Summary
Coronavirus disease 2019 (COVID-19) is associated with a wide spectrum of disease presentation, ranging from asymptomatic infection to acute respiratory distress syndrome (ARDS). Paradoxically, a direct relationship has been suggested between COVID-19 disease severity and the levels of circulating severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-specific antibodies, including virus-neutralizing titers. A serological analysis of 536 convalescent healthcare workers reveals that SARS-CoV-2-specific and virus-neutralizing antibody levels are elevated in individuals that experience severe disease. The severity-associated increase in SARS-CoV-2-specific antibody is dominated by immunoglobulin G (IgG), with an IgG subclass ratio skewed toward elevated receptor binding domain (RBD)- and S1-specific IgG3. In addition, individuals that experience severe disease show elevated SARS-CoV-2-specific antibody binding to the inflammatory receptor FcɣRIIIa. Based on these correlational studies, we propose that spike-specific IgG subclass utilization may contribute to COVID-19 disease severity through potent Fc-mediated effector functions. These results may have significant implications for SARS-CoV-2 vaccine design and convalescent plasma therapy.
Keywords: SARS-CoV-2, COVID-19, serology, Fc-effector functions, IgG subclass
Graphical abstract
Highlights
Levels of SARS-CoV-2 antibodies and neutralizing titers rise with COVID-19 severity
IgG1 and IgG3 dominate the SARS-CoV-2 convalescent antibody pool
Increases in spike-specific IgG3 subclass ratios correlate with severe COVID-19
Enhanced FcɣRIIIa binding is observed in severe COVID-19 cohorts
In a serological analysis of convalescent healthcare workers, Yates et al. explore antibody profiles associated with COVID-19 severity. Their analysis reveals an IgG subclass ratio skewed toward IgG3 that correlates with COVID-19 disease severity. The authors hypothesize a link between IgG3, Fc effector functions, and inflammation characteristic of severe COVID-19.
Introduction
The novel coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of coronavirus disease 2019 (COVID-19), a disease responsible for more than 2.6 million deaths in the span of approximately 1 year. The case incidence based on virus detection estimates over 164 million cases globally to date. However, if population-based serological surveys of SARS-CoV-2 are taken into account, the infection rate of SARS-CoV-2 is likely much higher.1,2 This discrepancy highlights the variability of COVID-19 disease presentation in the human population. The severity of disease caused by SARS-CoV-2 infection ranges from an asymptomatic presentation to acute respiratory distress syndrome (ARDS) and death. Risk factors, such as age, gender, and underlying disease, are known to be associated with COVID-19 disease severity; however, a subset of patients with severe disease are younger without obvious comorbidities. The immunological features associated with severe COVID-19 disease include high levels of inflammatory cytokines, low lymphocyte counts, high neutrophil to lymphocyte ratios, and increased serum proteins, such as C-reactive protein (CRP), ferritin, and D-dimer.3, 4, 5, 6 In addition, several studies have shown that SARS-CoV-2-specific antibodies and neutralizing titers are increased in patients who exhibit more severe disease.7, 8, 9, 10 Therefore, it is important to consider that SARS-CoV-2-specific antibodies may play multiple roles in COVID-19 pathogenesis, including control of viral infection, disease resolution, and immunopathology.
The humoral response to SARS-CoV-2 is primarily directed toward the viral nucleocapsid (N) protein and the spike protein that decorates the surface of the virus. The N protein is an RNA-binding protein composed of an N-terminal RNA binding domain and a C-terminal oligomerization domain that are essential for viral RNA transcription and replication.11 The spike protein is a multi-domain trimeric glycoprotein composed of two distinct subunits. The S1 subunit is composed of four domains, including the receptor binding domain (RBD) of the spike glycoprotein.12 The S2 domain forms the stalk-like portion of the full-length trimeric protein and is responsible for viral fusion with the host cell membrane. Antibody responses directed at the spike protein and RBD in particular have been identified as the main neutralizing component of the SARS-CoV-2 antibody response.13, 14, 15 A recent study found that distinct antibody signatures could be linked to different disease outcomes in hospitalized COVID-19 patients. Specifically, early spike-specific responses were associated with a positive outcome (convalescence), while early N-specific responses were associated with a negative outcome (death). Moreover, the Fc-associated functions of the antibody response, such as antibody-mediated phagocytosis, cytotoxicity, and complement deposition, were critical for disease resolution.16 In a follow-up study, Zohar et al.17 showed slow maturation of the SARS-CoV-2 antibody response, characterized by reduced levels of spike-specific immunoglobulin G (IgG) and Fc receptor (FcR)-mediated functions in non-survivors of COVID-19. However, more work is needed to characterize the antibody isotypes and subclasses generated in response to SARS-CoV-2 throughout the full spectrum of COVID-19 pathogenesis. In this study, we analyzed the humoral immune response to SARS-CoV-2 in a cohort of 536 convalescent healthcare workers that were stratified by COVID-19 disease severity. This cohort provided us with a snapshot of the SARS-CoV-2-specific antibody profile at convalescence as a window to previous disease pathogenesis. We found a significantly increased SARS-CoV-2-specific antibody response in severe COVID-19 patients when compared to patients who experienced mild and moderate disease symptoms. This severity-associated antibody increase was dominated by IgG, with a disproportionate IgG subclass response dominated by IgG3 and increased FcɣRIIIa binding.
Results
Study cohort
A total of 536 COVID-19 serum specimens from convalescent healthcare workers (HCWs) were received by the Wadsworth Center for SARS-CoV-2 serology testing. The sera were obtained from individuals who had tested positive by RT-PCR and had illness consistent with SARS-CoV-2 infection. Table 1 provides basic patient demographic information stratified by self-reported COVID-19 disease severity. The gender distribution of the study cohort was biased toward females (29% male and 69% female, with 2% gender unknown), reflecting the gender disparity within the HCWs. The mean age and days post-onset of symptoms (DPOs) were roughly the same in all gender categories. Approximately 10% of the study cohort experienced severe disease, 40% moderate disease, 40% mild disease, and 10% with uncharacterized disease. The average age of each group in our cohort increased with disease severity, as did the percentage of males within each group. In fact, the representation of males (65%) in the severe group was more than double that of the mild group (31%), illustrating a clear gender bias in COVID-19 disease severity (Table 1).
Table 1.
Study cohort of convalescent healthcare workers
| All | Male | Female | Unknown | |
|---|---|---|---|---|
| Count | 536 | 154 (29%) | 370 (69%) | 12 (2%) |
| Mean age (years) | 40 | 40 | 40 | 40 |
| Days post-onset | 41 | 40 | 41 | 42 |
| % mild disease | 40.5% | 44.2% | 38.9% | 41.7% |
| % moderate disease | 40.1% | 35.0% | 42.1% | 41.7% |
| % severe disease | 9.1% | 10.4% | 8.7% | 8.3% |
| % unknown severity | 10.3% | 10.4% | 10.3% | 8.3% |
Relationship of antibody production and virus neutralization capability with COVID-19 disease severity
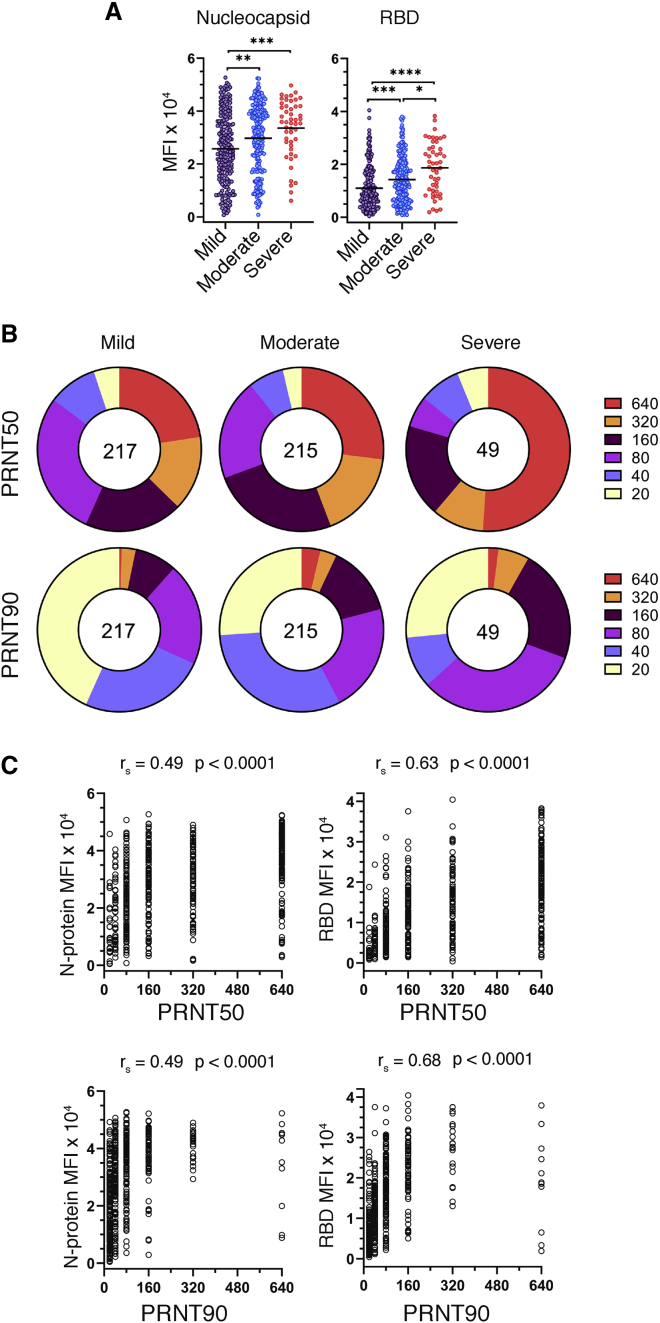
Serum samples were assessed for SARS-CoV-2-specific antibodies using a clinical microsphere immunoassay (MIA) to detect total antibody directed against N protein or RBD of SARS-CoV-2. We used the median fluorescence intensity (MFI) of phycoerythrin (PE)-labeled anti-human Ig to antigen-coupled beads as a qualitative measure of SARS-CoV-2-specific antibody abundance in donor serum. “Cutoff” values for each antibody isotype and antigen combination were defined as 3 standard deviations above the mean MFI, as determined by a panel of 94 pre-COVID-19 (2009) human serum specimens. Clinically positive antibody reactivity was defined as 6 standard deviations above the mean MFI (emergency use authorization: https://www.fda.gov/media/137541/download). We found that total serum antibody specific to the N protein and RBD of SARS-CoV-2 were increased with increasing disease severity (Figure 1A). To evaluate the relationship between overall antibody levels and protective antibody, we measured viral neutralization using a plaque reduction neutralization test (PRNT), where the highest dilution of sera providing 50% (PRNT50) or 90% (PRNT90) viral plaque reduction relative to a virus only control was reported as the neutralizing titer. Both PRNT50 and PRNT90 measurements identified a concomitant increase in virus-neutralizing titers with disease severity (Figure 1B). RBD-specific responses were strongly correlated with virus neutralization, as revealed through a Spearman’s correlation analysis with PRNT90 values and MFI values. The correlation with PRNT90 titers was greater for RBD-specific Ig than N-specific Ig (r = 0.68 and 0.53, respectively; Figure 1C). Because the function of the RBD is host cell attachment through ACE-2 binding, our data suggest that the neutralization activity of SARS-CoV-2-specific antibodies in our PRNT assay is primarily based on the ability to block viral attachment and uptake by host cells.13, 14, 15
Figure 1.
Relationship of antibody production and virus neutralization capability with COVID-19 disease severity
Serum specimens from convalescent COVID-19 donors were analyzed for reactivity to SARS-CoV-2 antigens and neutralization capacity.
(A) Median fluorescence intensity (MFI) of total Ig (IgM, -A, and -G) reactivity to SARS-CoV-2 nucleocapsid and RBD as determined by a microsphere immunoassay on convalescent COVID-19 serum specimens, grouped by disease severity (n = 481). Statistical significance was determined by the non-parametric Kruskal-Wallis test, where ∗p < 0.05, ∗∗p < 0.001, ∗∗∗p < 0.0001, and ∗∗∗∗p < 0.00001 adjusted for multiple comparisons by Dunn’s test.
(B) Reciprocal plaque reduction neutralization titer (PRNT) 50 and 90 dilutions based on a live virus assay on convalescent COVID-19 individuals, grouped by disease severity (n = 481). The relative size of each pie slice represents the percentage of specimens with a given titer. The center number represents the number of specimens in each group.
(C) PRNT50 and PRNT90 titers plotted against SARS-CoV-2 nucleocapsid or RBD MFI. Graphs and Spearman’s correlations are based on the full cohort (n = 536) patient specimens.
Isotype and antigen distribution of the SARS-CoV-2-specific antibody profile across COVID-19 disease severities
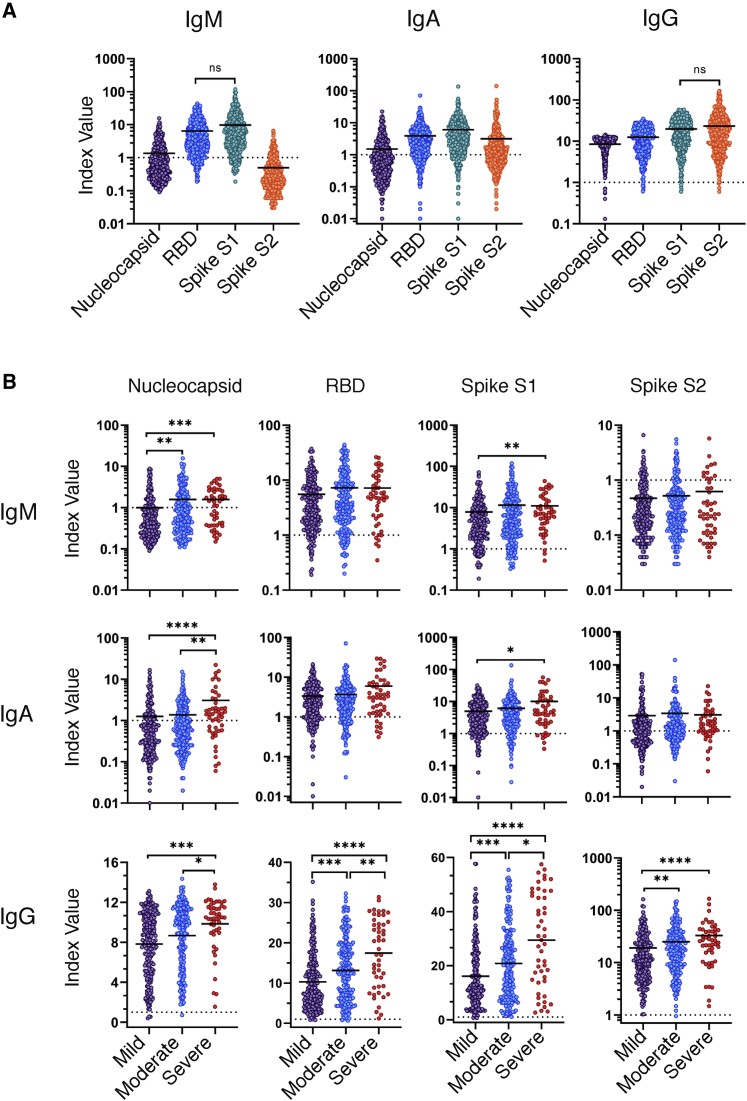
To better resolve the SARS-CoV-2 humoral immune response across COVID-19 severity groups, we tested additional antibody isotypes and specificities in our clinical MIA assay. To normalize our antibody measurements across antigens and isotypes, index value measurements were calculated by dividing the raw MFI by the cutoff value (3 standard deviations) as determined by the average MFI from a panel of pre-pandemic normal human serum specimens. In addition to N and RBD, we included the S1 and S2 subunit domains as antibody targets, allowing us to incorporate all the potential epitopes from the spike protein and to assess the contribution of each domain to the overall antibody response. In general, we found that antibody reactivity to SARS-CoV-2 antigens differed dramatically between isotypes. As expected, IgG was the dominant isotype generated in response to SARS-CoV-2 infection, with antibodies from 97% to 98% of serum specimens yielding positive recognition of all antigens and antigenic subunits tested (Figure 2A; Table S1). IgM and IgA were primarily reactive toward the spike S1 subunit, including the RBD—with 73% and 79% positivity, respectively. The presence of antigen-specific IgM at this convalescent time point is notable (∼day 40), as IgM is generally considered a biomarker of acute-phase infection. IgM and IgA with specificity to the nucleocapsid were rare, with only 22% of specimens testing positive for each isotype, respectively. IgM antibodies with specificity for the spike S2 subunit were even more rare (5%), while IgA responses displayed a 36% positivity rate. As shown in Figure 1, we observed that total SARS-CoV-2-specific antibody increased with disease severity. We next examined individual isotype distributions across disease categories. There was an increase in IgG generated in the severe COVID-19 group as compared to mild, against all antigens tested (N [1.3-fold; p < 0.001]; RBD [1.7-fold; p < 0.0001]; S1 [1.8-fold; p < 0.0001]; and S2 [1.7-fold; p < 0.0001]). In addition, IgM specific to the N protein had a 1.6-fold increase in the severe group, as compared to mild, with little or no change in other antigens. Finally, IgA generated against the N protein and the S1 subunit resulted in significant increases (2.4, p < 0.0001, and 2.0, p = 0.02, respectively)—the largest fold changes observed between mild and severe disease categories (Figure 2B; Table S2). The significant increase in N-specific IgA is consistent with the observation that exceptionally high levels of serum IgA were associated with ARDS.18 Furthermore, the observed correlation of IgM and IgA specific for the N protein increasing with disease severity is reminiscent of the early N-dominated response in deceased individuals reported by Atyeo et al.16 Our results illustrate that, although total SARS-CoV-2-specific antibody levels increase with COVID-19 severity, this change is not equal among antibody specificities or isotypes. The increase in total N-specific antibody and spike-specific IgG suggests there are enhanced levels of viral antigens and corresponding increases in T-cell-dependent B cell responses during severe COVID-19.
Figure 2.
Isotype and antigen distribution of the SARS-CoV-2-specific antibody response
Serum specimens from convalescent COVID-19 donors were analyzed for reactivity of IgM, IgA, and IgG specific to the SARS-CoV-2 nucleocapsid, RBD, S1 subunit, or S2 subunit. Index values represent the raw MFI divided by the cutoff value (3 standard deviations above the mean) determined by the average MFI of a panel of 94 pre-pandemic normal human serum specimens (cutoff = dashed line).
(A) Index values for IgM, IgA, and IgG reactivity to SARS-CoV-2 antigens on the full cohort (n = 536).
(B) Index values for the IgM, IgA, and IgG reactivity to SARS-CoV-2 antigens separated by COVID-19 disease severity (n = 481). Statistical significance was determined by the non-parametric Kruskal-Wallis test, where ∗p < 0.05, ∗∗p < 0.001, ∗∗∗p < 0.0001, and ∗∗∗∗p < 0.00001 adjusted for multiple comparisons by Dunn’s test.
See also Table S2.
IgG subclass and antigen distribution of the SARS-CoV-2-specific antibody profile across COVID-19 disease severities
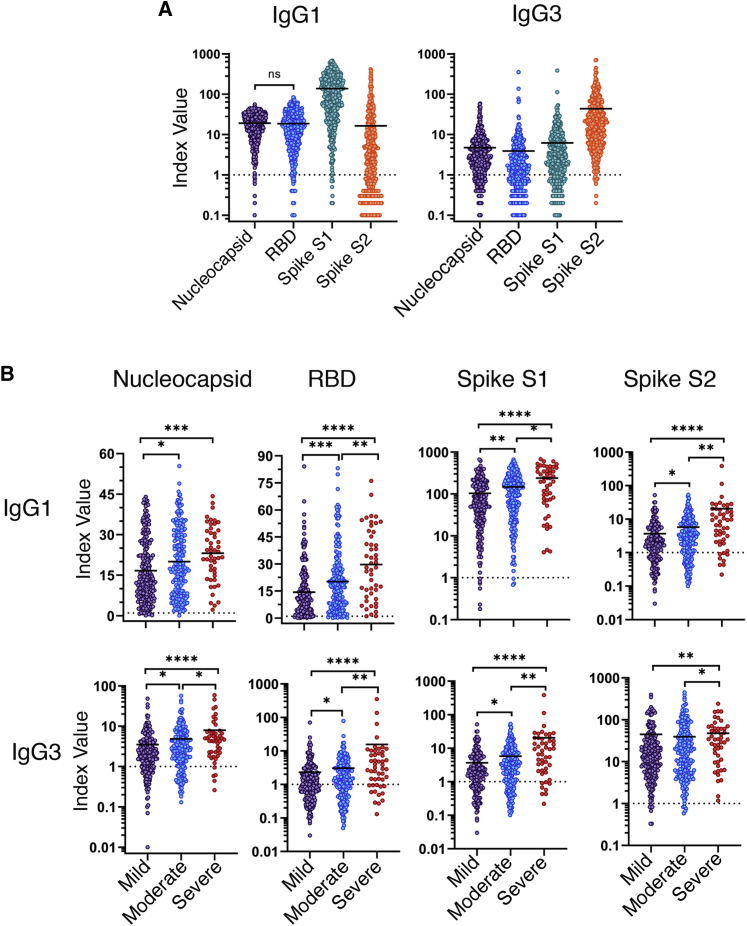
IgG is the classic antibody isotype involved in T-cell-dependent B cell responses, resulting in durable humoral memory. In humans, IgG can be further divided into four functional subclasses—IgG1, IgG2, IgG3, and IgG4. Each subclass has unique properties and effector functions that are primarily driven by the Fc portion of the antibody molecule, including complement activation, FcR binding, and serum half-life. In particular, the ability of an antibody to bind and signal through FcR on effector cells can have profound effects on disease resolution in many models of infectious disease. Therefore, we sought to characterize the IgG subclass usage of the SARS-CoV-2 antibody response. We found that IgG1 and IgG3 were the dominant IgG subclasses produced in response to SARS-CoV-2 infection (Figure 3A). The responses to nucleocapsid, RBD, and S1 subunit were each dominated by IgG1, with over 90% of specimens testing positive (Table S3). Strikingly, the IgG response toward the S2 subunit was dominated by the IgG3 subclass, with 94% of specimens yielding a positive clinical result. IgG2 responses were moderate, with a positivity rate ranging from 8% to 21%. Finally, IgG4 responses were especially rare with a clinical positivity rate of 0%–9% (Figure S1; Table S3). Next, we asked whether the IgG subclass representation changes in relation to COVID-19 disease severity. A small yet significant increase in N-specific IgG2 (1.5; p = 0.002) was associated with disease severity, while RBD-specific IgG2 trended downward with disease severity (Figure S1; Table S4). We observed significant increases in both IgG1 and IgG3 with increasing COVID-19 severity, specific for all antigens tested (N, RBD, S1, and S2). The largest difference was seen with the IgG3-specific response toward the RBD and S1 subunit with 6.8-fold (p < 0.0001) and 5.5-fold (p < 0.0001) increases in the mean index ratio between the mild and severe groups, respectively (Figure 3B; Table S4). Together, these results highlight distinct differences in the SARS-CoV-2-specific antibody profile among convalescent individuals that experienced mild, moderate, or severe symptoms of COVID-19.
Figure 3.
IgG subclass and antigen distribution of the SARS-CoV-2-specific antibody profile across COVID-19 disease severities
Serum specimens from convalescent COVID-19 donors were analyzed for reactivity of IgG1 and IgG3 specific to the SARS-CoV-2 nucleocapsid, RBD, S1 subunit, or S2 subunit. Index values represent the raw MFI divided by the cutoff value (3 standard deviations above the mean) determined by the average MFI of a panel of 94 pre-pandemic normal human serum specimens (cutoff = dashed line). Reactivity of IgG1, IgG2, and IgG3 to SARS-CoV-2 nucleocapsid, RBD, S1 subunit, or S2 subunit of the full patient cohort (A) or grouped by disease severity (B) is shown. Index value represents the raw MFI divided by the background cutoff value determined by a panel of 94 normal human serum specimens. Statistical significance was determined by the non-parametric Kruskal-Wallis test, where ∗p < 0.05, ∗∗p < 0.001, ∗∗∗p < 0.0001, and ∗∗∗∗p < 0.00001 adjusted for multiple comparisons by Dunn’s test. See also Figure S1 and Table S4.
Identification of variables associated with COVID-19 disease severity
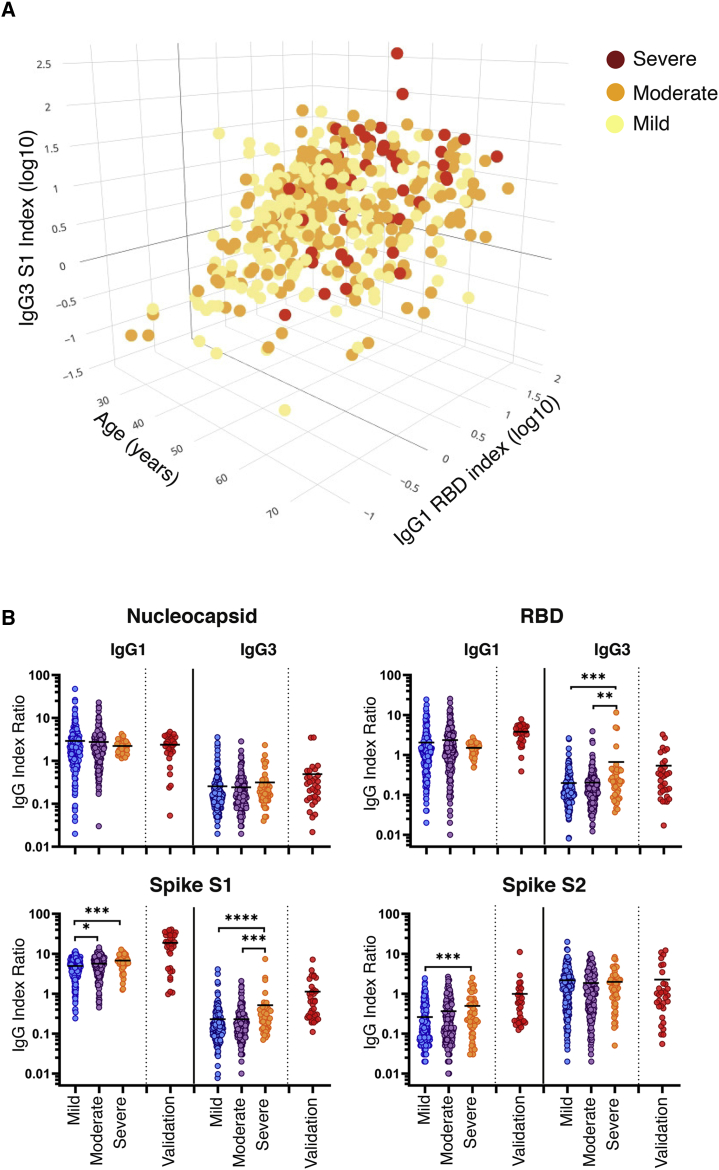
We sought to determine a minimal set of criteria that could best distinguish between individuals with different disease severities without overfitting. A training subset of the data was used to create an initial ordered probit regression model that included all potentially predictive variables, including antibody reactivities, gender, age, DPOs, and neutralizing antibody titers. Backward stepwise selection by Akaike information criterion (AIC) was performed on this model to determine the optimal set of features. When this model’s performance was measured on a testing subset of the data, it displayed higher accuracy (60%; Table S5) than both the initial all-inclusive model and any individual univariate model. The combined model that best discriminates between mild, moderate, and severe COVID-19 includes age, RBD-specific IgG1, and S1-specific IgG3. As shown in Figure 4A, the severe group clusters at high IgG1 RBD, IgG3 S1, and age measurements. In contrast, the mild and moderate groups cluster toward lower IgG1 RBD, IgG3 S1, and age measurements. In order to account for the confounding effects of age, a similar series of ordered probit regression models were created that included age as a covariate. All variables that were significantly associated with disease severity in the previous models retained their significant association with disease severity except S2-specific IgG1 (Table S6).
Figure 4.
Identification of variables associated with COVID-19 disease severity
(A) Three-dimensional scatterplot depicting the optimal feature set of disease-severity-associated features (age, log10-transformed index values [MFI/cutoff] for S1-specific IgG3, and RBD-specific IgG1) as determined by ordered probit regression and backward stepwise selection by Akaike information criterion. Data are displayed as the distribution of mild (yellow), moderate (orange), and severe (red) disease severities across variables in 478 patients.
(B) Serum sample index ratios for IgG1 and -3 specific to the N, RBD, S1, and S2 subunits were calculated by dividing the IgG subclass index value by the paired total IgG index value (IgGsubclass/IgGtotal) for each antigen. Index ratios were grouped by disease by disease severity (n = 481). Statistical significance was determined by the non-parametric Kruskal-Wallis test, where ∗p < 0.05, ∗∗p < 0.001, ∗∗∗p < 0.0001, and ∗∗∗∗p < 0.00001 adjusted for multiple comparisons by Dunn’s test.
See also Tables S5–S7.
Based on the results of the ordered probit regression model, we sought to define the relative contribution of both IgG1 and IgG3 to the overall SARS-CoV-2-specific pool of IgG. To this end, we calculated a ratio based on the index value of each IgG subclass divided by the total IgG index value of the same antigen (IgGsubclass/IgGtotal). No change was observed in N-specific IgG1 and IgG3 subclass ratios between COVID-19 disease severity groupings. We detected modest yet significant increases in the IgG1 subclass ratio to both the S1 and S2 subunits between mild and severe groups (1.4-fold, p = 0.0002 and 1.9-fold, p = 0.0004, respectively). A pronounced increase with the RBD-specific and S1 specific IgG3 subclass ratios was identified with increasing COVID-19 disease severity. Specifically, we measured 3.3-fold (p = 0.005) and 3.4-fold (p = 0.002) increases in the RBD-specific IgG3 subclass ratio between the moderate and severe groups as compared to the mild group. We also measured a 2.2-fold increase in the S1-specific IgG3 subclass ratio between the moderate and severe groups (p = 0.0002) and the mild and severe groups (p < 0.0001; Figure 4B; Table S7).
To confirm the severity-associated increase in IgG3, we tested a validation cohort (VC) of 32 acutely ill COVID-19 patients hospitalized due to the severity of their condition and were clinically positive for RBD-specific IgG. All serum samples were acquired at time of hospital admission (Table S8). The optimal three-variable ordered probit model predicted COVID-19 disease severity with 34.4% accuracy (95% confidence interval [CI] 0.186 and 0.532). The low predictive value of our model when tested on the VC is likely due to the sampling time point (convalescent versus acute) and significantly lower levels of IgG (Figure S3). In contrast, when we compared the RBD- and S1-specific index ratios of the VC to the convalescent cohort, we observed similar results. The mean IgG3 index ratio specific to the RBD (0.666) of the validation cohort was similar to the severe group (0.544), with a 2.7-fold increase (p = 0.004) over the mild group. Furthermore, the mean IgG3 subclass ratio of the VC specific to the S1 subunit (1.103) was significantly higher than all groups in the HCW cohort, with a 4.8-fold increase (p < 0.0001) over the mild group. In addition to high IgG3 index ratios to the spike proteins, we also observed significant increases in the IgG1 index ratio for the spike protein (RBD, S1, and S2), with 1.9- (p < 0.0001), 3.8- (p < 0.0001), and 3.8-fold (p < 0.0001) increases, respectively. These data highlight the utility of using index ratios where the IgG subclass is normalized to the level of total IgG and can be used across various stages during disease. In summary, these data show that a disease-severity-associated increase in total spike-specific IgG is accompanied by a proportional increase of IgG3 within the antibody pool.
Correlation of antibody measurements and COVID-19 clinical features
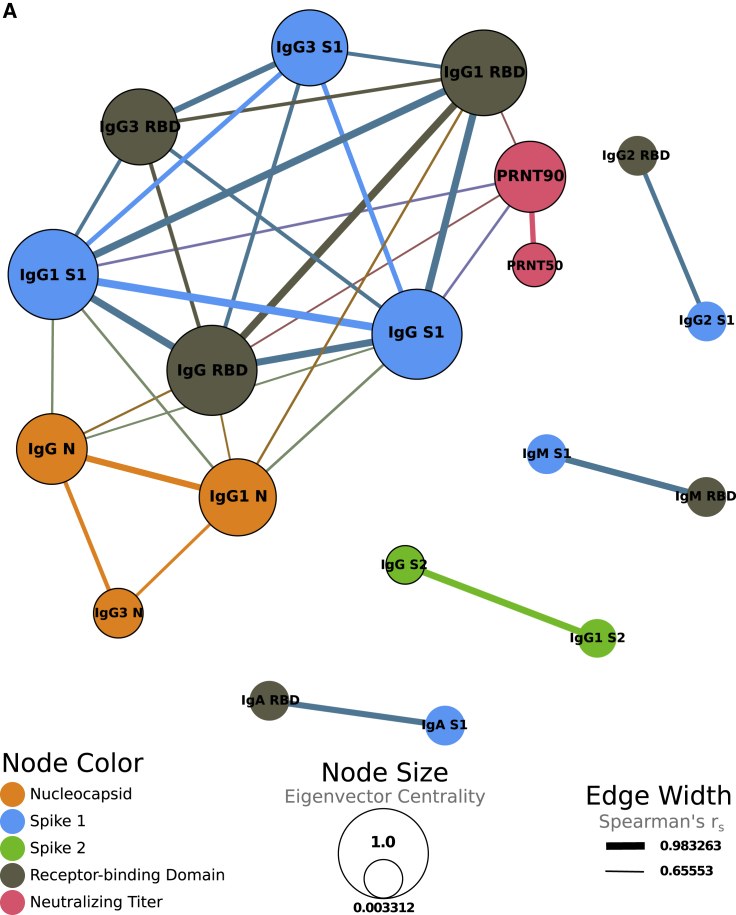
A correlation network was created to examine the relationship between antibody isotypes, subclasses, antigen specificity, viral neutralization, and clinical features of the HCW dataset. The network was based on a comprehensive correlation matrix (Figure S2) and stringently gated on only the strongest associations, with a Spearman’s coefficient above 0.65. As expected, S1 and RBD measurements were highly correlated, as were IgG and corresponding IgG subclasses. A dominant network cluster was formed between N-specific IgG, S1-specific IgG (including the RBD domain), and viral-neutralizing titers (Figure 5). N-specific IgG/IgG1/IgG3 formed its own mini-cluster on the edge of the highly inter-related IgG response to S1 and RBD. In addition, PRNT50 and PRNT90 measurements were very strongly correlated with each other (rs = 0.848), and the PRNT90 measurement was strongly correlated (0.5 ≤ rs ≤ 0.7) with IgG, IgG1, antibodies targeting the S1 domain, and RBD of the spike protein. This cluster was also significantly correlated with disease severity, as determined by ordered probit regression (Table S5). IgM, IgA, and IgG2 responses to S1 and RBD were also highly correlated yet separate from the central cluster. Taken together, our results reveal a shift in the proportional spike-specific IgG response toward the highly inflammatory IgG3 subclass19 that is linked to COVID-19 disease severity and not with increases in viral neutralizing activity.
Figure 5.
Correlation of antibody measurements and COVID-19 clinical features
(A) Correlation network displaying strongly correlated (Spearman’s rs > 0.65) variables. Edge thickness represents the magnitude of the correlation between variables. Node size represents eigenvector centrality, showing the influence of each node on the network. Node color represents whether that variable corresponds to an antibody targeting the nucleocapsid (orange), S1 (blue), S2 (green), or RBD (gray) regions or to neutralizing antibody titers (pink). Black borders around nodes correspond to variables with a significant correlation with severity determined by ordered probit regression modeling, while controlling for age as a confounding variable. All displayed correlations are statistically significant (Benjamini and Hochberg adjusted p < 0.05).
See also Figure S2.
Association of FcR binding to COVID-19 disease severity
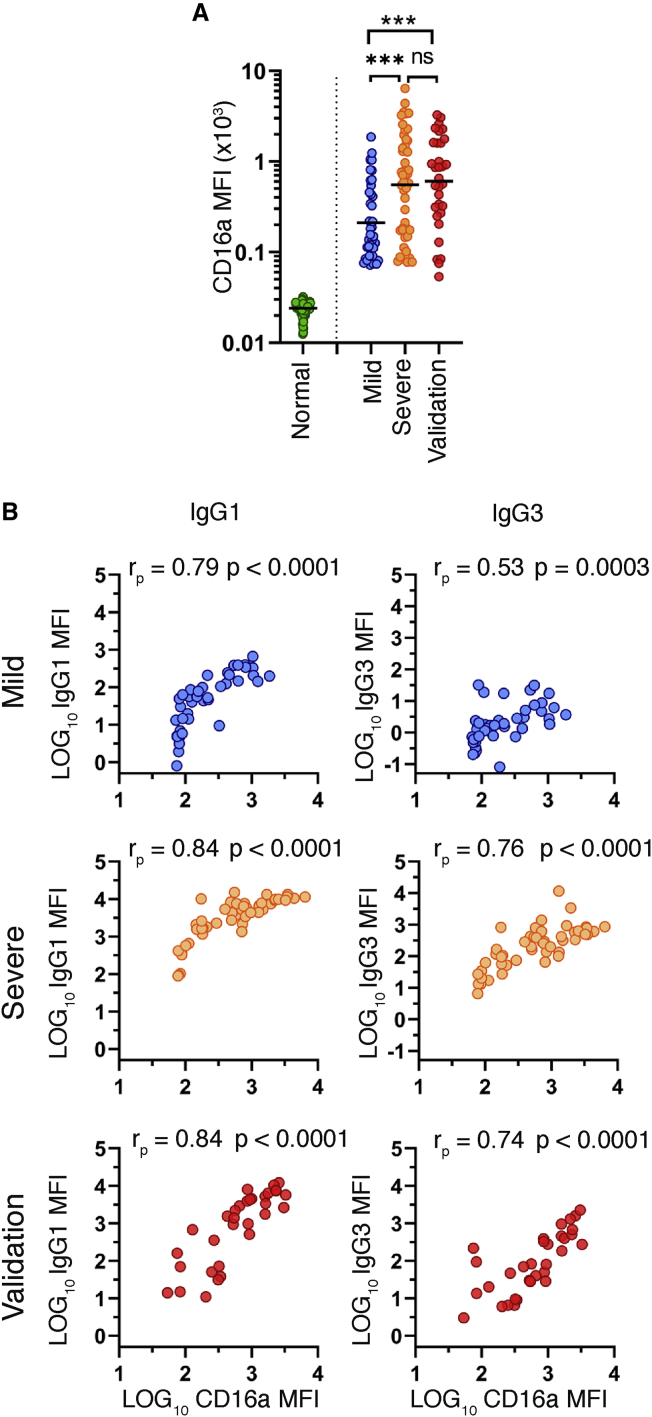
To demonstrate a functional relationship between proportional IgG subclass differences and disease severity, we developed a spike-specific microsphere-based FcR binding assay (STAR Methods). FcɣRIIIa (CD16a) is a type I activating FcR expressed on natural killer (NK) cells, subpopulations of macrophages and monocytes, and rare populations of T cells.20,21 IgG3 and specific glycosylation states of IgG1 (e.g., afucosylation) bind FcɣRIIIa with higher affinity than other subclasses.19 FcɣRIIIa is the sole FcR expressed on NK cells, where engagement of this receptor by immune complexes leads to inflammatory processes, such as antibody-dependent cellular cytotoxicity (ADCC) and downstream secretion of enzymes and inflammatory cytokines.22 As expected, a panel of 94 pre-pandemic normal human serum specimens showed minimal CD16a binding to full-length (FL)-spike-bound serum. In contrast, we detected a robust CD16a binding signal from COVID-19 patients, which included 43 randomly selected specimens from the mild HCW group, all 49 specimens from the severe HCW group, and the 32 VC specimens. We detected significantly increased FL-spike-specific CD16a binding in the severe (3.2-fold; p = 0.0009) and VC (2.9-fold; p = 0.001) groups, as compared to the mild group (Figure 6A). Pearson’s correlation analyses revealed strong, significant correlations between both S1-specific IgG1 and IgG3 with CD16a binding in all groups. In general, levels of IgG1 were better correlated with CD16a binding than IgG3 (Figure 6B). This difference was most noticeable in the “mild” group, where rp = 0.79 for IgG1 and rp = 0.53 for IgG3, which is likely due to the low levels of IgG3 in this group. In conclusion, the increased CD16a binding that we observed in the severe group and hospitalized validation cohort reveals a potential inflammatory component of the SARS-CoV-2 antibody response that correlates with COVID-19 disease severity.
Figure 6.
Association of Fc receptor binding to COVID-19 disease severity
Serum specimens from 94 pre-pandemic normal human serum specimens, 43 specimens from the mild group, all 49 specimens from the severe group, and all 32 acute hospitalized specimens from the validation cohort were tested for CD16a (FcɣRIIIa) binding to trimeric full-length spike (FL-spike)-specific antibody.
(A) MFI of FL-spike-specific CD16 binding as determined by a microsphere immunoassay. Statistical significance was determined by the non-parametric Kruskal-Wallis test, where ∗p < 0.05, ∗∗p < 0.001, ∗∗∗p < 0.0001, and ∗∗∗∗p < 0.00001 adjusted for multiple comparisons by Dunn’s test.
(B) Pearson’s correlation comparing the log10-transformed MFI of CD16a binding to the log10-transformed MFI of S1-specific IgG1 or IgG3.
Discussion
In viral infections, blockade of cell attachment and direct viral neutralization are thought to be the primary effector mechanisms through which antibodies contribute to host protection. However, antibodies also facilitate other mechanisms of pathogen clearance through the conserved Fc portion of the molecule. Fc effector functions include antibody-dependent complement deposition (ADCD), antibody-dependent cellular phagocytosis (ADCP), antibody-dependent neutrophil phagocytosis (ADNP), and ADCC and can be important for disease resolution. For example, ADCP and ADCC were shown to be critical mechanisms for vaccine-mediated protection against HIV.23,24 Antibody-mediated effector functions have also been shown for COVID-19 disease resolution, where spike-specific ADCP, ADNP, and ADCD were enhanced in COVID-19 survivors.16 In humans, IgG1 and IgG3 are the most common subclasses elicited by viral infections.25 Although IgG1 is the most abundant subclass in the serum, IgG3 is particularly effective at inducing effector functions through higher affinity interactions with complement proteins and activating type 1 Fc receptors.
In the present study, we performed a comprehensive analysis of the serum antibody response to SARS-CoV-2 in a cohort of 536 recovered HCWs who experienced varying degrees of COVID-19 severity. In general, we found that age, total SARS-CoV-2-specific antibody, and viral-neutralizing activity were increased in individuals who experienced severe disease. The serological profile for those who experienced severe COVID-19 was characterized by both a global increase in N-specific antibodies, as well as an increase in spike-specific IgG and SARS-CoV-2-neutralizing titers. Paradoxically, high-titer neutralizing antibodies are known to be protective and a biomarker of immunity in many viral infections.26,27 To rectify the disconnect between increased antibodies, increased neutralization, and increased disease severity, we performed a thorough analysis of the IgG subclass response to SARS-CoV-2. Our analysis revealed a substantial difference in the spike-specific IgG subclass composition, where a greater proportion of S1 and RBD-specific IgG3s were associated with COVID-19 severity in both a convalescent and acutely ill hospitalized cohort. Further analyses of the convalescent cohort found that spike-specific IgG1 and not IgG3 was most closely correlated with in vitro viral neutralization. Moreover, those with severe disease (including the validation cohort of hospitalized patients) showed increased spike-specific binding to FcɣRIIIa as compared to patients who experienced mild disease. Together, our data suggest that excess IgG3 may play an inflammatory role in the pathogenesis of COVID-19 in some individuals. Therefore, we hypothesize that an unbalanced IgG response enriched in inflammatory subclasses may promote excess inflammation, exacerbating the pathology of COVID-19.
An explanation for the overall increase in SARS-CoV-2-specific antibody in severe patients is simply increased viral loads and inflammatory signals, driving increased antigen presentation and stimulatory signals for humoral responses. However, many published studies support the hypothesis that imbalanced antibody responses to SARS-CoV-2 infection may contribute to severe disease. First, Liu et al.28 showed that anti-spike IgG promoted acute lung injury in humans and macaques infected with SARS-CoV-1 through macrophage polarization to an inflammatory phenotype. More recently, Garcia-Beltran et al.10 have shown the most severe COVID-19 patients have a significantly lower proportion of neutralizing titers within the total SARS-CoV-2 IgG antibody pool (neutralization potency; NT50/total IgG) than those with less severe disease—suggesting an accumulation of potentially disease enhancing, non-neutralizing antibodies. Increases in afucosylated IgG1 and a corresponding enhancement in FcɣRIIIa interactions have also been linked to COVID-19 disease severity.29,30 Increases in inflammatory forms of IgG1 may explain the significant proportional increases in IgG1 seen in the hospitalized validation cohort. In fact, Larsen et al.30 report that levels of IgG1 afucosylation changed as the antibody response matured, which highlights a key difference between our convalescent and acute validation cohort. Although some FcR-mediated effector functions, such as ADCP and ADCD, have been correlated with COVID-19 survival, spike-specific antibodies that promoted Fc-mediated NK cell activities were expanded in non-survivors16—highlighting the potential role for FcɣRIIIa-mediated functions in pathologic inflammation in severe COVID-19. Our data support this observation, as FcɣRIIIa is the only Fc receptor expressed on NK cells and has heightened affinity for IgG3. There is precedence for antibody and FcɣRIIIa-mediated inflammation causing detrimental effects in other viral infections. For example, secondary Dengue virus infection can lead to Dengue hemorrhagic fever or Dengue shock syndrome through antibody-dependent enhancement, where the presence of afucosylated IgG1 and FcɣRIIIa engagement was shown to be predictive of disease severity.31 In the context of these data, it is tempting to speculate that an increased proportion of spike-specific IgG3 may be predictive of long-term COVID-19 symptoms frequently seen during SARS-CoV-2 convalescence. Further studies are needed to address the true contribution of IgG subclasses and Fc effector functions to COVID-19 disease resolution or pathology. Although recent data suggest that ADCD plays a protective role in COVID-19,16 antibody-dependent pathways should not be ruled out as a potential driver of inflammation and disease pathology.32 In conclusion, we recommend that the potential contribution of S1 and RBD-specific IgG3 to COVID-19 disease severity should be a strong consideration for SARS-CoV-2 vaccine design, monoclonal antibody therapeutics, and convalescent plasma therapy.
Limitations of the study
The convalescent time point at which this study was conducted allowed us to capture a large cohort that was with well-associated disease severity classifications. The size of our patient cohort enabled a robust analysis of the antibody response to SARS-CoV-2 in individuals with self-reported mild, moderate, or severe disease. However, self-reporting disease severity can be subjective and may contribute to some overlap seen between the severity groupings. The late time point (day 40 post onset) of this study weakens the predictive capacity of our analysis for earlier points during infection. In addition, samples from the full spectrum of COVID-19 presentation, including asymptomatic individuals and non-survivors, would significantly strengthen our study. Overall, this study is correlational and would be strengthened by additional and comprehensive functional studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat Anti-Human IgM-PE | Southern Biotech | Cat# 2020-09; RRID: AB_2795606 |
| Goat Anti-Human IgA-PE | Southern Biotech | Cat# 2050-09; RRID: AB_2795707 |
| Goat Anti-Human IgG-PE | Southern Biotech | Cat# 2040-09; RRID: AB_2795648 |
| Mouse Anti-Human IgG1 Fc-PE | Southern Biotech | Cat# 9054-09; RRID: AB_2796628 |
| Mouse Anti-Human IgG2 Fc-PE | Southern Biotech | Cat# 9070-09; RRID: AB_2796639 |
| Mouse Anti-Human IgG3 Hinge-PE | Southern Biotech | Cat#9210-09; RRID: AB_2796701 |
| Mouse Anti-Human IgG4 Fc-PE | Southern Biotech | Cat# 9200-09; RRID: AB_2796693 |
| Mouse Anti-6X His | Southern Biotech | Cat# ab72467; RRID: AB_1267596 |
| Bacterial and virus strains | ||
| SARS-CoV-2, isolate USA-WA1/2020 | BEI Resources | Cat#NR18152281 |
| Biological samples | ||
| COVID-19 Human Serum Specimens | Wadsworth Center, New York State Department of Health | N/A |
| COVID-19 Human Serum Specimens | Albany Medical Center | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| MicroPlex Microspheres, Region 006 | Luminex Corp. | Cat#LC10006 |
| MicroPlex Microspheres, Region 019 | Luminex Corp. | Cat#LC10019 |
| MicroPlex Microspheres, Region 061 | Luminex Corp. | Cat#LC10061 |
| MicroPlex Microspheres, Region 071 | Luminex Corp. | Cat#LC10071 |
| SARS-CoV-2 Nucleoprotein, His-Tag (E.coli) | The Native Antigen Company | Cat#REC31812 |
| SARS-CoV-2 Spike S2, Sheep Fc-Tag (HEK293) | The Native Antigen Company | Cat#REC31807 |
| SARS-CoV-2 Spike protein S319-541-Myc-His (RBD Fragment) | Mass Biologics (https://www.umassmed.edu/massbiologics) | N/A |
| SARS-CoV-2 Spike protein S604-Myc-His (S1 Fragment) | Mass Biologics (https://www.umassmed.edu/massbiologics) | N/A |
| SARS-CoV-2 Spike protein S1208-Trimer-His | Mass Biologics (https://www.umassmed.edu/massbiologics) | N/A |
| Human Fc gamma RIIIA/CD16a (F176) Protein, His Tag (SPR & BLI verified) | Acro Biosystems | Cat#CDA-H5220 |
| xMAP Antibody Coupling (AbC) Kit | Luminex Corp. | Cat#40-50016 |
| ExpiFectamine™ 293 Transfection Kit | Thermo Fisher | Cat#A14525 |
| QuickChangeII XL Site-Directed Mutagenesis Kit | Agilent Technologies | Cat# 200521 |
| Experimental models: Cell lines | ||
| Vero E6 cells C1008 | ATCC | Cat#ATCC CRL-1586; RRID: CVCL_0574 |
| Expi293F™ cells | ThermoFisher | Cat#14527; RRID: CVCL_D615 |
| Recombinant DNA | ||
| pcDNA™ 3.1(+) Mammalian Expression Vector | ThermoFisher | Cat# V79020 |
| SARS-CoV-2 Spike Glycoprotein sequence | GenBank | GenBank: MN908947 |
| Software and algorithms | ||
| xPONENT 4.3 | Luminex Corp. | https://www.luminexcorp.com/?wpdmpro=xponent-software-version |
| GraphPad Prism 9 | GraphPad | RRID: SCR_002798 |
| R 4.0.2 | The R Foundation for Statistical Computing | https://www.r-project.org/ |
| Gephi 0.9.2 | Gephi | https://gephi.org/users/download/ |
| caret R package version 6.0-86 | The R Foundation for Statistical Computing | https://cran.r-project.org/web/packages/caret/index.html |
| MASS R package version 7.3-53.1 | The R Foundation for Statistical Computing | https://cran.r-project.org/web/packages/MASS/index.html |
| RANN R package version 2.6.1 | The R Foundation for Statistical Computing | https://cran.r-project.org/web/packages/RANN/index.html |
| corrplot R package version 0.84 | The R Foundation for Statistical Computing | https://cran.r-project.org/web/packages/corrplot/index.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Dr. William Lee (william.lee@health.ny.gov).
Materials availability
Resources used in this study will be made available to the scientific community upon request, and following execution of a material transfer agreement, by contacting Dr. William Lee (william.lee@health.ny.gov).
Data and code availability
The amino-acid sequence of the SARS-CoV-2 S glycoprotein sequence can be found on GenBank (MN908947). All other data generated are included in figures and tables in this published article.
Experimental model and subject details
Human subjects
Convalescent COVID-19 serum specimens
Studies were performed on sera from 536 clinical specimens submitted to the Wadsworth Center, New York State Department of Health for determination of antibody reactivity to SARS-CoV-2. The study population were recovered individuals who were all RT-PCR confirmed by Roche COVAS 6800 or Cepheid XPert for the presence of SARS-CoV-2 and who had a self-reported degree of disease severity (mild, moderate, or severe). All individuals had recovered from the disease and had defined days between the onset of symptoms and sample collection. Specimens were stored at 4°C until clinical tested was completed (> 1 week) and transferred to −80°C for long-term storage. Aliquots were made to minimize freeze-thaw. All testing and archiving of human specimens was approved by NYSDOH Institutional Review Board (IRB 20-021). See also Table 1.
Acute hospitalized COVID-19 serum specimens
The validation cohort included sera from 32 acute COVID-19 patients who were admitted to Albany Medical Center between October 2020 and January 2021. All patients had a positive for SARS-CoV-2 test result upon admission. All patients were hospitalized because of the severity of their illness as determined by their clinician with demographic data as reported in Table S8. Specimens were processed on the same day of collection and placed into −80C freezer for long term storage. The study was approved by the IRB at Albany Medical Center (protocol # 5929); all patients provided written informed consent.
Method details
Expression and purification of SARS-CoV-2 proteins
Spike glycoprotein expression and purification
The amino-acid sequence of the SARS-CoV-2 S glycoprotein sequence (GenBank: MN908947) were used to design a codon-optimized version for mammalian cell expression. The synthetic gene encoding the receptor binding domain (RBD) aa319- 541)) and S1 subunit (aa1-604) of the S glycoprotein were cloned into pcDNA 3.1 Myc/His in-frame with c-Myc and 6-histidine epitope tags that enabled detection and purification. The cloned genes were sequenced to confirm that no errors had accumulated during the cloning process. All constructs were transfected into Expi293 cells using ExpiFectamine 293 Transfection Kit (Thermo Fisher). Recombinant proteins were purified by immobilized metal chelate affinity chromatography using nickel-nitrilotriacetic acid (Ni-NTA) agarose beads, eluted from the columns using 250 mmol/L imidazole, and then dialyzed into phosphate-buffered saline (PBS), pH 7.2. Proteins were checked for size and purity by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
The stabilized trimer ectodomain of the SARS-CoV-2 spike glycoprotein sequence (GenBank: MN908947) residues 1-1208 was modified by adding two proline substitutions at residues 986 and 987, a “GSAS” substitution at residues 682-685, a C-terminal T4 fibritin trimerization otif, and HRV3C protease cleavage site, a TwinStrepTag and an 8x HisTag. The construct was cloned into the mammalian expression vector pcDNA3.1. The construct was then transfected into Expi293 cells using the ExpiFectamine Transfection Kit (ThermoFisher). Protein was purified using StrepTactin resin (IBA) followed by size-exlusion chromatography using a Superose 6 10/300 column (GE Healthcare).
SARS-CoV-2 specific microsphere immunoassay (MIA)
Specimens were assessed for the presence of antibodies reactive with SARS-CoV-2 using an MIA33. Recombinant SARS-CoV-2 nucleocapsid, RBD, S1, and S2 subunits were covalently linked to the surface of fluorescent microspheres (Luminex Corporation). Serum samples (25 μL at 1:100 dilution) and antigen-conjugated microspheres (25 μL at 5x104 microspheres/mL) were mixed and incubated 30 minutes at 37°C. Serum-bound microspheres were washed and incubated with phycoerythrin (PE)-conjugated secondary antibody. The PE-conjugated antibodies were chosen to specifically recognize, total Ig (Pan-Ig), IgM, IgA, IgG, IgG1, IgG2, IgG3, IgG4. After washing and final resuspension in buffer, the samples were analyzed on a FlexMap 3D analyzer using xPONENT software, version 4.3 (Luminex Corporation).
Fc receptor binding assay
SARS-CoV-2 Spike-specific FcR binding was determined using a modified MIA. Trimeric full-length spike protein (FL-Spike) was covalently coupled to the surface of fluorescent microspheres (Luminex Corporation). Serum samples (50 μL at 1:100 dilution) and antigen-conjugated microspheres (50 μL at 5x104 microspheres/mL) were mixed and incubated 30 minutes at 37°C. Serum-bound microspheres were washed and subsequently incubated with 50 μL recombinant CD16a-6XHis (Acro Biologics) at 20 μg/ml for 30 minutes at 37°C. Serum/CD16a-bound microspheres were washed and incubated with 50 μL PE-conjugated anti-6XHis tag antibody at 5 μg/ml for 30 minutes at 37°C. After washing and final resuspension in buffer, the samples were analyzed on a FlexMap 3D analyzer using xPONENT software, version 4.3 (Luminex Corporation), and signal was measured by median fluorescence intensity.
Plaque reduction neutralization assay (PRNT)
For the detection of SARS-CoV-2 neutralizing antibodies, 2-fold serially diluted test serum (100 μl) was mixed with 100 μL of 150-200 plaque forming units (PFUs) of SARS-CoV-2 (isolate USA-WA1/2020, BEI Resources, NR-52281) and incubated for 1 h at 37°C, 5% CO2. The virus:serum mixture (100 μl) was applied to VeroE6 cells grown to 95%–100% confluency in 6 well plates. Adsorption of the virus:serum mixture was allowed to proceed for 1 hour at 37°C, 5% CO2. Following the adsorption period, a 0.6% agar overlay prepared in cell culture medium (Eagle’s Minimal Essential Medium, 2% heat inactivated FBS, 100 μg/ml Penicillin G, 100 U/ml Streptomycin) was applied. Two days post-infection, a second agar overlay containing 0.2% neutral red prepared in cell culture medium was applied, and the number of plaques in each sample well were recorded after an additional 1-2 days incubation. Neutralizing titers were defined as the inverse of the highest dilution of serum providing 50% (PRNT50) or 90% (PRNT90) viral plaque reduction relative to a virus only control. This assay has been described and is considered the standard for determination of neutralizing virus-specific antibody titers34, 35, 36.
Quantification and statistical analysis
Ordered probit regression
Subjects with known disease severity and gender were included in ordered probit regression analysis. The data was randomly separated into training (70%) and testing (30%) subsets. Data was then centered by subtracting the mean of each predictor from that predictor’s values, and then scaled by dividing by the standard deviation. Imputation of missing values was performed using the k-nearest neighbors algorithm. Each created model’s accuracy was assessed by measuring its performance on the testing subset of data. One-sided binomial tests were performed to determine if the accuracy of each model was significantly better than the no-information rate. Variable selection for the final model was performed by backward stepwise selection by Akaike information criterion.
Correlation matrix
Spearman’s correlations were calculated using all complete pairs of variables in the dataset. All associated p values were then adjusted via the Benjamini and Hochberg method. The network created using these Spearman’s correlations was gated to only include those relationships with an rs > 0.65. Eigenvector centrality was calculated for each variable and is represented by the size of the respective node. Edge width corresponds with the strength of each correlation. R version 4.0.2 was used for the probit regression modeling and construction of the correlation matrix (https://www.R-project.org/).37 The caret,38 MASS,39 and RANN40 packages were also used for the probit regression. The corrplot package was used to create the correlation matrix.41 Gephi 0.9.2 was used in the construction of the correlation network.42
Acknowledgments
We acknowledge and thank the members of the following Wadsworth Center laboratories and core facilities for their expert technical assistance: Diagnostic Immunology, Steven Bush, Andrea Furuya, Theresa Lamson, Mary Marchewka, Randy Stone, Heidi Rose Tucker, Colleen Walsh, and Casey Warszycki, and the Tissue Culture Core. We thank Elizabeth Leadbetter and Gary Winslow for providing helpful discussions and critical reading of the manuscript. BioRender was used for the creation of the graphical abstract. This work was performed in part under a Project Award Agreement from the National Institute for Innovation in Manufacturing Biopharmaceuticals (NIIMBL), United States and financial assistance award 70NANB20H037 from the US Department of Commerce, National Institute of Standards and Technology. A portion of the work described in this publication was supported by Cooperative Agreement Number NU50CK000516 from Centers for Disease Control and Prevention, United States. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the Centers for Disease Control and Prevention. Research reported in this publication was supported, in part by the National Cancer Institute of the National Institutes of Health under Award Number U01CA260508. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
Conceptualization, J.L.Y.; methodology, J.L.Y., D.T.H., D.J.E., R.C.G., and A.P.D.; validation, J.L.Y., D.T.H., R.C.G., and A.P.D.; formal analysis, J.L.Y., D.J.E., M.S., and S.V.; investigation, J.L.Y., D.T.H., R.C.G., A.P.D., and A.F.P.; resources, K.M.H., K.C., M.H., M.E., Q.L., Y.W., R.P.-W., A.R., G.S., and K.S.; data curation, D.T.H., K.E.K., and V.L.D.; writing – original draft, J.L.Y. and D.J.E.; writing – review and editing, J.L.Y., D.J.E., N.J.M., and K.S.; visualization, J.L.Y. and D.J.E.; supervision, W.T.L., K.A.M., and N.J.M.
Declaration of interests
The authors declare no competing interests.
Published: June 14, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100329.
Contributor Information
Jennifer L. Yates, Email: jennifer.yates@health.ny.gov.
William T. Lee, Email: william.lee@health.ny.gov.
Supplemental information
References
- 1.McLaughlin C.C., Doll M.K., Morrison K.T., McLaughlin W.L., O’Connor T., Sholukh A.M., Bossard E.L., Phasouk K., Ford E.S., Diem K. High community SARS-CoV-2 antibody seroprevalence in a ski resort community, Blaine County, Idaho, US. preliminary results. medRxiv. 2020 doi: 10.1101/2020.07.19.20157198. [DOI] [Google Scholar]
- 2.Pollán M., Pérez-Gómez B., Pastor-Barriuso R., Oteo J., Hernán M.A., Pérez-Olmeda M., Sanmartín J.L., Fernández-García A., Cruz I., Fernández de Larrea N., ENE-COVID Study Group Prevalence of SARS-CoV-2 in Spain (ENE-COVID): a nationwide, population-based seroepidemiological study. Lancet. 2020;396:535–544. doi: 10.1016/S0140-6736(20)31483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D’Andrea K., UPenn COVID Processing Unit Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuri-Cervantes L., Pampena M.B., Meng W., Rosenfeld A.M., Ittner C.A.G., Weisman A.R., Agyekum R.S., Mathew D., Baxter A.E., Vella L.A. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020;5:eabd7114. doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.E., Katsaounou P. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27:992–1000.e3. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., Liao P., Qiu J.F., Lin Y., Cai X.F. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 8.Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J., Wu Q., Liu Z., Wang Q., Wu J., Hu Y., Bai T., Xie T., Huang M., Wu T. Spike-specific circulating T follicular helper cell and cross-neutralizing antibody responses in COVID-19-convalescent individuals. Nat. Microbiol. 2021;6:51–58. doi: 10.1038/s41564-020-00824-5. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Beltran W.F., Lam E.C., Astudillo M.G., Yang D., Miller T.E., Feldman J., Hauser B.M., Caradonna T.M., Clayton K.L., Nitido A.D. COVID-19-neutralizing antibodies predict disease severity and survival. Cell. 2021;184:476–488.e11. doi: 10.1016/j.cell.2020.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Z., Zhou Z., Chen Q. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B. 2020;10:1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suthar M.S., Zimmerman M.G., Kauffman R.C., Mantus G., Linderman S.L., Hudson W.H., Vanderheiden A., Nyhoff L., Davis C.W., Adekunle O. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell Rep. Med. 2020;1:100040. doi: 10.1016/j.xcrm.2020.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X., Li R., Pan Z., Qian C., Yang Y., You R., Zhao J., Liu P., Gao L., Li Z. Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell. Mol. Immunol. 2020;17:647–649. doi: 10.1038/s41423-020-0426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atyeo C., Fischinger S., Zohar T., Slein M.D., Burke J., Loos C., McCulloch D.J., Newman K.L., Wolf C., Yu J. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity. 2020;53:524–532.e4. doi: 10.1016/j.immuni.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zohar T., Loos C., Fischinger S., Atyeo C., Wang C., Slein M.D., Burke J., Yu J., Feldman J., Hauser B.M. Compromised humoral functional evolution tracks with SARS-CoV-2 mortality. Cell. 2020;183:1508–1519.e12. doi: 10.1016/j.cell.2020.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cervia C., Nilsson J., Zurbuchen Y., Valaperti A., Schreiner J., Wolfensberger A., Raeber M.E., Adamo S., Weigang S., Emmenegger M. Systemic and mucosal antibody responses specific to SARS-CoV-2 during mild versus severe COVID-19. J. Allergy Clin. Immunol. 2021;147:545–557.e9. doi: 10.1016/j.jaci.2020.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vidarsson G., Dekkers G., Rispens T. IgG subclasses and allotypes: from structure to effector functions. Front. Immunol. 2014;5:520. doi: 10.3389/fimmu.2014.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bournazos S., Gupta A., Ravetch J.V. The role of IgG Fc receptors in antibody-dependent enhancement. Nat. Rev. Immunol. 2020;20:633–643. doi: 10.1038/s41577-020-00410-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Sorge N.M., van der Pol W.L., van de Winkel J.G. FcgammaR polymorphisms: Implications for function, disease susceptibility and immunotherapy. Tissue Antigens. 2003;61:189–202. doi: 10.1034/j.1399-0039.2003.00037.x. [DOI] [PubMed] [Google Scholar]
- 22.Lu L.L., Suscovich T.J., Fortune S.M., Alter G. Beyond binding: antibody effector functions in infectious diseases. Nat. Rev. Immunol. 2018;18:46–61. doi: 10.1038/nri.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung A.W., Ghebremichael M., Robinson H., Brown E., Choi I., Lane S., Dugast A.S., Schoen M.K., Rolland M., Suscovich T.J. Polyfunctional Fc-effector profiles mediated by IgG subclass selection distinguish RV144 and VAX003 vaccines. Sci. Transl. Med. 2014;6:228ra38. doi: 10.1126/scitranslmed.3007736. [DOI] [PubMed] [Google Scholar]
- 24.Neidich S.D., Fong Y., Li S.S., Geraghty D.E., Williamson B.D., Young W.C., Goodman D., Seaton K.E., Shen X., Sawant S., HVTN 505 Team Antibody Fc effector functions and IgG3 associate with decreased HIV-1 risk. J. Clin. Invest. 2019;129:4838–4849. doi: 10.1172/JCI126391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferrante A., Beard L.J., Feldman R.G. IgG subclass distribution of antibodies to bacterial and viral antigens. Pediatr. Infect. Dis. J. 1990;9(8, Suppl):S16–S24. [PubMed] [Google Scholar]
- 26.Ng S., Nachbagauer R., Balmaseda A., Stadlbauer D., Ojeda S., Patel M., Rajabhathor A., Lopez R., Guglia A.F., Sanchez N. Novel correlates of protection against pandemic H1N1 influenza A virus infection. Nat. Med. 2019;25:962–967. doi: 10.1038/s41591-019-0463-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plotkin S.A. Correlates of protection induced by vaccination. Clin. Vaccine Immunol. 2010;17:1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H., Tang H., Nishiura K., Peng J., Tan Z. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4:e123158. doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chakraborty S., Gonzalez J., Edwards K., Mallajosyula V., Buzzanco A.S., Sherwood R., Buffone C., Kathale N., Providenza S., Xie M.M. Proinflammatory IgG Fc structures in patients with severe COVID-19. Nat. Immunol. 2021;22:67–73. doi: 10.1038/s41590-020-00828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larsen M.D., de Graaf E.L., Sonneveld M.E., Plomp H.R., Nouta J., Hoepel W., Chen H.-J., Linty F., Visser R., Brinkhaus M. Afucosylated IgG characterizes enveloped viral responses and correlates with COVID-19 severity. Science. 2021;371:eabc8378. doi: 10.1126/science.abc8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang T.T., Sewatanon J., Memoli M.J., Wrammert J., Bournazos S., Bhaumik S.K., Pinsky B.A., Chokephaibulkit K., Onlamoon N., Pattanapanyasat K. IgG antibodies to dengue enhanced for FcγRIIIA binding determine disease severity. Science. 2017;355:395–398. doi: 10.1126/science.aai8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ricklin D., Lambris J.D. Complement in immune and inflammatory disorders: pathophysiological mechanisms. J. Immunol. 2013;190:3831–3838. doi: 10.4049/jimmunol.1203487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong S.J., Furuya A., Zou J., Xie X., Dupuis A.P., 2nd, Kramer L.D., Shi P.Y. A multiplex microsphere immunoassay for Zika virus diagnosis. EBioMedicine. 2017;16:136–140. doi: 10.1016/j.ebiom.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindsey H.S., Calisher C.H., Mathews J.H. Serum dilution neutralization test for California group virus identification and serology. J. Clin. Microbiol. 1976;4:503–510. doi: 10.1128/jcm.4.6.503-510.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calisher C.H., Karabatsos N., Dalrymple J.M., Shope R.E., Porterfield J.S., Westaway E.G., Brandt W.E. Antigenic relationships between flaviviruses as determined by cross-neutralization tests with polyclonal antisera. J. Gen. Virol. 1989;70:37–43. doi: 10.1099/0022-1317-70-1-37. [DOI] [PubMed] [Google Scholar]
- 36.Shambaugh C., Azshirvani S., Yu L., Pache J., Lambert S.L., Zuo F., Esser M.T. Development of a high-throughput respiratory syncytial virus fluorescent focus-based microneutralization assay. Clin. Vaccine Immunol. 2017;24:e00225-17. doi: 10.1128/CVI.00225-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.R Development Core Team . 2020. R: A language and environment for statistical computing (R Foundation for Statistical Computing) [Google Scholar]
- 38.Kuhn M. 2020. Caret: classification and regression training. R package version 6.0-86.https://cran.r-project.org/web/packages/caret/index.html [Google Scholar]
- 39.Venables W.N., Ripley B.D. Fourth Edition. Springer; 2002. Modern Applied Statistics with S. [Google Scholar]
- 40.Arya S., Mount D., Kemp S.E., Jefferis G. 2019. RANN: fast nearest neighbor search (Wraps ANN Library) using L2 metric. R package version 2.6.1.https://cran.r-project.org/web/packages/RANN/index.html [Google Scholar]
- 41.Wei T., Simko V. 2017. R package “corrplot”: Visualization of a Correlation Matrix (Version 0.84)https://github.com/taiyun/corrplot [Google Scholar]
- 42.Bastian M., Heymann S., Jacomy M. Gephi: an open source software for exploring and manipulating networks. Procedings of the International AAAI Conference on Web and Social Media. 2009;3:361–362. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The amino-acid sequence of the SARS-CoV-2 S glycoprotein sequence can be found on GenBank (MN908947). All other data generated are included in figures and tables in this published article.