Abstract
Background: The Perilipin (PLIN) family of genes were previously shown to be involved in the formation and degradation of Lipid Droplets (LDs). In addition, they may play important roles in the development and progression of breast cancer. However, the prognostic value of PLIN family members in breast cancer patients remains unclear. Methods: Mutations and copy number alterations of PLIN family genes in breast cancer were examined using the cBioportal for Cancer Genomics. In addition, the expression patterns of PLIN family genes were explored using the UCSC Xena online tool. Finally, the Kaplan-Meier Plotter was used to investigate the prognostic value of PLIN family genes in breast cancer. Results: The findings revealed a low frequency of genetic alterations and amplification was the most frequent change in the PLIN family genes. Additionally, there was an increase in the expression of Perilipin 3 (PLIN3) in breast cancer tissues compared to normal breast tissues. However, expression of the other genes in the PLIN family was significantly lower in breast cancer tissues compared to normal breast tissues. Moreover, there was an increase in the expression levels of Perilipin 1 (PLIN1), PLIN3, Perilipin 4 (PLIN4) and Perilipin 5 (PLIN5) in the luminal A and luminal B subgroups. On the other hand, the expression of Perilipin 2 (PLIN2) was elevated in the human epidermal growth factor receptor 2 (HER2) positive and basal-like subgroups. Furthermore, Kaplan-Meier Plotter analysis demonstrated that high expression of PLIN1 might predict a longer Overall Survival (OS) in patients with breast cancer while overexpression of PLIN2 indicated poor OS of breast cancer patients. Conclusion: The findings from this study indicated that genes in the PLIN family were aberrantly expressed in breast cancer and may serve as novel therapeutic targets as well as prognostic biomarkers for the disease.
Keywords: Perilipin (PLIN) family genes, breast cancer, expression status, prognosis
Introduction
Initially, Lipid Droplets (LDs) were only considered to be storage organelles at the center of lipid and energy homeostasis. However, many studies over the recent years have shown that LDs are complicated, dynamic and multifunctional organelles that play an important role in processes such as membrane transport, protein degradation, signal transduction and regulation of gene expression [1].
In addition, cancer cells subjected to hypoxia or nutrient starvation have an outstanding ability to synthesize Fatty Acids (FAs) and have an increased accumulation of Lipid Droplets (LD) [2,3]. This accumulation of LD in turn contributes to the growth and survival of cancer cells under normoxia and hypoxia [4,5]. Moreover, previous studies showed that the presence of abundant LDs is a distinctive mark of cancer stem cells in colorectal cancer [6] and increased pulmonary metastases in glioblastoma [7].
Members of the Perilipin (PLIN) family are the most important LD associated proteins as they are involved in the formation and degradation of LDs. In addition, the PLIN family consists of 5 member proteins that share a conserved architecture and the ability to bind intracellular LDs [8,9]. The members include: Perilipin 1 (PLIN1), Perilipin 2 (PLIN2/ADFP), Perilipin 3 (PLIN3/Tip47), Perilipin 4 (PLIN4/S3-12) and Perilipin 5 (PLIN5//OXPAT/PAT1) [10]. Notably, PLIN2 and PLIN3 are ubiquitously expressed while PLIN1, PLIN4 and PLIN5 are mostly limited to adipocytes, skeletal muscle and cardiac muscle [11]. Moreover, recent studies demonstrated that aberrant expression of genes in the PLIN family may serve as a potential prognostic biomarker in various types of cancer including sarcomas [12], hepatocellular carcinoma [13], renal cancer [14] and breast cancer [15].
Breast cancer is the leading cause of cancer related deaths among females worldwide [16]. In addition, dysregulation of fatty acid metabolism has gained popularity as a warning sign of malignant transformation to breast cancer. Moreover, it was previously reported that accumulation of LDs may be closely related to increased aggressiveness in breast cancer [17]. Accumulating evidence also shows that genes in the PLIN family play a role in the invasion and metastasis of breast cancer through the regulation of lipid and energy metabolism. Furthermore, recent studies indicated that the PLIN family genes were strongly associated with clinical outcomes in breast cancer [15,18-21].
However, no study exists on the expression patterns and prognostic value of all the PLIN family genes in breast cancer. Therefore, the present study comprehensively explored the expression patterns and prognostic significance of the five PLIN family genes in breast cancer.
Material and methods
The study required neither ethical approval nor consent from patients since this was an integrative analysis of published data.
Mutations and copy number alterations in PLIN family genes
The cBioPortal for Cancer Genomics (https://www.cbioportal.org/) is an open source online tool that provides a visual analysis of multidimensional cancer genomics data [22]. In this study, mutations and copy number alterations in PLIN family genes in breast cancer were examined using TCGA data sets from the cBioportal for Cancer Genomics.
Expression profiles of PLIN family genes in breast cancer
UCSC Xena is an open-access exploration tool that allows researchers to analyze and visualize large-scale cancer genomics data sets, including TCGA and other public cancer genomics resources within the Xena Browser [23]. Therefore, the present study used UCSC Xena to explore the relationship between the expression levels of PLIN family genes and clinical data in breast cancer.
Analysis of the association between overall survival (OS) and PLIN family genes in breast cancer patients
The association between the mRNA expression levels of individual PLIN family members and Overall Survival (OS) in breast cancer patients was explored using the Kaplan-Meier Plotter (www.kmplot.com) [24]. The study further performed subgroup analyses of OS in breast cancer patients based on clinical pathological parameters, such as Estrogen Receptor (ER) status, Progesterone Receptor (PR) status, HER2 status, lymph node status, grade, molecular subtypes, and TP53 status. Briefly, the five PLIN family genes (PLIN1, PLIN2, PLIN3, PLIN4, and PLIN5) were respectively entered into the database to get the Kaplan-Meier survival plots based on the different clinical parameters. In addition, the breast cancer patients were divided into the ‘low’ and ‘high’ expression groups based on the mRNA expression levels of individual PLIN family genes, using the median value as the cutoff point. Thereafter, the Kaplan-Meier method along with the Log-rank test were used for univariate OS analysis. The Hazard ratio (HR) with 95% Confidence Intervals (CI) and the log-rank p values were also estimated. A p-value of <0.05 was considered to be statistically significant.
Results
Genetic alterations in PLIN family genes in breast cancer
Mutations and copy number alterations in PLIN family genes in breast cancer were examined by integrating 4 TCGA databases (TCGA, Cell 2015, TCGA Firehose Legacy, TCGA, Nature 2012, TCGA and PanCancer Atlas) from the cBioPortal for Cancer Genomics. The oncoprint in Supplementary Figure 1 presents the distribution of alterations in PLIN family genes in breast cancer. The results showed that the frequency of genetic alterations was low (1.3%~3%) and amplification was the most frequent change in the 5 genes from the PLIN family.
Expression patterns of PLIN family genes in breast cancer
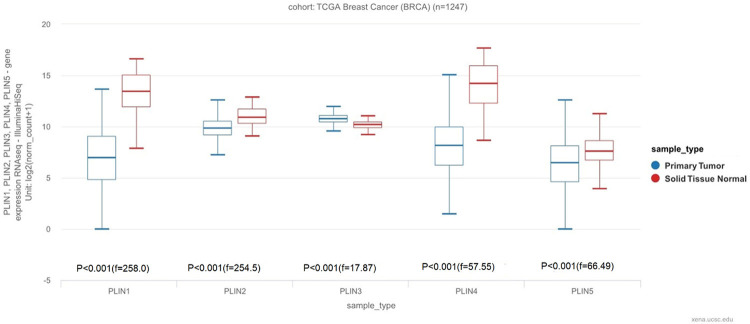
The UCSC database was used to analyze the mRNA expression levels of all the 5 PLIN family genes in breast cancer and normal tissues. The results indicated a decrease in the mRNA expression levels of PLIN1, PLIN2, PLIN4 and PLIN5 in breast cancer tissues compared to normal tissues. However, there was an increase in the mRNA expression levels of PLIN3 in breast cancer tissues compared to normal tissues. Moreover, PLIN3 had the highest expression levels in breast cancer tissues among the 5 PLIN family genes while PLIN5 had the lowest levels of expression. On the other hand, PLIN4 had the highest expression levels in normal breast tissues while PLIN5 had the lowest levels of expression (Figure 1).
Figure 1.
Expression patterns of the PLIN family genes in breast cancer using the UCSC database.
The relationship between PLIN family genes and clinicopathological parameters in breast cancer
The breast cancer patients were divided into various subgroups based on their clinicopathologic characteristics, including ER status, PR status, HER2 status, molecular subtypes, pathological types and nodal status. Thereafter, the UCSC Xena database was used to compare the mRNA expression levels of PLIN family genes in the different subgroups of breast cancer. The results revealed a significant increase in the mRNA expression levels of PLIN1, PLIN4 and PLIN5 in the ER (+) and PR (+) subgroups compared to the ER (-) and PR (-) categories. In contrast, expression of PLIN2 was negatively correlated with the ER and PR status in breast cancer. Additionally, PLIN1, PLIN4 and PLIN5 were overexpressed in the HER2 (-) subgroups compared to the HER2 (+) category. Nonetheless, the expression levels of PLIN2 and PLIN3 were not significantly correlated with the HER2 status. Moreover, there was an in increase in the expression levels of PLIN1, PLIN3, PLIN4 and PLIN5 in the luminal A and luminal B subgroups while the expression level of PLIN2 was elevated in the HER (+) and basal-like subgroups (Supplementary Figure 2). Furthermore, there was a decrease in the expression levels of PLIN1, PLIN4 and PLIN5 in the Infiltrating Ductal Carcinoma (IDC) subgroup compared to the Infiltrating Lobular Carcinoma (ILC) category. However, there was a significant increase in the expression of PLIN3 in the IDC subgroup compared to the ILC category. With regard to lymph node status, only PLIN2 was overexpressed in the N0 subgroup and there was no significant difference in the expression levels of the remaining genes between the N0 and N+ subgroups (Table 1).
Table 1.
The relationship between the expression levels of PLIN family genes and clinicopathological features
| Variables | No. | PLIN1 | PLIN2 | PLIN3 | PLIN4 | PLIN5 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|||||||
| mRNA | P value | mRNA | P value | mRNA | P value | mRNA | P value | mRNA | P value | ||
| ER | |||||||||||
| Positive | 601 | 7.2 | P<0.01 | 9.60 | P<0.01 | 10.7 | P = 0.67 | 8.22 | P<0.01 | 6.98 | P<0.01 |
| Negative | 179 | 5.18 | (t = 7.071) | 10.8 | (t = -13.08) | 10.7 | (t = -0.4287) | 6.73 | (t = 5.888) | 3.82 | (t = 15.60) |
| PR | |||||||||||
| Positive | 522 | 7.18 | P<0.01 | 9.62 | P<0.01 | 10.7 | P = 0.39 | 8.21 | P<0.01 | 7.12 | P<0.01 |
| Negative | 255 | 5.64 | (t = -5.412) | 10.4 | (t = 9.118) | 10.7 | (t = -0.8663) | 7.42 | (t = -4.946) | 4.43 | (t = -13.86) |
| HER2 | |||||||||||
| Positive | 114 | 5.75 | P<0.01 | 10.0 | P>0.05 | 10.7 | P = 0.78 | 6.58 | P<0.01 | 5.16 | P<0.01 |
| Negative | 652 | 6.82 | (t = 2.883) | 9.79 | (t = -1.951) | 10.7 | (t = -0.2747) | 8.03 | (t = 4.419) | 6.45 | (t = 4.456) |
| Molecular Subtypes | |||||||||||
| Luminal A | 231 | 7.75 | P<0.01 | 9.52 | P<0.01 | 10.8 | P<0.01 | 8.77 | P<0.01 | 7.16 | P<0.01 |
| Luminal B | 127 | 6.36 | (f = 27.11) | 9.59 | (f = 49.40) | 10.7 | (f = 7.025) | 7.76 | (f = 20.20) | 7.20 | (f = 73.93) |
| HER2+ | 58 | 5.75 | 10.1 | 10.8 | 6.64 | 4.57 | |||||
| Basal-like | 98 | 4.81 | 11.0 | 10.6 | 6.52 | 4.07 | |||||
| Tumor histology | |||||||||||
| IDC | 882 | 6.92 | P<0.01 | 10.0 | P=0.052 | 10.9 | P<0.01 | 8.21 | P<0.01 | 6.47 | P<0.01 |
| ILC | 210 | 9.29 | (t = 9.181) | 9.91 | (t = -1.948) | 10.7 | (t = 4.812) | 10.0 | (t = 8.048) | 7.94 | (t = 10.73) |
| Nodal status | |||||||||||
| N0 | 385 | 6.45 | P = 0.92 | 9.84 | P = 0.03 | 10.7 | P = 0.24 | 7.91 | P = 0.64 | 6.27 | P = 0.64 |
| N+ | 406 | 6.71 | (t = -0.0980) | 9.72 | (t = 2.144) | 10.7 | (t = -1.183) | 7.88 | (t = 0.4650) | 6.15 | (t = 0.4650) |
The prognostic value of PLIN family genes in breast cancer
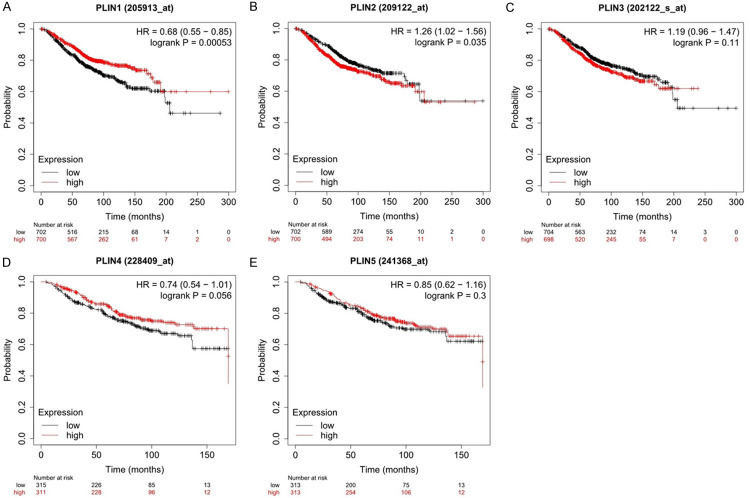
The prognostic value of the 5 PLIN family genes in breast cancer was examined using the Kaplan-Meier Plotter, which is an online tool. The Kaplan-Meier survival curves for breast cancer patients showed that high expression of PLIN1 mRNA was significantly correlated with a better OS. (Figure 2A: HR = 0.68, 95% CI: 0.55-0.85, log-rank P = 0.00053). However, high expression of PLIN2 had a significant association with worse OS. (Figure 2B: HR = 1.26, 95% CI: 1.02-1.56, log-rank P = 0.035). Moreover, there was no correlation between the mRNA expression levels of the other PLIN family genes (PLIN3, PLIN4 and PLIN5) and OS in breast cancer (Figure 2C-E).
Figure 2.
Prognostic value of the PLIN family genes in breast cancer patients using the Kaplan-Meier Plotter online database.
The prognostic value of PLIN family genes in breast cancer patients with different clinicopathological characteristics
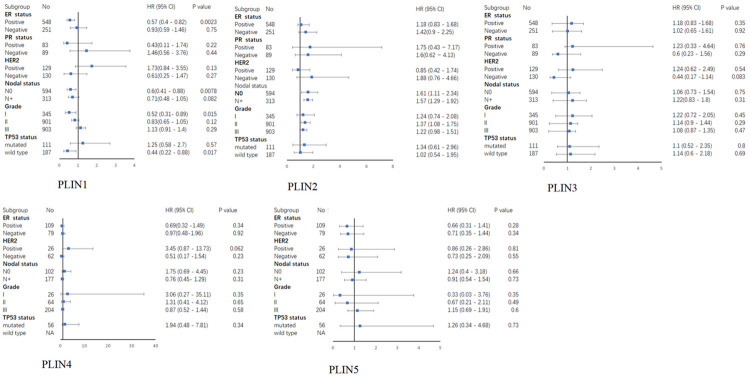
In order to evaluate the association between the expression levels of PLIN family genes and OS in breast cancer patients with different clinicopathological characteristics, subgroup analysis was performed based on the ER status, PR status, HER-2 status, lymph node status, grade and TP53 status (Figure 3). The results showed that high expression of PLIN1 was correlated with a long OS in breast cancer patients who were ER positive (Supplementary Figure 3A: HR = 0.57, 95% CI: 0.4-0.82, P = 0.0023), lymph node negative (Supplementary Figure 3B: HR = 0.6, 95% CI: 0.41-0.88, P = 0.0078) or in grade I (Supplementary Figure 3C: HR = 0.52, 95% CI: 0.31-0.89, P = 0.015). Additionally, high expression of PLIN2 was significantly related to a short OS in breast cancer patients with (Supplementary Figure 3D: HR = 1.61, 95% CI: 1.11-2.34, P = 0.012) or without (Supplementary Figure 3E: HR = 1.57, 95% CI: 1.29-1.92, P = 6.3e-06) lymph node metastasis or those who were in grade II (Supplementary Figure 3F: HR = 1.37, 95% CI: 1.08-1.75, P = 0.0094). However, the expression levels of PLIN3, PLIN4 and PLIN5 were not correlated with OS in all the subgroups of breast cancer patients.
Figure 3.
A Forest plot of the association between PLIN family genes and OS in breast cancer patients with different clinicopathological features.
The prognostic value of PLIN family genes in different molecular subtypes of breast cancer
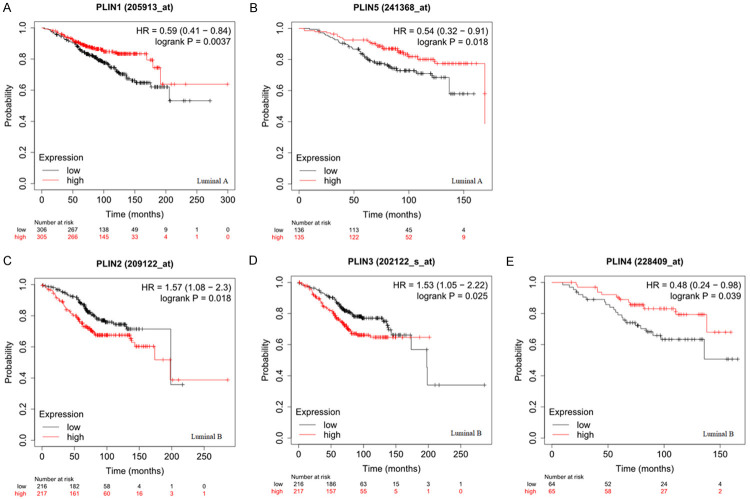
The study further investigated the prognostic value of PLIN family genes in different molecular subtypes of breast cancer, such as luminal A, luminal B, HER2 positive and basal-like. The results in Figure 4 show that high expression of PLIN1 (Figure 4A: HR = 0.59, 95% CI: 0.41-0.84, P = 0.0037) and PLIN5 (Figure 4B: HR = 0.54, 95% CI: 0.32-0.91, P = 0.018) was related to a longer OS in the luminal A type of breast cancer. However, the mRNA expression levels of PLIN2, PLIN3 and PLIN4 were not associated with OS in the luminal A subtype of breast cancer (Supplementary Table 1). On the other hand, high expression of PLIN2 (Figure 4C: HR = 1.57 95% CI: 1.08-2.3, P = 0.018) and PLIN3 (Figure 4D: HR = 1.53, 95% CI: 1.05-2.22, P = 0.025) was associated with worse OS in the luminal B subtype of breast cancer although high expression of PLIN4 (Figure 4E: HR = 0.48, 95% CI: 0.24-0.98, P = 0.039) was related to a longer OS. Moreover, the mRNA expression levels of PLIN1 and PLIN5 were not associated with OS in the luminal the B subtype of breast cancer (Supplementary Table 1). Nonetheless, none of PLIN family genes was significantly correlated with OS in the HER2-positive and basal-like subtypes of breast cancer (Supplementary Table 1).
Figure 4.
The prognostic value of PLIN family genes in breast cancer patients with different molecular subtypes using the Kaplan-Meier Plotter online database. A. Survival curves of PLIN1 (luminal A type). B. Survival curves of PLIN 5 (luminal A type). C. Survival curves of PLIN2 (luminal B type). D. Survival curves of PLIN3 (luminal B type). E. Survival curves of PLIN4 (luminal B type).
Discussion
This study comprehensively analyzed the expression status and prognostic value of PLIN family genes in breast cancer patients. The findings suggested that the expression levels of PLIN1, PLIN2, PLIN4 and PLIN5 were significantly lower in breast cancer tissues than in normal tissues. However, there was an increase in the expression of PLIN3 in breast cancer tissues compared to normal breast tissues. In addition, the results from survival analysis indicated that high expression of PLIN1 might predict longer OS in breast cancer patients although high expression of PLIN2 was linked to a worse OS in the cancer. Nonetheless, PLIN3, PLIN4 and PLIN5 had no significant effect on OS in breast cancer.
PLIN1 which is predominantly expressed in White Adipose Tissues (WAT), is located on the surface of LDs and involved in hormone-induced lipolysis as well as the formation of large LDs [25]. A recent study reported that PLIN1 was highly expressed in liposarcoma but was absent in non-lipomatous sarcomas [12]. In addition, a previous report showed that expression of PLIN1 was markedly downregulated in breast cancer [15]. Similarly, the results from the present study showed that expression of PLIN1 was lower in breast cancer tissues compared to normal breast tissues. Additionally, Kim et al. observed that expression of the PLIN1 protein was highest in HER2 (+) tumors and lowest in the Triple Begative Breast Cancer (TNBC) subtype [26]. However, the current study showed that the mRNA expression levels of PLIN1 were highest in the luminal A subtype and lowest in the basal-like subtype. This inconsistency in findings may be attributed to the small sample size and differences in unconformity between mRNA and proteins. The results from this study also showed that the mRNA expression levels of PLIN1 were higher in the ER (+) and PR (+) subgroups compared to the ER(-) and PR (-) subgroups of breast cancer. Notably, a recent study showed that estrogen could regulate the levels of PLIN1 and the Adipose Triglyceride Lipase (ATGL), controlling the size of LD and lipid accumulation by interacting with the estrogen receptor [27].
Few studies have reported on the correlation between PLIN1 and prognosis in breast cancer. In addition, the studies were conducted with relatively small sample sizes. For instance, Zhou et al. found that low expression of PLIN1 predicted a poorer overall metastatic relapse-free survival and PLIN1 was an independent predictor of OS in the ER positive and luminal A subtypes of breast cancer [15]. However, Jung et al. reported that high expression of PLIN1 was related to a shorter OS in metastatic breast cancer [20]. In the present study, the findings revealed that increased expression of PLIN1 predicted a longer OS in breast cancer patients. Similar to previous studies, the results also showed that high expression of PLIN1 was correlated with better OS in the luminal A and ER (+) subtypes of breast cancer. In addition, the present study showed that high expression of PLIN1 was associated with better OS in breast cancer patients with grade I and those who were lymph node negative. However, further studies are needed to validate these results.
On the other hand, PLIN2 is mainly expressed in the liver although it is also ubiquitously expressed [28]. Previous studies showed that PLIN2 regulates adipocyte differentiation and cellular LD accumulation under the control of the Peroxisome Proliferator-activated Receptors (PPARs) and the Retinoid X Receptor (RXR) [29,30]. Additionally, recent studies suggested that PLIN2 was overexpressed in a variety of tumors, including Burkitt Lymphoma [31], malignant melanoma [32], renal cell carcinoma [14] and lung adenocarcinoma [33]. PLIN2 was also shown to not only be expressed in breast cancer tissues but also in the non-neoplastic mammary ducts [21]. Herein, the results suggested that PLIN2 was expressed in breast cancer and normal breast tissues although its expression significantly decreased in tumor tissues compared to normal tissues. On the contrary, a previous study reported that expression of the PLIN2 protein was significantly increased in carcinoma tissues compared to non-tumor breast tissues [21]. This discrepancy may be attributed to differences in gene and protein expression or the statistical methods used.
Additionally, previous studies showed that expression of PLIN2 had a significant correlation with histological grade and HER2 status but was negatively associated with the ER status in breast cancer [21]. In this study, the results showed that PLIN2 expression was higher in the ER and PR negative subgroups compared to ER and PR positive subgroups. Moreover, previous studies suggested that the expression levels of PLIN family members in breast cancer were regulated by estrogen. Additionally, Pawlik et al. suggested that antiestrogen treatment resulted to an increase in the expression levels of PLIN2 in ER-positive breast cancer cells [34]. Furthermore, a previous study reported that there was in increase in the expression of PLIN2 in the HER2 positive and TNBC subtypes compared to the luminal subtypes [21]. The results in this study corroborated with those obtained previously since the expression of PLIN2 was significantly higher in the HER2 (+) and basal-like subtypes of breast cancer compared to the luminal A and luminal B subtypes.
To the best of our knowledge, no report exists on the relationship between PLIN2 expression and OS in breast patients. Previous studies suggested that PLIN2 might serve as an independent favorable prognostic factor for OS in clear cell renal cell carcinoma [14,35]. However, the present study showed that high expression of PLIN2 was a poor prognostic factor for OS in breast cancer. Moreover, existing evidence shows that PLIN2 is associated with more aggressive biological phenotypes in breast cancer [21]. Similar to this study, a previous study on lung adenocarcinoma also showed that overexpression of PLIN2 was associated with a shorter disease-free and OS. These results therefore indicate that PLIN2 has different tumor-specific roles in various types of cancer.
PLIN3 (also called TIP47) and PLIN2 have similar amino acid sequences and PLIN3 is also ubiquitously distributed among tissues [36]. Previous studies suggested that PLIN3 is involved in the transportation of lysosomal enzymes and plays a crucial role in the formation of LD and generation of Prostaglandin E2 (PGE2) [11]. PLIN3 was also reported to be widely expressed in various malignancies, including hepatocellular carcinoma, breast cancer, colon cancer and lung cancer [37]. Additionally, it was previously reported that the expression of PLIN3 was elevated in clear cell renal cell carcinoma (RCC) [38], cervical carcinoma [39] and liposarcoma [40]. Similarly, the present study revealed a significant increase in the expression of PLIN3 in breast cancer tissues compared to normal tissues. However, unlike in PLIN2, the results revealed that expression of PLIN3 was markedly lower in the basal-like breast cancer subtypes than in the other subtypes. The reason behind these differences is however currently unclear. Existing evidence shows that elevated levels of PLIN3 predicted poor DFS and OS in patients with RCC. However, the present study showed no correlation between PLIN3 and OS in patients with breast cancer. Nonetheless, this conclusion needs to be validated further.
PLIN4 is selectively expressed in WAT and is also present in skeletal muscles and human Mesenchymal Stem Cells (hMSCs) [41,42]. Although little is known on the biological functions of PLIN4, increasing evidence suggests that it may play a role in adipocyte differentiation. In addition, genomic variants can result in genomic instability leading to carcinogenesis. Notably, mutations in the PLIN4 gene have previously been reported in several primary tumors in humans, including gastric cancer [43] and lung cancer [44]. Additionally, a previous study showed that PLIN4 was expressed in some liposarcomas although it was absent in non-lipomatous sarcomas [40]. Moreover, reprogrammed lipid metabolism and accumulation of LDs were observed in TNBC resistant cells and clinically chemoresistant breast cancer. Interestingly, PLIN4, which coated the LDs was expressed in this resistant phenotype [19]. Similarly, the present study showed that there was a decrease in the expression levels of PLIN4 in the basal-like subtype of breast cancer.
PLIN5, also known as the Myocardial LD Protein (MLDP), was shown to be highly expressed in heart tissues, pancreatic islet β-cells and hepatic stellate cells [45,46]. PLIN5 may also have a role in stabilizing lipid homeostasis and providing fatty acids to the mitochondria [47,48]. According to a previous study, PLIN5 was significantly upregulated in the tumoral area of Hepatocellular Carcinoma (HCC) compared to the adjacent normal tissues, indicating a possible direct effect of PLIN5 in promoting tumor development [13]. Additionally, Burlaka et al. reported that expression of PLIN5 was higher in gastric cancer patients with distant metastasis compared to those without distant metastases [49]. In this study, there was a decrease in the mRNA expression levels of PLIN5 in breast cancer tissues compared to normal tissues. Moreover, a recent bioinformatics study revealed that high expression of PLIN5 was associated with better prognosis in lung adenocarcinoma [50]. However, the present study found no correlation between the expression of PLIN5 and OS in breast cancer.
The above findings therefore suggest that genes in the PLIN family may be involved in tumor development and are associated with prognosis in breast cancer. Although several mechanisms have been proposed, the exact mechanisms are still unclear. It was previously reported that fatty acids might exert various effects on cancer. For instance, fatty acids may function as signaling molecules, energy sources and necessary substrates for membrane synthesis during tumorigenesis and cancer progression [51]. In addition, recent studies showed that the PLIN family genes played essential roles in breast cancer by regulating lipid metabolism. Moreover, KEGG pathway analysis showed that the PLIN family genes were downstream targets for the PPARγ pathway. Notably, PPARγ is a member of the PPAR family and plays essential roles in adipocyte differentiation, formation of lipid droplets and lipid metabolism [52]. Furthermore, recent studies showed that PPARγ was down-regulated in breast cancer and inhibited breast cancer cell growth by regulating lipid metabolism [53-56]. Additionally, accumulating evidence indicates that genes in the PLIN family regulate the formation of lipid droplets by activating the PPARγ signaling pathway [57,58]. A previous study also showed that estradiol signals regulate the expression of PLIN1, controlling lipid droplet metabolism through the estrogen receptor in adipose tissues [27].
Conclusion
The present study comprehensively evaluated the expression profiles and prognostic value of PLIN family genes in breast cancer. The findings revealed that the PLIN family members were aberrantly expressed in breast cancer tissues, suggesting that they may play important roles in the progression of breast cancer by affecting the metabolism of LDs in the regulation of estrogen. In addition, survival analysis showed that PLIN1 could be a favorable prognostic marker for breast cancer although overexpression of PLIN2 was associated with poor prognosis. All in all, genes in the PLIN family may serve as novel therapeutic targets and prognostic biomarkers for breast cancer. However, further research is required to confirm the above results.
Acknowledgements
The present study was supported by the Medicine Health Science and Technology Program of Zhejiang Province (2020368427).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Olzmann JA, Carvalho P. Dynamics and functions of lipid droplets. Nat Rev Mol Cell Biol. 2019;20:137–155. doi: 10.1038/s41580-018-0085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wellen KE, Thompson CB. Cellular metabolic stress: considering how cells respond to nutrient excess. Mol Cell. 2010;40:323–332. doi: 10.1016/j.molcel.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bensaad K, Favaro E, Lewis CA, Peck B, Lord S, Collins JM, Pinnick KE, Wigfield S, Buffa FM, Li JL, Zhang Q, Wakelam MJO, Karpe F, Schulze A, Harris AL. Fatty acid uptake and lipid storage induced by HIF-1alpha contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 2014;9:349–365. doi: 10.1016/j.celrep.2014.08.056. [DOI] [PubMed] [Google Scholar]
- 5.Koizume S, Miyagi Y. Lipid Droplets: a key cellular organelle associated with cancer cell survival under normoxia and hypoxia. Int J Mol Sci. 2016;17:1430. doi: 10.3390/ijms17091430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tirinato L, Liberale C, Di Franco S, Candeloro P, Benfante A, La Rocca R, Potze L, Marotta R, Ruffilli R, Rajamanickam VP, Malerba M, De Angelis F, Falqui A, Carbone E, Todaro M, Medema JP, Stassi G, Di Fabrizio E. Lipid droplets: a new player in colorectal cancer stem cells unveiled by spectroscopic imaging. Stem Cells. 2015;33:35–44. doi: 10.1002/stem.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menard JA, Christianson HC, Kucharzewska P, Bourseau-Guilmain E, Svensson KJ, Lindqvist E, Indira Chandran V, Kjellen L, Welinder C, Bengzon J, Johansson MC, Belting M. Metastasis stimulation by hypoxia and acidosis-induced extracellular lipid uptake is mediated by proteoglycan-dependent endocytosis. Cancer Res. 2016;76:4828–4840. doi: 10.1158/0008-5472.CAN-15-2831. [DOI] [PubMed] [Google Scholar]
- 8.Kimmel AR, Sztalryd C. The perilipins: major cytosolic lipid droplet-associated proteins and their roles in cellular lipid storage, mobilization, and systemic homeostasis. Annu Rev Nutr. 2016;36:471–509. doi: 10.1146/annurev-nutr-071813-105410. [DOI] [PubMed] [Google Scholar]
- 9.Bickel PE, Tansey JT, Welte MA. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim Biophys Acta. 2009;1791:419–440. doi: 10.1016/j.bbalip.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okumura T. Role of lipid droplet proteins in liver steatosis. J Physiol Biochem. 2011;67:629–636. doi: 10.1007/s13105-011-0110-6. [DOI] [PubMed] [Google Scholar]
- 11.Itabe H, Yamaguchi T, Nimura S, Sasabe N. Perilipins: a diversity of intracellular lipid droplet proteins. Lipids Health Dis. 2017;16:83. doi: 10.1186/s12944-017-0473-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Straub BK, Witzel HR, Pawella LM, Renner M, Eiteneuer E, Hashani M, Schirmacher P, Roth W, Mechtersheimer G. Perilipin 1 expression differentiates liposarcoma from other types of soft tissue sarcoma. Am J Pathol. 2019;189:1547–1558. doi: 10.1016/j.ajpath.2019.04.017. [DOI] [PubMed] [Google Scholar]
- 13.Asimakopoulou A, Vucur M, Luedde T, Schneiders S, Kalampoka S, Weiss TS, Weiskirchen R. Perilipin 5 and lipocalin 2 expression in hepatocellular carcinoma. Cancers (Basel) 2019;11:385. doi: 10.3390/cancers11030385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao Q, Ruan H, Wang K, Song Z, Bao L, Xu T, Xiao H, Wang C, Cheng G, Tong J, Meng X, Liu D, Yang H, Chen K, Zhang X. Overexpression of PLIN2 is a prognostic marker and attenuates tumor progression in clear cell renal cell carcinoma. Int J Oncol. 2018;53:137–147. doi: 10.3892/ijo.2018.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou C, Wang M, Zhou L, Zhang Y, Liu W, Qin W, He R, Lu Y, Wang Y, Chen XZ, Tang J. Prognostic significance of PLIN1 expression in human breast cancer. Oncotarget. 2016;7:54488–54502. doi: 10.18632/oncotarget.10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 17.Abramczyk H, Surmacki J, Kopec M, Olejnik AK, Lubecka-Pietruszewska K, Fabianowska-Majewska K. The role of lipid droplets and adipocytes in cancer. Raman imaging of cell cultures: MCF10A, MCF7, and MDA-MB-231 compared to adipocytes in cancerous human breast tissue. Analyst. 2015;140:2224–2235. doi: 10.1039/c4an01875c. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, Frost S, Khushi M, Cantrill LC, Yu H, Arthur JW, Bright RK, Groblewski GE, Byrne JA. Delayed recruiting of TPD52 to lipid droplets - evidence for a “second wave” of lipid droplet-associated proteins that respond to altered lipid storage induced by Brefeldin A treatment. Sci Rep. 2019;9:9790. doi: 10.1038/s41598-019-46156-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sirois I, Aguilar-Mahecha A, Lafleur J, Fowler E, Vu V, Scriver M, Buchanan M, Chabot C, Ramanathan A, Balachandran B, Légaré S, Przybytkowski E, Lan C, Krzemien U, Cavallone L, Aleynikova O, Ferrario C, Guilbert MC, Benlimame N, Saad A, Alaoui-Jamali M, Saragovi HU, Josephy S, O’Flanagan C, Hursting SD, Richard VR, Zahedi RP, Borchers CH, Bareke E, Nabavi S, Tonellato P, Roy JA, Robidoux A, Marcus EA, Mihalcioiu C, Majewski J, Basik M. A unique morphological phenotype in chemoresistant triple-negative breast cancer reveals metabolic reprogramming and PLIN4 expression as a molecular vulnerability. Mol Cancer Res. 2019;17:2492–2507. doi: 10.1158/1541-7786.MCR-19-0264. [DOI] [PubMed] [Google Scholar]
- 20.Jung YY, Kim HM, Koo JS. Expression of lipid metabolism-related proteins in metastatic breast cancer. PLoS One. 2015;10:e0137204. doi: 10.1371/journal.pone.0137204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuniyoshi S, Miki Y, Sasaki A, Iwabuchi E, Ono K, Onodera Y, Hirakawa H, Ishida T, Yoshimi N, Sasano H. The significance of lipid accumulation in breast carcinoma cells through perilipin 2 and its clinicopathological significance. Pathol Int. 2019;69:463–471. doi: 10.1111/pin.12831. [DOI] [PubMed] [Google Scholar]
- 22.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldman M, Craft B, Hastie M, Repečka K, McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN, Zhu J, Haussler D. The UCSC Xena platform for public and private cancer genomics data visualization and interpretation. Nat Biotechnol. 2020;38:675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 25.Sztalryd C, Kimmel AR. Perilipins: lipid droplet coat proteins adapted for tissue-specific energy storage and utilization, and lipid cytoprotection. Biochimie. 2014;96:96–101. doi: 10.1016/j.biochi.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S, Lee Y, Koo JS. Differential expression of lipid metabolism-related proteins in different breast cancer subtypes. PLoS One. 2015;10:e0119473. doi: 10.1371/journal.pone.0119473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christian M. Nuclear receptor-mediated regulation of lipid droplet-associated protein gene expression in adipose tissue. Horm Mol Biol Clin Investig. 2013;14:87–97. doi: 10.1515/hmbci-2013-0028. [DOI] [PubMed] [Google Scholar]
- 28.Straub BK, Gyoengyoesi B, Koenig M, Hashani M, Pawella LM, Herpel E, Mueller W, Macher-Goeppinger S, Heid H, Schirmacher P. Adipophilin/perilipin-2 as a lipid droplet-specific marker for metabolically active cells and diseases associated with metabolic dysregulation. Histopathology. 2013;62:617–631. doi: 10.1111/his.12038. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki K, Takahashi K, Nishimaki-Mogami T, Kagechika H, Yamamoto M, Itabe H. Docosahexaenoic acid induces adipose differentiation-related protein through activation of retinoid x receptor in human choriocarcinoma BeWo cells. Biol Pharm Bull. 2009;32:1177–1182. doi: 10.1248/bpb.32.1177. [DOI] [PubMed] [Google Scholar]
- 30.Schaiff WT, Bildirici I, Cheong M, Chern PL, Nelson DM, Sadovsky Y. Peroxisome proliferator-activated receptor-gamma and retinoid X receptor signaling regulate fatty acid uptake by primary human placental trophoblasts. J Clin Endocrinol Metab. 2005;90:4267–4275. doi: 10.1210/jc.2004-2265. [DOI] [PubMed] [Google Scholar]
- 31.Ambrosio MR, Piccaluga PP, Ponzoni M, Rocca BJ, Malagnino V, Onorati M, De Falco G, Calbi V, Ogwang M, Naresh KN, Pileri SA, Doglioni C, Leoncini L, Lazzi S. The alteration of lipid metabolism in Burkitt lymphoma identifies a novel marker: adipophilin. PLoS One. 2012;7:e44315. doi: 10.1371/journal.pone.0044315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujimoto M, Matsuzaki I, Yamamoto Y, Yoshizawa A, Warigaya K, Iwahashi Y, Kojima F, Furukawa F, Murata SI. Adipophilin expression in cutaneous malignant melanoma. J Cutan Pathol. 2017;44:228–236. doi: 10.1111/cup.12868. [DOI] [PubMed] [Google Scholar]
- 33.Fujimoto M, Yoshizawa A, Sumiyoshi S, Sonobe M, Menju T, Hirata M, Momose M, Date H, Haga H. Adipophilin expression in lung adenocarcinoma is associated with apocrine-like features and poor clinical prognosis: an immunohistochemical study of 328 cases. Histopathology. 2017;70:232–241. doi: 10.1111/his.13048. [DOI] [PubMed] [Google Scholar]
- 34.Pawlik A, Słomińska-Wojewódzka M, Herman-Antosiewicz A. Sensitization of estrogen receptor-positive breast cancer cell lines to 4-hydroxytamoxifen by isothiocyanates present in cruciferous plants. Eur J Nutr. 2016;55:1165–1180. doi: 10.1007/s00394-015-0930-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tolkach Y, Lüders C, Meller S, Jung K, Stephan C, Kristiansen G. Adipophilin as prognostic biomarker in clear cell renal cell carcinoma. Oncotarget. 2017;8:28672–28682. doi: 10.18632/oncotarget.15639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu X, Gruia-Gray J, Copeland NG, Gilbert DJ, Jenkins NA, Londos C, Kimmel AR. The murine perilipin gene: the lipid droplet-associated perilipins derive from tissue-specific, mRNA splice variants and define a gene family of ancient origin. Mamm Genome. 2001;12:741–749. doi: 10.1007/s00335-01-2055-5. [DOI] [PubMed] [Google Scholar]
- 37.Straub BK, Herpel E, Singer S, Zimbelmann R, Breuhahn K, Macher-Goeppinger S, Warth A, Lehmann-Koch J, Longerich T, Heid H, Schirmacher P. Lipid droplet-associated PAT-proteins show frequent and differential expression in neoplastic steatogenesis. Mod Pathol. 2010;23:480–492. doi: 10.1038/modpathol.2009.191. [DOI] [PubMed] [Google Scholar]
- 38.Wang K, Ruan H, Song Z, Cao Q, Bao L, Liu D, Xu T, Xiao H, Wang C, Cheng G, Tong J, Meng X, Yang H, Chen K, Zhang X. PLIN3 is up-regulated and correlates with poor prognosis in clear cell renal cell carcinoma. Urol Oncol. 2018;36:343.e9–343.e19. doi: 10.1016/j.urolonc.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 39.Szigeti A, Minik O, Hocsak E, Pozsgai E, Boronkai A, Farkas R, Balint A, Bodis J, Sumegi B, Bellyei S. Preliminary study of TIP47 as a possible new biomarker of cervical dysplasia and invasive carcinoma. Anticancer Res. 2009;29:717–724. [PubMed] [Google Scholar]
- 40.Zhang Q, Zhang P, Li B, Dang H, Jiang J, Meng L, Zhang H, Zhang Y, Wang X, Li Q, Wang Y, Liu C, Li F. The expression of perilipin family proteins can be used as diagnostic markers of liposarcoma and to differentiate subtypes. J Cancer. 2020;11:4081–4090. doi: 10.7150/jca.41736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolins NE, Skinner JR, Schoenfish MJ, Tzekov A, Bensch KG, Bickel PE. Adipocyte protein S3-12 coats nascent lipid droplets. J Biol Chem. 2003;278:37713–37721. doi: 10.1074/jbc.M304025200. [DOI] [PubMed] [Google Scholar]
- 42.Nimura S, Yamaguchi T, Ueda K, Kadokura K, Aiuchi T, Kato R, Obama T, Itabe H. Olanzapine promotes the accumulation of lipid droplets and the expression of multiple perilipins in human adipocytes. Biochem Biophys Res Commun. 2015;467:906–912. doi: 10.1016/j.bbrc.2015.10.045. [DOI] [PubMed] [Google Scholar]
- 43.Zhang J, Huang JY, Chen YN, Yuan F, Zhang H, Yan FH, Wang MJ, Wang G, Su M, Lu G, Huang Y, Dai H, Ji J, Zhang J, Zhang JN, Jiang YN, Chen SJ, Zhu ZG, Yu YY. Whole genome and transcriptome sequencing of matched primary and peritoneal metastatic gastric carcinoma. Sci Rep. 2015;5:13750. doi: 10.1038/srep13750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qu LW, Zhou B, Wang GZ, Chen Y, Zhou GB. Genomic variations in paired normal controls for lung adenocarcinomas. Oncotarget. 2017;8:104113–104122. doi: 10.18632/oncotarget.22020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trevino MB, Machida Y, Hallinger DR, Garcia E, Christensen A, Dutta S, Peake DA, Ikeda Y, Imai Y. Perilipin 5 regulates islet lipid metabolism and insulin secretion in a cAMP-dependent manner: implication of its role in the postprandial insulin secretion. Diabetes. 2015;64:1299–1310. doi: 10.2337/db14-0559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin J, Chen A. Perilipin 5 restores the formation of lipid droplets in activated hepatic stellate cells and inhibits their activation. Lab Invest. 2016;96:791–806. doi: 10.1038/labinvest.2016.53. [DOI] [PubMed] [Google Scholar]
- 47.Bosma M, Minnaard R, Sparks LM, Schaart G, Losen M, de Baets MH, Duimel H, Kersten S, Bickel PE, Schrauwen P, Hesselink MK. The lipid droplet coat protein perilipin 5 also localizes to muscle mitochondria. Histochem Cell Biol. 2012;137:205–216. doi: 10.1007/s00418-011-0888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuramoto K, Sakai F, Yoshinori N, Nakamura TY, Wakabayashi S, Kojidani T, Haraguchi T, Hirose F, Osumi T. Deficiency of a lipid droplet protein, perilipin 5, suppresses myocardial lipid accumulation, thereby preventing type 1 diabetes-induced heart malfunction. Mol Cell Biol. 2014;34:2721–2731. doi: 10.1128/MCB.00133-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burlaka AP, Ganusevich II, Vovk AV, Burlaka AA, Gafurov MR, Lukin SN. Redox state of adipose tissue for patients with gastric cancer and its connection with the body mass index and distance from the tumor. Obes Res Clin Pract. 2020;14:34–38. doi: 10.1016/j.orcp.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 50.Ye J, Liu H, Xu ZL, Zheng L, Liu RY. Identification of a multidimensional transcriptome prognostic signature for lung adenocarcinoma. J Clin Lab Anal. 2019;33:e22990. doi: 10.1002/jcla.22990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Q, Luo Q, Halim A, Song G. Targeting lipid metabolism of cancer cells: a promising therapeutic strategy for cancer. Cancer Lett. 2017;401:39–45. doi: 10.1016/j.canlet.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 52.Ma L, Shi W, Ma X, Zou M, Chen W, Li W, Zou R, Chen X. Comprehensive analysis of differential immunocyte infiltration and the potential ceRNA networks during epicardial adipose tissue development in congenital heart disease. J Transl Med. 2020;18:111. doi: 10.1186/s12967-020-02279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rovito D, Gionfriddo G, Barone I, Giordano C, Grande F, De Amicis F, Lanzino M, Catalano S, Andò S, Bonofiglio D. Ligand-activated PPARγ downregulates CXCR4 gene expression through a novel identified PPAR response element and inhibits breast cancer progression. Oncotarget. 2016;7:65109–65124. doi: 10.18632/oncotarget.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, Wang Q, Dong L, Liu C, Sun Z, Gao L, Wang X. Morusin suppresses breast cancer cell growth in vitro and in vivo through C/EBPβ and PPARγ mediated lipoapoptosis. J Exp Clin Cancer Res. 2015;34:137. doi: 10.1186/s13046-015-0252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang Y, Zou L, Zhang C, He S, Cheng C, Xu J, Lu W, Zhang Y, Zhang H, Wang D, Shen A. PPARgamma and Wnt/beta-Catenin pathway in human breast cancer: expression pattern, molecular interaction and clinical/prognostic correlations. J Cancer Res Clin Oncol. 2009;135:1551–1559. doi: 10.1007/s00432-009-0602-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu YY, Liu H, Su L, Xu N, Xu DH, Liu HY, Spaner D, Bed-David Y, Li YJ. PPARγ inhibits breast cancer progression by upregulating PTPRF expression. Eur Rev Med Pharmacol Sci. 2019;23:9965–9977. doi: 10.26355/eurrev_201911_19563. [DOI] [PubMed] [Google Scholar]
- 57.Aranaz P, Navarro-Herrera D, Zabala M, Miguéliz I, Romo-Hualde A, López-Yoldi M, Martínez JA, Vizmanos JL, Milagro FI, González-Navarro CJ. Phenolic compounds inhibit 3T3-L1 adipogenesis depending on the stage of differentiation and their binding affinity to PPARγ. Molecules. 2019;24:1045. doi: 10.3390/molecules24061045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu L, Cui H, Xing S, Zhao G, Wen J. Effect of divergent selection for intramuscular fat content on muscle lipid metabolism in chickens. Animals (Basel) 2019;10:4. doi: 10.3390/ani10010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.