Abstract
Influenza D virus (IDV) is a novel type of influenza virus that infects and causes respiratory illness in bovines. Lack of host-specific in vitro model that can recapitulate morphology and physiology of in vivo airway epithelial cells has impeded the study of IDV infection. Here, we established and characterized bovine primary respiratory epithelial cells from nasal turbinate, soft palate, and trachea of the same calf. All three cell types showed characteristics peculiar of epithelial cells, polarized into apical-basolateral membrane, and formed tight junctions. Furthermore, these cells expressed both α−2,3- and α−2,6-linked sialic acids with α−2,3 linkage being more abundant. IDV strains replicated to high titers in these cells, while influenza A and B viruses exhibited moderate to low titers, with influenza C virus replication not detected. These findings suggest that bovine primary airway epithelial cells can be utilized to model infection biology and pathophysiology of IDV and other respiratory pathogens.
Keywords: bovine, primary cells, respiratory epithelial cells, influenza D virus, nasal turbinate, soft palate, trachea
1. Introduction
Influenza virus is an enveloped, single stranded, negative sense, RNA virus with a segmented genome, belonging to the Orthomyxoviridae family. Influenza virus is a respiratory pathogen with a wide host range and is further classified into four types: Influenza A (IAV), Influenza B (IBV), Influenza C (ICV), and Influenza D (IDV) (Krammer et al., 2018; Liu et al., 2020a). Of all four types, IAV has a wide range of vertebrate hosts with the capability of cross-species transmission. While IBV and ICV primarily infect humans, infections of swine and cattle have been reported for both IBV and ICV on some occasions (Ran et al., 2015; Sederdahl and Williams, 2020; Yan et al., 2020). The novel influenza D virus was initially isolated from swine and later from bovine species (Hause et al., 2014; Hause et al., 2013).
IDV primarily infects cattle with periodic spillover to other animal species. Antibodies against IDV has been detected in sheep, goat, horses, and camels (Murakami et al., 2019; Nedland et al., 2018; Oliva et al., 2019; Quast et al., 2015; Salem et al., 2017). So far, no evidence of human infection has been reported for IDV although antibody against IDV has been found in humans (Hause et al., 2014; Hause et al., 2013; Liu et al., 2020a; White et al., 2016; Yu et al., 2021). Emerging evidence has shown IDV as a key pathogen of the bovine respiratory disease. After experimental infection of ferrets, and pigs, IDV was isolated from nasal turbinates only (Dane et al., 2019; Flynn et al., 2018; Mitra et al., 2016; Nissly et al., 2020). However, IDV was detected from both upper and lower respiratory tracts after experimental infection of guinea pigs.
Experimental infection of native host cattle with IDV revealed the presence of viral RNA in both upper and lower respiratory tracts, including viral RNA detected in lungs of infected animals at 4 day post infection. Compared to the upper respiratory tract, relatively low amounts of viral RNA were found in the lower respiratory tract tissues (Ferguson et al., 2016; Hause et al., 2013; Hause et al., 2017; Salem et al., 2019; Sreenivasan et al., 2015; Su et al., 2017). The widespread distribution of IDV in North America, Europe, and Asia along with high prevalence in cattle farms has led to an emerging challenge to the livestock industry (Bailey et al., 2018; Dane et al., 2019). Although there has been significant improvement in the understanding of influenza virus biology, most of it is focused on IAV and IBV. The emergence of novel IDV type along with its associated disease burden in cattle has demanded continued efforts in IDV research including the development of in vitro physiologically relevant models specific to IDV.
Cell culture is a robust tool to study virus-host interactions and investigate viral protein functions in a controlled environment. Host-specific cell lines provide in vitro model that can mimic natural virus infection and spread in vitro. Currently available cell culture models for influenza D virus include the existing continuous cell lines that are extensively used for IAV, IBV, and ICV. A variety of cell lines like Madin-Darby canine kidney cell (MDCK), Madin-Darby bovine kidney (MDBK), and African green monkey kidney epithelial cells (Vero), have been used for both influenza research as well as for vaccine production (Milian and Kamen, 2015). These established cell lines are mostly transformed cells and were obtained from tissues that are not infected by the influenza virus under natural conditions. Existing evidence shows that tumor transformation during the development of these immortalized cell lines leads to hyper sialyation (Pearce and Laubli, 2016), and these transformed cell lines have defective antiviral innate immune responses (Seitz et al., 2010). These alterations, despite rendering the cell lines more permissive for influenza virus replication, may affect the physiological relevance of these cell lines in influenza research and the derived conclusions may require further validation in vivo. Bovine respiratory epithelium serves as the first line of defense against IDV infection and successful permissibility and replication of IDV depend on its unique ability in overcoming the physical and immunological barriers of the respiratory tract. Thus, host tissue specific primary cell culture from bovine is essential to model infection biology and pathogenesis of IDV. So far, only few respiratory cell lines of bovine origin are available, and these cell lines are either from fully undifferentiated respiratory tissue (Sobotta et al., 2017), or require sophisticated culture technique with cell differentiation that is heavily influenced by culture conditions (Cozens et al., 2018; Cozens et al., 2019; Lee and Chambers, 2019). More importantly, there are no cell lines derived from the upper and lower respiratory tract of same calf. Such bovine respiratory cell lines from both upper and lower airways share the same genetic background, which would be very useful in studying IDV tissue tropism, receptor utilization, and host-pathogen interaction.
Here we developed and characterized bovine primary respiratory cell lines from the upper and lower respiratory tracts of the same animal. Although IDV is primarily considered as an upper respiratory tract pathogen, experimental inoculation of cattle and mice demonstrated viral replication in the lower respiratory tract as well (Ferguson et al., 2016; Oliva et al., 2020; Salem et al., 2019). Bovine primary nasal turbinate and soft palate epithelial cells represent the upper respiratory tract, while bovine primary tracheal epithelial cells are from the lower respiratory tract. All three cell lines were characterized for epithelial cell-specific phenotypes including polarization potential and formation of tight junctions. The distribution pattern of glycans with terminal α- 2,3, and α−2,6 sialic acids on the cell surface were also studied for these cell types. Finally, the replication potential of all four types of influenza viruses was examined and compared in these cell lines with a hypothesis that IDV replicates more efficiently than other influenza types in bovine primary respiratory epithelial cells developed from this work.
2. Material and Methods
2.1. Establishment of primary epithelial cell cultures from the bovine respiratory tract
Nasal turbinate, soft palate and tracheal tissues were collected from one-year-old cattle slaughtered at South Dakota State University (SDSU) meat science laboratory. Tissues were collected immediately after slaughter in plain Dulbecco’s Modified Eagle’s Medium-F12 (DMEM/F-12; Invitrogen, Garland, NY) media containing 2X antibiotic/antimycotic (Gibco, catalogue number: 15240–062, Garland, NY). Blood was collected for obtaining serum for influenza D virus-specific antibodies testing. Haemagglutinin Inhibition (HI) assay showed the absence of antibodies against influenza D virus in this animal. We followed previously established protocol for isolation of cells from tissues (Katwal et al., 2019; Kaushik et al., 2008). Briefly, tissues were washed 3 times with 1X PBS and incubated for 2 h at 37°C in Collagenase II (200U/ml; Worthington Biochemical, Lakewood, NJ, catalogue number: LS004176). Cells were then strained with strainer (70 μm cell strainer), centrifuged at 500 x g for 5 minutes. The pellet was washed three times with DMEM/F-12 plain media containing antibiotic/antimycotic. Cells were then incubated at 37°C and 5% CO2 in a collagen coated flask. The DMEM/F-12 medium was used to culture these cells, which was supplemented with 5% Fetal Bovine Serum (Hyclone, catalogue number: SH30396–03, Logan, Utah), 1X antibiotic/antimycotic, 1% insulin transferrin selenium (ITS; Corning, Catalogue number: 354351), and 5 ng/ml of mouse epidermal growth factor (EGF; Corning, catalogue number: 4069007). The medium was changed every other day during cell culture. Once the flask was confluent, cells were detached using 0.25% Trypsin EDTA (Gibco; Garland, NY, Catalogue number: 25200–056) and split into two flasks for further passaging.
2.2. Enrichment and phenotypic characterization of cells
To remove fibroblasts from the culture, cell monolayers were treated with 0.05% trypsin EDTA every other day for 3 minutes followed by washing with 1X PBS. Immunocytochemistry was used to identify phenotypic characteristics of cells using the previously described protocol (Uprety et al., 2019). Approximately 0.5–1 × 105 cells were used to prepare cytospins. Acetone fixed cytospins were blocked for non-specific protein binding and endogenous peroxidase. Cytospins were then stained with one of the following four monoclonal antibodies (mAb); anti-cytokeratin mAb (C6909), anti-α-smooth muscle actin (α-SMA) mAb (A2547), anti-vimentin mAb (V5255), and anti-desmin mAb (D1033). These antibodies detect cell-specific marker proteins such as cytokeratin for epithelial cells, α-SMA for fibroblasts, vimentin for mesenchymal cells, and desmin for muscle cells, respectively. Monoclonal antibodies M5170, M9269, and M9144 were used as isotype controls for IgM, IgG1, and IgG2a, respectively. Primary and isotype control antibodies were purchased from Sigma-Aldrich. After 1 h incubation with primary antibody (0.1 μg), cytospins were stained with biotinylated goat anti mouse IgG2a or IgM or IgG1 antisera (1:2000 dilution, Caltag Laboratories) for 30 minutes. After incubation for 30 minutes with HRP-streptavidin, ready to use (RTU) diaminobenzene (DAB) substrate (Vector Laboratories) was used for chromogenic visualization of markers. Hematoxylin was used for counterstaining and images were taken using Olympus BX53 upright microscope.
2.3. Trans-electric epithelial resistance (TEER) and indirect immunofluorescence assay (IFA)
To identify the polarization potential of bovine respiratory epithelial cells, we seeded 0.5 × 106 cells (passages 4–6) in 6-well transwell membrane cell culture inserts (Corning; catalogue number: CLS3450). Two ml of medium was added on top and bottom of the insert. After 48 h of seeding, TEER was measured using Evom voltmeter (World Precision Instruments, Sarasota, FL). Indirect immunofluorescence assay (IFA) was used for detection of the tight junction proteins. For IFA, at day 10, the membrane was fixed in 4% paraformaldehyde. Triton X-100 (0.2%) was used for membrane permeabilization and 5% goat serum for blocking nonspecific binding. The membrane was incubated for 1 h with 5 μg/ml of rabbit anti-Claudin 3 (Invitrogen, catalogue number: 34–1700), rabbit anti-Occludin (Invitrogen, catalogue number: 71–1500), and normal rabbit serum (isotype control, 5 μg/ml). Membrane was incubated for 30 minutes with 1:200 dilution of goat anti-rabbit IgG Alexa-Fluor 488 antibody (Invitrogen, catalogue number: A11034). Propidium iodide was used for cell nuclear staining. Images were visualized using Olympus BX53 upright microscope.
2.4. Lectin binding assay
The expression of sialic acids in bovine respiratory cells was studied by staining with biotinylated lectins that bind to different sialic acid linkage. Approximately 5 × 105 cells (passages 5–7) were incubated for 1 h with biotinylated lectins MAL-I, MAL-II and SNA at 10 μg/ml final concentration. MAL-I binds to Siaα2–3Galβ1–4GlcNAc, MAL-II binds to Siaα2–3Galβ1–3GalNAc, and SNA binds to Siaα2–6Gal/GalNAc (Geisler and Jarvis, 2011; Shibuya et al., 1987). For MAL-II, inhibitor N-acetylneuramininc acid (NANA) was used, while lactose was used as an inhibitor for MAL-I and SNA. The specific inhibitor was added to cells at a final concentration of 200 μM, incubated for 10 minutes followed by the addition of lectin. Lectin and inhibitors were purchased from Vector Laboratories, Burlingame, CA. Cells were then incubated with Streptavidin-FITC (5 μg/ml) for 30 minutes. Cells were analyzed using FACS Calibur Cytometer (Becton Dickson, San Jose, CA) and the percentage of stained cells was calculated.
2.5. Viral replication kinetics and indirect fluorescence assay
Approximately 0.5 × 105 cells (passages 4–8) were seeded in 24-well plates and incubated for 72 h. After 72 h, cells were infected with 1 MOI of A/California/04/2009/H1N1 (CA04pdm09), B/Hongkong/286/2017, C/Johannesburg/1/66, D/bovine/Oklahoma/660/2013 (D/660), and D/swine/Oklahoma/1334/2011 (D/OK) viruses, and incubated at 37°C and 33°C, representing the core body temperature and relatively lower temperature in naso-pharyngeal cavity. All these viruses were propagated and titrated on MDCK cells. Initial titers of the virus inoculum (TCID50/ml) were 6.5 logs for CA04pdm09, D/OK, and B/Hongkong. Viral titers for D/660 was 6.6 logs, while viral titer for C/Johannesburg was 5.5 logs. The virus growth medium is DMEM/High Glucose media supplemented with 1μg/ml of TPCK trypsin (Thermo Fisher Scientific, Waltham, MA) and 1X antibiotic/antimycotic. Supernatants (100 μl) were collected every 24 h from infected cells and controls for virus titration. For virus titration, 1.5 ×104 MDCK cells were cultured overnight in 96-well plates. Plates were washed with 1 x PBS. Ten-fold serial dilutions of the virus infected cell culture supernatants were prepared in virus growth medium, which was followed by inoculation into pre-seeded 96-well plates with MDCK cells. Inoculated plates were incubated at 37°C or 33°C for 5 days. Hemagglutinin assay (HA) with 0.5% Turkey RBC (Lampire Biological Laboratories, Pipersville, PA) was used for determining virus infectivity. Reed and Muench method was used for calculation of virus titers (Reed and Muench, 1938).
On day 5, the wells were washed with 1 x PBS, fixed in acetone, blocked with 1% bovine serum albumin in 1 x PBS. Cells were then stained for the presence of viral protein. For CA04pdm09 infected cells, we used purified human IgG anti-influenza A/B antibody (1:400 dilutions, SAB biotherapeutics Inc) as primary antibody and mouse anti human FITC (1:500 dilutions, BETHYL) as the secondary antibody. For B/HK/286/2017, we used anti-IBV mouse monoclonal antibody pooled sera (1:400 dilutions, BEI, ATCC) as a primary antibody and goat anti-mouse FITC (1:400 dilutions, Thermo Fisher Scientific) as a secondary antibody. For C/Johannesburg/1/66 and the two strains of the influenza D virus, we used rabbit sera specific for each virus (1:100 dilution) as primary antibody and goat anti-rabbit Alexa Fluor 488 (1:400 dilutions, Invitrogen, catalogue number: A11034) as secondary antibody. Rabbit sera were generated for in-house purpose by commercial vendor. Virus infected cells were incubated with specific primary antibody for 1 h at 37°C and for 30 minutes with secondary antibody. Cells were counterstained with DAPI. Images were taken at 20X using Olympus BX53 upright microscope.
3. Results
3.1. Isolation and characterization of bovine primary respiratory cells.
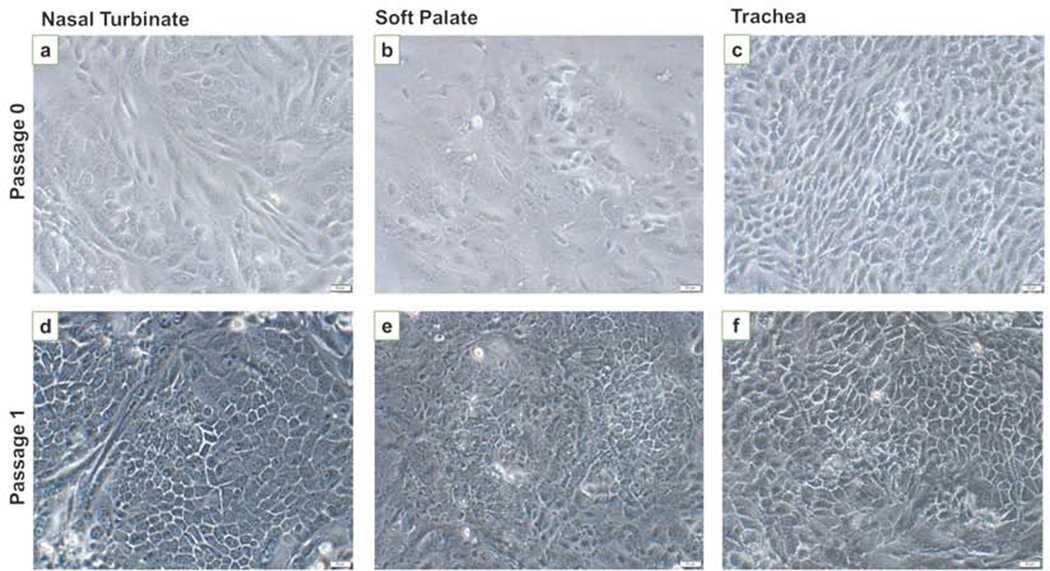
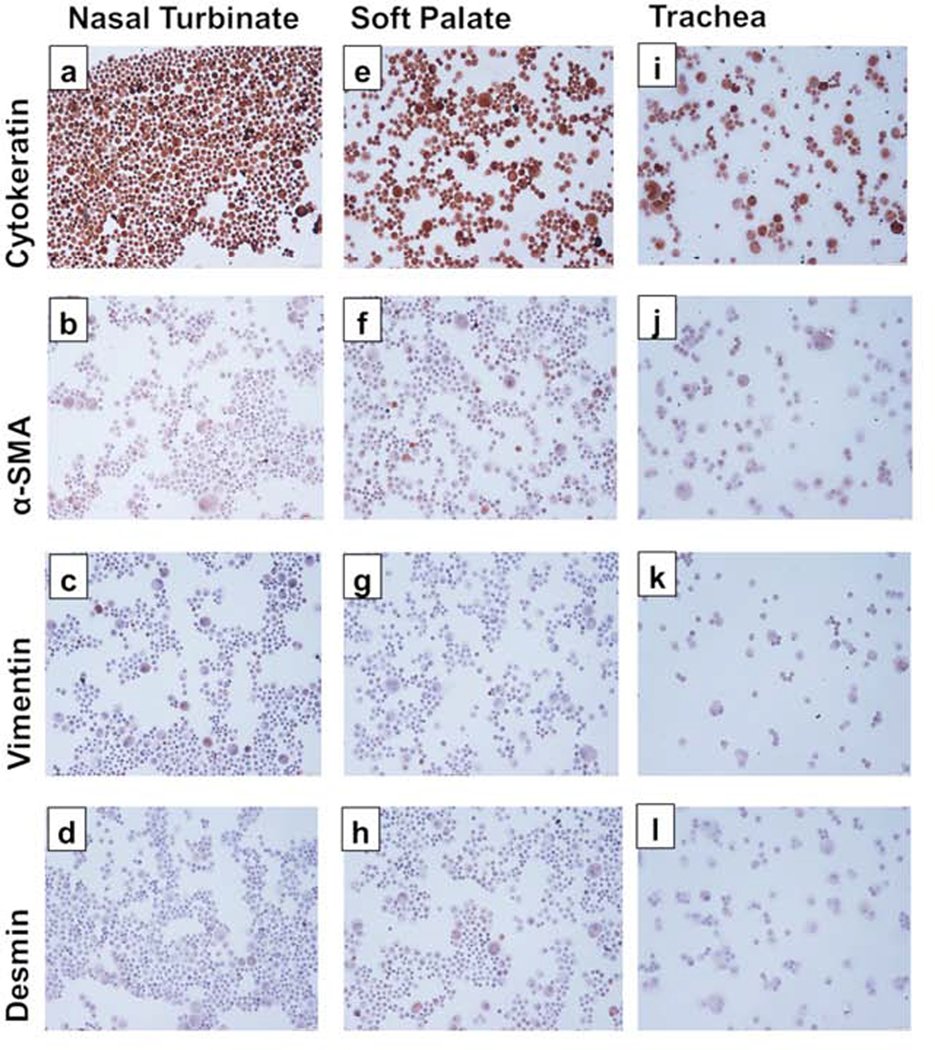
After 48 h of initial culture, cells began to form visible clusters. By day 5, all three cell types started to show clusters of epithelial cells (Fig. 1a-c). After 1st passage, most of the cells showed cobblestone morphology, typical of epithelial cells (Fig. 1d-f). Cells took 5–7 days for reaching confluency and were continued to be cultured on collagen-coated flasks. We were able to maintain these cells till passage 14 after which they started to undergo replicative senescence. After the first passage, cytospins were stained for anti-cytokeratin antibody only. The cytospins were positively stained for cytokeratin, indicative of the epithelial cell phenotype. Following trypsin treatment, cytospins were prepared on 3rd passage and stained for cell specific markers; cytokeratin, vimentin, α-SMA, and desmin. For all three cell types, cytokeratin staining was positive, indicating epithelial cell phenotype (Fig. 2 a, e, and i). All three cell types were mostly negative for α-SMA (Fig. 2 b, f, j), vimentin (Fig. 2 c, g, k), and desmin (Fig 2 d, h, l), which were indicative of the absence of fibroblast, and smooth muscle cell populations. Isotype specific antibodies stained negative for all three cell types, indicating the absence of non-specific binding of the primary antibodies used.
Fig. 1. Morphology of bovine nasal turbinate, soft palate, and tracheal cells.
Cell pellets obtained after collagenase digestion of tissues were seeded in collagen coated flasks. After 4–5 days cell clusters were visible with heterogenous cell populations (a-c). After the first passage and successive trypsin treatment, more homogenous cells with cobblestone morphology were observed (d-f). Images were taken on a phase contrast microscope at 20X. Scale bar represents 50 μM.
Fig. 2. Immunocytochemical staining of the primary bovine nasal turbinate, soft palate, and tracheal cells.
Cytospins from all three cell types were stained with monoclonal antibodies against marker proteins: cytokeratin (a, e, i), α-SMA (b, f, j), vimentin (c, g, k), and desmin (d, h, l). Brown staining indicates the presence of marker proteins. Bovine nasal turbinate, soft palate, and tracheal cells showed intense staining for cytokeratin but almost no staining for other marker proteins indicating epithelial phenotype of cells. Isotype control antibodies showed negative staining. Images were taken at 20X.
3.2. Presence of high TEER values and tight junctions in Bovine primary respiratory cells.
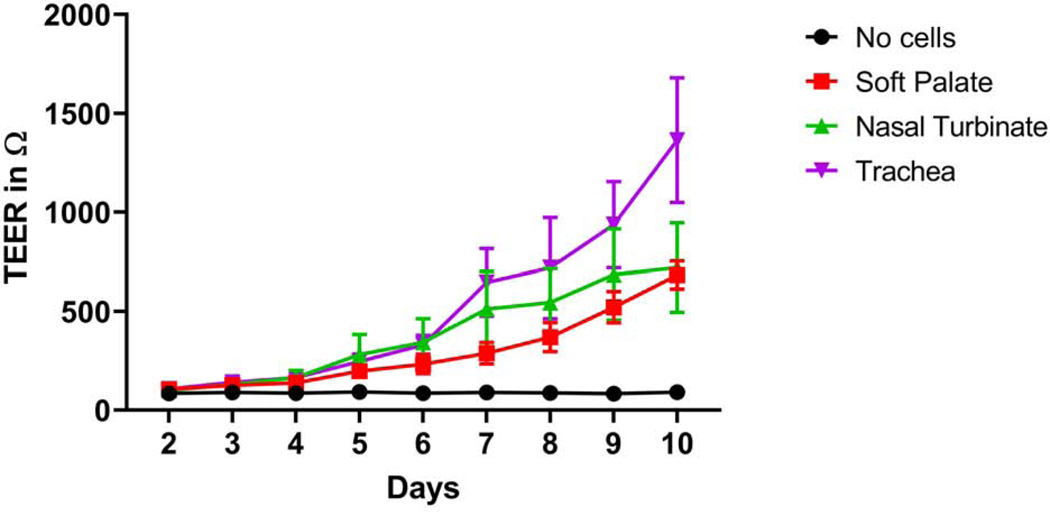
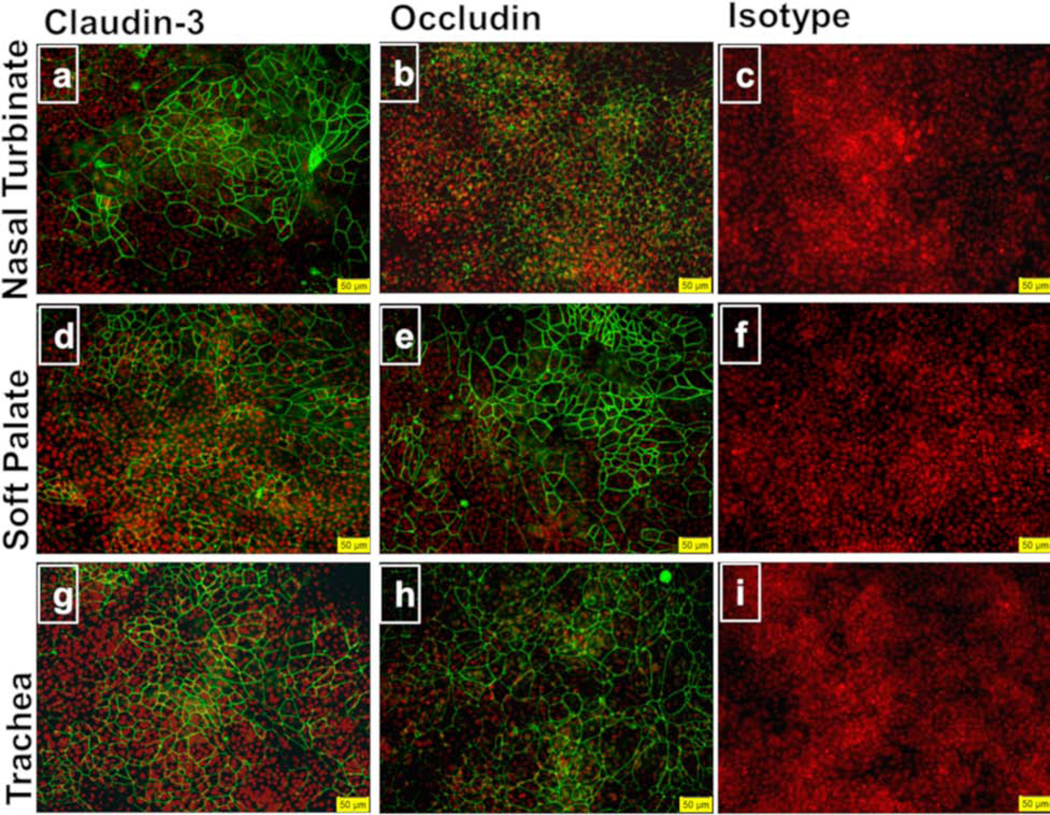
Bovine nasal turbinate, soft palate, and tracheal epithelial cells grown on transwell membranes polarized and formed tight junctions. Early passage cells (passages 4–7) were used for this experiment. By day 10, all three cell types polarized as indicated by high TEER values. Out of three cell types, bovine tracheal cells showed the highest TEER followed by nasal turbinate and soft palate epithelial cells, respectively (Fig. 3). To demonstrate the presence of tight junction proteins, we stained the transwell membrane with anti-occludin, and anti-claudin-3 antibodies. Bovine nasal turbinate, soft palate, and tracheal epithelial cells stained positive for claudin-3 (Fig. 4 a, d, g, respectively), and occludin (Fig. 4 b, e, h, respectively). Isotype controls stained negative for all three cell types (Fig. 4 c, f, i), indicating the lack of non-specific primary antibody binding.
Fig. 3. TEER measurements of the bovine nasal turbinate, soft palate, and tracheal epithelial cells.
Three different bovine respiratory epithelial cells were cultured on trans-well membranes and electric resistance was measured for 10 days using probe and volt-ohmmeter. A high TEER value indicates the polarization of epithelial cells. All three cell types showed medium to high TEER values indicating the polarization capacity of these cells. Data represents mean ± SEM of three independent experiments with the bars indicating standard error of mean.
Fig. 4. Tight junction proteins in bovine nasal turbinate, soft palate, and tracheal epithelial cells.
Cells grown on transwell membranes for TEER measurement were stained for the tight junction proteins claudin-3 (a, d, g), occludin (b, e, h), and isotype controls (c, f, i) in immunofluorescence assay. All three cell types showed the presence of tight junction proteins claudin-3 and occludin (stained as green). The cell nucleus is stained in red with DAPI. The data shown are representative of three independent experiments. Images were merged using Image J. Images were taken at 20X. Scale bar represents 50 μM.
3.3. Expression of α2–3 and α2–6 linked sialic acids in bovine primary respiratory cells.
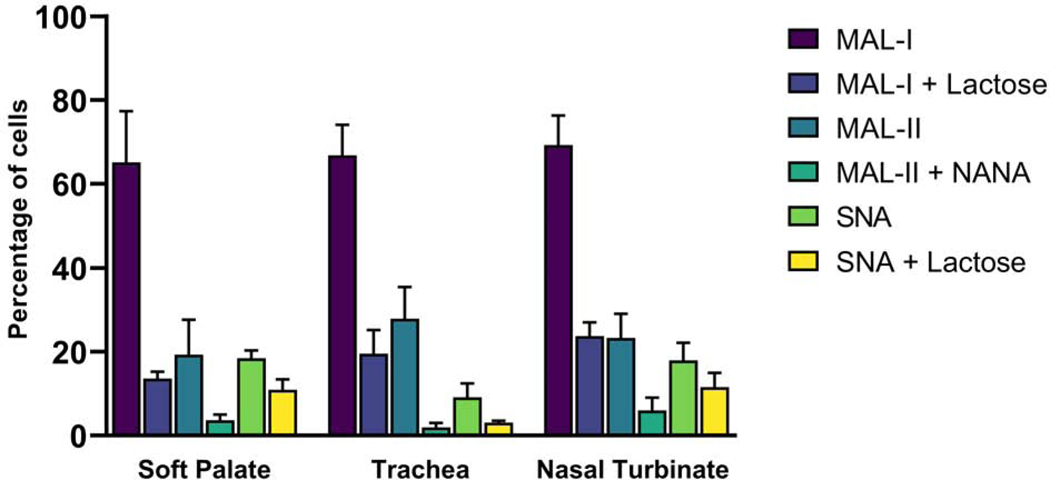
Biotinylated lectins MAL-I, MAL-II, and SNA were used to study sialic acid expression pattern in these bovine respiratory cells. The percentages of cells expressing Sia α2–3Galβ1–4GlcNAc based on MAL-I staining were 69.2%, 65.04%, and 66.7% for nasal turbinate, soft palate, and tracheal epithelial cells, respectively. The percentages of cells expressing Sia α2–3Gal-β13GalNAc based on MAL-II staining were 23.2%, 19.2%, 27.8%, respectively, for nasal turbinate, soft palate, and tracheal epithelial cells (Fig. 5). The percentages of cells expressing Siaα2–6Gal/GalNAc as demonstrated by positive SNA binding were 17.9%, 18.3%, and 9% for nasal turbinate, soft palate, and tracheal epithelial cells, respectively. Lactose was used as inhibitor for MAL-I and SNA, while N-acetylneuraminic acid (NANA) was used as inhibitor for MAL-II. Inhibitor Lactose reduced the percentage of positive cells for MAL-I to 23.7% for nasal turbinate (cf. 69.2% without the inhibitor), 13.5% for soft palate (cf. 65.04% without the inhibitor), and 19.4% for tracheal epithelial cells (cf. 66.7% without the inhibitor). NANA reduced the percentage positive cells for MAL-II to 5.9% for nasal turbinate (cf. 23.2% without the inhibitor), 3.6% for soft palate (cf. 19.2% without the inhibitor), and 1.8% for tracheal epithelial cells (cf. 27.8% without the inhibitor). Similarly, lactose inhibition reduced the percentage of cells positive for SNA binding to 11.5% for nasal turbinate (cf. 17.9% without the inhibitor), 10.9% for soft palate (cf. 18.3% without the inhibitor), and 3 % for tracheal epithelial cells (cf. 9.0% without the inhibitor). Reduction in the percentage of nasal turbinate, soft palate, and tracheal epithelial cells positive for MAL-I, MAL-II, and SNA binding indicated binding specificities of lectins (Fig. 5).
Fig. 5. Expression of glycans with α 2,3, or α 2,6 linked sialic acids in bovine primary respiratory epithelial cells.
Biotinylated lectins binding to sialic acids-containing glycans were used for staining of cells and the percentage of cells staining for each lectin was calculated by using flow cytometry. Lectin MAL-I and MAL-II bind to sialic acid linked to galactose by α 2,3 linkage, while SNA binds to sialic acid linked to galactose by α 2,6 linkage. The data represents the mean of three independent experiments. Error bar shows the standard error of the mean.
3.4. Susceptibility of bovine primary respiratory cells to IDV infection.
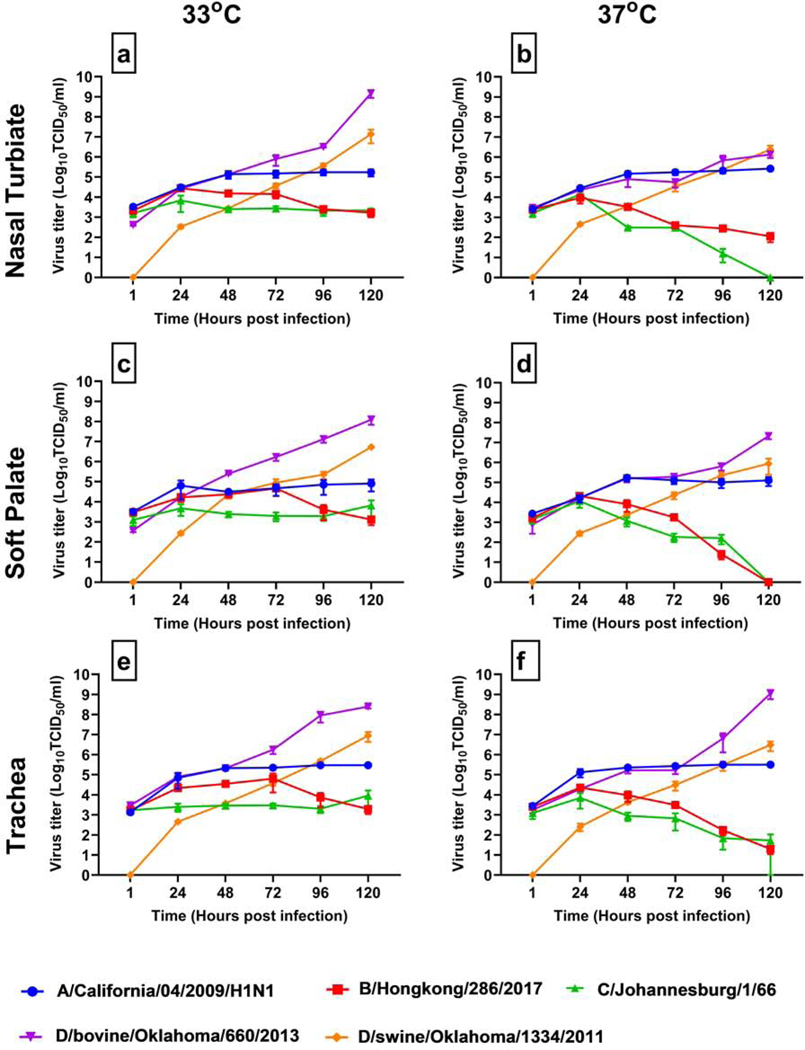
We infected bovine nasal turbinate, soft palate, and tracheal epithelial cells with 1.0 MOI for all four types of influenza viruses at two different temperatures, 33°C and 37°C. Of four types, influenza D viruses replicated more robustly in all three cell types at both temperatures than influenza A, influenza B, and influenza C viruses (Fig. 6). A/CA04/pdm09 virus replicated moderately in these cells reaching the peak titers of 5 logs, at least 3 logs lower than that of IDVs. Similarly, B/Hongkong/286/2017 virus showed its peak titers around 4 logs, one log lower than IAV. Interestingly, IDV-related C/Johannesburg/1/66 failed to replicate beyond the inoculum level in all three cell types over the course of 5-day experiments. However, the same inoculum of C/Johannesburg/1/66 replicated efficiently in MDCK cells especially at 33°C (data not shown). This indicated that ICV failed to replicate in bovine primary nasal turbinate, soft palate, and tracheal epithelial cells which was in marked contrast with other viruses that showed increased replication over time, regardless of high or low titers.
Fig. 6. Influenza virus replication kinetics at 33°C and 37°C.
All three cell types were infected with 1 MOI of the indicated viruses and were incubated at 33°C and 37°C. The supernatant was collected 1-hour post-infection and then every 24 hours during the course of five-day experiment. Ten-fold serial dilutions of supernatants were inoculated in 96-well culture plates. Viral infectivity endpoints were determined by the haemagglutinin assay with virus titers expressed in log10TCID50/ml were calculated using Reed and Muench method.
Of the two lineage-representative strains of influenza D virus, D/bovine/Oklahoma/660/2013 (D/660 lineage) with bovine origin replicated to the highest titer with the peak titer of 8 logs than D/swine/Oklahoma/1334/2011 (D/OK lineage) with swine-origin with the peak titer of 6.5 logs. At 33°C, D/bovine/Oklahoma/660/2013 (D/660 lineage) replicated better in nasal turbinate followed by tracheal and soft palate epithelial cells. At 37°C, D/bovine/Oklahoma/660/2013 (D/660 lineage) replicated to higher titers in tracheal cells followed by soft palate and nasal turbinate epithelial cells (Fig 6). D/swine/Oklahoma/1334/2011 (D/OK lineage) replicated to similar titers in all three cell types.
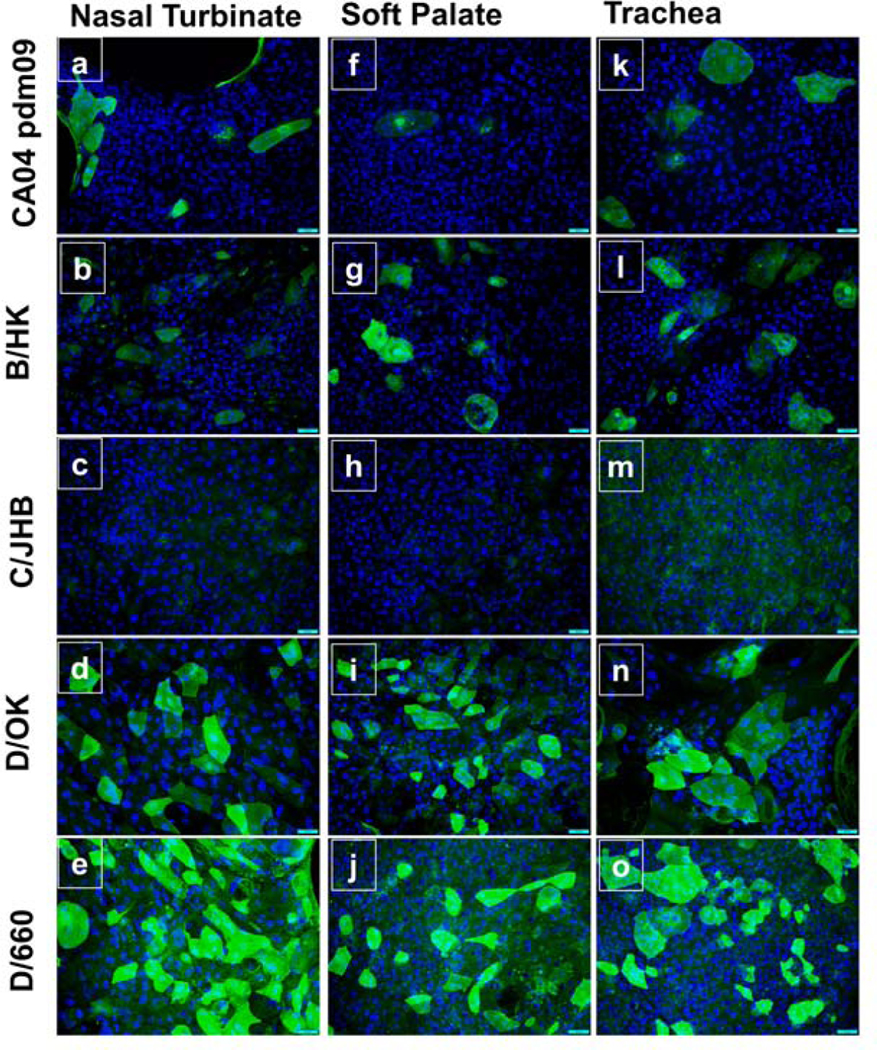
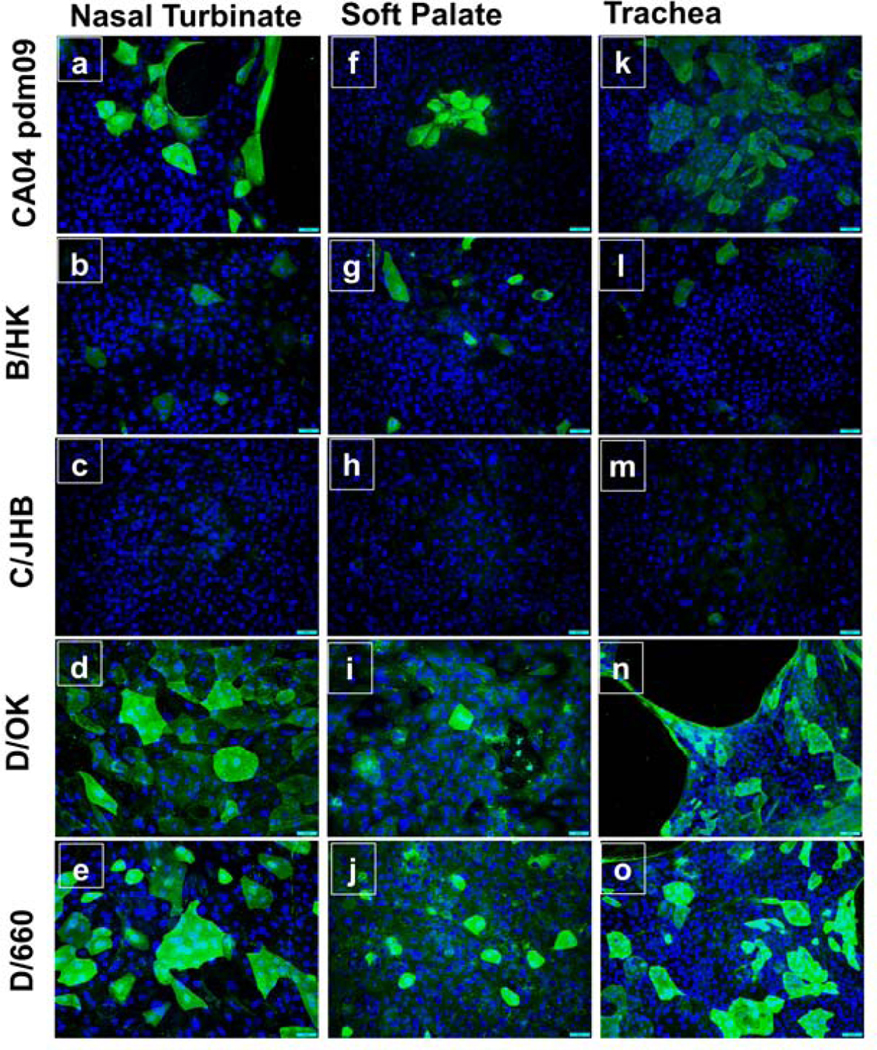
Immunofluorescence imaging analysis of virus-infected cells showed the presence of viral proteins. C/Johannesburg/1/66 infected bovine nasal turbinate, soft palate, and tracheal epithelial cells did not show any evident staining at both 33°C (Fig 7 c, h, m) and 37°C (Fig 8 c, h, m), indicating its replication incompetence, which was in good agreement with virus titration experiment above. All cell types that were infected with two influenza D strains i.e., D/bovine/Oklahoma/660/2013 (Fig 7 and 8 e, j, o), D/swine/Oklahoma/1334/2011 (Fig 7 and 8 d, i, n), showed intense staining at both temperatures in all three cell types. Further analysis of cells infected with IAV and IBV showed that cells infected with A/California/04/2009/H1N1 (CA04pdm09) exhibited the fluorescence intensity comparable to those infected with B/Hongkong/286/2017 at both temperatures. Cells infected with both viruses seemed to have more fluorescence signals at 37°C (Fig 8 a, f, k) than at 33°C (Fig 7 a, f, k).
Fig. 7. Immunofluorescence images of cells infected with different strains of four types of influenza viruses at 33°C.
At the end of the viral replication kinetics study (day 5 post-infection), cells were fixed in acetone and stained with rabbit polyclonal antibodies for ICV and IDV, mouse monoclonal antibody for IBV, or humanized cattle antibody for IAV. Alexa Fluor 488 conjugated (for ICV and IDV) or FITC conjugated (for IAV and IBV) secondary antibodies were used for visualization. High expression of the viral protein was detected in all three cell types for IDV strains, and moderate to low expression for IAV and IBV, respectively. We failed to detect the expression of ICV specific viral proteins. Images were taken at 20X magnification.
Fig. 8. Immunofluorescence images of cells infected with different strains of four types of influenza viruses at 37°C.
Indirect immunofluorescence assay as described in Fig. 7 was used to stain cells infected with different strains representative of four types of Influenza viruses. Consistent with findings in Fig 7, high expression of the viral protein was detected in all three cell types for IDV strains, and moderate to low expression was found for IAV and IBV, respectively. ICV specific viral protein could not be detected which is consistent with the results of viral replication kinetics assay. Images were taken at 20X magnification.
4. Discussion
Cell culture system provides a reliable in vitro model to study host-pathogen interactions and immune responses (Powell and Waters, 2017). There are various immortalized and well-established cell lines like MDCK, MDBK, Vero, and A549 cells that are routinely used to replicate influenza viruses. Primary human respiratory airway cell cultures have already been used for studying influenza A, and influenza B viruses. Recently identified influenza D virus primarily infects cattle and so far, a host-specific primary respiratory epithelial cell culture model is not available for this new virus (Hause et al., 2013). Lack of bovine primary respiratory epithelial cells also represents a hurdle for study of other respiratory pathogens that infect and cause diseases in this agricultural animal. In this study, we developed primary respiratory epithelial cell lines from both upper (Nasal turbinate and soft palate) and lower (trachea) respiratory tracts where IDV and other viral pathogens replicate and cause pathological effects. Recently soft palate was identified as the site of influenza virus adaptation (Lakdawala et al., 2015) and thus were included in the present study. We characterized these cell lines for epithelial cell phenotype and investigated the replication potential of influenza D as well as other influenza types.
Immunocytochemistry analysis demonstrated the presence of cytokeratin but the absence of vimentin, α-SMA, and desmin in all three types. Cytokeratin is an epithelial cell marker while vimentin is a marker for mesenchymal cells. Alpha-SMA and desmin respectively have been used as markers for myofibroblast and muscle cells. These markers were frequently used to differentiate one cell population from another (Council and Hameed, 2009; van der Velden et al., 1997). Here, we generated epithelial cell cultures devoid of fibroblast cells as demonstrated by an entirely cytokeratin positive population of cells. The primary respiratory epithelial cell cultures have a finite life span and often undergo replicative senescence as early as 4 passages (Rayner et al., 2019). We were able to culture them up to 14 passages without evident change in phenotypes after which cells started to show senescence.
To further investigate the polarization potential of these primary cells, we measured TEER and also stained the transwell membrane for demonstrating the presence of tight junction proteins. One of the characteristics of well-differentiated epithelium is cell polarity. The tight junction proteins help to maintain the polarity by keeping apical and basolateral domains intact and inhibiting intermixture (Cao et al., 2012; Riga et al., 2020). Maintenance of apical-basolateral polarity is essential for virus infection (Bergelson, 2009; Ruan et al., 2020) as viruses may have predilection to initiate infection from either apical or basolateral surface (Excoffon et al., 2008; Kotha et al., 2015; Tamhankar and Patterson, 2019). The TEER measurement has been used for the demonstration of the polarization potential of epithelial cells (Srinivasan et al., 2015). Here all three bovine primary respiratory cells showed moderate to high TEER values, thus indicating their ability to differentiate their surface membranes into apical-basolateral domains. Further, we demonstrated the presence of tight junction proteins, occludin, and claudin-3 in all three cell types. Thus, these cell lines have the differentiation ability and are mature respiratory epithelial cells with barrier integrity.
Influenza viruses use sialo-glycans with α−2,3 or α−2,6 linkage present in epithelial cells (Garcia-Sastre, 2010). Airway epithelial cells expressing the above receptors are the primary target of influenza virus although the distribution pattern of these receptors is unknown in many species (Ibricevic et al., 2006). Bovine primary respiratory cells established in this study showed higher expression of α−2,3 linked sialic acid than α−2,6 linked sialic acid. The distribution pattern, when compared for particular sialic acid (α−2,3 or α−2,6 linked), among nasal turbinate, soft palate, and tracheal epithelial cells was similar. Although all four influenza subtypes use glycans with terminal sialic acids as receptors, there are type-specific preferences towards engagement of different forms of sialic acids for entry and infection. The N and O substitution or modification of parent neuraminic acid leads to observed structural diversity of sialic acids in different mammalian species (Li and Chen, 2012). The 9-O- acetylation of Neu5Ac (prominent in humans) and Neu5Gc (prominent in animals) leads to the generation of Neu5,9Ac2 and Neu5Gc9Ac respectively (Baumann et al., 2015).
Although some reports have shown IAV infecting cattle, migratory birds or waterfowls are thought to be the primary reservoirs of IAV, and humans are primary hosts for IBV (Sreenivasan et al., 2019b). In this study, bovine primary respiratory cells supported the replication of IAV and IBV to moderate titers with IAV titer higher than IBV by 1 log. This observation appears to be in good agreement with previous studies showing that IBV has a narrow tropism, while IAV possesses a broad tropism (Long et al., 2019; Wang et al., 2007). The moderate to low titers of IAV and IBV could be due to differences in receptor binding between IAV and IBV. Influenza A, and B viruses demonstrate a binding preference to Neu5Ac over Neu5Gc. Sialic acid modifications like O-acetylation of Neu5Ac and Neu5Gc may alter the infectivity of IAV, and IBV either by reducing HA binding or by affecting the enzymatic ability of NA to cleave such modified sialic acids (Corfield et al., 1986; Wasik et al., 2017; Wasik et al., 2016). Interesingly, unlike IAV and IBV, ICV and IDV use 9-O-acetylated sialic acid especially 9-O-acetylated form of Neu5Ac and Neu5Gc (Liu et al., 2020b; Rogers et al., 1986; Song et al., 2016). The distribution pattern and diversity of sialic acids in different compartments along the respiratory tract, and more specifically in the lung, in humans is slowly unraveling (Jia et al., 2020). So far, no such comprehensive study on the distribution pattern of various sialic acids on the bovine respiratory tract is conducted, and thus it is a potential area for further study.
The infection of the bovine primary nasal turbinate, soft palate, and tracheal epithelial cells with four types of influenza viruses demonstrated that these cell lines are permissive to IDV as well as to influenza A and B virus infections. All three cell types supported IDV replication to high titers. Influenza D viruses diverged into five phylogenetic lineages, the D/660, D/OK, D/Yama2016, D/Yama/2019, and D/CA2019 (Huang et al., 2021; Murakami et al., 2020). Of the two strains, D/660 lineage virus propagated to a slightly higher titer than D/OK lineage. D/660 lineage virus strain used in the study is of bovine origin, which might be the reason for the higher replication fitness observed in these bovine respiratory cells compared to the D/OK lineage strain from swine. Congruent to this finding, swine D/OK had better replication fitness, than bovine D/660, in swine primary nasal turbinate cell (Sreenivasan et al., 2019a). A recent study agreed with our current finding where D/660 propagated better than D/OK in ovine respiratory tract explants. Additionally, they observed better replication in ciliated epithelial cells of the respiratory tract (trachea, nasal turbinate) (Holwerda et al., 2019; Mazzetto et al., 2020). We found higher replication titers for D/660 in nasal turbinate, and tracheal epithelial cells as well. It has been suggested that IDV replicates better at the upper respiratory tract than at the lower respiratory tract. Recombinant HEF from D660 lineage virus, however, showed increased staining in bovine lung tissues compared to those observed in nasal epithelium and pharyngeal tissues (Chiapponi et al., 2020). Future study is needed to determine whether receptor binding affinity relates to the level of viral staining in these tissues. Overall, in this study, the replication titers were slightly higher at 33°C than at 37°C for D/660 strain but had similar titers at both temperatures for D/OK strain. IDV replication in cell culture has shown no temperature-dependent restriction in vitro at above two temperatures and that IDV-HEF protein has been thought to be the basis for this thermal stability (Liu et al., 2020a; Yu et al., 2017).
At the amino acid level, ICV and IDV share 50% identity, with both viruses having single HEF in substitution of HA and NA proteins seen in IAV and IBV (Hause et al., 2013). Intriguingly, we found that human ICV failed to replicate in bovine primary epithelial cells, which was further confirmed by IFA. Bovine influenza C virus, detected in cattle with respiratory symptoms, shared 95% sequence identity to human ICV strains (Nissly et al., 2020; Zhang et al., 2018). Although ICV has been detected in cattle, and swine, it is mostly considered as a human pathogen (Sederdahl and Williams, 2020). Similar results were obtained when human airway epithelial cells were used to study replication of ICV and IDV (Holwerda et al., 2019). Although HEF protein of both ICV and IDV uses 9-O-acetylated sialic acid, IDV HEF uses both Neu5,9Ac2 and Neu5Gc9Ac while ICV prefers Neu5,9Ac2 for host cell entry (Liu et al., 2020b). The receptor-binding cavity of IDV-HEF is open and thus may account for its broad cell tropism when compared to ICV (Song et al., 2016). Using bovine primary cells developed from this work should enable further interrogation of viral determinants residing in the HEF protein that restricts human ICV infection in these bovine cells.
Current culture model systems for studying IDV utilizes transformed cell lines that are largely limited to translate the in vivo conditions of the bovine respiratory epithelium where IDV infection and spread occur. The bovine primary respiratory epithelial cell culture system described here represents a step forward in modeling IDV infection, some limitations, however, exist that can be further improved. For example, bovine immune cells are absent in these primary cells so the immune activation of infected epithelial cells and the impact of the communication involving epithelial and immune cells on infection outcome and pathogenesis of IDV cannot be measured. Thus, these bovine primary respiratory cells can be further improved by including immune cells, which can better model IDV infection biology and pathophysiology.
Conclusion
In this study, we have established and characterized bovine primary respiratory epithelial cell lines from nasal turbinate, soft palate, and trachea. Since all three cell lines were established from the same animal, these cell lines provide isogenous cell culture models to study novel influenza D virus as well as other microbial pathogens primarily infecting cattle through the respiratory tract. All three cell lines demonstrated a phenotype that was characteristic of well-differentiated epithelial cells, which can be readily preserved up to 14 passages (5–7 days each passage) in vitro. Moreover, these cells supported the efficient replication of IDV with variable susceptibility to other types of influenza viruses. Taken together, the results of our experiments collectively demonstrate that bovine primary respiratory epithelial cells can serve as a physiologically relevant in vitro system to model IDV replication and pathogenesis.
Research Highlights.
Established and characterized primary respiratory epithelial cells from nasal turbinate, soft palate, and trachea of same cattle.
All three cells showed epithelial cell phenotype, polarized into apicobasal surface, and formed tight junctions.
All three cell types supported replication of influenza D virus to high titers.
Isogenous bovine respiratory epithelial cell culture will be useful tool to study pathogenesis of influenza D virus as well as other respiratory virus of bovine respiratory disease complex.
Acknowledgments
This study was supported by NIH R01AI141889, Agricultural Experiment Station of the University of Kentucky, the William Robert Mills Chair Endowment Fund, and SDSU Agricultural Experiment Station Hatch grants # SD00H701-20 and 3AH-673. We would also like to acknowledge the SDSU meat lab for providing tissue samples.
Footnotes
Conflict of interests
The authors declare no conflict of interest.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bailey ES, Choi JY, Fieldhouse JK, Borkenhagen LK, Zemke J, Zhang D, Gray GC, 2018. The continual threat of influenza virus infections at the human-animal interface: What is new from a one health perspective? Evol Med Public Health 2018, 192–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann AM, Bakkers MJ, Buettner FF, Hartmann M, Grove M, Langereis MA, de Groot RJ, Muhlenhoff M, 2015. 9-O-Acetylation of sialic acids is catalysed by CASD1 via a covalent acetyl-enzyme intermediate. Nat Commun 6, 7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson JM, 2009. Intercellular junctional proteins as receptors and barriers to virus infection and spread. Cell Host Microbe 5, 517–521. [DOI] [PubMed] [Google Scholar]
- Cao X, Surma MA, Simons K, 2012. Polarized sorting and trafficking in epithelial cells. Cell Res 22, 793–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiapponi C, Ducatez M, Faccini S, Foni E, Gaudino M, Hägglund S, Luppi A, Meyer G, Moreno A, Näslund K, Nemanichvili N, Oliva J, Prosperi A, Rosignoli C, Renault V, Saegerman C, Sausy A, Snoeck C, Valarcher JF, Verheije H, Zohari S, 2020. Risk assessment for influenza D in Europe. EFSA Supporting Publications 17. [Google Scholar]
- Corfield AP, Sander-Wewer M, Veh RW, Wember M, Schauer R, 1986. The action of sialidases on substrates containing O-acetylsialic acids. Biol Chem Hoppe Seyler 367, 433–439. [DOI] [PubMed] [Google Scholar]
- Council L, Hameed O, 2009. Differential expression of immunohistochemical markers in bladder smooth muscle and myofibroblasts, and the potential utility of desmin, smoothelin, and vimentin in staging of bladder carcinoma. Mod Pathol 22, 639–650. [DOI] [PubMed] [Google Scholar]
- Cozens D, Grahame E, Sutherland E, Taylor G, Berry CC, Davies RL, 2018. Development and optimization of a differentiated airway epithelial cell model of the bovine respiratory tract. Sci Rep 8, 853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozens D, Sutherland E, Lauder M, Taylor G, Berry CC, Davies RL, 2019. Pathogenic Mannheimia haemolytica Invades Differentiated Bovine Airway Epithelial Cells. Infect Immun 87, e00078–00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dane H, Duffy C, Guelbenzu M, Hause B, Fee S, Forster F, McMenamy MJ, Lemon K, 2019. Detection of influenza D virus in bovine respiratory disease samples, UK. Transbound Emerg Dis 66, 2184–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffon KJ, Guglielmi KM, Wetzel JD, Gansemer ND, Campbell JA, Dermody TS, Zabner J, 2008. Reovirus preferentially infects the basolateral surface and is released from the apical surface of polarized human respiratory epithelial cells. J Infect Dis 197, 1189–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson L, Olivier AK, Genova S, Epperson WB, Smith DR, Schneider L, Barton K, McCuan K, Webby RJ, Wan XF, 2016. Pathogenesis of Influenza D Virus in Cattle. J Virol 90, 5636–5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn O, Gallagher C, Mooney J, Irvine C, Ducatez M, Hause B, McGrath G, Ryan E, 2018. Influenza D Virus in Cattle, Ireland. Emerg Infect Dis 24, 389–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sastre A, 2010. Influenza virus receptor specificity: disease and transmission. The American journal of pathology 176, 1584–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause BM, Collin EA, Liu R, Huang B, Sheng Z, Lu W, Wang D, Nelson EA, Li F, 2014. Characterization of a novel influenza virus in cattle and Swine: proposal for a new genus in the Orthomyxoviridae family. mBio 5, e00031–00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause BM, Ducatez M, Collin EA, Ran Z, Liu R, Sheng Z, Armien A, Kaplan B, Chakravarty S, Hoppe AD, Webby RJ, Simonson RR, Li F, 2013. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog 9, e1003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause BM, Huntimer L, Falkenberg S, Henningson J, Lechtenberg K, Halbur T, 2017. An inactivated influenza D virus vaccine partially protects cattle from respiratory disease caused by homologous challenge. Veterinary microbiology 199, 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holwerda M, Kelly J, Laloli L, Sturmer I, Portmann J, Stalder H, Dijkman R, 2019. Determining the Replication Kinetics and Cellular Tropism of Influenza D Virus on Primary Well-Differentiated Human Airway Epithelial Cells. Viruses 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Yu J, Hause BM, Park JY, Sreenivasan C, Uprety T, Sheng Z, Wang D, Li F, 2021. Emergence of New Phylogenetic Lineage of Influenza D virus with Broad Antigenicity in California, United States. Emerging Microbes & Infections, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibricevic A, Pekosz A, Walter MJ, Newby C, Battaile JT, Brown EG, Holtzman MJ, Brody SL, 2006. Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. J Virol 80, 7469–7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia N, Byrd-Leotis L, Matsumoto Y, Gao C, Wein AN, Lobby JL, Kohlmeier JE, Steinhauer DA, Cummings RD, 2020. The Human Lung Glycome Reveals Novel Glycan Ligands for Influenza A Virus. Sci Rep 10, 5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katwal P, Thomas M, Uprety T, Hildreth MB, Kaushik RS, 2019. Development and biochemical and immunological characterization of early passage and immortalized bovine intestinal epithelial cell lines from the ileum of a young calf. Cytotechnology 71, 127–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik RS, Begg AA, Wilson HL, Aich P, Abrahamsen MS, Potter A, Babiuk LA, Griebel P, 2008. Establishment of fetal bovine intestinal epithelial cell cultures susceptible to bovine rotavirus infection. J Virol Methods 148, 182–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotha PL, Sharma P, Kolawole AO, Yan R, Alghamri MS, Brockman TL, Gomez-Cambronero J, Excoffon KJ, 2015. Adenovirus entry from the apical surface of polarized epithelia is facilitated by the host innate immune response. PLoS Pathog 11, e1004696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F, Smith GJD, Fouchier RAM, Peiris M, Kedzierska K, Doherty PC, Palese P, Shaw ML, Treanor J, Webster RG, Garcia-Sastre A, 2018. Influenza. Nat Rev Dis Primers 4, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakdawala SS, Jayaraman A, Halpin RA, Lamirande EW, Shih AR, Stockwell TB, Lin X, Simenauer A, Hanson CT, Vogel L, Paskel M, Minai M, Moore I, Orandle M, Das SR, Wentworth DE, Sasisekharan R, Subbarao K, 2015. The soft palate is an important site of adaptation for transmissible influenza viruses. Nature 526, 122–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Chambers M, 2019. A co-culture model of the bovine alveolus. F1000Res 8, 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chen X, 2012. Sialic acid metabolism and sialyltransferases: natural functions and applications. Applied microbiology and biotechnology 94, 887–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Sheng Z, Huang C, Wang D, Li F, 2020a. Influenza D virus. Curr Opin Virol 44, 154–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Sreenivasan C, Yu H, Sheng Z, Newkirk SJ, An W, Smith DF, Chen X, Wang D, Li F, 2020b. Influenza D virus diverges from its related influenza C virus in the recognition of 9-O-acetylated N-acetyl- or N-glycolyl-neuraminic acid-containing glycan receptors. Virology 545, 16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JS, Mistry B, Haslam SM, Barclay WS, 2019. Host and viral determinants of influenza A virus species specificity. Nat Rev Microbiol 17, 67–81. [DOI] [PubMed] [Google Scholar]
- Mazzetto E, Bortolami A, Fusaro A, Mazzacan E, Maniero S, Vascellari M, Beato MS, Schiavon E, Chiapponi C, Terregino C, Monne I, Bonfante F, 2020. Replication of Influenza D Viruses of Bovine and Swine Origin in Ovine Respiratory Explants and Their Attachment to the Respiratory Tract of Bovine, Sheep, Goat, Horse, and Swine. Front Microbiol 11, 1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milian E, Kamen AA, 2015. Current and emerging cell culture manufacturing technologies for influenza vaccines. Biomed Res Int 2015, 504831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra N, Cernicchiaro N, Torres S, Li F, Hause BM, 2016. Metagenomic characterization of the virome associated with bovine respiratory disease in feedlot cattle identified novel viruses and suggests an etiologic role for influenza D virus. J Gen Virol 97, 1771–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S, Odagiri T, Melaku SK, Bazartseren B, Ishida H, Takenaka-Uema A, Muraki Y, Sentsui H, Horimoto T, 2019. Influenza D Virus Infection in Dromedary Camels, Ethiopia. Emerg Infect Dis 25, 1224–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S, Sato R, Ishida H, Katayama M, Takenaka-Uema A, Horimoto T, 2020. Influenza D Virus of New Phylogenetic Lineage, Japan. Emerg Infect Dis 26, 168–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedland H, Wollman J, Sreenivasan C, Quast M, Singrey A, Fawcett L, Christopher-Hennings J, Nelson E, Kaushik RS, Wang D, Li F, 2018. Serological evidence for the co-circulation of two lineages of influenza D viruses in equine populations of the Midwest United States. Zoonoses and public health 65, e148–e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissly RH, Zaman N, Ibrahim PAS, McDaniel K, Lim L, Kiser JN, Bird I, Chothe SK, Bhushan GL, Vandegrift K, Neibergs HL, Kuchipudi SV, 2020. Influenza C and D viral load in cattle correlates with bovine respiratory disease (BRD): Emerging role of orthomyxoviruses in the pathogenesis of BRD. Virology 551, 10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva J, Eichenbaum A, Belin J, Gaudino M, Guillotin J, Alzieu J-P, Nicollet P, Brugidou R, Gueneau E, Michel E, Meyer G, Ducatez MF, 2019. Serological Evidence of Influenza D Virus Circulation Among Cattle and Small Ruminants in France. Viruses 11, 516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva J, Mettier J, Sedano L, Delverdier M, Bourgès-Abella N, Hause B, Loupias J, Pardo I, Bleuart C, Bordignon PJ, Meunier E, Le Goffic R, Meyer G, Ducatez MF, 2020. Murine Model for the Study of Influenza D Virus. Journal of virology 94, e01662–01619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce OM, Laubli H, 2016. Sialic acids in cancer biology and immunity. Glycobiology 26, 111–128. [DOI] [PubMed] [Google Scholar]
- Powell JD, Waters KM, 2017. Influenza-Omics and the Host Response: Recent Advances and Future Prospects. Pathogens 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast M, Sreenivasan C, Sexton G, Nedland H, Singrey A, Fawcett L, Miller G, Lauer D, Voss S, Pollock S, Cunha CW, Christopher-Hennings J, Nelson E, Li F, 2015. Serological evidence for the presence of influenza D virus in small ruminants. Veterinary microbiology 180, 281–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran Z, Shen H, Lang Y, Kolb EA, Turan N, Zhu L, Ma J, Bawa B, Liu Q, Liu H, Quast M, Sexton G, Krammer F, Hause BM, Christopher-Hennings J, Nelson EA, Richt J, Li F, Ma W, 2015. Domestic pigs are susceptible to infection with influenza B viruses. J Virol 89, 4818–4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner RE, Makena P, Prasad GL, Cormet-Boyaka E, 2019. Optimization of Normal Human Bronchial Epithelial (NHBE) Cell 3D Cultures for in vitro Lung Model Studies. Sci Rep 9, 500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed LJ, Muench H, 1938. A Simple Method of Estimating Fifty Per Cent Endpoints12. American Journal of Epidemiology 27, 493–497. [Google Scholar]
- Riga A, Castiglioni VG, Boxem M, 2020. New insights into apical-basal polarization in epithelia. Curr Opin Cell Biol 62, 1–8. [DOI] [PubMed] [Google Scholar]
- Rogers GN, Herrler G, Paulson JC, Klenk HD, 1986. Influenza C virus uses 9-O-acetyl-N-acetylneuraminic acid as a high affinity receptor determinant for attachment to cells. Journal of Biological Chemistry 261, 5947–5951. [PubMed] [Google Scholar]
- Ruan T, Sun J, Liu W, Prinz RA, Peng D, Liu X, Xu X, 2020. H1N1 Influenza Virus Cross-Activates Gli1 to Disrupt the Intercellular Junctions of Alveolar Epithelial Cells. Cell Rep 31, 107801. [DOI] [PubMed] [Google Scholar]
- Salem E, Cook EAJ, Lbacha HA, Oliva J, Awoume F, Aplogan GL, Hymann EC, Muloi D, Deem SL, Alali S, Zouagui Z, Fevre EM, Meyer G, Ducatez MF, 2017. Serologic Evidence for Influenza C and D Virus among Ruminants and Camelids, Africa, 1991–2015. Emerg Infect Dis 23, 1556–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem E, Hagglund S, Cassard H, Corre T, Naslund K, Foret C, Gauthier D, Pinard A, Delverdier M, Zohari S, Valarcher JF, Ducatez M, Meyer G, 2019. Pathogenesis, Host Innate Immune Response, and Aerosol Transmission of Influenza D Virus in Cattle. J Virol 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sederdahl BK, Williams JV, 2020. Epidemiology and Clinical Characteristics of Influenza C Virus. Viruses 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz C, Frensing T, Hoper D, Kochs G, Reichl U, 2010. High yields of influenza A virus in Madin-Darby canine kidney cells are promoted by an insufficient interferon-induced antiviral state. J Gen Virol 91, 1754–1763. [DOI] [PubMed] [Google Scholar]
- Sobotta K, Bonkowski K, Liebler-Tenorio E, Germon P, Rainard P, Hambruch N, Pfarrer C, Jacobsen ID, Menge C, 2017. Permissiveness of bovine epithelial cells from lung, intestine, placenta and udder for infection with Coxiella burnetii. Vet Res 48, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Qi J, Khedri Z, Diaz S, Yu H, Chen X, Varki A, Shi Y, Gao GF, 2016. An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism. PLoS Pathog 12, e1005411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasan C, Thomas M, Sheng Z, Hause BM, Collin EA, Knudsen DE, Pillatzki A, Nelson E, Wang D, Kaushik RS, Li F, 2015. Replication and Transmission of the Novel Bovine Influenza D Virus in a Guinea Pig Model. J Virol 89, 11990–12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasan CC, Thomas M, Antony L, Wormstadt T, Hildreth MB, Wang D, Hause B, Francis DH, Li F, Kaushik RS, 2019a. Development and characterization of swine primary respiratory epithelial cells and their susceptibility to infection by four influenza virus types. Virology 528, 152–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasan CC, Thomas M, Kaushik RS, Wang D, Li F, 2019b. Influenza A in Bovine Species: A Narrative Literature Review. Viruses 11, 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan B, Kolli AR, Esch MB, Abaci HE, Shuler ML, Hickman JJ, 2015. TEER measurement techniques for in vitro barrier model systems. Journal of laboratory automation 20, 107–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S, Fu X, Li G, Kerlin F, Veit M, 2017. Novel Influenza D virus: Epidemiology, pathology, evolution and biological characteristics. Virulence 8, 1580–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamhankar M, Patterson JL, 2019. Directional entry and release of Zika virus from polarized epithelial cells. Virol J 16, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uprety T, Spurlin BB, Antony L, Sreenivasan C, Young A, Li F, Hildreth MB, Kaushik RS, 2019. Development and characterization of a stable bovine intestinal sub-epithelial myofibroblast cell line from ileum of a young calf. In vitro cellular & developmental biology. Animal 55, 533–547. [DOI] [PubMed] [Google Scholar]
- van der Velden LA, Schaafsma HE, Manni JJ, Ruiter DJ, Ramaekers FC, Kuijpers W, 1997. Cytokeratin and vimentin expression in normal epithelium and squamous cell carcinomas of the larynx. Eur Arch Otorhinolaryngol 254, 376–383. [DOI] [PubMed] [Google Scholar]
- Wang Q, Tian X, Chen X, Ma J, 2007. Structural basis for receptor specificity of influenza B virus hemagglutinin. Proceedings of the National Academy of Sciences of the United States of America 104, 16874–16879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasik BR, Barnard KN, Ossiboff RJ, Khedri Z, Feng KH, Yu H, Chen X, Perez DR, Varki A, Parrish CR, 2017. Distribution of O-Acetylated Sialic Acids among Target Host Tissues for Influenza Virus. mSphere 2, e00379–00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasik BR, Barnard KN, Parrish CR, 2016. Effects of Sialic Acid Modifications on Virus Binding and Infection. Trends Microbiol 24, 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SK, Ma W, McDaniel CJ, Gray GC, Lednicky JA, 2016. Serologic evidence of exposure to influenza D virus among persons with occupational contact with cattle. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology 81, 31–33. [DOI] [PubMed] [Google Scholar]
- Yan Y, Ou J, Zhao S, Ma K, Lan W, Guan W, Wu X, Zhang J, Zhang B, Zhao W, Wan C, Shi W, Wu J, Seto D, Yu Z, Zhang Q, 2020. Characterization of Influenza A and B Viruses Circulating in Southern China During the 2017–2018 Season. Front Microbiol 11, 1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hika B, Liu R, Sheng Z, Hause BM, Li F, Wang D, 2017. The Hemagglutinin-Esterase Fusion Glycoprotein Is a Primary Determinant of the Exceptional Thermal and Acid Stability of Influenza D Virus. mSphere 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Li F, Wang D, 2021. The first decade of research advances in influenza D virus. The Journal of general virology 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Porter E, Lohman M, Lu N, Peddireddi L, Hanzlicek G, Marthaler D, Liu X, Bai J, 2018. Influenza C Virus in Cattle with Respiratory Disease, United States, 2016–2018. Emerg Infect Dis 24, 1926–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]