Abstract
Objective: We aimed to investigate the mechanism of circular RNA circ_0084927 in the progression of breast cancer (BC). Methods: The levels of circ_0084927, miR-142-3p, and ELKS/RAB6-interacting/CAST family member-1 (ERC1) mRNA in the BC tissues and cells were detected by qRT-PCR. CCK8, colony formation, Transwell, and flow cytometry assays were performed to examine the cell proliferation, colony formation, cell invasion, and apoptosis, respectively, in the BC cells with regulated expressions of circ_0084927, miR-142-3p, and ERC1. RNase R treatment was employed to verify the circular structure of circ_0084927. Nucleocytoplasmic separation experiment, bioinformatics analysis, dual-luciferase reporter assay, and RNA immunoprecipitation were performed to investigate the ceRNA mechanism of circ_0084927. Results: High levels of circ_0084927 and ERC1 and low levels of miR-142-3p were detected in the BC tissues and cells. Knockdown of circ_0084927 promoted apoptosis and inhibited proliferation, colony formation, and invasion of BC cells (all P<0.05), whereas overexpression of circ_0084927 in the BC cells achieved the opposite effects. miR-142-3p is the target of circ_0084927. Overexpression of miR-142-3p could inhibit BC cell proliferation, colony formation, and cell invasion and induce apoptosis of the BC cells (all P<0.05), and the effects of miR-142-3p knockout on the BC cells could be reversed by silencing circ_0084927. miR-142-3p could target ERC1. Both ERC1 silencing and circ_0084927 knockout in the BC cells could achieve the tumor-suppressing effect, and this effect could be more remarkable under simultaneous ERC1 silencing and circ_0084927 knockout (all P<0.05). Conclusion: Circ_0084927 can promote the progression of BC by regulating the miR-142-3p/ERC1 axis.
Keywords: circ_0084927, miR-142-3p, ELKS/RAB6-interacting/CAST family member-1, breast cancer
Introduction
Breast cancer (BC) is one of the most fatal malignant tumors in women. Although great progress has been made in the diagnosis and treatment of BC, the recurrence rate and survival rate of patients are still not satisfactory [1-4]. With the change of lifestyle and environment, the incidence of BC in China shows an upward trend, and the disease can bring a huge burden to the patients’ families and the society [5,6]. Therefore, it is essential to explore the molecular mechanism of BC to improve the diagnosis and treatment of this cancer.
Circular RNA (circRNA) is a closed circular non-coding RNA without a 5’ cap and 3’ poly (A) tail. CircRNA is structurally stable, unaffected by RNA exonuclease, and widely exists in eukaryotes [7,8]. Many studies have found that circRNA is involved in the progression of various cancers at the transcriptional, post-transcriptional, and translational levels and can act as a miRNA sponge that affects the biological activity and function of miRNA target genes [9-11]. Xu et al. found that circRNA-UBAP2 can promote the proliferation and inhibit the apoptosis of ovarian cancer cells through the miR-382-5p/PRPF8 axis [12]. Other studies also found that circMET can regulate the miR-145-5p/CXCL3 axis to promote the proliferation, metastasis, and immune escape of non-small cell lung cancer cells [13]. It has been reported that circRNA also plays a key role in the development of BC. For instance, Gao et al. found that circ_0006528, as a competitive endogenous RNA, can promote the progression of human BC by activating miR-7-5p and MAPK/ERK signaling pathways [14]. However, the role of circ_0084927 in BC has not been revealed, but researchers demonstrated that overexpression of circ_0084927 can promote carcinogenesis through the miR-1179/CDK2 regulatory network in cervical cancer [15]. Circ_0084927 expression has also been confirmed to be increased in malignant pleural effusions related to lung cancer [16]. Interestingly, miR-142-3p, the potential target of circ_0084927 in our present study, can improve the chemosensitivity of BC cells and inhibit autophagy by targeting HMGB1 discovered by Liang et al. [17]. Other studies have also found that miR-142-3p plays the tumor-suppressing role in BC by targeting the RAC1/PAK1 pathway [18]. Also, ERC1, the potential target of miR-142-3p, has been found to act as an oncogene in BC development [19].
In the present study, we aimed to investigate the mechanism of circ_0084927 and its downstream signaling in the development of BC.
Materials and methods
Collection of primary tissue samples
The BC tissues and the adjacent normal tissues (2-5 cm away from the tumor) of 50 patients (age: 49.1±10.3 years) who received breast tumor resection surgery in our hospital from January 2018 to January 2020 were collected for this study. All tissue specimens were preserved at -80°C immediately after the resection. There were 19 cases with tumor diameter less than 2 cm, 20 with tumor diameter between 2 and 5 cm, and 11 with tumor diameter above 5 cm. Eighteen patients were in stage I, 22 in stage II, and 10 in stage III/IV. In terms of the pathological grade, 16 cases were in grade I, 26 in grade II, and 8 in grade III.
The patients were followed up for survival analysis using the Kaplan-Meier method. Informed consent was obtained from the patients, and the study was approved by the Ethics Committee of our hospital.
Cell culture
Normal human breast cells (MCF-10A) and human BC cell lines (MDA-MB-231, SUM-159, MCF-7, SK-BR-3, and MDA-MB-157) were purchased from ATCC, USA. The cells were cultured in RPMI1640 (31800, Solarbio, China) containing 10% fetal bovine serum. All the cells were incubated and sub-cultured in a humidified incubator containing 5% CO2 at 37°C.
qRT-PCR
Total RNA was extracted from the tissue samples and the cells using Trizol reagent (R0016, Beyotime Biotechnology, China). The purity and concentration of the RNAs were determined by ultraviolet spectrophotometer (Alpha1500, Lab-Spectrum Instruments, China). RNA was reversely transcribed to cDNA using the one-step reverse transcription kit (T2240, Solarbio., China). The PCR reaction system and the reaction conditions were prepared according to the manufacturer’s instructions. The relative expression levels of the genes were calculated using the 2ΔΔCt method with U6 as the internal reference of miRNA and GAPDH as the internal reference of circRNA and mRNA. The primer sequences are listed in Table 1.
Table 1.
Primer sequence for qRT-PCR
| Gene | Sequences (5’-3’) | |
|---|---|---|
| circ_0084927 | Forward | CGAAGAACGGAGAAGCTCT |
| Reverse | GTGCCCTGACTACGGTGTTA | |
| miR-142-3p | Forward | GGAGAGATCCCGAAATGTGTG |
| Reverse | GGTGTCCGAATGCTTGAAACCCT | |
| ERC1 | Forward | ATGGAAGTGCTCTTGAACC |
| Reverse | ATTCCATGTATGGCAAGTCAA | |
| GAPDH | Forward | ACCACAGTCCATGCCATCAC |
| Reverse | TCCACCACCCTGTTGCTGTA | |
| U6 | Forward | GCUUCGGCAGCACAUAUACUAAAAU |
| Reverse | CGCUUCACGAAUUUGCGUGUCAU |
Note: ERC1: ELKS/RAB6-interacting/CAST family member-1.
RNase R treatment
The total RNA was extracted from the BC cells. The RNA (10 μg) was incubated with 40U RNase R (R1030, Solarbio, China) at 37°C for 1 h. qRT-PCR was performed to quantify circ_0084927.
Cell transfection and grouping
According to the manufacturer’s instructions of LipofectamineTM 3000 kit, the BC cells were seeded into 96-well plates at a density of 104 cell/mL. LipofectamineTM 3000 (0.2 μL) was diluted in 5 μL of Opti MEM medium and mixed well. Master mix of DNA was prepared by diluting 0.3-3 μg of DNA (miRNA mimic, miRNA inhibitor, or siRNA) in 10 μL of Opti-MEM medium, followed by addition of 0.4 μL of P3000TM. Then, the diluted DNA was mixed with diluted LipofectamineTM 3000 at a ratio of 1:1 and incubated for 5 min. Afterward, the DNA-lipid complex was added to the BC cells with 70-90% confluence, and the cells were cultured for 24-48 h.
The grouping of the BC cells was as follows: the blank group (transfected with blank plasmid), the circ_0084927 group (transfected with circ_0084927), the si-NC group (transfected with si-NC), the si-circ group (transfected with si-circ), the miR-142-3p group (transfected with miR-142-3p mimic), the si-circ + miR-inhibitor group (transfected with si-circ and miR-inhibitor), the si-circ + NC inhibitor group (transfected with si-circ and NC inhibitor), and the ERC1 + si-circ group (transfected with pcDNA3.1-ERC1 overexpressing ERC1 and si-circ). NC inhibitor was the negative control of miR-inhibitor.
The supplier of miR-142-3p mimic and miR-142-3p inhibitor was Bioneer, South Korea; the supplier of pcDNA3.1 plasmid (V795-20) was Invitrogen, USA; and the supplier of LipofectamineTM 3000 kit (L3000015) was Thermo Fisher Scientific, USA. The si-RNA of cic_0084927 was designed and synthesized by Sango Biotech, China.
CCK-8 assay
The cells were inoculated into a 96-well plate at a density of 2*103 per well. The cells were added with 10 μL of CCK-8 (C0038, Beyotime Biotechnology, China) when being incubated at room temperature for 24 h, 48 h, and 72 h. Two hours later, the absorbance at 450 nm was measured by the microplate reader (HBS-1096C, DeTie, China) to analyze the cell proliferation. The experiment was repeated over three times in each group.
Colony formation assay
The colony formation ability of the cells was measured by the colony formation assay. The BC cells were inoculated into a 6-well plate at a density of 1*103 per well and incubated at 37°C for 15 days. Then, the cells were washed in PBS (C0221A, Beyotime Biotechnology, China), fixed with 4% paraformaldehyde (P0099, Beyotime Biotechnology, China), and stained with 0.1% crystal violet (C0121, Beyotime Biotechnology, China). Five fields were randomly picked to observe and photograph the colony formation under a microscope (CX23, Olympus, Japan). The number of clone in each field was calculated using the ImageJ software. The colony formation rate = (number of clone/number of cells inoculated) *100%. The experiment was repeated over three times in each group.
Transwell assay
The invasive ability of the cells was detected by the Transwell assay. The upper chamber of Transwell was pre-coated with Matrigel (354234, Solarbio, China) and added with 200 μL of serum-free medium (N6010, Solarbio, China) containing 1*105 cells. Meanwhile, 600 μL of RPMI1640 medium containing 10% fetal bovine serum (31800, Solarbio, China) was placed in the lower chamber. The cells were cultured at 37°C in 5% CO2 for 24 h. Afterward, the non-invading cells were removed from the upper chamber, and the invasive cells were fixed and stained with 4% paraformaldehyde (P0099, Beyotime Biotechnology, China) and 0.1% crystal violet (C0121, Beyotime Biotechnology, China), respectively. Five fields were randomly picked under an optical microscope (DSX100, Olympus, Japan) to observe the cell. The cells that had passed through the insert membrane were counted and the average value was calculated using the ImageJ software. The experiment was repeated over three times in each group, and the quantification of the invasive cells was performed using the ImageJ software.
Flow cytometry assay
The BC cells (5×105/well) were inoculated into a 96-well plate and cultured at 37°C for 24 h. Then, the cells were stained in the dark using the AnnexinV-FITC cell apoptosis detection kit (C1062S, Solarbio, China), and the apoptotic cells in each group were detected with the flow cytometer (Attune NxT, Thermo Fisher Scientific, USA). The experiment was repeated over three times in each group.
Western blot
RIPA (R0010, Solarbio, China) was used to lyse the BC cells to extract the total protein, and the protein concentration was determined using the BCA protein detection kit (PC0020, Solarbio, China). SDS-PAGE was performed, and the proteins were transferred to the PVDF membrane, sealed with skimmed milk for 2 h, and incubated with anti-ERC1 antibodies (1:500, K15450-JWP, Baiaolaibo Technology, China) at 4°C overnight. After removing the primary antibodies, the samples were incubated with the secondary antibodies (1:1,500, ab150077, Abcam, UK) at room temperature for 2 h. GAPDH (1:1,000, ab8245, Abcam, UK) was used as the internal reference, and the protein band was detected using the ECL chemiluminescence kit (P0018M, Beyotime Biotechnology, China). ImageJ software was used to analyze the gray value of the protein band. The relative expression level of the protein = The gray value of the target protein/the gray value of the internal control *100%. The experiment was repeated over three times in each group.
Nucleocytoplasmic separation assay
The nucleocytoplasmic separation assay was used to detect the subcellular localization of circ_0084927. The cells were digested with 0.25% trypsin (T1350, Solarbio, China) and centrifuged for 3 min at 1,000 g. According to the manufacturer’s instructions of the nucleocytoplasmic separation kit (KTP4002, AmyJet Scientific, China), the RNA was extracted from the nucleus and cytoplasm and was detected by qRT-PCR. GAPDH and U6 were used as the internal references for the RNAs from the nucleus and the cytoplasm, respectively.
Bioinformatic analysis
We used the web-based tool Circular RNA Interactome (https://circinteractome.nia.nih.gov) to predict the target of circ_0084927 and online databases RNA22 (https://cm.jefferson.edu/rna22/), TargetScan (http://www.targetscan.org/vert_71/), miRDB (http://www.mirdb.org/), and miRWalk (http://mirwalk.umm.uni-heidelberg.de/) to predict the target genes of miR-142-3p.
Dual-luciferase reporter assay
To verify the relationship between miR-142-3p and circ_0084927 and between miR-142-3p and ERC1, the full-length sequences of circ_0084927 and ERC1 3’-UTR with or without mutant miR-142-3p binding site were cloned into the pmir-Reporter plasmid. The wild and mutant reporter plasmids were cotransfecte 4 d with miR-142-3p mimic or control vectors into the BC cells. At 48 h after the transfection, the luciferase activity of the BC cells was analyzed using the dual-luciferase reporter kit (D0010, Solarbio, China). The experiment was repeated over three times in each group.
RNA-precipitation assay
The BC cells were lysed in lysis buffer (R0010, Solarbio, China) containing RNasin and protease inhibitor. The lysate was centrifuged at 12,000 g at 4°C for 30 min to obtain the supernatant. Magnetic beads coated with Ago-2 and IgG antibodies were added to the lysate for incubation at 4°C for 3 h followed by three wash in PBS. RNA was extracted from the magnetic beads using the Trizol reagent (R0016, Beyotime Biotechnology, China) for testing.
Statistical analysis
SPSS (version 18.0) software was used for data analysis. Measurement data are expressed as mean ± standard deviation (x̅ ± sd); count data are expressed as n and were tested by chi-square test or Fisher exact probability test. If the measurement data were in a normal distribution, the independent samples t-test was performed for the comparison between the two groups, and one-way analysis of variance was used for multi-group analysis. If the measurement data were not in a normal distribution, the Mann-Whitney U test was used for comparison between the two groups, and the Bonferroni method was used for pairwise comparisons. P<0.05 indicated that the difference was statistically significant.
Results
Circ_0084927 was up-regulated in the primary BC tissues and the BC cells
The heat map showed that the expression level of circ_0084927 was increased in the BC tissues compared with that in the normal adjacent tissues (n=50; Figure 1A, 1B). The mean value of circ_0084927 expression in the control group was used as the cutoff value, higher than the mean value was considered to be a high expression, and lower than the mean was considered to be a low expression. The median survival period of the patients with high circ_0084927 expression was 55 months, whereas that of the patients with low expression of circ_0084927 was 69 months, indicating that down-regulation of circ_0084927 was correlated with a longer survival period of patients with BC (P<0.05, Figure 1C). Also, the expression of circ_0084927 in the BC cell lines (MDA-MB-231, SUM-159, MCF-7, SK-BR-3, MDA-MB-157) was significantly higher than that in the normal cell lines (MCF-10A; Figure 1D). These results suggest that the dysregulation of circ_0084927 can be involved in the progression of BC.
Figure 1.
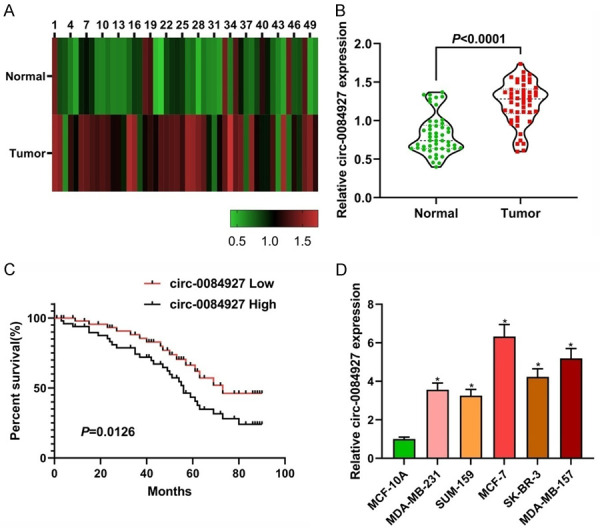
The expression levels of circ_0084927. A and B: The relative expression levels of circ_0084927 in primary BC tissues and adjacent normal tissues (n=50); C: The curve of Kaplan-Meier overall survival rate was calculated according to the expression level of circ_0084927; D: The expression level of circ_0084927 in the normal and BC cell lines. Compared with MCF-10A, *P<0.05. BC: breast cancer.
The relationship between circ_0084927 expression and clinicopathological features of the patients
As shown in Table 2, patients with high expression of circ_0084927 had higher TNM stage, more lymph node metastasis, and negative expression of PR (all P<0.05). There were no differences in age, ER, HER-2, and tumor size between the high expression group and the low expression group (all P>0.05).
Table 2.
Relationship between circ_0084927 expression and clinicopathological features in 50 patients with BC
| Variable | Circ_0084927 expression | P | ||
|---|---|---|---|---|
|
| ||||
| Low (n=25) | High (n=25) | |||
| Age (years) | ≤55 | 12 | 15 | 0.5709 |
| >55 | 13 | 10 | ||
| ER | Positive | 20 | 23 | 0.4174 |
| Negative | 5 | 2 | ||
| PR | Positive | 19 | 7 | 0.0016 |
| Negative | 6 | 18 | ||
| HER-2 | Positive | 15 | 11 | 0.3961 |
| Negative | 10 | 14 | ||
| TNM stage | I-II | 19 | 8 | 0.0041 |
| III-IV | 6 | 17 | ||
| Lymph node metastasis | Absent | 17 | 5 | 0.0014 |
| Present | 8 | 20 | ||
| Tumor size | <5 cm | 19 | 16 | 0.5380 |
| ≥5 cm | 6 | 9 | ||
Note: BC: breast cancer; ER: estrogen receptor; PR: progesterone receptor; HER-2: human epidermal growth factor receptor-2; TNM: tumor node metastasis.
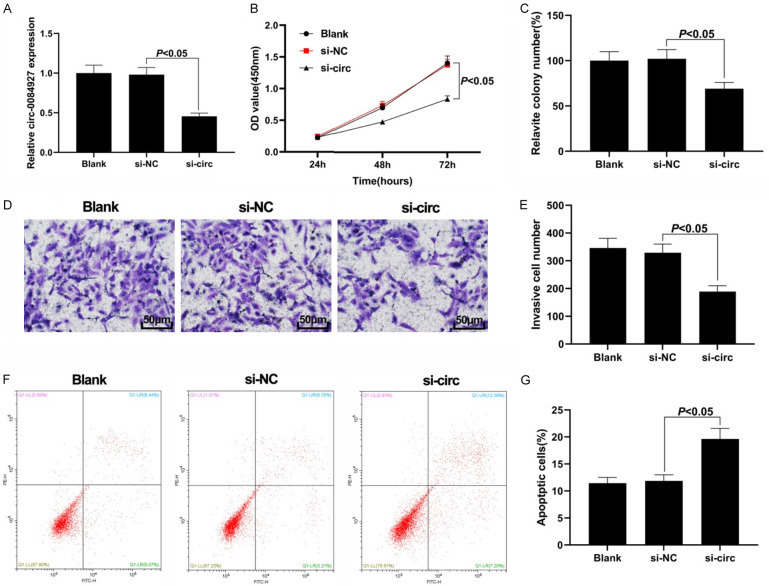
Knockout of circ_0084927 could inhibit the proliferation, colony formation, and invasion of the BC cells and could induce cellular apoptosis
We first verified the knockout efficiency of circ_0084927 (Figure 2A). The results of the CCK-8 assay showed that circ_0084927 knockout significantly decreased the proliferative ability of the BC cells compared with the si-NC group (Figure 2B). Moreover, the colony formation ability of the MCF-7 cells transfected with si-circ was significantly lower than that in the si-NC group (P<0.05, Figure 2C). Circ_0084927 knockout significantly reduced the number of invasive MCF-7 cells (Figure 2D, 2E). Flow cytometry analysis found that down-regulation of circ_0084927 in the MCF-7 cell line significantly promoted cellular apoptosis (P<0.05, Figure 2F, 2G). These findings demonstrate that knockout of circ_0084927 can inhibit the proliferation, colony formation, and invasion of BC cells and promote cellular apoptosis.
Figure 2.
Effect of circ_0084927 knockout on the BC cells. A: Knockout efficiency of circ_0084927; B: Cell proliferation results by CCK-8; C: Colony formation assay result; D, E: Cell invasion results by Transwell assay (200×); F, G: Cellular apoptosis results by flow cytometry assay. BC: breast cancer.
Circ_0084927 could sponge miR-142-3p
The results of the nucleocytoplasmic separation assay showed that the cir_0084927 was mainly located in the cytoplasm (Figure 3A). We also found that circ_0084927 could not be digested by RNase R, demonstrating the circular structure of circ_0084927 (Figure 3B). We then searched on the Circular RNA Interactome (https://circinteractome.nia.nih.gov) website to find the miR-142-3p binding site on circ_0084927 (Figure 3C). Dual-luciferase reporter assay showed that the dual-luciferase activity of the cells co-transfected with circ_0084927-WT and miR-142-3p was lower than that of the control group (P<0.05; Figure 3D). The results of the RIP assay showed that Ago2 could capture more circ_0084927 in the miR-142-3p group than in the NC group (P<0.05, Figure 3E). Moreover, the expression of circ_0084927 was suppressed after the overexpression of miR-142-3p compared with the NC group (Figure 3F). These results demonstrate that circ_0084927 can act as a ceRNA to sponge miR-142-3p. Compared with the normal tissues and the MCF-10A cells, the BC tissues and cells had lower expression levels of miR-142-3p (all P<0.05, Figure 3G and 3I). The expression of miR-142-3p was negatively correlated with the expression of circ_0084927 in BC (Figure 3H).
Figure 3.
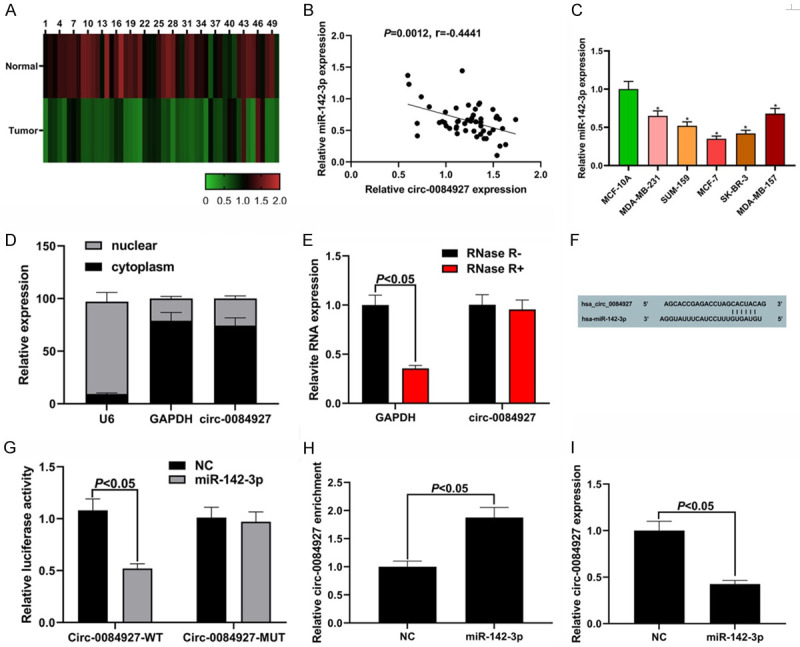
miR-142-3p is the target of circ_0084927. A: Circ_0084927 is mainly located in the cytoplasm; B: Resistance of circ_0084927 to RNase R treatment detected by qRT-PCR; C: The miR-142-3p binding site on circ_0084927; D: Dual-luciferase reporter assay result; E: RIP assay result; F: miR-142-3p suppressed the expression of circ_0084927; G: Expression level of miR-14-3p in the BC cells; H: Negative correlation between miR-142-3p and circ_0084927 in the BC tissues; I: Expression level of miR-142-3p in the BC cells. Compared with MCF-10A cells, *P<0.05. BC: breast cancer; RIP: RNA immunoprecipitation.
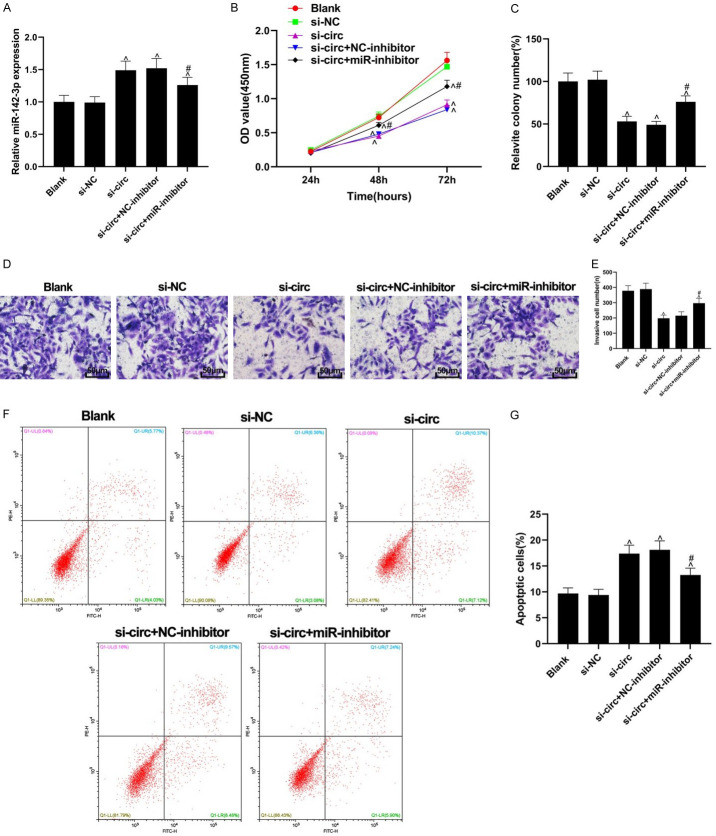
The effects of silencing circ_0084927 in the BC cells could be reversed by miR-142-3p knockdown
As shown in Figure 4A, the expression of miR-142-3p was increased following the knockout of circ_0084927. Compared with the si-NC group, the cell proliferation, colony formation, and invasion abilities in the si-circ group were inhibited (all P<0.05). The effects of si-circ transfection on cell proliferation, colony formation, and invasion abilities were partially reversed by miR-inhibitor transfection (all P<0.05, Figure 4B-E). Compared with the si-NC group, apoptosis was induced in the si-circ group, while the apoptosis-inducing effect of silencing circ_0084927 was partially reversed by miR-inhibitor transfection (P<0.05, Figure 4F, 4G).
Figure 4.
Effect of circ_0084927 on BC cells could be reversed by miR-142-3p. A: The expression levels of miR-142-3p in each group; B: Cell proliferation assessed by CCK-8 assay; C: The results of colony formation assay; D, E: Cell invasion detected by Transwell assay (200×); F, G: Cellular apoptosis assessed by flow cytometry assay. Compared with the si-NC group, ^P<0.05, compared with the si-circ group, #P<0.05.
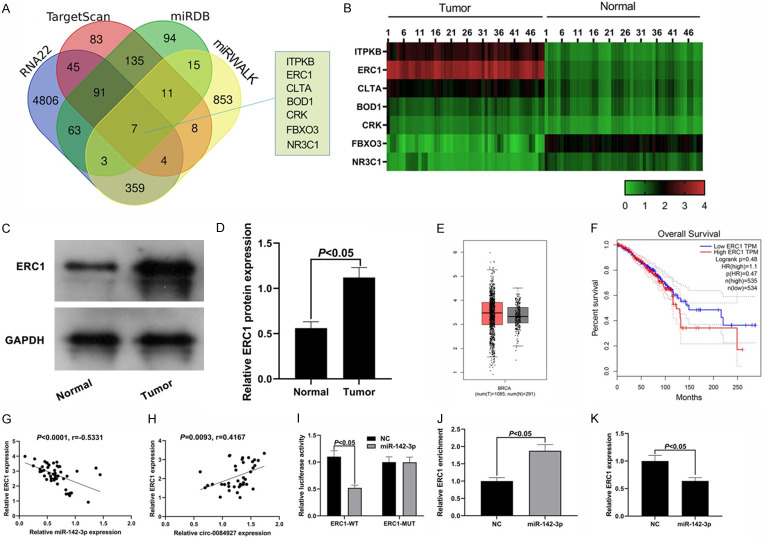
miR-142-3p could target ERC1
To clarify the regulatory mechanism of circ_0084927/miR-142-3p in BC, we investigated the potential targets of circ_0084927/miR-142-3p through the intersection of Wayne graph of RNA22, Targetscan, miRWalk, and miRDB. As shown in Figure 5A, FBXO3, ITPKB, CLTA, BOD1, ERC1, NR3C1, and CRK were identified as target genes. qRT-PCR was performed to detect the expression levels of the above target genes in the tumor tissues and normal tissues of the BC patients, and we found that five genes, ITPKB, ERC1, CLTA, BOD1, and CRK, were up-regulated in the cancer tissues, whereas the expression levels of FBXO3 and NR3C1 were downregulated in the cancer tissues (Figure 5B). After searching the previous studies, we chose to investigate ERC1 as the target gene of miR-142-3p in the subsequent experiments. We found that the protein expression level of ERC1 in the BC tissues was higher than that in the adjacent normal tissues (P<0.05, Figure 5C, 5D). Therefore, we speculated that ERC1 can act as an oncogene to regulate the apoptosis of cancer cells.
Figure 5.
ERC1 is the target of miR-142-3p. A: Prediction of miR-142-3p targets; B: The expression of FBXO3, ITPKB, CLTA, BOD1, ERC1, NR3C1, and CRK in the cancer tissues and the normal tissues. C, D: ERC1 expression level in the BC tissues; E, F: The expression levels of ERC1 in the cancer tissue and the adjacent tissues and the association between ERC1 level and patient survival period predicted by GEPIA website; G, H: ERC1 was negatively correlated with miR-142-3p and positively correlated with circ_0084927; I: Dual-luciferase reporter assay results; J: RIP assay results; K: The expression of ERC1 was inhibited by miR-142-3p. ERC1: ELKS/RAB6-interacting/CAST family member-1; BC: breast cancer; RIP: RNA immunoprecipitation.
After searching the GEPIA database (http://gepia.cancer-pku.cn/detail.php), we found that the expression level of ERC1 can increase in cancer tissues (Figure 5E), and the patients with high expression of ERC1 can have a shorter survival period than those with low expression of ERC1, even though the difference is not significant (Figure 5F). Correlation analysis showed that the expression of ERC1 in BC was positively correlated with circ_0084927 and negatively correlated with miR-142-3p (Figure 5G, 5H). Dual-luciferase reporter assay showed that miR-142-3p could significantly reduce the luciferase activity of the cells with wild-type ERC1 3’UTR, but had no significant effect on the cells with mutant ERC1 3’UTR, indicating that there was a targeting relationship between ERC1 and miR-142-3p (Figure 5I). The results of the RIP assay showed that the Ago2 antibody could capture more ERC1 in the miR-142-3p group than in the NC group (Figure 5J). As shown in Figure 5K, overexpression of miR-142-3p suppressed the expression of ERC1. These results suggest that ERC1 can be negatively regulated by miR-142-3p.
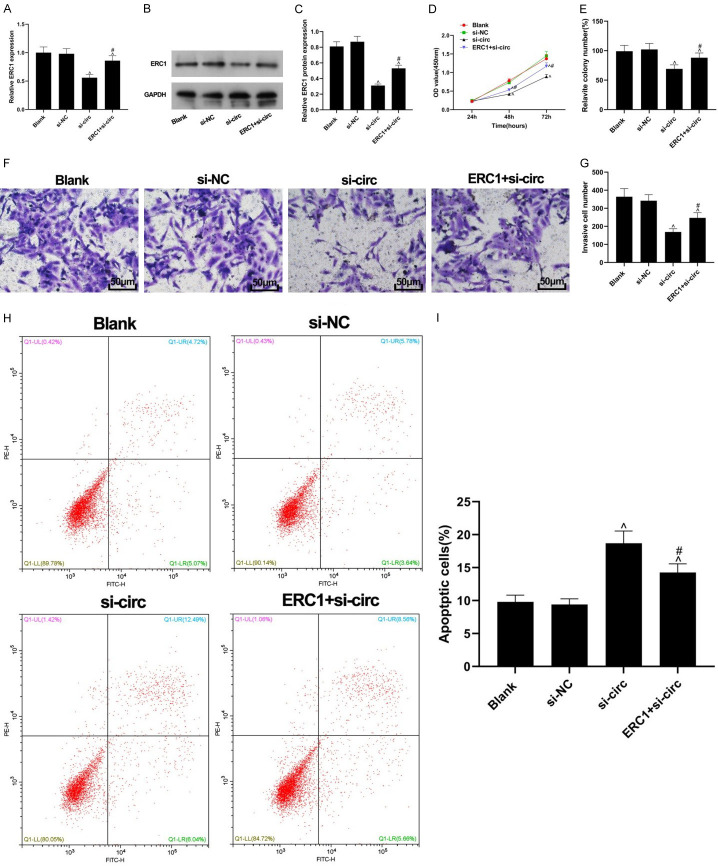
The inhibitory effect of circ_0084927 knockout on BC development could be partially reversed by overexpression of ERC1
As shown in Figure 6A-C, compared with the si-NC group, the expression level of ERC1 decreased significantly in the si-circ group, and decreased expression of ERC1 could be partially reversed by transfection of ERC1 to the cells with si-circ (both P<0.05). The inhibitory effect of silencing cir_0084927 on the cell proliferative ability was also partially reversed by up-regulation of ERC1 (P<0.05, Figure 6D). The colony-forming ability of the ERC1 + si-circ group was greater than that in the si-circ group (P<0.05, Figure 6E). Transwell assay showed that the weakened invasiveness of the cells with circ_0084927 knockout could be restored by the up-regulation of ERC1 (P<0.05, Figure 6F, 6G). The apoptosis induced by silencing circ_0084927 was inhibited by the upregulation of ERC1 (P<0.05, Figure 6H, 6I). These results suggest that the inhibitory effect of circ_0084927 knockout on the BC cells can be partially reversed by the up-regulation of ERC1.
Figure 6.
ERC1 partially reversed the effects of circ_0084927 silencing on the BC cells. A: The mRNA level of ERC1 in the BC cells; B: The protein band of ERC1; C: Quantification of the protein levels of ERC1; D: BC cell proliferation assessed by CCK-8 assay; E: BC cell colony formation results; F, G: BC cell invasion results detected by Transwell assay; H, I: Cellular apoptosis assessed by flow cytometry assay. Compared with the si-NC group, ^P <0.05, compared with the si-circ group, #P<0.05. ERC1: ELKS/RAB6-interacting/CAST family member-1; BC: breast cancer.
Discussion
BC is a heterogeneous tumor with different clinical features, disease processes, and responses to specific treatment [20-22]. However, the potential molecular mechanism regulating the progression of BC is still unclear. It is essential to discover the effective molecular biomarkers and identify the targets for a better therapeutic intervention of BC.
Increasing evidence has shown that multiple circRNAs play important roles in BC development [23,24]. For example, it has been found that circRNA_103809 is highly expressed in BC tissues and cells, and knockout of circRNA_103809 can effectively inhibit the growth and metastasis of BC cells [25]. Circ_DCAF6 can up-regulate GLI1 through sponging miR-616-3p, thereby promoting the growth of BC cells [26]. In this study, we found that circ_0084927 was highly expressed in BC tissues and cells; circ_0084927 knockout could inhibit the proliferation, invasion, and colony formation of BC cells and could induce BC cell apoptosis, whereas overexpression of circ_0084927 could achieve the opposite effects. A high level of circ_0084927 was highly correlated with a poor survival rate of patients with BC. These findings suggest that the dysregulation of circ_0084927 is involved in the progression of BC.
Through the bioinformatics analysis, we found that circ_0084927 could sponge various miRNAs. Among these miRNAs, we selected miR-142-3p as the target of circ_0084927 in this study. MiR-142-3p has been proven to inhibit the proliferation of colorectal cancer cells, and miR-491-5p also inhibits the proliferation, migration, and invasion of prostate cancer cells [27,28]. The role of miR-142-3p in BC has been preliminarily clarified, and this miRNA has a low expression in BC and has anti-tumor effect. In this study, it was found that miR-142-3p was decreased in BC tissues. Through dual-luciferase reporter assay, we verified the targeting relationship between circ_0084927 and miR-142-3p and found a negative correlation between the expression levels of these two. After measuring the BC cell proliferation, invasion, and apoptosis, we found that the effect of circ_0084927 knockout on BC cells could be partially reversed by miR-142-3p inhibitor.
To find the potential target of circ_0084927/miR-142-3p in BC, we screened 7 target genes using several prediction tools. In the ceRNA mechanism, the expression of lncRNA is consistent with the regulated expression of mRNA, we thus focused on the five up-regulated genes, ITPKB, ERC1, CLTA, BOD1, and CRK. Studies have found that the expression of ITPKB is increased in endometrial carcinoma, but the role of ITPKB in BC is not clear [29]. Other studies have found that BOD1, CRK, and CLTA-4, a family member of CLTA are dysregulated or mutated in BC, but there is no evidence of how they affect the progression of BC [30-32]. ERC1 is a protein-coding gene. The main diseases associated with ERC1 are thyroid carcinoma and distal monomer 12p. It has been proven that ERC1 is involved in the progression of thyroid papillary carcinoma and non-small cell lung cancer [33,34]. Besides, some studies have found that ERC1 is a factor related to BC cell invasion and metastasis, and the inhibition or knockout of ERC1 can disable the basic activity of carcinogenic transcription factor NF- κB [19]. Therefore, we focused on ERC1 in this study and found the targeting relationship between ERC1 and miR-142-3p. ERC1 was highly expressed in the BC tissues and cells and promoted the malignant progression of BC cells. A negative correlation between the expression of ERC1 and expression of miR-142-3p and a positive correlation between the expression of ERC1 and expression of circ_0084927 in BC were observed. miR-142-3p could induce BC cell apoptosis and inhibit BC cell proliferation, invasion, and colony formation, while these effects could be partially reversed by up-regulation of ERC1. The inhibitory effects of circ_0084927 knockout on the proliferation, cloning, and invasion of BC cells and the promotion effect of circ_0084927 knockout on BC cellular apoptosis could be partially reversed by up-regulation of ERC1. The above results suggest that circ_0084927 can regulate ERC1 through sponging miR-142-3p, thereby promoting the development of BC.
In conclusion, circ_0084927 upregulation can suppress the expression of miR-142-3p and increase the expression of ERC1, thus promoting the progression of BC. However, since this study only made a simple analysis of circ_0084927 in the tumor transplantation model, more studies need to be conducted in the future to explore the mechanism of circ_0084927/miR-142-3p/ERC1 to further clarify its role in BC. Circ_0084927/miR-142-3p/ERC1 can be a potential therapeutic target for BC. The possible mechanism of this study is listed in Figure 7.
Figure 7.
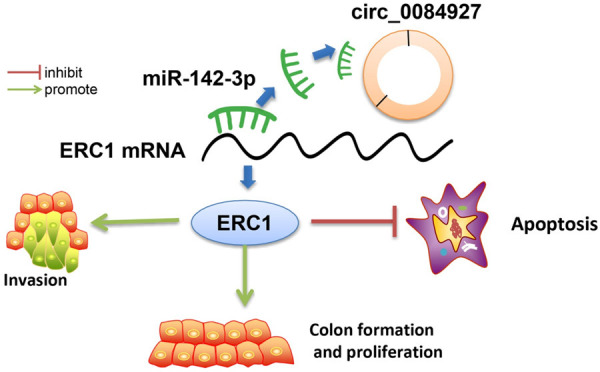
Circ_0084927 can promote BC cell proliferation and invasion and inhibit BC cell apoptosis by regulating the miR-142-3p/ERC1 axis. BC: breast cancer; ERC1: ELKS/RAB6-interacting/CAST family member-1.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81873373) and Young Innovative and Entrepreneurial Talents of “Grassland Talents” Project of Inner Mongolia Autonomous Region (Q2017042).
Disclosure of conflict of interest
None.
References
- 1.Liu X, Bi L, Wang Q, Wen M, Li C, Ren Y, Jiao Q, Mao JH, Wang C, Wei G, Wang Y. miR-1204 targets VDR to promotes epithelial-mesenchymal transition and metastasis in breast cancer. Oncogene. 2018;37:3426–3439. doi: 10.1038/s41388-018-0215-2. [DOI] [PubMed] [Google Scholar]
- 2.Luo ML, Zheng F, Chen W, Liang ZM, Chandramouly G, Tan J, Willis NA, Chen CH, Taveira MO, Zhou XZ, Lu KP, Scully R, Wulf GM, Hu H. Inactivation of the prolyl isomerase pin1 sensitizes BRCA1-proficient breast cancer to PARP inhibition. Cancer Res. 2020;80:3033–3045. doi: 10.1158/0008-5472.CAN-19-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yao Y, Saw PE, Nie Y, Wong PP, Jiang L, Ye X, Chen J, Ding T, Xu L, Yao H, Hu H, Xu X. Multifunctional sharp pH-responsive nanoparticles for targeted drug delivery and effective breast cancer therapy. J Mater Chem B. 2019;7:576–585. doi: 10.1039/c8tb02600a. [DOI] [PubMed] [Google Scholar]
- 4.Zong Y, Zhang Y, Hou D, Xu J, Cui F, Qin Y, Sun X. The lncRNA XIST promotes the progression of breast cancer by sponging miR-125b-5p to modulate NLRC5. Am J Transl Res. 2020;12:3501–3511. [PMC free article] [PubMed] [Google Scholar]
- 5.Choi J, Cha YJ, Koo JS. Adipocyte biology in breast cancer: from silent bystander to active facilitator. Prog Lipid Res. 2018;69:11–20. doi: 10.1016/j.plipres.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Chandler BC, Moubadder L, Ritter CL, Liu M, Cameron M, Wilder-Romans K, Zhang A, Pesch AM, Michmerhuizen AR, Hirsh N, Androsiglio M, Ward T, Olsen E, Niknafs YS, Merajver S, Thomas DG, Brown PH, Lawrence TS, Nyati S, Pierce LJ, Chinnaiyan A, Speers C. TTK inhibition radiosensitizes basal-like breast cancer through impaired homologous recombination. J Clin Invest. 2020;130:958–973. doi: 10.1172/JCI130435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, Lu N, Wang L, Wang Y, Li M, Zhou Y, Yan H, Cui M, Zhang M, Zhang L. Circular RNAs and esophageal cancer. Cancer Cell Int. 2020;20:362. doi: 10.1186/s12935-020-01451-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo Y, Wang L, Gou R, Tang L, Liu P. Noncoding RNAs in peritoneal fibrosis: background, mechanism, and therapeutic approach. Biomed Pharmacother. 2020;129:110385. doi: 10.1016/j.biopha.2020.110385. [DOI] [PubMed] [Google Scholar]
- 9.Dai J, Zhuang Y, Tang M, Qian Q, Chen JP. CircRNA UBAP2 facilitates the progression of colorectal cancer by regulating miR-199a/VEGFA pathway. Eur Rev Med Pharmacol Sci. 2020;24:7963–7971. doi: 10.26355/eurrev_202008_22479. [DOI] [PubMed] [Google Scholar]
- 10.Wu Q, Yuan ZH, Ma XB, Tang XH. Low expression of CircRNA HIPK3 promotes osteoarthritis chondrocyte apoptosis by serving as a sponge of miR-124 to regulate SOX8. Eur Rev Med Pharmacol Sci. 2020;24:7937–7945. doi: 10.26355/eurrev_202008_22476. [DOI] [PubMed] [Google Scholar]
- 11.Xu Y, Yao Y, Gao P, Cui Y. Upregulated circular RNA circ_0030235 predicts unfavorable prognosis in pancreatic ductal adenocarcinoma and facilitates cell progression by sponging miR-1253 and miR-1294. Biochem Biophys Res Commun. 2019;509:138–142. doi: 10.1016/j.bbrc.2018.12.088. [DOI] [PubMed] [Google Scholar]
- 12.Xu Q, Deng B, Li M, Chen Y, Zhuan L. circRNA-UBAP2 promotes the proliferation and inhibits apoptosis of ovarian cancer though miR-382-5p/PRPF8 axis. J Ovarian Res. 2020;13:81. doi: 10.1186/s13048-020-00685-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pei X, Chen SW, Long X, Zhu SQ, Qiu BQ, Lin K, Lu F, Xu JJ, Zhang PF, Wu YB. circMET promotes NSCLC cell proliferation, metastasis, and immune evasion by regulating the miR-145-5p/CXCL3 axis. Aging (Albany NY) 2020;12:13038–13058. doi: 10.18632/aging.103392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao D, Qi X, Zhang X, Fang K, Guo Z, Li L. hsa_circRNA_0006528 as a competing endogenous RNA promotes human breast cancer progression by sponging miR-7-5p and activating the MAPK/ERK signaling pathway. Mol Carcinog. 2019;58:554–564. doi: 10.1002/mc.22950. [DOI] [PubMed] [Google Scholar]
- 15.Qu X, Zhu L, Song L, Liu S. circ_0084927 promotes cervical carcinogenesis by sponging miR-1179 that suppresses CDK2, a cell cycle-related gene. Cancer Cell Int. 2020;20:333. doi: 10.1186/s12935-020-01417-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wen Y, Wang Y, Xing Z, Liu Z, Hou Z. Microarray expression profile and analysis of circular RNA regulatory network in malignant pleural effusion. Cell Cycle. 2018;17:2819–2832. doi: 10.1080/15384101.2018.1558860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang L, Fu J, Wang S, Cen H, Zhang L, Mandukhail SR, Du L, Wu Q, Zhang P, Yu X. MiR-142-3p enhances chemosensitivity of breast cancer cells and inhibits autophagy by targeting HMGB1. Acta Pharm Sin B. 2020;10:1036–1046. doi: 10.1016/j.apsb.2019.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu T, He BS, Pan B, Pan YQ, Sun HL, Liu XX, Xu XN, Chen XX, Zeng KX, Xu M, Wang SK. MiR-142-3p functions as a tumor suppressor by targeting RAC1/PAK1 pathway in breast cancer. J Cell Physiol. 2020;235:4928–4940. doi: 10.1002/jcp.29372. [DOI] [PubMed] [Google Scholar]
- 19.Alpay M, Backman LR, Cheng X, Dukel M, Kim WJ, Ai L, Brown KD. Oxidative stress shapes breast cancer phenotype through chronic activation of ATM-dependent signaling. Breast Cancer Res Treat. 2015;151:75–87. doi: 10.1007/s10549-015-3368-5. [DOI] [PubMed] [Google Scholar]
- 20.Pu S, Chen H, Zhou C, Yu S, Liao X, Zhu L, He J, Wang B. Major postoperative complications in esophageal cancer after minimally invasive esophagectomy compared with open esophagectomy: an updated meta-analysis. J Surg Res. 2020;257:554–5. doi: 10.1016/j.jss.2020.08.011. [DOI] [PubMed] [Google Scholar]
- 21.Brooks JD, Boice JD Jr, Shore RE, Reiner AS, Smith SA, Bernstein L, Knight JA, Lynch CF, John EM, Malone KE, Mellemkjaer L, Langballe R, Liang X, Woods M, Tischkowitz M, Concannon P, Stram DO. A case-control study of the joint effect of reproductive factors and radiation treatment for first breast cancer and risk of contralateral breast cancer in the WECARE study. Breast. 2020;54:62–69. doi: 10.1016/j.breast.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gokce B, Akcok I, Cagir A, Pesen-Okvur D. A new drug testing platform based on 3D tri-culture in lab-on-a-chip devices. Eur J Pharm Sci. 2020;155:105542. doi: 10.1016/j.ejps.2020.105542. [DOI] [PubMed] [Google Scholar]
- 23.Wang JM, Li XJ, Wang J. Circular RNA circ_0067934 functions as an oncogene in breast cancer by targeting Mcl-1. Eur Rev Med Pharmacol Sci. 2019;23:9499–9505. doi: 10.26355/eurrev_201911_19444. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L, Dong X, Yan B, Yu W, Shan L. CircAGFG1 drives metastasis and stemness in colorectal cancer by modulating YY1/CTNNB1. Cell Death Dis. 2020;11:542. doi: 10.1038/s41419-020-2707-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang W, Lu Y, Wang F, Huang X, Yu Z. Circular RNA circRNA_103809 accelerates bladder cancer progression and enhances chemo-resistance by activation of miR-516a-5p/FBXL18 axis. Cancer Manag Res. 2020;12:7561–7568. doi: 10.2147/CMAR.S263083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye G, Pan R, Zhu L, Zhou D. Circ_DCAF6 potentiates cell stemness and growth in breast cancer through GLI1-Hedgehog pathway. Exp Mol Pathol. 2020;116:104492. doi: 10.1016/j.yexmp.2020.104492. [DOI] [PubMed] [Google Scholar]
- 27.Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, Chen C, Chang W, Ping Y, Ji P, Wu J, Quan W, Yao Y, Zhou Y, Sun Z, Li D. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-β1 axis. Mol Cancer. 2020;19:117. doi: 10.1186/s12943-020-01235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan YF, Chen ZY, Wang L, Wang M, Liu XH. MiR-142-3p functions as an oncogene in prostate cancer by targeting FOXO1. J Cancer. 2020;11:1614–1624. doi: 10.7150/jca.41888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeda A, Witek A, Janikowska G, Cwynar G, Janikowski T, Ciałon M, Orchel J, Mazurek U. Expression profile of genes associated with the histaminergic system estimated by oligonucleotide microarray analysis HG-U133A in women with endometrial adenocarcinoma. Ginekol Pol. 2014;85:172–179. doi: 10.17772/gp/1709. [DOI] [PubMed] [Google Scholar]
- 30.Navarrete-Bernal MGC, Cervantes-Badillo MG, Martínez-Herrera JF, Lara-Torres CO, Gerson-Cwilich R, Zentella-Dehesa A, Ibarra-Sánchez MJ, Esparza-López J, Montesinos JJ, Cortés-Morales VA, Osorio-Pérez D, Villegas-Osorno DA, Reyes-Sánchez E, Salazar-Sojo P, Tallabs-Utrilla LF, Romero-Córdoba S, Rocha-Zavaleta L. Biological landscape of triple negative breast cancers expressing CTLA-4. Front Oncol. 2020;10:1206. doi: 10.3389/fonc.2020.01206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Troschel FM, Böhly N, Borrmann K, Braun T, Schwickert A, Kiesel L, Eich HT, Götte M, Greve B. miR-142-3p attenuates breast cancer stem cell characteristics and decreases radioresistance in vitro. Tumour Biol. 2018;40:1010428318791887. doi: 10.1177/1010428318791887. [DOI] [PubMed] [Google Scholar]
- 32.Davra V, Saleh T, Geng K, Kimani S, Mehta D, Kasikara C, Smith B, Colangelo NW, Ciccarelli B, Li H, Azzam EI, Kalodimos CG, Birge RB, Kumar S. Cyclophilin a inhibitor debio-025 targets crk, reduces metastasis, and induces tumor immunogenicity in breast cancer. Mol Cancer Res. 2020;18:1189–1201. doi: 10.1158/1541-7786.MCR-19-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou X, Shou J, Sheng J, Xu C, Ren S, Cai X, Chu Q, Wang W, Zhen Q, Zhou Y, Li W, Pan H, Li H, Sun T, Cheng H, Wang H, Lou F, Rao C, Cao S, Pan H, Fang Y. Molecular and clinical analysis of Chinese patients with anaplastic lymphoma kinase (ALK)-rearranged non-small cell lung cancer. Cancer Sci. 2019;110:3382–3390. doi: 10.1111/cas.14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee YC, Chen JY, Huang CJ, Chen HS, Yang AH, Hang JF. Detection of NTRK1/3 rearrangements in papillary thyroid carcinoma using immunohistochemistry, fluorescent in situ hybridization, and next-generation sequencing. Endocr Pathol. 2020;31:348–358. doi: 10.1007/s12022-020-09648-9. [DOI] [PubMed] [Google Scholar]