Abstract
Background:
Hepatic stellate cell (HSC) differentiation/activation is central to liver fibrosis and is innately linked to the immune response to liver injury. Exosomes (EXOs) are important means of communication between cell populations. This study sought to characterize EXO release from HSCs and the effect of HSC-EXOs on macrophage cytokine release/function.
Methods.
Liver from a rat fibrosis model was analyzed for EXO expression and localization. Quiescent and culture-activated rat and mouse HSCs and activated human HSCs were analyzed for microRNA expression. Mouse, rat, and human HSCs were culture-activated and EXOs purified from culture medium prior to addition to macrophages, and interleukin-6 (IL-6) and tumor necrosis factor-α (TNFα) mRNA and protein measured. The effect of activated HSC-EXOs on macrophage migration was assayed.
Results.
Activation of rat HSCs led to increased EXO production in vivo, an effect mirrored by in vitro rat HSC culture-activation. Culture activation of mouse and rat HSCs led to altered EXO microRNA profiles, with a similar microRNA profile detected in activated human HSCs. Addition of activated HSC-EXOs to macrophages stimulated IL-6 and TNFα mRNA expression and protein secretion in mouse and human macrophages, but not for rat HSC-EXO-macrophages. Addition of human EXOs to macrophages stimulated migration, effects mirrored by the direct addition of rhIL-6 and rhTNFα.
Conclusions.
HSC-EXOs associate with macrophages and stimulate cytokine synthesis-release and macrophage migration. Constructing a comprehensive understanding of EXOs interactions between liver cell populations in the setting of inflammation/fibrosis increases the potential for developing new diagnostic/therapeutic approaches.
Keywords: Hepatic stellate cell, Exosome, Macrophage, Interleukin-6, Tumor necrosis factor-α
LAY SUMMARY.
Exosomes (EXOs) are small vesicles released from cells that enter neighboring cells to deliver proteins, lipids, mRNA, and microRNA cargos. Hepatic fibrosis is a worsening liver pathology characterized by inflammation, hepatic stellate cell activation (HSC), and increased tissue scarring. In this study we report EXOs derived from activated-HSCs stimulates pro-fibrotic cytokine release in macrophages and increase macrophage migration, events associated with liver fibrosis progression.
INTRODUCTION
Liver cirrhosis most commonly arises due to sustained hepatic injury and is most commonly caused by chemical (alcohol) [1], viral (hepatitis B and/or C virus [HBV/HCV]) infection) [2], and/or metabolic (obesity) [3] events. Within these etiologies coordinated local and systemic immune responses are invoked to initiate tissue repair [4]. During disease progression repeat hepatocyte damage leads to necrotic and apoptotic cell death and the release of soluble mediators, apoptotic bodies, microvesicles (MVs), and exosomes (EXOs), factors that regulate hepatic stellate cell (HSC) and hepatic macrophage (Kupffer cell [KC]) activation [5, 6].
Extracellular vesicles (ECVs) are classified by their secretory pathway and size as being either membrane-derived MVs (100–1000nm) or EXOs (30–100nm; endosomal multi-vesicular bodies released on fusion with the cell membrane) [7, 8]. Increasing evidence suggests EXOs can be produced by a range of hepatic cell populations and contain diverse cargos that may include lipids, proteins, mRNAs, and microRNAs (miRs) [9–11]. Following release, EXOs are transferred to other cells via receptor interaction, membrane fusion, and/or endocytosis/phagocytosis as a means of intercellular communication [12]. While some cells spontaneously release EXOs, EXO secretion can also be induced by changes in the extracellular and/or intracellular environment. For example, changes in the rate of MV release and cargo profile can occur due to changes in inflammatory status across a range of cell types [13–15], as well as in renal [16] and metabolic disease states [17]. Similarly, increases in intracellular calcium [18] and cytoskeletal remodeling [19] induce MV release, events also reported to be central during HSC activation [6].
The primary function of EXOs appears to be via direct interaction with nearby cells to alter target cell function [12]. For example, many mRNAs characterized within EXOs consist of 3’UTR sequences capable of binding miRs to inhibit their activity [20]. Equally, the transfer of mature miRs can target mRNA expression to inhibit protein synthesis to modify target cell function [21]. This has important implications during hepatic injury, where changes in EXO release and/or changes content can function to alter hepatic cell repopulation and differentiation, the hepatic immune response, and neoangiogenesis [11].
Within the fibrotic nodes that form in the cirrhotic liver, EXOs represent an emerging means of intercellular signaling during the coordinated immune responses that occur during liver repair. Previous studies report significant changes in miR profiles between freshly isolated (quiescent) and cultured activated HSCs [22]. Similarly, HSC-derived EXOs (HSC-EXOs) contain unique “miR signatures” that vary depending on the underlying liver pathologies in which they are released [23, 24]. While previous studies have addressed the role of hepatocyte and macrophage-derived EXOs in modifying HSC function [25, 26], relatively few studies have addressed how HSC-derived EXOs may alter macrophage function. The aims of this study were to examine potential mechanisms whereby HSC-EXOs may coordinate macrophage responses.
MATERIALS AND METHODS
Assurances.
Animal studies were approved by our Institutional Animal Care and Use Committee and conform to the NIH Guide for the Care and Use of Laboratory Animals.
Rat bile duct ligation (BDL) model.
A 4-week rat common bile duct ligation (BDL) and sham-operated [control] model (4 weeks) was employed in male Sprague-Dawley rats (250–300g, Charles River Laboratories, Wilmington, MA) [27].
Immunohistochemistry (IHC).
Liver sections (6μm) were stained using an anti-CD63 antibody (AbCam, Cambridge, MA) and detected using a fluorescein isothiocyanate (FITC) labeled anti-mouse secondary antibody. Sections were countered-stained with 4′,6-diamidino-2-phenylindole (DAPI) for nuclear localization [27].
Primary rat and mouse hepatic stellate cell (HSC) isolation.
Male Sprague-Dawley rats (>500g; Charles River Laboratories, N=10) or male C57BL/6 mice (>30g; Charles River Laboratories, N=30) were used. Isolation of HSCs was performed by in situ pronase-collagenase perfusion followed by differential centrifugation. Cell purity/viability was confirmed by autofluorescence and trypan blue exclusion [28].
Cell culture.
Rodent hepatic stellate cells (HSCs).
Freshly isolated rat or mouse HSCs were resuspended in Dulbecco’s modified Eagle medium (DMEM; ThermoFisher Scientific, Waltham, MA) supplemented with 10% (v/v) exosome-free fetal bovine serum (EXO-free FBS; System Biosciences, Palo Alto, CA), and 100U/mL penicillin/100μg/mL streptomycin (P/S) in culture flasks [28]. For day-1 HSCs, 4-hours after initial cell seeding/attachment, culture medium was replaced with DMEM supplemented with 1% (v/v) EXO-free FBS and P/S. For day-14 (culture-activated) HSCs, 4-hours after initial cell seeding/attachment, culture medium was replaced with DMEM supplemented with 10% (v/v) EXO-free FBS and P/S, with culture medium replaced every 48-hours thereafter. Twenty-four hours prior to EXO collection, culture medium was replaced with DMEM supplemented with 1% (v/v) EXO-free FBS and P/S.
Human hepatic stellate cells (HSCs).
Human TWNT4 and LX2 HSCs (American Type Culture Collection; ATCC, Manassas, VA) were cultured in DMEM supplemented with 10% (v/v) EXO-free FBS and P/S. Four hours after seeding/attachment culture medium was replaced with 10% (v/v) EXO-free FBS and P/S, and culture medium replaced every 48-hours thereafter [9]. Twenty-four hours prior to EXO collection, culture medium was replaced with DMEM supplemented with 1% (v/v) EXO-free FBS and P/S medium.
Macrophages.
The RAW 264.7 mouse macrophage cell line (ATCC) was grown in DMEM high-glucose medium containing 10% (v/v) EXO-free FBS, P/S, glutamine (2mM/L) and sodium pyruvate (1mM/mL)). The NR8383 rat macrophage cell line was grown in Ham’s F12K medium (Thermo Fisher Scientific) containing 15% (v/v) EXO-free FBS, P/S, glutamine (2mM/mL) and sodium bicarbonate (1.5g/L). The human promyelotic leukemic HL-60 cell line (ATCC) was grown in RPMI-1640 medium (Thermo Fisher Scientific) containing 20% (v/v) EXO-free FBS, P/S, glutamine (2mM/L), and sodium pyruvate (1mM/mL). To induce HL-60 cells to a macrophage phenotype cells were treated with 12–0-tetradecanoylphorbol 13-acetate (160nM/mL, 48-hours).
Exosome isolation from hepatic stellate cells (HSCs).
For mouse and rat HSC day-1 EXO isolation culture medium was collected 24Hrs following medium replacement with 1% (v/v) EXO-free FBS medium. For day-14 HSCs culture medium was replaced with 1% (v/v) EXO-free FBS medium on day-13, and culture medium collected 24-hours later. For human HSCs, cells were cultured for 10-days (10% (v/v) EXO-free FBS) and, 24-hours prior to EXO collection, culture medium was replaced with 1% (v/v) EXO-free FBS medium. Exosomes were isolated and purified using an EXOQuick-TC kit (System Bioscience) and re-suspended in 100µL PBS. Exosome suspensions were quantified using an EXOCET Exosome Quantitation Kit (System Biosciences) prior to storage (−80° C). To determine EXO production in HSCs, following culture medium collection/EXO quantification, HSCs were detached from the culture flask and counted (Countess Automatic Cell Counter, Thermo-Fisher Scientific), and data expressed as EXOs/HSC.
To confirm/characterize HSC-EXOs, lysates from HSCs and HSC-EXOs were analyzed by Western blot for membrane-bound markers using antibodies against CD63 and CD81, a non-membrane bound marker using an antibody against Tsg101, and a marker not expressed in EXOs using an antibody against calnexin (all antibodies purchased from Santa Cruz Biotechnology, Dallas,TX) (Supplemental Figure 1).
Electron Microscopy.
Rat HSCs were isolated from normal liver, resuspended in DMEM supplemented with 10% (v/v) EXO-free FBS and P/S, and seeded into tissue culture flasks. For day 1 (quiescent) HSCs, culture medium was replaced 4 hours after initial seeding with DMEM containing 1% (v/v) EXO-free FBS and P/S. After 24 hours, culture medium was collected, EXOs isolated, fixed with Karnovsky’s Fixative, and washed with Cacodylate buffer. For day 14 (activated) HSCs, culture medium was replaced 4-hours after initial seeding with DMEM containing 10% (v/v) EXO-free FBS and P/S and replaced every 48-hours thereafter. Twenty-four hours prior to EXO collection, culture medium was replaced with DMEM containing 1% (v/v) Exo-FBS and P/S. Culture medium was collected 24-hours later and EXO isolation, fixation (Karnovsky’s Fixative) and washing (Cacodylate buffer) performed. Formvar carbon-coated copper mesh grids were prepared by placing them in Alcian Blue (5-minutes), followed by rinsing (deionized water x3) and air drying (room temperature). Exosome samples were transferred to grids, air-dried, and the grid placed on a drop of 2% Phosphotungstic acid (pH 7.0, 30-seconds). Image capture was performed using a Philips CM10 Transmission Electron Microscope.
Immunofluorescent cytochemistry (IFC).
Exosome localization was visualized by IFC using RAW 264.7 macrophages exposed to EXOs collected from quiescent (day-1) or culture-activated (day-14) HSCs isolated from normal rat liver. Briefly, HSCs were isolated as previously described and seeded in 8-well chamber slides. Following fixation, the EXO cargo was stained with carboxy-fluorescein succinimidyl diacetate and detected at 494/521nm (excitation/emission). Cell membranes were stained with 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate. Representative images were captured and merged to determine EXO-membrane localization.
miRNA analysis in HSC-EXOs.
Culture medium was collected from mouse and rat HSCs (day-1 and day-14) and activated human HSCs (grown in 10% (v/v) EXO-FBS), and EXOs purified and quantified as previously described. RNA isolation was performed using a Quick-RNA MiniPrep Kit (Zymo Research Corp, Irvine, CA) and an iScript cDNA kit (Bio-Rad, Hercules, CA) was used for cDNA synthesis. Individual miR expression was determined by polyadenylation of total RNA (250ng) followed by cDNA synthesis (NCode VILO miRNA cDNA synthesis kit, Life Technologies, Grand Island, NY). Quantitative real time PCR (qRT-PCR) was performed on cDNA using an Express SYBR-Green miR qRT-PCR kit (Life Technologies) using miR specific primers (Supplemental Table 1) with universal reverse primer. Fold-changes were calculated using comparative Ct analysis and normalized to snU6 (control).
Macrophage mRNA analysis.
Macrophages were exposed to HSC-EXOs (1010 EXO/mL macrophage culture medium), LPS (20ng/mL), or HSC-EXO + LPS (1010 EXO/mL + 20ng/mL LPS) for 24 or 48 hours. Culture medium was removed and total RNA isolated using a Quick-RNA MiniPrep Kit (Zymo Research Corp, Irvine, CA) and reverse-transcribed to cDNA (iScript cDNA kit, Biorad, Hercules, CA). A qRT-PCR was performed using iQ SYBR Green Supermix (Biorad) for interleukin-6 (IL-6) and tumor necrosis factor-α (TNFα) mRNA and normalized to glyceraldehyde-3-phosphatedehydrogenase (GAPDH) (Supplemental Table 1).
Macrophage IL-6 and TNFα protein production.
Culture medium was collected from mouse, rat, and human macrophages following exposure to HSC-EXOs, LPS, or HSC-EXO + LPS for 24 or 48 hours. Levels of IL-6 and TNF-α protein in the culture medium were determined using species-specific enzyme-linked immunosorbent assay (ELISA; Thermo Fisher Scientific).
Cell Migration.
Cell migration of human macrophages (HL-60) was performed using transwell Boyden chambers (8µm pore, Corning Inc., Lowell, MA) as previously reported [29]. Following overnight attachment (upper chamber) the culture medium in the lower chamber was replaced with culture medium containing human HSC (TWNT4)-EXOs (1x1010/mL), IL-6 (1µg/mL), TNF-α (1µg/mL), or IL-6 (1µg/mL) + TNF-α (1µg/mL). In parallel experiments culture medium in the lower chamber was replaced with 0 or 10% (v/v) Exo-FBS (negative/positive controls). After capturing representative images (48-hours), membranes were placed in 10% (w/v) sodium dodecyl sulfate and quantification performed by measuring the absorbance of crystal violet at 592nm [29].
Statistical analyses.
Data is presented as mean ± SEM. All experiments were performed a minimum of three independent times unless otherwise stated. Statistical analyses were performed using one-way ANOVA for grouped comparisons, with Tukey’s multiple comparison test or paired Student’s t test. p < 0.05 was considered significant.
RESULTS
Hepatic stellate cell (HSC) activation leads to increased exosome (EXO) production.
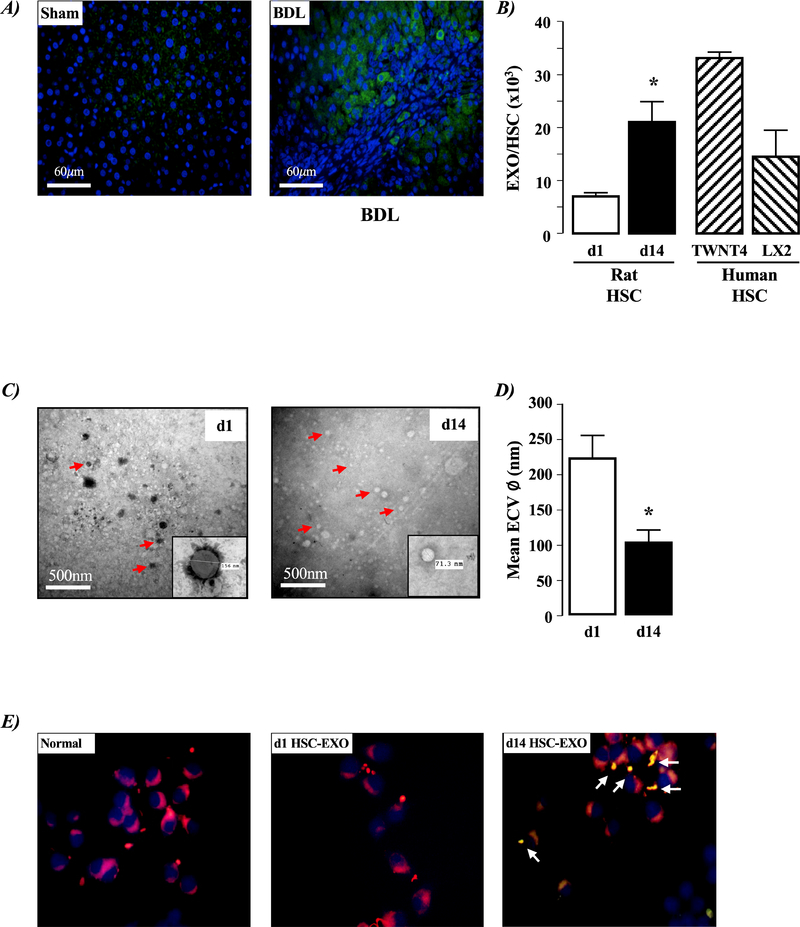
Increased EXO detection was identified using immunohistochemistry in hepatic tissue from BDL versus sham operated rats (Figure 1A). Using culture medium from quiescent (day-1) and culture-activated (day-14) HSCs isolated from normal rat liver, and culture-activated human HSC cells (TWNT4, LX2) we detected increased ECV production in day-14 versus day-1 rat HSCs, and the level of ECV production in cultured human HSCs was of the same order of magnitude as that detected in day-14 rat HSCs (Figure 1B, P<0.05 day 14 versus day1 rat HSC-ECVs, n=4 independent experiments). Using SEM, we identified the majority of ECVs collected from day-1 rat HSCs were in the 125–250nm size range, while those from day-14 rat HSCs were in the 50–100nm size range (Figures 1C& D, P<0.05 day-14 versus day-1 rat HSC ECV size, n=4 independent experiments).
Figure 1. Liver injury and hepatic stellate cell (HSC) exosome (EXO) production.
A) Representative liver sections from sham operated and 4-week bile duct ligated (BDL) rat model of liver injury. Sections were stained with anti-CD63/FITC-labeled secondary antibody (green), and counter-stained with 4′,6-diamidino-2-phenylindole (DAPI) to identify nuclei (blue). B) Number of exosomes (EXOs) released from hepatic stellate cells (HSCs) isolated from normal liver tissue after 1 (d1; quiescent) or 14 (d14; culture-activated) days in culture, and activated human HSCs (TWNT4 and LX2). Data expressed as EXOs released/HSC. *P<0.05 d14 versus d1, n=3 independent experiments. C) Representative electron micrograph images of extracellular vesicles (ECVs) collected from hepatic stellate cells (HSCs) isolated from normal liver (d1; quiescent, d14; culture-activated). Insert illustrates measurement tool for size quantification. D) Quantification of ECV size from d1 and d14 rat HSCs. *P<0.05 d14 versus d1, n=3 independent experiments. E) Representative immunocytochemistry images of untreated macrophages (RAW 264.7) and macrophages following exposure to d1 or d14 EXOs collected from HSCs isolated from normal rat liver. White arrows indicate region of macrophage membrane-EXO colocalization.
Exosomes collected from activated HSCs colocalize to macrophage cell membranes.
Exosomes were not readily detected by immunofluorescent cytochemistry in mouse RAW 264.7 macrophages (Figure 1E). Similarly, following exposure of RAW 264.7 macrophages to day-1 rat HSC-EXOs failed to detect EXO localization in macrophages (Figure 1E). However, when RAW 264.7 macrophages were exposed to day-14 rat HSC-EXOs, clustering of EXOs was evidenced at macrophage cell membranes (Figure 1E).
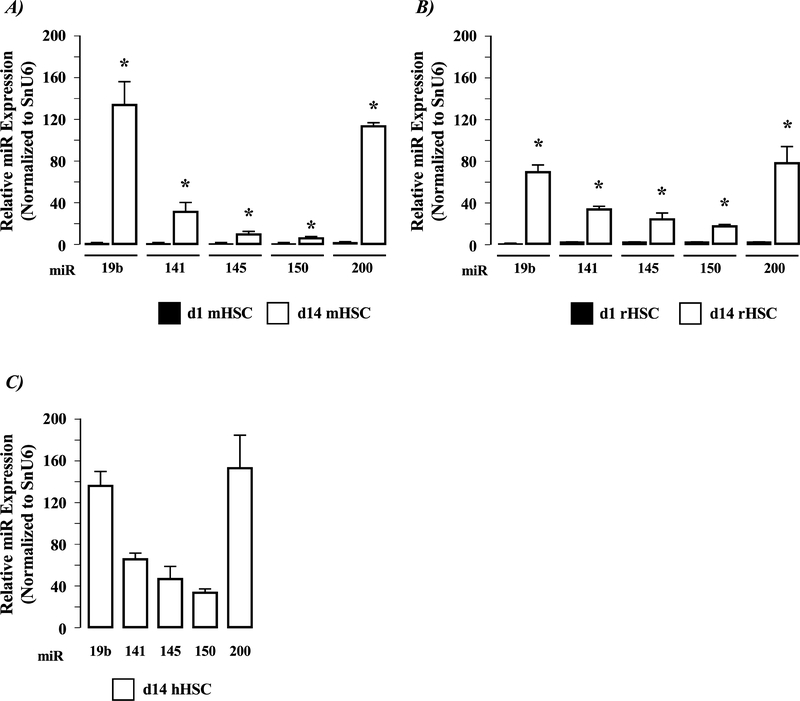
Altered micro-RNA (miR) profiles in rat HSCs following culture.
Analysis of miRs 19b, 141, 145, 150 and 200) demonstrated low detection levels in day-1 mouse and rat HSC-EXOs (Figure 2A& B). Conversely, all of the miRs analyzed were robustly expressed in EXOs collected from day-14 mouse and rat HSCs (Figure 2A& B, P<0.05 day-14 versus day-1 miR expression, n=3 independent experiments). Analysis of these miRs in human HSC-EXOs revealed all of the miR species detected in culture-activated rodent EXOs were also readily detectable in human-EXOs, the most abundantly detected being miR19b and miR200 (Figure 2c).
Figure 2. HSC activation alters exosome (EXO) microRNA profiles.
Exosomes (EXO) were collected, and purified from A) Mouse HSCs isolated from normal liver after 1 (d1 mHSC) or 14 (d14 mHSC) days in culture; B) Rat HSCs isolated from normal liver after 1 (d1 rHSC) or 14 (d14 rHSC) days in culture; C) Human HSCs (hHSC) isolated from cultured TWNT4 cells. EXOs were analyzed for miR19b, 141, 145, 150 and 200 expression. *P<0.05 d14 versus d1 HSC-EXOs, n=3 independent experiments.
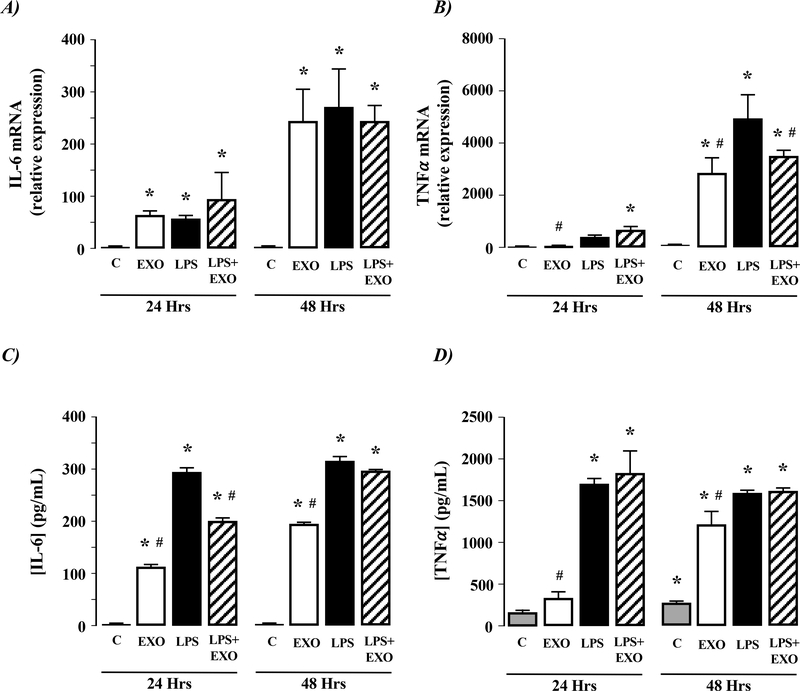
Day-14 (activated) mouse HSC-EXOs stimulate IL-6 and TNFα production in macrophages.
Exposure of mouse macrophages (RAW 264.7) to LPS revealed significant IL-6 and TNFα mRNA expression within 24-hours (Figure 3A& B, P<0.05 versus control [unstimulated] cells, n=4 independent experiments). Similarly, exposure of RAW 264.7cells to EXOs from 14-day mouse HSCs demonstrated increased IL-6 and TNFα mRNA (Figure 3A& B, P<0.05 versus control [unstimulated] cells, n=4 independent experiments). Of note, the effect of EXOs on IL-6 mRNA expression increased between 24 and 48 hours, while changes in TNFα mRNA were only evidenced following 48-hours EXO exposure, and the effect of HSC-EXOs on TNFα mRNA expression were not as great as that obtained following LPS exposure (Figure 3A& B). Combined exposure of RAW 264.7 cells to EXOs and LPS did not alter IL-6 or TNFα mRNA expression compared to EXOs alone (Figure 3A& B). Both LPS and EXOs increased IL-6 and TNFα protein detection from macrophages, with expression profiles mirroring that of the respective changes in mRNA detection (Figure 3C& D). Exposure of RAW 264.7 cells to EXO and LPS revealed that after 24-hours exposure IL-6 protein levels detected were between those for EXO or LPS alone (Figure 3C). However, by the 48-hour time point, the effects of EXO and LPS on IL-6 were equivalent to that of LPS alone (Figure 3C& D).
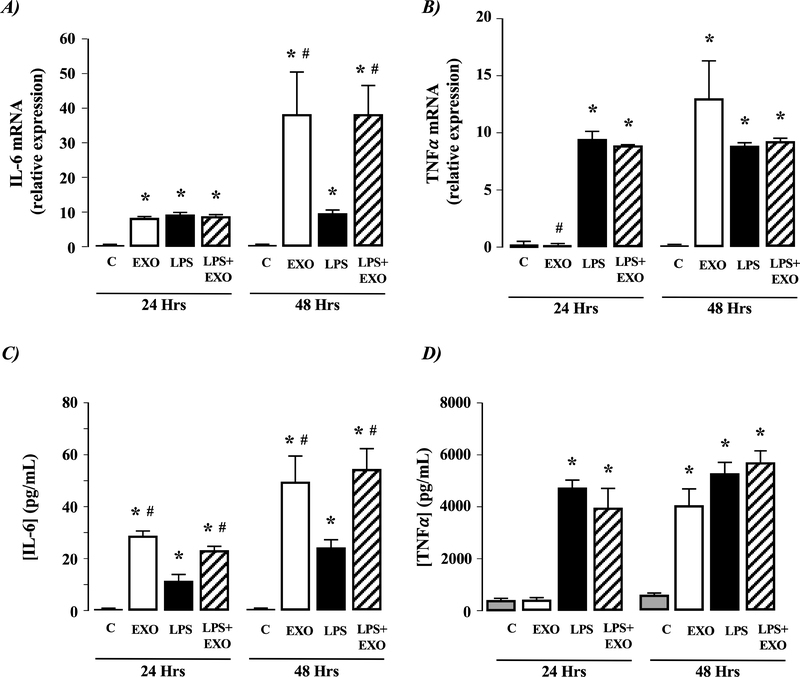
Figure 4. Exosomes from culture-activated rat HSCs do not stimulate IL-6 or TNFα production in macrophages.
Exosomes (EXOs) from day 14 culture-activated rat HSCs were added to culture medium of NR8383 macrophages for 24 or 48 hours. Cells were analyzed for A) IL-6 mRNA expression or B) TNF-α mRNA expression. Culture medium was analyzed for C) IL-6 protein secretion or D) TNF-α protein secretion. *P<0.05 versus control (C), #P<0.05 versus LPS only, n=4 independent experiments.
Activated rat HSC-EXOs do not stimulate IL-6 and TNFα production in macrophages.
Using the rat (NR8383) macrophage cell line LPS significantly stimulated IL-6 and TNFα mRNA and protein detection (Figure 4A-D). Exposure of NR8383 cells to day-14 rat HSC-EXOs failed to affect IL-6 or TNFα mRNA expression or protein secretion or the effect of LPS on macrophage IL-6 or TNFα mRNA expression or protein secretion (Figure 4A-D).
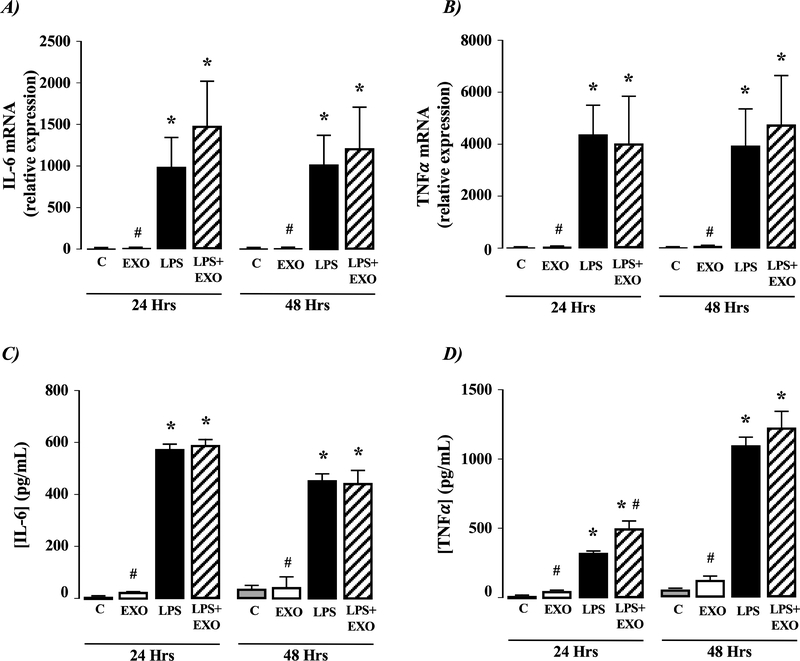
Figure 5. Exosomes from activated human HSCs stimulate IL-6 and TNFα production in macrophages.
Exosomes (EXOs) from activated human TWNT4 HSCs were added to culture medium of HL-60 macrophages for 24 or 48 hours. Cells were analyzed for A) IL-6 mRNA expression or B) TNF-α mRNA expression. Culture medium was analyzed for C) IL-6 protein secretion and D) TNF-α protein secretion. *P<0.05 versus control (C), #P<0.05 versus LPS only, n=4 independent experiments.
Activated human HSC-EXOs stimulate IL-6 and TNFα production in macrophages.
Exposure of HL60 macrophages to LPS induced increased IL-6 and TNFα mRNA expression within 24 hours, effects that were sustained at 48 hours (Figure 5A& B, P<0.05 versus control [unstimulated] cells, n=4 independent experiments). Similarly, exposure of HL60 cells to EXOs from cultured TWNT-4 HSCs demonstrated increased IL-6 and TNFα mRNA expression (Figure 5A& B, P<0.05 versus control [unstimulated] cells, n=4 independent experiments). Of note, the effect of EXOs on IL-6 mRNA expression increased significantly between 24–48 hours, while changes in TNFα mRNA were only evidenced following 48 hours HSC-EXO exposure (Figure 5A and B, P<0.05 versus control [unstimulated] cells, n=4 independent experiments). Combined exposure of HL60 cells to EXOs and LPS did not significantly alter IL-6 or TNFα mRNA expression compared to EXOs alone( Figure 5A& B). Analysis of HL-60 culture medium following exposure to LPS, EXOs, or LPS and EXOs revealed both LPS and EXOs increased IL-6 and TNα protein secretion with expression profiles mirroring that of respective changes in mRNA detection (Figure 5A-D).
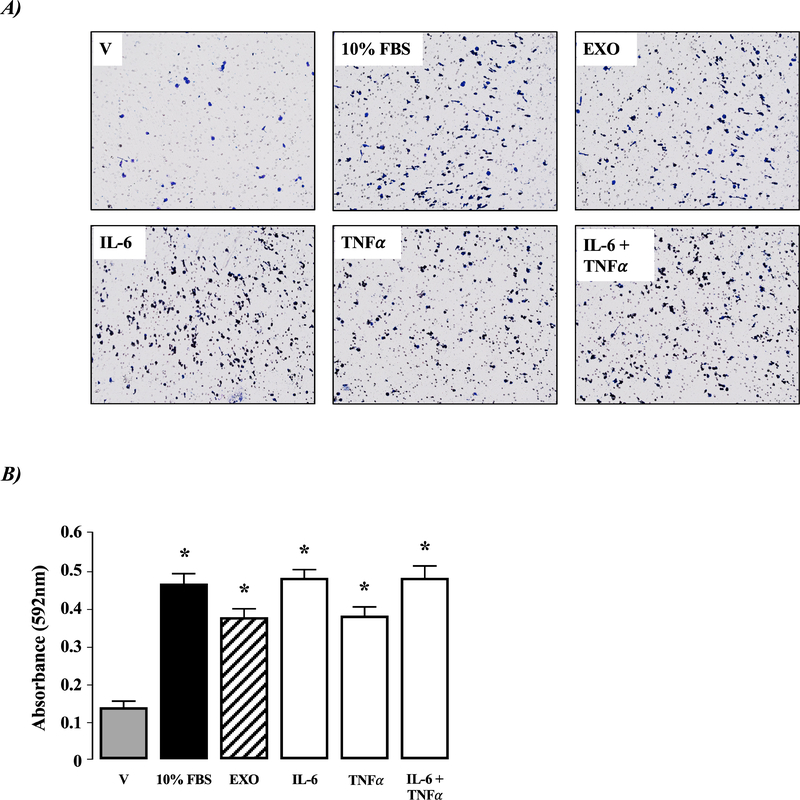
Figure 6. Activated human HSC-EXOs stimulate macrophage migration.
Exosomes (EXO) from activated human TWNT4 HSCs were added to culture medium of HL-60 macrophages for 48 hours. In parallel, TWNT4 HSCs were exposed to vehicle (V; 0.1% (v/v) FBS), 10% (v/v) FBS, IL-6 (1μg/mL), TNF-α (1μg/mL) or IL-6 (1μg/mL) +TNF-α (1μg/mL). A) Representative micrographs of transwell membranes following crystal violet staining. B) Quantification of migration data following cell solubilization and measurement of absorbance at λ = 592nm). *P<0.05 versus V, n=4 independent experiments.
IL-6 does not stimulate TNFα production in macrophages.
To investigate whether HSC-EXO stimulation of IL-6 is involved in TNFα production, HL60 macrophages were treated with rhIL-6 (48Hrs). This approach demonstrated rhIL-6 did not stimulate TNFα mRNA production (Supplemental Figure 2).
Activated HSC-derived EXOs stimulate macrophage migration.
Exposure of HL-60 macrophages to HSC-EXOs stimulated cell migration (Figure 6 A& B, P<0.05 versus vehicle, n=4 independent experiments). Similarly, both IL-6 and TNFα stimulated HL-60 migration, and there was no synergistic effect of IL-6 and TNFα (Figure 6 A& B, P<0.05 versus vehicle, n=4 independent experiments).
Figure 3. Exosomes from culture-activated mouse HSCs stimulate IL-6 and TNFα production in macrophages.
Exosomes (EXOs) from day 14 culture-activated mouse HSCs were added to culture medium of mouse (RAW 264.7) macrophages for 24 or 48 hours. Macrophages were analyzed for A) IL-6 mRNA expression and B) TNF-α mRNA expression. Culture medium was analyzed for C) IL-6 protein secretion and D) TNF-α protein secretion. *P<0.05 versus control (C), #P<0.05 versus LPS only, n=4 independent experiments.
DISCUSSION
Liver fibrosis most commonly arises during regeneration and repair in the setting of injury and inflammation [5]. A critical event during fibrosis is HSC activation, in which HSCs transform from a quiescent, vitamin A rich state to a proliferative, contractile, myofibroblast-like state [6]. While much work has focused on the activity of specific cell signaling pathways during HSC activation, increasing evidence implies multicellular intrahepatic and systemic responses to liver injury are important in defining disease progression [30]. In this study we focused on the role of EXOs as paracrine signaling entities, and the potential role of HSC-EXOs in regulating macrophage function. Our data demonstrate HSC culture-activation altered the profile of extracellular vesicle production from predominantly larger EVs (day 1) to smaller EXOs (day 14). Furthermore, the EXO miR profile changed following HSC culture-activation such that activated HSC-EXOs co-localized with macrophages and stimulated macrophage IL-6 and TNFα synthesis and release and macrophage migration.
Following EXO release, the effect of EXOs on target cells depends on the specific components contained within the EXO, factors that can comprise of lipids, miRs, mRNAs, and proteins [7, 8]. Previous studies report significant roles for EXOs released from hepatocytes [31, 32], macrophages [25], cholangiocytes [33], and hepatoma cells [34] during the initiation and acceleration of HSC activation [35]. To our knowledge, our data represent the first reports of a functional role for HSC-EXOs in regulating macrophage function, implying a dynamic interaction within hepatic cell populations during HSC activation/fibrosis. Of specific interest was the demonstration of altered miR profiles between EXOs collected from quiescent versus activated HSCs. Previous studies report HSC-EXOs contain unique “miR signatures” based on the underlying hepatic pathology [36–38], with miRs 18–22 being [predominately] inhibitors of proteins coding gene expression. Functionally, miRs bind the 3’ untranslated region (through varying degrees of specificity) to regulate protein expression. A single miR thus has the potential to interact with hundreds of mRNAs which could in turn have wide-spread effects on gene expression and intracellular signaling. Previous studies report loss of miR19b in HSCs influences TGFβ signaling during hepatic injury through interaction with the TGFβ-receptor-II to increase expression [27]. This decrease in miR19b occurred despite increased expression of the miR17–92 cluster (which contains miR19b and five other miRs) in models of hepatic injury, and expression of individual miRs in the cluster varied between cells and HSC-EXOs [9]. Additionally, manipulation of miR19b (via HSC over-expression) alters cellular and EXO expression of miRs within the miR17–92 cluster [9, 39].
In addition to miRs, EXOs contain a range of mRNAs and proteins that can alter target cell function [35]. Given the possible combinations, and relative levels of expression within EXOs, it seems likely that the effect of EXOs are dependent on both the specific contents, and the relative amount of each component. Thus, it is important to highlight that while our study focused on specific miR species, further analysis is required to determine the potential presence of other miRs (as well as mRNAs and proteins) in EXOs derived from quiescent and activated HSC to provide a comprehensive view of HSC-EXO function.
Of note within our study, unlike culture-activated mouse and human HSCs, EXOs derived from culture-activated rat HSCs failed to induce cytokine production in rat macrophages. To investigate this, additional studies were performed in which macrophages from one species were exposed to HSC-EXOs from another species. Using this approach, we determined both human and mouse HSC-EXOs stimulated IL-6 and TNFα production in the rat macrophage cell line, but rat HSC-EXOs were incapable of stimulating IL-6 or TNFα in human or mouse macrophages (Supplemental Figure 3). These findings were unexpected given our data indicating the differences in miR profiles between EXOs collected from quiescent versus activated rat HSCs. One explanation may be the possibility of damage to the structural integrity of the rat EXOs during collection/storage, although this seems unlikely given that mouse and human HSC-EXOs were collected in an identical manner. Similarly, it is unlikely the differences we observed were due to cell contamination resulting from individual animals since EXOs were pooled following collection. Alternative explanations may relate to changes in the formation/releases processes of the rat HSC-EXOs in culture. That is, in addition to the EXO contents it is equally important that the EXO-membrane is recognized/able to fuse with the target cell membrane. Given our ability to demonstrate cross-species macrophage activation for human and mouse HSC-EXOs, and the inability of rat HSC-EXOs to stimulate rat, human or mouse macrophage cytokine production, the possibility exists that the rat HSC-EXOs contain the content profile of activated HSCs, but were unable to enter into the macrophages.
In considering our results, it is important to highlight several limitations when interpreting these data. Our studies utilized EXOs derived from culture-activated HSCs, as opposed to EXOs derived from disease-state activated HSCs. This may be of particular relevance given the dynamic interaction between different hepatic and non-hepatic cell types that occurs during injury-wound healing, and the cellular remodeling-repopulation that occurs in vivo. Similarly, our studies addressed the direct effect of HSC-EXOs on macrophage-cytokine production, yet it is equally likely other pathways are regulated by other miR species contained within the HSC-EXOs. While several candidate pathways exist, the role of Notch-signaling may prove particularly fruitful for future investigation. Previous studies report important roles for Notch signaling during both HSC activation [40] and macrophage polarization, changes in macrophage polarization being an important event during macrophage infiltration [41]. Given the significance of Notch signaling in HSCs and macrophages during the progression of hepatic fibrosis [42], the intriguing possibility is raised that miR species involved in HSC activation may also play a role in regulating macrophage polarization during fibrosis. Finally, our studies focused on changes in a specific group of miRs previously identified as being altered during HSC transition [24–27]. However, this does not mean other miRs, or combinations of miRs (and/or mRNA species), may be equally important in regulating macrophage-cytokine production, and a comprehensive gene array analyses is required to identify such changes.
CONCLUSION
Increasing evidence suggests EXOs play a significant role in the tissue microenvironment as carriers of miRs, RNA species, and proteins that influence cell function in recipient cells. In this study we report HSC-EXOs associate with macrophages in vivo and HSC-EXOs stimulate cytokine synthesis-release and migration in macrophages in vitro. These effects may be mediated, at least in part, by changes in HSC-EXO miR profiles that occur as a result of HSC trans-differentiation from a quiescent to activated state. Constructing a comprehensive understanding of HSC-EXO cargos, and how HSC-EXOs interact during hepatic inflammation and fibrosis, raises the potential for developing innovative diagnostic or therapeutic approaches to monitor and/or treat liver injury.
Supplementary Material
Supplemental Figure 1. Exosome characterization. A) To assess EXO characteristics/purity Western blot analysis was performed using EXO and hepatic stellate cell (HSC) lysates with antibodies against membrane-bound markers (anti-CD63 and anti-CD81), a non-membrane bound marker (anti-Tsg101), and a marker not expressed in EXOs (anti-calnexin). [N.B. Insufficient mouse HSC lysates were available for analysis following collection for RNA analysis. All EXOs were purified from culture medium using identical techniques (Exoquick, Systems Bioscience, Paolo Alto, CA)]. B) To assess range of HSC-derived EXOs for use in macrophage studies, EXOs were collected and purified from human HSC culture medium and counted. Human macrophages (HL60) were seeded at 1 × 106 cells/well and exposed to either LPS (20ng/mL) or 1 × 103, 10 × 103 or 20 × 103 HSC-EXOs. 24Hrs later HL-60 culture medium was collected and analyzed for IL-6 and TNF-α expression by ELISA. n=3 independent experiments, *P versus control (untreated).
Supplemental Figure 2. Interleukin-6 does not stimulate TNFα production in macrophages. Human HL60 macrophages were treated with either rhIL-6, exosomes (EXO) isolated from culture activated human HSCs, or LPS (48Hrs) and analyzed for TNF-α protein expression by ELISA. n=4 independent experiments. *P<0.05 versus untreated control (C), #P<0.05 versus rhIL-6.
Supplemental Figure 3. Activated rat HSC-EXOs do not stimulate IL-6 and TNFα production in mouse or human macrophages yet activated mouse and human HSC-EXOs stimulate IL-6 and TNFα production in rat macrophages. Exosomes (EXO) from day 14 culture activated rat HSCs were added to culture medium of mouse (RAW 264.7) macrophages for 48 hours. Culture medium was analyzed for A) IL-6 and B) TNF-α protein levels by ELISA. Exosomes (EXO) from day 14 culture activated rat HSCs were added to culture medium of human (HL-60) macrophages for 48 hours. Culture medium was analyzed for C) IL-6 and D) TNF-α protein levels by ELISA. *P<0.05 versus Control (C), n=4 independent experiments. EXO from day 14 culture activated mouse HSCs were added to culture medium of rat macrophages (NR8383) for 48 hours. Culture medium was analyzed for E) IL-6 and F) TNF-α protein levels by ELSIA. Exosomes (EXO) from activated human HSCs (TWNT4) were added to culture medium of rat macrophages (NR8383) for 48 hours. Culture medium was analyzed for G) IL-6 and H) TNF-α protein levels by ELISA. *P<0.05 versus Control (C), n=4 independent experiments.
Financial Support
This study was supported by a grant from the National Institutes of Health (AA022702).
Abbreviations:
- HSC
Hepatic stellate cell
- EXO
Exosome
- MV
microvessicle
- KC
Kupffer cell
- miR
MicroRNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: The authors do not have any disclosures to report.
DISCLOSURES
Declaration of conflict of interest: None of the authors declare any conflicts of interest.
Conflict of interest:
The authors do not have any disclosures to report.
REFERENCES
- [1].Neuman MG, Malnick S, Maor Y, Nanau RM, Melzer E, Ferenci P, Seitz HK, Mueller S, Mell H, Samuel D, Cohen LB, Kharbanda KK, Osna NA, Ganesan M, Thompson KJ, McKillop IH, Bautista A, Bataller R, French SW, Alcoholic liver disease: Clinical and translational research, Exp Mol Pathol 99 (2015) 596–610. [DOI] [PubMed] [Google Scholar]
- [2].Razavi H, Global Epidemiology of Viral Hepatitis, Gastroenterol Clin North Am 49 (2020) 179–189. [DOI] [PubMed] [Google Scholar]
- [3].Kumar R, Priyadarshi RN, Anand U, Non-alcoholic Fatty Liver Disease: Growing Burden, Adverse Outcomes and Associations, J Clin Transl Hepatol 8 (2020) 76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wan M, Han J, Ding L, Hu F, Gao P, Novel Immune Subsets and Related Cytokines: Emerging Players in the Progression of Liver Fibrosis, Front Med (Lausanne) 8 (2021) 604894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lech M, Anders HJ, Macrophages and fibrosis: How resident and infiltrating mononuclear phagocytes orchestrate all phases of tissue injury and repair, Biochim Biophys Acta 1832 (2013) 989–997. [DOI] [PubMed] [Google Scholar]
- [6].Thompson KJ, McKillop IH, Schrum LW, Targeting collagen expression in alcoholic liver disease, World J Gastroenterol 17 (2011) 2473–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Crescitelli R, Lasser C, Szabo TG, Kittel A, Eldh M, Dianzani I, Buzas EI, Lotvall J, Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes, J Extracell Vesicles 2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Vlassov AV, Magdaleno S, Setterquist R, Conrad R, Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials, Biochim Biophys Acta 1820 (2012) 940–948. [DOI] [PubMed] [Google Scholar]
- [9].Brandon-Warner E, Feilen NA, Culberson CR, Field CO, deLemos AS, Russo MW, Schrum LW, Processing of miR17–92 Cluster in Hepatic Stellate Cells Promotes Hepatic Fibrogenesis During Alcohol-Induced Injury, Alcohol Clin Exp Res 40 (2016) 1430–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Han W, Duan Z, Roles of exosomes in liver metastases: Novel diagnosis and treatment choices, J Cell Physiol 234 (2019) 21588–21600. [DOI] [PubMed] [Google Scholar]
- [11].Malhi H, Emerging role of extracellular vesicles in liver diseases, Am J Physiol Gastrointest Liver Physiol 317 (2019) G739–G749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Colombo M, Raposo G, Thery C, Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles, Annu Rev Cell Dev Biol 30 (2014) 255–289. [DOI] [PubMed] [Google Scholar]
- [13].Harting MT, Srivastava AK, Zhaorigetu S, Bair H, Prabhakara KS, Toledano Furman NE, Vykoukal JV, Ruppert KA, Cox CS Jr., Olson SD, Inflammation-Stimulated Mesenchymal Stromal Cell-Derived Extracellular Vesicles Attenuate Inflammation, Stem Cells 36 (2018) 79–90. [DOI] [PubMed] [Google Scholar]
- [14].Lanyu Z, Feilong H, Emerging role of extracellular vesicles in lung injury and inflammation, Biomed Pharmacother 113 (2019) 108748. [DOI] [PubMed] [Google Scholar]
- [15].Yamamoto S, Niida S, Azuma E, Yanagibashi T, Muramatsu M, Huang TT, Sagara H, Higaki S, Ikutani M, Nagai Y, Takatsu K, Miyazaki K, Hamashima T, Mori H, Matsuda N, Ishii Y, Sasahara M, Inflammation-induced endothelial cell-derived extracellular vesicles modulate the cellular status of pericytes, Sci Rep 5 (2015) 8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nakaoka H, Hirono K, Yamamoto S, Takasaki I, Takahashi K, Kinoshita K, Takasaki A, Nishida N, Okabe M, Ce W, Miyao N, Saito K, Ibuki K, Ozawa S, Adachi Y, Ichida F, MicroRNA-145–5p and microRNA-320a encapsulated in endothelial microparticles contribute to the progression of vasculitis in acute Kawasaki Disease, Sci Rep 8 (2018) 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dini L, Tacconi S, Carata E, Tata AM, Vergallo C, Panzarini E, Microvesicles and exosomes in metabolic diseases and inflammation, Cytokine Growth Factor Rev 51 (2020) 27–39. [DOI] [PubMed] [Google Scholar]
- [18].Messenger SW, Woo SS, Sun Z, Martin TFJ, A Ca(2+)-stimulated exosome release pathway in cancer cells is regulated by Munc13–4, J Cell Biol 217 (2018) 2877–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Speziali G, Liesinger L, Gindlhuber J, Leopold C, Pucher B, Brandi J, Castagna A, Tomin T, Krenn P, Thallinger GG, Olivieri O, Martinelli N, Kratky D, Schittmayer M, Birner-Gruenberger R, Cecconi D, Myristic acid induces proteomic and secretomic changes associated with steatosis, cytoskeleton remodeling, endoplasmic reticulum stress, protein turnover and exosome release in HepG2 cells, J Proteomics 181 (2018) 118–130. [DOI] [PubMed] [Google Scholar]
- [20].O’Brien J, Hayder H, Zayed Y, Peng C, Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation, Front Endocrinol (Lausanne) 9 (2018) 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tetta C, Ghigo E, Silengo L, Deregibus MC, Camussi G, Extracellular vesicles as an emerging mechanism of cell-to-cell communication, Endocrine 44 (2013) 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Coll M, El Taghdouini A, Perea L, Mannaerts I, Vila-Casadesus M, Blaya D, Rodrigo-Torres D, Affo S, Morales-Ibanez O, Graupera I, Lozano JJ, Najimi M, Sokal E, Lambrecht J, Gines P, van Grunsven LA, Sancho-Bru P, Integrative miRNA and Gene Expression Profiling Analysis of Human Quiescent Hepatic Stellate Cells, Sci Rep 5 (2015) 11549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bala S, Szabo G, MicroRNA Signature in Alcoholic Liver Disease, Int J Hepatol 2012 (2012) 498232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Szabo G, Sarnow P, Bala S, MicroRNA silencing and the development of novel therapies for liver disease, J Hepatol 57 (2012) 462–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chen L, Yao X, Yao H, Ji Q, Ding G, Liu X, Exosomal miR-103–3p from LPS-activated THP-1 macrophage contributes to the activation of hepatic stellate cells, FASEB J 34 (2020) 5178–5192. [DOI] [PubMed] [Google Scholar]
- [26].Devhare PB, Sasaki R, Shrivastava S, Di Bisceglie AM, Ray R, Ray RB, Exosome-Mediated Intercellular Communication between Hepatitis C Virus-Infected Hepatocytes and Hepatic Stellate Cells, J Virol 91 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lakner AM, Steuerwald NM, Walling TL, Ghosh S, Li T, McKillop IH, Russo MW, Bonkovsky HL, Schrum LW, Inhibitory effects of microRNA 19b in hepatic stellate cell-mediated fibrogenesis, Hepatology 56 (2012) 300–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lakner AM, Walling TL, McKillop IH, Schrum LW, Altered aquaporin expression and role in apoptosis during hepatic stellate cell activation, Liver Int 31 (2011) 42–51. [DOI] [PubMed] [Google Scholar]
- [29].Attal N, Sullivan MT, Girardi CA, Thompson KJ, McKillop IH, Fatty acid binding protein-4 promotes alcohol-dependent hepatosteatosis and hepatocellular carcinoma progression, Transl Oncol 14 (2020) 100975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Albillos A, de Gottardi A, Rescigno M, The gut-liver axis in liver disease: Pathophysiological basis for therapy, J Hepatol 72 (2020) 558–577. [DOI] [PubMed] [Google Scholar]
- [31].Lee YS, Kim SY, Ko E, Lee JH, Yi HS, Yoo YJ, Je J, Suh SJ, Jung YK, Kim JH, Seo YS, Yim HJ, Jeong WI, Yeon JE, Um SH, Byun KS, Exosomes derived from palmitic acid-treated hepatocytes induce fibrotic activation of hepatic stellate cells, Sci Rep 7 (2017) 3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Khatun M, Ray RB, Mechanisms Underlying Hepatitis C Virus-Associated Hepatic Fibrosis, Cells 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Liu R, Li X, Zhu W, Wang Y, Zhao D, Wang X, Gurley EC, Liang G, Chen W, Lai G, Pandak WM, Robert Lippman H, Bajaj JS, Hylemon PB, Zhou H, Cholangiocyte-Derived Exosomal Long Noncoding RNA H19 Promotes Hepatic Stellate Cell Activation and Cholestatic Liver Fibrosis, Hepatology 70 (2019) 1317–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhou Y, Ren H, Dai B, Li J, Shang L, Huang J, Shi X, Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts, J Exp Clin Cancer Res 37 (2018) 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zivko C, Fuhrmann G, Luciani P, Liver-derived extracellular vesicles: A cell by cell overview to isolation and characterization practices, Biochim Biophys Acta Gen Subj (2020) 129559. [DOI] [PubMed] [Google Scholar]
- [36].Bala S, Szabo G, MicroRNA Signature in Alcoholic Liver Disease, International Journal of Hepatology 2012 (2012) 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bala S, Marcos M, Szabo G, Emerging role of microRNAs in liver diseases, World J Gastro 15 (2009) 5633–5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Szabo G, Sarnow P, Bala S, MicroRNA silencing and the development of novel therapies for liver disease, J Hepatol 57 462–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Brandon-Warner E, Benbow JH, Swet JH, Feilen NA, Culberson CR, McKillop IH, deLemos AS, Russo MW, Schrum LW, Adeno-Associated Virus Serotype 2 Vector-Mediated Reintroduction of microRNA-19b Attenuates Hepatic Fibrosis, Hum Gene Ther 29 (2018) 674–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ezhilarasan D, MicroRNA interplay between hepatic stellate cell quiescence and activation, Eur J Pharmacol 885 (2020) 173507. [DOI] [PubMed] [Google Scholar]
- [41].Zheng S, Zhang P, Chen Y, Zheng S, Zheng L, Weng Z, Inhibition of Notch Signaling Attenuates Schistosomiasis Hepatic Fibrosis via Blocking Macrophage M2 Polarization, PLoS One 11 (2016) e0166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bansal R, van Baarlen J, Storm G, Prakash J, The interplay of the Notch signaling in hepatic stellate cells and macrophages determines the fate of liver fibrogenesis, Sci Rep 5 (2015) 18272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Exosome characterization. A) To assess EXO characteristics/purity Western blot analysis was performed using EXO and hepatic stellate cell (HSC) lysates with antibodies against membrane-bound markers (anti-CD63 and anti-CD81), a non-membrane bound marker (anti-Tsg101), and a marker not expressed in EXOs (anti-calnexin). [N.B. Insufficient mouse HSC lysates were available for analysis following collection for RNA analysis. All EXOs were purified from culture medium using identical techniques (Exoquick, Systems Bioscience, Paolo Alto, CA)]. B) To assess range of HSC-derived EXOs for use in macrophage studies, EXOs were collected and purified from human HSC culture medium and counted. Human macrophages (HL60) were seeded at 1 × 106 cells/well and exposed to either LPS (20ng/mL) or 1 × 103, 10 × 103 or 20 × 103 HSC-EXOs. 24Hrs later HL-60 culture medium was collected and analyzed for IL-6 and TNF-α expression by ELISA. n=3 independent experiments, *P versus control (untreated).
Supplemental Figure 2. Interleukin-6 does not stimulate TNFα production in macrophages. Human HL60 macrophages were treated with either rhIL-6, exosomes (EXO) isolated from culture activated human HSCs, or LPS (48Hrs) and analyzed for TNF-α protein expression by ELISA. n=4 independent experiments. *P<0.05 versus untreated control (C), #P<0.05 versus rhIL-6.
Supplemental Figure 3. Activated rat HSC-EXOs do not stimulate IL-6 and TNFα production in mouse or human macrophages yet activated mouse and human HSC-EXOs stimulate IL-6 and TNFα production in rat macrophages. Exosomes (EXO) from day 14 culture activated rat HSCs were added to culture medium of mouse (RAW 264.7) macrophages for 48 hours. Culture medium was analyzed for A) IL-6 and B) TNF-α protein levels by ELISA. Exosomes (EXO) from day 14 culture activated rat HSCs were added to culture medium of human (HL-60) macrophages for 48 hours. Culture medium was analyzed for C) IL-6 and D) TNF-α protein levels by ELISA. *P<0.05 versus Control (C), n=4 independent experiments. EXO from day 14 culture activated mouse HSCs were added to culture medium of rat macrophages (NR8383) for 48 hours. Culture medium was analyzed for E) IL-6 and F) TNF-α protein levels by ELSIA. Exosomes (EXO) from activated human HSCs (TWNT4) were added to culture medium of rat macrophages (NR8383) for 48 hours. Culture medium was analyzed for G) IL-6 and H) TNF-α protein levels by ELISA. *P<0.05 versus Control (C), n=4 independent experiments.