Abstract
The relevance of stem cell-derived exosomes has been implicated in necrotizing enterocolitis, while the involvement of serum-derived exosomes from children with Hirschsprung-associated enterocolitis (HAEC) in pathogenesis of HAEC remains unclear. This study set to identify the roles of exosomal microRNA (miR)-18a-5p from sera of HAEC patients in human-derived colonic epithelial NCM460 cells and in mice with HAEC. Exosomes were isolated from the sera of healthy children (Healthy-exo), patients with Hirschsprung’s disease (HSCR) (HSCR-exo) or HAEC (HAEC-exo). A microarray analysis of miRNAs was implemented to assess the enrichment of miRNAs in these exosomes. HAEC-exo was significantly enriched in miR-18a-5p. HAEC-exo led to the generation of a pro-inflammatory microenvironment, inhibition of cellular DNA synthesis, and promotion of apoptosis in NCM460 cells. Mechanistically, miR-18a-5p targeted and repressed retinoid-related orphan receptor α (RORA) expression, thereby regulating the Sirtuin 1 (SIRT1)/nuclear factor-kappa B (NFκB) pathway. Overexpression of RORA ameliorated inflammatory damage in NCM460 cells caused by exosomal miR-18a-5p. HAEC-exo exacerbated inflammatory damage in HAEC mice, and this facilitation was reversed after RORA overexpression. Collectively, exosomal miR-18a-5p was a promoter of HAEC, which induces the intestine cell apoptosis and inflammatory responses through the inhibition of SIRT1/NFκB pathway by targeting RORA.
Keywords: Hirschsprung-associated enterocolitis, miR-18a-5p, RORA, SIRT1/NFκB, inflammation
Introduction
Hirschsprung’s disease (HSCR), showing the characteristics of a lack of enteric (intrinsic) neurons from variable lengths of the most distal bowel, has an incidence of approximately 1:5,000 live births, with a 4:1 gender bias (male:female) [1]. Hirschsprung-associated enterocolitis (HAEC), the commonest and most severe complication of HSCR, happens despite the surgical methods for HSCR and may be fatal if timely and effective treatment is not in place [2]. Risk factors for HAEC development consist of Trisomy 21, long-segment disease, obstruction, and previous history of HAEC [3]. Since the cause of HAEC is largely unknown, treatment regimens are empiric and directed toward easing acute symptoms and managing the factors that may led to pathogenesis, including rectal irrigations and the use of antibiotics [4]. Therefore, much progress should be made in recognizing the causes of HAEC and developing effective treatment strategies.
Exosomes, extracellular vesicles (Evs) with a size range of ~40 to 160 nm in diameter with an endosomal origin, are associated with various diseases, and proteins, metabolites, and nucleic acids shuttled by exosomes into recipient cells powerfully alter their biological response, which can be disease-promoting or restraining [5]. After injury and the ensuing inflammation, immune cell- and stroma cell-derived Evs can change the production of cytokines in wounded mucosa to mediate cell migration, proliferation, as well as differentiation, making them either an inhibitor and a contributor of inflammation and tissue repair [6]. Moreover, serum EVs in a mouse model of colitis were discovered to promote inflammatory responses of gut macrophages and increased production of tumor necrosis factor alpha (TNF-α) [7]. Therefore, we postulated that the serum exosomes are associated with intestinal inflammation in HAEC. As we mentioned, exosomes modulate intercellular communication between different cell types by bearing proteins, lipids, and RNAs, thus altering normal and pathological conditions [8]. Interestingly, hsa-microRNA (miR)-18a-5p was significantly differentially expressed in the serum of patients with mild traumatic brain injury, which was correlated with pathways of inflammatory response, neurological disease, and cell development [9]. As a consequence, we assumed that miR-18a-5p, at least partially, is responsible for the pro-inflammatory effects of serum exosomes in HAEC. Ednrb mutant (Ednrb-/-) mouse is an acknowledged animal model of colorectal aganglionosis which displays several features of HSCR in human, including the progression of HAEC [10]. The aim of this work was to expound the functional role of exosomal miR-18a-5p from HAEC in Ednrb-/- mice, and how it can influence the progression of HAEC by mediation of inflammation and proliferation of colonic epithelial cells, which may offer a novel therapeutic strategy for HAEC.
Materials and methods
Clinical samples
An ethical approval was obtained from the Institutional Ethics Committee of Jiaxing Maternal and Child Health Hospital, and written informed consents were acquired from legal guardian(s) of participants. Thirty health children, 30 children with HSCR and 10 children with HAEC from June 2019 to June 2022 in Jiaxing Maternal and Child Health Hospital were randomly selected. Fresh blood samples were collected between 8 and 9 a.m. locally (at least 6 hours after fasting). After discarding the first 1 mL, blood samples were collected from the anterior elbow vein into 5 mL serum tubes by venipuncture with a 21-gauge needle. All test tubes were maintained upright for a 30 min clotting. The serum was centrifuged at 1500×g for 15 min at room temperature and left for one hour, and the supernatant was taken. All samples (1.5 mL each) were frozen in liquid nitrogen and then preserved in a -80°C refrigerator.
Isolation and characterization of exosomes
Exosomes were collected and extracted from physically examined healthy children, HSCR patients, and HAEC patients using ultracentrifugation (160,000×g, 16 h, 4°C) and named Health-exo, HSCR-exo, and HAEC-exo, respectively. The exosomes were resuspended using PBS, and their concentration was determined by BCA kits (Thermo Fisher Scientific Inc., Waltham, MA, USA).
The exosome dilution was added dropwise to a 2-mm copper grid and allowed to stand for 5 min at ambient temperature before excess liquid was gently aspirated off with filter paper. With the 3% (w/v) sodium phosphotungstate solution supplement, the samples were dried at room temperature and observed and photographed by TEM (Carl Zeiss, Oberkochen, Germany). Western blot experiments were applied to determine the expression patterns of the exosomal marker proteins CD63 and TSG101. The particle size distribution of exosome samples was assessed by a Nananosight NS300 particle size analyzer (NTA) equipped with a 450 nm laser.
Western blot analysis
Total proteins were isolated from the cells using RIPA (Beyotime Biotechnology Co., Ltd., Shanghai, China). Protein sample concentrations were assessed by the BCA Protein Assay Kit (Thermo Scientific). After electrophoresis in 15% SDS-polypropylene gels, the proteins were transferred to PVDF membranes. The membranes were sealed with 5% skim milk, cut according to the molecular weight of the pre-stained marker, probed with the primary antibody overnight on a shaker at 4°C, and reacted with horseradish peroxidase-labeled secondary antibody for 60 min at ambient temperature. Protein signals were visualized by BeyoECL Plus (Ultrasensitive ECL Chemiluminescence Kit, Beyotime), and GAPDH was utilized as a housekeeping gene to measure the relative expression of the protein. The antibodies used in the western blot are demonstrated in Table 1.
Table 1.
Antibodies used for western blot
| Primary antibodies | Dilution | Catalog No. | Manufacturer |
|---|---|---|---|
| CD63 | 1:1000 | ab134045 | Abcam |
| TSG101 | 1:1000 | ab125011 | Abcam |
| RORA | 1:500 | #TA803200 | Thermo Fisher Scientific |
| SIRT1 | 1:1000 | ab110304 | Abcam |
| NF-κB | 1:1000 | #8242 | Cell signaling technology |
| p-NF-κB p65 (Ser536) | 1:1000 | #3033 | Cell signaling technology |
| GAPDH | 1:1000 | #5174 | Cell signaling technology |
| Secondary antibody | Dilution | Catalog No. | Manufacturer |
| Goat anti-mouse IgG | 1:10000 | ab205719 | Abcam |
| Goats against-rabbit IgG | 1:10000 | ab205718 | Abcam |
Note: RORA, retinoid-related orphan receptor α; SIRT1, Sirtuin 1; NF-κB, nuclear factor-kappa B; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IgG, immunoglobulin G.
RT-qPCR
Total RNA was isolated from tissues or cells with the help of TRIzol (Invitrogen, Carlsbad, CA, USA). RNA was isolated from exosomes with the help of a SeraMir Exosome RNA Purification Kit (System Biosciences, Palo Alto, CA, USA). The extracted RNA was reversely transcribed to cDNA (miRNA/total) with the help of the Prime Script miRNA cDNA Synthesis Kit (Qiagen company, Hilden, Germany) or PrimeScript RT Kit (Qiagen), followed by RT-qPCR. qPCR was performed on a Q6 real-time PCR System (Applied Biosystems, Carlsbad, CA, USA) with the help of SYBR Green Master Mix (4309155, Applied Biosystems) or the miScript PCR Kit (Qiagen). The relative expression was normalized to GAPDH or U6, using the 2-ΔΔCt method. The primers are listed in Table 2.
Table 2.
Primer used for RT-qPCR
| Gene | Forward primer (5’-3’) | Reverse primer (5’-3’) |
|---|---|---|
| Human | ||
| miR-18a-5p | AGGTGCATCTAGTGCAG | GAACATGTCTGCGTATCTC |
| RORA | CACCAGCATCAGGCTTCTTTCC | GTATTGGCAGGTTTCCAGATGCG |
| ATM | TGTTCCAGGACACGAAGGGAGA | CAGGGTTCTCAGCACTATGGGA |
| HIF1A | TATGAGCCAGAAGAACTTTTAGGC | CACCTCTTTTGGCAAGCATCCTG |
| SMAD2 | GGGTTTTGAAGCCGTCTATCAGC | CCAACCACTGTAGAGGTCCATTC |
| Mouse | ||
| miR-18a-5p | AAGGTGCATCTAGTGCAGA | GAACATGTCTGCGTATCTC |
| RORA | CAGAGCAATGCCACCTACTCCT | CTGCTTCTTGGACATCCGACCA |
| Internal reference | ||
| GAPDH | GTCTCCTCTGACTTCAACAGCG | ACCACCCTGTTGCTGTAGCCAA |
| U6 | CGCGCTTCGGCA GCACATATACT | ACGCTTCACGAATTTGCGTGTC |
Note: miR, microRNA; RORA, retinoid-related orphan receptor α; ATM, ataxia-telangiectasia-mutated; HIF1A, hypoxia-inducible factor-1α.
miRNA microarray
Total miRNA of exosomes was linearly amplified using the Global miRNA Amplification Kit (System Bioscience), followed by labeling using the miRCURY™ Hy3/Hy5 Power Labeling Kit (Exiqon, Vedbaek, Denmark). Hybri-dization was performed on the miRCURY™ LNA miRNA Array (Exiqon). An Axon GenePix 4000B microarray scanner (Axon Instruments, Foster City, CA, USA) was then utilized for scanning. Grid alignment and data extraction were then implemented using the genepix pro software (version 6.0, Axon Instruments). In at least one exosome sample, miRNAs with a foreground signal intensity two times higher than the background signal were considered to be expressed, and ten times higher than the background signal were considered to be enriched miRNAs.
Cell culture and treatment
The colonic epithelial cell line NCM460 (ATCC, Manassas, VA, USA) were grown in DMEM containing 10% FBS, 100 U/mL penicillin and 100 µg/mL streptomycin and maintained in a 37°C incubator with 5% CO2.
Cells were co-cultured with HAEC-exo at different concentrations (5, 10, 15, 20 µg/mL), while the control cells were treated with an equal volume of PBS. The miR-18a-5p mimic, RORA overexpression plasmid (oe-RORA) and their respective controls used for cell transfection were purchased from VectorBuilder (Guangzhou, Guangdong, China). Lipofectamine 3000 (Invitrogen) was utilized for all transfections following the manufacturer’s protocol.
Exosome internalization
Exosomes were labeled with PKH26 (Sigma-Aldrich Chemical Company, St Louis, MO, USA). Briefly, exosomes were resuspended in diluent C (Sigma) and incubated with PKH26 at 37°C for 5 min. Excess staining solution was discarded by the centrifugation. The PKH26-labeled exosomes were co-cultured with NCM460 in medium free of FBS for 24 h. Nuclei were stained with DAPI (Sigma) and viewed under a ZEISS inverted fluorescence microscope (ZEISS).
Methylthiazolyldiphenyl-tetrazolium bromide (MTT) assay
The effect of exosomes at different concentrations on the viability of NCM460 was examined using the MTT kit (Thermo Fisher). The exosome-treated cells were plated into 96-well plates (5000 cells per well). After the addition of 10 µL MTT solution, the incubation continued for 4 h. The cells were incubated with 100 µL Formazan lysis buffer for another 3-4 h or so until all the formazan purple crystals were found to be dissolved when observed under an ordinary light microscope. The optical density at 570 nm was read with a spectrophotometer (Shanghai Metash Instruments Co., Ltd., Shanghai, China).
ELISA
The levels of pro-inflammatory factors in the supernatant of NCM460 cells were examined by Human ELISA Kits for IL-6 (ab178013) and TNF-α (ab181421). The levels of pro-inflammatory factors in mouse serum were assessed using mouse ELISA Kits for IL-6 (ab100712) and TNF-α (ab208348) as per the manufacturer’s instructions, and the concentrations of relevant factors were measured according to the plotted standard curves.
EdU assay
EdU kits (RiboBio, Guangzhou, Guangdong, China) were utilized to evaluate the DNA synthesis of cells. The transfected NCM460 were cultured with 50 μmol/L EdU for 120 min at 37°C, fixed with 4% paraformaldehyde for 0.5 h and stained with 1× Apollo reaction cocktail for 0.5 h. After a 30-min reaction with 100 μL Hoechst 33342 (5 μg/mL), a fluorescent microscope (ZEISS) was utilizd to examine the percentage of EdU-positive cells.
Immunofluorescence staining
After a 10-min ice-bath with 2% paraformaldehyde, a 15-min permeabilization with 0.1% Triton ×100 (Thermo Fischer Scientific) in 1× PBS, and a 1 h sealing at ambient temperature with 1% BSA (Gibco, Carlsbad, CA, USA), the cells were incubated overnight at 4°C with the primary antibody to Mucin 1 (MUC1, 1:1000, ab109185, Abcam). Afterwards, cells were incubated with secondary goat anti-rabbit antibody against IgG H&L (Alexa Fluor® 594, 1:200, ab150080, Abcam) for 1 h. The slides were mounted using a VECTASHIELD® containing DAPI aqueous mounting medium (Vector Laboratories, Inc., Burlingame, CA, USA) and viewed by the fluorescent microscope (ZEISS). The Image J 1.48 version software (NIH, Bethesda, MD, USA) was utilized for quantification (at least 200 cells from 5 images).
Luciferase reporter assay
Potential binding sites for RORA to miR-18a-5p were downloaded from Starbase (http://starbase.sysu.edu.cn/), and corresponding mutation binding sites were designed. RORA 3’UTR containing these sites was sub-cloned into the pmirGLO vectors (Promega, Madison, WI, USA). The NCM460 cells were co-transfected with the luciferase reporter plasmid, pRL-TK Renilla plasmid and NC mimic or miR-18a-5p with the help of Lipofectamine 3000 (Invitrogen). Following 2 d, a dual-luciferase assay system (Promega) was utilized to assess the luciferase activity, and Renilla luciferase activity was used as a loading control.
RNA immunoprecipitation (RIP)
Magna RIP Kit (Millipore, Billerica, MA, USA) was utilized as per the manufacturer’s protocol. The RNA magnetic beads were conjugated to antibodies to Argonaute2 (AGO2) (Millipore) or isotype control antibody anti-IgG (Millipore). Then, the relative expression of miR-18a-5p and RORA was measured using RT-qPCR.
Experimental animals and sample collection
Ednrb-/- mice (Ednrbtm1Ywa/J on a hybrid C57BL/6J-129Sv background; 003295) were from Jackson Laboratory (Sacramento, CA, USA). Approval for this study was received from the Animal Care and Use Committee in Jiaxing Maternal and Child Health Hospital. The Chinese National Institute of Health Guide for the Care and Use of Laboratory Animals was followed to implement all animal experimental procedures. Randomly selected 2-week-old Ednrb-/- homozygote mice were used for the experiment, and wild-type littermates (Ednrb+/+) served as normal controls. The mice were randomly assigned into four groups (6 mice in each, Table 3). Lentivirus LV-RORA was from GenePharma (Shanghai, China) with a virus titer of 1×108 TU/mL. Mice were euthanized the day after the last injection, and blood and colonic tissue were collected.
Table 3.
Animal grouping
| Group | Number | Description |
|---|---|---|
| Control | 6 | Wild-type littermates (Ednrb+/+) |
| Model | 6 | Ednrb-/- homozygote mice without any other treatment |
| HAEC-exo | 6 | Ednrb-/- homozygote mice were injected with 20 µg HAEC-exo via tail vein every 3 days for 3 times |
| HAEC-exo + LV-RORA | 6 | Ednrb-/- homozygote mice were injected with 20 µg HAEC-exo along with 20 µL RORA overexpressing lentivirus LV-RORA via tail vein for 3 times |
Note: HAEC, Hirschsprung-associated enterocolitis; RORA, retinoid-related orphan receptor α.
Evaluation of bacterial counts in colonic tissues
Collected colonic tissues were homogenized with sterile saline. Following centrifugation at 13,000 g for 10 min at 4°C, supernatant of the tissue lysate (100 μL) containing 100 μg colonic tissue extract was spread on blood agar plates in duplicate and cultured at 37°C for 2 d. Bacterial counts were expressed as the mean number of bacterial colony-forming units in one gram of colonic tissues (wet weight).
Immunohistochemical staining
Colonic tissues were fixed in 4% paraformaldehyde, paraffin-embedded, and cut. The 5-μm-thick sections were dewaxed using xylene and hydrated with alcohol. Antigen repair was then carried out for 0.5 h. Next, sections were reacted with 3% H2O2 to eliminate endogenous peroxidase and reacted with primary antibody antibodies against MUC1 (1:100, ab15481, Abcam) and p-NFκB (S536) (1:50, ab28856, Abcam) overnight at 4°C. The sections were the hybridized with the secondary antibody to IgG (1:1000, ab205718, Abcam) at 37°C for 60 min, visualized with diaminobenzidine (56990, Abcam), counter-stained using hematoxylin, and dehydrated in alcohol and xylene. Finally, the sections were fixed using neutral gum and viewed under a microscope. Five fields of view were randomly chosen by Image J software to determine the positive rate of genes.
Histopathological evaluation
The 5-μm-thick colonic samples were sectioned and stained with hematoxylin-eosin (HE) (Sigma). Based on a previous report [11], histopathological scores were evaluated by two pathologists who were unaware of grouping on the basis of the degree of inflammation, neutrophilic and lymphoid tissue infiltration, crypt damage, crypt abscess formation, submucosal edema, loss of goblet cells, and reactive epithelial hyperplasia. Each item was scored from 0 to 4 according to the severity of pathological damage from the lowest to the highest, and the final score was the sum of all scores (0 to 28 points).
Data analysis
GraphPad Prism 8.0 (GraphPad, San Diego, CA, USA) was utilized for processing statistical analyses, and measurement data were exhibited as SD. Data between two groups were compared with unpaired t-test, while data among multiple groups were compared using one-way or two-way ANOVA, along with Tukey’s post-hoc tests. A value of P < 0.05 was indicative of a statistical significance.
Results
Exosome characterization
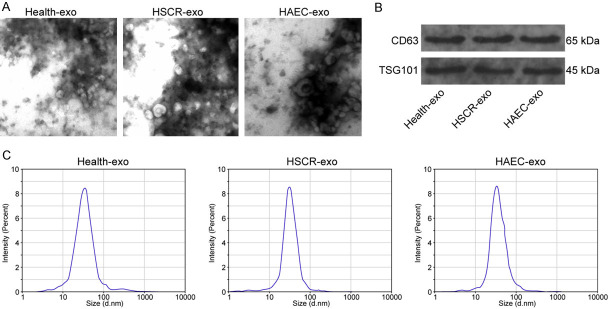
Health-exo, HSCR-exo, and HAEC-exo were isolated from the sera of healthy children, children with HSCR or HAEC. The structure of exosomes was observed by TEM, and all three groups of exosomes exhibited a round or oval structure (Figure 1A). Western blot detection of exosomal marker proteins showed that all three groups of exosomes expressed CD63 and TSG101 to different degrees (Figure 1B). By analyzing the exosome diameters using NTA, we observed that the diameters of the three groups of exosomes were around 100 nm (Figure 1C).
Figure 1.
Exosomes were extracted from the serum of healthy children and children with HSCR or HAEC. A. Exosomes analyzed by TEM. B. Exosomal markers CD63 and TSG101 determined by western blot. C. The exosome size distribution analyzed by NTA.
miR-18a-5p is significantly elevated in HAEC-exo
Serum exosomes from five healthy children, five children with HSCR, and five children with HAEC were randomly selected for exosomal miRNA microarray analysis. A total of eight significantly differentially expressed miRNAs were identified (Figure 2A) by setting the threshold of |log2 Fold change| > 2 for each two groups. miR-18a-5p is the miRNA with the most significant foldchange. Moreover, a notable increase in miR-18a-5p expression in HSCR-exo and HAEC-exo was observed compared with Health-exo, with a particularly significant increase in HAEC-exo. The miR-18a-5p expression in the remaining exosome samples was detected by RT-qPCR, and we again observed the same results (Figure 2B).
Figure 2.
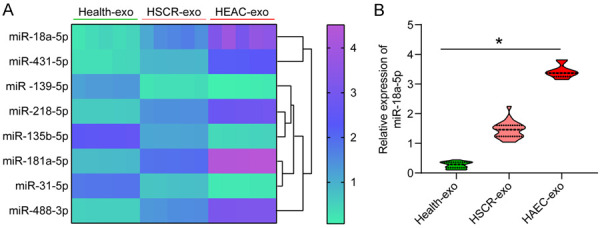
The aberrantly expressed miRNAs among the Healthy-exo, HSCR-exo and HAEC-exo. A. Enrichment of miRNAs in exosomes from different sources (|log2 Fold change| > 2 between each two groups). B. miR-18a-5p expression in exosomes of different origins determined by RT-qPCR. The data are exhibited as mean ± SD. The significant differences among the groups were compared by one-way ANOVA (Tukey’s test). *P < 0.05. Data are representative of 3 independent experiments.
HAEC-exo treatment promotes the formation of a pro-inflammatory microenvironment in colonic epithelial cells
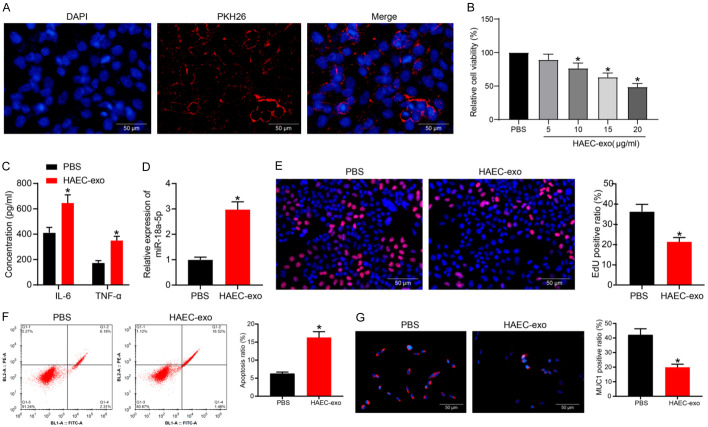
To examine the effect of HAEC-exo on the colonic epithelial cell line, we first co-cultured NCM460 cells with PKH26-labeled HAEC-exo for 24 h. A significant uptake of HAEC-exo by NCM460 was noted (Figure 3A). We then treated the cells with different concentrations of HAEC-exo and detected cell viability changes by MTT (Figure 3B). HAEC-exo inhibited the proliferation of NCM460 cells in a concentration-dependent fashion. By the time the concentration of HAEC-exo reached 20 μg/mL, the viability of the cells had been reduced by more than half. Therefore, we chose the concentration of 20 µg/mL HAEC-exo for the subsequent experiments.
Figure 3.
HAEC-exo exacerbate the inflammation and NCM460 cell apoptosis. (A) The uptake of HAEC-exo by NCM460 cells. (B) Cell viability examined by MTT assay. (C) The concentration of pro-inflammatory factors in cell supernatants assessed by ELISA assay. (D) The miR-18a-5p expression in cells by RT-qPCR. (E) Cellular DNA synthesis activity determined by EdU staining. (F) Apoptosis rate determined by flow cytometry analysis. (G) MUC1 expression in cells examined by immunofluorescence staining. The data are exhibited as mean ± SD. Significant differences between two groups were compared using an unpaired t-test (D-G). The significant differences among the groups were compared by one-way (B) or two-way (C) ANOVA, followed by Tukey’s test. *P < 0.05. Data are representative of 3 independent experiments.
The supernatant of exosome-treated NCM460 cell culture medium was harvested, and ELISA was implemented to detect the release of IL-6 and TNF-α (Figure 3C). HAEC-exo treatment significantly increased the IL-6 and TNF-α levels in the supernatant, indicating the formation of a pro-inflammatory microenvironment. By RT-qPCR, we detected a significant elevation of intracellular miR-18a-5p expression (Figure 3D). DNA synthesis in the cells was assessed by EdU staining, and we observed a remarkable decline in the rate of EdU-positive cells after HAEC-exo treatment (Figure 3E). Flow cytometry results showed that exosome treatment also promoted apoptosis (Figure 3F). We then detected the expression of MUC1 in cells by immunofluorescence. HAEC-exo treatment significantly inhibited the expression of MUC1 as well (Figure 3G).
miR-18a-5p binds to and negatively modulates RORA
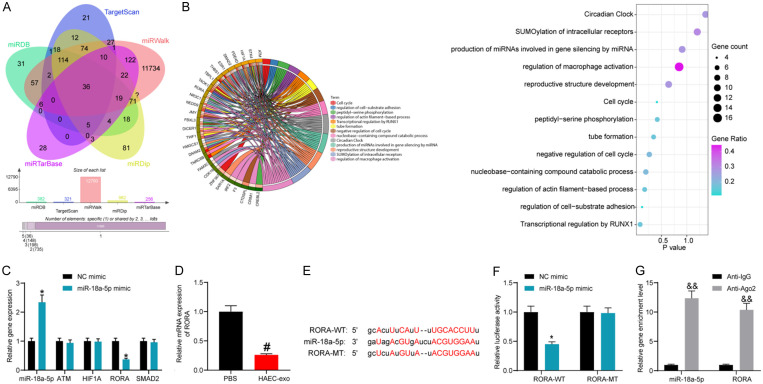
To investigate the downstream mechanism of miR-18a-5p, we predicted the downstream target genes of hsa-miR-18a-5p in miRDB (http://mirdb.org/), TargetScan (http://www.targetscan.org/vert_72/), miRWalk (http://mirwalk.umm.uni-heidelberg.de/), miRDip (http://ophid.utoronto.ca/mirDIP/), and miRTarBase (http://mirtarbase.cuhk.edu.cn/php/search.php), with a total of 36 intersections (Figure 4A). Pathway enrichment analysis was performed for these potential target genes (Figure 4B). Among the five pathways with the highest enrichment, there were four common potential target genes: ATM, HIF1A, RORA, and SMAD2. We transfected miR-18a-5p mimic and its control into NCM460 cells and measured the expression patterns of miR-18a-5p and these target genes by RT-qPCR. miR-18a-5p mimic successfully augmented the miR-18a-5p levels in cells and repressed the expression of RORA, without significant effects on the expression of other genes (Figure 4C). The expression of RORA in HAEC-exo-treated NCM460 cells was detected by RT-qPCR, and a significant reduction in its expression was also observed (Figure 4D).
Figure 4.
RORA is a potential target of miR-18a-5p. (A) Potential downstream targets of miR-18a-5p. (B) Pathway enrichment analysis of target genes. (C) The expression profiles of miR-18a-5p and its target genes in response to miR-18a-5p mimic examined by RT-qPCR. (D) RORA expression in response to HAEC-exo or PBS treatment by RT-qPCR. (E) Potential binding sites of RORA to miR-18a-5p and its mutation sites. (F) The effect of miR-18a-5p mimic on RORA-WT/RORA-MT luciferase activity determined by dual-luciferase assays. (G) The binding ability of miR-18a-5p to RORA mRNA examined by RIP assay. The data are exhibited as mean ± SD. Significant differences between two groups were assessed using an unpaired t-test (D-G). The significant differences among the groups were compared by two-way (C, F and G) ANOVA, followed by Tukey’s test. *, #P < 0.05, &&P < 0.01. Data are representative of 3 independent experiments.
Potential binding sites for RORA to miR-18a-5p were downloaded in StarBase, and mutant binding sites were designed (Figure 4E). miR-18a-5p mimic significantly repressed the luciferase activity of RORA using the dual-luciferase assay (Figure 4F). In the RIP experiment, anti-Ago2 significantly enriched miR-18a-5p and RORA, again demonstrating the targeting relationship between miR-18a-5p and RORA mRNA (Figure 4G).
Overexpression of RORA rescues the damaging effects of HAEC-exo on cells by mediating the SIRT1/NFκB pathway
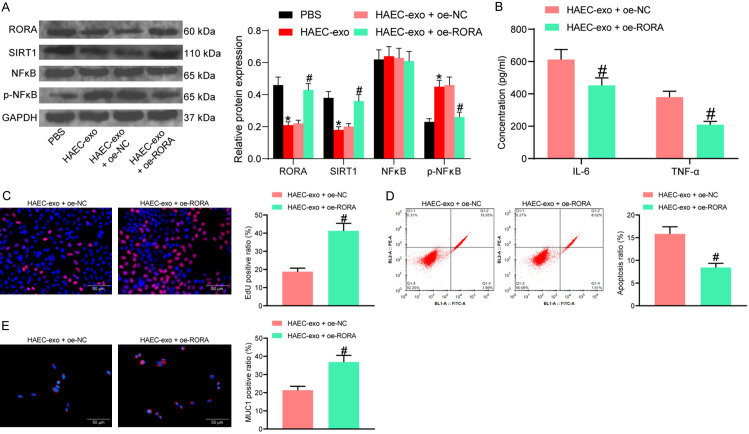
To explore whether exosomal miR-18a-5p from HAEC serum exerts pro-inflammatory effects by targeting RORA, we transfected the RORA overexpression plasmid oe-RORA into HAEC-exo-treated NCM460 cells. According to a previous report [12], RORA can regulate inflammatory damage by mediating the SIRT1/NFκB axis. We, therefore, examined the effects of HAEC-exo treatment and transfection with oe-RORA on protein expression of RORA, SIRT1, and NFκB as well as the extent of NFκB phosphorylation in cells by western blot experiments. We observed that HAEC-exo drastically inhibited the expression of RORA and SIRT1, while promoting the extent of NFκB phosphorylation. The expression of RORA and SIRT1 was significantly restored after transfection with oe-RORA, while the phosphorylation level of NFκB was significantly reduced (Figure 5A).
Figure 5.
RORA restoration represses intestinal epithelial cell apoptosis and inflammation. (A) RORA, SIRT1 protein expression and NFκB phosphorylation examined by western blot. (B) The concentration of pro-inflammatory factors in cell supernatants assessed by ELISA assay. (C) Cellular DNA synthesis activity determined by EdU staining. (D) Apoptosis rate determined by flow cytometry analysis. (E) MUC1 expression in cells examined by immunofluorescence staining. The data are exhibited as mean ± SD. Significant differences between two groups were assessed using an unpaired t-test (C-E). The significant differences among the groups were compared by two-way (A and B) analysis of variance, followed by Tukey’s test. *, #P < 0.05. Data are representative of 3 independent experiments.
Next, we examined whether overexpression of RORA affects the pro-inflammatory effect of HAEC-exo on NCM460 cells. By detecting the release of pro-inflammatory factors in the cell culture medium supernatant with ELISA assay, we observed that overexpression of RORA significantly reduced the concentration of IL-6 and TNF-α (Figure 5B). EdU staining revealed that overexpression of RORA rescued the inhibitory effect of HAEC-exo on cellular DNA synthesis (Figure 5C). Flow cytometry results exhibited that overexpression of RORA inhibited apoptosis caused by HAEC-exo (Figure 5D). Immunofluorescence staining, consistently, exhibited that RORA overexpression stimulated the MUC1 expression in the cells (Figure 5E).
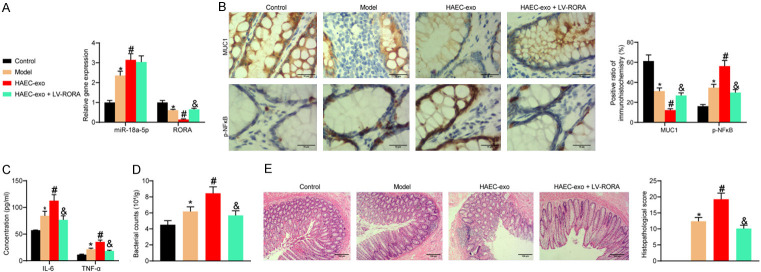
HAEC-exo exacerbates inflammatory damage in HAEC mice
Colonic tissues from mice were harvested, and the miR-18a-5p and RORA mRNA expression was detected by RT-qPCR assay. miR-18a-5p expression was much higher and RORA expression was lower in the colonic tissue of the HAEC mice than the control mice, and HAEC-exo treatment exacerbated the expression difference of the two genes. LV-RORA treatment drastically elevated the expression of RORA in tissues but had no notable effect on the miR-18a-5p expression (Figure 6A). Immunohistochemical assay revealed that MUC1 expression was significantly lower and that of p-NFκB was significantly higher in the colonic tissues of the HAEC mice compared with the control mice. HAEC-exo treatment further suppressed MUC1 expression and promoted p-NFκB expression. By contrast, MUC1 expression was significantly higher and p-NFκB expression was significantly lower after RORA overexpression (Figure 6B).
Figure 6.
HAEC-exo exacerbates the inflammatory responses and apoptosis in HEAC mice. (A) miR-18a-5p and RORA expression in the colonic tissues of mice by RT-qPCR. (B) Expression of MUC1 and p-NFκB in the colonic tissue of mice examined by immunohistochemistry. (C) The concentration of pro-inflammatory factors in the serum of mice assessed by ELISA assay. (D) Bacterial counts in colonic tissue of mice. (E) Pathological changes in colonic tissue examined by HE staining. There are 6 mice in each group, and the images shown are representative ones. The significant differences among the groups were compared by one-way (D and E) or two-way (A-C) ANOVA, followed by Tukey’s test. *, #, &P < 0.05.
The serum from each group of mice was collected for ELISA assay. The levels of IL-6 and TNF-α were much higher in the HAEC mice versus the control mice. HAEC-exo exacerbated the inflammatory response in the serum of mice, while LV-RORA significantly alleviated the release of pro-inflammatory factor by HAEC-exo (Figure 6C). Bacterial counts were then measured on colonic tissues of mice. Colonic tissue in the HAEC mice exhibited significant bacterial invasion compared to the control mice. Treatment with HAEC-exo accelerated bacterial invasion of colonic tissue, while overexpression of RORA significantly reduced bacterial invasion (Figure 6D). HE staining showed that mice with HAEC exhibited pathological changes, such as intestinal structural disruption, crypt loss and inflammatory cell infiltration, and HAEC-exo significantly exacerbated the pathological changes in the colon of mice. Still, overexpression of RORA significantly reduced the damaging effects of HAEC-exo on colonic tissues (Figure 6E).
Discussion
Conditions of premature babies that necessitate medical treatment frequently affect the central nervous system, the cardiorespiratory system, and the gastrointestinal tract, among which HAEC represents a common complication of HSCR with substantial morbidity and mortality, and its etiology and pathophysiology remain to be elucidated [13]. Therefore, more effective therapeutic interventions are called for, among which Evs are garnering great attention since they can regulate genes in target cells through the delivery of RNA species, such as miRNAs [14]. The current study set to probe the possible involvement of exosomal miR-18a-5p/RORA/SIRT1/NFκB signaling in HAEC. Our findings suggested that exosomal miR-18a-5p downregulated RORA, and consequently activated the SIRT1/NFκB signaling pathway, thereby encouraging the inflammatory response in HEAC mice.
Previously, McCulloh et al. reported that stem cell-derived exosomes reduced the incidence and severity of necrotizing enterocolitis as effectively as the stem cells, supporting the potential for exosome therapy for necrotizing enterocolitis [15]. The function of exosome cargoes is strongly influenced by the exosome origins, and exosomes derived from different sources exert enormously different effects on recipient cells [16]. Our in vitro evidence provided that HAEC-exo remarkably enhanced the inflammation and NCM460 apoptosis. The release of TNF-α is the initial step for inflammatory responses, which facilitates immune cell recruitment, while IL-6 levels are promoted as a consequence of secondary secretion in response to TNF-α and IL-1β [17]. Meanwhile, the mechanistical studies underlying HAEC-exo using miRNA-based microarray analysis found that miR-18a-5p was the most drastically enriched miRNA in the serum-derived exosomes of patients with HAEC. In addition, Cho et al. reported that miR-18a-5p was overexpressed in exosomes derived from hepatocellular carcinoma cells and also highly linked to prognosis of hepatocellular carcinoma [18]. Five serum miRNAs, including miR-25, miR-92a, miR-133a, miR-218-1, and miR-483-5p have been identified to be associated with HSCR development, signifying potential non-invasive diagnostic options for HSCR screening [19]. Nevertheless, the involvement of serum miRNAs in HAEC is largely unknown. Still, the upregulation of miR-18a-5p has been indicated in the serum of women with endometriosis and patients with colorectal cancer [20,21]. Besides, suppression of the miR-17~92 cluster has been documented to repress the podocyte apoptosis, inflammation, and fibrosis induced by high-glucose culture [22]. Since miR-18a is a part of that family, the possible interaction between miR-18a-5p and inflammation is thus convincible.
Furthermore, another key finding of the current study revealed the ability of miR-18a-5p to induce the SIRT1/NFκB signaling pathway via RORA suppression. RORA, located at 15q22.2, has four domains: N-terminal (A/B) domain, zinc finger DNA-binding domain, hinge domain, and a C-terminal ligand-binding domain [23]. RORA has the capacity to halt the inflammation and ease inflammation injuries, therefore playing a fundamental role in the development of diabetes, asthma, and allergic rhinitis [24-26]. Likewise, our rescue experiments provided that overexpression of RORA curbed the cell apoptosis and inflammation in the presence of HAEC-exo. Under the context of periodontitis, the pro-inflammatory and pro-apoptotic effects of miR-498 overexpression on human periodontal ligament cells were weakened by RORA overexpression [27]. As mentioned above, RORA has been indicated to protect macrophages against lipopolysaccharide-induced inflammation by impairing the SIRT1/NFκB pathway [12]. Therefore, we next tested whether this pathway was also involved in the exosomal miR-18a-5p/RORA axis-mediated inflammation. Western blot results substantiated their involvement. Quiet in line with our findings, the intestine epithelial cells were also under the protection of porcine milk-derived exosomes against injury evoked by lipopolysaccharide through the action of exosomal miRNAs and the TLR4/NFκB pathway [28]. On top of that, lipopolysaccharide upregulated miR-132/212 expression in HAEC, facilitating pyroptosis by targeting SIRT1 [29]. On the basis of these evidences, we are reasonable to conclude that exosomal miR-18a-5p enhanced the inflammatory responses and colonic epithelial cell apoptosis in HAEC via RORA-dependent SIRT1/NFκB signaling pathway activation.
Conclusion
In conclusion, the current study identified RORA as a putative target of miR-18a-5p in colonic epithelial cells, and further demonstrated the role of the exosomal miR-18a-5p/RORA regulatory axis in mediating inflammatory signals and colonic epithelial cell apoptosis via the SIRT1/NFκB signaling (Figure 7). Our findings on the pro-inflammatory role of exosomal miR-18a-5p from patients’ serum provide fresh insights into the mechanism of HAEC and offers potential targets for applications. Nonetheless, additional studies are necessary to corroborate these findings and to expand the translational potential of this direction. A potential pitfall of this work may be the application of single dose of HAEC-exo in vivo, which might affect the clinical translation.
Figure 7.
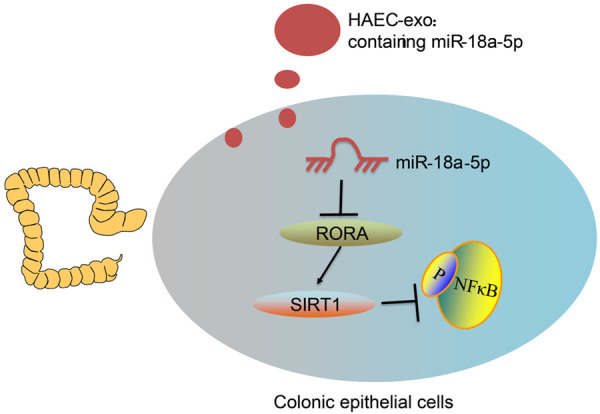
A diagram for the molecular mechanism. Serum-derived exosomes from children with HAEC exacerbate inflammatory damage in HAEC by delivering miR-18a-5p to inhibit RORA expression in colonic epithelial cells, thereby suppressing SIRT1 expression and promoting NFκB phosphorylation.
Acknowledgements
The authors would like to thank 2019 Zhejiang Medical and Health Science and Technology Plan Project (2019KY220) and 2020 Zhejiang Medical and Health Science and Technology Plan Project (2020KY960). This work was supported by 2019 Zhejiang Medical and Health Science and Technology Plan Project (2019KY220) and 2020 Zhejiang Medical and Health Science and Technology Plan Project (2020KY960).
Disclosure of conflict of interest
None.
References
- 1.McKeown SJ, Stamp L, Hao MM, Young HM. Hirschsprung disease: a developmental disorder of the enteric nervous system. Wiley Interdiscip Rev Dev Biol. 2013;2:113–129. doi: 10.1002/wdev.57. [DOI] [PubMed] [Google Scholar]
- 2.Jiao CL, Chen XY, Feng JX. Novel insights into the pathogenesis of hirschsprung’s-associated enterocolitis. Chin Med J (Engl) 2016;129:1491–1497. doi: 10.4103/0366-6999.183433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Svetanoff WJ, Dekonenko C, Osuchukwu O, Oyetunji TA, Aguayo P, Fraser JD, Juang D, Snyder CL, Hendrickson R, Peter SS, Rentea RM. Inpatient management of Hirschsprung’s associated enterocolitis treatment: the benefits of standardized care. Pediatr Surg Int. 2020;36:1413–1421. doi: 10.1007/s00383-020-04747-4. [DOI] [PubMed] [Google Scholar]
- 4.Gosain A, Frykman PK, Cowles RA, Horton J, Levitt M, Rothstein DH, Langer JC, Goldstein AM American Pediatric Surgical Association Hirschsprung Disease Interest Group. Guidelines for the diagnosis and management of Hirschsprung-associated enterocolitis. Pediatr Surg Int. 2017;33:517–521. doi: 10.1007/s00383-017-4065-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bui TM, Mascarenhas LA, Sumagin R. Extracellular vesicles regulate immune responses and cellular function in intestinal inflammation and repair. Tissue Barriers. 2018;6:e1431038. doi: 10.1080/21688370.2018.1431038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong WY, Lee MM, Chan BD, Kam RK, Zhang G, Lu AP, Tai WC. Proteomic profiling of dextran sulfate sodium induced acute ulcerative colitis mice serum exosomes and their immunomodulatory impact on macrophages. Proteomics. 2016;16:1131–1145. doi: 10.1002/pmic.201500174. [DOI] [PubMed] [Google Scholar]
- 8.Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 9.Devoto C, Lai C, Qu BX, Guedes VA, Leete J, Wilde E, Walker WC, Diaz-Arrastia R, Kenney K, Gill J. Exosomal microRNAs in military personnel with mild traumatic brain injury: preliminary results from the chronic effects of neurotrauma consortium biomarker discovery project. J Neurotrauma. 2020;37:2482–2492. doi: 10.1089/neu.2019.6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng Z, Dhall D, Zhao L, Wang HL, Doherty TM, Bresee C, Frykman PK. Murine model of Hirschsprung-associated enterocolitis. I: phenotypic characterization with development of a histopathologic grading system. J Pediatr Surg. 2010;45:475–482. doi: 10.1016/j.jpedsurg.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang R, Liao Y, Wang L, He P, Hu Y, Yuan D, Wu Z, Sun X. Exosomes derived from M2b macrophages attenuate DSS-induced colitis. Front Immunol. 2019;10:2346. doi: 10.3389/fimmu.2019.02346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han S, Li Z, Han F, Jia Y, Qi L, Wu G, Cai W, Xu Y, Li C, Zhang W, Hu D. ROR alpha protects against LPS-induced inflammation by down-regulating SIRT1/NF-kappa B pathway. Arch Biochem Biophys. 2019;668:1–8. doi: 10.1016/j.abb.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Demehri FR, Halaweish IF, Coran AG, Teitelbaum DH. Hirschsprung-associated enterocolitis: pathogenesis, treatment and prevention. Pediatr Surg Int. 2013;29:873–881. doi: 10.1007/s00383-013-3353-1. [DOI] [PubMed] [Google Scholar]
- 14.Matei AC, Antounians L, Zani A. Extracellular vesicles as a potential therapy for neonatal conditions: state of the art and challenges in clinical translation. Pharmaceutics. 2019;11:404. doi: 10.3390/pharmaceutics11080404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCulloh CJ, Olson JK, Wang Y, Zhou Y, Tengberg NH, Deshpande S, Besner GE. Treatment of experimental necrotizing enterocolitis with stem cell-derived exosomes. J Pediatr Surg. 2018;53:1215–1220. doi: 10.1016/j.jpedsurg.2018.02.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcus ME, Leonard JN. FedExosomes: engineering therapeutic biological nanoparticles that truly deliver. Pharmaceuticals (Basel) 2013;6:659–680. doi: 10.3390/ph6050659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hussain MT, Iqbal AJ, Norling LV. The role and impact of extracellular vesicles in the modulation and delivery of cytokines during autoimmunity. Int J Mol Sci. 2020;21:7096. doi: 10.3390/ijms21197096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho HJ, Eun JW, Baek GO, Seo CW, Ahn HR, Kim SS, Cho SW, Cheong JY. Serum exosomal MicroRNA, miR-10b-5p, as a potential diagnostic biomarker for early-stage hepatocellular carcinoma. J Clin Med. 2020;9:281. doi: 10.3390/jcm9010281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang W, Li H, Tang J, Wu W, Qin J, Lei H, Cai P, Huo W, Li B, Rehan V, Xu X, Geng Q, Zhang H, Xia Y. Specific serum microRNA profile in the molecular diagnosis of Hirschsprung’s disease. J Cell Mol Med. 2014;18:1580–1587. doi: 10.1111/jcmm.12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alves Dos Santos K, Clemente Dos Santos IC, Santos Silva C, Gomes Ribeiro H, de Farias Domingos I, Nogueira Silbiger V. Circulating exosomal miRNAs as biomarkers for the diagnosis and prognosis of colorectal cancer. Int J Mol Sci. 2020;22:346. doi: 10.3390/ijms22010346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cosar E, Mamillapalli R, Ersoy GS, Cho S, Seifer B, Taylor HS. Serum microRNAs as diagnostic markers of endometriosis: a comprehensive array-based analysis. Fertil Steril. 2016;106:402–409. doi: 10.1016/j.fertnstert.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 22.Fan X, Hao Z, Li Z, Wang X, Wang J. Inhibition of miR-17~92 cluster ameliorates high glucose-induced podocyte damage. Mediators Inflamm. 2020;2020:6126490. doi: 10.1155/2020/6126490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eftekharian MM, Noroozi R, Sayad A, Sarrafzadeh S, Toghi M, Azimi T, Komaki A, Mazdeh M, Inoko H, Taheri M, Mirfakhraie R. RAR-related orphan receptor A (RORA): a new susceptibility gene for multiple sclerosis. J Neurol Sci. 2016;369:259–262. doi: 10.1016/j.jns.2016.08.045. [DOI] [PubMed] [Google Scholar]
- 24.Acevedo N, Saaf A, Soderhall C, Melen E, Mandelin J, Pietras CO, Ezer S, Karisola P, Vendelin J, Gennas GB, Yli-Kauhaluoma J, Alenius H, von Mutius E, Doekes G, Braun-Fahrlander C, Riedler J, van Hage M, D’Amato M, Scheynius A, Pershagen G, Kere J, Pulkkinen V. Interaction between retinoid acid receptor-related orphan receptor alpha (RORA) and neuropeptide S receptor 1 (NPSR1) in asthma. PLoS One. 2013;8:e60111. doi: 10.1371/journal.pone.0060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang HS, Okamoto K, Takeda Y, Beak JY, Gerrish K, Bortner CD, DeGraff LM, Wada T, Xie W, Jetten AM. Transcriptional profiling reveals a role for RORalpha in regulating gene expression in obesity-associated inflammation and hepatic steatosis. Physiol Genomics. 2011;43:818–828. doi: 10.1152/physiolgenomics.00206.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Xue K, Zheng Y, Wang Y, Xu C. RORA overexpression alleviates nasal mucosal injury and enhances red blood cell immune adhesion function in a mouse model of allergic rhinitis via inactivation of the wnt/beta-catenin signaling pathway. Int Arch Allergy Immunol. 2019;180:79–90. doi: 10.1159/000500637. [DOI] [PubMed] [Google Scholar]
- 27.Huang N, Li C, Sun W, Wu J, Xiao F. Long non-coding RNA TUG1 participates in LPS-induced periodontitis by regulating miR-498/RORA pathway. Oral Dis. 2020;27:600–610. doi: 10.1111/odi.13590. [DOI] [PubMed] [Google Scholar]
- 28.Xie MY, Hou LJ, Sun JJ, Zeng B, Xi QY, Luo JY, Chen T, Zhang YL. Porcine milk exosome MiRNAs attenuate LPS-induced apoptosis through inhibiting TLR4/NF-kappaB and p53 pathways in intestinal epithelial cells. J Agric Food Chem. 2019;67:9477–9491. doi: 10.1021/acs.jafc.9b02925. [DOI] [PubMed] [Google Scholar]
- 29.Li H, Zhou L, Zhi Z, Lv X, Wei Z, Zhang X, Tang W, Tong M. Lipopolysaccharide upregulates miR-132/212 in Hirschsprung-associated enterocolitis, facilitating pyroptosis by activating NLRP3 inflammasome via targeting Sirtuin 1 (SIRT1) Aging (Albany NY) 2020;12:18588–18602. doi: 10.18632/aging.103852. [DOI] [PMC free article] [PubMed] [Google Scholar]