Abstract
Advances in laboratory testing have significantly increased the detection of rare genetic etiologies of neurodevelopmental psychiatric disorders (NPD), particularly developmental delay / intellectual disability, autism spectrum disorder, and schizophrenia. Establishing a genetic diagnosis has important medical and personal utility for individuals with these conditions. Diagnostic genetic tests for NPD are clinically available but under-utilized outside of medical genetics settings. Without clear multidisciplinary consensus recommendations, active involvement of medical specialists working with NPD patients, and practical education and training, the implementation of genetic testing for NPD will continue to lag behind other areas of medicine. In the long-term, collaborative efforts to address educational, logistical, and workforce obstacles will improve patient care and pave the way for targeted, effective NPD treatments.
Keywords: neurodevelopmental, psychiatric, genetic disorders, diagnostic genetic testing, copy number variants, single nucleotide variants, Autism, Schizophrenia, developmental delay, intellectual disability, genetic counseling, implementation, child neurology, child psychiatry, developmental pediatrics, precision medicine, Genomics, genomic sequencing
Introduction
Advances in clinical laboratory testing have significantly increased our ability to detect rare genetic etiologies of developmental and psychiatric conditions, including developmental delay / intellectual disability (DD/ID), autism spectrum disorder (ASD) and schizophrenia (SCZ). These and other disorders, such as attention deficit and hyperactivity disorder (ADHD) and Tourette’s disorder, show overlap in symptomatologies and frequently occur as co-morbid conditions. As such, they can be categorized under the broad umbrella of neurodevelopmental psychiatric disorders (NPD). While NPD refers to this full continuum of pediatric and adult brain disorders, our primary focus here is on DD/ID, ASD, and SCZ, as these NPD have the most compelling evidence in support of routine clinical genetic testing.
Various diagnostic genetic tests to identify the underlying etiologies of NPD have been clinically available for decades. Beginning with low resolution karyotypes as far back as the 1960s, and advancing through today’s next generation sequencing technologies, these tests have been almost exclusively employed by a small cadre of medical geneticists, limiting their clinical reach in NPD populations. A major breakthrough in genetic diagnosis came with the introduction of chromosomal microarray analysis (CMA) in the early 2000s, allowing clinical identification of many submicroscopic copy number variants (CNVs) (i.e., microdeletions and microduplications) which had previously gone undetected [1,2]. Over a decade of experience with CMA has revealed dozens of pathogenic CNVs that confer large effects on brain function and cut across a broad range of clinical NPD [3–5]. These CNVs have primary neurodevelopmental and psychiatric manifestations but vary with regard to the presence of congenital anomalies and overt facial dysmorphism [6,7].
Currently, the clinical availability of exome sequencing (ES) has significantly increased the diagnostic yield of genetic testing [8–10]. ES detects rare, pathogenic sequence variations in the coding regions of the genome, allowing the diagnosis of single gene etiologies of NPD. Many commercial laboratories have now expanded their ES analyses to also detect CNVs, eliminating the need for separate CMA in most cases [11]. At the same time, the cost of ES has dropped dramatically, while coverage by health insurance plans in some countries has increased, particularly for pediatric developmental indications [12]. The result has been an exponential rise over the past decade in the number of known genetic etiologies of NPD. Several large research surveys have reported pathogenic copy number and single gene variants for a combined yield of at least 40% of DD/ID and 25% of ASD [9,13–15]. Emerging data from SCZ cohorts has identified CNVs in up to 8% and sequence variants in 1–2% [16–21], with higher percentages among SCZ subgroups having significant cognitive disabilities and/or congenital anomalies [22,23]. Further SCZ studies are required to confirm these associations at a level beyond chance expectations and will likely increase the diagnostic yield. The high prevalence of rare causative variants in DD/ID and ASD has been corroborated by data from years of clinical testing in medical genetics practice. By contrast, diagnostic testing for SCZ etiologies is rarely offered in clinical settings. While preliminary research suggests more modest diagnostic yields for CMA and ES in SCZ – pointing to a predominantly polygenic etiology - our understanding of the contributory role of rare genetic variants in this population is still evolving [24,25].
Diagnostic Genetic Testing for NPD in Clinical Practice
Studies of rare genetic NPD etiologies have confirmed extensive phenotypic variability, even within families. The 1q21.1 recurrent microdeletion, for example, may manifest as ASD in a child, bipolar disorder and epilepsy in her mother, and a variety of different cognitive and behavioral symptoms in the extended family [26]. These observations have called into question the biological validity of current clinical diagnostic approaches, given their lack of consistency with the shared underlying biology and dimensional nature of neuropsychiatric traits [27,28]. They also highlight the important family implications of genetic diagnosis [3,29] and the potential for new treatment discoveries through cross-disorder research strategies [30]. Against this dynamic backdrop of gene discovery and dimensional models of brain disorders, however, the adoption of widely available diagnostic testing by non-geneticist NPD specialists (e.g., psychiatrists, developmental pediatricians, neurologists) has been slow to take root [31–33]. Currently, clinical genetic testing for NPD remains almost entirely focused on young children, mainly targeting DD/ID and/or ASD, and implemented through referral to medical genetics specialists. This contrasts with other areas of medicine, such as Cardiology and Oncology where diagnostic genetic testing is already well-integrated into routine clinical care [34,35].
Published consensus guidelines from various expert panels and professional societies are in place for NPD commonly encountered by pediatric and adult brain specialists. For example, ES, CMA, and fragile X analysis are recommended by several groups for the evaluation of children with DD/ID and ASD, regardless of the presence of physical anomalies [2,9,36–39]. Similar guidelines for adults with these disorders have been slower to emerge, although evidence is building in support of the clinical and personal utility of diagnostic testing beyond childhood [3,40,41]. In both pediatric and adult medicine, genetic testing recommendations for the same clinical disorder may vary, depending on the particular professional group cited, the recency of its review, and the strength and clarity of its published statement. For example, the American College of Medical Genetics (ACMG) has long recommended fragile X and CMA for the evaluation of all children with DD/ID and ASD [36,38]. ACMG has not yet issued similar guidance for ES [42], although a separate consensus statement by multiple expert groups strongly recommends ES as a first-tier clinical test, citing evidence for high diagnostic yields in DD/ID and ASD [9]. The United Kingdom-based National Institute for Health and Care Excellence advocates a more restricted, traditional approach to genetic testing, based on input from Medical Genetics and weighted on the presence of dysmorphic features and congenital anomalies [43]. Some individual research groups have urged genetic testing for patients with SCZ [22,40], given reports of clinically relevant CNVs in this population; however, recommendations from professional psychiatric organizations have been more tepid. Practice guidelines from the American Psychiatric Association mention “genetic testing” without further description in a table of “suggested assessments” for individuals with SCZ [44]. The Canadian Psychiatry Association endorses consideration of “genetic testing based on the history and physical examination of the patient, especially at the time of the first episode of psychosis” [45]. The International Society of Psychiatric Genetics suggests that diagnostic testing “may have value” for patients with DD/ID and ASD, while offering no specific recommendations on SCZ [46].
In principal, best practices in clinical care should be agnostic to reimbursement and access issues, yet short-term concerns about the regional availability of certain genetic tests can weaken guidance. Such reticence is reflected in vague position statements that fail to adequately inform clinical decision-making, and in some cases, inappropriately advise the continued use of outdated technologies, such as karyotyping [37]. The lack of consistent, cross-disciplinary recommendations has slowed clinical implementation of diagnostic genetic testing for NPD, making it difficult to assess its diagnostic yield and utility outside of a research context. At the same time, the absence of clinical testing data, particularly for SCZ, has hampered the development of consensus guidelines, which would in turn drive insurance reimbursement. As a result of this circular dilemma, genetic testing for SCZ has so far failed to gain traction toward widespread implementation outside of medical genetics settings.
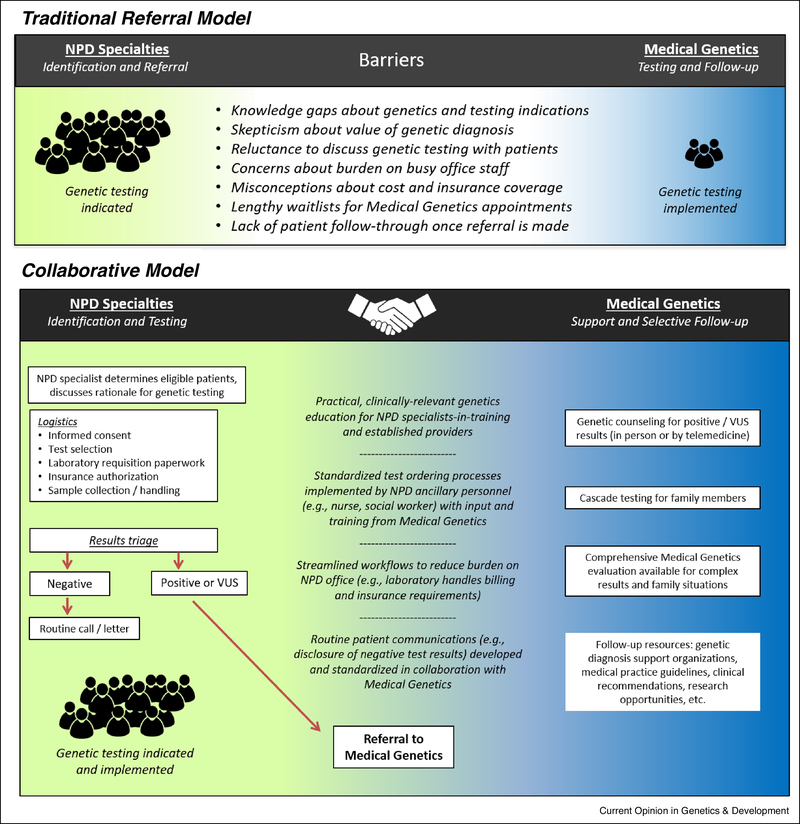
Surveys of NPD specialists reveal that they rarely initiate diagnostic testing, even in patients with ASD or ID/DD where the recommendations are strongest [33]. This is confirmed in studies of patient experiences, including a recent finding that only 3% of children with ASD had undergone both fragile X testing and CMA [31]. Reported barriers to implementation include: clinicians’ lack of training in medical genetics; a scarcity of genetic counselors to manage consenting, results disclosure, and family follow-up; uncertainty about which test to choose; and difficulty interpreting and explaining test results. Additional obstacles include concerns about insurance coverage, as well as skepticism about the clinical utility of genetic diagnosis. It appears that NPD specialists are largely aware of genetic testing recommendations but rarely implement them [33]. Those who do most often refer patients for Medical Genetics specialty appointments without directly ordering recommended laboratory testing (Figure 1). Given the long waiting lists and limited availability of medical geneticists, this continued reliance on inefficient referral practices greatly reduces the chance of testing follow-through by patients with NPD and their families. Psychiatry, Neurology, and Developmental Pediatrics currently trail other areas of specialty medicine in integrating genetic testing into patient care [34,35], despite relatively high diagnostic yields and robust evidence for clinical utility in NPD. This lag also has important downstream implications for NPD research, as advances in precision medicine hinge on the identification of rare genomic variants which, when paired with clinical correlation, can fuel treatment breakthroughs [47,48].
Fig. 1.
A traditional approach of referring patients to Medical Genetics is widely used by NPD clinicians but fails to achieve adequate implementation of genetic testing recommendations. A collaborative model facilitates consistent genetic testing and follow-up for patients with NPD and their families.
Closing the Implementation Gap
We propose here three practical steps to address the current obstacles in the way of a successful evidence-based implementation of genetic testing for NPD in the clinic (Table 1.):
Table 1:
Strategies to Increase Implementation of Diagnostic Genetic Testing for NPD
| Goal | Current Status | Proposed Strategy |
|---|---|---|
| Establishment of cross-disciplinary consensus recommendations on NPD genetic testing | • Competing and conflicting guidelines by multiple professional groups • Siloed dissemination and inconsistent compliance within NPD specialty areas |
• Cross-disciplinary consensus and publication of a single, authoritative source of genetic testing guidance for children and adults with NPD • Regular updates to reflect changing laboratory technology and research evidence • Widespread dissemination across NPD clinical specialties and professional groups |
| Streamlined clinical workflows that minimize the burden of genetic test ordering and follow-up | • High administrative and logistical burden on NPD clinicians and office personnel • Low rates of implementation despite awareness of clinical recommendations • Reliance on traditional model of referral to Medical Genetics |
• Collaboration and training from regional Medical Genetics resources on test selection, consent procedures, and results triage • NPD office staff and testing laboratory develop shared processes for billing, specimen collection, sample shipping, etc. • Standardized protocols for disclosure of negative results to patients, with positive and ambiguous results referred to Medical Genetics |
| Practical genetics education for trainees and established NPD clinicians | • Inconsistent knowledge base in medical genetics across NPD specialists • Need for NPD-specific training on test selection, diagnostic yields, results interpretation, and communication with patients and families |
• Development of practical curricula for NPD trainees and established clinicians, with a focus on essential knowledge related to diagnostic genetic testing • Increased clinician confidence in genetic test implementation reinforced through training and illustrative positive experiences of patients diagnosed with genetic etiologies of NPD |
1. Establishment of cross-disciplinary consensus recommendations
Starting with diagnostic genetic testing for DD/ID, ASD, and SCZ, efforts are needed to synthesize competing recommendations, guidelines, and position statements into a single authoritative source of clear, prescriptive guidance. Recommendations should encompass both children and adults with DD/ID, ASD and SCZ, regardless of whether these diagnoses present as the primary referral indication or an observed secondary comorbidity. Consistent expert opinion, regularly updated to reflect changing laboratory technologies and relevant NPD discoveries, should continually inform test selection, including the future clinical use of polygenic risk scores [24,49]. Ideally, a joint practice guideline authored by representatives from multiple professional societies and stakeholders would ensure broad cross-disciplinary dissemination to NPD medical care providers. As the professional discipline most often on the front lines of diagnostic laboratory testing, Medical Genetics is an obvious choice to lead such an initiative, in close collaboration with relevant clinical NPD groups. In the United States, there is precedent for such an approach in Oncology, where genetic testing and surveillance recommendations are consolidated under widely implemented guidelines of the National Comprehensive Cancer Network [50]. As a secondary benefit, the establishment of national testing recommendations for cancer informed US healthcare policy and insurance coverage efforts, areas that remain to be addressed for NPD.
2. Creation of streamlined clinical workflows that minimize the burden of genetic test ordering and follow-up
For busy clinicians unfamiliar with genetic test selection and ordering in the age of next generation sequencing, the prospect of directly handling this aspect of NPD care can seem daunting [32,33,51]. However, the traditional practice of simply referring patients to a medical geneticist is no longer an acceptable way to ensure that testing recommendations are followed, given the volume of eligible patients and the scarcity of medical genetics providers. Fortunately, genetic test selection for DD/ID and ASD is relatively straightforward, in most cases involving a tiered strategy of fragile X analysis reflexing, if negative, to ES with CNV detection on the same patient sample. Local practices may vary but should be made transparent and available to ordering NPD clinicians. CMA may be the diagnostic test of choice for SCZ, rather than ES, based on the current low yield of detectable sequence variants. In North America, commercial genetics laboratories typically employ genetic counselors who serve as a technical resource for ordering providers, ensuring appropriate test selection. DNA can now be reliably extracted for analysis from saliva or oral buccal samples in most cases, facilitating specimen collection and shipping. Many labs also provide solutions for some of the most time-consuming aspects of genetic testing, including full-service handling of insurance pre-authorization requirements and patient billing.
Aside from these logistical considerations, there are more nuanced aspects of the genetic testing process that NPD clinicians can efficiently manage through careful workflow planning in collaboration with a regional genetics group. For example, genetic testing has the potential to reveal unanticipated findings with implications beyond the original testing indication, including the discovery of sensitive information about family relationships (e.g., non-paternity), as well as medically-actionable secondary genomic findings [52]. For this reason, an informed consent process is recommended prior to testing to alert patients to the types of results that might be revealed, while offering options for limited disclosure of secondary findings. Obtaining consent for genetic testing does not have to be an onerous process [53,54] and with appropriate education can be efficiently carried out by NPD specialists themselves or delegated to trained office staff.
NPD clinicians may not feel adequately prepared to interpret genetic laboratory test reports and may be reluctant to take on responsibility for ordering an unfamiliar test [32]. By establishing a mutually beneficial shared workflow with regional genetics providers, test results can be efficiently triaged, with a Medical Genetics referral generated only for those patients with positive or ambiguous results, as illustrated in figure 1. Cross-disciplinary collaborations such as these can increase adherence to genetic testing recommendations while advancing clinicians’ knowledge about rare NPD etiologies. In the long-term, positive outcomes related to genetically-informed medical care and patient satisfaction will further reinforce the value of clinical genetic testing in the eyes of NPD specialists.
3. Development of practical genetics education strategies for new NPD trainees and established clinicians
At present, many established and future NPD clinicians, particularly psychiatrists, lack the necessary training that would allow them to routinely consider genetic testing as part of their diagnostic armamentarium [55]. Given the evidence for an increasing proportion of NPD patients for whom a genetic etiology can be identified, and the clinical importance of elucidating this aspect of their diagnosis, the inclusion of basic genetic knowledge in the training curricula for NPD clinicians is both logical and necessary. The extent of information offered during training does not have to cover a deep understanding of genetic architecture or the technical workings of various genetic testing methods, but should be sufficient to provide the clinician with confidence to consider genetic testing as part of the diagnostic work-up in his or her own practice. At the very minimum, basic knowledge should include a broad understanding of the current evidence from genetic studies which provides the rationale for testing; the different genetic testing methods and their specific yields; which pros and cons of testing should be discussed with patients; the practical aspects of ordering genetic testing; how workflows are organized locally (see previous section), and how test results are interpreted and communicated with the family. Recently, Nurnberger et al. [55] have proposed an inventory of themes in genetics along with recommendations to include these as essential knowledge components in the training of psychiatrists.
Conclusions
While individually rare, genomic copy number and sequence variants collectively account for a significant proportion of DD/ID, ASD, and SCZ etiologies. Establishing an underlying genetic diagnosis has important medical and personal utility for individuals with these conditions, while also unlocking rare disorder research opportunities that will drive precision medicine breakthroughs in NPD treatment. Powerful diagnostic genetic tests for NPD are clinically available but currently under-utilized outside of medical genetics settings. Without clear multidisciplinary consensus recommendations, a collaborative workflow regarding genetic testing across the NPD and genetic disciplines, and educational strategies about genetic testing for future NPD clinicians, the implementation of diagnostic genetic testing will continue to lag behind other areas of medicine. In the long-term, collaborative efforts to address educational, logistical, and workforce obstacles will improve patient care and pave the way for targeted, effective NPD treatments.
Acknowledgments
2. Funding
X Funding was received for this work.
All of the sources of funding for the work described in this publication are acknowledged below:
Funding: This work was supported by the National Institute of Mental Health of the National Institutes of Health [grant number U01 MH 119705]
Funding supported an overall aim of the project to increase diagnosis of rare genetic etiologies of neurodevelopmental and psychiatric disorders
□No funding was received for this work.
Footnotes
Author declaration
1. Conflict of Interest
Potential conflict of interest exists:
We wish to draw the attention of the Editor to the following facts, which may be considered as potential conflicts of interest, and to significant financial contributions to this work:
The nature of potential conflict of interest is described below:
Brenda Finucane serves as a consultant for the National Fragile X Foundation
Jacob A.S. Vorstman serves as a consultant for NoBias Therapeutics Inc.
David H. Ledbetter serves as a scientific consultant to Natera, Inc. and X-Therma, Inc.
We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
3. Intellectual Property
X We confirm that we have given due consideration to the protection of intellectual property associated with this work and that there are no impediments to publication, including the timing of publication, with respect to intellectual property. In so doing we confirm that we have followed the regulations of our institutions concerning intellectual property.
4. Research Ethics Not applicable
□We further confirm that any aspect of the work covered in this manuscript that has involved human patients has been conducted with the ethical approval of all relevant bodies and that such approvals are acknowledged within the manuscript.
□IRB approval was obtained (required for studies and series of 3 or more cases)
□Written consent to publish potentially identifying information, such as details or the case and photographs, was obtained from the patient(s) or their legal guardian(s).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rauch A, Rüschendorf F, Huang J, Trautmann U, Becker C, Thiel C, Jones KW, Reis A, Nürnberg P. Molecular karyotyping using an SNP array for genomewide genotyping. J Med Genet 2004. 41:916–22. doi: 10.1136/jmg.2004.022855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, Church DM, Crolla JA, Eichler EE, Epstein CJ, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet 2010. 86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin CL, Wain KE, Oetjens MT, Tolwinski K, Palen E, Hare-Harris A, Habegger L, Maxwell EK, Reid JG, Walsh LK, et al. Identification of Neuropsychiatric Copy Number Variants in a Health Care System Population. JAMA Psychiatry 2020. 22:e202159. doi: 10.1001/jamapsychiatry.2020.2159. Erratum in: JAMA Psychiatry. 2020 Sep 2: e202861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chawner SJRA, Owen MJ, Holmans P, Raymond FL, Skuse D, Hall J, van den Bree MBM. Genotype-phenotype associations in children with copy number variants associated with high neuropsychiatric risk in the UK (IMAGINE-ID): a case-control cohort study. Lancet Psychiatry 2019. 6:493–505. doi: 10.1016/S2215-0366(19)30123-3. [DOI] [PubMed] [Google Scholar]
- 5.Kirov G CNVs in neuropsychiatric disorders. Hum Mol Genet 2015. 24(R1):R45–49. doi: 10.1093/hmg/ddv253. [DOI] [PubMed] [Google Scholar]
- 6.Owen D, Bracher-Smith M, Kendall KM, Rees E, Einon M, Escott-Price V, Owen MJ, O’Donovan MC, Kirov G. Effects of pathogenic CNVs on physical traits in participants of the UK Biobank. BMC Genomics 2018. 19:867. doi: 10.1186/s12864-018-5292-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, Goldstein A, Filipink RA, McConnell JS, Angle B, Meschino WS, et al. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N Engl J Med 2012. 367:1321–1331. doi: 10.1056/NEJMoa1200395. Erratum in: N Engl J Med, 2012 Dec 13;367(24):2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stefanski A, Calle-López Y, Leu C, Pérez-Palma E, Pestana-Knight E, Lal D. Clinical sequencing yield in epilepsy, autism spectrum disorder, and intellectual disability: A systematic review and meta-analysis. Epilepsia 2020. doi: 10.1111/epi.16755. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srivastava S, Love-Nichols J A, Dies KA, Ledbetter DH, Martin CL, Chung WK, Firth HV, Frazier T, Hansen RL, Prock L, et al. Meta-analysis and multidisciplinary consensus statement: exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet Med 2019. 21:2413–2421. doi: 10.1038/s41436-019-0554-6 Erratum in Genet Med 2020 22:1731–1732.** Based on a meta-analysis of existing studies, the authors conclude that exome sequencing (ES) consistently outperforms chromosomal microarray analysis in terms of diagnostic yield for the etiological evaluation of neurodevelopmental psychiatric disorders (NPD). They recommend elevating ES to a first-tier clinical test for individuals presenting with autism, ID, and other NPD.
- 10.Waggoner D, Wain KE, Dubuc AM, Conlin L, Hickey SE, Lamb AN, Martin CL, Morton CC, Rasmussen K, Schuette JL, et al. Yield of additional genetic testing after chromosomal microarray for diagnosis of neurodevelopmental disability and congenital anomalies: a clinical practice resource of the American College of Medical Genetics and Genomics (ACMG). Genet Med 2018, 20:1105–1113. doi: 10.1038/s41436-018-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quenez O, Cassinari K, Coutant S, Lecoquierre F, Le Guennec K, Rousseau S, Richard AC, Vasseur S, Bouvignies E, Bou J et al. Detection of copy-number variations from NGS data using read depth information: a diagnostic performance evaluation. Eur J Hum Genet 2020. doi: 10.1038/s41431-020-0672-2. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trosman JR, Weldon CB, Slavotinek A, Norton ME, Douglas MP, Phillips KA. Perspectives of US private payers on insurance coverage for pediatric and prenatal exome sequencing: Results of a study from the Program in Prenatal and Pediatric Genomic Sequencing (P3EGS). Genet Med 2020. 22:283–291. doi: 10.1038/s41436-019-0650-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossi M, El-Khechen D, Black MH, Farwell Hagman KD, Tang S, Powis Z. Outcomes of diagnostic exome sequencing in patients with diagnosed or suspected autism spectrum disorders. Pediatr Neurol 2017. 70:34–43.e2. doi: S0887-8994(16)30572-0 [pii]. [DOI] [PubMed] [Google Scholar]
- 14.Tammimies K, Marshall CR, Walker S, Kaur G, Thiruvahindrapuram B, Lionel AC, Yuen RK, Uddin M, Roberts W, Weksberg R et al. Molecular diagnostic yield of chromosomal microarray analysis and whole-exome sequencing in children with autism spectrum disorder. JAMA 2015. 314:895–903. doi: 10.1001/jama.2015.10078 [doi]. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, Braxton A, Beuten J, Xia F, Niu Z et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med 2013. 369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howrigan DP, Rose SA, Samocha KE, Fromer M, Cerrato F, Chen WJ, Churchhouse C, Chambert K, Chandler SD, Daly MJ et al. Exome sequencing in schizophrenia-affected parent-offspring trios reveals risk conferred by protein-coding de novo mutations. Nat Neurosci 2020. 23:185–193. doi: 10.1038/s41593-019-0564-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, Antaki D, Shetty A, Holmans PA, Pinto D, et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet 2017. 49:27–35. doi: 10.1038/ng.3725. Epub 2016 Nov 21. Erratum in: Nat Genet. 2017 49:651. Erratum in: Nat Genet 2017. 49:1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh T, Walters JTR, Johnstone M, Curtis D, Suvisaari J, Torniainen M, Rees E, Iyegbe C, Blackwood D, McIntosh AM, et al. The contribution of rare variants to risk of schizophrenia in individuals with and without intellectual disability. Nat Genet 2017. 49:1167–1173. doi: 10.1038/ng.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genovese G, Fromer M, Stahl EA, Ruderfer DM, Chambert K, Landén M, Moran JL, Purcell SM, Sklar P, Sullivan PF et al. Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nat Neurosci 2016. 19:1433–1441. doi: 10.1038/nn.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rees E, Walters JT, Georgieva L, Isles AR, Chambert KD, Richards AL, Mahoney-Davies G, Legge SE, Moran JL, McCarroll SA, et al. Analysis of copy number variations at 15 schizophrenia-associated loci. Br J Psychiatry 2014. 204:108–114. doi: 10.1192/bjp.bp.113.131052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costain G, Lionel AC, Merico D, Forsythe P, Russell K, Lowther C, Yuen T, Husted J, Stavropoulos DJ, Speevak M et al. Pathogenic rare copy number variants in community-based schizophrenia suggest a potential role for clinical microarrays. Hum Mol Genet 2013. 22:4485–4501. doi: 10.1093/hmg/ddt297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foley C, Heron EA, Harold D, Walters J, Owen M, O’Donovan M, Sebat J, Kelleher E, Mooney C, Durand A, et al. Identifying schizophrenia patients who carry pathogenic genetic copy number variants using standard clinical assessment: retrospective cohort study. Br J Psychiatry 2020. 216:275–279. doi: 10.1192/bjp.2019.262.** The authors describe benefits to clinical identification of copy number variants (CNVs) in individuals with schizophrenia. They propose that patients at highest risk for having a CNV may be identified through careful documentation of neurodevelopmental features.
- 23.Lowther C, Merico D, Costain G, Waserman J, Boyd K, Noor A, Speevak M, Stavropoulos DJ, Wei J, Lionel AC et al. Impact of IQ on the diagnostic yield of chromosomal microarray in a community sample of adults with schizophrenia. Genome Med 2017. 9:105 doi: 10.1186/s13073-017-0488-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davies RW, Fiksinski AM, Breetvelt EJ, Williams NM, Hooper SR, Monfeuga T, Bassett AS, Owen MJ, Gur RE, Morrow BE et al. Using common genetic variation to examine phenotypic expression and risk prediction in 22q11.2 deletion syndrome. Nat Med. 2020. doi: 10.1038/s41591-020-1103-1. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szatkiewicz JP, Fromer M, Nonneman RJ, Ancalade N, Johnson JS, Stahl EA, Rees E, Bergen SE, Hultman CM, Kirov G et al. Characterization of Single Gene Copy Number Variants in Schizophrenia. Biol Psychiatry 2020. 87:736–744. doi: 10.1016/j.biopsych.2019.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernier R, Steinman KJ, Reilly B, Wallace AS, Sherr EH, Pojman N, Mefford HC, Gerdts J, Earl R, Hanson E et al. Clinical phenotype of the recurrent 1q21.1 copy-number variant. Genet Med 2016. 18:341–349. doi: 10.1038/gim.2015.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smoller JW, Andreassen OA, Edenberg HJ, Faraone SV, Glatt SJ, Kendler KS. Psychiatric genetics and the structure of psychopathology. Mol Psychiatry 2019. 24:409–420. doi: 10.1038/s41380-017-0010-4. Erratum in: Mol Psychiatry. 2019. 24:471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moreno-De-Luca A, Myers SM, Challman TD, Moreno-De-Luca D, Evans DW, Ledbetter DH: Developmental brain dysfunction: revival and expansion of old concepts based on new genetic evidence. Lancet Neurol 2013, 12:406–414. doi: 10.1016/S1474-4422(13)70011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finucane B, Myers SM. Genetic counseling for autism spectrum disorder in an evolving theoretical landscape. Curr Genet Med Rep 2016. 4:147–153. doi: 10.1007/s40142-016-0099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bodkin JA, Coleman MJ, Godfrey LJ, Carvalho CMB, Morgan CJ, Suckow RF, Anderson T, Öngür D, Kaufman MJ, Lewandowski KE et al. Targeted Treatment of Individuals with Psychosis Carrying a Copy Number Variant Containing a Genomic Triplication of the Glycine Decarboxylase Gene. Biol Psychiatry 2019. 86:523–535. doi: 10.1016/j.biopsych.2019.04.031.* The authors describe the efficacy of targeted interventions using glycine and D-cycloserine in a mother and son with psychosis, both of whom had a known genomic variant involving a glycine decarboxylase gene. This case report illustrates the potential for successful precision medicine approaches to the treatment of genetic etiologies of psychiatric disorders.
- 31.Moreno-De-Luca D, Kavanaugh BC, Best CR, Sheinkopf SJ, Phornphutkul C, Morrow EM. Clinical Genetic Testing in Autism Spectrum Disorder in a Large Community-Based Population Sample. JAMA Psychiatry 2020. 77:979–981. doi: 10.1001/jamapsychiatry.2020.0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vorstman JAS, Parr JR, Moreno-De-Luca D, Anney RJL, Nurnberger JI Jr, Hallmayer JF. Autism genetics: opportunities and challenges for clinical translation. Nat Rev Genet 2017. 18:362–376. doi: 10.1038/nrg.2017.4. [DOI] [PubMed] [Google Scholar]
- 33.Tchaconas A, Adesman A. Diagnostic Evaluation of Children with Autism Spectrum Disorders: Clinician Compliance with Published Guidelines. J Dev Behav Pediatr 2017. 38:29–38. doi: 10.1097/DBP.0000000000000349. [DOI] [PubMed] [Google Scholar]
- 34.Wilkinson B, George E, Horton S, Bellaby J, Min SS, Gama R. A service evaluation: impact of nurse-led regional familial hypercholesterolaemia service on a hospital adult lipid clinic. Br J Nurs 2020. 29:1206–1208. doi: 10.12968/bjon.2020.29.20.1206. [DOI] [PubMed] [Google Scholar]
- 35.Malone ER, Oliva M, Sabatini PJB, Stockley TL, Siu LL. Molecular profiling for precision cancer therapies. Genome Med 2020. 12:8. doi: 10.1186/s13073-019-0703-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaefer GB, Mendelsohn NJ, Professional Practice and Guidelines Committee. Clinical genetics evaluation in identifying the etiology of autism spectrum disorders: 2013 guideline revisions. Genet Med 2013. 2013:399–407. doi: 10.1038/gim.2013.32. [DOI] [PubMed] [Google Scholar]
- 37.Filipek PA, Accardo PJ, Ashwal S, Baranek GT, Cook EH, Dawson G, Gordon B, Gravel JS, Johnson CP, Kallen RJ et al. Practice parameter: screening and diagnosis of autism: report of the quality standards subcommittee of the American Academy of Neurology and the Child Neurology Society. Neurology 2000. 55:468–479. doi: 10.1212/WNL.55.4.468. [DOI] [PubMed] [Google Scholar]
- 38.Manning M, Hudgins L, Professional Practice and Guidelines Committee. Array-based technology and recommendations for utilization in medical genetics practice for detection of chromosomal abnormalities. Genet Med 2010. 12:742–745. doi: 10.1097/GIM.0b013e3181f8baad. Addendum in: Genet Med. 2020 Jun 8. doi: 10.1038/s41436-020-0848-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson CP, Myers SM, American Academy of Pediatrics Council on Children with Disabilities. Identification and evaluation of children with autism spectrum disorders. Pediatrics 2007. 120:1183–1206. doi: 10.1542/peds.2007-2361. [DOI] [PubMed] [Google Scholar]
- 40.Finucane BM, Myers SM, Martin CL, Ledbetter DH. Long overdue: including adults with brain disorders in precision health initiatives. Curr Opin Genet Dev 2020. 65:47–52. doi: 10.1016/j.gde.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minardi R, Licchetta L, Baroni MC, Pippucci T, Stipa C, Mostacci B, Severi G, Toni F, Bergonzini L, Carelli V et al. Whole-exome sequencing in adult patients with developmental and epileptic encephalopathy: It is never too late. Clin Genet 2020. 98:477–485. doi: 10.1111/cge.13823. [DOI] [PubMed] [Google Scholar]
- 42.Malinowski J, Miller DT, Demmer L, Gannon J, Pereira EM, Schroeder MC, Scheuner MT, Tsai AC, Hickey SE, Shen J et al. Systematic evidence-based review: outcomes from exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability. Genet Med 2020. 22:986–1004. doi: 10.1038/s41436-020-0771-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.National Institute for Health and Care Excellence. Autism spectrum disorder in under 19s: recognition, referral and diagnosis. September 2011. Epub: https://www.nice.org.uk/guidance/cg128/resources/autism-spectrum-disorder-in-under-19s-recognition-referral-and-diagnosis-pdf-35109456621253 [PubMed]
- 44.American Psychiatric Association. The American Psychiatric Association Practice Guideline for the Treatment of Patients with Schizophrenia, Third Edition. December 2019. Epub: https://DOI.ORG/10.1176/APPI.BOOKS.9780890424841
- 45.Canadian Psychiatric Association. Clinical Practice Guideline for Schizophrenia. CPA website; 2020. https://www.cpa-apc.org/clinical-resources/clinical-practice-guidelines/ [Google Scholar]
- 46.Genetic Testing Committee of the International Society of Psychiatric Genetics (ISPG). Genetic Testing Statement: Genetic Testing and Psychiatric Disorders. ISPG Website; 2019. https://ispg.net/genetic-testing-statement/ [Google Scholar]
- 47.Schwaederle M, Zhao M, Lee JJ, Eggermont AM, Schilsky RL, Mendelsohn J, Lazar V, Kurzrock R. Impact of precision medicine in diverse cancers: A meta-analysis of phase II clinical trials. J Clin Oncol 2015. 33:3817–3825. doi: 10.1200/JCO.2015.61.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Endo A A historical perspective on the discovery of statins. Proc Jpn Acad Ser B Phys Biol Sc 2010. 86:484–493. doi: 10.2183/pjab.86.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bergen SE, Ploner A, Howrigan D, CNV Analysis Group and the Schizophrenia Working Group of the Psychiatric Genomics Consortium, O’Donovan MC, Smoller JW, Sullivan PF, Sebat J, Neale B, Kendler KS. Joint contributions of rare copy number variants and common SNPs to risk for schizophrenia. Am J Psychiatry 2019. 176:29–35. doi: 10.1176/appi.ajp.2018.17040467.** Based on data from over 21,000 control-matched individuals with schizophrenia, the authors found evidence for an additive model whereby rare pathogenic copy number variants and common, small-effect SNPs interact to influence schizophrenia risk.
- 50.National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology. NCCN Website; 2020. https://www.nccn.org/professionals/physician_gls/default.aspx [Google Scholar]
- 51.Sullivan PF, Owen MJ . Increasing the Clinical Psychiatric Knowledge Base About Pathogenic Copy Number Variation. Am J Psychiatry 2020. 177:204–209. doi: 10.1176/appi.ajp.2019.19040335.*In this commentary, the authors take their case in support of genetic testing directly to clinical psychiatrists, providing practical information and offering compelling arguments in favor of diagnosing rare genetic etiologies of psychiatric disorders.
- 52.Kalia SS, Adelman K, Bale SJ, Chung WK, Eng C, Evans JP, Herman GE, Hufnagel SB, Klein TE, Korf BR. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med 2017. 19:249–255. doi: 10.1038/gim.2016.190. Erratum in: Genet Med 2017. 19:484. [DOI] [PubMed] [Google Scholar]
- 53.Kraft SA, Porter KM, Duenas DM, Guerra C, Joseph G, Lee SS, Shipman KJ, Allen J, Eubanks D, Kauffman TL et al. Participant Reactions to a Literacy-Focused, Web-Based Informed Consent Approach for a Genomic Implementation Study. AJOB Empir Bioeth 2020. 26:1–11. doi: 10.1080/23294515.2020.1823907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riggs ER, Azzariti DR, Niehaus A, Goehringer SR, Ramos EM, Rodriguez LL, Knoppers B, Rehm HL, Martin CL; Clinical Genome Resource Education Working Group. Development of a consent resource for genomic data sharing in the clinical setting. Genet Med 2019. 21:81–88. doi: 10.1038/s41436-018-0017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nurnberger JI Jr, Austin J, Berrettini WH, Besterman AD, DeLisi LE, Grice DE, Kennedy JL, Moreno-De-Luca D, Potash JB, Ross DA et al. What Should a Psychiatrist Know About Genetics? Review and Recommendations from the Residency Education Committee of the International Society of Psychiatric Genetics. J Clin Psychiatry 2018. 80:17nr12046. doi: 10.4088/JCP.17nr12046. [DOI] [PMC free article] [PubMed] [Google Scholar]