Abstract
Background:
Characterizing the complexity of environmental exposures in relation to human health is critical to advancing our understanding of health and disease throughout the life span. Extant cohort studies open the door for such investigations more rapidly and inexpensively than launching new cohort studies and the Human Health Exposure Analysis Resource (HHEAR) provides a resource for implementing life-stage exposure studies within existing study populations. Primary challenges to incorporation of environmental exposure assessment in health studies include: (1) lack of widespread knowledge of biospecimen and environmental sampling and storage requirements for environmental exposure assessment among investigators; (2) lack of availability of and access to laboratories capable of analyzing multiple environmental exposures throughout the life-course; and (3) studies lacking sufficient power to assess associations across life-stages. HHEAR includes a consortium of researchers with expertise in laboratory analyses, statistics and logistics to overcome these limitations and enable inclusion of exposomics in human health studies.
Objective:
This manuscript describes the structure and strengths of implementing the harmonized HHEAR resource model, and our approaches to addressing challenges. We describe how HHEAR incorporates analyses of biospecimens and environmental samples and human health studies across the life span - serving as a model for incorporating environmental exposures into national and international research. We also present program successes to date.
Discussion:
HHEAR provides a full-service laboratory and data analysis exposure assessment resource, linking scientific, life span, and toxicological consultation with both laboratory and data analysis expertise. HHEAR services are provided without cost but require NIH, NCI, NHLBI, or ECHO funding of the original cohort; internal HHEAR scientific review and approval of a brief application; and adherence to data sharing and publication policies. We describe the benefits of HHEAR’s structure, collaborative framework and coordination across project investigators, analytical laboratories, biostatisticians and bioinformatics specialists; quality assurance/quality control (QA/QC) including integrated sample management; and tools that have been developed to support the research (exposure information pages, ontology, new analytical methods, common QA/QC approach across laboratories, etc.). This foundation supports HHEAR’s inclusion of new laboratory and statistical analysis methods and studies that are enhanced by including targeted analysis of specific exposures and untargeted analysis of chemicals associated with phenotypic endpoints in biological and environmental samples.
Conclusion:
HHEAR is an interdisciplinary teams of toxicologists, epidemiologists, laboratory and data scientists across multiple institutions to address broad and complex questions that benefit from integrated laboratory and data analyses. HHEAR’s processes, features, and tools include all life stages and analysis of biospecimens and environmental samples. They are available to the wider scientific community to augment studies by adding state of the art environmental analyses to be linked to human health outcomes.
Introduction
The etiology of health and disease is increasingly recognized to result from a complex interplay of environmental influences with the genome and biological processes across time. While genomics is a well-established field, the exposome is frequently a key missing component. This is significant as it is estimated that 70 to 90% of chronic disease risks are affected to some extent by the environment (Rappaport et al. 2010). Systematically analyzing multiple exposures and their associations with health endpoints throughout the life-course and across cohorts is the 21st century challenge facing human health researchers (Wild 2005, Cui et al. 2016). The time varying nature of the environment makes exposomics an extraordinarily complex science. DNA sequence is static but environment changes with time and requires multiple assessments. Furthermore, the life-stage at the time of assessment is important.
Specific challenges facing exposomics include the lack of sufficient resources to measure the multiple environmental exposures at life-stage specific windows of susceptibility, while including a sufficient number of subjects to attain the statistical power required to assess associations across developmental life-stages (fetal, infancy, childhood, pre-adolescence, adolescence, etc.). To overcome the latter challenge, evidence accumulated from independently conducted studies is required - especially data from longitudinal cohorts. Combining such cohorts with newer laboratory analysis technologies, such as untargeted metabolomics analyses, allow for simultaneous analysis of thousands of biomarkers (i.e., endogenous metabolites) and exogenous chemical exposures; however, these analysis methods are complex to apply and interpret, and are even more challenging when comparing results across laboratories (Rappaport 2012; Uppal et al. 2016; Athersuch 2016). Cross-validation of these discovery-driven approaches with traditional targeted biomonitoring and data analysis methodologies require organization and standard assessments for comparisons and integration. In addition, familiarity with environmental assessment tools is still lacking for many health researchers, particularly those in fields outside environmental health. By partnering with a harmonized exposure analysis resource, the interdisciplinary research team (neurosciences, infectious disease/immunology, endocrinology, etc. with exposure science) is more likely to discover new insights that will drive the field of health research forward. This kind of inter-disciplinary cross-talk is often the seed that leads to breakthroughs in science.
To address these issues, the National Institute of Environmental Health Sciences (NIEHS), the National Cancer Institute (NCI), the National Heart, Lung, and Blood Institute (NHLBI), and Environmental Influences on Child Health Outcomes (ECHO) established the Human Health Exposure Analysis Resource (HHEAR) in 2019. HHEAR is a consortium of environmental health researchers and laboratories which provides eligible investigators access to expert laboratory and data analyses that add or expand environmental exposures in health studies, at no cost to the investigator (Balshaw et al. 2017). HHEAR is a continuation of the NIEHS Children’s Health Exposure Analysis Resource (CHEAR) that was expanded to include environmental sample analysis and health outcomes at all life stages. In addition, HHEAR has the capability for analyses of environmental samples linked to health outcomes to examine the sources of environmental exposures. The goal is to provide the research community access to laboratory and statistical analyses to add or expand the inclusion of environmental exposures in their research and to make that data publicly available. This will serve as a means to improve our knowledge of the comprehensive effects of environmental exposures on human health throughout the life course. In addition, the data will add to our understanding of the processes that occur between exposure and disease, which can help health officials determine possible intervention points to prevent disease and improve chemical risk assessment.
This manuscript describes the structure and strengths of implementing the harmonized HHEAR model and our approaches to addressing challenges. We describe how HHEAR incorporates analyses of biospecimens and environmental samples and data for human health studies across the life span - serving as a model for incorporating environmental exposures into national and international research. We also present program achievements to date.
Discussion
Governance and Structure
HHEAR governance includes a deliberative body – the Steering Committee, and a decision-making entity – the Executive Committee. The resource focuses on consensus building for decision making, with input from working groups established to address specific facets of the program. The HHEAR Scientific Expert Panel, which reviews proposals, is composed of subject matter experts outside the HHEAR Program with relevant scientific expertise. A major strength of HHEAR is the transdisciplinary expertise and years of experience in health research among the consortium members (Wright et al. 2018). As the research community’s portal to HHEAR services, the Coordinating Center is particularly important in this initiative, organizing the many administrative activities, and allowing the laboratory and data analysis experts to focus on their specific technical work.
Outreach
An outreach effort is ongoing to inform the human health research community about HHEAR’s available services. Because many researchers have not previously included environmental exposures or expanded their exposure assessments in their study designs, education on the potential of HHEAR to enhance their research is needed. The public website is the primary communication vehicle. The website encourages researchers to consider whether and how their research could utilize the services and incorporate environmental exposures into their studies. This is accomplished via Frequently Asked Questions about HHEAR and Exposure Information Pages which explains why environmental exposures are relevant to health, provide descriptions of specific chemical classes that could be analyzed and the kinds of analyses provided including media (e.g., blood, urine, serum, dust, sediment/soil, or silicone wristbands), and quality and quantity of samples necessary for laboratory analysis. As the exposome and new, highly sophisticated untargeted exposomic and metabolomic analyses are not well known among health researchers, a separate information page has been included on the public website to explain these terms.
Over the four years of CHEAR, the website had over 7,600 new visitors and over 26,700 page views, with over 300 people subscribed to receive regular email updates about CHEAR services. Since the application process for the program’s continuation as HHEAR opened on January 31st, 2020, the HHEAR website has had over 6,000 new visitors and over 21,132 page views. The top five pages visited have been the HHEAR Homepage, How to Apply, HHEAR Application and Review Schedule, Targeted Analysis, and the Sample Matrix table (https://hhearprogram.org/).
Outreach activities also include presentations by HHEAR resource investigators to other NIH institutes and initiatives and at professional meetings, and publications in relevant journals. This is building on the outreach efforts by CHEAR, notably a series in Current Opinions in Pediatrics (Balshaw et al. 2017; Peterson and Hecht 2017; Stingone et al 2017; Wolff et al 2017; Wright 2017).
Overall, efforts from CHEAR resulted in the completion of over 73,000 sample analyses across more than 45 projects, exceeding the program goals to support 20-40 studies (20,000 sample analyses) in 2018 and an additional 10 studies (10,000 sample analyses) in 2019. As HHEAR, the resource continues to further contribute to these harmonized analyses of studies across the life span. Notably, the studies have focused on a wide variety of health endpoints (Fig. 1) and analysis groups (Fig. 2). The proportion of health outcomes will change as studies of adult populations are added to the resource, e.g., cancer outcomes will increase through funding from NCI.
Fig. 1.
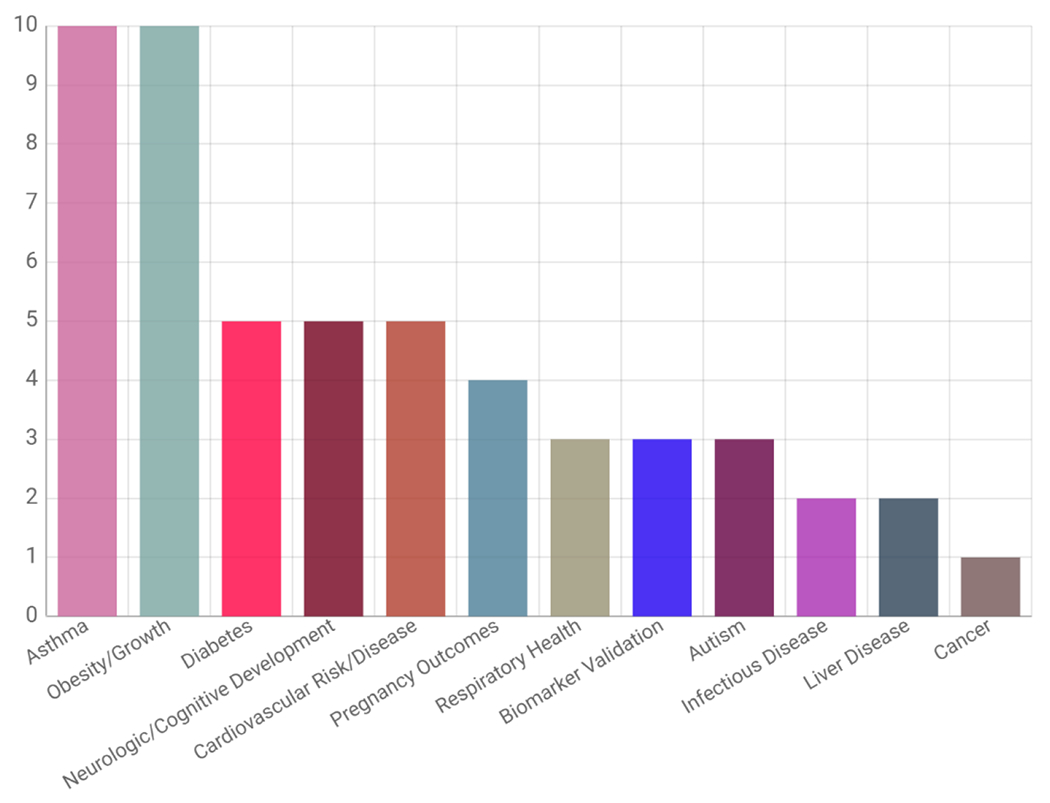
Studies by Health Outcome
Fig. 2. Studies by Lab Analysis Panel.
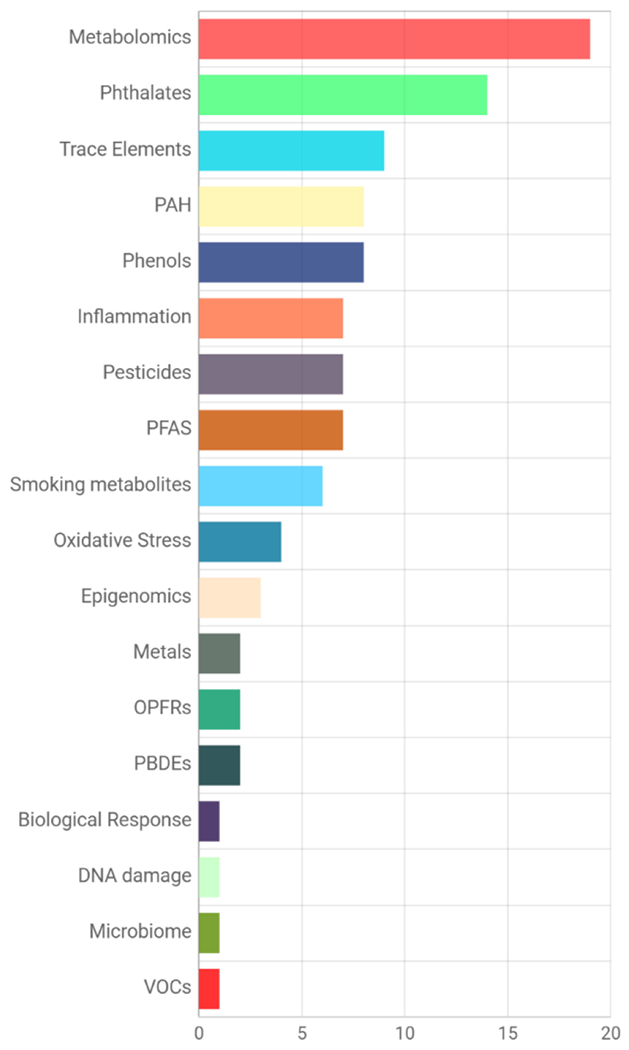
(PAH, Polycyclic Aromatic Hydrocarbons; PFAS, Per- and Polyfluoroalkyl Substances; OPFRs, Organophosphorus Flame Retardants; PBDEs, Polybrominated Diphenyl Ether Flame Retardants; VOCs, Volatile Organic Compounds)
Application process
Because many applicants had not previously considered environmental exposures in their research, a system of collaboration was developed to provide early and expert consultative services, including proposal interest review and feasibility. HHEAR laboratory hub network scientists discuss with the applicant the feasibility of the proposed laboratory analyses and suitability of the available biospecimens and/or environmental samples. The HHEAR Data Center consults with the applicant on the feasibility of the proposed data analysis plan. This process results in greatly improved final applications to HHEAR and expands opportunities for including environmental assessment in the applicant’s studies.
The HHEAR consortium has also developed a reference table with quantified measures for longitudinal surveillance of the total United States population. The most current National Health and Nutrition Examination Study (NHANES) data (Centers for Disease Control and Prevention 2019) have been integrated into the HHEAR laboratories analytes table to support feasibility assessments for biomonitoring projects. NHANES has collected biological specimens (i.e., blood, serum, plasma, urine) continuously since 1999 and reports known exposures across the United States population. Chemicals of potential concern (e.g., arsenic, environmental phenols) continue to be added to NHANES, with the most recent report including data on more than 350 chemicals. Including the NHANES exposure data with the HHEAR laboratory analytes supports assessment of the technical requirements for measuring potential toxicants (i.e., acceptable level of detection), for determining the likely prevalence of detectable levels in the population, identifying of unique analytes needed, and providing information on the distribution of exposure in the population. This document is used by both the HHEAR Lab Hubs and Data Center to support their feasibility assessments.
The HHEAR Scientific Expert Panel assigns an overall priority level to each project, based on a variety of considerations, including scientific merit, study type, health endpoints, populations, exposures, and the ability and willingness to share data, as well as the ability of a specific study to contribute to the overall assessment of human health impacts and relationship with respect to HHEAR objectives and the research priorities of the funding agencies. The funding agency makes the final decision for project approval based on the panel’s recommendation, requested funding, and other considerations.
Each investigator agrees to public use of the complete de-identified dataset (epidemiologic and biomarker data) after an embargo period. Investigators also obtain a letter from their Institutional Review Board (IRB) attesting that the original study consent permits the use of samples and data for the HHEAR project as well as unspecified future use that could arise from the publicly available data.
Workflow management
The Coordinating Center has implemented a systematic proposal review and project management process via proposal tracking and service request applications. These systems allow for monitoring of the progress of each proposal from its initial submission to final resolution, and each approved project from approval to laboratory and data analysis, and finally to publications that result from the work. These systems provide proposal and project progress in real-time.
Laboratory analysis services
HHEAR offers targeted and untargeted analyses of human biospecimens and environmental samples (Table 1). Targeted analyses are directed by hypotheses about potential associations between a health-related endpoint and specific exposures [including environmental phenols, per- and polyfluoroalkyl substances (PFAS), pesticides, phthalates, polycyclic aromatic hydrocarbons (PAH), trace elements, volatile organic compounds (VOCs), and tobacco markers]. Untargeted analyses are hypothesis-free exploratory analyses to discover new associations between chemicals or metabolites, either endogenous metabolites or exogenous xenobiotics and their metabolites, and health. One of the attractive features of untargeted exposure assessment by metabolomics is the direct linkage of indicators of environmental exposure with alterations in endogenous molecules, which can be mapped onto biological pathways to provide a simultaneous assessment of both exposure and response.
Table 1. Examples of laboratory analyses provided by HHEAR.
For sample matrices noted with an * or +, applicants consult with resource Lab Hubs about availability and utility of analyses using the sample matrix. An expanded table of laboratory analyses is available on the HHEAR website.
| Lab Analysis Panel | Exposure/Preferred Biospecimen | Environmental/Personal Samplers | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Urine | Plasma/Serum | Whole Blood | Experimental Research Biomarkers | Air Samplers | Water | House Dust | Soil/Sediment | Silicone Wristbands | |
| Targeted Analyses | |||||||||
| Brominated Flame Retardants | at least 1 mL | X* | Breast milk, placenta, adipose tissue | X* | 100 mg | 1 gram | 1/5 of band | ||
| Per- and Polyfluoroalkyl Substances | 0.1-1.0 mL | X* | Amniotic fluid, breast milk, dried blood spots, seminal plasma | X* | 50 mL wastewater, 200 mL surface water, 1000 mL drinking water | 100 mg | 1 gram | 1/5 of band | |
| Polycyclic Aromatic Hydrocarbons | 0.1-3.0 mL | X+ | 100 mg | 1 gram | 1/5 of band | ||||
| Phthalates | 01.-1.0 mL | Amniotic fluid | X+ | 100 mg | 1 gram | 1/5 of band | |||
| Untargeted Analyses | |||||||||
| Untargeted | 10 to 250 μl | 10 to 250 μl | 10 to 250 μl | Feces, saliva, tissue extracts | 50 mL wastewater, 200 mL surface water, 1000 mL drinking water | 100 mg | 1 gram | 1/5 of band | |
Initially, the resource provided analyses of biological response indicators. These included a broad range of methods from epigenomic analyses to assays for molecular markers of common biological pathways such as inflammatory signaling and oxidative stress markers. HHEAR now focuses on exposure measurements in biospecimens and environmental samples. In association with the NIEHS Superfund Research Program, HHEAR provides targeted and untargeted analysis to comprehensively measure exposures in air samplers, silicone wristbands, and environmental media, including water, soil, sediment, and dust, collected from human health studies (Table 1). The silicone wristbands provide additional information on cumulative exposures (Hammel et al. 2018). The type of biospecimen or environmental sample, the volume available, and how the samples were collected and stored will affect the feasibility of requested analyses. Applicant consultations with the resource Lab Hubs about the availability and utility of analyses for sample matrices are important to determine appropriateness in proposed projects.
To ensure the most accurate and precise data, the HHEAR Laboratory Network has designed and implemented quality assurance/quality control (QA/QC) schemes for laboratory analyses and reporting.
Targeted analysis is a fairly “mature” field of environmental exposure assessment and typically includes a relatively stringent QA/QC scheme for each method. As such, targeted analysis include rigorous intra- and inter-laboratory quality control procedures and regular participation in international proficiency testing programs. In addition, internal “HHEAR” QC materials (pooled urine, whole blood, or plasma/serum) have been developed to be characterized by each laboratory and included in each analytical run. Kannan et al. (2021 in issue) and Galusha et al. (2021) further elaborate on the QC and harmonization for organic and inorganic chemical analyses across the CHEAR/HHEAR laboratory network. This enables the Data Center to conduct direct comparisons between laboratory analyses and help to identify any inter-laboratory biases (Mazzella et al. 2021). All Lab Hubs participate in common external Proficiency Testing (PT) programs as appropriate. The PT programs include trace elements in whole blood, serum, and urine (NYS DOH Biomonitoring); organics in urine and serum (German EQUAS and/or Centre de Toxicologie du Québec); targeted metals analyses in drinking water and soil (ERA #590, ERA #551, ERA #620); and targeted organic analyses in drinking water and soils (ERA #960, ERA #625). If there is not an external PT program that covers a HHEAR project analytes in an exposure group for a given matrix, the HHEAR Lab Hubs agree on approaches such as splitting samples with other labs – either within or external to HHEAR or other review and QC measures for these unique situations. These efforts provide assurances that the offered analyses adhere to strict performance metrics and are comparable not only within HHEAR but with services provided by other laboratories around the world.
The untargeted approaches also undergo rigorous QC measures and take advantage of common quality control materials (pooled urine or plasma) that are analyzed across all HHEAR laboratories and for each study being analyzed to facilitate the harmonization of results from each study. The HHEAR untargeted analyses are also benchmarked against other metabolomics programs, such as the NIH Common Fund metabolomics program, to ensure that the highest quality services are being provided. Transparency in the HHEAR Lab Hubs reporting confidence and evidence of identification and annotation is critical for data harmonization.
In addition, the resource includes a component to support development of methods either to measure emerging environmental exposures or to utilize new biological and environmental matrices that do not currently have existing, well-validated monitoring methods in place. Examples of these studies from CHEAR include a rapid method for analysis of perfluoralkyl substances in serum by hybrid solid-phase extraction (Honda et al. 2018); determination of iodide in dried blood spots (Kim et al. 2018); operationalizing a differential ion-mobility platform (DMS) for untargeted metabolomics analysis (Wernisch et al. 2018), and others (https://hhearprogram.org/methods-development).
Data Analysis Services.
In addition to the comprehensive laboratory analyses, the HHEAR resource contains a full-service data center that serves as the data conduit between the laboratories and the HHEAR investigators (https://hheardatacenter.mssm.edu/). The Data Center provides a secure venue to access the data generated by the HHEAR laboratories, as well as linked data provided by the study investigators, which include data on health-related outcomes and phenotypes as well as additional exposure data if available.
HHEAR studies are heterogeneous and include a wide variety of scientific data including exposures, outcomes and demographic variables. To facilitate the harmonization of this disparate data into a cohesive data repository, an ontology for HHEAR was developed (https://hheardatacenter.mssm.edu/resources.asp). This ontology leverages several established open-source ontologies to create a standard vocabulary of environmental, perinatal, pediatric and public health concepts. The ontology is then used to map individual study terms to concepts within the HHEAR standard vocabulary, identifying commonalities across studies and opportunities for data pooling and/or meta-analysis. The creation of a HHEAR-specific ontology also enables the development of technologies that allow the HHEAR data repository to be browsed and queried using standardized terms and concepts, rather than individual study variable names. This greatly enhances the user experience and facilitates the users’ ability to identify and download similar data across HHEAR studies.
The HHEAR Data Center leverages a group of statistical analysts, biostatisticians and bioinformatics experts to insure the optimal analysis of HHEAR researchers’ data. The statistical team has a wide variety and depth of expertise in environmental health research including environmental exposure mixtures analysis, high dimensional data analysis, and metabolomics data analysis. The Data Center analysts’ access to the HHEAR lab personnel allows in-depth, multidisciplinary discussions of the biomarker results (both targeted and untargeted data) that are necessary to fully understand the data produced and to appropriately include them in the statistical analyses. The HHEAR Data Center provides the statistical analysis code to the HHEAR investigator so they can better understand their study results and have a template for running future analyses.
The Data Center provides consultation to statistical methods and informatics tools for investigators using HHEAR laboratory analysis services as well as for meta- and pooled-analysis across studies supported by HHEAR or added by individual investigators. As the HHEAR data repository grows, these analyses will include linkages across developmental windows and multiple environmental factors; it will also include the direct linkage and cross-validation of targeted, untargeted, biological, and environmental response data.
Additional HHEAR Services
In addition to supporting the addition of environmental exposure measure to human health studies, HHEAR provides a number of other services. HHEAR works to assist potential collaborating researchers in planning and developing proposals. HHEAR periodically supports a Pilot and Feasibility Program to provide an opportunity for investigators to study novel environmental exposures and outcomes on a small scale or to generate preliminary data to apply for research funding from NIH or other granting agencies. There were seven Pilot and Feasibility Program projects in CHEAR. These included pilot studies looking at immunometabolic biomarkers of tobacco exposure in children with cystic fibrosis; environmental exposures associated with Kawasaki disease; body size and cholesterol fractions of perfluoralkyl substances; health outcomes associated with maternal gut microbiome and environmental contaminants; arsenic exposure in children with eosinophilic esophagitis; pesticide exposure and childhood acute myeloid leukemia; and metabolomics profiles of multiple endocrine disruptor exposures during adolescence.
Some projects were also approved to develop and test CHEAR operational and quality control procedures for submission and review of proposals, and coordination and standardization of laboratory and data exposure analyses. Examples of these studies include investigating urinary polycyclic aromatic hydrocarbon metabolite associations with biomarkers of inflammation, angiogenesis, and oxidative stress in pregnant women (Ferguson et al. 2017); and urinary trace metals individually and in mixtures in association with preterm birth (Kim et al. 2018) (https://hhearprogram.org/chear-process-and-procedures-development).
Publications
One of HHEAR’s measures of success is the number of scientific publications that have and will come as a result of the supported studies (Fig. 3). These initially addressed methods development, i.e., new targeted and untargeted exposure and biological response measures (as described above) and statistical methods developed and validated by the resource laboratories and the Data Center. HHEAR investigators will continue to publish their findings for studies of exposure and health outcomes. Research highlights and publications are posted on the public website (https://hhearprogram.org/chear-publications).
Fig. 3.

Number of resource publications by publication year
In addition, as data are made public in the HHEAR data repository, in accordance with the HHEAR publication policy, other investigators will be able to access and utilize this data. To facilitate analysis of the data, particularly high dimensional data, HHEAR publications document various analytic approaches that are established as well as novel in the field of metabolomics (CHEAR Metabolomics Analysis Team et al. 2020).HHEAR publications also describe statistical methods to advance approaches for mixture analyses and for the identification of susceptible windows of exposure (Bello et al. 2017; Liu et al. 2018).
Evaluation
An evaluation plan contains ten outcomes or key indicators and associated metrics of the success of the HHEAR program. Outcomes include inter lab data comparability; data harmonization; capacity to measure multiple exposures; dissemination of findings; team science; and coordination with ECHO.
As HHEAR is relatively new, an evaluation has not yet been conducted. Some key metrics from the CHEAR evaluation have been presented above, i.e., numbers of studies funded by health outcome and analysis panel, number of publications, and number of website visits. Additional metrics from the CHEAR evaluation include numbers of requests for services submitted (75) and approved (32), percent of clients who are new to NIEHS (50%), numbers of presentations at professional meeting (65), number of new processes for analyzing exposures (35), number of databases, software or statistical models developed (10), number of new tools to measure exposures (10), and number of new biological assays developed (3). The CHEAR evaluation was used to improve the proposal review process for HHEAR. This included revising the Scientific Expert Panel process for external review to include a focus on project contributions to the field and adding feasibility confirmation to evaluate investigator responsiveness and final assessment of project feasibility.
Conclusions
HHEAR is a unique resource that provides comprehensive services in laboratory and statistical analyses allowing researchers to add environmental exposures to their studies of human health. The program expands the number of studies that include environmental exposure analysis and creates a public database of human exposures and their health consequences which improves our knowledge of the comprehensive effects of environmental exposures on human health throughout the life course.
In addition, HHEAR’s implementation of rigorous quality control for laboratory analyses, development of an exposure ontology, and establishment of a data repository allow for harmonization of data and provide the research community with the opportunity and tools to integrate complex exposure data across multiple studies. Ultimately, HHEAR promotes the exploration of the complicated interactions between multiple environmental and other factors as determinants of human health and development.
A similar exposome framework has been adopted by the broader national and international environmental health research community (Gilbertson et al. 2021; Finch and Kulminski 2019; Maitre et al. 2018; Wright et al. 2018; Buck Louis et al. 2017; Vineis et al. 2017; Kawamoto et al. 2014). While each of these programs, including HHEAR, is distinct, they provide an opportunity for coordination, cross-study harmonization, data integration, and demonstration of the power of the exposome. Such coordination has the potential to revolutionize our understanding of the importance of multiple environmental factors along the life course trajectory. Disease-based studies allow HHEAR and partner clients to assess the role of environment in complex diseases, such as diabetes, and the potential for toxic environmental chemicals to exacerbate the impact of infections, such as malaria. HHEAR, through assessing chemical signatures of biological and environmental response may, in turn, enable a greater understanding of the emerging concept of biological resilience. The accumulated burden of damage resulting from life-stage specific multiple environmental exposures may decrease the ability of the individual to appropriately compensate for the next exposure. Understanding the role of exposure across time can help to distinguish resilient and susceptible responses of individuals and populations (Dennis et al. 2016; Miller and Jones 2014).
Acknowledgments
The authors recognize the other principal investigators of the CHEAR and HHEAR components who are not co-authors here: Lee Ferguson and Heather Stapleton (Duke University Lab Hub); Gary Miller and Lance Waller (Emory Lab Hub); Chuck Burant, Dana Dolinoy, and John Meeker (Michigan Lab Hub); Stephen Hecht (Minnesota Lab Hub); Manish Arora (Mount Sinai Lab Hub); Xiuxia Du, Timothy Fennell, Keith Levine, and Susan Sumner (Research Triangle Institute); Kenneth Aldous, Kurunthachalam Kannan, and Patrick Parsons (Wadsworth Lab Hub); Chris Gennings and Susan Teitelbaum (Data Center).
This work was supported by the National Institute of Environmental Health Sciences through grants with each CHEAR (U2CES026560, U2CES026561, U2CES026544, U2CES026553, U2CES026533, U2CES026542, U2CES026555, and U24ES026539) and HHEAR (U2CES026561, U2CES026533, U2CES026542, U2CES030859, U2CES030857, U2CES030851, U2CES026555, and U24ES026539) component. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the National Cancer Institute, or the National Heart, Lung, and Blood Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare they have no actual or potential competing financial interests.
References
- Athersuch T, 2016. Metabolome analyses in exposome studies: Profiling methods for a vast chemical space. Archives of Biochemistry and Biophysics 589, 177–186. 10.1016/j.abb.2015.10.007 [DOI] [PubMed] [Google Scholar]
- Balshaw DM, Collman GW, Gray KA, Thompson CL, 2017. The Children’s Health Exposure Analysis Resource: enabling research into the environmental influences on children’s health outcomes. Curr Opin Pediatr 29, 385–389. 10.1097/MOP.0000000000000491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello GA, Arora M, Austin C, Horton MK, Wright RO, Gennings C, 2017. Extending the Distributed Lag Model framework to handle chemical mixtures. Environ Res 156, 253–264. 10.1016/j.envres.2017.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck Louis GM, Smarr MM, Patel CJ, 2017. The Exposome Research Paradigm: an Opportunity to Understand the Environmental Basis for Human Health and Disease. Curr Envir Health Rpt 4, 89–98. 10.1007/s40572-017-0126-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Fourth Report on Human Exposure to Environmental Chemicals, Updated Tables, (January 2019). Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, https://www.cdc.gov/exposurereport/ [Google Scholar]
- CHEAR Metabolomics Analysis Team, Mazzella M, Sumner SJ, Gao S, Su L, Diao N, Mostofa G, Qamruzzaman Q, Pathmasiri W, Christiani DC, Fennell T, Gennings C, 2020. Quantitative methods for metabolomic analyses evaluated in the Children’s Health Exposure Analysis Resource (CHEAR). J Expo Sci Environ Epidemiol 30, 16–27. 10.1038/s41370-019-0162-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Balshaw DM, Kwok RK, Thompson CL, Collman GW, Birnbaum LS, 2016. The Exposome: Embracing the Complexity for Discovery in Environmental Health. Environmental Health Perspectives 124. 10.1289/EHP412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis KK, Auerbach SS, Balshaw DM, Cui Y, Fallin MD, Smith MT, Spira A, Sumner S, Miller GW, 2016. The Importance of the Biological Impact of Exposure to the Concept of the Exposome. Environmental Health Perspectives 124, 1504–1510. 10.1289/EHP140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson KK, McElrath TF, Pace GG, Weller D, Zeng L, Pennathur S, Cantonwine DE, Meeker JD, 2017. Urinary Polycyclic Aromatic Hydrocarbon Metabolite Associations with Biomarkers of Inflammation, Angiogenesis, and Oxidative Stress in Pregnant Women. Environ Sci Technol 51, 4652–4660. 10.1021/acs.est.7b01252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch CE, Kulminski AM, 2019. The Alzheimer’s Disease Exposome. Alzheimer’s & Dementia 15, 1123–1132. 10.1016/j.jalz.2019.06.3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galusha AL, Merrill L, Palmer CD, Amarasiriwardena C, Parsons PJ, 2021. Measurement harmonization and traceability for trace element analyses across the Children’s Health Exposure Analysis Resource laboratory network. Environ Res 193, 110302. 10.1016/j.envres.2020.110302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbertson PK, Forrester S, Andrews L, McCann K, Rogers L, Park C, Moye J, 2021. The National Children’s Study Archive Model: A 3-Tier Framework for Dissemination of Data and Specimens for General Use and Secondary Analysis. Front. Public Health 9, 526286. 10.3389/fpubh.2021.526286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel SC, Phillips AL, Hoffman K, Stapleton HM, 2018. Evaluating the Use of Silicone Wristbands To Measure Personal Exposure to Brominated Flame Retardants. Environ. Sci. Technol acs.est.8b03755. 10.1021/acs.est.8b03755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda M, Robinson M, Kannan K, 2018. A rapid method for the analysis of perfluorinated alkyl substances in serum by hybrid solid-phase extraction. Environ. Chem 15, 92. 10.1071/EN17192 [DOI] [Google Scholar]
- Kannan K, Stathis A, Mazzella MJ, Andra SS, Barr DB, Hecht SS, Merrill LS, Galusha AL, Parsons PJ, 2021. Quality assurance and harmonization for targeted biomonitoring measurements of environmental organic chemicals across the Children’s Health Exposure Analysis Resource laboratory network. International Journal of Hygiene and Environmental Health 234, 113741. 10.1016/j.ijheh.2021.113741 (in issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto T, Nitta H, Murata K, Toda E, Tsukamoto N, Hasegawa M, Yamagata Z, Kayama F, Kishi R, Ohya Y, Saito H, Sago H, Okuyama M, Ogata T, Yokoya S, Koresawa Y, Shibata Y, Nakayama S, Michikawa T, Takeuchi A, Satoh H, Working Group of the Epidemiological Research for Children’s Environmental Health, 2014. Rationale and study design of the Japan environment and children’s study (JECS). BMC Public Health 14, 25. 10.1186/1471-2458-14-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SS, Meeker JD, Carroll R, Zhao S, Mourgas MJ, Richards MJ, Aung M, Cantonwine DE, McElrath TF, Ferguson KK, 2018. Urinary trace metals individually and in mixtures in association with preterm birth. Environ Int 121, 582–590. 10.1016/j.envint.2018.09.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim U-J, Kannan K, 2018. Method for the Determination of Iodide in Dried Blood Spots from Newborns by High Performance Liquid Chromatography Tandem Mass Spectrometry. Anal. Chem 90, 3291–3298. 10.1021/acs.analchem.7b04827 [DOI] [PubMed] [Google Scholar]
- Liu SH, Bobb JF, Lee KH, Gennings C, Claus Henn B, Bellinger D, Austin C, Schnaas L, Tellez-Rojo MM, Hu H, Wright RO, Arora M, Coull BA, 2018. Lagged kernel machine regression for identifying time windows of susceptibility to exposures of complex mixtures. Biostatistics 19, 325–341. 10.1093/biostatistics/kxx036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitre L, de Bont J, Casas M, Robinson O, Aasvang GM, Agier L, Andrušaitytė S, Ballester F, Basagaña X, Borràs E, Brochot C, Bustamante M, Carracedo A, de Castro M, Dedele A, Donaire-Gonzalez D, Estivill X, Evandt J, Fossati S, Giorgis-Allemand L, R Gonzalez J, Granum B, Grazuleviciene R, Bjerve Gützkow K, Småstuen Haug L, Hernandez-Ferrer C, Heude B, Ibarluzea J, Julvez J, Karachaliou M, Keun HC, Hjertager Krog N, Lau C-HE, Leventakou V, Lyon-Caen S, Manzano C, Mason D, McEachan R, Meltzer HM, Petraviciene I, Quentin J, Roumeliotaki T, Sabido E, Saulnier P-J, Siskos AP, Siroux V, Sunyer J, Tamayo I, Urquiza J, Vafeiadi M, van Gent D, Vives-Usano M, Waiblinger D, Warembourg C, Chatzi L, Coen M, van den Hazel P, Nieuwenhuijsen MJ, Slama R, Thomsen C, Wright J, Vrijheid M, 2018. Human Early Life Exposome (HELIX) study: a European population-based exposome cohort. BMJ Open 8, e021311. 10.1136/bmjopen-2017-021311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzella MJ, Barr DB, Kannan K, Amarasiriwardena C, Andra SS, Gennings C, 2021. Evaluating inter-study variability in phthalate and trace element analyses within the Children’s Health Exposure Analysis Resource (CHEAR) using multivariate control charts. J Expo Sci Environ Epidemiol 31, 318–327. 10.1038/s41370-021-00293-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GW, Jones DP, 2014. The nature of nurture: refining the definition of the exposome. Toxicol Sci 137, 1–2. 10.1093/toxsci/kft251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson LA, Hecht SS, 2017. Tobacco, e-cigarettes, and child health. Curr Opin Pediatr 29, 225–230. 10.1097/MOP.0000000000000456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport SM, Smith MT, 2010. Epidemiology. Environment and disease risks. Science 330, 460–461. 10.l126/science.1192603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport SM, 2012. Biomarkers intersect with the exposome. Biomarkers 17, 483–489. 10.3109/1354750X.2012.691553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingone JA, Mervish N, Kovatch P, McGuinness DL, Gennings C, Teitelbaum SL, 2017. Big and disparate data: considerations for pediatric consortia. Curr Opin Pediatr 29, 231–239. 10.1097/MOP.0000000000000467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal K, Walker DI, Liu K, Li S, Go Y-M, Jones DP, 2016. Computational Metabolomics: A Framework for the Million Metabolome. Chem Res Toxicol 29, 1956–1975. 10.1021/acs.chemrestox.6b00179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vineis P, Chadeau-Hyam M, Gmuender H, Gulliver J, Herceg Z, Kleinjans J, Kogevinas M, Kyrtopoulos S, Nieuwenhuijsen M, Phillips DH, Probst-Hensch N, Scalbert A, Vermeulen R, Wild CP, EXPOsOMICS Consortium, 2017. The exposome in practice: Design of the EXPOsOMICS project. Int J Hyg Environ Health 220, 142–151. 10.1016/j.ijheh.2016.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernisch S, Afshinnia F, Rajendiran T, Pennathur S, 2018. Probing the application range and selectivity of a differential mobility spectrometry-mass spectrometry platform for metabolomics. Anal Bioanal Chem 410, 2865–2877. 10.1007/s00216-018-0978-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild CP, 2005. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev 14, 1847–1850. 10.1158/1055-9965.EPI-05-0456 [DOI] [PubMed] [Google Scholar]
- Winett L, Wallack L, Richardson D, Boone-Heinonen J, Messer L, 2016. A Framework to Address Challenges in Communicating the Developmental Origins of Health and Disease. Curr Environ Health Rep 3, 169–177. 10.1007/s40572-016-0102-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff MS, Buckley JP, Engel SM, McConnell RS, Barr DB, 2017. Emerging exposures of developmental toxicants. Curr Opin Pediatr 29, 218–224. 10.1097/MOP.0000000000000455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization, 2006. Principles for evaluating health risks in children associated with exposure to chemicals. Environmental health criteria; 237. [Google Scholar]
- Wright RO, 2017. Environment, susceptibility windows, development, and child health. Curr Opin Pediatr 29, 211–217. 10.1097/MOP.0000000000000465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright RO, Teitelbaum S, Thompson C, Balshaw D, CHEAR Network, 2018. The child health exposure analysis resource as a vehicle to measure environment in the environmental influences on child health outcomes program. Curr Opin Pediatr 30, 285–291. 10.1097/MOP.0000000000000601 [DOI] [PMC free article] [PubMed] [Google Scholar]


