) and Malaysia (Malaysian, allele frequency = 0.361, n = 1,974) (Gonzalez-Galarza et al., 2020) (Table S1), where both the number of confirmed cases and COVID-19 mortality are relatively lower than those in European countries and the USA (https://coronavirus.jhu.edu; as of May 22, 2021).
There are some limitations in this study. First, we hypothesized that the emergence of the L452R and Y453F mutations might be attributed to the immune pressure triggered by HLA-A24-mediated cellular immunity. However, since the frequency of HLA-A24-positive individuals in California and Denmark is inaccessible, the magnitude of HLA-A24-mediated immunity against SARS-CoV-2 in these states and countries remains unclear. Second, the magnitude of NF9-mediated HLA-A24-restricted antiviral cellular immunity was not revealed. To clarify this, further investigations with larger cohorts of COVID-19 patients are essential. Third, the effects of the L452 and Y453 substitutions on viral pathogenicity, disease severity, mortality, and transmissibility remain unaddressed. To fully characterize the virological features of these mutants, further investigations using animal models and epidemiological data will be required. Nevertheless, we provide direct evidence suggesting that mutations in the RBM, including L452R (in the B.1.427/429 lineage) and Y453F (in the B1.1.298 lineage), may confer escape from HLA-A24-restricted cellular immunity, and furthermore, that the L452R mutant has increased replication capacity. Accordingly, variants, particularly those possessing the L452R mutation, such as the B.1.427/429 lineage, are a potential threat in countries and regions with a predominance of HLA-A24 individuals, and deep surveillance and tracing of the epidemic related to these variants are urgently required.
Finally, in May 2021, a novel variant of concern, the B.1.617 lineage, emerged that was closely associated with a massive COVID-19 surge in India. Notably, in addition to the B.1.427/429 lineage, the L452R mutation is a hallmark of the B.1.617 lineage. Therefore, the L452R mutation would be a mutation of concern and should be targeted by deep surveillance.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FITC-conjugated anti-human CD3 antibody | Biolegend | Cat# 300440; RRID: AB_2562046 |
| BV510-conjugated anti-human CD3 antibody | Biolegend | Cat# 317331; RRID: AB_2561376 |
| APC-Cy7-conjugated anti-human CD8 antibody | Biolegend | Cat# 301016; RRID: AB_314134 |
| PerCP-Cy5.5-conjugated anti-human CD14 antibody | Biolegend | Cat# 325622; RRID: AB_893250 |
| PerCP-Cy5.5-conjugated anti-human CD19 antibody | Biolegend | Cat# 302230; RRID: AB_2073119 |
| PE-Cy7-conjugated anti-human CD25 antibody | Biolegend | Cat# 356107; RRID: AB_2561974 |
| APC-conjugated anti-human CD137 antibody | Biolegend | Cat# 309809; RRID: AB_830671 |
| BV421-conjugated anti-human CD107a antibody | Biolegend | Cat# 328625; RRID: AB_10899581 |
| PE-conjugated anti-human IFN-γ antibody | BD Biosciences | Cat# 554552; RRID: AB_395474 |
| PE-Cy7-conjugated anti-human TNF-α antibody | Biolegend | Cat# 502930; RRID: AB_2204079 |
| APC-conjugated anti-human IL-2 antibody | Biolegend | Cat# 500310; RRID: AB_315097 |
| Anti-ACE2 antibody | R&D Systems | Cat# AF933; RRID: AB_355722 |
| APC-conjugated anti-goat IgG | R&D Systems | Cat# F0108; RRID: AB_573124 |
| Goat normal IgG | R&D Systems | Cat# AB-108-C; RRID: AB_354267 |
| Anti-SARS-CoV-2 S antibody | GeneTex | Cat# GTX635654; RRID: AB_2888548 |
| APC-conjugated anti-rabbit IgG | Jackson ImmunoResearch | Cat# 111-136-144; RRID: AB_2337987 |
| Rabbit normal IgG | SouthernBiotech | Cat# 0111-01; RRID: AB_2732899 |
| Bacterial and virus strains | ||
| SARS-CoV-2 (strain WK-521) | Matsuyama et al., 2020 | GISAID ID: EPI_ISL_408667 |
| Biological samples | ||
| Human PBMCs | This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Ficoll-Paque Plus | GE Healthcare Life Sciences | Cat# 17-1440-03 |
| RPMI1640 medium | Thermo Fisher Scientific | Cat# 11875101 |
| Fetal calf serum (FCS) | Sigma-Aldrich | Cat# 172012-500ML |
| Penicillin streptomycin (PS) | Sigma-Aldrich | Cat# P4333-100ML |
| Dulbecco’s modified Eagle’s medium (high glucose) | Wako | Cat# 044-29765 |
| Eagle’s minimum essential medium (with L-glutamine and phenol red) | Wako | Cat# 051-07615 |
| Dulbecco’s modified Eagle’s medium (low glucose) | Wako | Cat# 041-29775 |
| G418 | Nacalai Tesque | Cat# G8168-10ML |
| Dulbecco’s modified Eagle’s medium (high glucose) | Sigma-Aldrich | Cat# R8758-500ML |
| Blasticidin | InvivoGen | Cat# ant-bl-1 |
| Ham’s F-12K medium | Wako | Cat# 080-08565 |
| Expi293 expression medium | Thermo Fisher Scientific | Cat# A1435101 |
| SARS-CoV-2 PepTivator peptide pools | Miltenyi Biotec | Cat# 130-126-700 |
| Recombinant human IL-2 | Peprotec | Cat# 200-02 |
| NF9 peptide (NYNYLYRLF, residues 448-456 of the SARS-CoV-2 S protein) | This study | N/A |
| L452R peptide (NYNYRYRLF, L5R in NF9) | This study | N/A |
| Y453F peptide (NYNYLFRLF, Y6F in NF9) | This study | N/A |
| 7-aminoactinomycin D | Biolegend | Cat# 420404 |
| Paraformaldehyde | Nacalai Tesque | Cat# 09154-85 |
| Brefeldin A | Sigma-Aldrich | Cat# B7651 |
| Monensin | Biolegend | Cat# 420701 |
| PrimeSTAR GXL DNA polymerase | Takara | Cat# R050A |
| KpnI | New England Biolab | Cat# R0142S |
| NotI | New England Biolab | Cat# R1089S |
| MluI-HF | New England Biolab | Cat# R3198L |
| SmaI | New England Biolab | Cat# R0141L |
| HpaI | New England Biolab | Cat# R0105L |
| XhoI | New England Biolab | Cat# R0146S |
| Bilirubin | Sigma-Aldrich | Cat# 14370-1G |
| CF640R succinimidyl ester | Biotium | Cat# 92108 |
| Lipofectamine 3000 | Thermo Fisher Scientific | Cat# L3000015 |
| Lipofectamine 2000 | Thermo Fisher Scientific | Cat# 11668019 |
| DNase I | Sigma-Aldrich | Cat# 11284932001 |
| TransIT-LT1 | Takara | Cat# MIR2300 |
| Puromycin | InvivoGen | Cat# ant-pr-1 |
| Doxycycline | Takara | Cat# 1311N |
| TURBO DNase | Thermo Fisher Scientific | Cat# AM2238 |
| Minimum essential medium (2 × ), no phenol red | Thermo Fisher Scientific | Cat# 11935046 |
| Nonessential amino acid | Thermo Fisher Scientifc | Cat# 11140-050 |
| Carboxymethyl cellulose | Sigma-Aldrich | Cat# C9481-500G |
| Formaldehyde | Nacalai Tesque | Cat# 37152-51 |
| Crystal violet | Nacalai Tesque | Cat# 09804-52 |
| Methylene blue | Nacalai Tesque | Cat# 22412-14 |
| Recombinant RNase inhibitor | Takara | Cat# 2313B |
| Transcriptor reverse transcriptase | Roche | Cat# 03531317001 |
| Critical commercial assays | ||
| Cytofix/Cytoperm Fixation/Permeabilization solution kit | BD Biosciences | Cat# 554714 |
| QIAamp RNA blood mini kit | QIAGEN | Cat# 52304 |
| SuperScript III reverse transcriptase | Thermo Fisher Scientific | Cat# 18080085 |
| ExpiFectamine 293 transfection kit | Thermo Fisher Scientific | Cat# A14525 |
| KAPA HiFi HotStart ReadyMix kit | Roche | Cat# KK2601 |
| One-Glo luciferase assay system | Promega | Cat# E6130 |
| GENEART site-directed mutagenesis system | Thermo Fisher Scientific | Cat# A13312 |
| QIAamp viral RNA mini kit | QIAGEN | Cat# 52906 |
| One Step TB Green PrimeScript PLUS RT-PCR kit | Takara | Cat# RR096A |
| SARS-CoV-2 direct detection RT-qPCR kit | Takara | Cat# RC300A |
| GeneJET gel extraction kit | Thermo Fisher Scientific | Cat# K0692 |
| EnduRen live cell substrate | Promega | Cat# E6481 |
| Experimental models: Cell lines | ||
| Human: C1R-A2402 cells | Karaki et al., 1993 | N/A |
| Human: HEK293 cells | ATCC | CRL-1573 |
| Human: HEK293T cells | ATCC | CRL-3216 |
| African green monkey (Chlorocebus sabaeus): Vero cells | JCRB | JCRB0111 |
| African green monkey (Chlorocebus sabaeus): VeroE6/TMPRSS2 cells | JCRB | JCRB1819 |
| Mink (Neovison vison): Mv.1.Lu cells | ATCC | CCL-64 |
| Human: HEK293-C34 cells | Torii et al., 2021 | N/A |
| Human: HEK293-ACE2 cells | This study | N/A |
| Human: A549-ACE2 cells | This study | N/A |
| Human: Expi293F cells | Thermo Fisher Scientific | Cat# A14527 |
| Yeast (Saccharomyces cerevisiae): strain EBY100 | ATCC | MYA-4941 |
| Oligonucleotides | ||
| Forward primer for pC-SARS2-S: TTG GGTACC ATG TTT GTG TTC CTG GTG CTG | Ozono et al., 2021 | N/A |
| Reverse primer for pC-SARS2-S: GTG GCGGCCGC TCT AGA TTC AGG TGT AGT GCA GTT T | Ozono et al., 2021 | N/A |
| Forward primer for S L452R: GTG GGA GGC AAC TAC AAC TAC CGT TAC | This study | N/A |
| Forward primer for S Y453F: GTG GGA GGC AAC TAC AAC TAC CTC TTC AGA | This study | N/A |
| Reverse primer for S L452R/L453F: GTT GTA GTT GCC TCC CAC CTT | This study | N/A |
| Forward primer for S N501Y: TCC TAT GGC TTC CAA CCA ACC TAT GGA | This study | N/A |
| Reverse primer for S N501Y: TGG TTG GAA GCC ATA GGA TTG | This study | N/A |
| Forward primer for S S13I: TGC CAC TGG TGT CCA TCC AGT GTG TGA ACC T | This study | N/A |
| Reverse primer for S S13I: AGG TTC ACA CAC TGG ATG GAC ACC AGT GGC A | This study | N/A |
| Forward primer for S W152C: GAA CAA CAA GTC CTG TAT GGA GTC TGA GTT C | This study | N/A |
| Reverse primer for S W152C: GAA CTC AGA CTC CAT ACA GGA CTT GTT GTT C | This study | N/A |
| Forward primer for S D614G: CTG TGC TCT ACC AGG GTG TGA ACT GTA CTG A | This study | N/A |
| Reverse primer for S D614G: TCA GTA CAG TTC ACA CCC TGG TAG AGC ACA G | This study | N/A |
| Forward primer for S HV69-70del: GGT TCC ATG CCA TCT CTG GCA CCA ATG GCA | This study | N/A |
| Reverse primer for S HV69-70del: TGC CAT TGG TGC CAG AGA TGG CAT GGA ACC | This study | N/A |
| Mink ACE2 forward primer for cloning: ATG TTA GGC TCT TCC TGG CTC CTT | This study | N/A |
| Mink ACE2 reverse primer for cloning: CTA AAA TGA CGT CTG AAC ATC ATC GAC | This study | N/A |
| Mink ACE2 forward primer for the preparation of mink ACE2 expression: TTG GGT ACC ATG TTA GGC TCT TCC TGG CTC C | This study | N/A |
| Mink ACE2 reverse primer for the preparation of mink ACE2 expression: AAA GAT CTC GAG CTA AAA TGA CGT CTG AAC ATC | This study | N/A |
| Primer for the preparation of RBD L452R expression plasmid (RBD L452R):GGA CAG CAA GGT GGG AGG CAA CTA CAA CTA CAG ATA CAG ACT GTT CAG GAA GAG CAA C | This study | N/A |
| Primer for the preparation of RBD Y453F expression plasmid (RBD Y453F): CTC AAA TGG TTT CAG GTT GCT CTT CCT GAA CAG TCT GAA GAG GTA GTT GTA GTT GCC TCC C | This study | N/A |
| Primer for the preparation of RBD N501Y expression plasmid (RBD N501Y): GTA TGG TTG GTA GCC CAC TCC ATA GGT TGG TTG GAA GCC ATA GGA TTG | This study | N/A |
| Primer for the preparation of the RBD expression plasmid CAT GGG AAA ACA TGT TGT TTA CGG AG | This study | N/A |
| Primer for the preparation of the RBD expression plasmid preparation: GCA GCC CCA TAA ACA CAC AGT AT | This study | N/A |
| Forward primer for the preparation of Fragment 8, S L452R: CTA AGG TTG GTG GTA ATT ATA ATT ACC GGT ATA GAT TGT TTA GGA AGT CTA ATC | This study | N/A |
| Reverse primer for the preparation of Fragment 8, S L452R: GAT TAG ACT TCC TAA ACA ATC TAT ACC GGT AAT TAT AAT TAC CAC CAA CCT TAG | This study | N/A |
| Forward primer for the preparation of Fragment 8, S Y453F: GGT TGG TGG TAA TTA TAA TTA CCT GTT TAG ATT GTT TAG GAA GTC TAA TCT C | This study | N/A |
| Reverse primer for the preparation of Fragment 8, S Y453F: GAG ATT AGA CTT CCT AAA CAA TCT AAA CAG GTA ATT ATA ATT ACC ACC AAC C | This study | N/A |
| Forward primer for the preparation of Fragment 8, S N501Y: CAA TCA TAT GGT TTC CAA CCC ACT TAT GGT GTT GGT TAC CAA CCA TAC AG | This study | N/A |
| Reverse primer for the preparation of Fragment 8, S N501Y: CTG TAT GGT TGG TAA CCA ACA CCA TAA GTG GGT TGG AAA CCA TAT GAT TG | This study | N/A |
| SARS-CoV-2 S forward primer for the mutation verification (WK-521, 22607-22630 forward): GCA TCT GTT TAT GCT TGG AAC AGG | Torii et al., 2021 | N/A |
| SARS-CoV-2 S reverse primer for the mutation verification (WK-521, 23342-23367 reverse): CCT GGT GTT ATA ACA CTG ACA CCA CC | Torii et al., 2021 | N/A |
| SARS-CoV-2 N forward primer for real-time RT-PCR: AGC CTC TTC TCG TTC CTC ATC AC | This study | N/A |
| SARS-CoV-2 N reverse primer for real-time RT-PCR: CCG CCA TTG CCA GCC ATT C | This study | N/A |
| Forward primer for competition assay: CAG GGC AAA CTG GAA AGA TT | This study | N/A |
| Reverse primer for competition assay: TCA GCA ATG TCT CTG CCA AA | This study | N/A |
| Sequencing primer for competition assay: CAG GGC AAA CTG GAA AGA TT | This study | N/A |
| Random hexamer | Sigma-Aldrich | N/A |
| Recombinant DNA | ||
| Plasmid: pC-SARS2-S | Ozono et al., 2021 | N/A |
| Plasmid: pCAGGS | Niwa et al., 1991 | N/A |
| Plasmid: pC-SARS2-S-L452R | This study | N/A |
| Plasmid: pC-SARS2-S-Y453F | This study | N/A |
| Plasmid: pC-SARS2-S-N501Y | This study | N/A |
| Plasmid: pC-SARS2-S-D614G | Ozono et al., 2021 | N/A |
| Plasmid: pC-SARS2-S-B.1.429 [S13I/W152C/L452R/D614G] | This study | N/A |
| Plasmid: pC-SARS2-S-B.1.1.298 [HV69-70del/Y453F/D614G] | This study | N/A |
| Plasmid: pTargeT-human ACE2 | Fukushi et al., 2007 | N/A |
| Plasmid: pLV-EF1a-IRES-Puro | Addgene | Cat# #85132 |
| Plasmid: pLV-EF1a-human ACE2-IRES-Puro | This study | N/A |
| Plasmid: pC-ACE2 | Ozono et al., 2021 | N/A |
| Plasmid: pC-mink ACE2 | This study | N/A |
| Plasmid: pHL-sec | Addgene | Cat# 99845 |
| Plasmid: pHL-sec-human ACE2 (residues 18-740) | Zahradník et al., 2021a | N/A |
| Plasmid: pJYDC1 | Addgene | Cat# 162458 |
| Plasmid: pJYDC1-SARS-CoV-2 S RBD (residues 336-528) | Zahradník et al., 2021a | N/A |
| Plasmid: pJYDC1-SARS-CoV-2 S RBD L452R | This study | N/A |
| Plasmid: pJYDC1-SARS-CoV-2 S RBD Y453F | This study | N/A |
| Plasmid: pJYDC1-SARS-CoV-2 S RBD N501Y | This study | N/A |
| Plasmid: psPAX2-IN/HiBiT | Ozono et al., 2020 | N/A |
| Plasmid: pWPI-Luc2 | Ozono et al., 2020 | N/A |
| Plasmid: pC-TMPRSS2 | Ozono et al., 2021 | N/A |
| Plasmid: pΔ-NRF | Naldini et al., 1996 | N/A |
| Plasmid: pVSV-G | Addgene | Cat# 138479 |
| Plasmid: template plasmids for the preparation of the partial SARS-CoV-2 genome fragments for CPER, see Table S6 | Torii et al., 2021 | N/A |
| Plasmid: pDSP1-7 | Kondo et al., 2011 | N/A |
| Plasmid: pDSP8-11 | Kondo et al., 2011 | N/A |
| Software and algorithms | ||
| NetMHCpan (v 4.1) |
Reynisson et al., 2020; Katoh and Standley, 2013 |
http://www.cbs.dtu.dk/services/NetMHCpan/ |
| NetMHC (v 4.0) | Andreatta and Nielsen, 2016; Lundegaard et al., 2008; Nielsen et al., 2003 | http://www.cbs.dtu.dk/services/NetMHC/ |
| SMM tool | Andreatta and Nielsen, 2016 | http://tools.iedb.org/mhci/help/ |
| PickPocket (v 1.1) | Zhang et al., 2009 | http://www.cbs.dtu.dk/services/PickPocket/ |
| NetMHCcons (v 1.1) | Karosiene et al., 2012 | http://www.cbs.dtu.dk/services/NetMHCcons/ |
| MAFFT suite (v7.467) | Katoh and Standley, 2013 | https://mafft.cbrc.jp/alignment/software/ |
| ModelTest-NG (v0.1.5) | Darriba et al., 2020 | https://github.com/ddarriba/modeltest |
| RAxML-NG (v1.0.0) | (Kozlov et al., 2019) | https://github.com/amkozlov/raxml-ng |
| FigTree (v1.4.4) | Andrew Rambaut | http://tree.bio.ed.ac.uk/software/figtree |
| trimAl (v1.4.rev22) | Capella-Gutiérrez et al., 2009 | http://trimal.cgenomics.org |
| Python (v3.7) | Python Software Foundation | https://www.python.org |
| Sequencher (v5.1) | Gene Codes Corporation | N/A |
| PyMOL (v2.3.0) | Schrödinger | https://pymol.org/2/ |
| UCSF Chimera (v1.13) | Pettersen et al., 2004 | N/A |
| Prism | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Tracy | Rausch et al., 2020 | https://www.gear-genomics.com/teal/ |
| EditR | Kluesner et al., 2018 | http://baseeditr.com |
| Other | ||
| GISAID | Freunde von GISAID e.V. | https://www.gisaid.org |
| Pangolin | Rambaut et al., 2020 | https://cov-lineages.org/pangolin.html |
| 0.45-μm pore size filter | Thermo Fisher Scientific | Cat# 09-740-114 |
| 40-μm cell strainer | SPL Life Sciences | Cat# 93040 |
| HisTrap Fast Flow column | Cytiva | Cat# 17-5255-01 |
| Ultracel-3 regenerated cellulose membrane | Merck | Cat# UFC900324 |
| 0.45-μm pore size filter | Merck | Cat# SLGVR33RB |
| 96-well black plate | PerkinElmer | Cat# 6005225 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kei Sato (KeiSato@g.ecc.u-tokyo.ac.jp).
Materials availability
All unique reagents generated in this study are listed in the Key Resources Table and available from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability
N/A
Experimental model and subject details
Ethics statement
All protocols involving human subjects recruited at Kyushu University Hospital, Japan, National Hospital Organization Kyushu Medical Center, Japan, and Tokyo Metropolitan Cancer and Infectious Diseases Center Komagome Hospital, Japan, were reviewed and approved by the Ethics Committee for Epidemiological and General Research at the Faculty of Life Science, Kumamoto University (approval numbers 2066 and 461). All human subjects provided written informed consent.
Cell culture
Human PBMCs were obtained from three HLA-A∗24:02-positive seronegative donors (average age: 44, range: 36-56, 100% male), sixteen HLA-A∗24:02-positive COVID-19 convalescent donors (average age: 53, range: 27-83, 81% male, median sampling day post PCR positivity: 20, day range: 3-173), and six HLA-A24-negative COVID-19 convalescent donors (average age: 44, range: 24-67, 67% male, median sampling day post PCR positivity: 15, day range: 2-171) (Table S2). The PBMCs were purified by a density gradient centrifugation using Ficoll-Paque Plus (GE Healthcare Life Sciences, cat# 17-1440-03) and stored in liquid nitrogen until further use. C1R cells expressing HLA-A∗24:02 (C1R-A2402) (Karaki et al., 1993) were maintained in RPMI 1640 medium (Thermo Fisher Scientific, cat# 11875101) containing 10% fetal calf serum (FCS) and 1% antibiotics (penicillin and streptomycin; PS). HEK293 cells (a human embryonic kidney cell line; ATCC CRL-1573) and HEK293T cells (a human embryonic kidney cell line; ATCC CRL-3216) were maintained in Dulbecco’s modified Eagle’s medium (high glucose) (Wako, cat# 044-29765) containing 10% FCS and 1% PS. Vero cells [an African green monkey (Chlorocebus sabaeus) kidney cell line; JCRB0111] were maintained in Eagle’s minimum essential medium (Wako, cat# 051-07615) containing 10% FCS and 1% PS. VeroE6/TMPRSS2 cells [an African green monkey (Chlorocebus sabaeus) kidney cell line; JCRB1819] (Matsuyama et al., 2020) and Mv.1.Lu cells [a mink (Neovison vison) lung epithelial cell line; ATCC CCL-64] (Sato et al., 2009) were maintained in Dulbecco’s modified Eagle’s medium (low glucose) (Wako, cat# 041-29775) containing 10% FCS, G418 (1 mg/mL; Nacalai Tesque, cat# G8168-10ML) and 1% PS. HEK293-C34 cells, the IFNAR1 KO HEK293 cells expressing human ACE2 and TMPRSS2 by doxycycline treatment (Torii et al., 2021), were maintained in Dulbecco’s modified Eagle’s medium (high glucose) (Sigma-Aldrich, cat# R8758-500ML) containing 10% FCS, blasticidin (10 μg/mL; InvivoGen, cat# ant-bl-1) and 1% PS. A549 cells (a human lung epithelial cell line; ATCC CCL-185) were maintained in Ham’s F-12K medium (Wako, cat# 080-08565) containing 10% FCS and 1% PS. Expi293F cells (Thermo Fisher Scientific, cat# A14527) were maintained in Expi293 expression medium (Thermo Fisher Scientific, cat# A1435101).
Method details
Viral genomes and phylogenetic analyses
All viral genome sequences and annotation information used in this study were downloaded from GISAID (https://www.gisaid.org) as of March 15, 2021 (750,243 sequences). We used viral nucleotide sequences that did not contain any undetermined nucleotides in the region coding the S protein for our analysis (581,367 sequences). SARS-CoV-2 variants containing the L452R or Y453F mutation were sorted from the verified 581,367 sequences (Tables S3, S4, and S5). To infer the phylogeny of the B.1.1.298 lineage (Figure S2), we collected 657 sequences belonging to the B.1.1.298 lineage that do not contain any undetermined nucleotides. For the phylogenetic analysis, we used 345 representative SARS-CoV-2 sequences by removing identical ones. We aligned entire genome sequences by using the FFT-NS-2 program in MAFFT suite v7.467 (Katoh and Standley, 2013) and removed gapped regions using trimAl v1.4.rev22 with the gappyout option (Capella-Gutiérrez et al., 2009). We selected GTR+I as the best-fit nucleotide substitution model using ModelTest-NG v0.1.5 (Darriba et al., 2020). Based on this model, we generated a maximum-likelihood-based phylogenetic tree using RAxML-NG v1.0.0 (Kozlov et al., 2019) with a bootstrap test (n = 100).
Activation induced marker assay
Expansion of antigen-specific human CD8+ T cells and the analysis of the surface expression levels of the activation markers CD25 and CD137 were performed as previously described (Wolfl et al., 2007). Briefly, human PBMCs were pulsed with 1 μg/mL SARS-CoV-2 PepTivator peptide pools (“S overlap peptides”) (Miltenyi Biotec, cat# 130-126-700) and maintained in RPMI 1640 medium (Thermo Fisher Scientific, cat# 11875101) containing 10% FCS and 30 U/mL recombinant human IL-2 (Peprotec, cat# 200-02) for 10-14 days. The in-vitro expanded CD8+ T cells (i.e., CTL lines) were stimulated with or without the NF9 peptide (NYNYLYRLF, residues 448-456 of the SARS-CoV-2 S protein; synthesized by Scrum Inc.). After incubation at 37°C for 1 h, the cells were washed, and surface proteins (CD3, CD8, CD14, CD19, CD25 and CD137) were stained with the antibodies listed in Key Resources Table. Dead cells were stained with 7-aminoactinomycin D (Biolegend, cat# 420404). After incubation for 20 min on ice, the cells were fixed with 1% paraformaldehyde (Nacalai Tesque, cat# 09154-85), and the levels of protein surface expression were analyzed by flow cytometry using a FACS Canto II (BD Biosciences). The data obtained by flow cytometry were analyzed with FlowJo software (Tree Star).
Analysis of the multifunctionality and cytotoxic potential of CD8+ T cells
C1R-A2402 cells were pulsed with or without the NF9 peptide or its derivatives [the NF9-L452R peptide (NYNYRYRLF, L5R in NF9) and the NF9-Y453F peptide (NYNYLFRLF, Y6F in NF9); synthesized by Scrum Inc.] at concentrations from 0.1 to 10 nM at 37°C for 1 h. The cells were washed twice with PBS, mixed with the CTL lines generated from COVID-19 convalescent samples (see above) and incubated with RPMI 1640 medium (Thermo Fisher Scientific, cat# 11875101) containing 10% FCS, 5 μg/mL brefeldin A (Sigma-Aldrich, cat# B7651), 2 μM monensin (Biolegend, cat# 420701) and a BV421-anti-CD107a antibody (Biolegend, cat# 420404) in a 96-well U plate at 37°C for 5 h. The cells were washed, and surface proteins (CD3, CD8, CD14 and CD19) were stained with the antibodies listed in Key Resources Table. Dead cells were stained with 7-aminoactinomycin D (Biolegend, cat# 420404). After incubation at 37°C for 30 min, the cells were fixed and permeabilized with a Cytofix/Cytoperm Fixation/Permeabilization solution kit (BD Biosciences, cat# 554714); intracellular proteins (IFN-γ, TNF-α and IL-2) were stained with the antibodies listed in Key Resources Table. After incubation at room temperature for 30 min, the cells were washed, and levels of protein expression were analyzed by flow cytometry using a FACS Canto II (BD Biosciences) followed by analysis using FlowJo software (Tree Star).
In silico HLA binding assay
The binding affinity of NF9 and its derivatives to HLA-A24 was predicted using the following five tools: NetMHCpan (v 4.1) (Reynisson et al., 2020), NetMHC (v 4.0) (Andreatta and Nielsen, 2016; Lundegaard et al., 2008; Nielsen et al., 2003), SMM tool (Andreatta and Nielsen, 2016), PickPocket (v 1.1) (Zhang et al., 2009), and NetMHCcons (v 1.1) (Karosiene et al., 2012).
Plasmid construction
Plasmids expressing the SARS-CoV-2 S protein mutants (pC-S-L452R, pC-SARS2-S-Y453F, pC-SARS2-S-N501Y, pC-SARS2-S-B.1.429 [S13I/W152C/L452R/D614G] and pC-SARS2-S-B.1.1.298 [HV69-70del/Y453F/D614G]) were generated by site-directed mutagenesis PCR using pC-SARS2-S (kindly provided by Kenzo Tokunaga) (Ozono et al., 2021) as the template and the following primers: S forward, 5′-TTG GGTACC ATG TTT GTG TTC CTG GTG CTG-3′; S reverse, 5′-GTG GCGGCCGC TCT AGA TTC AGG TGT AGT GCA GTT T-3′; S Y453F forward, 5′-GTG GGA GGC AAC TAC AAC TAC CTC TTC AGA-3′; and S L452R/Y453F reverse, 5′-GTT GTA GTT GCC TCC CAC CTT-3′; S N501Y forward, 5′-TCC TAT GGC TTC CAA CCA ACC TAT GGA-3′; S N501Y reverse, 5′-TGG TTG GAA GCC ATA GGA TTG-3′; S S13I forward, 5′-TGC CAC TGG TGT CCA TCC AGT GTG TGA ACC T-3′; S S13I reverse, 5′-AGG TTC ACA CAC TGG ATG GAC ACC AGT GGC A-3′; S W152C forward, 5′-GAA CAA CAA GTC CTG TAT GGA GTC TGA GTT C-3′; S W152C reverse, 5′-GAA CTC AGA CTC CAT ACA GGA CTT GTT GTT C-3′; S D614G forward, 5′-CTG TGC TCT ACC AGG GTG TGA ACT GTA CTG A-3′; S D614G reverse, 5′-TCA GTA CAG TTC ACA CCC TGG TAG AGC ACA G-3′; S HV69-70del forward, 5′-GGT TCC ATG CCA TCT CTG GCA CCA ATG GCA-3′; and S HV69-70del reverse, 5′-TGC CAT TGG TGC CAG AGA TGG CAT GGA ACC-3′. The resulting PCR fragment was digested with KpnI and NotI and inserted into the KpnI-NotI site of the pCAGGS vector (Niwa et al., 1991).
To construct the expression plasmid for human ACE2 (GenBank: NM_021804.3) (pLV-EF1a-human ACE2-IRES-Puro), the MluI-SmaI fragment pTargeT-human ACE2 (kindly provided by Shuetsu Fukushi) (Fukushi et al., 2007) was inserted into the MluI-HpaI site of pLV-EF1a-IRES-Puro (Addgene #85132).
To construct the expression plasmid for mink ACE2 (GenBank: MW269526.1) (pC-mink ACE2), mink cDNA was used as the template. To prepare mink cDNA, cellular mRNA was extracted from Mv.1.Lu cells using a QIAamp RNA blood mini kit (QIAGEN, cat# 52304), and mink mRNA was reversed transcribed using SuperScript III reverse transcriptase (Thermo Fisher Scientific, cat# 18080085) according to the manufacturers’ protocols. The DNA fragment including the mink ACE2 open reading frame was obtained by RT-PCR using PrimeSTAR GXL DNA polymerase (Takara, cat# R050A) and the following primers: Mink ACE2 forward, 5′-ATG TTA GGC TCT TCC TGG CTC CTT-3′; and Mink ACE2 reverse, 5′-CTA AAA TGA CGT CTG AAC ATC ATC GAC-3′. To add the linker sequence, additional PCR was performed using PrimeSTAR GXL DNA polymerase (Takara, cat# R050A). The DNA fragment including the mink ACE2 open reading frame was used as the template, and two primers, Kpn1 mink ACE2 forward, 5′-TTG GGT ACC ATG TTA GGC TCT TCC TGG CTC C-3′; and Xho1 mink ACE2 reverse: 5′-AAA GAT CTC GAG CTA AAA TGA CGT CTG AAC ATC-3′, were used. The resultant DNA fragment was digested with KpnI and XhoI and inserted into the KpnI-XhoI site of pC-ACE2 (a human ACE2 expression plasmid kindly provided by Kenzo Tokunaga) (Ozono et al., 2021). Nucleotide sequences were determined by a DNA sequencing service (Fasmac), and the sequence data were analyzed by Sequencher v5.1 software (Gene Codes Corporation).
Preparation of soluble human ACE2
To prepare soluble human ACE2, the expression plasmid for the extracellular domain of human ACE2 (residues 18-740) based on pHL-sec (Addgene, cat# 99845) (Zahradník et al., 2021a) was transfected into Expi293F cells using an ExpiFectamine 293 transfection kit (Thermo Fisher Scientific, cat# A14525) according to the manufacturer’s protocol. At three days posttransfection, the culture medium was harvested, centrifuged, and filtered through a 0.45-μm pore size filter (Thermo Fisher Scientific, cat# 09-740-114). The filtered medium was applied to a 5 mL HisTrap Fast Flow column (Cytiva, cat# 17-5255-01) equilibrated with phosphate-buffered saline (PBS) using a ÄKTA pure chromatography system (Cytiva). The column was washed with PBS, and the pure human ACE2 protein (residues 18-740) was eluted using PBS supplemented with 300 mM imidazole (pH 7.4). The buffer was exchanged with PBS by using an Ultracel-3 regenerated cellulose membrane (Merck, cat# UFC900324), and the purified protein was concentrated. The purity of the prepared protein was assessed using a Tycho NT.6 system (NanoTemper).
Preparation of a yeast-based SARS-CoV-2 RBD expression system
The labeling-free yeast surface display plasmid pJYDC1 (Addgene, cat# 162458) (Zahradník et al., 2021b) encoding the SARS-CoV-2 S RBD (residues 336-528) (pJYDC1-RBD) (Zahradník et al., 2021a) was modified by the restriction enzyme-free cloning procedure (Peleg and Unger, 2014). To prepare the plasmids with the mutated RBD, megaprimers were amplified by PCR using the KAPA HiFi HotStart ReadyMix kit (Roche, cat# KK2601) with the following primers according to the manufacturer’s protocol: RBD L452R forward:5′-GGA CAG CAA GGT GGG AGG CAA CTA CAA CTA CAG ATA CAG ACT GTT CAG GAA GAG CAA C-3′; RBD Y453F reverse: 5′-CTC AAA TGG TTT CAG GTT GCT CTT CCT GAA CAG TCT GAA GAG GTA GTT GTA GTT GCC TCC C-3′; RBD N501Y reverse: 5′-GTA TGG TTG GTA GCC CAC TCC ATA GGT TGG TTG GAA GCC ATA GGA TTG-3′; pCT_seq reverse: 5′-CAT GGG AAA ACA TGT TGT TTA CGG AG-3′; and pCTCON_seq forward: 5′-GCA GCC CCA TAA ACA CAC AGT AT-3′. The PCR products were integrated into pJYDC1 by integration PCR as previously described (Peleg and Unger, 2014).
Analysis of the binding affinity of the SARS-CoV-2 S RBD variants for human ACE2 by yeast surface display
The pJYDC1-based yeast display plasmids expressing SARS-CoV-2 RBD and its mutants were transformed into yeast (Saccharomyces cerevisiae; strain EBY100, ATCC MYA-4941) and selected by growth on SD-W plates (Peleg and Unger, 2014). Single colonies were grown in 1 mL liquid SD-CAA medium (Zahradník et al., 2021a) overnight at 30°C (220 rpm) and used to inoculate 1/9 medium (Zahradník et al., 2021a) with 1 nM bilirubin (Sigma-Aldrich, cat# 14370-1G) for expression cultures. The cells were washed with PBS-B buffer [PBS supplemented with bovine serum albumin (1 g/l)] and aliquoted to solutions consisting of PBS-B buffer with 14 different concentrations (ranging from 100 nM to 1 pM) of the human ACE2 protein (residues 18-740) labeled with CF640R succinimidyl ester (Biotium, cat# 92108). The volume of the analysis solution was adjusted (1-100 ml) to reduce the effect of ligand depletion (Zahradník et al., 2021b). Yeasts expressing the SARS-CoV-2 S RBDs were incubated with the analysis solution overnight to allow for equilibrium. Subsequently, the yeast cells were washed with PBS-B buffer and passed through a 40-μm cell strainer (SPL Life Sciences, cat# 93040), after which binding affinity to the CF640R-labeled human ACE2 protein (residues 18-740) was examined using an Accuri C6 flow cytometer (BD Biosciences). The fluorescent signal was processed as previously described (Zahradník et al., 2021b), and the standard noncooperative Hill equation was fitted by nonlinear least-squares regression using Python v3.7 (https://www.python.org).
Pseudovirus assay
To prepare pseudoviruses, lentivirus (HIV-1)-based, luciferase-expressing reporter viruses pseudotyped with the SARS-CoV-2 S protein and its derivatives, HEK293T cells (1 × 106 cells) were cotransfected with 1 μg of psPAX2-IN/HiBiT (Ozono et al., 2020), 1 μg of pWPI-Luc2 (Ozono et al., 2020), and 500 ng of plasmids expressing parental S or its derivatives (L452R, Y453F, N501Y, D614G, B.1.429 and B.1.1.298) using Lipofectamine 3000 (Thermo Fisher Scientific, cat# L3000015) according to the manufacturer’s protocol. At two days posttransfection, the culture supernatants were harvested, centrifuged, and treated with 37.5 U/mL DNase I (Sigma-Aldrich, cat# 11284932001) at 37°C for 30 min. The amount of pseudoviruses prepared was quantified using the HiBiT assay, and the measured value was normalized to the level of HIV-1 p24 antigen as previously described (Ozono et al., 2021; Ozono et al., 2020). The pseudoviruses prepared were stored at –80°C until use.
To prepare target cells for pseudovirus infection, HEK293T cells (1 × 106 cells) were cotransfected with 250 ng of pC-TMPRSS2 (Ozono et al., 2021) and 500 ng of either pC-ACE2 or pC-mink ACE2 using Lipofectamine 2000 (Thermo Fisher Scientific, cat# 11668019) according to the manufacturer’s protocol. At two days posttransfection, the cells (22,000 cells/100 μl) were seeded in 96-well plates and infected with 100 μL of the pseudoviruses prepared at 4 different doses (1, 3, 5 and 10 ng of p24 antigen). At two days postinfection, the infected cells were lysed with a One-Glo luciferase assay system (Promega, cat# E6130), and the luminescent signal was measured using a CentroXS3 plate reader (Berthhold Technologies).
Lentiviral transduction
Lentiviral transduction was performed as described previously (Anderson et al., 2018; Ikeda et al., 2019). Briefly, the VSV-G-pseudotyped lentivirus vector expressing human ACE2 was generated by transfecting 2.5 μg of pLV-EF1a-human ACE2-IRES-Puro plasmid with 1.67 μg of pΔ-NRF (expressing HIV-1 gag, pol, rev, and tat genes) (Naldini et al., 1996) and 0.83 μg of pVSV-G (expressing VSV-G; Addgene, cat# 138479) into HEK293T cells (3 × 106 cells) using TransIT-LT1 (Takara, cat# MIR2300) according to the manufacturer’s protocol. At two days posttransfection, the culture supernatants were harvested and centrifuged, and the supernatants were filtered with a 0.45-μm pore size filter (Millipore, cat# SLGVR33RB) and collected as the lentiviral vector. The lentivirus vectors were concentrated by centrifugation (at 22,000 × g for 2 h at 4°C), and the concentrated vectors were inoculated into target cells (HEK293 cells and A549 cells) and incubated at 37°C. At two days posttransduction, the cells were selected using culture medium containing 1 μg/mL puromycin (InvivoGen, cat# ant-pr-1). Puromycin-selected cells with relatively higher ACE2 expression were sorted by a FACS Aria II (BD Biosciences) and expanded. After expansion, the expression level of surface ACE2 was verified by a FACS Canto II (BD Biosciences). A goat anti-ACE2 polyclonal antibody (R&D Systems, cat# AF933) and an APC-conjugated donkey anti-goat IgG (R&D Systems, cat# F0108) were used for surface ACE2 staining. Normal goat IgG (R&D Systems, cat# AB-108-C) was used as the negative control for this assay.
Protein structure
PyMOL v2.3 (https://pymol.org/2/) was used for 3D visualization of the SARS-CoV-2 S and human ACE2 proteins (Figures 2A and 2G) with the cocrystal structure of the SARS-CoV-2 S and human ACE2 proteins (PDB: 6M17) (Yan et al., 2020). The L452R substitution (Figure 2G) was prepared using UCSF Chimera v1.13 (Pettersen et al., 2004).
SARS-CoV-2 reverse genetics
Recombinant SARS-CoV-2 was generated by circular polymerase extension reaction (CPER) as previously described (Torii et al., 2021). In brief, 9 DNA fragments encoding the partial genome of SARS-CoV-2 (strain WK-521; GISAID ID: EPI_ISL_408667) (Matsuyama et al., 2020) were prepared by PCR using PrimeSTAR GXL DNA polymerase (Takara, cat# R050A). A linker fragment encoding hepatitis delta virus ribozyme (HDVr), bovine growth hormone (BGH) polyA signal and cytomegalovirus (CMV) promoter was also prepared by PCR. The corresponding SARS-CoV-2 genomic region and the templates and primers of this PCR are summarized in Table S6. The 10 obtained DNA fragments were mixed and used for CPER (Torii et al., 2021).
To produce recombinant SARS-CoV-2, the CPER products were transfected into HEK293-C34 cells using TransIT-LT1 (Takara, cat# MIR2300) according to the manufacturer’s protocol. At one day posttransfection, the culture medium was replaced with Dulbecco’s modified Eagle’s medium (high glucose) (Sigma-Aldrich, cat# R8758-500ML) containing 2% FCS, 1% PS and doxycycline (1 μg/mL; Takara, cat# 1311N). At six days posttransfection, the culture medium was harvested and centrifuged, and the supernatants were collected as the seed virus. To remove the CPER products (i.e., SARS-CoV-2-related DNA), 1 mL of the seed virus was treated with 2 μL TURBO DNase (Thermo Fisher Scientific, cat# AM2238) and incubated at 37°C for 1 h. Complete removal of the CPER products (i.e., SARS-CoV-2-related DNA) from the seed virus was verified by PCR. To prepare the working virus of recombinant SARS-CoV-2 for virological experiments (Figures 2H–2L), 100 μL of the seed virus was inoculated into VeroE6/TMPRSS2 cells (5,000,000 cells in a T-75 flask). At one h after infection, the culture medium was replaced with Dulbecco’s modified Eagle’s medium (low glucose) (Wako, cat# 041-29775) containing 2% FCS and 1% PS; at two days postinfection, the culture medium was harvested and centrifuged, and the supernatants were collected as the working virus.
To generate recombinant SARS-CoV-2 mutants, mutations were inserted in fragment 8 (Table S6) using the GENEART site-directed mutagenesis system (Thermo Fisher Scientific, cat# A13312) according to the manufacturer’s protocol with the following primers: Fragment 8_S L452R forward, 5′-CTA AGG TTG GTG GTA ATT ATA ATT ACC GGT ATA GAT TGT TTA GGA AGT CTA ATC-3′; Fragment 8_S L452R reverse, 5′-GAT TAG ACT TCC TAA ACA ATC TAT ACC GGT AAT TAT AAT TAC CAC CAA CCT TAG-3′; Fragment 8_S Y453F forward, 5′-GGT TGG TGG TAA TTA TAA TTA CCT GTT TAG ATT GTT TAG GAA GTC TAA TCT C-3′; Fragment 8_S Y453F reverse, 5′-GAG ATT AGA CTT CCT AAA CAA TCT AAA CAG GTA ATT ATA ATT ACC ACC AAC C-3′; Fragment 8_S N501Y forward, 5′-CAA TCA TAT GGT TTC CAA CCC ACT TAT GGT GTT GGT TAC CAA CCA TAC AG-3′; and Fragment 8_S N501Y reverse, 5′-CTG TAT GGT TGG TAA CCA ACA CCA TAA GTG GGT TGG AAA CCA TAT GAT TG-3′, according to the manufacturer’s protocol. Nucleotide sequences were determined by a DNA sequencing service (Fasmac), and the sequence data were analyzed by Sequencher version 5.1 software (Gene Codes Corporation). The CPER for the preparation of SARS-CoV-2 mutants was performed using mutated fragment 8 instead of parental fragment 8. Subsequent experimental procedures correspond to the procedure for parental SARS-CoV-2 preparation (described above). To verify insertion of the mutation in the working viruses, viral RNA was extracted using a QIAamp viral RNA mini kit (QIAGEN, cat# 52906) and reverse transcribed using SuperScript III reverse transcriptase (Thermo Fisher Scientific, cat# 18080085) according to the manufacturers’ protocols. DNA fragments including the mutations inserted were obtained by RT-PCR using PrimeSTAR GXL DNA polymerase (Takara, cat# R050A) and the following primers: WK-521 22607-22630 forward: 5′-GCA TCT GTT TAT GCT TGG AAC AGG-3′; and WK-521 23342-23367 reverse: 5′-CCT GGT GTT ATA ACA CTG ACA CCA CC-3′. Nucleotide sequences were determined as described above, and sequence chromatograms (Figure 2H) were visualized using the web application Tracy (https://www.gear-genomics.com/teal/) (Rausch et al., 2020).
Plaque assay
One day prior to infection, 200,000 Vero cells were seeded into a 12-well plate. The virus was diluted with serum-free virus dilution buffer [1 × minimum essential medium (Temin’s modification) (Thermo Fisher Scientific, cat# 11935046) with 20 mM HEPES, nonessential amino acids (Thermo Fisher Scientific, cat# 11140-050) and antibiotics]. After removing the culture medium, the cells were infected with 500 μl of diluted virus at 37°C. At two h postinfection, 1 mL of mounting solution [1 × minimum essential medium containing 3% FCS and 1.5% carboxymethyl cellulose (Sigma, cat# C9481-500G)] was overlaid, followed by incubation at 37°C. At three days postinfection, the culture medium was removed, and the cells were washed with PBS three times and fixed with 10% formaldehyde (Nacalai Tesque, cat# 37152-51) or 4% paraformaldehyde (Nacalai Tesque, cat# 09154-85). The fixed cells were washed with tap water, dried, and stained with staining solution [2% crystal violet (Nacalai Tesque, cat# 09804-52) or 0.1% methylene blue (Nacalai Tesque, cat# 22412-14) in water]. The stained cells were washed with city water and dried, and the number of plaques was counted to calculate PFU.
SARS-CoV-2 infection
One day prior to infection, 10,000 HEK293-ACE2, A549-ACE2 and VeroE6/TMPRSS2 cells were seeded into a 96-well plate. Recombinant SARS-CoV-2 (10, 100 or 1,000 PFU) was inoculated and incubated at 37°C for 1 h. The infected cells were washed, and 180 μl of culture medium was added. The culture supernatant (10 μl) was harvested at indicated time points and used for real-time RT-PCR to quantify the viral RNA copy number.
Real-time RT-PCR
The amount of viral RNA in culture supernatant was quantified by real-time RT-PCR as previously described (Shema Mugisha et al., 2020), with some modifications. In brief, 5 μL of culture supernatant was mixed with 5 μL of 2 × RNA lysis buffer [2% Triton X-100, 50 mM KCl, 100 mM Tris-HCl (pH 7.4), 40% glycerol, 0.8 U/μl recombinant RNase inhibitor (Takara, cat# 2313B)] and incubated at room temperature for 10 min. RNase-free water (90 μl) was added, and the diluted sample (2.5 μl) was used as the template for real-time RT-PCR performed according to the manufacturer’s protocol using the One Step TB Green PrimeScript PLUS RT-PCR kit (Takara, cat# RR096A) and the following primers: Forward N, 5′-AGC CTC TTC TCG TTC CTC ATC AC-3′; and Reverse N, 5′-CCG CCA TTG CCA GCC ATT C-3′. The copy number of viral RNA was standardized with a SARS-CoV-2 direct detection RT-qPCR kit (Takara, cat# RC300A). The fluorescent signal was acquired using a QuantStudio 3 Real-Time PCR system (Thermo Fisher Scientific), a CFX Connect Real-Time PCR Detection system (Bio-Rad) or a 7500 Real Time PCR System (Applied Biosystems).
Competition assay
One day prior to infection, 100,000 HEK293-ACE2 cells were seeded into 12-well plates. Parental virus and the L452R mutant were mixed at a 1:1 ratio based on the PFU. The virus mixture was inoculated at a final concentration of 1,000 PFU (i.e., 500 PFU for each virus) and incubated at 37°C for 1 h. The infected cells were washed, and 1 mL of culture medium was added. The culture supernatant (140 μl) was harvested at the indicated time points, and 140 μl culture medium was supplied. Viral RNAs were isolated from the harvested culture supernatant using a QIAamp viral RNA mini kit (QIAGEN, cat# 52906), and cDNA was synthesized using reverse transcriptase (Roche, cat# 03531317001) with a random hexamer (Sigma-Aldrich). The resultant cDNAs were amplified with a primer set (forward, 5′-CAG GGC AAA CTG GAA AGA TT-3′ and reverse, 5′-TCA GCA ATG TCT CTG CCA AA-3′). The PCR products were purified using a GeneJET gel extraction kit (Thermo Fisher Scientific, cat# K0692) and subjected to Sanger sequencing with the following primer: 5′-CAG GGC AAA CTG GAA AGA TT-3′. The nucleotide variation at position 22,917 (i.e., T for parental virus, G for the L452R mutant) was quantified by using EditR (Kluesner et al., 2018).
SARS-CoV-2 S-based fusion assay
The SARS-CoV-2 S-based fusion assay was performed according to the HIV-1 envelope protein-based fusion assay (Ikeda et al., 2018; Kondo et al., 2011) with some modifications. This assay utilizes a dual split protein (DSP) encoding Renilla luciferase (RL) and GFP genes, and the respective split proteins, DSP1-7 and DSP8-11, are expressed in effector and target cells by transfection (Ikeda et al., 2018; Kondo et al., 2011).
On day 1, effector cells (i.e., S-expressing cells) and target cells (i.e., ACE2-expressing cells) were prepared. To prepare effector cells, HEK293 cells were cotransfected with the expression plasmids for parental S or its derivatives (L452R, Y453F and N501Y) (400 ng) and pDSP1-7 (400 ng) using TransIT-LT1 (Takara, cat# MIR2300). To prepare target cells, HEK293 cells were cotransfected with pC-ACE2 (1,000 ng) and pDSP8-11 (400 ng). On day 2 (24 h posttransfection), 16,000 effector cells were detached and reseeded into 96-well black plates (PerkinElmer, cat# 6005225), and target cells were reseeded at a density of 1,000,000 cells/2 mL/well in 6-well plates. On day 4 (72 h posttransfection), target cells were incubated with EnduRen live cell substrate (Promega, cat# E6481) for three h and then detached, and 32,000 target cells were applied to a 96-well plate with effector cells. RL activity was measured at the indicated time points by using a Centro XS3 LB960 (Berthhold Technologies). The S proteins expressed on the surface of effector cells were stained with a rabbit anti-SARS-CoV-2 monoclonal antibody (GeneTex, cat# GTX635654) and an APC-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch, cat# 111-136-144). Normal rabbit IgG (SouthernBiotech, cat# 0111-01) was used as a negative control. Eexpression levels of surface S proteins were analyzed using a FACS Canto II (BD Biosciences). RL activity was normalized to the mean fluorescence intensity (MFI) of surface S proteins, and the normalized values are shown as fusion activity.
Quantification and statistical analysis
Data analyses were performed using Prism 7 (GraphPad Software). In Figures 1B and 1F, data are presented as median with SD, and n represents the number of PBMC donor. In the other figure panels, data are presented as average with SD, and n represents the number of technical replicate. In Figures 1B and 1F, statistically significant differences are determined by the Wilcoxon signed-rank test. In Figure 1I, statistically significant differences are determined by ANOVA, with multiple comparisons by Bonferroni correction. In Figure 2C, statistically significant difference are determined by the Mann-Whitney U test. In Figures 2E, 2F, and 2J–2N, statistically significant difference are determined by Student’s t test.
Consortia
The Genotype to Phenotype Japan (G2P-Japan) Consortium: Mika Chiba, Mai Fujimi, Hirotake Furihata, Haruyo Hasebe, Kazuko Kitazato, Naoko Misawa, Mikari Motomura, Akiko Oide, Sachiko Sakata, Mai Suganami, Yoshitaka Suzuki, Yutaka Suzuki, Miyoko Takahashi, Jiaqi Wu, and Miyabishara Yokoyama.
Acknowledgments
We would like to thank all members belonging to the Genotype to Phenotype Japan (G2P-Japan) Consortium. We thank Drs. Sho Fujiwara, Kazuaki Fukushima, Masaru Tanaka, and Akifumi Imamura (Tokyo Metropolitan Cancer and Infectious Diseases Center Komagome Hospital, Japan) for supporting the collection of COVID-19 convalescent samples; Dr. Mizuki Kitamatsu and Mr. Yoshiki Aritsu (Kindai University, Japan) for supporting the preparation of synthetic peptides; Drs. Hiroyuki Kishi and Hiroshi Hamana (University of Toyama, Japan) for helpful suggestions; and Dr. Kenzo Tokunaga (National Institute of Infectious Diseases, Japan), Dr. Shuetsu Fukushi (National Institute of Infectious Diseases, Japan), Dr. Masafumi Takiguchi (Kumamoto University, Japan), and Dr. Jin Gohda (The University of Tokyo, Japan) for providing plasmids, reagents, and cells. The super-computing resource was provided by Human Genome Center at The University of Tokyo and the NIG supercomputer at ROIS National Institute of Genetics.
This study was supported in part by AMED Research Program on Emerging and Re-emerging Infectious Diseases 20fk0108163 (to A.S.), 20fk0108539 (to T.U.), 20fk0108146 (to K.S.), 19fk0108171 (to S.N. and K.S.), 20fk0108270 (to K.S.), and 20fk0108413 (to T.I., S.N., and K.S.); AMED Research Program on HIV/AIDS 21fk0410046 (to C.M.), 20fk0410019 (to T.U. and K.S.), 20fk0410014 (to K.S.), and 21fk0410039 (to K.S.); AMED Japan Program for Infectious Diseases Research and Infrastructure 20wm0325009 (to A.S.); JST A-STEP JPMJTM20SL (to T.I.); JST J-RAPID JPMJJR2007 (to K.S.); JST SICORP (e-ASIA) JPMJSC20U1 (to K.S.); JST SICORP JPMJSC21U5 (to K.S.); JST CREST JPMJCR20H6 (to S.N,) and JPMJCR20H4 (to K.S); JSPS KAKENHI Grant-in-Aid for Scientific Research B 18H02662 (to K.S.) and 21H02737 (to K.S.); JSPS KAKENHI Grant-in-Aid for Scientific Research on Innovative Areas 16H06429 (to S.N. and K.S.), 16K21723 (to S.N. and K.S.), 17H05823 (to S.N.), 17H05813 (to K.S.), 19H04843 (to S.N.), and 19H04826 (to K.S.); JSPS Fund for the Promotion of Joint International Research (Fostering Joint International Research) 18KK0447 (to K.S.); JSPS Core-to-Core Program JPJSCCB20190009 (to T.U.) and JPJSCCA20190008 (A. Advanced Research Networks) (to K.S.); JSPS Research Fellow DC1 19J20488 (to I.K.); JSPS Leading Initiative for Excellent Young Researchers (LEADER) (to T.I.); ONO Medical Research Foundation (to K.S.); Ichiro Kanehara Foundation (to K.S.); Lotte Foundation (to K.S.); Mochida Memorial Foundation for Medical and Pharmaceutical Research (to K.S.); Daiichi Sankyo Foundation of Life Science (to K.S.); Sumitomo Foundation (to K.S.); Uehara Foundation (to K.S.); Takeda Science Foundation (to C.M., T.I., and K.S.); The Tokyo Biochemical Research Foundation (to K.S.); Mitsubishi Foundation (to T.I.); Shin-Nihon Foundation of Advanced Medical Research (to T.I.); an intramural grant from Kumamoto University COVID-19 Research Projects (AMABIE) (to C.M., T.I., and T.U.); Kumamoto University International Collaborative Research Grants (to T.U.); Intercontinental Research and Educational Platform Aiming for Eradication of HIV/AIDS (to T.I. and T.U.); 2020 Tokai University School of Medicine Research Aid (to S.N.); and Joint Usage/Research Center program of Institute for Frontier Life and Medical Sciences, Kyoto University (to K.S.). T.S.T and I.N. are the recipients of the doctoral course scholarship from Japanese Government.
Author contributions
C.M., M.T., J.Z., A.S., H.N., T.S.T., I.N., I.K., K.U., Y.Y., R.S., T.I., and K.S. performed the experiments. S.T., T.F., G.S., and Y.M. prepared experimental materials. J.Z. and Y.K. performed structural analysis. S.N. performed molecular phylogenetic analysis. A.Y., N. Shimoto, Y.N., R.M., T.T., and N. Sekiya performed clinical analysis and collected clinical samples. C.M., M.T., J.Z., A.S., J.I., T.I., S.N., T.U., and K.S. designed the experiments and interpreted the results. K.S. wrote the original manuscript. C.M., J.Z., A.S., T.I., S.N, and T.U. modified the manuscript. All authors reviewed and proofread the manuscript. The Genotype to Phenotype Japan (G2P-Japan) Consortium contributed to the project administration.
Declaration of interests
The authors declare no competing interests.
Published: June 15, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.chom.2021.06.006.
Supplemental information
References
- Anderson B.D., Ikeda T., Moghadasi S.A., Martin A.S., Brown W.L., Harris R.S. Natural APOBEC3C variants can elicit differential HIV-1 restriction activity. Retrovirology. 2018;15:78. doi: 10.1186/s12977-018-0459-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreatta M., Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics. 2016;32:511–517. doi: 10.1093/bioinformatics/btv639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Mossman K., Baker M.L. Zooanthroponotic potential of SARS-CoV-2 and implications of reintroduction into human populations. Cell Host Microbe. 2021;29:160–164. doi: 10.1016/j.chom.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayarri-Olmos R., Rosbjerg A., Johnsen L.B., Helgstrand C., Bak-Thomsen T., Garred P., Skjoedt M.-O. The SARS-CoV-2 Y453F mink variant displays a pronounced increase in ACE-2 affinity but does not challenge antibody neutralization. J. Biol. Chem. 2021;296:100536. doi: 10.1016/j.jbc.2021.100536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC Birth: final data for 2018. 2019. https://www.cdc.gov/nchs/data/nvsr/nvsr68/nvsr68_13-508.pdf
- CDC Emerging SARS-CoV-2 variants (updated January 28, 2021) 2020. https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-brief-emerging-variants.html
- Challen R., Brooks-Pollock E., Read J.M., Dyson L., Tsaneva-Atanasova K., Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ. 2021;372:n579. doi: 10.1136/bmj.n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar A., Liu J., Martinot A.J., McMahan K., Mercado N.B., Peter L., Tostanoski L.H., Yu J., Maliga Z., Nekorchuk M., et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier D.A., De Marco A., Ferreira I.A.T.M., Meng B., Datir R.P., Walls A.C., Kemp S.A., Bassi J., Pinto D., Silacci-Fregni C., et al. CITIID-NIHR BioResource COVID-19 Collaboration. COVID-19 Genomics UK (COG-UK) Consortium Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593:136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damas J., Hughes G.M., Keough K.C., Painter C.A., Persky N.S., Corbo M., Hiller M., Koepfli K.P., Pfenning A.R., Zhao H., et al. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc. Natl. Acad. Sci. USA. 2020;117:22311–22322. doi: 10.1073/pnas.2010146117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D., Posada D., Kozlov A.M., Stamatakis A., Morel B., Flouri T. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 2020;37:291–294. doi: 10.1093/molbev/msz189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Jarvis C.I., CMMID COVID-19 Working Group. Edmunds W.J., Jewell N.P., Diaz-Ordaz K., Keogh R.H. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021;593:270–274. doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Garcia-Knight M.A., Khalid M.M., Servellita V., Wang C., Morris M.K., Sotomayor-González A., Glasner D.R., Reyes K.R., Gliwa A.S., et al. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. medRxiv. 2021 doi: 10.1101/2021.03.07.21252647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S., Shackelton L.A., Holmes E.C. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- Fryer H.R., Frater J., Duda A., Palmer D., Phillips R.E., McLean A.R. Cytotoxic T-lymphocyte escape mutations identified by HLA association favor those which escape and revert rapidly. J. Virol. 2012;86:8568–8580. doi: 10.1128/JVI.07020-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushi S., Mizutani T., Sakai K., Saijo M., Taguchi F., Yokoyama M., Kurane I., Morikawa S. Amino acid substitutions in the s2 region enhance severe acute respiratory syndrome coronavirus infectivity in rat angiotensin-converting enzyme 2-expressing cells. J. Virol. 2007;81:10831–10834. doi: 10.1128/JVI.01143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao A., Chen Z., Amitai A., Doelger J., Mallajosyula V., Sundquist E., Segal F.P., Carrington M., Davis M.M., Streeck H., et al. Learning from HIV-1 to predict the immunogenicity of T cell epitopes in SARS-COV-2. iScience. 2021;24:102311. doi: 10.1016/j.isci.2021.102311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383.e9. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Galarza F.F., McCabe A., Santos E.J.M.D., Jones J., Takeshita L., Ortega-Rivera N.D., Cid-Pavon G.M.D., Ramsbottom K., Ghattaoraya G., Alfirevic A., et al. Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res. 2020;48(D1):D783–D788. doi: 10.1093/nar/gkz1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grint D.J., Wing K., Williamson E., McDonald H.I., Bhaskaran K., Evans D., Evans S.J., Walker A.J., Hickman G., Nightingale E., et al. Case fatality risk of the SARS-CoV-2 variant of concern B.1.1.7 in England, 16 November to 5 February. Euro Surveill. 2021;26:2100256. doi: 10.2807/1560-7917.ES.2021.26.11.2100256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann P.J., Hatta M., Chiba S., Maemura T., Fan S., Takeda M., Kinoshita N., Hattori S.I., Sakai-Tagawa Y., Iwatsuki-Horimoto K., et al. Transmission of SARS-CoV-2 in domestic cats. N. Engl. J. Med. 2020;383:592–594. doi: 10.1056/NEJMc2013400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Arora P., Groß R., Seidel A., Hörnich B.F., Hahn A.S., Krüger N., Graichen L., Hofmann-Winkler H., Kempf A., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Zhang L., Krüger N., Graichen L., Kleine-Weber H., Hofmann-Winkler H., Kempf A., Nessler S., Riggert J., Winkler M.S., et al. SARS-CoV-2 mutations acquired in mink reduce antibody-mediated neutralization. Cell Rep. 2021;35:109017. doi: 10.1016/j.celrep.2021.109017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., 3rd, Leist S.R., Schäfer A., Nakajima N., Takahashi K., et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370:1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C., Shen M., Han X., Chen Q., Li L., Chen S., Zhang J., Gao F., Wang W., Wang Y., et al. Identification of cross-reactive CD8+ T cell receptors with high functional avidity to a SARS-CoV-2 immunodominant epitope and its natural mutant variants. bioRxiv. 2020 doi: 10.1101/2020.11.02.364729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda T., Symeonides M., Albin J.S., Li M., Thali M., Harris R.S. HIV-1 adaptation studies reveal a novel Env-mediated homeostasis mechanism for evading lethal hypermutation by APOBEC3G. PLoS Pathog. 2018;14:e1007010. doi: 10.1371/journal.ppat.1007010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda T., Molan A.M., Jarvis M.C., Carpenter M.A., Salamango D.J., Brown W.L., Harris R.S. HIV-1 restriction by endogenous APOBEC3G in the myeloid cell line THP-1. J. Gen. Virol. 2019;100:1140–1152. doi: 10.1099/jgv.0.001276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaki S., Kariyone A., Kato N., Kano K., Iwakura Y., Takiguchi M. HLA-B51 transgenic mice as recipients for production of polymorphic HLA-A, B-specific antibodies. Immunogenetics. 1993;37:139–142. doi: 10.1007/BF00216838. [DOI] [PubMed] [Google Scholar]
- Kared H., Redd A.D., Bloch E.M., Bonny T.S., Sumatoh H., Kairi F., Carbajo D., Abel B., Newell E.W., Bettinotti M.P., et al. SARS-CoV-2-specific CD8+ T cell responses in convalescent COVID-19 individuals. J. Clin. Invest. 2021;131:e145476. doi: 10.1172/JCI145476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karosiene E., Lundegaard C., Lund O., Nielsen M. NetMHCcons: a consensus method for the major histocompatibility complex class I predictions. Immunogenetics. 2012;64:177–186. doi: 10.1007/s00251-011-0579-8. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.I., Kim S.G., Kim S.M., Kim E.H., Park S.J., Yu K.M., Chang J.H., Kim E.J., Lee S., Casel M.A.B., et al. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe. 2020;27:704–709.e2. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyotani K., Toyoshima Y., Nemoto K., Nakamura Y. Bioinformatic prediction of potential T cell epitopes for SARS-Cov-2. J. Hum. Genet. 2020;65:569–575. doi: 10.1038/s10038-020-0771-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluesner M.G., Nedveck D.A., Lahr W.S., Garbe J.R., Abrahante J.E., Webber B.R., Moriarity B.S. EditR: a method to quantify base editing from Sanger sequencing. The CRISPR Journal. 2018;1:239–250. doi: 10.1089/crispr.2018.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo N., Miyauchi K., Matsuda Z. Monitoring viral-mediated membrane fusion using fluorescent reporter methods. Curr Protoc Cell Biol. 2011 doi: 10.1002/0471143030.cb2609s50. Chapter 26, Unit 26.9. [DOI] [PubMed] [Google Scholar]
- Koopmans M. SARS-CoV-2 and the human-animal interface: outbreaks on mink farms. Lancet Infect. Dis. 2021;21:18–19. doi: 10.1016/S1473-3099(20)30912-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Sheffield COVID-19 Genomics Group Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov A.M., Darriba D., Flouri T., Morel B., Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35:4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Gruta N.L., Gras S., Daley S.R., Thomas P.G., Rossjohn J. Understanding the drivers of MHC restriction of T cell receptors. Nat. Rev. Immunol. 2018;18:467–478. doi: 10.1038/s41577-018-0007-5. [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Le Bert N., Clapham H.E., Tan A.T., Chia W.N., Tham C.Y.L., Lim J.M., Kunasegaran K., Tan L.W.L., Dutertre C.A., Shankar N., et al. Highly functional virus-specific cellular immune response in asymptomatic SARS-CoV-2 infection. J. Exp. Med. 2021;218:e20202617. doi: 10.1084/jem.20202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie A.J., Pfafferott K.J., Chetty P., Draenert R., Addo M.M., Feeney M., Tang Y., Holmes E.C., Allen T., Prado J.G., et al. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 2004;10:282–289. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Nie J., Wu J., Zhang L., Ding R., Wang H., Zhang Y., Li T., Liu S., Zhang M., et al. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell. 2021;184:2362–2371.e9. doi: 10.1016/j.cell.2021.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Hu G., Wang Y., Ren W., Zhao X., Ji F., Zhu Y., Feng F., Gong M., Ju X., et al. Functional and genetic analysis of viral receptor ACE2 orthologs reveals a broad potential host range of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2025373118. e2025373118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu J., Xia H., Zhang X., Fontes-Garfias C.R., Swanson K.A., Cai H., Sarkar R., Chen W., Cutler M., et al. Neutralizing activity of BNT162b2-elicited serum. N. Engl. J. Med. 2021;384:1466–1468. doi: 10.1056/NEJMc2102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., VanBlargan L.A., Bloyet L.M., Rothlauf P.W., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Liebeskind M.J., et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29:477–488.e44. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundegaard C., Lamberth K., Harndahl M., Buus S., Lund O., Nielsen M. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8-11. Nucleic Acids Res. 2008;36 doi: 10.1093/nar/gkn202. W509–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Hernández F., Isaak-Delgado A.B., Alfonso-Toledo J.A., Muñoz-García C.I., Villalobos G., Aréchiga-Ceballos N., Rendón-Franco E. Assessing the SARS-CoV-2 threat to wildlife: Potential risk to a broad range of mammals. Perspectives in Ecology and Conservation. 2020;18:223–234. doi: 10.1016/j.pecon.2020.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F., et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. USA. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy K.R., Rennick L.J., Nambulli S., Robinson-McCarthy L.R., Bain W.G., Haidar G., Duprex W.P. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science. 2021;371:1139–1142. doi: 10.1126/science.abf6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagutelli X., Prot M., Levillayer L., Salazar E.B., Jouvion G., Conquet L., Donati F., Albert M., Gambaro F., Behillil S., et al. The B1.351 and P.1 variants extend SARS-CoV-2 host range to mice. bioRxiv. 2021:436013. doi: 10.1101/2021.03.18.436013. [DOI] [Google Scholar]
- Moore E., Grifoni A., Weiskopf D., Schulten V., Arlehamn C.S.L., Angelo M., Pham J., Leary S., Sidney J., Broide D., et al. Sequence-based HLA-A, B, C, DP, DQ, and DR typing of 496 adults from San Diego, California, USA. Hum. Immunol. 2018;79:821–822. doi: 10.1016/j.humimm.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V.J., Feldmann F., Williamson B.N., van Doremalen N., Pérez-Pérez L., Schulz J., Meade-White K., Okumura A., Callison J., Brumbaugh B., et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585:268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldini L., Blömer U., Gallay P., Ory D., Mulligan R., Gage F.H., Verma I.M., Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- Nelde A., Bilich T., Heitmann J.S., Maringer Y., Salih H.R., Roerden M., Lübke M., Bauer J., Rieth J., Wacker M., et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 2021;22:74–85. doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- Nielsen M., Lundegaard C., Worning P., Lauemøller S.L., Lamberth K., Buus S., Brunak S., Lund O. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12:1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Yamamura K., Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- OIE Infection with SARS-CoV-2 in animals (January 2021) 2021. https://www.oie.int/fileadmin/Home/MM/EN_Factsheet_SARS-CoV-2.pdf
- Oude Munnink B.B., Sikkema R.S., Nieuwenhuijse D.F., Molenaar R.J., Munger E., Molenkamp R., van der Spek A., Tolsma P., Rietveld A., Brouwer M., et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371:172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozono S., Zhang Y., Tobiume M., Kishigami S., Tokunaga K. Super-rapid quantitation of the production of HIV-1 harboring a luminescent peptide tag. J. Biol. Chem. 2020;295:13023–13030. doi: 10.1074/jbc.RA120.013887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozono S., Zhang Y., Ode H., Sano K., Tan T.S., Imai K., Miyoshi K., Kishigami S., Ueno T., Iwatani Y., et al. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat. Commun. 2021;12:848. doi: 10.1038/s41467-021-21118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish C.R., Holmes E.C., Morens D.M., Park E.C., Burke D.S., Calisher C.H., Laughlin C.A., Saif L.J., Daszak P. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol. Mol. Biol. Rev. 2008;72:457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg Y., Unger T. Application of the Restriction-Free (RF) cloning for multicomponents assembly. Methods Mol. Biol. 2014;1116:73–87. doi: 10.1007/978-1-62703-764-8_6. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J.A., Mitchell B.M., Plante K.S., Debbink K., Weaver S.C., Menachery V.D. The variant gambit: COVID-19's next move. Cell Host Microbe. 2021;29:508–515. doi: 10.1016/j.chom.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O’Toole Á., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T., Fritz M.H., Untergasser A., Benes V. Tracy: basecalling, alignment, assembly and deconvolution of sanger chromatogram trace files. BMC Genomics. 2020;21:230. doi: 10.1186/s12864-020-6635-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynisson B., Alvarez B., Paul S., Peters B., Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48(W1):W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Yamamoto S.P., Misawa N., Yoshida T., Miyazawa T., Koyanagi Y. Comparative study on the effect of human BST-2/Tetherin on HIV-1 release in cells of various species. Retrovirology. 2009;6:53. doi: 10.1186/1742-4690-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulien I., Kemming J., Oberhardt V., Wild K., Seidel L.M., Killmer S., Sagar, Daul F., Salvat Lago M., Decker A., et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8+ T cells. Nat. Med. 2021;27:78–85. doi: 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- Selzer T., Albeck S., Schreiber G. Rational design of faster associating and tighter binding protein complexes. Nat. Struct. Biol. 2000;7:537–541. doi: 10.1038/76744. [DOI] [PubMed] [Google Scholar]
- Shema Mugisha C., Vuong H.R., Puray-Chavez M., Bailey A.L., Fox J.M., Chen R.E., Wessel A.W., Scott J.M., Harastani H.H., Boon A.C.M., et al. A simplified quantitative real-time PCR assay for monitoring SARS-CoV-2 growth in cell culture. MSphere. 2020;5 doi: 10.1128/mSphere.00658-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Tang H., McDanal C., Wagh K., Fischer W., Theiler J., Yoon H., Li D., Haynes B.F., Sanders K.O., et al. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe. 2021;29:529–539.e3. doi: 10.1016/j.chom.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z., et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supasa P., Zhou D., Dejnirattisai W., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Nutalai R., Tuekprakhon A., et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell. 2021;184:2201–2211.e7. doi: 10.1016/j.cell.2021.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii S., Ono C., Suzuki R., Morioka Y., Anzai I., Fauzyah Y., Maeda Y., Kamitani W., Fukuhara T., Matsuura Y. Establishment of a reverse genetics system for SARS-CoV-2 using circular polymerase extension reaction. Cell Rep. 2021;35:109014. doi: 10.1016/j.celrep.2021.109014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxlmayr M.W., Obinger C. Directed evolution of proteins for increased stability and expression using yeast display. Arch. Biochem. Biophys. 2012;526:174–180. doi: 10.1016/j.abb.2012.04.022. [DOI] [PubMed] [Google Scholar]
- Volz E., Hill V., McCrone J.T., Price A., Jorgensen D., O’Toole A., Southgate J., Johnson R., Jackson B., Nascimento F.F., et al. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;184:64–75.e11. doi: 10.1016/j.cell.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C., Muecksch F., Rutkowska M., Hoffmann H.H., Michailidis E., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife. 2020 doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Coronavirus disease 2019. 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- WHO SARS-CoV-2 mink-associated variant strain – Denmark (November 6, 2020) 2020. https://www.who.int/csr/don/06-november-2020-mink-associated-sars-cov2-denmark/en/
- Wilson E.A., Hirneise G., Singharoy A., Anderson K.S. Total predicted MHC-I epitope load is inversely associated with population mortality from SARS-CoV-2. Cell Rep. Med. 2021;2:100221. doi: 10.1016/j.xcrm.2021.100221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfl M., Kuball J., Ho W.Y., Nguyen H., Manley T.J., Bleakley M., Greenberg P.D. Activation-induced expression of CD137 permits detection, isolation, and expansion of the full repertoire of CD8+ T cells responding to antigen without requiring knowledge of epitope specificities. Blood. 2007;110:201–210. doi: 10.1182/blood-2006-11-056168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H., Nkolola J.P., Liu J., Li Z., Chandrashekar A., et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183:739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahradník J., Dey D., Marciano S., Schreiber G. An enhanced yeast display platform demonstrates the binding plasticity under various selection pressures. bioRxiv. 2021 doi: 10.1101/2020.12.16.423176. [DOI] [Google Scholar]
- Zahradník J., Marciano S., Shemesh M., Zoler E., Chiaravalli J., Meyer B., Rudich Y., Dym O., Elad N., Schreiber G. SARS-CoV-2 RBD in vitro evolution follows contagious mutation spread yet generates an able infection inhibitor. bioRxiv. 2021 doi: 10.1101/2021.01.06.425392. [DOI] [Google Scholar]
- Zhang H., Lund O., Nielsen M. The PickPocket method for predicting binding specificities for receptors based on receptor pocket similarities: application to MHC-peptide binding. Bioinformatics. 2009;25:1293–1299. doi: 10.1093/bioinformatics/btp137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Davis B.D., Chen S.S., Sincuir Martinez J.M., Plummer J.T., Vail E. Emergence of a novel SARS-CoV-2 variant in Southern California. JAMA. 2021;325:1324–1326. doi: 10.1001/jama.2021.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Chen D., Szabla R., Zheng M., Li G., Du P., Zheng S., Li X., Song C., Li R., et al. Broad and differential animal angiotensin-converting enzyme 2 receptor usage by SARS-CoV-2. J. Virol. 2020 doi: 10.1128/JVI.00940-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Thao T.T.N., Hoffmann D., Taddeo A., Ebert N., Labroussaa F., Pohlmann A., King J., Steiner S., Kelly J.N., et al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature. 2021;592:122–127. doi: 10.1038/s41586-021-03361-1. [DOI] [PubMed] [Google Scholar]
- Zuo J., Dowell A.C., Pearce H., Verma K., Long H.M., Begum J., Aiano F., Amin-Chowdhury Z., Hoschler K., Brooks T., et al. Robust SARS-CoV-2-specific T cell immunity is maintained at 6 months following primary infection. Nat. Immunol. 2021;22:620–626. doi: 10.1038/s41590-021-00902-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
N/A