Abstract
Elongation of RNA polymerase II (Pol II) is affected by many factors including DNA damage. Bulky damage, such as lesions caused by ultraviolet (UV) radiation, arrests Pol II and inhibits gene transcription, and may lead to genome instability and cell death. Cells activate transcription-coupled nucleotide excision repair (TC-NER) to remove Pol II-impeding damage and allow transcription resumption. TC-NER initiation in humans is mediated by Cockayne syndrome group B (CSB) protein, which binds to the stalled Pol II and promotes assembly of the repair machinery. Given the complex nature of the TC-NER pathway and its unique function at the interface between transcription and repair, new approaches are required to gain in-depth understanding of the mechanism. Advances in genomic approaches provide an important opportunity to investigate how TC-NER is initiated upon damage-induced Pol II stalling and what factors are involved in this process. In this Review, we discuss new mechanisms of TC-NER revealed by genome-wide DNA damage mapping and new TC-NER factors identified by high-throughput screening. As TC-NER conducts strand-specific repair of mutagenic damage, we also discuss how this repair pathway causes mutational strand asymmetry in the cancer genome.
Keywords: DNA damage, RNA Pol II, CPD-seq, CSB, Mutagenesis
1. Introduction
Gene transcription by RNA polymerase II (Pol II) is essential for all cellular processes, yet the journey of Pol II along transcribed genes is not as smooth as expected. DNA in living cells can be damaged by a number of agents [1], and the resulting damage can interfere with Pol II function. Endogenous damaging sources, including cellular reactive oxygen specifies (ROS), alkylating agents (e.g., S-adenosylmethionine), spontaneous base hydrolysis and deamination, and replication errors [1], can generate up to 105 DNA lesions in each mammalian genome per day [2]. These lesions are mostly chemical modifications to the base and they can block Pol II elongation to different extents. For example, in vitro data indicates that 8-oxo-7,8-dihydroguanine (8-oxo-G), one of the most frequent endogenous oxidative damage, does not arrest Pol II transcription [3]. However, 8-oxo-G can be further oxidized to form 5-guanidinohydantoin (Gh) and spiroiminodihydantoin (Sp) [4]. Gh and Sp have been shown to strongly block Pol II elongation and the blockage cannot be resolved by transcription elongation factors [5]. Additionally, endogenous base lesions are primarily removed from the genome by base excision repair (BER), in which the first step is the cleavage of the damaged base by glycosylases and generation of apurinic/apyrimidinic sites (AP sites) [6]. AP sites have been shown to block Pol II elongation [7]. AP sites also arise in the cell by spontaneous depurination at high frequency (~104 lesions per cell per day) [2]. Hence, endogenous DNA damage (e.g., AP sites, oxidative damage, and others) can inhibit Pol II elongation.
‘Bulky’ DNA damage, frequently induced by environmental damaging chemicals and radiation, can be even more harmful to Pol II elongation than endogenous base damage. In this regard, it has long been known that solar ultraviolet (UV) radiation induces bulky photolesions that distort the DNA double helix and block DNA replication and gene transcription [8]. UV-A and residual UV-B comprise the major damaging wavelengths in solar UV and they can produce significant amount of photolesions in each exposed cell [9], mainly including cyclobutane pyrimidine dimers (CPDs) and (6–4) photoproducts (6–4PPs) [10]. RNA Pol II stalling at a UV lesion creates a stable ternary complex consisting of stalled Pol II, UV-damaged DNA template, and the nascent transcript. The half-life of an arrested Pol II at a CPD is ~ 20 hours in vitro in the absence of relevant repair proteins, and Pol II causes a footprint of approximately 40 nt covering the damage nearly symmetrically [11]. The strong Pol II stalling at a CPD lesion is problematic for the cell, as transcription cannot continue and repair may be inhibited by Pol II. Therefore, repair of UV damage to rescue the stalled Pol II is vital for maintenance of genomic stability and cell survival [12]. Bulky lesions are also caused by the damaging chemical benzo[a]pyrene (BaP) in tobacco smoke. BaP is metabolized in the cell to form a potent mutagen, (+)benzo[a]pyrene-7,8-dihydrodiol-9,10-epoxide (BPDE), which covalently binds to deoxyguanosines (dGs) in DNA and causes formation of BPDE-dG adducts [13]. The BPDE-dG adduct has been shown to block Pol II elongation [14]. Both UV and BaP are well-known carcinogens of the skin and lung, respectively. Therefore, the cellular response to remove DNA lesions induced by UV and BaP is important for normal cell functions and cancer prevention.
Transcription-coupled nucleotide excision repair (TC-NER), a subpathway of nucleotide excision repair (NER), is a critical cellular mechanism that repairs transcription-stalling DNA damage. Different from global genomic NER (GG-NER), which relies on surveillance proteins such as Xeroderma pigmentosum complementation group C protein (XPC) to recognize damage [15], TC-NER utilizes elongating Pol II to scan the transcribed strand and ‘recognize’ transcription-stalling damage [16]. After damage recognition, both GG-NER and TC-NER recruit transcription factor IIH (TFIIH), a ten-subunit protein complex consisting of two important DNA helicases, XPD and XPB, to the damage site [17]. TFIIH, together with the scaffolding protein XPA and replication protein A (RPA), unwinds the two strands around the damage and verifies the presence of damage [18], thus creating a preincision DNA bubble that is recognized by repair endonucleases ERCC1-XPF and XPG [15]. XPF and XPG cleave the damaged strand on the 5’ and 3’ sides relative to the damage, respectively, releasing a single-stranded DNA fragment of ~30 nucleotide (nt) containing the lesion. The gap is then filled by DNA polymerases using undamaged DNA strand as the template. DNA ligase I or ligase IIIα-XRCC1 is then recruited to seal the DNA backbone [15].
TC-NER was first reported in the mid 1980’s in the study of UV damage repair in mammalian cells [19,20]. Early studies revealed that repair of CPDs in the actively expressing dihydrofolate reductase (Dhfr) gene is faster on the transcribed strand (TS) relative to the non-transcribed strand (NTS), exhibiting a clear repair asymmetry between the two strands [19]. These pioneering studies on TC-NER were followed by investigations in other model organisms such as Escherichia coli [21], Saccharomyces cerevisiae [22], Arabidopsis thaliana [23], and Drosophila melanogaster [24]. Altogether, these studies established TC-NER as a conserved mechanism that conducts strand-biased and TS-preferential repair in active genes [16]. It is important to note that TC-NER repairs a wide range of bulky lesions, including UV damage, BPDE-dG adducts [25], and DNA crosslinks caused by the anti-cancer drug cisplatin [26,27]. Additionally, published data also suggest a role for TC-NER in the repair of non-bulky lesions such as alkylation damage [28] and AP sites [29].
2. TC-NER mechanisms revealed by structural and biochemical studies
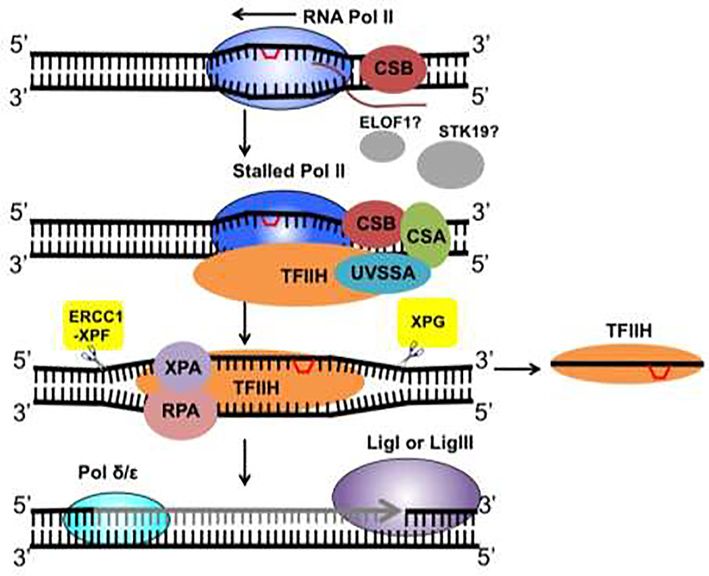
Previous studies have identified important TC-NER factors such as Cockayne syndrome group B protein (CSB), Cockayne syndrome group A protein (CSA), and UV Stimulated Scaffold Protein A (UVSSA) [30]. Recent data suggests that assembly of the TC-NER complex occurs in a cooperative way in which CSB binds to the stalled Pol II first and assists the recruitment of CSA and UVSSA [31]. CSA functions in an E3 ubiquitin ligase complex to facilitate CSB and Pol II ubiquitylation [30], while UVSSA recruits the NER core complex transcription factor IIH (TFIIH) [31]. As mentioned earlier, damage recognition in GG-NER is dependent on XPC, which binds to DNA damage and recruits TFIIH to the damage site in GG-NER [15]. The steps following TFIIH recruitment, including DNA unwinding, repair excision, DNA synthesis and ligation (Figure 1), are believed to be the same between GG-NER and TC-NER [15].
Figure 1.
TC-NER in mammalian cells. RNA Pol II stalling at a DNA lesion leads to the binding of CSB, which recruits CSA and UVSSA. UVSSA then promotes TFIIH recruitment. The elongation factor ELOF1 and the serine/threonine kinase STK19 are also important for TC-NER, but their roles remain elusive. The helicases in TFIIH unwind the two strands of damaged DNA to facilitate dual incision by ERCC1-XPF and XPG on the 5’ and 3’ sides of the lesion, respectively. The excised DNA fragment containing the lesion is bound by TFIIH and released. The gap on the damaged strand is filled by DNA polymerases and DNA ligases. Transcription can be resumed after repair of the damage.
It has long been known Pol II stalling is not only caused by DNA damage, but also triggered by genomic features such as specific DNA sequences and nucleosomes [32], yet the cell can distinguish between different transcription barriers and only commits DNA lesions to TC-NER. How does the cell ‘know’ if the obstacle is a genuine DNA lesion? Recent studies of the CSB ortholog in Saccharomyces cerevisiae, Rad26, provide new insights into the underlying mechanism [33]. CSB and Rad26 are known to function as elongation factors during normal Pol II transcription [34,35]. The structure of the yeast Pol II-Rad26 complex indicates that Rad26 binds to DNA upstream of elongating Pol II, and utilizes its ATP-dependent 3’-to-5’ DNA translocase activity to pull the transcribed strand away from Pol II [33], which promotes the forward movement of Pol II. This directional DNA translocase activity of Rad26 allows Pol II to bypass small barriers (e.g., poly(A) DNA sequence, non-bulky lesions, and nucleosomes [33,36]) without the need of invoking TC-NER. However, if the barrier is strong and cannot be bypassed by Pol II, such as CPDs, Rad26 moves toward the arrested Pol II and eventually binds to it [37]. The structural data shows that Rad26 binds directly to Pol II, through interactions with regions of the Pol II complex that serve as docking sites for the elongation factor Spt4-Spt5 [33,37], which has been shown as a TC-NER suppressor in yeast [38,39]. The overlapping binding sites between Rad26 and Spt4-Spt5 suggest that CSB/Rad26 may compete against Spt4-Spt5 and displace it from stalled Pol II, thus switching Pol II from elongation to repair mode [37]. CSB/Rad26 may also create new binding sites for NER factors such as TFIIH. Therefore, the DNA translocase activity of CSB/Rad26 plays an important role in deciding whether TC-NER is required by testing if the DNA obstacle can be bypassed by Pol II [12]. In addition to CSB/Rad26, emerging evidence indicates that Pol II itself and components in the Pol II transcription complex also affect TC-NER. For example, studies in yeast have shown important roles for the non-essential Pol II subunits Rpb9 and Rpb4 [40], Pol II-associated factor (PAF) complex [41], and transcription termination factor Sen1 [42], in regulating TC-NER. Among these factors, Rpb9 and Sen1 can facilitate repair, while Rpb4 and PAF function as TC-NER suppressors [43], suggesting complex regulation of TC-NER by Pol II and Pol II-associated proteins.
Although published data have revealed intriguing structural and biochemical mechanisms for TC-NER, important questions still exist. For example, CSB and Rad26 are not always required for TC-NER in cells [44–46], which raises questions as to how TC-NER initiates without CSB/Rad26 in some genes or genic regions. It is also unknown how TC-NER copes with the chromatin structure in eukaryotes. Furthermore, no CSB homolog has been identified in the Drosophila genome, yet strong TC-NER activity was found in Drosophila cells [24], suggesting some species may employ factors not belonging to the CSB family to initiate TC-NER. Recent studies using genomic approaches provide new information that helps us address these important questions. In the subsequent sections, we discuss how data obtained with genome-wide damage mapping uncovers common but variable requirements for CSB/Rad26 in different chromatin regions (e.g., transcription initiation vs. elongation) and how high-throughput screens identify new TC-NER regulators.
3. TC-NER mechanisms revealed by genome-wide mapping of DNA damage and repair
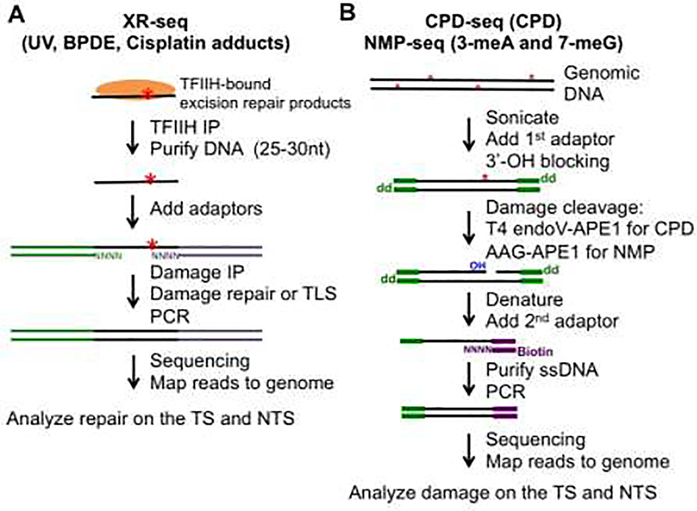
Several genomic methods have been developed in recent years to study formation and repair of different types of DNA damage [47,48]. Of particular interest, methods utilizing next-generation sequencing (NGS) for strand-specific analysis of DNA damage have offered an important opportunity to analyze genome-wide TC-NER. Here we focus on studies using three damage/repair mapping methods: XR-seq, CPD-seq, and NMP-seq (Figure 2).
Figure 2.
Schematic representation of DNA damage/repair mapping methods. (A) XR-seq is a method that maps excision repair products (~30 nt), which are bound by TFIIH in the cell. These short fragments are extracted from cell lysates and ligated to sequencing adaptors. Fragments containing damage are purified by damage immuneprecipitation (IP) using a DNA damage-specific antibody. After damage repair or bypass with a translesion synthesis (TLS) DNA polymerase, the DNA fragment is used for PCR amplification. (B) CPD-seq and NMP-seq are used for mapping UV damage (i.e., CPDs) and alkylation lesions such as 3-meA and 7-meG. Red asterisks represent DNA damage. Damage is cleaved with DNA repair enzymes to generate a new 3’-OH group, which is ligated to a sequencing DNA adaptor (2nd adaptor).
XR-seq:
Excision repair sequencing (XR-seq) was published in 2015 to analyze excision repair of UV damage in the human genome [49]. XR-seq utilizes TFIIH co-immunoprecipitation to capture the ~30 nt fragments that are excised during NER, which are then ligated to sequencing adaptors and subjected to NGS [49,50] (Figure 2A). As the NER intermediates are single stranded and generated from the damaged strand, XR-seq data can distinguish repair between the TS and NTS of transcribed genes, and thus can be utilized for simultaneous analysis of GG-NER and TC-NER. Importantly, XR-seq data obtained in normal human fibroblasts irradiated by UV presented a clear TC-NER signature for CPDs at the genome scale, with faster repair on the TS relative to the NTS [49]. In contrast, 6–4PPs repair is evenly distributed between the two strands, due to recognition of 6–4PPs by XPC and efficient repair through the GG-NER subpathway [49]. Additionally, TC-NER of CPDs is significantly diminished across the genome in CSB- or UVSSA-deficient cells [31,49], consistent with the important roles of CSB and UVSSA in TC-NER initiation. XR-seq was also applied to repair studies of BPDE-dG and DNA crosslink damage caused by tobacco smoke and cisplatin, respectively. Similar to CPDs, XR-seq data indicates that both BPDE-dGs and cisplatin-induced 1,2-GpG crosslinks are preferentially repaired on the TS of active genes by TC-NER [25,26], although the TC-NER effect on BPDE-dGs is relatively weak. The low TC-NER is likely because BPDE-dG adducts can be efficiently repaired by GG-NER, similar to the repair of 6–4PPs.
As the NER dual incision step is highly conserved, the XR-seq methodology can be used for repair mapping in other organisms. Indeed, an XR-seq study conducted in E. coli showed abundant NER intermediates following UV treatment, and the excision product is ~13 nt [51], shorter than that in humans. A strong TC-NER signature, shown by fast repair on the TS, was found for almost all E. coli genes, and this signature is dependent on the Mutation Frequency Decline (Mfd) protein [51], the counterpart of CSB in E. coli, thus confirming the crucial role for Mfd in bacterial TC-NER. Furthermore, XR-seq data generated in UV-irradiated Drosophila cells identified clear TC-NER activity that is comparable to the level observed in mammalian cells [24], even though a search of the Drosophila genome failed to identify orthologs to CSB, CSA, or UVSSA. The XR-seq data suggests that Drosophila (and potentially other organisms lacking canonical TC-NER factors) may utilize non-canonical TC-NER factors to initiate repair upon transcription stalling; however, the identity of Drosophila TC-NER initiation factor is unclear.
CPD-seq:
Cyclobutane Pyrimidine Dimer sequencing (CPD-seq) was initially developed to analyze CPDs in the yeast genome [52], and it has been extended to human cells for genome-wide CPD mapping [53,54]. This method uses a CPD-specific glycosylase (T4 endonuclease V) and the AP endonuclease (APE1) to cleave CPD damage and generate a nick on the damaged stand. The resulting 3’-OH group on the 5’ side of the CPD damage is then ligated to a sequencing adaptor (Figure 2B, purple adaptor), which allows precise mapping of the CPD damage at single-nucleotide resolution in a strand-specific manner [52,55]. Using CPD-seq, genome-wide CPDs can be mapped to analyze initial damage as well as remaining damage at different repair time points. Normalization of CPDs after repair to the initial damage generates fraction of remaining (i.e., unrepaired) damage, which conversely correlates with CPD repair [55]. By analyzing the remaining damage on the TS and NTS of active genes, repair of CPDs by TC-NER and GG-NER can be compared to each other.
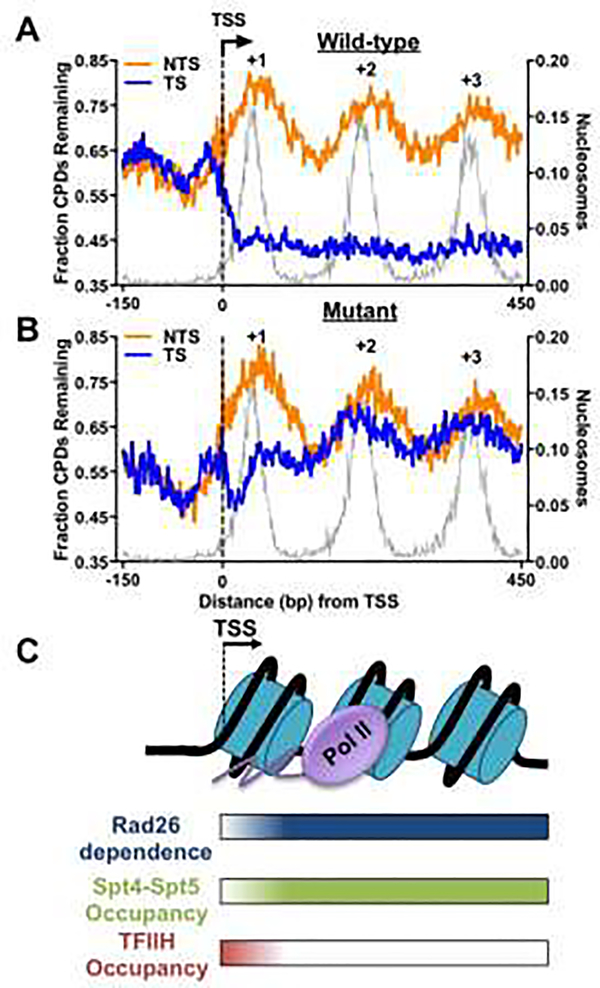
Analysis of CPD-seq data in wild-type yeast cells shows faster repair on the TS than that on the NTS, and the TC-NER efficiency is correlated with gene transcription frequency, with highly expressing genes showing the most prominent TC-NER activity [52]. Transcribed DNA is packaged into regularly spaced nucleosomes [56]. Although biochemical studies have shown an inhibitory effect of the nucleosome on GG-NER [57], how nucleosomes affect TC-NER is less clear. Intriguingly, analysis of the CPD-seq data in wild-type yeast cells has revealed periodic unrepaired CPD peaks near central dyads of nucleosomes on the NTS [58,59] (Figure 3A), consistent with the known function of nucleosomes in preventing the access of GG-NER factors to damage [57]. In stark contrast, repair by TC-NER (i.e., repair on the TS) is not inhibited by nucleosomes and robust TC-NER is observed both at the nucleosomal dyad axis and in linker DNA (Figure 3A), which is likely due to transient disruption of nucleosomes by elongating Pol II during TC-NER [16]. Therefore, nucleosomes display strikingly different effects on GG-NER and TC-NER in yeast, and the difference is correlated with distinct nucleosome structures encountered by the two NER subpathways (i.e., intact nucleosomes for GG-NER but disrupted nucleosome structure for TC-NER).
Figure 3.
Role of Rad26 in yeast TC-NER revealed by CPD-seq analysis. (A) Repair of CPDs in wild-type cells at 2h. Yeast genes (n = 5,205) [74] were aligned at their transcription-start site (TSS) and the fraction of remaining CPDs at 2h (CPD-2h normalized to CPDs-0h) was plotted in DNA regions around the TSS. Transcribed strand (TS) and non-transcribed strand (NTS) are analyzed separately. CPD-seq data was downloaded from Gene Expression Omnibus (accession code GSE145911) [59] and reanalyzed in transcribed regions near the TSS (from −150bp upstream to 450bp downstream of the TSS). The gray background depicts yeast nucleosome occupancy generated with published MNase-seq data [75]. (B) Repair of CPDs in Rad26-deficient cells (i.e., rad26Δ) at 2h in yeast genes. (C) Model depicting the variable requirements for Rad26 in yeast TC-NER in different chromatin regions (e.g., +1 and downstream nucleosomes), and its correlation with the occupancy of transcription elongation factor Spt4-Spt5 and initiation/repair factor TFIIH.
CPD-seq analysis indicates that TC-NER is significantly diminished among most genes in a Rad26-deficient yeast strain (i.e., rad26Δ) [59], confirming a key role for Rad26 in promoting TC-NER. However, a careful examination of the repair data indicates that some yeast genes and some specific genic regions do not require Rad26 to conduct TC-NER. First, a small number of highly transcribed yeast genes can activate TC-NER in a Rad26-independent manner. These Rad26-independent genes comprise the most actively expressing genes in yeast and they may utilize Rpb9, a non-essential subunit of the Pol II complex to perform TC-NER [40]. Although this subset of genes is independent of Rad26, TC-NER in these genes still depends on TFIIH [59], suggesting that Rad26-independent TC-NER relies on the same set of core NER enzymes for repair. Second, TC-NER in the +1 nucleosome (the first nucleosome downstream of the transcription start site [TSS]) of almost all yeast genes is largely independent of Rad26, particularly for the first 30-nt on the TSS-proximal side (Figure 3B). In contrast, Rad26 is essential for TC-NER downstream of the +1 nucleosome, suggesting variable Rad26 requirements in different chromatin regions. Interestingly, Rad26-independent TC-NER on the TSS-proximal side of the +1 nucleosome is correlated with high TFIIH occupancy in this specific region, revealed by high-resolution ChIP-exonuclease (ChIP-exo) data [59,60]. Biochemical data have shown that TFIIH is associated with Pol II in this region to promote transcription initiation and early elongation [61]. The high TFIIH occupancy suggests that TFIIH is readily available for DNA unwinding and damage verification upon Pol II stalling in the TSS-proximal region, which allows TC-NER in the absence of Rad26 (Figure 3C). In the downstream nucleosomes where Rad26 is needed for TC-NER, the TFIIH occupancy is low but the occupancy of the elongation factor Spt4-Spt5 is high [59] (Figure 3C), suggesting a role for Rad26 in removing Spt4-Spt5, a known yeast TC-NER suppressor [38,39], from the stalled Pol II. Indeed, CPD-seq analysis in a rad26Δspt4Δ double mutant shows significant recovery of TC-NER in the downstream nucleosomes [59], further confirming that eviction of Spt4-Spt5 by Rad26 is an important step that switches Pol II from elongation to repair. Hence, the high-resolution CPD-seq data, coupled with the genome-wide occupancy analyses of transcription factors (i.e., TFIIH and Spt4-Spt5), uncovers the common but variable requirements for Rad26 in different transcribed regions, and reveals the interplay between Rad26 and the transcription machinery to initiate repair when Pol II encounters DNA damage.
NMP-seq:
The CPD-seq methodology was modified to develop a new method, NMP-seq (N-methylpurine sequencing), to map alkylation damage such as 7-methyguanine (7meG) and 3-methyladenine (3meA) (Figure 2B) [28]. The alkylation damage can be induced by exposing cells to alkylating agents such as methyl methanesulfonate (MMS). The damage mapping strategy of NMP-seq is similar to CPD-seq, except a different DNA repair enzyme is used for the initial damage cleavage. In NMP-seq, alkylation damage is cleaved by the glycosylase alkyladenine glycosylase (AAG), followed by APE1, to generate the damage-associated 3’-OH group for sequencing adaptor ligation [28] (Figure 2B). NMP-seq data confirmed that BER is the primary repair pathway for alkylation damage in wild-type yeast cells, consistent with previous studies [6]. However, analysis of the NMP-seq data in a BER-deficient yeast strain, mag1Δ, which lacks the glycosylase Mag1 (ortholog of human AAG), shows preferential repair of 3meA damage on the TS relative to the NTS. Consistent with the repair data, genome-scale sequencing of MMS-induced mutations in the mag1Δ mutant also showed significantly enriched adenine mutations on the NTS [28]. These data suggest that 3meA lesions in the BER mutant (i.e., mag1Δ) may inhibit RNA Pol II elongation and invoke TC-NER, resulting in reduction of 3meA damage and adenine mutations on the TS [28]. As a comparison, the strand asymmetry in repair and mutagenesis is significantly diminished in wild-type cells, conceivably due to rapid removal of the damage by BER. As alkylation damage such as 3meA is non-bulky, the NMP-seq data suggests that TC-NER may also target some DNA base lesions for strand-specific repair. Consistent with this finding, data generated in yeast cells has also shown that AP sites, which are not bulky lesions, are repaired by TC-NER and fewer mutations associated with AP sites are found on the TS relative to the NTS [29].
4. New TC-NER regulators identified by high-throughput screens
Identification of new TC-NER factors and characterization of their cooperative actions with the known repair factors are important for understanding this repair pathway, particularly for organisms lacking canonical TC-NER factors such as CSB. The CRISPR-Cas9 technology, coupled with synthesized genome-scale single-guide RNA library (e.g., [62]), provides a powerful system to introduce mutations in gene coding regions that cause loss of gene functions. This strategy has been used to identify gene candidates whose loss affects cellular response to drug treatment [62]. The same strategy can be utilized to screen for new genes involved in different DNA repair pathways. A recent study conducting whole-genome CRISPR-Cas9 screens in human cells identified a number of new genes that affect cellular sensitivity to DNA damaging agents. Of particular interest to NER research, the screens identified two new TC-NER factors: elongation factor 1 homolog (ELOF1) and serine/threonine kinase 19 (STK19) [63]. The DNA damage sensitivity profile in ELOF1 knockout cells is similar to cells lacking CSA, CSB, or UVSSA: high sensitivity to UV, cisplatin, and illudin S, but resistance to trabectedin (also known as ecteinascidin 743) [63]. DNA damage induced by UV, cisplatin, or illudin S is repaired by TC-NER, while trabectedin, a natural marine product, requires active TC-NER in the cell to exert its cytotoxicity and resistance to trabectedin is correlated with defective TC-NER [64]. Hence, the screening data suggests that ELOF1 is a likely TC-NER factor required for normal TC-NER activity. ELOF1 is a conserved transcription elongation factor [65] and its yeast ortholog, Elf1, interacts with Spt4-Spt5 to increase Pol II transcription through nucleosome barriers [66]. STK19 is a CSB-interacting protein and promotes transcription recovery post UV irradiation [67], suggesting a potential role for STK19 in the response to UV-induced Pol II stalling. Similar to ELOF1, loss of STK19 sensitizes human cells to illudin S but increases resistance to trabectedin [63]. The detailed role for ELOF1 and STK19 in TC-NER is currently unclear, and more studies are needed to understand their functions.
A yeast genetic screen was performed using a set of yeast knockout mutants in the absence of the GG-NER factor Rad7 [68]. These yeast mutants were exposed to UV light to screen for candidates showing significantly increased UV sensitivity compared to the rad7 single mutant. This screen found that mutants of the PAF and Ccr4-Not complexes exhibit high UV sensitivity, suggesting their potential roles in TC-NER. The TC-NER impairment in these mutants was confirmed by a Southern blot-based DNA repair assay [68]. Notably, PAF and Ccr4-Not complexes are both transcription elongation factors, suggesting TC-NER in yeast (and potentially in other eukaryotes) is broadly regulated by factors participating in transcription elongation. The role of PAF in yeast TC-NER was further examined using a high-resolution gel-based repair assay [41]. However, analyses of CPD repair at the actively transcribed RPB2 gene revealed that PAF only marginally promotes Rad26-dependent TC-NER but significantly suppresses Rad26-independent TC-NER [41]. Additionally, PAF appears to cooperate with the elongation factor Spt4-Spt5 to exert its suppressive role in repair.
Proteomic screens have also been utilized to identify new TC-NER factors. As Pol II and CSB are the central factors in TC-NER, a previous study using purified CSB and Pol II identified proteins interacting with each of them by mass spectrometry [67]. The interactomes were then compared between cells with and without UV treatment, and proteins that specifically interact with CSB or Pol II upon UV irradiation were recognized as candidates for TC-NER or other Pol II rescue pathways. The proteomic screens identified STK19 as a CSB-interacting protein [67], which was later confirmed as a TC-NER factor in the CRISPR-Cas9 screen [63]. Additional factors that bind to CSB or Pol II upon DNA damage were also identified; however, whether and how they play a role in TC-NER have not been characterized.
5. TC-NER and cancer: mutational strand asymmetry in the cancer genome
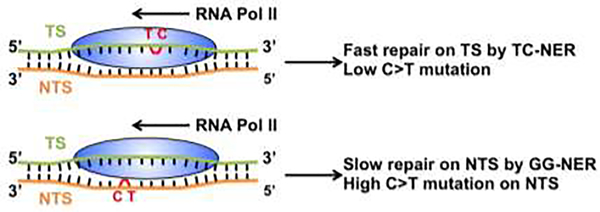
Due to the strand-specific repair by TC-NER, DNA damage on the TS of active genes is repaired more rapidly than damage on the NTS. As a result, the unrepaired damage on the NTS may cause mutations if they are replicated by DNA polymerases, and thus lead to more mutations on the NTS relative to the TS (Figure 4). Indeed, consistent with the known function of TC-NER in the repair of UV and BPDE adducts, UV-induced C>T mutations in skin cancer and smoking-induced G>T mutations in lung cancer, respectively, are significantly enriched on the NTS [69].
Figure 4.
Role of TC-NER in causing mutational strand asymmetry. Upper panel: Damage (e.g., UV-induced TC dimer) located on the transcribed strand (TS) stalls Pol II elongation and is repaired by TC-NER. The fast repair leads to low C>T mutation frequency on the TS. Lower panel: A TC dimer located on the non-transcribed strand (NTS) cannot be repaired by TC-NER and is left for repair by GG-NER. The slow repair by GG-NER causes high C>T mutation frequency on the NTS when the damage is replicated by a DNA polymerase.
The mutational strand asymmetry can also be used to investigate cancer mutations with no clear knowledge of causative DNA damage. For example, skin cancer such as cutaneous melanoma contains non-canonical mutations such as T>C and T>A that do not fit the UV signature mutation (i.e., C>T) [70], but it is not clear what causes these non-canonical mutations. A recent study sequenced the genome of UV-treated yeast cells and identified many UV signature C>T mutations as well as a large number of T>C and T>A transitions [71]. Interestingly, T>C mutations are enriched in TTN as well as CTN trinucleotide contexts, and thus mainly occur at the 3’ position of a dipyrimidine (i.e., TT or CT with mutations on the underlined T), suggesting they likely originate from known UV photolesions such as CPDs or 6–4PPs. T>A mutations are mainly found in NTA trinucleotides and are not specifically associated with dipyrimidines. Further analysis in yeast genes indicates that T>C mutations are elevated on the NTS (non-transcribed strand) relative to the TS (transcribed strand), similar to the UV signature C>T mutations [69], confirming T>C mutations are likely caused by CPDs or 6–4PPs. In contrast, T>A mutations in the NTA context are elevated on the TS relative to the NTS. Analysis in melanomas also shows elevated T>A mutations in the NTA context on the TS relative to NTS [71]. Thus, the strand information (i.e., TS vs. NTS) suggests that T>A mutations (in the NTA context) may originate from a DNA lesion on the opposite strand, at a corresponding TAN sequence context. The lesion is likely formed between TA and that the central adenine is mutated to thymine (i.e., A>T mutation) during replication [71]. Indeed, studies using UV-irradiated oligonucleotides containing tandem TA repeats and genome-wide mapping of atypical UV damage in yeast cells collectively indicate formation of TA lesions by UV irradiation that can be cleaved by UV DNA endonuclease (UVDE) [71]. The TA lesion likely inhibits Pol II elongation and invokes TC-NER for strand-specific repair on the TS, and thus leads to reduced A>T mutations on the TS. Therefore, the strand information of non-canonical UV mutations is useful to determine the underlying DNA lesions.
Similarly, the strand information of A>G mutations in liver cancer also suggests that TC-NER may play a role. The A>G mutations are found in some liver patients but the causative DNA damage is unclear. By analyzing the distribution of A>G mutations between the TS and NTS, it has been shown that the mutation frequency is significantly higher on the NTS than on the TS in actively transcribed genes [69]. The strand asymmetry suggests that A>G mutations in liver cancer are likely caused by an unknown bulky lesion formed on adenines. Interestingly, A>G mutations in liver cancer are also significantly increased on the NTS in the transcribed region relative to the flanking intergenic regions such as promoters, suggesting a potential effect of ‘transcription-coupled damage’ that boosts damage formation on the NTS [69]. Hence, these analyses suggest that both transcription-coupled repair and damage formation contribute to significantly elevated A>G mutations on the NTS.
6. Discussion and future directions
In this Review, we have discussed new findings in TC-NER revealed by genomic approaches. First, genome-wide damage mapping methods have provided an unprecedented opportunity to study TC-NER at the whole genome scale and at high resolution. These data not only confirmed the critical role for CSB and its counterparts (e.g., Rad26 and Mfd) in coupling transcription and repair, but they further uncovered new insights into how these TC-NER factors interact with the transcription machinery to initiate repair upon DNA damage-induced transcription stalling. For example, analysis of CPD-seq data obtained in yeast mutants indicates that Rad26 is specifically required in TSS-distal transcribed regions (e.g., downstream of the +1 nucleosome) in order to displace Spt4-Spt5, but not in the TSS-proximal region due to high TFIIH occupancy [59]. An open question associated with this finding is whether this is a yeast-specific phenomenon or a common mechanism shared by other organisms. Further studies in mammalian cells will likely uncover if CSB plays a similar role. Mapping of the mutagenic 3meA lesions using the NMP-seq method revealed a surprising role for TC-NER to repair non-bulky base damage when BER is deficient. This finding suggests that TC-NER may repair both bulky and non-bulky lesions, and plays a broader role than previously expected. Although in vitro data have shown weak or lack of inhibition to RNA polymerases for some non-bulky damage, these studies were performed with naked DNA templates without the chromatin structure. It is possible that the transcription-impeding effect will be amplified when non-bulky damage is embedded in chromatin, which by itself imposes restrictions to Pol II elongation [72]. Second, genome-scale screens have allowed identification of new TC-NER factors in human cells and this strategy can be potentially utilized in organisms lacking CSB orthologs. Investigation of how the newly identified factors such as ELOF1 and STK19 [63] cooperate with the known TC-NER factors will further improve our understanding of this repair pathway. Finally, we discussed how the TC-NER activity is associated with mutational strand asymmetry in human cancers. While a large number of mutational signatures have been identified mainly by analyzing the trinucleotide contexts [73], the cause for many of them is still unclear. Future studies incorporating mutational strand information into the sequence contexts will offer new insights into the causative DNA damage, which is important for cancer etiology and prevention.
Highlights.
Genome-wide DNA damage mapping methods reveal new insights into TC-NER
Genome-scale CRISPR-Cas9 screens identify new TC-NER factors
Mutational strand asymmetry provides important information to the causative DNA damage in human cancers
Acknowledgement
We thank Dr. John Wyrick for reading the manuscript. DNA repair studies related to this review are supported by NIH grants (R21ES029302 to P.M. and R01ES030993 to K.J.L.), a pilot grant from UNM Center for Metals in Biology and Medicine (P20GM130422), UNM Comprehensive Cancer Center Support Grant NCI P30CA118100, and UNM Analytical and Translational Genomics Shared Resource.
Abbreviations:
- NER
nucleotide excision repair
- TC-NER
transcription-coupled NER
- GG-NER
global genomic NER
- BER
base excision repair
- Pol II
RNA polymerase II
- CS
Cockayne syndrome
- CSB
Cockayne syndrome group B protein
- XPC
Xeroderma pigmentosum complementation group C protein
- NGS
next-generation sequencing
- CPD
cyclobutane pyrimidine dimer
- 6–4PPs
(6–4) photoproducts
- BaP
benzo[a]pyrene
- BPDE
(+)benzo[a]pyrene-7,8-dihydrodiol-9,10-epoxide
- BPDE-dGs
BPDE-deoxyguanosines
- CPD-seq
cyclobutane pyrimidine dimer sequencing
- XR-seq
excision repair sequencing
- NMP-seq
N-methylpurine sequencing
- AP sites
apurinic/apyrimidinic sites
- TS
transcribed strand
- NTS
non-transcribed strand
- TSS
transcription start site
- UVSSA
UV Stimulated Scaffold Protein A
- TFIIH
transcription factor IIH
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Tiwari V, Wilson DM, DNA Damage and Associated DNA Repair Defects in Disease and Premature Aging, The American Journal of Human Genetics. 105 (2019) 237–257. 10.1016/j.ajhg.2019.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ciccia A, Elledge SJ, The DNA damage response: making it safe to play with knives, Mol Cell. 40 (2010) 179–204. 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tornaletti S, Maeda LS, Kolodner RD, Hanawalt PC, Effect of 8-oxoguanine on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II, DNA Repair (Amst). 3 (2004) 483–494. 10.1016/j.dnarep.2004.01.003. [DOI] [PubMed] [Google Scholar]
- [4].Cadet J, Davies KJA, Medeiros MH, Di Mascio P, Wagner JR, Formation and repair of oxidatively generated damage in cellular DNA, Free Radic Biol Med. 107 (2017) 13–34. 10.1016/j.freeradbiomed.2016.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Oh J, Fleming AM, Xu J, Chong J, Burrows CJ, Wang D, RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair–π and CH–π interactions, PNAS. 117 (2020) 9338–9348. 10.1073/pnas.1919904117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Krokan HE, Bjørås M, Base Excision Repair, Cold Spring Harb Perspect Biol. 5 (2013). 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wang W, Walmacq C, Chong J, Kashlev M, Wang D, Structural basis of transcriptional stalling and bypass of abasic DNA lesion by RNA polymerase II, Proc Natl Acad Sci U S A. 115 (2018) E2538–E2545. 10.1073/pnas.1722050115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mao P, Wyrick JJ, Roberts SA, Smerdon MJ, UV-Induced DNA Damage and Mutagenesis in Chromatin, Photochemistry and Photobiology. 93 (2017) 216–228. 10.1111/php.12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cadet J, Douki T, Formation of UV-induced DNA damage contributing to skin cancer development, Photochem. Photobiol. Sci. 17 (2018) 1816–1841. 10.1039/C7PP00395A. [DOI] [PubMed] [Google Scholar]
- [10].Sinha RP, Häder DP, UV-induced DNA damage and repair: a review, Photochem Photobiol Sci. 1 (2002) 225–236. 10.1039/b201230h. [DOI] [PubMed] [Google Scholar]
- [11].Selby CP, Drapkin R, Reinberg D, Sancar A, RNA polymerase II stalled at a thymine dimer: footprint and effect on excision repair., Nucleic Acids Res. 25 (1997) 787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gregersen LH, Svejstrup JQ, The Cellular Response to Transcription-Blocking DNA Damage, Trends in Biochemical Sciences. 43 (2018) 327–341. 10.1016/j.tibs.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kozack R, Seo KY, Jelinsky SA, Loechler EL, Toward an understanding of the role of DNA adduct conformation in defining mutagenic mechanism based on studies of the major adduct (formed at N(2)-dG) of the potent environmental carcinogen, benzo[a]pyrene, Mutat Res. 450 (2000) 41–59. 10.1016/s0027-5107(00)00015-4. [DOI] [PubMed] [Google Scholar]
- [14].Perlow RA, Kolbanovskii A, Hingerty BE, Geacintov NE, Broyde S, Scicchitano DA, DNA adducts from a tumorigenic metabolite of benzo[a]pyrene block human RNA polymerase II elongation in a sequence- and stereochemistry-dependent manner, J Mol Biol. 321 (2002) 29–47. 10.1016/s0022-2836(02)00593-4. [DOI] [PubMed] [Google Scholar]
- [15].Schärer OD, Nucleotide Excision Repair in Eukaryotes, Cold Spring Harb Perspect Biol. 5 (2013). 10.1101/cshperspect.a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hanawalt PC, Spivak G, Transcription-coupled DNA repair: two decades of progress and surprises, Nature Reviews Molecular Cell Biology. 9 (2008) 958–970. 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- [17].Okuda M, Nakazawa Y, Guo C, Ogi T, Nishimura Y, Common TFIIH recruitment mechanism in global genome and transcription-coupled repair subpathways, Nucleic Acids Res. 45 (2017) 13043–13055. 10.1093/nar/gkx970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li C-L, Golebiowski FM, Onishi Y, Samara NL, Sugasawa K, Yang W, Tripartite DNA Lesion Recognition and Verification by XPC, TFIIH, and XPA in Nucleotide Excision Repair, Molecular Cell. 59 (2015) 1025–1034. 10.1016/j.molcel.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mellon I, Spivak G, Hanawalt PC, Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene, Cell. 51 (1987) 241–249. 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- [20].Bohr VA, Smith CA, Okumoto DS, Hanawalt PC, DNA repair in an active gene: Removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall, Cell. 40 (1985) 359–369. 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- [21].Mellon I, Hanawalt PC, Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand, Nature. 342 (1989) 95–98. 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- [22].Smerdon MJ, Thoma F, Site-specific DNA repair at the nucleosome level in a yeast minichromosome, Cell. 61 (1990) 675–684. 10.1016/0092-8674(90)90479-x. [DOI] [PubMed] [Google Scholar]
- [23].Oztas O, Selby CP, Sancar A, Adebali O, Genome-wide excision repair in Arabidopsis is coupled to transcription and reflects circadian gene expression patterns, Nat Commun. 9 (2018) 1503. 10.1038/s41467-018-03922-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Deger N, Yang Y, Lindsey-Boltz LA, Sancar A, Selby CP, Drosophila, which lacks canonical transcription-coupled repair proteins, performs transcription-coupled repair, J. Biol. Chem. 294 (2019) 18092–18098. 10.1074/jbc.AC119.011448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Li W, Hu J, Adebali O, Adar S, Yang Y, Chiou Y-Y, Sancar A, Human genome-wide repair map of DNA damage caused by the cigarette smoke carcinogen benzo[a]pyrene, PNAS. (2017) 201706021. 10.1073/pnas.1706021114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hu J, Lieb JD, Sancar A, Adar S, Cisplatin DNA damage and repair maps of the human genome at single-nucleotide resolution, Proc. Natl. Acad. Sci. U.S.A. 113 (2016) 11507–11512. 10.1073/pnas.1614430113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Slyskova J, Sabatella M, Ribeiro-Silva C, Stok C, Theil AF, Vermeulen W, Lans H, Base and nucleotide excision repair facilitate resolution of platinum drugs-induced transcription blockage, Nucleic Acids Res. 46 (2018) 9537–9549. 10.1093/nar/gky764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mao P, Brown AJ, Malc EP, Mieczkowski PA, Smerdon MJ, Roberts SA, Wyrick JJ, Genome-wide maps of alkylation damage, repair, and mutagenesis in yeast reveal mechanisms of mutational heterogeneity, Genome Res. (2017). 10.1101/gr.225771.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kim N, Jinks-Robertson S, Abasic Sites in the Transcribed Strand of Yeast DNA Are Removed by Transcription-Coupled Nucleotide Excision Repair, Mol Cell Biol. 30 (2010) 3206–3215. 10.1128/MCB.00308-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Vermeulen W, Fousteri M, Mammalian Transcription-Coupled Excision Repair, Cold Spring Harbor Perspectives in Biology. 5 (2013) a012625–a012625. 10.1101/cshperspect.a012625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].van der Weegen Y, Golan-Berman H, Mevissen TET, Apelt K, González-Prieto R, Goedhart J, Heilbrun EE, Vertegaal ACO, van den Heuvel D, Walter JC, Adar S, Luijsterburg MS, The cooperative action of CSB, CSA, and UVSSA target TFIIH to DNA damage-stalled RNA polymerase II, Nature Communications. 11 (2020) 2104. 10.1038/s41467-020-15903-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Noe Gonzalez M, Blears D, Svejstrup JQ, Causes and consequences of RNA polymerase II stalling during transcript elongation, Nature Reviews Molecular Cell Biology. 22 (2021) 3–21. 10.1038/s41580-020-00308-8. [DOI] [PubMed] [Google Scholar]
- [33].Xu J, Lahiri I, Wang W, Wier A, Cianfrocco MA, Chong J, Hare AA, Dervan PB, DiMaio F, Leschziner AE, Wang D, Structural basis for the initiation of eukaryotic transcription-coupled DNA repair, Nature. 551 (2017) 653–657. 10.1038/nature24658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Selby CP, Sancar A, Cockayne syndrome group B protein enhances elongation by RNA polymerase II, Proc. Natl. Acad. Sci. U.S.A. 94 (1997) 11205–11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lee S-K, Yu S-L, Prakash L, Prakash S, Requirement for Yeast RAD26, a Homolog of the HumanCSB Gene, in Elongation by RNA Polymerase II, Mol. Cell. Biol. 21 (2001) 8651–8656. 10.1128/MCB.21.24.8651-8656.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Xu J, Wang W, Xu L, Chen J-Y, Chong J, Oh J, Leschziner AE, Fu X-D, Wang D, Cockayne syndrome B protein acts as an ATP-dependent processivity factor that helps RNA polymerase II overcome nucleosome barriers, PNAS. 117 (2020) 25486–25493. 10.1073/pnas.2013379117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wang W, Xu J, Chong J, Wang D, Structural Basis of DNA Lesion Recognition for Eukaryotic Transcription-Coupled Nucleotide Excision Repair, DNA Repair (Amst) 71 (2018) 43–55. 10.1016/j.dnarep.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jansen LET, Spt4 modulates Rad26 requirement in transcription-coupled nucleotide excision repair, The EMBO Journal. 19 (2000) 6498–6507. 10.1093/emboj/19.23.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ding B, LeJeune D, Li S, The C-terminal Repeat Domain of Spt5 Plays an Important Role in Suppression of Rad26-independent Transcription Coupled Repair, Journal of Biological Chemistry. 285 (2010) 5317–5326. 10.1074/jbc.M109.082818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Li S, Smerdon MJ, Rpb4 and Rpb9 mediate subpathways of transcription-coupled DNA repair in Saccharomyces cerevisiae, EMBO J. 21 (2002) 5921–5929. 10.1093/emboj/cdf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tatum D, Li W, Placer M, Li S, Diverse roles of RNA polymerase II-associated factor 1 complex in different subpathways of nucleotide excision repair, J Biol Chem. 286 (2011) 30304–30313. 10.1074/jbc.M111.252981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Li W, Selvam K, Rahman SA, Li S, Sen1, the yeast homolog of human senataxin, plays a more direct role than Rad26 in transcription coupled DNA repair, Nucleic Acids Research. 44 (2016) 6794–6802. 10.1093/nar/gkw428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Li W, Li S, Facilitators and Repressors of Transcription-coupled DNA Repair in Saccharomyces cerevisiae, Photochemistry and Photobiology. 93 (2017) 259–267. 10.1111/php.12655. [DOI] [PubMed] [Google Scholar]
- [44].Tu Y, Bates S, Pfeifer GP, Sequence-specific and Domain-specific DNA Repair in Xeroderma Pigmentosum and Cockayne Syndrome Cells, J. Biol. Chem. 272 (1997) 20747–20755. 10.1074/jbc.272.33.20747. [DOI] [PubMed] [Google Scholar]
- [45].Tijsterman M, Verhage RA, van de Putte P, Tasseron-de Jong JG, Brouwer J, Transitions in the coupling of transcription and nucleotide excision repair within RNA polymerase II-transcribed genes of Saccharomyces cerevisiae, Proc. Natl. Acad. Sci. U.S.A. 94 (1997) 8027–8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Li S, Transcription coupled nucleotide excision repair in the yeast Saccharomyces cerevisiae: The ambiguous role of Rad26, DNA Repair. 36 (2015) 43–48. 10.1016/j.dnarep.2015.09.006. [DOI] [PubMed] [Google Scholar]
- [47].Li W, Sancar A, Methodologies for detecting environmentally induced DNA damage and repair, Environmental and Molecular Mutagenesis. 61 (2020) 664–679. 10.1002/em.22365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wyrick JJ, Roberts SA, Genomic approaches to DNA repair and mutagenesis, DNA Repair. 36 (2015) 146–155. 10.1016/j.dnarep.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hu J, Adar S, Selby CP, Lieb JD, Sancar A, Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution, Genes Dev. 29 (2015) 948–960. 10.1101/gad.261271.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hu J, Li W, Adebali O, Yang Y, Oztas O, Selby CP, Sancar A, Genome-wide mapping of nucleotide excision repair with XR-seq, Nature Protocols. 14 (2019) 248–282. 10.1038/s41596-018-0093-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Adebali O, Chiou Y-Y, Hu J, Sancar A, Selby CP, Genome-wide transcription-coupled repair in Escherichia coli is mediated by the Mfd translocase, PNAS. 114 (2017) E2116–E2125. 10.1073/pnas.1700230114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Mao P, Smerdon MJ, Roberts SA, Wyrick JJ, Chromosomal landscape of UV damage formation and repair at single-nucleotide resolution, PNAS. 113 (2016) 9057–9062. 10.1073/pnas.1606667113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Mao P, Brown AJ, Esaki S, Lockwood S, Poon GMK, Smerdon MJ, Roberts SA, Wyrick JJ, ETS transcription factors induce a unique UV damage signature that drives recurrent mutagenesis in melanoma, Nature Communications. 9 (2018) 2626. 10.1038/s41467-018-05064-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Elliott K, Boström M, Filges S, Lindberg M, den Eynden JV, Ståhlberg A, Clausen AR, Larsson E, Elevated pyrimidine dimer formation at distinct genomic bases underlies promoter mutation hotspots in UV-exposed cancers, PLOS Genetics. 14 (2018) e1007849. 10.1371/journal.pgen.1007849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Mao P, Wyrick JJ, Genome-Wide Mapping of UV-Induced DNA Damage with CPD-Seq, in: Hancock R (Ed.), The Nucleus, Springer US, New York, NY, 2020: pp. 79–94. 10.1007/978-1-0716-0763-3_7. [DOI] [PubMed] [Google Scholar]
- [56].Jiang C, Pugh BF, Nucleosome positioning and gene regulation: advances through genomics, Nat Rev Genet. 10 (2009) 161–172. 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hara R, Mo J, Sancar A, DNA Damage in the Nucleosome Core Is Refractory to Repair by Human Excision Nuclease, Mol Cell Biol. 20 (2000) 9173–9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Mao P, Smerdon MJ, Roberts SA, Wyrick JJ, Asymmetric repair of UV damage in nucleosomes imposes a DNA strand polarity on somatic mutations in skin cancer, Genome Res. 30 (2020) 12–21. 10.1101/gr.253146.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Duan M, Selvam K, Wyrick JJ, Mao P, Genome-wide role of Rad26 in promoting transcription-coupled nucleotide excision repair in yeast chromatin, Proc Natl Acad Sci USA. (2020) 202003868. 10.1073/pnas.2003868117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Vinayachandran V, Reja R, Rossi MJ, Park B, Rieber L, Mittal C, Mahony S, Pugh BF, Widespread and precise reprogramming of yeast protein–genome interactions in response to heat shock, Genome Res. 28 (2018) 357–366. 10.1101/gr.226761.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zawel L, Kumar KP, Reinberg D, Recycling of the general transcription factors during RNA polymerase II transcription, Genes Dev. 9 (1995) 1479–1490. [DOI] [PubMed] [Google Scholar]
- [62].Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F, Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells, Science. 343 (2014) 84–87. 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Olivieri M, Cho T, Álvarez-Quilón A, Li K, Schellenberg MJ, Zimmermann M, Hustedt N, Rossi SE, Adam S, Melo H, Heijink AM, Sastre-Moreno G, Moatti N, Szilard RK, McEwan A, Ling AK, Serrano-Benitez A, Ubhi T, Feng S, Pawling J, Delgado-Sainz I, Ferguson MW, Dennis JW, Brown GW, Cortés-Ledesma F, Williams RS, Martin A, Xu D, Durocher D, A Genetic Map of the Response to DNA Damage in Human Cells, Cell. 0 (2020). 10.1016/j.cell.2020.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Takebayashi Y, Pourquier P, Zimonjic DB, Nakayama K, Emmert S, Ueda T, Urasaki Y, Kanzaki A, Akiyama SI, Popescu N, Kraemer KH, Pommier Y, Antiproliferative activity of ecteinascidin 743 is dependent upon transcription-coupled nucleotide-excision repair, Nat Med. 7 (2001) 961–966. 10.1038/91008. [DOI] [PubMed] [Google Scholar]
- [65].Prather D, Krogan NJ, Emili A, Greenblatt JF, Winston F, Identification and Characterization of Elf1, a Conserved Transcription Elongation Factor in Saccharomyces cerevisiae, Mol Cell Biol. 25 (2005) 10122–10135. 10.1128/MCB.25.22.10122-10135.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Ehara H, Kujirai T, Fujino Y, Shirouzu M, Kurumizaka H, Sekine S, Structural insight into nucleosome transcription by RNA polymerase II with elongation factors, Science. 363 (2019) 744–747. 10.1126/science.aav8912. [DOI] [PubMed] [Google Scholar]
- [67].Boeing S, Williamson L, Encheva V, Gori I, Saunders RE, Instrell R, Aygün O, Rodriguez-Martinez M, Weems JC, Kelly GP, Conaway JW, Conaway RC, Stewart A, Howell M, Snijders AP, Svejstrup JQ, Multiomic Analysis of the UV-Induced DNA Damage Response, Cell Rep. 15 (2016) 1597–1610. 10.1016/j.celrep.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Gaillard H, Tous C, Botet J, González-Aguilera C, Quintero MJ, Viladevall L, García-Rubio ML, Rodríguez-Gil A, Marín A, Ariño J, Revuelta JL, Chávez S, Aguilera A, Genome-Wide Analysis of Factors Affecting Transcription Elongation and DNA Repair: A New Role for PAF and Ccr4-Not in Transcription-Coupled Repair, PLOS Genetics. 5 (2009) e1000364. 10.1371/journal.pgen.1000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Haradhvala NJ, Polak P, Stojanov P, Covington KR, Shinbrot E, Hess J, Rheinbay E, Kim J, Maruvka Y, Braunstein LZ, Kamburov A, Hanawalt PC, Wheeler DA, Koren A, Lawrence MS, Getz G, Mutational strand asymmetries in cancer genomes reveal mechanisms of DNA damage and repair, Cell. 164 (2016) 538–549. 10.1016/j.cell.2015.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, Patch A-M, Kakavand H, Alexandrov LB, Burke H, Jakrot V, Kazakoff S, Holmes O, Leonard C, Sabarinathan R, Mularoni L, Wood S, Xu Q, Waddell N, Tembe V, Pupo GM, De Paoli-Iseppi R, Vilain RE, Shang P, Lau LMS, Dagg RA, Schramm S-J, Pritchard A, Dutton-Regester K, Newell F, Fitzgerald A, Shang CA, Grimmond SM, Pickett HA, Yang JY, Stretch JR, Behren A, Kefford RF, Hersey P, Long GV, Cebon J, Shackleton M, Spillane AJ, Saw RPM, López-Bigas N, Pearson JV, Thompson JF, Scolyer RA, Mann GJ, Whole-genome landscapes of major melanoma subtypes, Nature. 545 (2017) 175–180. 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
- [71].Laughery MF, Brown AJ, Bohm KA, Sivapragasam S, Morris HS, Tchmola M, Washington AD, Mitchell D, Mather S, Malc EP, Mieczkowski PA, Roberts SA, Wyrick JJ, Atypical UV Photoproducts Induce Non-canonical Mutation Classes Associated with Driver Mutations in Melanoma, Cell Reports. 33 (2020) 108401. 10.1016/j.celrep.2020.108401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Li B, Carey M, Workman JL, The Role of Chromatin during Transcription, Cell. 128 (2007) 707–719. 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- [73].Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Ng AWT, Wu Y, Boot A, Covington KR, Gordenin DA, Bergstrom EN, Islam SMA, Lopez-Bigas N, Klimczak LJ, McPherson JR, Morganella S, Sabarinathan R, Wheeler DA, Mustonen V, Getz G, Rozen SG, Stratton MR, The repertoire of mutational signatures in human cancer, Nature. 578 (2020) 94–101. 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Park D, Morris AR, Battenhouse A, Iyer VR, Simultaneous mapping of transcript ends at single-nucleotide resolution and identification of widespread promoter-associated noncoding RNA governed by TATA elements, Nucleic Acids Res. 42 (2014) 3736–3749. 10.1093/nar/gkt1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Weiner A, Hsieh T-HS, Appleboim A, Chen HV, Rahat A, Amit I, Rando OJ, Friedman N, High-Resolution Chromatin Dynamics during a Yeast Stress Response, Molecular Cell. 58 (2015) 371–386. 10.1016/j.molcel.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]