Abstract
Objective: To explore the role and of miR-132, HMGA2 and PI3K/AKT pathway in mice with Alzheimer’s disease (AD). Methods: The mice were divided into 7 groups: the normal group, the model group (AD model mice), the NC group (AD mice injected with negative control (NC) vector), the miR-132 mimic group (AD mice injected with miR-132 mimics), the miR-132 inhibitor group (AD mice injected with miR-132 inhibitor), the si-HMGA2 group (AD mice injected with HMGA2 silencing vector), and the miR-132 inhibitor + si-HMGA2 group (model mice treated with miR-132 inhibitor and si-HMGA2). Y-maze experiment and related molecular biology experiments were performed. Results: The double-luciferase reporter assay verified that miR-132 could target and inhibit the expression of HMGA2A. Compared with the NC group, model mice had decreased learning and memory ability, reduced miR-132, p-PI3K/PI3K, p-AKT/AKT, AQP4 expression as well as GFAP GSH-Px, SOD, ATP, and T-AOC levels, but increased expression of HMGA2 and the levels of TNF-α, IL-6, NO, IL-1β, MAO, and MDA (P<0.017). Up-regulation of miR-132 or silencing HMGA2 could partly reverse the changes, but inhibition of miR-132 would exaggerate the brain injury and these molecular changes (P<0.017). The combination uses of si-HMGA2 and miR-132 inhibitor could reverse the changes caused by miR-132 inhibitor (P<0.017). Conclusion: miR-132 could downregulate the expression of HMGA2 and promote the expression of the PI3K/AKT pathway, so as to achieve a protective effect on brain in AD mice.
Keywords: miR-132, HMGA2, PI3K/AKT pathway, Alzheimer’s disease
Introduction
Alzheimer’s disease (AD) is the most common neurodegenerative disease in the elderly [1-3]. It mainly occurs among individuals over 60 years old. AD patients show progressive memory loss, cognitive impairment, resulting in behavioral abnormalities. The occurrence of AD seriously endangers the health of the elderly and results in a burden to the family and society [4-6].
The expression of high-mobility group AT-hook 2 (HMGA2) is reported to be upregulated in different malignant tumors, including gliomas [7-9]. HMGA2 is mainly involved in regulating the transcriptional process of genes, thereby regulating the life activities of organism [10-13].
The PI3K/Akt signaling pathway regulates the permeability of the mitochondrial membrane and participates in the metabolic processes of amyloid protein, which is also involved in the formation of dendritic spines and development of dendritic neurons. A previous study also reports that miR-132 alleviates brain injury by protecting the blood-brain barrier when ischemic stroke occurs. The upregulation of miR-132 could also regulate neuronal maturation and morphogenesis [14-18].
However, there is no clear literature about the expression changes of miR-132 and HMGA2 in AD and the regulatory relationship between them. Therefore, we constructed mouse models of AD and treated them with miR-132 and HMGA2 with two objectives: to explore the relationship between miR-132 and HMGA2, and to study the mechanism of miR-132 and HMGA2 on hippocampal injury in mice with AD.
Materials and methods
Experimental materials
A total of 180 male Kunming mice (10 weeks old, SPF grade, weighing 21.42±2.08 g) were purchased from the Experimental Animal Center of Shandong University. All the mice were maintained in an environment of 12 h light and 12 h dark, at 22°C, with 45-65% humidity. They ate freely for a week before model establishment. This study has been reviewed and approved by the ethics committee of our hospital.
Establishment of mouse models of AD
Among 180 mice, 20 mice were randomly selected as normal group and the rest were used to construct an AD model. Briefly, mice were anesthetized with 0.4% pentobarbital sodium (40 mg/kg) and disinfected routinely, then AD model was induced by injecting 3 mg/kg of scopolamine (0.3 mg/mL; Xuzhou Lai’en Pharmaceutical Co., Ltd., Jiangsu, China) into the subcutaneous occipital area of the posterior brain, once a day for 2 weeks [19]. The mice in the normal group were injected with the same dose of normal saline by the same approach. The preliminary criteria for the successful establishment of AD were as follows: slow exercise, reduced food intake, no response to external stimuli, dry hair, quadriplegia, overbalance, and rotation to the right during tail rise [20]. There were 145 mice successfully modeled (success rate 90.63%), and 120 mice were selected and divided into groups for follow-up analysis. The rest were euthanized by rapid cervical dislocation.
Experimental grouping and processing
The model mice were divided into 6 groups (n=20 per group): the AD model group (AD mice), the negative control (NC) group (AD mice injected with NC vector into dentate gyrus), the miR-132 mimic group (AD mice injected with miR-132 mimics into dentate gyrus), the miR-132 inhibitor group (AD mice injected with miR-132 inhibitor into dentate gyrus), the Si-HMGA2 group (AD mice injected with HMGA2 silencing vector into dentate gyrus), and the miR-132 inhibitor + Si-HMGA2 group (AD mice injected with miR-132 inhibitor and HMGA2 silencing vector into dentate gyrus). miR-132 mimic, miR-132 inhibitor and Si-HMGA2 were purchased from Tiangen Biotech (Beijing) Co., Ltd., China. The mice were anesthetized with 2% pentobarbital acid. The 3 mM of corresponding vectors was injected into the dentate gyrus of mice under stereotaxic apparatus, perpendicular to the dorsal longitudinal axis of the hippocampus. The injection coordinates were 2.9 mm deep to the dura mater, 2 mm lateral toward the right side, and 3 mm posterior to bregma. The plane of incision was 2.4 mm lower than that of the interaural line [21]. The injection was carried out once a week. The follow-up experiment was performed after 8 weeks of continuous injection [22]. The operation was carried out in strict accordance with the instructions.
Double-luciferase reporter experiment
The biological prediction website (www.targetscan.org) screened out that there may be a binding site between miR-132 and HMGA2, which was verified by the double-luciferase reporting system. The double-luciferase reporter gene vectors, PGL3-HMGA2 wt and PGL3-HMGA2 mut, were constructed, respectively. Rellina plasmid and two kinds of reporter plasmids were co-transfected into HEK 293T cells with miR-132 plasmid and NC plasmid, respectively. Following a period of 24 h, a double-luciferase detection was performed.
Y-maze tests
All the mice underwent the Y-maze test. The Y-maze test device is composed of three wooden arms with an angle of 120°C between them. Each arm was 40 cm long, 12 cm high, 10 cm wide at the top, and 5 cm wide at the bottom. The Y-maze test was carried out on day 3 after completion of the interference injection. In the experiment, the mice were placed at the end of one arm and were allowed to enter and leave the 3 arms freely. Over a period of 8 min, we recorded the total number and order of each mouse entering the 3 arms. We recorded the correct alternating reaction times and the total alternating reaction times. Entering the 3 different arms continuously was defined as one correct alternating reaction. The test results were expressed by the spontaneous alternating reaction rate (%) = correct alternating reaction times/total times - 2 * 100%.
Immunofluorescence
Paraffin sections of the hippocampal tissues were made from three mice from each group, respectively, by fixing them with 10% neutral buffer formalin and embedding them in paraffin. Aquaporin 4 (AQP4) and glial fibrillary acidic protein (GFAP) were used as markers of astrocytes [23]. Detection of the expression of AQP4 and GFAP was used to determine the activation of glial cells in each group. The sections were primarily sealed with rabbit monoclonal antibody AQP4 (1:200, ab46182, Abcam, USA) and goat polyclonal antibody GFAP (1:1,000, ab4674, Abcam, USA) at room temperature for 1 h. The secondary goat anti-rabbit IgG-FITC (1:1,000, ab6717, Abcam, USA) and monkey anti-goat IgG-Cy5 (1:1,000, ab6566, Abcam, USA) were blocked for 30 min. The nuclei were stained with DAPI (Invitrogen Molecular Probes, Carlsbad, CA, USA) after the second antibody was sealed. Anti-fluorescence quenching agent was used to mount slices, which were observed under a fluorescence microscope (XSP-BM22AY, Shanghai Optical Instrument Factory, Shanghai, China).
Quantitative reverse transcription PCR (qRT-PCR)
Hippocampal tissues of 3 mice from each group were taken to extract the total RNA by Trizol method (Invitrogen, Carlsbad, CA, USA). According to the TaqMan MicroRNA Assays Reverse Transcription primer (4427975, Applied Biosystems, USA), reverse transcription was performed to obtain cDNA and the reaction condition was 37°C for 30 min and then at 85°C for 5 s. Primers were synthesized by Beijing Tsingke Biotechnology Co., Ltd. (Table 1). Quantitative PCR (7500, ABI, USA) was carried out with the following conditions: pre-denaturation at 95°C for 10 min, denaturation at 95°C for 10 s and annealing at 60°C for 20 s, for a total of 35 cycles. The PCR reaction μL of qPCR forward primer (10 μM), 0.8 μL of qPCR reverse primer (10 μM), μL of ROX reference dye II, 10 μL of SYBR Premix Ex TaqTM II, 2.0 μL of cNDA template, and 6.0 μL of sterilized distilled water. U6 was used as the internal reference for the relative expression of miR-132. The expression of GAPDH was used as the internal reference for the relative expression of mRNA of HMGA2. 2-ΔΔCt indicated the relative expression level of the target gene.
Table 1.
Primer sequences
| Primer | Sequence (5’-3’) |
|---|---|
| miR-132 | F: GGGAACCGTGGCTTTCGAT |
| R: GTGCAGGGTCCGAGGT | |
| HMGA2 | F: GCCCAGAAGAAAGCAGAGACC |
| R: TCTGAACGACTTGTTGTGGC | |
| U6 | F: CTCGCTTCGGCAGCACATA |
| R: AACGATTCACGAATTTGCGT | |
| GAPDH | F: TGGCCTTCCGTGTTCCTAC |
| R: GAGTTGCTGTTGAAGTCGCA |
Western blot (WB)
Total protein was extracted from hippocampal tissues of 5 mice from each group and was separated by SDS-PAGE gel electrophoresis and transferred to a Polyvinylidene fluoride (PVDF)-imprinted membrane at a constant voltage of 80 V. After incubating with sealing solution for 1 h, rabbit anti-HMGA2 (1:3,000, ab97267, Abcam, USA), p-PI3K (1:3,000, ab154589, Abcam, USA), p-AKT (1:1,000, ab38449, Abcam, USA), PI3K (1:1,000, ab151549, Abcam, USA), AKT (1:10,000, ab179463, Abcam, USA), and GAPDH (1:10,000, ab181603, Abcam, USA) were added onto the membrane overnight at 4°C. Goat anti-rabbit IgG (1:20,000, ab97051, Abcam, USA) labeled with HRP was incubated with membranes at 37°C for 2 h. The membrane was developed. The relative expression of protein = Gray value of the protein band/the gray value of the GAPDH band in the same sample.
Enzyme-linked immunosorbent assay (ELISA)
The hippocampal tissues of 6 mice from each group were washed with normal saline, dried with neutral filter paper, and placed in a homogenizer. The 10% tissue homogenate buffer was prepared by mixing 0.25 mol/L sucrose and 0.01 mol/L Tris with homogenized tissue. The buffer was centrifuged at 10,000 g at 4°C for 30 min. The supernatant was then collected. The levels of tumor necrosis factor-α (TNF-α, ab208348, Abcam, USA), interleukin-6 (IL-6, ab203360, Abcam, USA), nitric oxide (NO, ab272517, Abcam, USA) and interleukin-1 beta (IL-1β, ab197742, Abcam, USA) in the brain tissue of each group were measured. According to the operation instructions of the ELISA kit, the optical density value of each sample was detected at 450 nm by an enzyme marker (BioTek Synergy 2).
Detection of oxidative stress damage index
The supernatant of the 10% tissue homogenate of 3 mice from each group was prepared as described above. The operation was performed in strict accordance with the kit scheme. Colorimetry was conducted to detect the level of monoamine oxidase (MAO), malondialdehyde (MDA), glutathione peroxidase (GSH-Px), superoxide dismutase (SOD), adenosine triphosphate (ATP), and total antioxidant capacity (TAOC). All the kits were purchased from Shanghai Enzyme-linked Biotechnology Co., Ltd., Shanghai, China.
Statistical analysis
Data analysis and processing were carried out through SPSS version 21.0 (SPSS Inc., Chicago, IL, USA) software and all measurement data were in normal distribution and were represented as mean ± standard deviation (x̅ ± sd). For the comparison between two independent groups, t test was used and P<0.05 indicated a significant difference. For comparison among multiple groups, one-way ANOVA combined with post hoc Bonferroni test was used f, and P<0.017 indicated a significant difference.
Results
miR-132 targeted HMGA2 and downregulation of the expression
The bioinformatic analysis website screened out that there may be a binding site between miR-132 and HMGA2 (Figure 1A). The dual-luciferase reporter system was used to verify the relationship between miR-132 and HMGA2 (Figure 1B). The results showed that there was no significant difference in the HMGA2 mut fluorescence value between the miR-132 group and the NC group. However, the fluorescence value of HMGA2 wt in the miR-132 group was significantly decreased versus the NC group (P<0.05).
Figure 1.
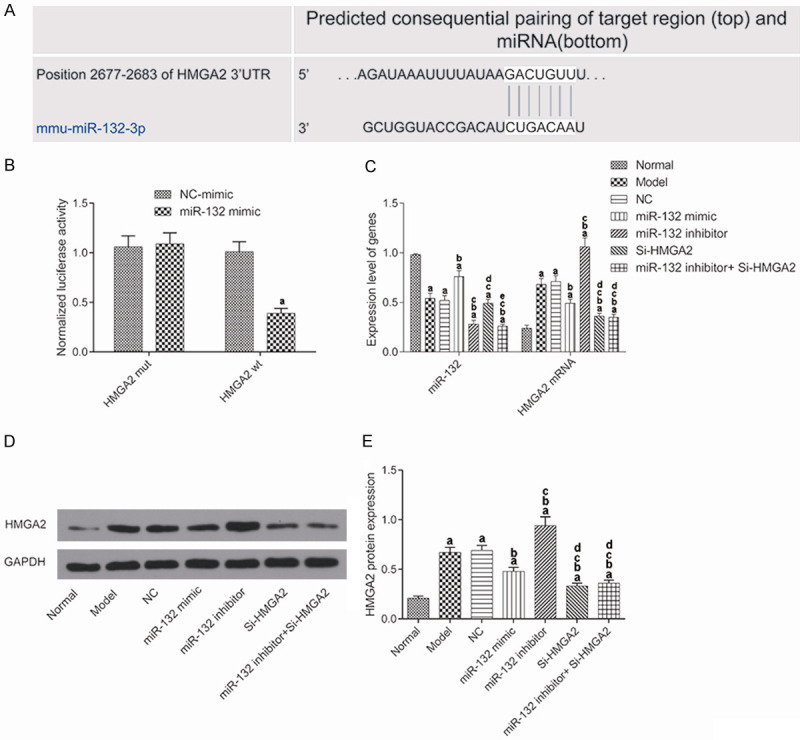
miR-132 targeted HMGA2 and downregulated its expression. (A) Predicted binding sites between miR-132 and HMGA2 from a bioinformatic analysis website; (B) The binding relationship between miR-132 and HMGA2 confirmed by dual luciferase reporting system. aP<0.05, compared with negative control mimic group. (C) Expression of miR-132 and HMGA2 in the brain tissues detected by qRT-PCR; (D) Protein expression of HMGA2 in the brain tissues detected by western blot; (E) HMGA2 protein expression. Both in (C and E), aP<0.017, compared with normal group. bP<0.017, compared with model group. cP<0.017, compared with miR-132 mimic group. dP<0.017, compared with miR-132 inhibitor group. eP<0.017, compared with Si-HMGA2 group.
To further confirm the regulatory relationship between miR-132 and HMGA2, the expression of miR-132 and HMGA2 in the hippocampal tissue of each group was detected by qRT-PCR (Figure 1C) and WB (Figure 1D and 1E). The results showed that compared with the normal mice, AD mice had significantly lower expression of miR-132 but higher expression of HMGA2 (P<0.017). In the miR-132 mimic group, AD mice had significantly increased miR-132 expression but significantly decreased HMGA2 expression, while miR-132 inhibitor had an opposite effect. In the si-HMGA2 group, AD mice had no significantly changed expression of miR-132, but their HMGA2 expression was significantly decreased (P<0.017). In the miR-132 inhibitor + Si-HMGA2 group, the expression of miR-132 and HMGA2 was both significantly decreased (P<0.017). These results suggested that miR-132 could negatively regulate the expression of HMGA2.
Learning and memory ability of the mice
The learning and memory ability of the mice in each group was measured by the Y-maze (Figure 2). The spontaneous alternating response rate of the model group was significantly decreased when compared to that of the normal group (P<0.017). Compared with AD model group, the miR-132 mimic group and the Si-HMGA2 group showed a significant increase in the spontaneous alternating response rate (P<0.017), while the miR-132 inhibitor group obtained a lower rate (P<0.017). The additional use of Si-HMGA2 could reverse the low rate caused by miR-132 inhibitor.
Figure 2.
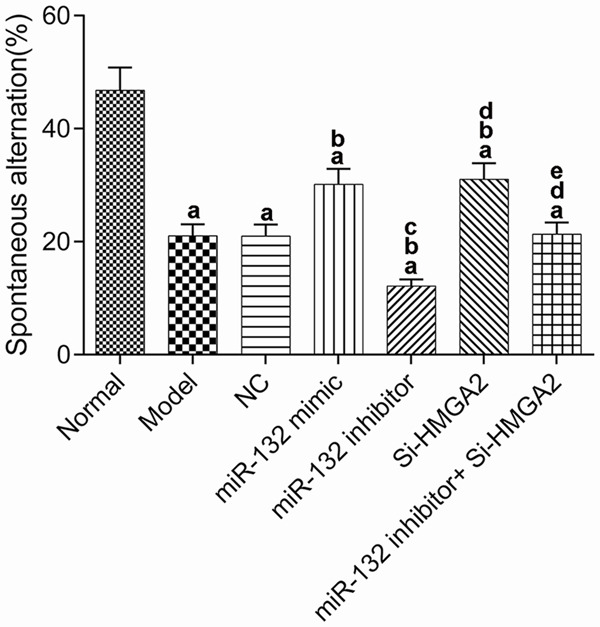
Learning and memory ability of mice detected by Y maze. aP<0.017, compared with normal group. bP<0.017, compared with model group. cP<0.017, compared with miR-132 mimic group. dP<0.017, compared with miR-132 inhibitor group. eP<0.017, compared with Si-HMGA2 group.
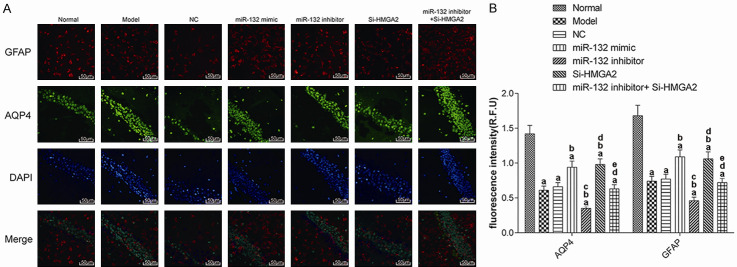
Brain injury in mice
To explore the impact of injury to the hippocampus in mice from each group, we detected the expression of AQP4 and GFAP in hippocampus tissue by immunofluorescence (Figure 3). Compared to the normal group, the fluorescence intensity and breadth of AQP4 and GFAP in the model group decreased in varying degrees (P<0.017). Compared with AD model group, the intensity and breadth of AQP4 and GFAP fluorescence in the miR-132 mimic group and the Si-HMGA2 group were increased significantly (P<0.017), while in the miR-132 inhibitor group, it decreased significantly (P<0.017). The additional use of Si-HMGA2 could reverse the effect of miR-132 inhibitor. These results suggest that miR-132 can promote the activation of glial cells by targeting and negatively regulating the expression of HMGA2, thereby protecting the brains of mice with AD.
Figure 3.
The expression of AQP4 and GFAP (200×). A: Immunofluorescence; B: Fluorescence intensity. aP<0.017, compared with normal group. bP<0.017, compared with model group. cP<0.017, compared with miR-132 mimic group. dP<0.017, compared with miR-132 inhibitor group. eP<0.017, compared with Si-HMGA2 group. AQP4: Aquaporin 4; GFAP: glial fibrillary acidic protein.
Changes in the expression of inflammatory factors in mice
TNF-α, IL-6, NO, and IL-1β levels in the hippocampus of mice from each group were measured using ELISA (Figure 4). The levels of TNF-α, IL-6, NO, and IL-1β in the model group were significantly higher than those in the normal group (P<0.017). Compared with the model group, the levels of TNF-α, IL-6, NO and IL-1β were decreased significantly in the miR-132 mimic group and the Si-HMGA2 group (P<0.017). Conversely, these indexes were increased significantly in the miR-132 inhibitor group (P<0.017). The additional use of Si-HMGA2 could reverse the effects of miR-132 inhibitor. These results suggest that miR-132 can inhibit the inflammatory response in the hippocampus of AD mice by inhibition of HMGA2 expression.
Figure 4.
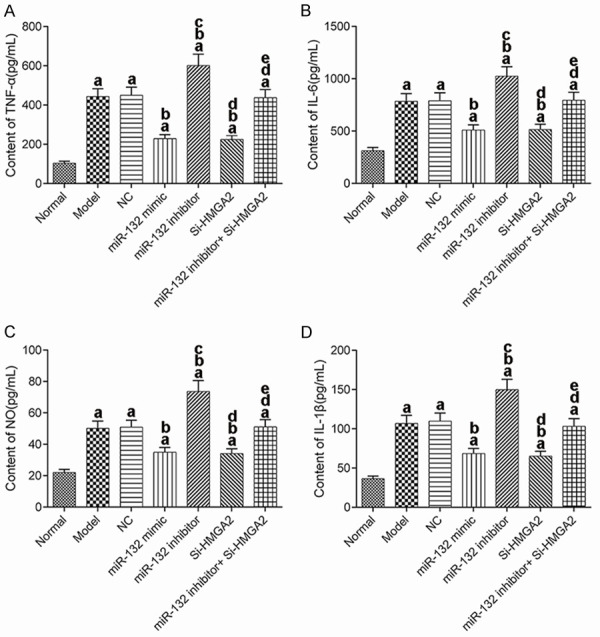
Contents of inflammatory factors in hippocampal tissue of mice. A: TNF-α; B: IL-6; C: NO; D: IL-1β. aP<0.017, compared with normal group. bP<0.017, compared with model group. cP<0.017, compared with miR-132 mimic group. dP<0.017, compared with miR-132 inhibitor group. eP<0.017, compared with Si-HMGA2 group. TNF-α: tumor necrosis factor-α; IL: interleukin; NO: nitric oxide.
Oxidative stress injury in mice
We detected the levels of MAO, MDA, GSH-Px, SOD, ATP, and T-AOC in the hippocampus of the mice (Figure 5). Compared with the normal group, the levels of MAO and MDA in the model groups were significantly increased, while the levels of GSH-Px, SOD, ATP, and T-AOC significantly decreased (P<0.017). Compared with the AD model group, the levels of MAO and MDA in the miR-132 mimic group and the Si-HMGA2 group decreased significantly, while the levels of GSH-Px, SOD, ATP and T-AOC increased significantly (P<0.017). These indexes showed an opposite trend in the miR-132 inhibitor group (P<0.017). The additional use of Si-HMGA2 could reverse the effects of miR-132 inhibitor. These results suggest that miR-132 can inhibit oxidative stress injury in hippocampus of AD mice by targeting and negatively regulating the expression of HMGA2.
Figure 5.
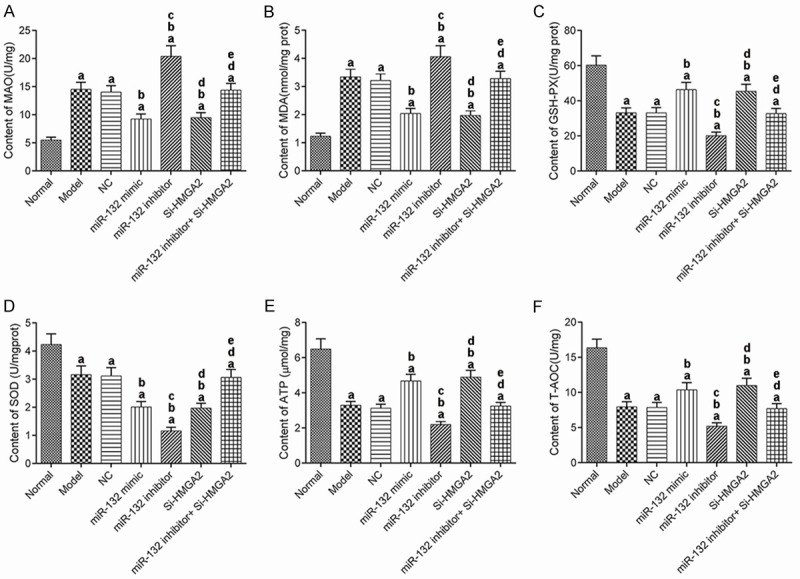
The contents of indices for oxidative stress injury in mice. A: MAO; B: MDA; C: GSH-Px; D: SOD; E: ATP; F: T-AOC. aP<0.017, compared with normal group. bP<0.017, compared with model group. cP<0.017, compared with miR-132 mimic group. dP<0.017, compared with miR-132 inhibitor group. eP<0.017, compared with Si-HMGA2 group. MAO: monoamine oxidase; MDA: malondialdehyde; GSH-Px: glutathione peroxidase; SOD: superoxide dismutase; ATP: adenosine triphosphate; T-AOC: total antioxidant capacity.
Changes in expression of PI3K/AKT pathway in mice hippocampus
WB was used to detect the expression of p-PI3K/PI3K and p-AKT/AKT in the hippocampal tissue from mice in each group (Figure 6). The expressions of p-PI3K/PI3K and p-AKT/AKT in the model group were significantly lower than those in the normal group (P<0.017). Compared with AD model group, the expression of p-PI3K/PI3K and p-AKT/AKT in the miR-132 mimic group and the Si-HMGA2 group was significantly higher, which was significantly lower than that in the miR-132 inhibitor group (P<0.017). This suggested that the expression of PI3K/AKT pathway was significantly decreased in AD mice. Moreover, both miRNA-132 and HMGA2 can effectively regulate the expression of the PI3K/AKT pathway.
Figure 6.
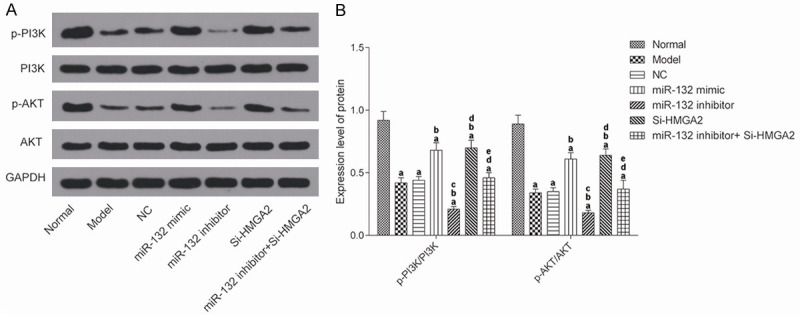
The expression of PI3K/AKT pathway in the hippocampus of mice by western blot detection. A: Protein blot; B: The results of protein expression. aP<0.017, compared with normal group. bP<0.017, compared with model group. cP<0.017, compared with miR-132 mimic group. dP<0.017, compared with miR-132 inhibitor group. eP<0.017, compared with Si-HMGA2 group.
Discussion
There is presently no treatment available for AD [24,25]. Brain damage may be one of the causes of the occurrence and development of AD. Therefore, exploring the protective mechanisms of the brain tissue is of great significance for the prevention and treatment of AD [26-28].
In normal mice, the level of expression of HMGA2 is very low. However, it is significantly upregulated in several malignant tumors, ischemic strokes, and other diseases [29]. Studies have shown that HMGA2 is involved in the senescence of bone marrow mesenchymal stem cells in aged rats. It can also inhibit phosphorylation of the PI3K/AKT pathway. Activation of the PI3K/AKT pathway has a protective effect on neurons, which can improve AD [30]. After detecting the expression of HMGA2 in the brain tissue, we found that the expression of HMGA2 mRNA and protein in the brain tissue of mice with AD was significantly higher than that of normal mice. Tang et al. also found that HMGA2 expression was abnormal in AD model mice, which is consistent with our results [31].
Therefore, we injected Si-HMGA2 vector into mouse models of AD and tested the learning and memory ability of mice by the Y-maze test to further explore the effect of HMGA2 on AD in mice. We detected the expression of glial cell markers AQP4 and GFAP to observe the brain injury of mice and detected the levels of indicators related to inflammation and oxidative stress reaction in the hippocampal tissues of mice. Our results indicate that AD mice treated by silencing HMGA2 had increased expression of the PI3K/AKT signal pathway, improved learning and memory ability, alleviated brain injury, and decreased inflammatory and oxidative stress reactions. All these results indicated that HMGA2 may have a negative role in mice with AD.
A previous study reported that overexpression of miR-132 in primary hippocampal neuronal culture had a strong protective effect on ischemia-induced neuronal death, which be a new therapeutic target for improving neurodegeneration and cognitive deficits associated with ischemic stroke [32]. In the bioinformatic website, we found that there is a binding site between miR-132 and HMGA2, which had been verified by double-luciferase reporter assay, thus we further speculate that miR-132 may be an upstream regulatory element of HMGA2 and regulate its expression. Our results showed that AD model mice had significantly lower expression of miR-132 than the normal ones. Moreover, up-regulation of miR-132 in AD mice could inhibit their expression of HMGA2, inhibit the inflammatory and oxidative stress response in the brain, inhibit brain injury, and improve learning and memory ability. Therefore, miR-132 had similar treatment effect to Si-HMGA2, indicating that miR-132 up-regulation may play a protective role in brain damage of AD mice by targeting inhibition of HMGA2 expression.
We also noted that there was no significant difference in HMGA2 expression between the Si-HMGA2 group and the miR-132 inhibitor + Si-HMGA2 group. However, learning and memory ability, nerve injury, inflammation, and stress indexes in the miR-132 inhibitor + Si-HMGA2 group were significantly worse than those in the Si-HMGA2 group, and these indicators were also at significantly different levels compared to the model group. At present, we have not been able to explain this phenomenon. We speculated that miR-132 may act on other targets and then affect mice with AD. As the research is in its initial stages, further study including clinical effects and dosage variation is required.
In conclusion, miR-132 can target and inhibit the expression of HMGA2 and upregulate expression of the PI3K/AKT pathway to alleviate brain damage in AD mice, which may be a new treatment target for AD.
Disclosure of conflict of interest
None.
References
- 1.Zenaro E, Piacentino G, Constantin G. The blood-brain barrier in Alzheimer’s disease. Neurobiol Dis. 2017;107:41–56. doi: 10.1016/j.nbd.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones DT, Graff-Radford J, Lowe VJ, Wiste HJ, Gunter JL, Senjem ML, Botha H, Kantarci K, Boeve BF, Knopman DS, Petersen RC, Jack CR Jr. Tau, amyloid, and cascading network failure across the Alzheimer’s disease spectrum. Cortex. 2017;97:143–159. doi: 10.1016/j.cortex.2017.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cummings J, Lee G, Ritter A, Sabbagh M, Zhong K. Alzheimer’s disease drug development pipeline: 2017. Alzheimers Dement (N Y) 2017;3:367–384. doi: 10.1016/j.trci.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen CT, Chang CC, Chang WN, Tsai NW, Huang CC, Chang YT, Wang HC, Kung CT, Su YJ, Lin WC, Cheng BC, Su CM, Hsiao SY, Hsu CW, Lu CH. Neuropsychiatric symptoms in Alzheimer’s disease: associations with caregiver burden and treatment outcomes. QJM. 2017;110:565–570. doi: 10.1093/qjmed/hcx077. [DOI] [PubMed] [Google Scholar]
- 5.Sarraf S, Tofighi G. Deep learning-based pipeline to recognize Alzheimer’s disease using fMRI data. 2016 Future Technologies Conference (FTC) 2016;31:1–5. [Google Scholar]
- 6.Lourenço CF, Ledo A, Barbosa RM, Laranjinha J. Neurovascular uncoupling in the triple transgenic model of Alzheimer’s disease: impaired cerebral blood flow response to neuronal-derived nitric oxide signaling. Exp Neurol. 2017;291:36–43. doi: 10.1016/j.expneurol.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 7.Strell C, Norberg KJ, Mezheyeuski A, Schnittert J, Kuninty PR, Moro CF, Paulsson J, Schultz NA, Calatayud D, Löhr JM, Frings O, Verbeke CS, Heuchel RL, Prakash J, Johansen JS, Östman A. Stroma-regulated HMGA2 is an independent prognostic marker in PDAC and AAC. Br J Cancer. 2017;117:65–77. doi: 10.1038/bjc.2017.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun J, Sun B, Sun R, Zhu D, Zhao X, Zhang Y, Dong X, Che N, Li J, Liu F, Zhao N, Wang Y, Zhang D. HMGA2 promotes vasculogenic mimicry and tumor aggressiveness by upregulating Twist1 in gastric carcinoma. Sci Rep. 2017;7:2229. doi: 10.1038/s41598-017-02494-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong J, Wang R, Ren G, Li X, Wang J, Sun Y, Liang J, Nie Y, Wu K, Feng B, Shang Y, Fan D. HMGA2-FOXL2 axis regulates metastases and epithelial-to-mesenchymal transition of chemoresistant gastric cancer. Clin Cancer Res. 2017;23:3461–3473. doi: 10.1158/1078-0432.CCR-16-2180. [DOI] [PubMed] [Google Scholar]
- 10.Li W, Wang Z, Zha L, Kong D, Liao G, Li H. HMGA2 regulates epithelial-mesenchymal transition and the acquisition of tumor stem cell properties through TWIST1 in gastric cancer. Oncol Rep. 2017;37:185–192. doi: 10.3892/or.2016.5255. [DOI] [PubMed] [Google Scholar]
- 11.Li X, Wang S, Li Z, Long X, Guo Z, Zhang G, Zu J, Chen Y, Wen L. The lncRNA NEAT1 facilitates cell growth and invasion via the miR-211/HMGA2 axis in breast cancer. Int J Biol Macromol. 2017;105:346–353. doi: 10.1016/j.ijbiomac.2017.07.053. [DOI] [PubMed] [Google Scholar]
- 12.Tan L, Wei X, Zheng L, Zeng J, Liu H, Yang S, Tan H. Amplified HMGA2 promotes cell growth by regulating Akt pathway in AML. J Cancer Res Clin Oncol. 2016;142:389–399. doi: 10.1007/s00432-015-2036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleemann M, Schneider H, Unger K, Bereuther J, Fischer S, Sander P, Marion Schneider E, Fischer-Posovszky P, Riedel CU, Handrick R, Otte K. Induction of apoptosis in ovarian cancer cells by miR-493-3p directly targeting AKT2, STK38L, HMGA2, ETS1 and E2F5. Cell Mol Life Sci. 2019;76:539–559. doi: 10.1007/s00018-018-2958-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie J, Ubango J, Ban Y, Chakravarti D, Kim JJ, Wei JJ. Comparative analysis of AKT and the related biomarkers in uterine leiomyomas with MED12, HMGA2, and FH mutations. Genes Chromosomes Cancer. 2018;57:485–494. doi: 10.1002/gcc.22643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen KF, Sakamoto K, Wayman GA, Impey S, Obrietan K. Transgenic miR132 alters neuronal spine density and impairs novel object recognition memory. PLoS One. 2010;5:e15497. doi: 10.1371/journal.pone.0015497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ucar A, Vafaizadeh V, Jarry H, Fiedler J, Klemmt PA, Thum T, Groner B, Chowdhury K. miR-212 and miR-132 are required for epithelial stromal interactions necessary for mouse mammary gland development. Nat Genet. 2010;42:1101–1108. doi: 10.1038/ng.709. [DOI] [PubMed] [Google Scholar]
- 17.Luikart BW, Bensen AL, Washburn EK, Perederiy JV, Su KG, Li Y, Kernie SG, Parada LF, Westbrook GL. miR-132 mediates the integration of newborn neurons into the adult dentate gyrus. PLoS One. 2011;6:e19077. doi: 10.1371/journal.pone.0019077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jimenez-Mateos EM, Bray I, Sanz-Rodriguez A, Engel T, McKiernan RC, Mouri G, Tanaka K, Sano T, Saugstad JA, Simon RP, Stallings RL, Henshall DC. miRNA expression profile after status epilepticus and hippocampal neuroprotection by targeting miR-132. Am J Pathol. 2011;179:2519–2532. doi: 10.1016/j.ajpath.2011.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma Y, Ma B, Shang Y, Yin Q, Wang D, Xu S, Hong Y, Hou X, Liu X. Flavonoid-rich ethanol extract from the leaves of diospyros kaki attenuates D-galactose-induced oxidative stress and neuroinflammation-mediated brain aging in mice. Oxid Med Cell Longev. 2018;2018:8938207. doi: 10.1155/2018/8938207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui B, Zhang S, Wang Y, Guo Y. Farrerol attenuates β-amyloid-induced oxidative stress and inflammation through Nrf2/Keap1 pathway in a microglia cell line. Biomed Pharmacother. 2019;109:112–119. doi: 10.1016/j.biopha.2018.10.053. [DOI] [PubMed] [Google Scholar]
- 21.Tang Y, Bao JS, Su JH, Huang W. MicroRNA-139 modulates Alzheimer’s-associated pathogenesis in SAMP8 mice by targeting cannabinoid receptor type 2. Genet Mol Res. 2017;16:gmr16019166. doi: 10.4238/gmr16019166. [DOI] [PubMed] [Google Scholar]
- 22.Grazyna R, Craig AS. Astrocyte pathology in major depressive disorder: insights from human postmortem brain tissue. Curr Drug Targets. 2013;14:1225–1236. doi: 10.2174/13894501113149990156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lepelletier FX, Mann DM, Robinson AC, Pinteaux E, Boutin H. Early changes in extracellular matrix in Alzheimer’s disease. Neuropathol Appl Neurobiol. 2017;43:167–182. doi: 10.1111/nan.12295. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Liu M, Le A, Gao Y, Shen D. Alzheimer’s disease diagnosis using landmark-based features from longitudinal structural MR images. IEEE J Biomed Health Inform. 2017;21:1607–1616. doi: 10.1109/JBHI.2017.2704614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sims R, van der Lee SJ, Naj AC, Bellenguez C, Badarinarayan N, Jakobsdottir J, Kunkle BW, Boland A, Raybould R, Bis JC, Martin ER, Grenier-Boley B, Heilmann-Heimbach S, Chouraki V, Kuzma AB, Sleegers K, Vronskaya M, Ruiz A, Graham RR, Olaso R, Hoffmann P, Grove ML, Vardarajan BN, Hiltunen M, Nöthen MM, White CC, Hamilton-Nelson KL, Epelbaum J, Maier W, Choi SH, Beecham GW, Dulary C, Herms S, Smith AV, Funk CC, Derbois C, Forstner AJ, Ahmad S, Li H, Bacq D, Harold D, Satizabal CL, Valladares O, Squassina A, Thomas R, Brody JA, Qu L, Sánchez-Juan P, Morgan T, Wolters FJ, Zhao Y, Garcia FS, Denning N, Fornage M, Malamon J, Naranjo MCD, Majounie E, Mosley TH, Dombroski B, Wallon D, Lupton MK, Dupuis J, Whitehead P, Fratiglioni L, Medway C, Jian X, Mukherjee S, Keller L, Brown K, Lin H, Cantwell LB, Panza F, McGuinness B, Moreno-Grau S, Burgess JD, Solfrizzi V, Proitsi P, Adams HH, Allen M, Seripa D, Pastor P, Cupples LA, Price ND, Hannequin D, Frank-García A, Levy D, Chakrabarty P, Caffarra P, Giegling I, Beiser AS, Giedraitis V, Hampel H, Garcia ME, Wang X, Lannfelt L, Mecocci P, Eiriksdottir G, Crane PK, Pasquier F, Boccardi V, Henández I, Barber RC, Scherer M, Tarraga L, Adams PM, Leber M, Chen Y, Albert MS, Riedel-Heller S, Emilsson V, Beekly D, Braae A, Schmidt R, Blacker D, Masullo C, Schmidt H, Doody RS, Spalletta G, Longstreth WT Jr, Fairchild TJ, Bossù P, Lopez OL, Frosch MP, Sacchinelli E, Ghetti B, Yang Q, Huebinger RM, Jessen F, Li S, Kamboh MI, Morris J, Sotolongo-Grau O, Katz MJ, Corcoran C, Dunstan M, Braddel A, Thomas C, Meggy A, Marshall R, Gerrish A, Chapman J, Aguilar M, Taylor S, Hill M, Fairén MD, Hodges A, Vellas B, Soininen H, Kloszewska I, Daniilidou M, Uphill J, Patel Y, Hughes JT, Lord J, Turton J, Hartmann AM, Cecchetti R, Fenoglio C, Serpente M, Arcaro M, Caltagirone C, Orfei MD, Ciaramella A, Pichler S, Mayhaus M, Gu W, Lleó A, Fortea J, Blesa R, Barber IS, Brookes K, Cupidi C, Maletta RG, Carrell D, Sorbi S, Moebus S, Urbano M, Pilotto A, Kornhuber J, Bosco P, Todd S, Craig D, Johnston J, Gill M, Lawlor B, Lynch A, Fox NC, Hardy J ARUK Consortium; Albin RL, Apostolova LG, Arnold SE, Asthana S, Atwood CS, Baldwin CT, Barnes LL, Barral S, Beach TG, Becker JT, Bigio EH, Bird TD, Boeve BF, Bowen JD, Boxer A, Burke JR, Burns JM, Buxbaum JD, Cairns NJ, Cao C, Carlson CS, Carlsson CM, Carney RM, Carrasquillo MM, Carroll SL, Diaz CC, Chui HC, Clark DG, Cribbs DH, Crocco EA, DeCarli C, Dick M, Duara R, Evans DA, Faber KM, Fallon KB, Fardo DW, Farlow MR, Ferris S, Foroud TM, Galasko DR, Gearing M, Geschwind DH, Gilbert JR, Graff-Radford NR, Green RC, Growdon JH, Hamilton RL, Harrell LE, Honig LS, Huentelman MJ, Hulette CM, Hyman BT, Jarvik GP, Abner E, Jin LW, Jun G, Karydas A, Kaye JA, Kim R, Kowall NW, Kramer JH, LaFerla FM, Lah JJ, Leverenz JB, Levey AI, Li G, Lieberman AP, Lunetta KL, Lyketsos CG, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Morris JC, Murrell JR, Myers AJ, O’Bryant S, Olichney JM, Pankratz VS, Parisi JE, Paulson HL, Perry W, Peskind E, Pierce A, Poon WW, Potter H, Quinn JF, Raj A, Raskind M, Reisberg B, Reitz C, Ringman JM, Roberson ED, Rogaeva E, Rosen HJ, Rosenberg RN, Sager MA, Saykin AJ, Schneider JA, Schneider LS, Seeley WW, Smith AG, Sonnen JA, Spina S, Stern RA, Swerdlow RH, Tanzi RE, Thornton-Wells TA, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Van Eldik LJ, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Wilhelmsen KC, Williamson J, Wingo TS, Woltjer RL, Wright CB, Yu CE, Yu L, Garzia F, Golamaully F, Septier G, Engelborghs S, Vandenberghe R, De Deyn PP, Fernadez CM, Benito YA, Thonberg H, Forsell C, Lilius L, Kinhult-Stählbom A, Kilander L, Brundin R, Concari L, Helisalmi S, Koivisto AM, Haapasalo A, Dermecourt V, Fievet N, Hanon O, Dufouil C, Brice A, Ritchie K, Dubois B, Himali JJ, Keene CD, Tschanz J, Fitzpatrick AL, Kukull WA, Norton M, Aspelund T, Larson EB, Munger R, Rotter JI, Lipton RB, Bullido MJ, Hofman A, Montine TJ, Coto E, Boerwinkle E, Petersen RC, Alvarez V, Rivadeneira F, Reiman EM, Gallo M, O’Donnell CJ, Reisch JS, Bruni AC, Royall DR, Dichgans M, Sano M, Galimberti D, St George-Hyslop P, Scarpini E, Tsuang DW, Mancuso M, Bonuccelli U, Winslow AR, Daniele A, Wu CK GERAD/PERADES, CHARGE, ADGC, EADI. Peters O, Nacmias B, Riemenschneider M, Heun R, Brayne C, Rubinsztein DC, Bras J, Guerreiro R, Al-Chalabi A, Shaw CE, Collinge J, Mann D, Tsolaki M, Clarimón J, Sussams R, Lovestone S, O’Donovan MC, Owen MJ, Behrens TW, Mead S, Goate AM, Uitterlinden AG, Holmes C, Cruchaga C, Ingelsson M, Bennett DA, Powell J, Golde TE, Graff C, De Jager PL, Morgan K, Ertekin-Taner N, Combarros O, Psaty BM, Passmore P, Younkin SG, Berr C, Gudnason V, Rujescu D, Dickson DW, Dartigues JF, DeStefano AL, Ortega-Cubero S, Hakonarson H, Campion D, Boada M, Kauwe JK, Farrer LA, Van Broeckhoven C, Ikram MA, Jones L, Haines JL, Tzourio C, Launer LJ, Escott-Price V, Mayeux R, Deleuze JF, Amin N, Holmans PA, Pericak-Vance MA, Amouyel P, van Duijn CM, Ramirez A, Wang LS, Lambert JC, Seshadri S, Williams J, Schellenberg GD. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat Genet. 2017;49:1373–1384. doi: 10.1038/ng.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alzheimer’s Association Calcium Hypothesis Workgroup. Calcium hypothesis of Alzheimer’s disease and brain aging: a framework for integrating new evidence into a comprehensive theory of pathogenesis. Alzheimers Dement. 2017;13:178–182. e17. doi: 10.1016/j.jalz.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 27.Bagyinszky E, Giau VV, Shim K, Suk K, An SSA, Kim S. Role of inflammatory molecules in the Alzheimer’s disease progression and diagnosis. J Neurol Sci. 2017;376:242–254. doi: 10.1016/j.jns.2017.03.031. [DOI] [PubMed] [Google Scholar]
- 28.Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, Harvey D, Jack CR Jr, Jagust W, Morris JC, Petersen RC, Saykin AJ, Shaw LM, Toga AW, Trojanowski JQ. Recent publications from the Alzheimer’s disease neuroimaging initiative: reviewing progress toward improved AD clinical trials. Alzheimers Dement. 2017;13:e1–e85. doi: 10.1016/j.jalz.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fedele M, Battista S, Kenyon L, Baldassarre G, Fidanza V, Klein-Szanto AJ, Parlow AF, Visone R, Pierantoni GM, Outwater E, Santoro M, Croce CM, Fusco A. Overexpression of the HMGA2 gene in transgenic mice leads to the onset of pituitary adenomas. Oncogene. 2002;21:3190–3198. doi: 10.1038/sj.onc.1205428. [DOI] [PubMed] [Google Scholar]
- 30.Cui H, Song R, Wu J, Wang W, Chen X, Yin J. MicroRNA-337 regulates the PI3K/AKT and Wnt/β-catenin signaling pathways to inhibit hepatocellular carcinoma progression by targeting high-mobility group AT-hook 2. Am J Cancer Res. 2018;8:405–421. [PMC free article] [PubMed] [Google Scholar]
- 31.Tang L, Liu L, Li G, Jiang P, Wang Y, Li J. Expression profiles of long noncoding RNAs in intranasal LPS-mediated Alzheimer’s disease model in mice. Biomed Res Int. 2019;2019:9642589. doi: 10.1155/2019/9642589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hwang JY, Kaneko N, Noh KM, Pontarelli F, Zukin RS. The gene silencing transcription factor REST represses miR-132 expression in hippocampal neurons destined to die. J Mol Biol. 2014;426:3454–3466. doi: 10.1016/j.jmb.2014.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]