Abstract
Objectives: This study analyzed the effect of dexmedetomidine (DEX) on biological behavior of osteosarcoma cells through expression of miR-1307. Methods: We performed routine culture of human osteosarcoma cells MG-63 and randomly divided into control group, low-dose DEX group (25 ng/ml), medium-dose DEX group (50 ng/ml) and high-dose DEX group (100 ng/ml). Subsequently, we detected the cell proliferation (by CCK8 method), cell apoptosis (flow cytometry), mir-1307 expression (qRT-PCR), cell invasion (Transwell), and cell migration (scratch test) respectively. Results: The growth rate of osteosarcoma cells MG-63 slowed down with the increase of DEX concentration. Compared with the control group, the cellular absorbance in groups with different DEX dose decreased remarkably after 72 hours of culture (P<0.05). The proportion of apoptotic cells increased as well with the uplifting of DEX concentration, and the apoptotic rate in medium and high dosed DEX groups were remarkably higher than which in control group (P<0.05). Compared with the control group, the invasive ability of MG-63 cells after DEX treatment decreased significantly, and with the increase of DEX concentration, the number of invasive cells declined more obviously (P<0.05). Compared with the control group, the mobility rate of MG-63 cells after DEX treatment decreased significantly, and with the increase of DEX concentration, the cell mobility rate decreased more remarkably (P<0.05). In addition, the relative expression of miR-1307 in MG-63 cells after DEX treatment decreased significantly comparing to the control group, and the decline was more noteworthy with the increase of DEX concentration (P<0.05). Conclusion: DEX can effectively inhibit the proliferation, invasion, metastasis, and apoptosis of osteosarcoma cells in a dose-dependent manner, and its efficacy may be related to its regulation of miR-1307 expression.
Keywords: Dexmedetomidine (Dex), miR-1307, osteosarcoma, cellar biological behavior
Introduction
Osteosarcoma is a primary bone cancer, which accounts for around 20% of all bone tumors. Osteosarcoma originates from mesenchymal progenitor cells and can differentiate into bone, cartilage and fibers. Most patients with osteosarcoma are diagnosed within 20 years after birth [1,2]. The common complications of osteosarcoma are pulmonary metastasis and pathological fracture. For patients with metastatic osteosarcoma, a further aggravation of condition may occur with typical clinical symptoms including pain, limited joint activity, and local swelling [3]. Although the survival time of patients can be prolonged after treatment, it still imposes serious impact on patients’ physical and mental health. The oncogenesis and progression of osteosarcoma are affected by multiple factors and cumulative effects in stages, including environmental factors and genetic factors [4,5]. Dexmedetomidine (DEX) is a kind of α2-adrenoceptor agonist, and has multiple effects of sedation, anti-anxiety, anti-sympathetic and stress reducing. DEX can be used in postoperative sedation and analgesia in patients with osteosarcoma. Recent studies by scholars have shown that DEX also has biological activities such as anti-inflammatory, anti-oxidant and anti-cancer biological activities. It can promote the survival of ovarian cancer model rats by enhancing its immunologic function, and can substantially improve the prognosis of patients undergoing gastric cancer surgery [6,7]. This study explored and analyzed the effect of DEX on biological behavior of osteosarcoma cells through the expression of miR-1307.
Materials and methods
Cell lines and reagents
The human osteosarcoma cells MG-63 were purchased from Representative Culture Preservation Committee of Chinese Academy of Sciences. We adopted DMEM cell culture medium containing 10% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin for culture. The culture was carried out under the condition of 5% CO2 at 37°C, the incubator humidity was kept at 95%, the medium was changed once every other day, and the subculture was carried out in 2-3 days.
The CCK8 kit was purchased from Jiangsu Biyuntian Biotechnology Research Institute; Australian Fetal Bovine Serum was purchased from Gbico, USA; Penicillin (100 U/mL)/streptomycin (100 g/ml) were purchased from Sigma Company, USA; DMEM culture medium was purchased from Gbico, USA; Annexin V-FITC/PI double-staining cell apoptosis detection kit was purchased from Shanghai Meiji Biotechnology Co., Ltd.; DEX was purchased from Nanjing Laifuse Biotechnology Co., Ltd.
Methods
Cell grouping and treatment
We inoculated the cells in a culture plate, randomly classified into control group, low-dose DEX group (25 ng/ml), medium-dose DEX group (50 ng/ml) and high-dose DEX group (100 ng/ml), and treated with corresponding concentration of DEX for 24 h.
CCK8 assay
We inoculated the cells in a 96-well plate, and treated with 0, 25, 50 and 100 ng/ml DEX for 0 h, 24 h, 48 h and 72 h, respectively after the cells adherent to the plate wall. The activity of cells in each group was detected in line with the CCK8 kit instructions. CCK8 solution of 20 μL was added and the cell were incubated at 37°C for 4 h. The OD value in each group at 490 nm was detected by a microplate reader.
Apoptosis detected by flow cytometry
The MG-63 cells in good growth condition in each group were selected for trypsinization (without EDTA). The cells were centrifuged at 250 × g for 5 min, washed twice with pre-cooled PBS × 10 min, and transferred to a flow cytometry tube. The cells were re-suspended with 100 μl × binding buffer, added with 5 μl Annexin V-FITC and PI and incubated for 10 min at room temperature. 400 μl 1 × binding buffer was then added to the cells and placed within 1 h to detect the cell apoptosis rate by flow cytometry. The experiment was repeated 3 times.
qRT PCR experiment
The total RNA was extracted by Trizol, and its concentration was detected by NanoDrop. We took 2 ng RNA for reverse transcription, and calculated the RNA volume required for reverse transcription according to RNA concentration. cDNA was synthesized according to the reverse transcription kit instructions, and the reaction system of qRT-PCR was configured with cDNA as the template. The reaction conditions of qRT-PCR were 95°C for 10 s, 95°C for 30 s, 60°C for 30 s and 72°C for 30 s (a total of 40 sets of circulation were completed), and the operation was carried out in accordance with the kit instructions. ABI 7500 real-time PCR was used for detection, and the relative expression of miR-1307 was calculated with 2-ΔΔCt method by regarding U6 as internal reference gene. The primer sequences of miR-1307 was the upstream primer 5’-aactcGGCGTGGC-3’ and downstream primer 5’-gagcagGCTGGAGAA-3’; The primer sequences of U6 was the upstream primer 5’-ATTGGAACGATACAGAGAAGATT-3’ and the downstream primer 5’-GGAACGCTTCACGAATTTG-3’.
Detection of cell invasion by transwell assay
Spread the Matrige on the filter membrane in advance, added 2 ml cell suspension to Transwell upper chamber, and processed with the above method for different groups. Added DMEM medium containing 10% fetal bovine serum to the lower chamber, and carefully removed the upper chamber of Transwell after 24 hours of incubation. Carefully removed the tumor cells in the upper chamber of the polycarbonate filter, fixed the tumor cells in the lower chamber of the polycarbonate filter with 950 ml/L ethanol, and stained with hematoxylin for around 10 minutes. Subsequently, we randomly selected 5 fields from the polycarbonate filter membrane under a light microscope, and calculated the number of cells in each field, and took the average value.
Detection of cell migration by scratch test
Adjusted the density of cells in logarithmic growth phase to 2 × 105 cells/ml. Added each well of the 6-well plate with 1 ml cell suspension. After the monolayer growth of the cells covered the bottom of the plate, a sterile pipette tip was used to carefully scratch the bottom of the plate, and the cell debris was gently washed away with PBS. After the above groups were cultured for 24 h, we observed the cells and photographed under an inverted microscope, and calculated its mobility. Three parallel holes were set in each group, and the experiment was repeated for 3 times.
Mobility (%) = (initial scratch width - scratch width of corresponding point)/initial scratch width × 100%.
Statistical analysis
The statistical analysis was conducted by software tool SPSS 23.0. The comparison between the two groups was conducted by t-test, the comparison between multiple groups was performed by one-way analysis of variance (ANOVA), and the experimental results were expressed as x̅ ± sd. P<0.05 indicated that the difference of data was statistically significant.
Results
Detection of cell proliferation by CCK8
The growth rate of osteosarcoma cells MG-63 slowed down with the increased concentration of DEX; and compared with the control group, the cellular absorbance in groups with different DEX dose decreased remarkably after 72 hours of culture (P<0.05), as illustrated in Figure 1.
Figure 1.
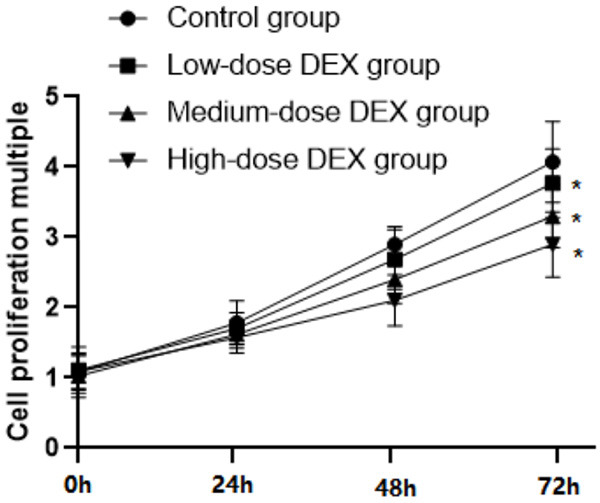
Comparison of each group’s OD value of at each time period. Note: Compared with the control group, *P<0.05.
Comparison of apoptosis rate
We stained the cells with Annexin V-FITC and PI, and subsequently analyzed apoptosis by flow cytometry. The proportion of apoptotic cells increased with the uplifting of DEX concentration, and the apoptotic rates in medium and high dosed DEX groups were remarkably higher than which in control group (P<0.05), as shown in Table 1 and Figure 2.
Table 1.
Comparison of apoptosis rate in each group (%, x̅ ± sd)
| Group | Apoptosis Rate | F | P |
|---|---|---|---|
| Control group | 8.17±2.54 | 30.974 | 0.000 |
| Low-dose DEX group | 9.10±1.89 | ||
| Medium-dose DEX group | 12.03±2.65* | ||
| High-dose DEX group | 14.11±3.10* |
P<0.05 compared with the control group.
Figure 2.
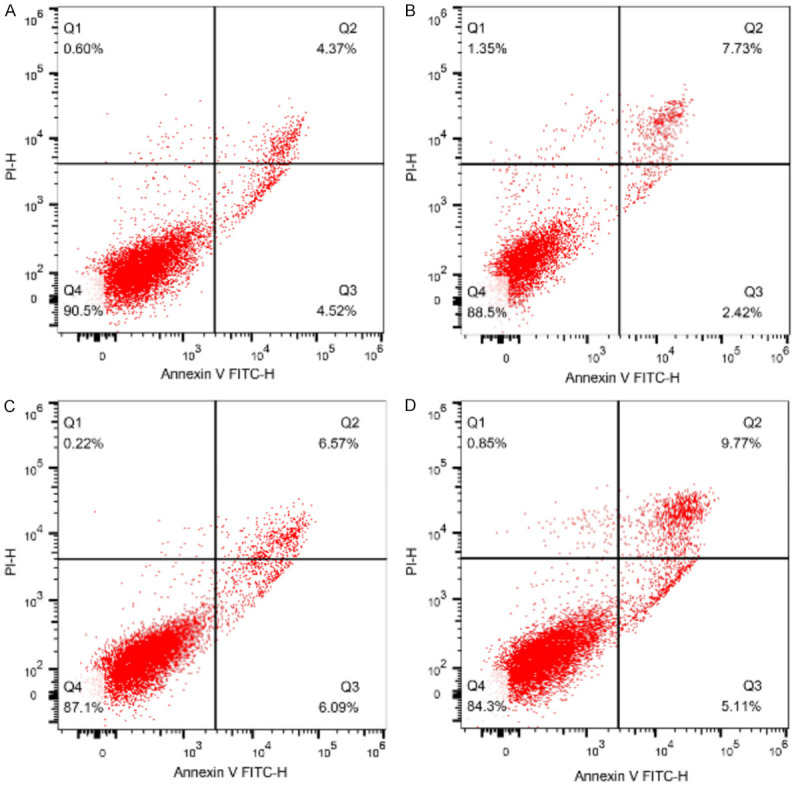
Comparison of apoptosis rate in each group. Note: A: Control group; B: Low-dose DEX group; C: Medium-dose DEX group; D: High-dose DEX group.
Detection of cell invasion by Transwell assay
Compared with the control group, the invasive ability of MG-63 cells decreased significantly after DEX treatment, and the number of invasive cells has decreased more obviously with the increase of DEX concentration (P<0.05), as shown in Table 2 and Figure 3.
Table 2.
Comparison of quantity of invasive cells in each group (number, x̅ ± sd)
| Group | Number of invasive cells per well | F | P |
|---|---|---|---|
| Control group | 216.48±25.94 | 25.931 | 0.000 |
| Low-dose DEX group | 189.64±20.28* | ||
| Medium-dose DEX group | 162.36±24.03*,# | ||
| High-dose DEX group | 143.41±23.49*,#,Δ |
P<0.05 compared with the control group;
P<0.05 compared with the low-dose DEX group;
P<0.05 compared with the medium-dose DEX group.
Figure 3.
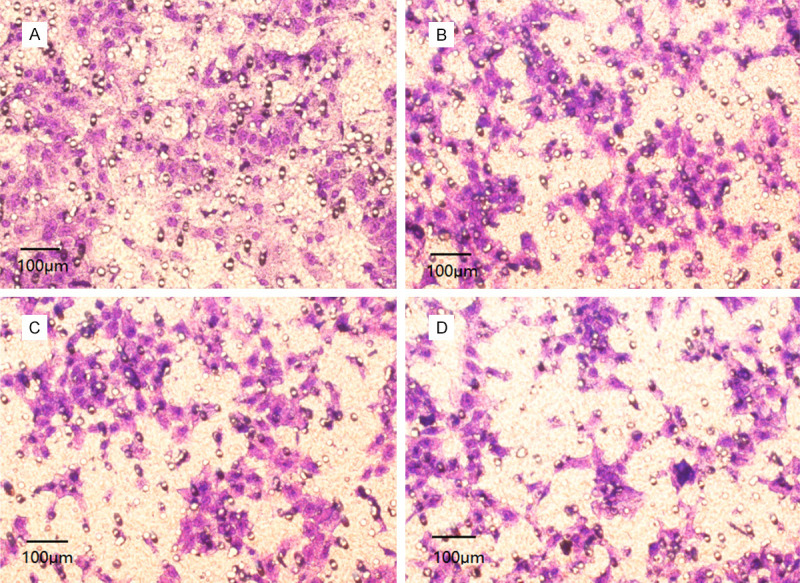
Comparison of quantity of invasive cells in each group. Note: A: Control group; B: Low-dose DEX group; C: Medium-dose DEX group; D: High-dose DEX group.
Cell migration detected by scratch test
Compared with the control group, the mobility rate of MG-63 cells after DEX treatment decreased significantly, and with the increase of DEX concentration, the cell mobility rate decreased more remarkably (P<0.05), as illustrated in Table 3 and Figure 4.
Table 3.
Comparison of cell mobility rate in each group (%, x̅ ± sd)
| Group | Cell mobility rate | F | P |
|---|---|---|---|
| Control group | 76.02±5.12 | 19.895 | 0.000 |
| Low-dose DEX group | 62.30±7.20* | ||
| Medium-dose DEX group | 56.33±6.11*,# | ||
| High-dose DEX group | 51.08±4.63*,#,Δ |
P<0.05 compared with the control group;
P<0.05 compared with the low-dose DEX group;
P<0.05 compared with the medium-dose DEX group.
Figure 4.
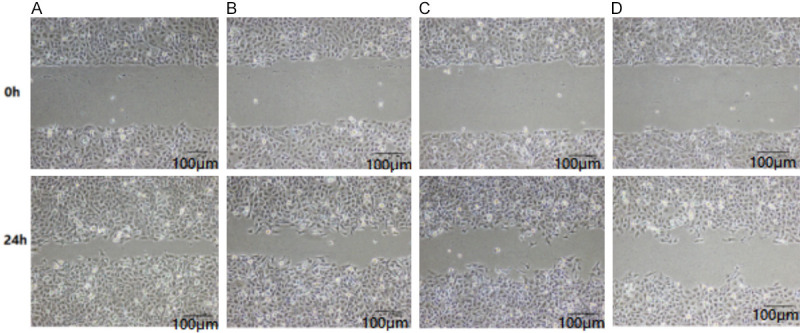
Comparison of cell migration rate in each group (scale: 100 μm). Note: A. Control group; B. Low-dose DEX group; C. Medium-dose DEX group; D. High-dose DEX group.
The relative expression of miR-1307 detected by qRT-PCR
The relative expression of miR-1307 in MG-63 cells after DEX treatment decreased dramatically comparing to the control group, and the decline was more noteworthy with the increase of DEX concentration (P<0.05), as shown in Table 4.
Table 4.
The relative expression levels of miR-1307 in each group (x̅ ± sd)
| Group | The relative expression of miR-1307 | F | P |
|---|---|---|---|
| Control group | 1.000±0.000 | 12.083 | 0.000 |
| Low-dose DEX group | 0.847±0.124* | ||
| Medium-dose DEX group | 0.622±0.170*,# | ||
| High-dose DEX group | 0.506±0.109*,#,Δ |
P<0.05 compared with the control group;
P<0.05 compared with the low-dose DEX group;
P<0.05 compared with the medium-dose DEX group.
Discussion
According to the latest reports, anesthetics such as propofol and morphine have impact on the development of malignant tumors [8]. DEX, as a novel receptor agonist, is highly selective for α2-adrenergic receptors in brain and spinal cord. Therefore, DEX can produce sedative, anti-anxiety and analgesic effects in the application of anesthetics without bringing respiratory depression [9-11]. Studies have shown that DEX can effectively inhibit the cell proliferation of glioma, lung carcinoma and breast cancer. Meanwhile, it can also inhibit the growth of ovarian cancer by inhibiting the p38MAPK/NF-κB signaling pathway [12-14]. Recent studies have also revealed that DEX can notably inhibit the proliferation, migration and invasion of breast tumor cell line MDA-MB-231 in a dose-dependent manner by activating the α2-adrenerin-receptor/ERK signaling pathway [15,16]. In addition, DEX can restrain the proliferation and migration of osteosarcoma cells and promote cellular apoptosis by up-regulating the expression degree of miR-520a-3p [17]. This study explored and analyzed the effect of DEX on the biological behavior of osteosarcoma cells via expression of miR-1307.
miRNA is tightly related to the biological behaviors of tumors, such as tumor occurrence and progression, metastasis and invasion. Current researches have shown that miR-1307 plays a crucial role in tumor progression and can be treated as a tumor marker to affect treatment and prognosis of tumor [18-20]. miR-1307, which is located on human chromosome 10, is a highly conserved miRNA with its function very limited to the public at present. Studies have suggested that miR-1307 may be related to chemotherapy resistance of tumors [21-23]. Meanwhile, there were reports indicating that the expression of miR-1307 in osteosarcoma cells increased dramatically than which in normal osteocytes, and such expression was closely connected to the proliferation, invasion, apoptosis, and other biological behaviors of osteosarcoma cells [24,25].
The results of this study showed that the growth rate of osteosarcoma cells MG-63 slowed slow down with the increase of DEX concentration, and compared with the control group, the cellular absorbance in groups with different DEX dose decreased remarkably after 72 hours of culture. The proportion of apoptotic cells increased along with the uplifting of DEX concentration, and the apoptotic rate in medium and high-dose DEX groups were remarkably higher than which in control group. Compared with the control group, the invasive ability and the mobility rate of MG-63 cells after DEX treatment decreased significantly, and the number of invasive cells and the rate of mobility decreased even more dramatically with the increase of DEX concentration. The above experiments explained that DEX has a certain anti-tumor effect and can effectively inhibit the proliferation, apoptosis, invasion and mobility of osteosarcoma cells in a dose-dependent manner, which was similar to the related research outcomes of scholars [26]. Besides, the relative expression of miR-1307 in MG-63 cells after DEX treatment decreased significantly comparing to the control group, and the decline was more obvious with the increase of DEX concentration. According to scholars’ studies [27], the relative expression of miR-1307 in osteosarcoma cells and tissues was significantly increased, and correlated with the biological behavior of osteosarcoma cells. DEX may affect such biological behavior of osteosarcoma cells by inhibiting the expression of the gene.
The results of this study were basically consistent with the reports of other scholars [28], that DEX may have a certain influence on the biological behaviors such as proliferation and invasion of osteosarcoma cells, and such influencing mechanism may be related to the regulation of miR-1307 expression in cells. While miR-1307 can regulate tumor cell proliferation, differentiation, and apoptosis through a variety of signaling pathways, its specific regulatory mechanism still under further research and analysis.
In summary, DEX can efficaciously inhibit the proliferation, invasion, metastasis, and apoptosis of osteosarcoma cells in a dose-dependent manner, and its effect may be connected to the regulation of miR-1307 expression.
Disclosure of conflict of interest
None.
References
- 1.Wang WT, Qi Q, Zhao P, Li CY, Yin XY, Yan RB. miR-590-3p is a novel microRNA which suppresses osteosarcoma progression by targeting SOX9. Biomed Pharmacother. 2018;107:1763–1769. doi: 10.1016/j.biopha.2018.06.124. [DOI] [PubMed] [Google Scholar]
- 2.He M, Shen P, Qiu C, Wang J. miR-627-3p inhibits osteosarcoma cell proliferation and metastasis by targeting PTN. Aging (Albany NY) 2019;11:5744–5756. doi: 10.18632/aging.102157. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Shekhar R, Priyanka P, Kumar P, Ghosh T, Khan MM, Nagarajan P, Saxena S. The microRNAs miR-449a and miR-424 suppress osteosarcoma by targeting cyclin A2 expression. J Biol Chem. 2019;294:4381–4400. doi: 10.1074/jbc.RA118.005778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao H, Yan P, Wang J, Zhang Y, Zhang M, Wang Z, Fu Q, Liang W. Clinical significance of tumor miR-21, miR-221, miR-143, and miR-106a as biomarkers in patients with osteosarcoma. Int J Biol Markers. 2019;34:184–193. doi: 10.1177/1724600819843537. [DOI] [PubMed] [Google Scholar]
- 5.Zhang W, Wei L, Sheng W, Kang B, Wang D, Zeng H. miR-1225-5p functions as a tumor suppressor in osteosarcoma by targeting Sox9. DNA Cell Biol. 2020;39:78–91. doi: 10.1089/dna.2019.5105. [DOI] [PubMed] [Google Scholar]
- 6.Li Q, Song S, Ni G, Li Y, Wang X. Serum miR-542-3p as a prognostic biomarker in osteosarcoma. Cancer Biomark. 2018;21:521–526. doi: 10.3233/CBM-170255. [DOI] [PubMed] [Google Scholar]
- 7.Li M, Shen Y, Wang Q, Zhou X. MiR-204-5p promotes apoptosis and inhibits migration of osteosarcoma via targeting EBF2. Biochimie. 2019;158:224–232. doi: 10.1016/j.biochi.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Jiang W, Liu J, Xu T, Yu X. MiR-329 suppresses osteosarcoma development by downregulating Rab10. FEBS Lett. 2016;590:2973–2981. doi: 10.1002/1873-3468.12337. [DOI] [PubMed] [Google Scholar]
- 9.Ji Q, Xu X, Song Q, Xu Y, Tai Y, Goodman SB, Bi W, Xu M, Jiao S, Maloney WJ, Wang Y. miR-223-3p inhibits human osteosarcoma metastasis and progression by directly targeting CDH6. Mol Ther. 2018;26:1299–1312. doi: 10.1016/j.ymthe.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu K, Huang J, Ni J, Song D, Ding M, Wang J, Huang X, Li W. MALAT1 promotes osteosarcoma development by regulation of HMGB1 via miR-142-3p and miR-129-5p. Cell Cycle. 2017;16:578–587. doi: 10.1080/15384101.2017.1288324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang DZ, Jing SF, Hao SB, Huang XY, Miao QT, Gao JF. MiR-218 promotes apoptosis of U2OS osteosarcoma cells through targeting BIRC5. Eur Rev Med Pharmacol Sci. 2018;22:6650–6657. doi: 10.26355/eurrev_201810_16140. [DOI] [PubMed] [Google Scholar]
- 12.Zhang RM, Tang T, Yu HM, Yao XD. LncRNA DLX6-AS1/miR-129-5p/DLK1 axis aggravates stemness of osteosarcoma through Wnt signaling. Biochem Biophys Res Commun. 2018;507:260–266. doi: 10.1016/j.bbrc.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 13.Wang K, Qi XJ, Liu HZ, Su H. MiR-361 inhibits osteosarcoma cell lines invasion and proliferation by targeting FKBP14. Eur Rev Med Pharmacol Sci. 2018;22:79–86. doi: 10.26355/eurrev_201801_14103. [DOI] [PubMed] [Google Scholar]
- 14.Chen H, Gao S, Cheng C. MiR-323a-3p suppressed the glycolysis of osteosarcoma via targeting LDHA. Hum Cell. 2018;31:300–309. doi: 10.1007/s13577-018-0215-0. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Zhang J, Xing C, Wei S, Guo N, Wang Y. miR-486 inhibited osteosarcoma cells invasion and epithelial-mesenchymal transition by targeting PIM1. Cancer Biomark. 2018;23:269–277. doi: 10.3233/CBM-181527. [DOI] [PubMed] [Google Scholar]
- 16.Wang SN, Luo S, Liu C, Piao Z, Gou W, Wang Y, Guan W, Li Q, Zou H, Yang ZZ, Wang D, Wang Y, Xu M, Jin H, Xu CX. miR-491 inhibits osteosarcoma lung metastasis and chemoresistance by targeting alphaB-crystallin. Mol Ther. 2017;25:2140–2149. doi: 10.1016/j.ymthe.2017.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, Ma W, Ma J, Xiao L, Hao D. Upregulation of miR-95-3p inhibits growth of osteosarcoma by targeting HDGF. Pathol Res Pract. 2019;215:152492. doi: 10.1016/j.prp.2019.152492. [DOI] [PubMed] [Google Scholar]
- 18.Shabani P, Izadpanah S, Aghebati-Maleki A, Baghbani E, Baghbanzadeh A, Fotouhi A, Bakhshinejad B, Aghebati-Maleki L, Baradaran B. Role of miR-142 in the pathogenesis of osteosarcoma and its potential as therapeutic approach. J Cell Biochem. 2019;120:4783–4793. doi: 10.1002/jcb.27857. [DOI] [PubMed] [Google Scholar]
- 19.Ren D, Zheng H, Fei S, Zhao JL. MALAT1 induces osteosarcoma progression by targeting miR-206/CDK9 axis. J Cell Physiol. 2018;234:950–957. doi: 10.1002/jcp.26923. [DOI] [PubMed] [Google Scholar]
- 20.Cao J, Han X, Qi X, Jin X, Li X. TUG1 promotes osteosarcoma tumorigenesis by upregulating EZH2 expression via miR-144-3p. Int J Oncol. 2017;51:1115–1123. doi: 10.3892/ijo.2017.4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong D, Zhang Z. NAIF1 suppresses osteosarcoma progression and is regulated by miR-128. Cell Biochem Funct. 2018;36:443–449. doi: 10.1002/cbf.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He M, Wang G, Jiang L, Qiu C, Li B, Wang J, Fu Y. miR-486 suppresses the development of osteosarcoma by regulating PKC-delta pathway. Int J Oncol. 2017;50:1590–1600. doi: 10.3892/ijo.2017.3928. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Wang X, Peng L, Gong X, Zhang X, Sun R, Du J. miR-423-5p inhibits osteosarcoma proliferation and invasion through directly targeting STMN1. Cell Physiol Biochem. 2018;50:2249–2259. doi: 10.1159/000495085. [DOI] [PubMed] [Google Scholar]
- 24.Lv T, Liu Y, Li Z, Huang R, Zhang Z, Li J. miR-503 is down-regulated in osteosarcoma and suppressed MG63 proliferation and invasion by targeting VEGFA/Rictor. Cancer Biomark. 2018;23:315–322. doi: 10.3233/CBM-170906. [DOI] [PubMed] [Google Scholar]
- 25.Shi YK, Guo YH. MiR-139-5p suppresses osteosarcoma cell growth and invasion through regulating DNMT1. Biochem Biophys Res Commun. 2018;503:459–466. doi: 10.1016/j.bbrc.2018.04.124. [DOI] [PubMed] [Google Scholar]
- 26.Fu D, Lu C, Qu X, Li P, Chen K, Shan L, Zhu X. LncRNA TTN-AS1 regulates osteosarcoma cell apoptosis and drug resistance via the miR-134-5p/MBTD1 axis. Aging (Albany NY) 2019;11:8374–8385. doi: 10.18632/aging.102325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Zeng X, Wang N, Zhao W, Zhang X, Teng S, Zhang Y, Lu Z. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. 2018;17:89. doi: 10.1186/s12943-018-0837-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujii R, Osaka E, Sato K, Tokuhashi Y. MiR-1 Suppresses proliferation of osteosarcoma cells by up-regulating p21 via PAX3. Cancer Genomics Proteomics. 2019;16:71–79. doi: 10.21873/cgp.20113. [DOI] [PMC free article] [PubMed] [Google Scholar]


