Abstract
Background: The new S100 protein family member S100A16 is functionally expressed in various cancers. This study explored the prognostic value and potential role of S100A16 in pancreatic cancer (PC). Methods: RNA-seq and clinical data were obtained from The Cancer Genome Atlas-Pancreatic Adenocarcinoma (TCGA-PAAD) dataset to compare the expression level of S100A16 between groups. The genes co-expressed with S100A16 in TCGA-PAAD were analyzed using cBioPortal. Gene Ontology and Kyoto Encyclopedia of Genes and genomes enrichment analyses were also performed on these genes. Pathways related to S100A16 expression dysregulation were explored using gene set enrichment analysis. The Tumor Immune Estimation Resource was used to analyze the correlation between S100A16 and infiltrating immune cells. The Kaplan-Meier method and Cox analyses were used to assess the prognostic significance of S100A16 for PC. Results: The S100A16 expression level was high in PC and increased with the degree of malignancy. The S100A16 functions in PC were mainly enriched in the immune modules, but negatively correlated with the immune activity (T-cell, cytokine, immune, co-receptor, signaling adaptor, cell adhesion molecule, chemokine, and JAK/STAT signaling) and infiltration level (T cells and macrophages). The strongest negative correlation was observed between the expression of CD8+ T cells and S100A16. Furthermore, high S100A16 expression also indicated worse overall survival and, therefore, worse prognosis of PC. Conclusion: S100A16 is a potential independent prognostic marker and immunotherapy target for PC. Mechanistically, S100A16 potentially affects prognosis by extensive immunosuppression, including the inhibition of the anti-tumor immune response of CD8+ T cells.
Keywords: S100A16, pancreatic cancer, prognostic marker, immune infiltration, immunosuppression, CD8+ T cells
Introduction
Pancreatic cancer (PC) is a devastating malignancy with an overall 5-year survival of less than 10%. The most common form of PC is pancreatic ductal adenocarcinoma (PDAC) accounting for more than 90% of all cases. PC has an insidious onset and there is currently a lack of effective tools for early diagnosis. Once diagnosed, radical resection remains the only treatment. Unfortunately, only 10-20% of patients are eligible for surgery, and even those diagnosed with localized and resectable tumors has 5-year survival rate of only 20% [1]. Current research on the etiology, detection and treatment of PC is lagging behind other tumors, increasing the need to advance related research for the benefit of patients [2]. In recent years, there have been some studies using high-throughput sequencing technology to analyze the potential molecular mechanisms, and identify new diagnostic markers and therapeutic targets for PC [3]. This also provides an opportunity for analyzing the role of specific genes in the prognosis of PC.
The highly conserved S100 family of proteins has been implicated in various stages of tumor progression. It consists of an EF-hand superfamily of Ca2+-binding proteins with more than 25 members [4]. S100 proteins have a wide range of biological functions and are involved in processes such as proliferation, apoptosis, migration, inflammation, and differentiation [5]. It has been reported that the expression of S100 proteins can be used to assist diagnosis, predict prognosis, guide treatment decisions, and supervise treatment effects [6]. Noteworthy, the expression and function of various S100 proteins vary in different tumor types. For instance, S100 calcium-binding protein A16, also known as S100A16, is a new member of the S100 family originally isolated from an astrocytoma tumor [7]. Furthermore, in lung adenocarcinoma [8], breast cancer [9], and ovarian cancer [10], patients with high expression of S100A16 have poor prognosis, while in colorectal cancer [11] and oral squamous cell carcinoma [12], high expression predicts a better prognosis. However, its prognostic value and role in PC remain to be fully understood. Therefore, in the current study we obtained expression profile for S100A16 gene in PC using The Cancer Genome Atlas (TCGA) database. We further screened genes co-expressed with S100A16 and speculated the related functions and pathways of S100A16. Based on these results, we evaluated the role of S100A16 in immune regulation and analyzed its ability to predict the prognosis of patients with PC.
Materials and methods
Data download
All datasets analyzed in this study are publicly available. The RNA-Seq data (FPKM format) and clinical data (normal = 4, tumor = 178) in the TCGA-Pancreatic Adenocarcinoma (PAAD) project were retrieved and downloaded through the Genomic Data Commons (GDC) Data Portal (https://portal.gdc.cancer.gov). The obtained data collection of the clinically relevant information was accessed on November 30, 2020.
Comparative analysis of S100A16 expression levels among different groups
The expression profile of S100A16 was extracted. The expression levels under different grouping situations (normal tissue vs. tumor tissue; ductal adenocarcinoma vs. others; pathological T1/2 vs. T3/4; histological grade 1 vs. grade 2/3/4; and clinical stage I vs. stage II/III/IV) were compared using a Student’s t-test (two-tailed) performed by SPSS 25.0 (IBM Corporation, Chicago, Illinois, USA) (P < 0.05).
Functional analysis of S100A16 co-expressed genes
The cBioPortal platform (http://www.cbioportal.org/) was utilized to analyze the co-expression of S100A16 and other genes in PC. Genes with |R| ≥ 0.6 and P < 0.05 were designated as co-expressed genes of S100A16. The clusterProfiler R package [13] was used to conduct Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses on the genes co-expressed with S100A16 to infer the associated functions and pathways of S100A16 in PC.
Gene set enrichment analysis (GSEA) of high and low S100A16-expression groups
Samples in the TCGA-PAAD dataset were classified into high and low expression groups based on their S100A16 expression level. Afterwards, the related pathways in the groups were identified by GSEA software 4.0 (https://www.gsea-msigdb.org/gsea/index.jsp).
Correlation analysis between S100A16 and infiltrating immune cells
The ESTIMATE R package (https://bioinformatics.mdanderson.org/estimate/rpackage.html) was used to calculate the immune score of each sample, and all samples were then classified into a high-immune or low-immune group according to their respective immune scores. The S100A16 expression was then compared between these two immune groups. Spearman rank correlation was performed to assess the association between S100A16 and immune markers with the Tumor Immune Estimation Resource (TIMER) (http://cistrome.shinyapps.io/timer/).
Prognostic valuation of S100A16 for PC
Univariate and multivariate Cox analysis was performed using SPSS 25.0 (Chicago, Il, USA) to evaluate the prognostic values of S100A16 and other clinical parameters for PC. The Kaplan-Meier plotter (KM plotter) (http://kmplot.com/analysis/) was applied to analyze the survival status of the high and low S100A16-expression groups.
Statistical analysis
The clinical data and S100A16 expression information were downloaded from the TCGA database, and then analyzed with R studio (https://cran.r-project.org/;version 1.2.1335) and SPSS 25.0. The correlation between clinicopathological characteristics and S100A16 expression was analyzed using a Student’s t-test (two-tailed). Univariate Cox analysis was used to evaluate the risk factors related to the survival of pancreatic cancer patients. Subsequently, clinical parameters of P < 0.05 were used in the multivariate Cox analysis to assess prognostic factors. The Kaplan-Meier method was used to analyze the difference in overall survival (OS) between the S100A16 high and low expression groups. Spearman analysis was used to analyze the correlation between S100A16 expression level and immune infiltration. All statistical tests were two-sided tests, and P < 0.05 was regarded as statistically significant.
Results
S100A16 was highly expressed in PC and positively correlated with the degree of malignancy
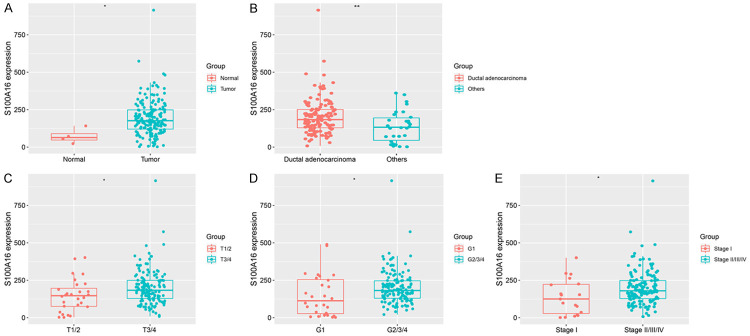
We evaluated mRNA expression of S100A16 and found differences between PC samples and at different clinical stages. Compared with normal tissues, the expression level of S100A16 in pancreatic tumor tissues was significantly upregulated (P < 0.05; Figure 1A). Furthermore, the expression level of S100A16 in PDAC was significantly higher than that in other pancreatic tumors (P < 0.01; Figure 1B). The level of S100A16 expression also significantly increased with increase of malignant degree (P < 0.05 for all; Figure 1C-E).
Figure 1.
Comparison of S100A16 expression between groups in PC. A. Normal tissue vs. tumor. B. Ductal adenocarcinoma vs. others. C. Pathological T1/2 vs. pathological T3/4. D. Histological grade 1 vs. histological grade 2/3/4. E. Stage I vs. stage II/III/IV. *P < 0.05, **P < 0.01. PC, pancreatic cancer.
The functions of S100A16 co-expressed genes were mainly enriched in immune modules
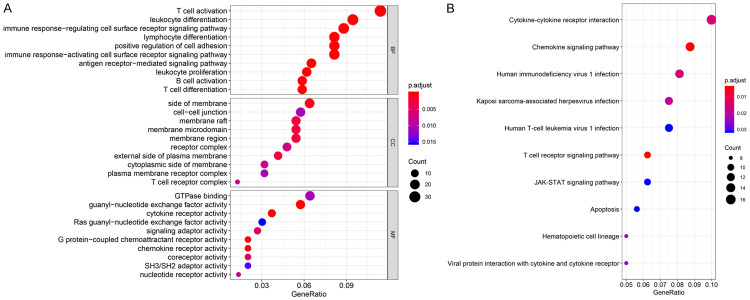
To explore the functions of S100A16 and pathways involved in PC, we screened the genes co-expressed with S100A16 according to |R| ≥ 0.6 and P < 0.05. Three terms were elucidated in GO enrichment analysis-biological processes (BP), cellular components (CC), and molecular functions (MF). The analysis revealed that the most enriched GO-BP were closely related to immune system functions, including T-cell activation, leukocyte differentiation, and the immune response-regulating cell surface receptor signaling pathway, among others. The most enriched GO-CC were related to the cell membrane structure and receptors, including the side of membranes, membrane raft, receptor complex, and others. The most enriched GO-MF were related to the activity of receptors and adaptors, including cytokine receptor activity, chemokine receptor activity, and signaling adaptor activity (all P < 0.05, Figure 2A). KEGG enrichment analysis revealed that the main enrichment pathways were cytokine-cytokine receptor interaction, the chemokine signaling pathway, T-cell receptor signaling pathway, and JAK-STAT signaling pathway (all P < 0.05, Figure 2B).
Figure 2.
GO and KEGG enrichment analyses of S100A16 co-expressed genes. A. Top 10 bubble plot of GO enrichment analysis. B. Top 10 bubble plot of KEGG enrichment analysis. Gene Ratio: the ratio of the number of genes associated with a certain term to the total number of differentially expressed genes. The size and color of the bubbles indicate the corresponding gene counts and P. adjust values, respectively. GO, gene ontology; BP, biological process; CC, cellular component; MF, molecular function; KEGG, Kyoto Encyclopedia of Genes and Genomes.
S100A16 is negatively correlated with immune activity
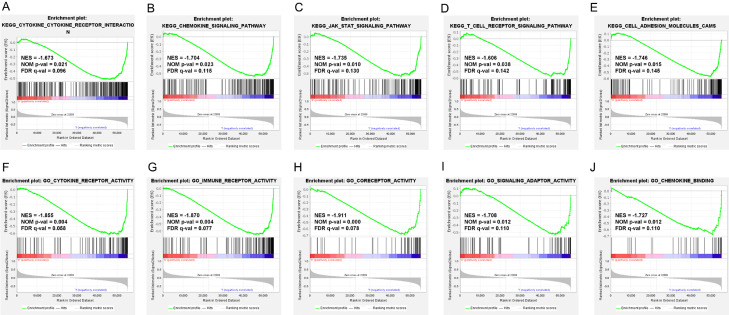
We further explored the potential functions of S100A16 in PC using GSEA. The GO and KEGG pathways were analyzed by GSEA, and differential gene sets were screened according to a false detection rate (FDR) q-value ≤ 0.25, absolute value of normalized enrichment score (|NES|) ≥ 1.0, and nominal (NOM) P-value ≤ 0.05. The results indicated that S100A16 expression was negatively correlated with immune activity (Figure 3A, 3B). When S100A16 was highly expressed, the activity of a variety of cell surface receptors and ligands decreased, including the T-cell receptor, cytokine receptor, immune receptor, coreceptor, and signaling adaptor (Figure 3D, 3F-I). Furthermore, the activity of cell adhesion molecules (CAMs), along with the chemokine signaling pathway and JAK-STAT signaling pathway, also declined (Figure 3C, 3E, 3J). These results together suggested that S100A16 may be involved in inhibiting immune cell recruitment, immune signal transduction, and promoting tumor metastasis.
Figure 3.
GSEA of the high and low S100A16-expression groups in PC. GSEA results showing the Top 5 downregulated KEGG pathways (A-E) and GO classification (F-J) associated with immune activity in the PC samples with high S100A16 expression. NES, normalized enrichment score; NOM p-val, nominal P-value; FDR q-val, false discovery rate q-value; GSEA, gene set enrichment analysis; PC, pancreatic cancer; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
S100A16 is negatively correlated with immune-cell infiltration in PC
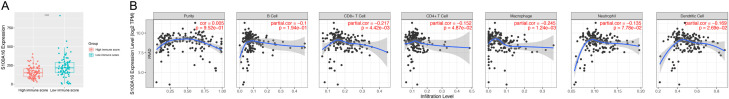
Based on the above analyses, we further explored the relationship between S100A16 and individual immune status in PC. S100A16 expression in the low-immune group was significantly higher than that in the high-immune group (P < 0.0001, Figure 4A), suggesting that S100A16 had an immunosuppressive effect. We also explored the correlation between S100A16 and different infiltrating immune cells, including B cells, T cells, dendritic cells (DCs), macrophages, monocytes, natural killer cells (NK cells), and neutrophils in the TIMER database. Additionally, the relationship of S100A16 and the subsets of T cells and macrophages, such as T-cell exhaustion, follicular helper T cells (Tfh), T helper (Th)1, Th2, Th17, regulatory T (Treg) cells, M1 macrophages, M2 macrophages, and tumor-associated macrophages (TAM), were also investigated. The results demonstrated that almost all immune-cell infiltrations in PC were inversely related to the expression of S100A16 (all P < 0.05), except for B cells and M1 macrophages (Figure 4A; Table 1). And among these immune cells, the strongest negative correlation occurred between the expression of CD8+ T cells and S100A16 (Figure 4B; Table 1).
Figure 4.
S100A16 expression is associated with immune infiltrating cells in PC. A. Immune scores were determined for the samples, which were applied to classify the samples into high-immune or low-immune groups according to the scores. The expression of S100A16 was higher in samples with low-immune scores compared to that in samples with high-immune scores. B. S100A16 expression is significantly negatively correlated with the level of immune-cell infiltration. PC, pancreatic cancer.
Table 1.
Spearman’s correlation analysis between S100A16 and markers of immune cells in PC
| Description | Gene markers | S100A16 | |
|---|---|---|---|
|
| |||
| Cor | P | ||
| B cell | CD19 | -0.1199302 | 0.11081159 |
| CD79A | -0.1327914 | 0.07722656 | |
| CD8+ T cell | CD8A | -0.3943527 | 5.13E-08 |
| CD8B | -0.2975552 | 5.49E-05 | |
| Dendritic cell | HLA-DPA1 | -0.3618189 | 6.96E-07 |
| HLA-DRA | -0.3453255 | 2.35E-06 | |
| HLA-DPB1 | -0.3415427 | 3.08E-06 | |
| NRP1 (BDCA-4) | -0.2836567 | 1.25E-04 | |
| CD1C (BDCA-1) | -0.2488582 | 8.09E-04 | |
| HLA-DQB1 | -0.2182153 | 0.00343139 | |
| ITGAX (CD11c) | -0.1448548 | 0.05370906 | |
| M1 Macrophage | IRF5 | -0.0237307 | 0.75320173 |
| PTGS2 (COX2) | 0.13740492 | 0.06740264 | |
| NOS2 (INOS) | -0.0691574 | 0.35899967 | |
| M2 Macrophage | MS4A4A | -0.3240856 | 1.02E-05 |
| CD163 | -0.2929803 | 7.23E-05 | |
| VSIG4 | -0.2186021 | 0.00337322 | |
| Monocyte | CD115 (CSF1R) | -0.3841369 | 1.20E-07 |
| CD86 | -0.2585095 | 4.94E-04 | |
| Natural killer cell | KIR3DL3 | 0.04545805 | 0.54682486 |
| KIR3DL2 | -0.2373857 | 0.00142008 | |
| KIR3DL1 | -0.2348641 | 0.00160134 | |
| KIR2DL4 | -0.1087693 | 0.14839317 | |
| KIR2DL1 | -0.2152592 | 0.00390682 | |
| KIR2DS4 | -0.1710711 | 0.02242149 | |
| KIR2DL3 | -0.1495663 | 0.04630219 | |
| Neutrophils | CEACAM8 (CD66b) | -0.0941052 | 0.21150061 |
| ITGAM (CD11b) | -0.1907858 | 0.01074319 | |
| CCR7 | -0.1720206 | 0.02167506 | |
| T cell (general) | CD2 | -0.3435207 | 2.67E-06 |
| CD3E | -0.2636532 | 3.77E-04 | |
| CD3D | -0.2023324 | 0.00675873 | |
| T cell exhaustion | HAVCR2 (TIM-3) | -0.2295206 | 0.00205711 |
| CTLA4 | -0.2249975 | 0.00253188 | |
| PDCD1 (PD-1) | -0.215586 | 0.00385149 | |
| LAG3 | -0.2129971 | 0.00430985 | |
| GZMB | -0.2070566 | 0.00555192 | |
| TAM | IL10 | -0.2304933 | 0.00196624 |
| CD68 | -0.0541709 | 0.4726548 | |
| CCL2 | -0.1914834 | 0.01045379 | |
| Tfh | BCL6 | -0.0384306 | 0.6105392 |
| IL21 | -0.2219269 | 0.00290865 | |
| Th1 | STAT4 | -0.3854338 | 1.08E-07 |
| STAT1 | -0.0504258 | 0.50384675 | |
| TBX21 (T-bet) | -0.3615366 | 7.11E-07 | |
| TNF (TNF-α) | 0.08238303 | 0.27428733 | |
| IFNG (IFN-γ) | -0.1645449 | 0.02817663 | |
| Th17 | IL17A | -0.1131636 | 0.13258416 |
| STAT3 | -0.2754626 | 1.98E-04 | |
| Th2 | IL13 | -0.2523209 | 6.79E-04 |
| STAT5A | -0.223672 | 0.00268873 | |
| STAT6 | 0.09434578 | 0.21033029 | |
| GATA3 | 0.02288401 | 0.76173782 | |
| Treg | STAT5B | -0.5321526 | 2.10E-14 |
| CCR8 | -0.3242737 | 1.01E-05 | |
| FOXP3 | -0.2399763 | 0.00125356 | |
| TGFB1 (TGFβ) | 0.19671208 | 0.00849485 | |
Cor, R value of Spearman’s correlation; TAM, tumor-associated macrophage; Tfh, T follicular helper cell; Th, T helper cell; Treg, regulatory T cell.
S100A16 was a potential prognostic marker for PC
KM survival analysis showed that high S100A16 expression was associated with poor prognosis of PC based on OS (log-rank P = 0.0022; Figure 5). Univariate Cox analysis showed that histology, tumor, node, grade, stage, and S100A16 expression were significantly correlated with the prognosis of patients with PC (all P < 0.05, Table 2). Multivariate Cox analysis revealed that S100A16 was an independent prognostic factor of PC (hazard ratio, HR = 1.828; 95% CI: 1.171-2.854; P = 0.008) with higher S100A16 expression predicting a worse prognosis for PC. These results consistently indicated that S100A16 could be a potential prognostic marker for PC.
Figure 5.
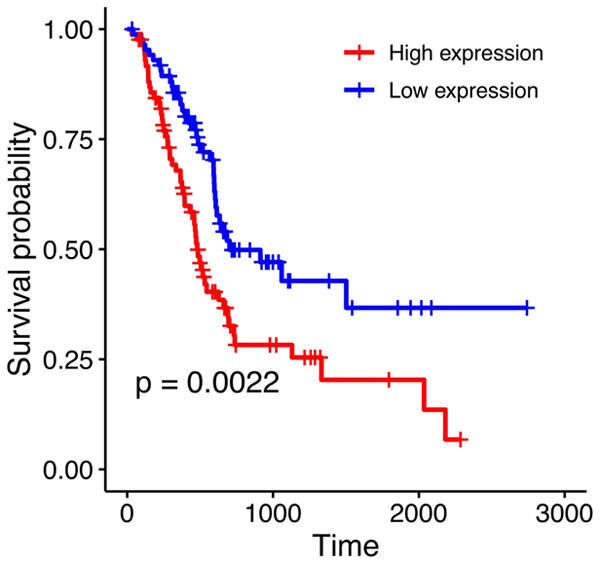
The OS curves of high and low S100A16-expression groups in PC. Log-rank test was used to evaluate the expression level of S100A16 in PC with 50% being the cutoff value. KM-plot analysis was used to produce the OS curve. OS, overall survival; PC, pancreatic cancer; KM, Kaplan-Meier.
Table 2.
Associations of clinicopathological factors and S100A16 with overall survival
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
|
|
|
|||
| HR (95% CI) | P | HR (95% CI) | P | |
| Histology | 0.293 (0.141-0.608) | 0.001 | 0.461 (0.192-1.108) | 0.083 |
| G1 vs. G2/3/4 | 2.160 (1.137-4.104) | 0.019 | 1.497 (0.774-2.898) | 0.231 |
| M | 1.028 (0.246-4.295) | 0.97 | ||
| N | 2.083 (1.239-3.503) | 0.006 | 2.300 (1.197-4.420) | 0.012 |
| T1/2 vs. T3/4 | 2.021 (1.071-3.815) | 0.03 | 1.634 (0.602-4.434) | 0.335 |
| Stage I vs. II/III/IV | 2.283 (1.046-4.980) | 0.038 | 0.402 (0.098-1.641) | 0.204 |
| S100A16 | 1.913 (1.252-2.923) | 0.003 | 1.828 (1.171-2.854) | 0.008 |
G, grade of pancreatic cancer; M, metastases prevalence; N, lymph node; T, tumor size. HR, hazard ratio (HR = 1: No effect, HR < 1: reduction in hazard, HR > 1: increase in hazard). CI, confidence interval, the narrower the confidence interval, the higher the credibility.
Discussion
PC is a deadly cancer with a grave prognosis. As is well known, abnormal expression of genes is involved in tumorigenesis and affects prognosis [14]. Previous studies have indicated the relation of the S100 protein family to the occurrence of tumors [6]. As a new member of the S100 protein family, S100A16 has recently been shown to be differentially expressed in multiple cancers [8-12] (Figure S1), suggesting that it may be involved in tumorigenesis. In this study, we analyzed the expression of S100A16 based on the RNA-seq profile of samples available in TCGA-PAAD (Figure S2) and found that high expression of S100A16 was positively correlated with the malignant degree of PC, including tumor histological grade and pathological stage. However, the potential role of S100A16 in regulating PC was still obscure. To this end, we explored the molecular function and potential mechanism of S100A16 in PC with different analysis software from multiple perspectives.
Through GO and KEGG enrichment analysis, it was clear that the genes co-expressed with S100A16 were mainly enriched in immune-related functional modules. The results of GSEA further demonstrated that when S100A16 was highly expressed, the activities of multiple cell receptors (T-cell receptor, cytokine receptor, immune receptor, coreceptor, and signaling adaptor), CAMs, and signaling pathways such as the chemokine signaling pathway and JAK-STAT signaling pathway were down-regulated. From the perspective of tumor immunity, the activity of the above receptors and adaptors is necessary for both T-cell activation and the chemokine signaling pathway to function [15,16]. CAMs play a key role in almost every step of the anti-tumor immune response, including the uptake of tumor antigens, activation of tumor-specific cytotoxic T cells, recruitment of leukocytes into tumor sites, and the killing of tumor cells [17]. Furthermore, it has been reported that up-regulation of the JAK-STAT signaling pathway is involved in tumorigenesis [18], whereas its down-regulation is also linked to some tumors [19]. At present, it is uncertain whether the inhibitory tendency of S100A16 on the JAK-STAT pathway is definitive or whether crosstalk occurs with other pathways. This requires experimental verification in the future. Overall, these results provided insight into the roles of S100A16 in PC and its effect on suppressing anti-tumor immunity.
It is well known that the immune system plays a crucial role in tumor progression [20]. Immune cells infiltrating tumors can be regulated by various cytokines and chemokines to inhibit or drive tumor progression, which exhibit potential prognostic significance [21]. Accordingly, we specifically conducted Spearman correlation analysis for S100A16 and immune-related markers. The results showed that S100A16 was significantly negatively associated with most immune markers, suggesting that it had a potential regulatory role in the anti-tumor immunity of PC. In particular, the expression of CD8+ T cell gene markers (CD8A and CD8B) had a strong negative correlation with S100A16 (Table 1). The significant inhibitory effect of S100A16 on CD8+ T cells (R = -0.217, P = 4.42e-3, Figure 4B) confirmed that high CD8+ T-cell infiltration was the most favorable prognostic factor for PC (pertaining to OS, disease-free survival, progression-free survival, and conditional survival) [22]. This suggests that S100A16 could inhibit the anti-tumor immune response mediated by CD8+ T cells [23], thereby affecting the prognosis of PC. Furthermore, while M1 macrophage gene markers (IRF5, PTGS2, and NOS2) had no significant correlation with S100A16 (all P > 0.05, Table 1), M2 macrophage markers (MS4A4A, CD163, and VSIG4) had a strong correlation with S100A16 (all P < 0.005, Table 1), indicating that S100A16 may play a regulatory role in the TAMs polarization. In DCs, the gene markers HLA-DPA1, HLA-DRA, HLA-DPB1, NRP1, CD1C, HLA-DQB1, and ITGAX were strongly negatively correlated with the expression of S100A16, suggesting that S100A16 can inhibit the innate immune memory response mediated by DCs [24], and thus suppress anti-tumor immunity. Moreover, the markers KIR3DL2, KIR3DL1, KIR2DL1, KIR2DS4, and KIR2DL3 of NK cells were also negatively correlated with the expression of S100A16, suggesting that S100A16 can inhibit the cytotoxicity of NK cells. We also observed a negative correlation between S100A16 and the overall expression level of T cells, as well as noting an inhibitory trend for each T-cell subgroup (Table 1). Taken together, the above results suggest an immunosuppressive role of S100A16, and also a primary anti-tumor immune function by inhibiting CD8+ T cells, DCs, and NK cells. It is known that high levels of immune infiltrating cells, especially CD8+ T cells, are characteristic of immunogenic warm tumors, which have a better response to immunotherapy. To transform immunogenic cold tumors into hot tumors, it is necessary to therapeutically stimulate the infiltration of active cytotoxic T cells into tumors [25,26]. Therefore, it is reasonable to speculate that S100A16 may be useful in this manner as a potential immunotherapy target.
S100A16 is highly expressed in lung adenocarcinoma, breast cancer, ovarian cancer, bladder cancer [27], and prostate cancer [28]. Some in vitro experiments have confirmed the involvement of S100A16 in tumorigenesis, including proliferation, epithelial-mesenchymal transition, and metastasis of tumor cells [29-31]. At present, its immune-related functions have not been well investigated [32], whereas its prognostic value has received full attention in many tumors, especially lung adenocarcinoma [8,33,34]. Our Kaplan-Meier survival analysis also revealed that the high S100A16 expression subgroup had a lower OS, indicating a worse prognosis. Subsequent Cox analysis further showed that S100A16 is a potential independent prognostic factor for PC. However, additional future research is needed to strengthen the clinical relevance of these findings.
In summary, this study highlights the significance of S100A16 in the prognosis of PC and its potential regulatory role in tumor-immune interactions. Our analyses showed that high S100A16 expression had a negative impact on the prognosis and immune-cell infiltration in patients with PC. However, this study also has some limitations. Using database-backed bioinformatics allowed access to large sample sizes, together with inexpensive and convenient analysis. However, as the analysis relied on public databases, the specific molecular biological mechanism of S100A16 involved in regulating immune infiltration still needs further verification. In future research, traditional experiments are necessary to explore the specific mechanism by which S100A16 affects immune infiltration and clinically validate its predictive value for the prognosis of PC.
Conclusions
S100A16 is a potential independent prognostic marker for PC and can be utilized to assess the level of immune-cell infiltration in PC tumor tissues. In addition, S100A16 exhibits extensive immunosuppressive activity, in turn affecting the prognosis of PC, mainly by inhibiting the anti-tumor immune response of CD8+ T cells, DCs, and NK cells. These findings indicate that S100A16 may be a potential immunotherapy target of PC.
Acknowledgements
We thank MD. Leilei Xia at the Department of Obstetrics and Gynecology (Changhai Hospital) for her kind bioinformatics support.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic cancer. Lancet. 2020;395:2008–2020. doi: 10.1016/S0140-6736(20)30974-0. [DOI] [PubMed] [Google Scholar]
- 2.Grossberg AJ, Chu LC, Deig CR, Fishman EK, Hwang WL, Maitra A, Marks DL, Mehta A, Nabavizadeh N, Simeone DM, Weekes CD, Thomas CR Jr. Multidisciplinary standards of care and recent progress in pancreatic ductal adenocarcinoma. CA Cancer J Clin. 2020;70:375–403. doi: 10.3322/caac.21626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network. Electronic address: andrew_aguirre@dfci.harvard.edu; Cancer Genome Atlas Research Network. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017;32:185–203. e13. doi: 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalez LL, Garrie K, Turner MD. Role of S100 proteins in health and disease. Biochim Biophys Acta Mol Cell Res. 2020;1867:118677. doi: 10.1016/j.bbamcr.2020.118677. [DOI] [PubMed] [Google Scholar]
- 5.Bresnick AR, Weber DJ, Zimmer DB. S100 proteins in cancer. Nat Rev Cancer. 2015;15:96–109. doi: 10.1038/nrc3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allgower C, Kretz AL, von Karstedt S, Wittau M, Henne-Bruns D, Lemke J. Friend or foe: S100 proteins in cancer. Cancers (Basel) 2020;12:2037. doi: 10.3390/cancers12082037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marenholz I, Heizmann CW. S100A16, a ubiquitously expressed EF-hand protein which is up-regulated in tumors. Biochem Biophys Res Commun. 2004;313:237–244. doi: 10.1016/j.bbrc.2003.11.115. [DOI] [PubMed] [Google Scholar]
- 8.Saito K, Kobayashi M, Nagashio R, Ryuge S, Katono K, Nakashima H, Tsuchiya B, Jiang SX, Saegusa M, Satoh Y, Masuda N, Sato Y. S100A16 is a prognostic marker for lung adenocarcinomas. Asian Pac J Cancer Prev. 2015;16:7039–7044. doi: 10.7314/apjcp.2015.16.16.7039. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka M, Ichikawa-Tomikawa N, Shishito N, Nishiura K, Miura T, Hozumi A, Chiba H, Yoshida S, Ohtake T, Sugino T. Co-expression of S100A14 and S100A16 correlates with a poor prognosis in human breast cancer and promotes cancer cell invasion. BMC Cancer. 2015;15:53. doi: 10.1186/s12885-015-1059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bai Y, Li LD, Li J, Lu X. Prognostic values of S100 family members in ovarian cancer patients. BMC Cancer. 2018;18:1256. doi: 10.1186/s12885-018-5170-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun X, Wang T, Zhang C, Ning K, Guan ZR, Chen SX, Hong TT, Hua D. S100A16 is a prognostic marker for colorectal cancer. J Surg Oncol. 2018;117:275–283. doi: 10.1002/jso.24822. [DOI] [PubMed] [Google Scholar]
- 12.Sapkota D, Bruland O, Parajuli H, Osman TA, Teh MT, Johannessen AC, Costea DE. S100A16 promotes differentiation and contributes to a less aggressive tumor phenotype in oral squamous cell carcinoma. BMC Cancer. 2015;15:631. doi: 10.1186/s12885-015-1622-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu G, Wang LG, Han Y, He QY. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hausser J, Alon U. Tumour heterogeneity and the evolutionary trade-offs of cancer. Nat Rev Cancer. 2020;20:247–257. doi: 10.1038/s41568-020-0241-6. [DOI] [PubMed] [Google Scholar]
- 15.Chen L, Flies DB. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol. 2013;13:227–242. doi: 10.1038/nri3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borroni EM, Savino B, Bonecchi R, Locati M. Chemokines sound the alarmin: the role of atypical chemokine in inflammation and cancer. Semin Immunol. 2018;38:63–71. doi: 10.1016/j.smim.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Harjunpaa H, Llort Asens M, Guenther C, Fagerholm SC. Cell adhesion molecules and their roles and regulation in the immune and tumor microenvironment. Front Immunol. 2019;10:1078. doi: 10.3389/fimmu.2019.01078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ihara S, Kida H, Arase H, Tripathi LP, Chen YA, Kimura T, Yoshida M, Kashiwa Y, Hirata H, Fukamizu R, Inoue R, Hasegawa K, Goya S, Takahashi R, Minami T, Tsujino K, Suzuki M, Kohmo S, Inoue K, Nagatomo I, Takeda Y, Kijima T, Mizuguchi K, Tachibana I, Kumanogoh A. Inhibitory roles of signal transducer and activator of transcription 3 in antitumor immunity during carcinogen-induced lung tumorigenesis. Cancer Res. 2012;72:2990–2999. doi: 10.1158/0008-5472.CAN-11-4062. [DOI] [PubMed] [Google Scholar]
- 19.Kohanbash G, Carrera DA, Shrivastav S, Ahn BJ, Jahan N, Mazor T, Chheda ZS, Downey KM, Watchmaker PB, Beppler C, Warta R, Amankulor NA, Herold-Mende C, Costello JF, Okada H. Isocitrate dehydrogenase mutations suppress STAT1 and CD8+ T cell accumulation in gliomas. J Clin Invest. 2017;127:1425–1437. doi: 10.1172/JCI90644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. Nat Rev Cancer. 2006;6:24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- 21.Fan JQ, Wang MF, Chen HL, Shang D, Das JK, Song J. Current advances and outlooks in immunotherapy for pancreatic ductal adenocarcinoma. Mol Cancer. 2020;19:32. doi: 10.1186/s12943-020-01151-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orhan A, Vogelsang RP, Andersen MB, Madsen MT, Hölmich ER, Raskov H, Gögenur I. The prognostic value of tumour-infiltrating lymphocytes in pancreatic cancer: a systematic review and meta-analysis. Eur J Cancer. 2020;132:71–84. doi: 10.1016/j.ejca.2020.03.013. [DOI] [PubMed] [Google Scholar]
- 23.van der Leun AM, Thommen DS, Schumacher TN. CD8(+) T cell states in human cancer: insights from single-cell analysis. Nat Rev Cancer. 2020;20:218–232. doi: 10.1038/s41568-019-0235-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eisenbarth SC. Dendritic cell subsets in T cell programming: location dictates function. Nat Rev Immunol. 2019;19:89–103. doi: 10.1038/s41577-018-0088-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Golstein P, Griffiths GM. An early history of T cell-mediated cytotoxicity. Nat Rev Immunol. 2018;18:527–535. doi: 10.1038/s41577-018-0009-3. [DOI] [PubMed] [Google Scholar]
- 26.Ochoa de Olza M, Navarro Rodrigo B, Zimmermann S, Coukos G. Turning up the heat on non-immunoreactive tumours: opportunities for clinical development. Lancet Oncol. 2020;21:e419–e430. doi: 10.1016/S1470-2045(20)30234-5. [DOI] [PubMed] [Google Scholar]
- 27.Yao R, Lopez-Beltran A, Maclennan GT, Montironi R, Eble JN, Cheng L. Expression of S100 protein family members in the pathogenesis of bladder tumors. Anticancer Res. 2007;27:3051–3058. [PubMed] [Google Scholar]
- 28.Wang R, Wu Y, Yu J, Yang G, Yi H, Xu B. Plasma messenger RNAs identified through bioinformatics analysis are novel, non-invasive prostate cancer biomarkers. Onco Targets Ther. 2020;13:541–548. doi: 10.2147/OTT.S221276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Yang Y, Ma X, Xin W, Fan X. S100A16 regulates HeLa cell through the phosphatidylinositol 3 kinase (PI3K)/AKT signaling pathway. Med Sci Monit. 2020;26:e919757. doi: 10.12659/MSM.919757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou W, Pan H, Xia T, Xue J, Cheng L, Fan P, Zhang Y, Zhu W, Xue Y, Liu X, Ding Q, Liu Y, Wang S. Up-regulation of S100A16 expression promotes epithelial-mesenchymal transition via Notch1 pathway in breast cancer. J Biomed Sci. 2014;21:97. doi: 10.1186/s12929-014-0097-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang D, Zhang C, Xu P, Liu Y, Mo X, Sun Q, Abdelatty A, Hu C, Xu H, Zhou G, Xia H, Lan L. S100A16 promotes metastasis and progression of pancreatic cancer through FGF19-mediated AKT and ERK1/2 pathways. Cell Biol Toxicol. 2021 doi: 10.1007/s10565-020-09574-w. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 32.Sun L, Zhang Z, Yao Y, Li WY, Gu J. Analysis of expression differences of immune genes in non-small cell lung cancer based on TCGA and ImmPort data sets and the application of a prognostic model. Ann Transl Med. 2020;8:550. doi: 10.21037/atm.2020.04.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi M, Nagashio R, Saito K, Aguilar-Bonavides C, Ryuge S, Katono K, Igawa S, Tsuchiya B, Jiang SX, Ichinoe M, Murakumo Y, Saegusa M, Satoh Y, Sato Y. Prognostic significance of S100A16 subcellular localization in lung adenocarcinoma. Hum Pathol. 2018;74:148–155. doi: 10.1016/j.humpath.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 34.Katono K, Sato Y, Kobayashi M, Nagashio R, Ryuge S, Igawa S, Ichinoe M, Murakumo Y, Saegusa M, Masuda N. S100A16, a promising candidate as a prognostic marker for platinum-based adjuvant chemotherapy in resected lung adenocarcinoma. Onco Targets Ther. 2017;10:5273–5279. doi: 10.2147/OTT.S145072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.