Abstract
Objective: This study aimed to explore the clinical characteristics of 742 patients with re-current Corona Virus Disease in 2019 (COVID-19), so as to provide relevant evidence for clinical diagnosis and treatment of re-infected patients. Methods: Altogether 742 discharged COVID-19 patients were analyzed retrospectively and were divided into re-infected patients (n=60) and non-re-infected patients (n=682) according to whether they became nucleic acid positive again after discharge. The time form leaving the hospital to re-infection and the time form the first nucleic acid negative test results to being re-infected were recorded. The clinical characteristics of the two groups were compared when they were admitted to the hospital. Logistic regression analysis was carried out on disease indicators with statistical differences between the two groups. Results: Compared with non-re-infected patients, there were statistical differences in age, contact history, fatigue, chills, nasal congestion and runny nose, lung CT observations, clinical classification and lymphocyte count of re-infected patients (P<0.05). Logistic regression analysis showed that nasal congestion and a runny nose, a lymphocyte count less than 0.93×109 cells/L, and age ≥65 years were the risk factors of being re-infected. The ROC curve showed that the cut-off value of lymphocyte count was 0.847×109 cells/L, and the AUC of predicted re-infection was 0.867. Conclusion: The symptoms of nasal congestion and runny nose, lymphocyte count less than 0.93×109 cells/L and, aged more than 65 years are the risk factors for the recurrent positive rates for COVID-19 patients, and lymphocyte count has certain clinical value in predicting recurrent patients.
Keywords: Corona Virus Disease 2019, recurrent positive, risk factors
Introduction
Corona Virus Disease 2019 (COVID-19) has broken out all over the world and has been deemed by the World Health Organization as a public health emergency of international concern. COVID-19 is an acute respiratory infection [1]. According to statistics, as of September 19th, COVID-19 has infected more than 30 million people and caused nearly one million deaths. Most COVID-19 patients have fever, cough, myalgia and dyspnea [2]. It has not only damaged the health care system, but also has had a damaging impact on the economy [3,4]. It has been reported that older age, increased complications and being male may related to the increased risk of poor prognosis for COVID-19 [5-7].
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the key virus causing COVID-19 [8,9]. It can not only spread through respiratory droplets, contact and aerosol, but it also exist in the feces in vitro for a long time [10]. The re-infection of patients via testing of nucleic acids may still occur in cured patients for a period of time [11,12]. At present, there are few reports about the re-infection of discharged COVID-19 patients. This paper retrospectively analyzed 742 discharged patients with COVID-19, and compared the clinical symptoms, biological indicators and the time from leaving hospital to becoming re-infected in patients, so as to explore the potential diagnostic indicators and provide relevant basis for predicting re-infection.
Materials and methods
COVID-19 cases
According to “Guidelines for Diagnosis and Treatment of Corona Virus Disease 2019” (the 7th Edition) issued by the Health Commission of the People’s Republic of China, 742 patients confirmed with COVID-19 were included in this study from February 20, 2020 to April 3, 2020 [13]. This study was approved by the Ethics Committee of Xiangyang No. 1 People’s Hospital, Hubei University of Medicine and also obtained the written informed consent of the participating patients.
Discharge criteria of COVID patients: the body temperature had returned to normal for more than 3 days, chest CT showed that the lung lesions were obviously reduced, dyspnea was obviously improved, and the nucleic acid test results of nasopharyngeal swabs were negative for at least 2 consecutive times (the interval of nucleic acid test was 24 hours). Patients discharged from hospital were observed in isolation for 14 days. Re-infection criteria: Patients with positive nucleic acid detection in the digestive tract or respiratory tract were regarded as re-infected [13]. The discharged patients were gived a patient health card in the Healthy Wuhan APP, and re-infected patients were screened through the nucleic acid detection and regularly tested after discharge (the 2nd week and 4th week after discharge). Special patients (patients with nucleic acid detection of re-infection and patients with prolonged course of re-infection) were followed up by telephone. The follow-up time of discharged patients was 4 weeks after discharge. Re-admission and isolation were carried out for the patients with re-infection, and those who were in close contact with them were followed up.
Data collection
The medical records of 742 discharged COVID-19 patients were analyzed retrospectively, including 60 re-infected patients and 682 non-re-infected patients. Routine blood work data, age, sex, admission symptoms, CT results, time from leaving hospital to re-infection, and time from first negative nucleic acid test to being re-infected were collected.
Statistical analysis
SPSS 26.0 (IBM, USA) was used for statistical analysis. Before analyzing the measurement data, the normal distribution test was carried out, and the non-normally distribution data were expressed by quartile (M (Q1, Q3)). Wilcoxon rank sum test was used for comparison between the two groups. The counting data were expressed by the number of cases (proportion), and the comparison was made by χ2 test. Logistic regression analysis was carried out for the indicators with statistical differences in univariate analysis by using progressive forward LR. Clinical value of lymphocyte count in predicting re-infection was evaluated by ROC. When P<0.05, the difference was statistically significant.
Results
Comparison of clinical features between the two groups
From February 20, 2020 to April 3, 2020, a total of 742 patients with COVID-19 were discharged from the hospital, including 60 (8.1%) cases of re-infected patients and 682 (91.9%) cases of non-re-infected patients after discharge. Among 60 patients with re-infected, 2 cases were mild (3.3%), 58 cases were moderate (96.7%), and there were no serious cases. There were significant differences in age, contact history, fatigue, chills, nasal congestion and runny nose, chest CT, clinical classification and lymphocyte count between the two groups (P<0.05), as shown in Table 1.
Table 1.
Clinical characteristics of re-infected patients and non-re-infected patients (n (%), median (min, max))
| Clinical characteristics | RP (n=60) | NRP (n=682) | P |
|---|---|---|---|
| Age | 0.030 | ||
| <65 years | 30 (50.0) | 437 (64.1) | |
| ≥65 years | 30 (50.0) | 245 (35.9) | |
| Sex | 0.919 | ||
| Male | 23 (38.3) | 266 (39.0) | |
| Female | 37 (61.7) | 416 (61.0) | |
| Contact information | 0.034 | ||
| Unidentified source of infection | 45 (75.0) | 582 (85.3) | |
| Contact with confirmed case | 15 (25.0) | 100 (14.7) | |
| Coexisting disorder | |||
| Hypertension | 16 (26.7) | 198 (29.0) | 0.698 |
| Diabetes | 7 (11.7) | 89 (13.0) | 0.760 |
| Hyperlipoidemia | 2 (3.3) | 11 (1.6) | 0.645 |
| Liver cyst | 1 (1.7) | 32 (4.7) | 0.445 |
| Fatty liver | 1 (1.7) | 4 (0.6) | 0.345 |
| Gastritis | 2 (3.3) | 10 (1.5) | 0.252 |
| Bronchiectasis | 1 (1.7) | 10 (1.5) | 0.607 |
| Cerebral infarction | 1 (1.7) | 15 (2.2) | 1.000 |
| Cancer* | 3 (5.0) | 13 (1.9) | 0.263 |
| Total with ≥2 symptoms | 13 (21.7) | 153 (22.4) | 0.891 |
| Signs and symptoms | |||
| Fever | 37 (61.7) | 416 (61.0) | 0.919 |
| Fatigue | 35 (58.3) | 306 (44.9) | 0.045 |
| Chills | 34 (56.7) | 244 (35.8) | <0.001 |
| Cough | 31 (51.7) | 360 (52.8) | 0.868 |
| Expectoration | 9 (15.0) | 105 (15.4) | 0.935 |
| Myalgia | 15 (25.0) | 145 (21.3) | 0.500 |
| Stuffy nose or Runny | 11 (18.3) | 16 (2.3) | <0.001 |
| Chest distress or Chest pain | 11 (18.3) | 133 (19.5) | 0.826 |
| Asthma or panting | 10 (16.7) | 164 (24.0) | 0.196 |
| Diarrhea | 4 (6.7) | 19 (2.8) | 0.203 |
| Nausea and vomitin | 3 (5.0) | 27 (4.0) | 0.960 |
| Anorexia | 2 (3.3) | 28 (4.1) | 1.000 |
| Sore throat | 1 (1.7) | 34 (5.0) | 0.398 |
| Dyspnea | 1 (1.7) | 12 (1.8) | 1.000 |
| Asymptomatic | 8 (13.3) | 73 (10.7) | 0.531 |
| Lung CT | <0.001 | ||
| Unilateral | 40 (66.7) | 276 (40.5) | |
| Bilateral | 18 (30.0) | 384 (56.3) | |
| Nodule shadow | 2 (3.2) | 22 (3.2) | |
| Predominantly CT patter | |||
| Ground-glass opacities | 12 (20.0) | 190 (27.9) | 0.190 |
| Atelectasis | 3 (5.0) | 19 (2.8) | 0.567 |
| Pulmonary fibrosis | 4 (6.7) | 66 (9.7) | 0.444 |
| Thickening of the adjacent pleura | 20 (33.3) | 169 (24.8) | 0.145 |
| Pleural effusion | 2 (3.3) | 18 (2.6) | 1.000 |
| Clinical classification | 0.029 | ||
| Light type | 2 (3.3) | 22 (3.2) | |
| Popular type | 58 (96.7) | 588 (86.2) | |
| Severe type | 0 (0.0) | 66 (9.7) | |
| Critical type | 0 (0.0) | 6 (0.9) | |
| Highest temperature during hospitalization | 0.613 | ||
| <37.5°C | 54 (90.0) | 582 (85.3) | |
| 37.5-38.0°C | 5 (8.3) | 83 (12.2) | |
| 38.1-39.0°C | 1 (1.7) | 17 (2.5) | |
| White-cell count (×109/liter) | 6.0 (4.4, 7.3) | 5.7 (4.7, 6.7) | 0.141 |
| Lymphocyte count (×109/liter) | 0.93 (0.82, 1.24) | 1.37 (1.21, 2.02) | 0.008 |
| Monocyte count (×109/liter) | 0.39 (0.31, 0.47) | 0.38 (0.30, 0.47) | 0.328 |
| Neutrophil count (×109/liter) | 3.67 (2.77, 4.63) | 3.41 (2.65, 4.24) | 0.129 |
| Platelet count (×109/liter) | 213.5 (172.3, 251.5) | 222.0 (181.0, 257.0) | 0.727 |
| Lymp/Mono | 4.5 (3.5, 5.9) | 4.3 (3.3, 5.6) | 0.530 |
| Neut/Lymp | 2.1 (1.7, 2.7) | 2.0 (1.6, 2.8) | 0.840 |
| Plt/Lymp | 121.8 (89.8, 170.6) | 130.6 (101.8, 171.7) | 0.238 |
| Alanine aminotransferase (U/L; normal range 0-55) | 19.7 (11.5, 27.1) | 20.8 (13.4, 34.6) | 0.227 |
| Aspartate aminotransferase (U/L; normal range 5-34) | 15.2 (12.2, 18.4) | 16.3 (12.9, 22.4) | 0.074 |
| Total bilirubin (μmol/L; normal range 3.4-20.5) | 9.8 (8.1, 13.7) | 9.5 (7.4, 12.5) | 0.172 |
| Direct bilirubin (μmol/L; normal range 0-8.6) | 4.1 (3.3, 6.5) | 4.1 (3.2, 5.2) | 0.242 |
| Albumin (g/L; normal range 35-52) | 39.7 (37.1, 41.1) | 39.2 (36.7, 41.6) | 0.709 |
| Alkaline phosphatase (IU/L; normal range 40-150) | 67.0 (57.3, 80.0) | 70.0 (58.3, 82.0) | 0.416 |
| Creatine kinase (U/L; normal range <190) | 58.0 (51.5, 79.0) | 56.0 (42.0, 81.0) | 0.325 |
| Myoglobin (ng/mL; normal range 0-106) | 32.1 (26.8, 45.8) | 31.8 (25.0, 42.5) | 0.257 |
| Hypersensitive troponin I (pg/mL; normal range 0-34.2) | 2.8 (1.5, 4.8) | 2.5 (1.4, 5.1) | 0.926 |
| Creatine kinase isoenzymes (ng/mL; normal range 0-3.1) | 0.8 (0.6, 1.0) | 0.6 (0.4, 0.9) | 0.055 |
| Hypersensitive C-reactive protein (mg/L; normal range 0-10) | 1.4 (0.6, 3.3) | 1.3 (0.5, 3.0) | 0.496 |
| Erythrocyte sedimentation rate for 30 minutes (mm/H; normal range 0-15) | 31.0 (13.5, 66.5) | 26.0 (12.0, 62.5) | 0.886 |
| Prothrombin time (s; normal range 9.2-15) | 11.4 (11.0, 12.1) | 11.3 (10.9, 11.8) | 0.252 |
| D-dimer (mg/L; normal range 0-0.55) | 0.3 (0.1, 0.4) | 0.3 (0.2, 0.6) | 0.420 |
| Urea nitrogen (mmol/L; normal range 3.2-7.4) | 4.0 (3.6, 5.2) | 4.5 (3.7, 5.5) | 0.067 |
| Serum creatinine (μmol/L; normal range 64-104) | 61.9 (54.4, 77.7) | 63.3 (55.2, 75.3) | 0.721 |
| Interleukin-6 (pg/mL; normal range 0-10) | 1.5 (1.5, 1.5) | 1.5 (1.5, 1.5) | 0.814 |
| Oxygen therapy | 0.152 | ||
| No | 25 (41.7) | 350 (51.3) | |
| Yes | 35 (58.3) | 332 (48.7) |
Included in this category is any type of cancer.
RP: re-infected patients; NRP: non-re-infected patients.
Comparison of the first cure time of re-infected patients
The results showed that there were 2 cases with the time from the first negative nucleic acid test to re-infection less than 7 days, all of which were moderate cases. There were 22 cases with 8-14 days, 1 case was mild and 21 cases were moderate. There were 36 cases with more than 14 days, 1 case was mild and 35 cases were moderate. There was no statistical difference between mild cases and moderate cases in the time from initial cure to re-infection (P=0.903), as shown in Table 2.
Table 2.
Time from the first negative nucleic acid of discharge index to returning to be virally positive (n (%), range 15.7 days)
| Days | Light type (n=2) | Popular type (n=58) | P |
|---|---|---|---|
| 0.903 | |||
| <7 days | 0 (0.0) | 2 (3.4) | |
| 8-14 days | 1 (50.0) | 21 (36.2) | |
| >14 days | 1 (50.0) | 35 (60.4) |
Two mild re-infected patients were found within 8-14 days after discharge. Among the moderate cases, 10 cases were re-infected within 7 days after discharge, 47 cases 8-14 days after discharge and 1 case 14 days after discharge. There was no statistical difference between mild cases and moderate cases in the time from discharge to re-infection (P=0.830), as shown in Table 3.
Table 3.
Time from leaving hospital to returning the be virally positive (n (%), range 9.3 days)
| Light type (n=2) | Popular type (n=58) | P | |
|---|---|---|---|
| 0.830 | |||
| <7 days | 0 (0.0) | 10 (17.2) | |
| 8-14 days | 2 (100.0) | 47 (81.1) | |
| >14 days | 0 (0.0) | 1 (1.7) |
Note: In table, there was one moderate re-infected patient who has been discharged from hospital for more than 14 days, and one patient who entered the isolation point since he was discharged from hospital and did not leave the isolation point until re-infection.
Risk factors of re-infection
Logistic regression was used to analyze the risk factors with P<0.05 in single factor analysis in Table 1. The assignment result was shown in Table 4.
Table 4.
Assignment table
| Variable | Variable assignment |
|---|---|
| Result | non-re-positive=1, re-positive=2 |
| Age | <65 years=0, ≥65 years=1 |
| Severity of illness | Light, normal type=0; heavy, critical type=1 |
| Contact history | No=0, Yes=1 |
| Fatigue | No=0, Yes=1 |
| Chills | No=0, Yes=1 |
| Nasal congestion and runny nose | No=0, Yes=1 |
| Lung CT | No nodule shadow=0; nodule shadow=1 |
| Lymphocyte count* | ≥0.93×109/L=0, <0.93×109/L=1 |
The average lymphocyte count of re-positive group was taken as the boundary.
Logistic regression analysis showed that nasal congestion and runny nose, lymphocyte count less than 0.93×109 cells /L, and aged more than 65 years were the risk factors of re-infection (P<0.05 or P<0.001), as shown in Table 5.
Table 5.
Logistic multivariate analysis of independent risk factors for re-infection
| B | S.E. | Wald | Variance | P | Exp (B) | 95% CI | |
|---|---|---|---|---|---|---|---|
| Nasal congestion and runny nose | 2.324 | 0.509 | 20.819 | 1 | 0.000 | 0.376 | 0.198-0.715 |
| Age ≥65 years old | 0.806 | 0.357 | 5.098 | 1 | 0.024 | 2.239 | 1.112-4.508 |
| Lymphocytes <0.93×109/L | -1.188 | 0.462 | 6.609 | 1 | 0.010 | 0.305 | 0.123-0.754 |
ROC results
ROC results are shown in Figure 1 and Table 6. The cut-off value of lymphocyte count was 0.847×109 cells/L, and the AUC of predicted re-infection was 0.867 (P<0.001).
Figure 1.
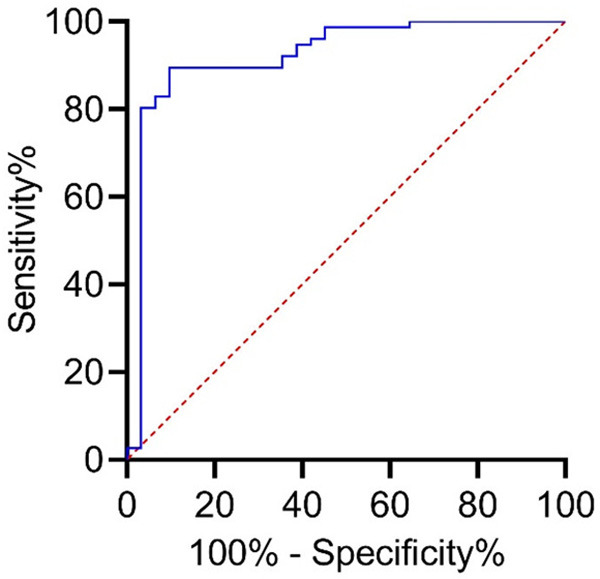
ROC curve (area under the curve is 0.760, 95% CI: 0.685-0.836, progressive significance is 0.000).
Table 6.
ROC curve
| Cut off value | Sensitivity | Specificity | Youden index | |
|---|---|---|---|---|
| Lymphocyte count (×109/ liter) | 0.847 | 0.917 | 0.815 | 0.732 |
Discussion
The latent period of COVID-19 is mostly 3-7 days, and some patients progress rapidly. In severe cases, they may develop into acute respiratory distress syndrome, bleeding/coagulation dysfunction, septic shock, and metabolic acidosis hard to correct, thus increasing the risk of death [14-18]. Previous studies have detected nucleic acid positivity in the sputum of patients cured from COVID, which means that the cured patients may still be virus carriers [11,19]. Some studies have found that among the COVID patients, the symptoms of the re-infection patients after cure were mild or moderate [12]. Although the above studies all point out that cured patients with COVID-19 still have the possibility of being nucleic acid positive again, the current clinical research on patients with re-infection is still under exploration, and the relationship between clinical data and re-infection needs to be deeply explored [20-22].
After exploring the time form discharge to re-infection, it was found that no matter the type severity, the discharged patients are likely to recover within 8-14 days after discharge. There was a re-infected patient after discharge which occurred after more than 14 days. The patient was held in isolation (the results of nucleic acid detection showed that one section was positive and two sections were negative) and they never left the isolation area. Therefore, it is still necessary to follow up the discharged patients for a certain period of time, pay close attention to the nucleic acid changes of patients, and strengthen the prevention and control of isolation areas.
This paper also compared the differences in clinical features between re-infected patients and non-re-infected patients. We found that 50.0% re-infected patients were more than 65 years old, and logistic regression analysis suggested that being aged more than 65 years was an independent risk factor for re-infection after discharge. Therefore, it is necessary to focus on the re-infection of discharged patients older than 65 years. The symptoms of nasal congestion and runny nose were also independent risk factors for re-infection. However, as nasal congestion and runny nose are common cold symptoms, the specific clinical significance of these indexes needs further study. The results also showed that lymphocyte count less than 0.93×109 cells/L may also increase the risk of re-infection. Viral infection may have a certain impact on the immune system. Leukomonocytes are an important immune cell in human body, and the decrease of lymphocyte level may be due to immune consumption caused by excessive immune response. Therefore, we should pay attention to the lymphocyte level of COVID-19 patients after they were discharged from hospital. In addition, we further discussed the clinical value of lymphocyte count in predicting re-infection. The results showed that when the cut-off value of lymphocyte count was 0.847×109 cells/L, the AUC for predicting re-positive was 0.867, suggesting that lymphocyte count has certain predictive value.
To sum up, nasal congestion and runny nose, lymphocyte count less than 0.93×109 cells/L, and age more than 65 years were the risk factors of re-infection in patients with COVID-19, and lymphocyte count has certain clinical value in predicting reactivation.
However, this study still has some limitations. Although age, nasal congestion and runny nose, and lymphocyte count less than 0.93×109 cells/L may be the risk factors of re-infection, this paper lacks dynamic data and fails to deeply understand the dynamic changes of other biological indicators in different periods of re-infected patients. According to the clinical data provided in this paper, the number of COVID patients younger than 18 years old was only 3 cases, as such we failed to fully study the possibility of re-infection in minor patients. Future studies will focus on young patients and study the potential relationship between youthand re-infection. In this study, it is considered that patients with moderate cases have high possibility of re-infection, so we can focus on the physical indicators of patients with moderate cases in the future research, in order to obtain more clinical information related to re-infection.
Disclosure of conflict of interest
None.
References
- 1.Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao G, Tan W China Novel Coronavirus Investigating and Research Team. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang TH, Wu JL, Chang LY. Clinical characteristics and diagnostic challenges of pediatric COVID-19: a systematic review and meta-analysis. J Formos Med Assoc. 2020;119:982–989. doi: 10.1016/j.jfma.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H, Li X, Li T, Zhang S, Wang L, Wu X, Liu J. The genetic sequence, origin, and diagnosis of SARS-CoV-2. Eur J Clin Microbiol Infect Dis. 2020;39:1629–1635. doi: 10.1007/s10096-020-03899-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X, Chen X, Chen L, Deng C, Zou X, Liu W, Yu H, Chen B, Sun X. The evidence of SARS-CoV-2 infection on ocular surface. Ocul Surf. 2020;18:360–362. doi: 10.1016/j.jtos.2020.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 8.Niu S, Tian S, Lou J, Kang X, Zhang L, Lian H, Zhang J. Clinical characteristics of older patients infected with COVID-19: a descriptive study. Arch Gerontol Geriatr. 2020;89:104058. doi: 10.1016/j.archger.2020.104058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H, Guo J, Wang C, Luo F, Yu X, Zhang W, Li J, Zhao D, Xu D, Gong Q, Liao J, Yang H, Hou W, Zhang Y. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. Lancet. 2020;395:809–815. doi: 10.1016/S0140-6736(20)30360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu S, Guo X, Geary K, Zhang D. Emerging therapeutic strategies for COVID-19 patients. Discoveries (Craiova) 2020;8:e105. doi: 10.15190/d.2020.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elfiky AA. Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253:117592. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Health Commission of the People’s Republic of China and National Administration of Traditional Chinese Medicine. Diagnosis and Treatment Protocol for Novel Coronavirus Infection-Induced Pneumonia version 7 (trial) Quan Ke Yi Xue Lin Chuang Yu Jiao Yu. 2020;18:100–105. [Google Scholar]
- 14.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong KH, Choi JP, Hong SH, Lee J, Kwon JS, Kim SM, Park SY, Rhee JY, Kim BN, Choi HJ, Shin EC, Pai H, Park SH, Kim SH. Predictors of mortality in Middle East respiratory syndrome (MERS) Thorax. 2018;73:286–289. doi: 10.1136/thoraxjnl-2016-209313. [DOI] [PubMed] [Google Scholar]
- 16.Gorbalenya A, Baker S, Baric R, de Groot R, Drosten C, Gulyaeva A, Haagmans B, Lauber C, Leontovich A, Neuman B, Penzar D, Perlman S, Poon L, Samborskiy D, Sidorov I, Sola I, Ziebuhr J. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Z, Shi L, Wang YJ, Zhang JY, Huang L, Zhang C, Liu SH, Zhao P, Liu HX, Zhu L, Tai YH, Bai CQ, Gao TT, Song JW, Xia P, Dong JH, Zhao JM, Wang FS. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henry BM, Vikse J. Clinical characteristics of COVID-19 in China. N Engl J Med. 2020;382:1860–1861. doi: 10.1056/NEJMc2005203. [DOI] [PubMed] [Google Scholar]
- 19.Qu YM, Kang EM, Cong HY. Positive result of Sars-Cov-2 in sputum from a cured patient with COVID-19. Travel Med Infect Dis. 2020;34:101619. doi: 10.1016/j.tmaid.2020.101619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang CJ, Ng CY, Brook RH. Response to COVID-19 in Taiwan: big data analytics, new technology, and proactive testing. JAMA. 2020;323:1341–1342. doi: 10.1001/jama.2020.3151. [DOI] [PubMed] [Google Scholar]
- 21.Shen C, Wang Z, Zhao F, Yang Y, Li J, Yuan J, Wang F, Li D, Yang M, Xing L, Wei J, Xiao H, Yang Y, Qu J, Qing L, Chen L, Xu Z, Peng L, Li Y, Zheng H, Chen F, Huang K, Jiang Y, Liu D, Zhang Z, Liu Y, Liu L. Treatment of 5 critically Ill patients with COVID-19 with convalescent plasma. JAMA. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chakraborty I, Maity P. COVID-19 outbreak: migration, effects on society, global environment and prevention. Sci Total Environ. 2020;728:138882. doi: 10.1016/j.scitotenv.2020.138882. [DOI] [PMC free article] [PubMed] [Google Scholar]


