Abstract
Treatment of aggressive glioblastoma multiforme (GBM) must be based on very precise histological and molecular diagnostic of GBM type. According to the WHO guidelines, only tissue biopsy is a relevant source of cellular material evaluated in the diagnostic process to specify the tumor features. Nevertheless, obtaining a GBM biopsy is complicated and relies mostly on resection surgery. Evaluating circulating free DNA and/or circulating tumor cells (CTCs) in the clinic, using a liquid biopsy could represent a non-invasive cancer care optimization. In the present study, the peripheral blood of patients undergoing GBM resection (n = 18) was collected and examined for CTCs. The feasibility of GBM molecular diagnostics from a simple non-invasive peripheral blood withdrawal was evaluated. The size-based enriched CTCs were analyzed using cytomorphology and their origin confirmed based on mutational analysis. In addition, shared DNA mutations in CTCs and in primary tumor tissue were searched. For the identification of CTCs, next generation sequencing (NGS) was used. The GeneReader™ sequencing platform enables targeted sequencing of a 12-gene panel and direct evaluation of detected gene variations using QIAGEN Clinical Insight Analyze (QCI-A) software with a special algorithm for liquid biopsy sequencing analysis. Herein, we present a standard operating procedure for CTC enrichment in GBM patients, CTC in vitro culture, CTC cytomorphological evaluation, and NGS analysis of CTCs using the QIAGEN Actionable Insights Tumor (ATP) Panel. CTCs were present in all tested patients (18/18). The NGS data generated for formalin-fixed paraffin-embedded (FFPE) primary tumor tissues and CTCs reached significantly high-quality parameters. The comparisons between different sample types (CTCs vs. primary tumors) and sampling area (different primary tumor regions) showed a significant level of concordance, indicating CTC testing could be used for patient monitoring and recurrence awareness. Notably, more mutations were detected when analyzing CTC samples compared with the paired primary tumors (n = 3). The results confirm the feasibility of using CTCs as a source of tumor DNA in a diagnostic process, especially when evaluating the molecular characteristics of GBMs. A major advantage of the presented NGS approach for detecting CTCs is the simultaneous identification of several markers relevant for GBM diagnostics, allowing molecular diagnostics on cytological specimens and potential administration of innovative targeted therapies.
Keywords: CTCs, liquid biopsy, metacell, glioblastoma, culturing, in vitro, gene expression, sequencing
Introduction
Malignant primary brain tumors are the third leading cause of death in adults 15-34 years of age [1]. One of the most common and aggressive primary malignant brain tumors is glioblastoma multiforme (GBM). The current median overall survival (OS) of GBM patients is approximately 16 months [2,3]. Despite the aggressive and highly invasive nature of GBM cells, the tumor rarely develops extracranial metastasis. However, clinical descriptions of GBM metastases have become more frequent. These observations indicate GBM can disseminate not only via the cerebrospinal fluid (CSF), but also through the circulatory system such as bloodstream and lymphatic vessels [4]. Analyses of cerebrum autopsy specimens showed the incidence of extracranial glioma metastases has an incidence of 0.4% [5]. These data indirectly indicate the existence of circulating tumor cells (CTCs) and disseminated tumor cells (DTCs) in glioblastoma.
CTCs are cells that have been shed from the site of tumor origin. CTCs could be used for disease monitoring in GBM patients if the protocol for CTC isolation and identification is standardized. In the present study, a CTC enrichment protocol using peripheral blood of GBM patients undergoing surgery was proposed. Based on the study model, not only standard GBM tissue was obtained but also size-based enriched CTCs, and their relationship to the primary tumor was confirmed on a molecular level. The simplest way to confirm the association between the primary tumor and CTCs is the detection of known mutations. Because many new genes have been discovered in GBM research, next generation sequencing (NGS) could provide significantly more information leading to new clinical trials and targeted drug administration for both the primary GBM and recurrent tumors. There is a narrow therapeutic window when clinicians can determine the disease progression and identifying CTCs with a simple blood test during suspicious recurrences would be advantageous.
Evidence shows that CTC count and their characteristics have prognostic validity in epithelial cancers and is associated with progression-free survival (PS), OS, and stage disease. The clinical utility of CTCs in GBM is limited due to the technical obstacles associated with their isolation and detection.
A major revision of the WHO classification for tumors of the central nervous system was implemented in 2016 [6]. The main adjustment was the incorporation of molecular criteria to the diagnostic classification. In previous studies, molecular characteristics were shown to not only hold a diagnostic value but also provide more detailed information regarding prognosis [7,8].
Targeted panels in NGS allow parallel detection of several markers relevant for glioma diagnostics and yield assay information that would otherwise require several tests. The cost of NGS is rapidly decreasing and the test can be performed on very limited tissue material (minimal requirement for our protocol is 10 ng of DNA from approximately 1,500 cells consisting of at least 30% neoplastic cells) independent of the method by which the tissue was obtained (e.g., resection, biopsy, or cytology).
In the present study, CTCs were detected in GBMs and analyzed using NGS. The obtained sequencing data were used with clinical data to improve the therapy outcome for patients.
Materials and methods
Patients
A total of 18 patients diagnosed with GBM and who underwent surgical resection were enrolled in the present study. Testing for the presence of CTCs in peripheral blood was performed to characterize the level of tumor dissemination. For each patient, two samples of approximately 8 mL of venous blood were drawn from the antecubital vein prior to surgery and placed into S-Monovette tubes (Sarstedt AG & Co., Numbrecht, Germany) containing 1.6 mg EDTA/mL blood as an anticoagulant. The samples were processed at room temperature using an isolation procedure and completed within 24 hours after the blood draw. Clinical data were collected from all participating patients. The patient characteristics are shown in Table 1.
Table 1.
Patient characteristics and paired CTCs and NGS results (CTCs, circulating tumor cells; NGS, next generation sequencing)
| Sex | Code | Age | Surgery type | Histology | Tumor special characteristics if any | Survival | CTC present | Mutation detected in CTCs | Primary tumor (FFPE) | Mutation detected in primary tumor (FFPE) - 1st, 2nd, 3rd … Sample | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| F | 159116 | 51 | resection | GBM | alive | YES | PIK3CA | 3 samples | ALK, PIK3CA, KIT | PIK3CA | |
| F | 164616 | 46 | resection | LGG WHO II | Ki 67 10-15%, without codeletion 13p19q | alive | YES | 1 sample | none | ||
| M | 168716 | 51 | resection | GBM | X | YES | 2 samples | EGFR, PIK3CA | EGFR, PIK3CA | ||
| F | 192816 | 54 | resection | GBM | X | YES | 2 samples | PIK3CA | PIK3CA | ||
| F | 202516 | 65 | resection | GBM | X | YES | 1 sample | KIT, EBBR2 | |||
| M | 203516 | 52 | resection | GBM | Ki 67 20% | X | YES | 1 sample | KIT | ||
| M | 220216 | 32 | resection | LGG WHO II | LGG- astrocytoma grade II | alive | YES | 2 samples | none | PDGFR (second run) | |
| M | 230916 | 65 | resection | LGG-GBM | x | YES | NO | ||||
| M | 231816 | 70 | biopsy | GBM | Ki 67 20% | x | YES | NO | |||
| M | 249516 | 39 | resection | GBM | Ki 67 20% | x | YES | 2 samples | PDGFR, EGFR | PDGFR, EGDR | |
| F | 274216 | 37 | resection | GBM | Ki67 30%, secondary GBM | x | YES | ||||
| M | 7818 | 75 | resection | GBM | x | YES | ALK, BRAF, EGFR, PDGRA, PIK3CA | 1 sample | ALK, BRAF, EGFR, PDGRA, PIK3CA | ||
| M | 11518 | 63 | resection | GBM | ATRX +, Ki67 25% | x | YES | ALK, EGFR, PDGFR, PIK3CA | 1 sample | ALK, EGFR, PDGFR | |
| M | 86818 | 74 | resection | GBM | x | YES | ALK, EGFR, KRAS, PDGFR, PIK3CA | ||||
| M | 37419 | 57 | resection | GBM | Ki67 15% | alive | YES | EGFR, PDGFR | |||
| F | 41119 | 34 | resection | LGG | without codeletion, 1p36/19q13 | alive | YES | EGFR | |||
| M | 47519 | 66 | resection | GBM | Ki 67 20% | alive | YES | EGFR, KIT PDGFR | |||
| F | 59919 | 34 | biopsy | GBM | alive | YES | EGFR, KIT KRAS, PDGFR, PIK3CA | ||||
The protocol for this study was approved by the Ethical Committee of Krajska zdravotní a.s. in Usti and Labem. Written informed consent was obtained from all study subjects and the study was conducted in accordance with the principles of the Declaration of Helsinki.
CTCs enrichment and culture
The presence of CTCs was determined based on single cell cytomorphology followed by molecular testing (qPCR analysis).
To enrich CTCs, a size-based separation protocol and MetaCell® tubes were used (MetaCell s.r.o., Prague, Czech Republic) [9-11]. The process is based on the filtration of peripheral blood through a porous polycarbonate membrane (pores 8 μM in diameter). The minimum and maximum volumes of the filtered peripheral blood may be adjusted up to 50 mL with fluid. The enriched CTCs were directly transferred onto a separation membrane in a 6-well culture plate. Next, 4 mL RPMI media was added to the filter top and CTCs were cultured on the membrane in vitro under standard cell culture conditions (37°C, 5% CO2 atmosphere) and observed using an inverted microscope. CTCs were grown in FBS-enriched RPMI medium (10%) for a minimum of 14 days on the membrane. The cultured cells were analyzed using vital fluorescent staining microscopy after 3-5 days of in vitro culture (Celltracker™, NucBlue™, MitoTracker™, Thermo Fisher Scientific, Waltham, MA, USA). Several cytomorphological parameters were evaluated. The viable cells (stained on the separation membrane) were examined using fluorescence microscopy at 20 × magnification to locate the cells and at 60 × magnification for detailed cytomorphological analysis. Isolated cells and/or clusters of cells of interest were selected, digitized, and examined by an experienced researcher and/or pathologist. CTCs were defined as cells presenting the following characteristics: (i) nuclear size ≥ 10 μm; (ii) irregularity of the nuclear contour; (iii) presence of a visible cytoplasm; (iv) prominent nucleoli (v) high nuclear-cytoplasmic ratio; (vi) “fatty” cytoplasm; (vii) mitochondrial network presence.
NGS analysis
Sample and DNA isolation
Formalin-fixed paraffin-embedded (FFPE) tumor material from primary tumor tissue samples were collected. The following tumor tissue types were considered: glioblastoma, low-grade glioma, astrocytoma, and metastatic bladder carcinoma (Table 1). All samples were processed using the routine diagnostic pipeline from 2017 thru 2019. Simultaneously, CTCs were collected, analyzed, and stored in RLT buffer (-20°C) for DNA isolation. DNA was isolated for both sample types (primary tumors and CTCs) using the DNA mini-blood separation protocol using QIAcube (Qiagen, Hilden, Germany). The purity and concentration of DNA were determined using Nanodrop and Qubit (Thermo Fisher Scientific).
GeneReader assay and sequencing
DNA (40 ng) from each sample was used as template for the QIAGEN Actionable Insights Tumor (ATP) Panel. For each sample, the QIAGEN ATP Panel amplifies 330 amplicons covering 16.7 kb, interrogating 773 unique variant positions in 12 genes of high prognostic and therapeutic relevance in solid tumors (KRAS, NRAS, KIT, BRAF, PDGFRA, ALK, EGFR, ERBB2, PIK3CA, ERBB3, ESR1, and RAF1).
Targeted amplicons were further processed to generate a library for sequencing. Libraries were prepared using the QIAGEN GeneRead DNA Library Kit and an automated protocol on a QIAcube according to the manufacturer’s instructions. Both PCR-enriched DNA and GeneRead libraries were qualified and quantified externally using a capillary electrophoresis system according to the manufacturer’s instructions. The clonal amplification was provided via emulsion PCR. Bead enrichment steps were conducted using the GeneRead Clonal Amp Q Kit and an automated protocol on GeneRead QIAcube according to the manufacturer’s instructions. Following clonal amplification, amplicon libraries were sequenced using the QIAGEN GeneRead Sequencing Q Kit and an automated protocol on the GeneReader instrument (all protocols available at http://www.qiagen.com).
GeneReader data processing
QIAGEN Clinical Insight Analyze (QCI-A) software was used to perform the secondary analysis of FASTQ reads generated automatically by the GeneReader and align the read data to the hg19 reference genome sequence, call sequence variants, and generate an interactive report for visualization and quality control of the sequencing results as well as a summary of the data (Supplementary File).
All comparable variants were identified using QCI-A secondary analysis pipeline for the ATP on FFPE material (ATPf), where a 3% allelic fraction cut-off was used to call variants for FFPE samples. The pipeline for liquid biopsies (ATPp), which allows a 1% allelic fraction cut-off was not applied for CTCs as expected because we also wanted the CTCs to receive the same, strong conditions as FFPE.
After reviewing data validity, variants were imported as industry standard variant call format (VCF) into the QCI-Interpret (QCI-I) web interface, which enables data interpretation for the previously identified variants. QCI-I then generated a per sample report for each detected variant based on the curated content of QIAGEN Knowledge Base including summary of findings, direct link to the data source, and the eventual recommended treatment.
Direct clinical relevance of GeneReader sequencing data
The ATP NGS assay was developed to focus on clinically relevant mutations which are included in approved therapeutic labels, professional practice guidelines, and active late-stage clinical trials. This approach results in a selection of genes and variants with an unparalleled level of direct clinical relevance. The integrated bioinformatics pipeline QCI-A allows analysis of the sequences and identification of genetic aberrations while providing intrinsic quality control measures to ensure confidence in the call sequence of variants. QCI-I analyzes direct clinical relevance and reports relevant therapy or clinical trials.
Results
Herein, we present a standard operating procedure for CTC enrichment in GBM patients, CTC in vitro culture, CTC cytomorphological evaluation, and NGS analysis of CTCs using the QIAGEN ATP Panel.
First, CTCs were detected preoperatively in peripheral blood of patients (n = 18) who underwent GBM resection. Size-based enriched CTCs were cultured in vitro and analyzed using cytomorphology. Second, the feasibility of GBM molecular diagnostics from a simple non-invasive peripheral blood withdrawal was evaluated.
Cytomorphological analysis
CTCs were evaluated in duplicate (2 × 18) and detected independently in all blood samples tested (18/18). CTCs were present in patients harboring tumors of different histological origin (GBM, low-grade glioma, astrocytoma). Detailed clinical data associated with the presence and characteristics of the CTCs are shown in Table 1.
In general, cytomorphological analysis of CTCs consisted of two microscopy evaluations at two different time points (days 3 and 5 after enrichment and in vitro culture) performed by two independent evaluators.
Enriched CTCs are shown in Figure 1. Typically, cells of blast-like phenotype with smooth nucleus and rather pale cytoplasm were observed. The cytoplasm was often enriched by lipids, which is also typical for CTCs in testicular cancer. The mitochondrial network was usually very tightly organized. In more differentiated cell subtypes, several nucleoli (e.g., 5) were typically present. CTCs were not large in size on average (< 15 µm).
Figure 1.
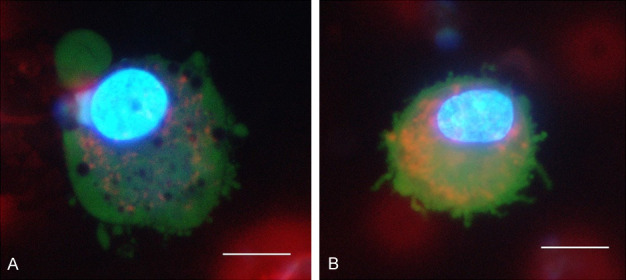
Circulating tumor cells (CTCs) in glioblastoma multiforme (GBM) patients are shown after short term in vitro culture and vital fluorescent staining (Celltracker™, NucBlue™, Mitotracker™) to enable evaluation of cytological GBM features. Typically, CTCs in GBM appear as blast-like cells with a smooth nucleus in lipid-enriched cytoplasm (A) with pale cytoplasm in combination with typical cancerous nucleoli signs (B). Cells with several nucleoli (e.g., >5) are often present.
NGS of CTCs and primary tumors
For the clinical application of CTCs in GBM management, the origin of the enriched CTCs was confirmed using mutational analysis. Shared DNA mutations in CTCs and in primary tumor tissue samples were searched.
NGS was used for efficient CTC identification. The GeneReader™ sequencing platform enables targeted sequencing of an entire gene group (n = 12) and direct evaluation of detected gene variants using the QCI-A software with a special algorithm for liquid biopsy sequencing analysis. Then, the QCI-A results were interpreted using the QCI-I software.
The NGS data generated for FFPE primary tumor tissues and CTCs reached significantly high-quality parameters. The comparisons between different sample types (CTCs vs. primary tumors) and sampling areas (different primary tumor regions) showed a significant level of concordance (Figure 2). The reported quality of NGS indicates CTC testing could be used for patient monitoring and recurrence awareness. Notably, more mutations were detected when analyzing CTC samples compared with the paired primary tumors (n = 3).
Figure 2.
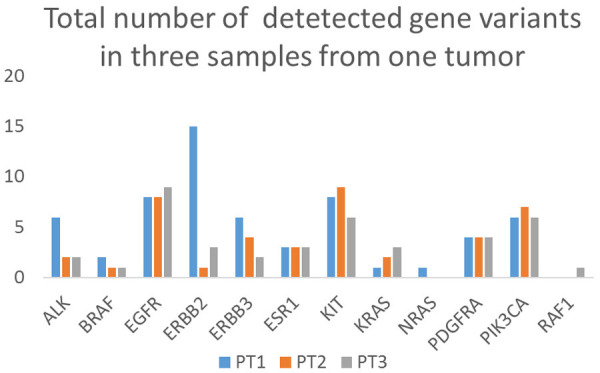
Total number of gene variants detected in three tissue samples from the same primary tumor. High concordance between the three samples in number of NGS-detected gene variants was found for BRAF, EGFR, KIT, PDGFRA, and PIK3CA genes.
Targeted NGS using the semi-automatic workflow of the GeneReader System showed the possibility of decreasing the time necessary for preparing and sequencing samples while maintaining high accuracy and run performance.
For each of the 8 CTC and 16 primary tumor samples tested, 1,279,108 reads on average were detected (1,362,147 reads for primary tumors and 1,149,711 for CTCs) and the samples were determined valid. The results confirm the feasibility of using CTCs as a source of tumor DNA in the diagnostic process, especially when evaluating the molecular characteristics of GBMs (Figures 3, 4).
Figure 3.
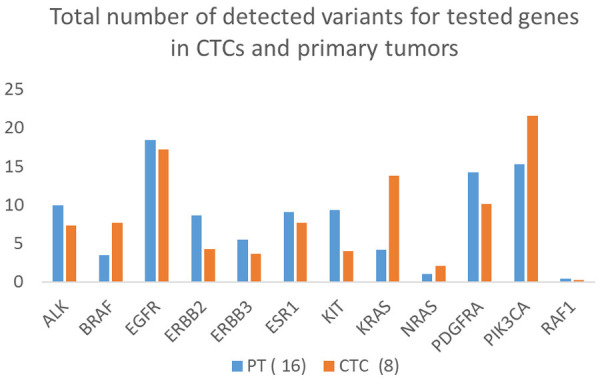
Total number of gene variants detected in CTCs and primary tumors. The data were evaluated for the CTC and primary tumor samples and averaged to minimize errors. As shown in the Figure, several of the mutations are found more frequently in CTCs (e.g., more mutations in BRAF, KRAS, and PIK3CA genes), indicating that only cell populations with these mutations are circulating in the blood and may be prognostic factors for more aggressive disease.
Figure 4.
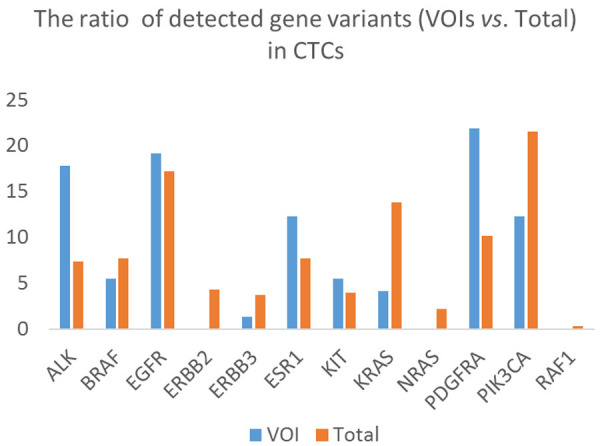
Non-synonymous variants of interest (VOIs) and total number of gene variants detected in CTCs. The data show that most mutations detected in CTCs were found in EGFR, PDFGR, and PIK3CA genes.
When duplicates and triplicates of primary tumor tissue samples were compared, significant differences in read quantity were not observed. Similarly, if CTCs and primary tumors from the same patient were compared, the efficiency of the NGS reactions generated similar read volumes, read quality, and above-average coverage (≥ 500). More detail can be found in Supplementary File which include completed automatic reports for every sample.
Based on the basic sequencing quality score, NGS results for all tested samples were determined valid. This indicates the implemented Standard Operating Protocol (SOP) for CTC sequencing (NGS) and generated data (sequences) can be used for further software analysis and clinical interpretation (Table 2). Several examples of detected gene variants and their frequencies are shown for CTCs and primary tumor sample types.
Table 2.
Comparison of the frequency of non-synonymous variants of interest (VOIs) and total number of detected variants in CTC and primary tumor samples
| Non-synonymous VOIs | Total number of detected variants | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| % Avg | % CTC | % Primary tumor | % Avg | % CTC | % Primary tumor | |
| ALK | 20 | 20 | 1,836,735 | 878,187 | 9,774436 | 8,181818182 |
| BRAF | 7,5 | 8,57,1429 | 6,122449 | 5,382436 | 7,518797 | 4,090909091 |
| EGFR | 17,5 | 14,28571 | 18,36735 | 15,58074 | 15,03759 | 15,90909091 |
| ERBB2 | 0 | 0 | 0 | 7,932011 | 5,263158 | 9,545454545 |
| ERBB3 | 0 | 0 | 0 | 6,232295 | 3,007519 | 8,181818182 |
| ESR1 | 16,25 | 11,42857 | 18,36735 | 7,082153 | 6,766917 | 7,272727273 |
| KIT | 13,75 | 5,714286 | 18,36735 | 10,1983 | 7,518797 | 11,81818182 |
| KRAS | 1,25 | 2,857143 | 0 | 7,365439 | 8,270677 | 6,818181818 |
| NRAS | 0 | 0 | 0 | 2,266289 | 2,255639 | 2,272727273 |
| PDGFRA | 16,25 | 20 | 12,2449 | 8,78187 | 9,022556 | 8,636363636 |
| PIK3CA | 12,5 | 17,14286 | 8,163265 | 19,83003 | 25,56391 | 16,36363636 |
| RAF1 | 0 | 0 | 0 | 0,566572 | 0 | 0,909090909 |
As shown in the Table, certain gene variants were found more frequently in CTCs or primary tumors (shown in bold; the number is higher than the average of CTC and primary tumor). Notably, the frequency of several mutations is doubled in CTCs (e.g., PDGFRA and PIK3CA genes) compared with primary tumors.
Reporting of NGS-detected sequence variants to the clinics
The mutation (variant) calling process was successfully based on connection of an automatic data transfer, analysis, and interpretation. At the end of the analysis process, a clinically relevant report summarizing the mutations with pathogenic impact was obtained (according to the American Molecular Pathologists classification) [12].
The transparency of the variant separation into the four main categories (tiers) renders the data easier to interpret for clinicians before enqueuing patients into the running clinical studies. Sequence variants in somatic conditions are divided into four categories based on their clinical impact: tier I, variants with strong clinical significance (level A and B evidence); tier II, variants with potential clinical significance (level C or D evidence); tier III, variants with unknown clinical significance; and tier IV, variants that are benign or likely benign. Detected somatic variants of the tested genes included single nucleotide variants (SNVs), missense mutations, and indels.
The clinical impact of a detected gene variant is determined according to currently available evidence in genomic databases. In the GeneReader system, this categorization process is provided by the QCI-I software. We show some of the comparisons for tumor duplicates (samples BC4 vs. BC5, BC6 vs. BC7, BC11 vs. BC12) made on the QCI-I level that analyzing duplicates may reveal significant differences in the detected mutation frequency on the primary tumor level (Table 3). These differences are then interpreted for clinical practice.
Table 3.
Comparison of the QCI-I results for chosen primary tumor duplicates with reported data on gene variant pathogenicity
| Gene | Variant | Tier | Pathogenicity | Effect | Allelic frequency (%) | |
|
| ||||||
| Sample BC4 FFPE | Sample BC5 FFPE | |||||
|
| ||||||
| EGFR | c.2125G>A, p.E709K | 2C | ü | missense | 4,81 | - |
| PIK3CA | c.3073A>G, p.T1025A | 2C | ü | missense | - | 11 |
| KIT | c.1737T>C, p.D579D | 4 | possibly not | missense | 4,17 | - |
|
| ||||||
| Sample BC6 FFPE | Sample BC7 FFPE | |||||
|
| ||||||
| PIK3CA | c.1049A>G, p.D350G | 2C | ü | missense | 4,49 | - |
|
| ||||||
| Sample BC11 FFPE | Sample BC12 FFPE | |||||
|
| ||||||
| EGFR | c.2508C>T, p.R836R | 4 | possibly not | synonymous | 26 | 42 |
| PDGFRA | c.661C>T, p.L221F | 4 | possibly not | missense | 36 | 40 |
Hypothetically, this primary tumor heterogeneity may be overcome by using CTC populations, which statistically could be more mixed and carry all the mutations from the primary tumor inside the CTCs from various tumors.
Comparison of the QCI-I results for chosen CTCs and paired primary tumor samples with reported data on gene variant pathogenicity show more mutations are generally detected in CTC than in primary tumor samples, however, the variants were present at very low frequency. This is an issue because only mutations with ≥ 1% frequency are reported to the clinic. If the frequency is < 1%, the quality of NGS analysis for the specific gene has to be re-evaluated. As shown in Table 4, the above-mentioned frequency is significant; however, several mutations in the PIK3CA gene occurred.
Table 4.
Comparisons of the QCI-I results for chosen CTCs and paired primary tumor samples with reported data on gene variant pathogenicity
| Gene | Variants | Tier | Pathogenicity | Effect | Allelic frequency (%) | |
|---|---|---|---|---|---|---|
|
| ||||||
| CTC | BC8 FFPE | |||||
| BRAF | c.1750C>T, p.L584F | 1A | ü | missense | 0,59 | - |
| PIK3CA | c.241G>A, p.E81K | 2C | ü | missense | 1,01 | - |
| ALK | c.3833A>G, p.Y1278C | 2C | Very Likely | missense | 0,52 | - |
| PIK3CA | c.328G>A, p.E110K | 2C | Very Likely | missense | 1,3 | - |
| PIK3CA | c.3025G>C, p.G1009R | 2C | Very Likely | missense | - | 4,03 |
More mutations were detected in the CTC samples than in the primary tumor samples (BC8), however, variants were generally present at very low frequency (only mutations with ≥ 1% are reported to the clinic). If the frequency is < 1%, the quality of NGS analysis for the specific gene has to be re-evaluated. As shown in the Table, several mutations in the PIK3CA gene occurred.
Discussion
Brain tumors may be difficult to access and surgery may result in morbidity. Processing of tissue samples, including fixation and paraffin embedding, may limit subsequent molecular analysis. The use of liquid biopsies as a complement to tumor biopsy analysis offers advantages in confirming diagnosis, identifying mutations present, monitoring tumor evolution, and response to therapy.
Circulating biomarkers, including circulating tumor nucleic acids, CTCs, and extracellular vesicles (EVs), which contain tumor DNA as well as other macromolecules, have shown tremendous promise as a type of liquid biopsy in oncology [11,12]. Although the technical aspects of biomarker detection require further optimization, these tools have already demonstrated diagnostic, prognostic, and predictive value in several tumor types.
Several independent groups have reported the presence of CTCs in blood samples from GBM patients.
Sullivan demonstrated that CTCs were identified in at least one blood specimen from 13 of 33 patients (39%; 26 of 87 samples) with GBM [13]. In one study, only 29 (20%) of the cohort including 141 patients with GBM had detectable CTCs, and in many cases, only one cell was found per 10-mL of blood sample per patient [14]. MacArthur and colleagues described the detection of GBM CTCs using nestin and human telomerase as markers in a novel telomerase assay. CTCs were detected in 71% of patients and were predictive of disease progression [15].
Possible candidates for clinical applications in glioblastoma, the challenging isolation, and the low number of CTCs detected restrict the number of available studies. Traditionally, antibody-mediated capture (positive selection) was used to isolate CTCs by targeting extracellular membrane proteins such as EpCAM. While successful for most solid tumors, for non-epithelial cancers such as brain gliomas, identifying a tumor-specific membrane protein is extremely difficult. Furthermore, only CTCs with the target protein are isolated using this approach.
Glioma cells frequently undergo EMT to support tumor propagation and invasiveness [16]. Mesenchymal GBM appears to be the most aggressive and regularly self-renews [17,18]. The mesenchymal GBM is not the only subtype that contributes to metastases. GBM is heterogeneous and all four subtypes can coexist in the same tumor [19]. In a study by Sullivan, all the GBM CTCs detected in patient samples as well as patient-derived xenografts shared a mesenchymal expression profile. Similarly, in the index GBM patient with multiple systemic metastases, all extracranial lesions were predominantly mesenchymal [13].
We recently introduced a size-based separation method which efficiently achieves isolation of CTCs, depletion of leucocytes without lysis, and culturing of CTCs. CTCs have never been isolated and directly cultured in patients with GBM. The majority of patients with glioblastoma have circulating brain tumor cells within the peripheral blood. Because these cells are very rare and express a subset of markers present in primary GBMs, their identification was made possible using the size-based separation method. We were the first to report on the successive long-term culturing of GBM CTCs and sequencing comparison.
Glioblastoma is a highly malignant brain tumor with limited treatment options. However, pseudo-progression is observed in approximately 20% of patients [20,21] and can be difficult to distinguish from true progression. Magnetic resonance imaging (MRI) is often the only alternative for clinicians, but its effectiveness is limited due to multiple factors. Only surgery followed by pathological confirmation of vital tumor cells in the lesion can verify the progressive state. However, the surgery often leaves the patient clinically unfit for further treatment. Therefore, a less time-consuming and non-invasive method for treatment monitoring is needed and a blood-based biopsy appears promising.
A liquid biopsy has several advantages compared with tissue biopsy. Liquid biopsy is a simple and less invasive procedure that can provide similar information from certain body fluids (mainly blood) than what is usually obtained from a tissue biopsy sample [22]. In addition, a liquid biopsy has potential clinical utility that could facilitate early detection of cancer as well as improve patient follow-up by managing tumor progression and monitoring therapy response [23]. Liquid biopsy, and the associated ease of monitoring treatment response, would allow significantly more discovered (targeted) drugs to be tested in clinical trials and eventually used in the clinic. Once established, using liquid biopsy with NGS can become an easily used routine in molecular pathology laboratories.
Acknowledgements
This research was financed through grant Nr 217104002 provided by Krajska zdravotni a.s.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Buckner JC, Brown PD, O’Neill BP, Meye FB, Wetmore CJ, Uhm JH. Central nervous system tumors. Mayo Clin Proc. 2007;82:1271–86. doi: 10.4065/82.10.1271. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R. Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: a randomized clinical trial. JAMA. 2017;318:2306–16. doi: 10.1001/jama.2017.18718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawton CD, Nagasawa DT, Yang I, Fessler RG, Smith ZA. Leptomeningeal spinal metastases from glioblastoma multiforme: treatment and management of an uncommon manifestation of disease. J Neurosurg Spine. 2012;17:438–448. doi: 10.3171/2012.7.SPINE12212. [DOI] [PubMed] [Google Scholar]
- 5.Smith DR, Hardman JM, Earle KM. Contiguous glioblastoma multiforme and fibrosarcoma with extracranial metastasis. Cancer. 1969;24:270–6. doi: 10.1002/1097-0142(196908)24:2<270::aid-cncr2820240210>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 6.Kolostova K, Pinkas M, Jakabova A, Pospisilova E, Svobodova P, Spicka J, Cegan M, Matkowski R, Bobek V. Molecular characterization of circulating tumor cells in ovarian cancer. Am J Cancer Res. 2016;6:973–80. [PMC free article] [PubMed] [Google Scholar]
- 7.Motzer RJ, Bander NH, Nanus DM. Renal-cell carcinoma. N Engl J Med. 1996;335:865–875. doi: 10.1056/NEJM199609193351207. [DOI] [PubMed] [Google Scholar]
- 8.de Bono JS, Scher HI, Montgomery RB. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 9.Kolostova K, Matkowski M, Jędryka M, Soter K, Cegan M, Pinkas M, Pavlasek J, Spicka J, Bobek V. The added value of circulating tumor cells examination in ovarian cancer staging of ovarian cancer. Am J Cancer Res. 2015;5:3363–75. [PMC free article] [PubMed] [Google Scholar]
- 10.Kolostova K, Matkowski R, Gürlich R, Grabowski K, Soter K, Lischke R, Schützner J, Bobek V. Detection and cultivation of circulating tumor cells in gastric cancer. Cytotechnology. 2016;68:1095–102. doi: 10.1007/s10616-015-9866-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cegan M, Kolostova K, Matkowski R, Broul M, Schraml J, Fiutowski M, Bobek V. In vitro culturing of viable circulating tumor cells of urinary bladder cancer. Int J Clin Exp Pathol. 2014;7:7164–7171. [PMC free article] [PubMed] [Google Scholar]
- 12.Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, Tsimberidou AM, Vnencak-Jones CL, Wolff DJ, Younes A, Nikiforova MN. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the association for molecular pathology, American society of clinical oncology, and college of American pathologists. J Mol Diagn. 2017;19:4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sullivan JP, Nahed BV, Madden MW, Oliveira SM, Springer S, Bhere D, Chi AS, Wakimoto H, Rothenberg SM, Sequist LV, Kapur R, Shah K, Iafrate AJ, Curry WT, Loeffler JS, Batchelor TT, Louis DN, Toner M, Maheswaran S, Haber DA. Brain tumor cells in circulation are enriched for mesenchymal gene expression. Cancer Discov. 2014;4:1299–1309. doi: 10.1158/2159-8290.CD-14-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller C, Holtschmidt J, Auer M. Hematogenous dissemination of glioblastoma multiforme. Sci Transl Med. 2014;6:247–301. doi: 10.1126/scitranslmed.3009095. [DOI] [PubMed] [Google Scholar]
- 15.Macarthur KM, Kao GD, Chandrasekaran S. Detection of brain tumor cells in the peripheral blood by a telomerase promoter-based assay. Cancer Res. 2014;74:2152–2159. doi: 10.1158/0008-5472.CAN-13-0813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ortensi B, Setti M, Osti D, Pelicci G. Cancer stem cell contribution to glioblastoma invasiveness. Stem Cell Res Ther. 2013;4:18. doi: 10.1186/scrt166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, de Tribolet N, Regli L, Wick W, Kouwenhoven MC, Hainfellner JA, Heppner FL, Dietrich PY, Zimmer Y, Cairncross JG, Janzer RC, Domany E, Delorenzi M, Stupp R, Hegi ME. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance. J. Clin. Oncol. 2008;26:3015–24. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 18.Colman H, Zhang L, Sulman EP, McDonald JM, Shooshtari NL, Rivera A, Popoff S, Nutt CL, Louis DN, Cairncross JG, Gilbert MR, Phillips HS, Mehta MP, Chakravarti A, Pelloski CE, Bhat K, Feuerstein BG, Jenkins RB, Aldape K. A multigene predictor of outcome in glioblastoma. Neuro Oncol. 2010;12:49–57. doi: 10.1093/neuonc/nop007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sottoriva A, Spiter I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavaré S. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taal W, Brandsma D, de Bruin HG, Bromberg JE, Swaak-Kragten AT, Smitt PA, van Es CA, van den Bent MJ. Incidence of early pseudo-progression in a cohort of malignant glioma patients treated with chemoirradiation with temozolomide. Cancer. 2008;113:405–10. doi: 10.1002/cncr.23562. [DOI] [PubMed] [Google Scholar]
- 21.Gerstner ER, McNamara MB, Norden AD, Lafrankie D, Wen PY. Effect of adding temozolomide to radiation therapy on the incidence of pseudo-progression. J Neurooncol. 2009;94:97–101. doi: 10.1007/s11060-009-9809-4. [DOI] [PubMed] [Google Scholar]
- 22.Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, Douville C, Javed AA, Wong F, Mattox A. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019;20:71–88. doi: 10.1038/s41576-018-0071-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


