Abstract
Fluid shear stress provided by blood flow instigates a transition from active blood vessel network expansion during development, to vascular homeostasis and quiescence that is important for mature blood vessel function. Here we show that SMAD6 is required for endothelial cell flow-mediated responses leading to maintenance of vascular homeostasis. Concomitant manipulation of the mechanosensor Notch1 pathway and SMAD6 expression levels revealed that SMAD6 functions downstream of ligand-induced Notch signaling and transcription regulation. Mechanistically, full-length SMAD6 protein was needed to rescue Notch loss-induced flow misalignment. Endothelial cells depleted for SMAD6 had defective barrier function accompanied by upregulation of proliferation-associated genes and down regulation of junction-associated genes. The vascular protocadherin PCDH12 was upregulated by SMAD6 and required for proper flow-mediated endothelial cell alignment, placing it downstream of SMAD6. Thus, SMAD6 is a required transducer of flow-mediated signaling inputs downstream of Notch1 and upstream of PCDH12, as vessels transition from an angiogenic phenotype to maintenance of a homeostatic phenotype.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10456-021-09777-7.
Keywords: Endothelial cells, Blood vessels, Blood flow, Notch, SMAD, BMP, PCDH12
Introduction
Blood vessel formation intersects with tissue metabolic needs and architecture in multiple ways, including remodeling of the initial vascular plexus via shear stress provided by blood flow [1–3]. Endothelial cells orchestrate this remodeling and eventual transition to vascular homeostasis, as elevated heart pumping and blood volume increase fluid shear stress [4–7]. In contrast to the sprouting and blood vessel remodeling that characterize the “activated” endothelium of development, endothelial cells become quiescent under laminar flow at steady state. This transition is an important prerequisite for endothelial cell functions involved in homeostasis, including formation of a vascular barrier that regulates oxygen and nutrient exchange [8–11]. As homeostasis is induced, endothelial cells become less proliferative and remodel their cytoskeleton and junctions to align parallel to the flow vector, presumably to reduce flow-induced cellular stress [12–16]. The importance of this response is highlighted by the association of arterial laminar flow with protection from atherosclerosis, and the correlation between disturbed flow and atheroprone regions of vessels [17, 18]. Although several cell-surface protein complexes function as direct endothelial cell mechanosensors [19], and even more signaling pathways have flow-responsive components, how these sensors and pathways transduce flow-mediated inputs to set up and maintain vessel homeostasis remains poorly understood.
Notch and BMP are two pathways involved in endothelial cell flow responses. Notch1 was recently identified as a direct flow mechanosensor, and endothelial deletion of Notch1 leads to flow misalignment, increased permeability, and atherosclerotic plaque formation [20–22]. More generally, Notch signaling is upregulated in arteries, which are exposed to higher shear stress than veins of equivalent diameter, and Notch is thought to be responsible for maintaining arterial differentiation [23–25]. Although BMP pathway components are not direct mechanosensors, some are flow-regulated [4]; BMP signaling regulates blood vessel formation and function in complex and context-dependent ways [26–29], with proangiogenic signaling downstream of ligands such as BMP2 and BMP6 countered by antiangiogenic signaling downstream of BMP9 and/or BMP10. Human mutations in the Type 1 receptor Alk1/ACVRL1, the co-receptor Endoglin, or a common signaling component SMAD4 lead to Hereditary Hemmorrhagic Telangiectasia (HHT) that is characterized by arterio-venous malformations (AVMs) and hemorrhage [30–32]. AVM formation requires genetic loss of BMP signaling, blood flow, and a third proangiogenic or pro-inflammatory stimulus [33–36], suggesting an intimate relationship between BMP signaling and flow responses in blood vessels.
There are several intersection points between Notch and BMP signaling in endothelial cells. For example, transcriptional responses are altered in complex and interdependent ways when both Notch and BMP signaling are activated, leading to cooperative effects on signaling and endothelial cell quiescence [37–39]. We showed that proangiogenic endothelial cell BMP signaling in vitro and in vivo is regulated in part by an intracellular negative regulator of BMP signaling, SMAD6 [40]. SMAD6 expression is upregulated by laminar blood flow and genetic loss leads to vascular hemorrhage [41, 42], but whether it functions in the transition to stable homeostasis is not known. Here, we reveal a requirement for SMAD6 in the flow-mediated alignment of endothelial cells, homeostatic quiescence, and barrier function downstream of the mechanosensor Notch1. Flow-induced endothelial cell-cell junction gene expression required SMAD6, and a SMAD6-regulated protocadherin affected homeostatic flow responses. Thus, SMAD6 is a required transducer of endothelial flow-mediated responses instigated by Notch signaling and required for maintenance of vascular homeostasis.
Results
SMAD6 is required for homeostatic laminar flow-induced endothelial cell alignment and polarization
To begin to understand how endothelial cell flow-mediated responses that characterize vascular homeostasis are maintained, we asked whether an inhibitory SMAD, SMAD6, regulates endothelial cell responses to extended periods of laminar shear stress. Smad6−/− mutant embryos and pups with a knock-in lacZ reporter in the Smad6 locus strongly express lacZ in endothelial cells of embryonic and early post-natal arteries, but not veins of similar diameter and stage, after the onset of blood flow [42, 43]. Since shear stress induced by laminar flow is higher in arteries, this suggests that endothelial SMAD6 expression is induced by homeostatic laminar flow. Thus, we chose conditions of 15 dynes/cm2 (d/cm2) for 72 h for analysis of SMAD6 function in homeostatic arterial flow responses. Under these conditions, both human umbilical vein endothelial cells (HUVEC) and human arterial endothelial cells (HAEC) showed a two to threefold increase in Smad6 RNA (Supp. Fig 1A, B), consistent with a previous report [41].
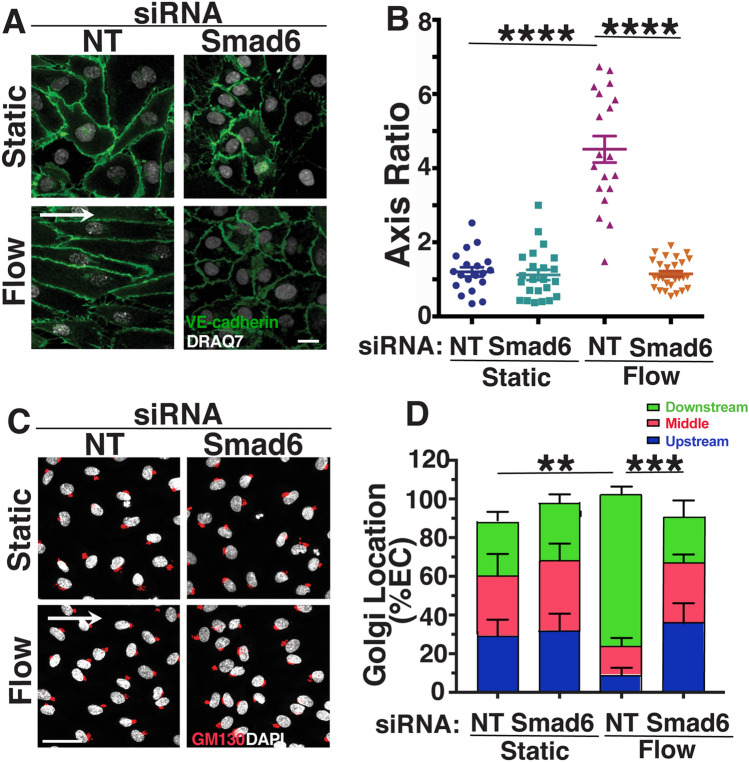
We next examined the function of SMAD6 in endothelial cell responses to homeostatic laminar flow, by interrogating HUVEC and HAEC depleted for Smad6 RNA via knockdown (KD) and exposed to flow. We found that both venous and arterial endothelial cells with reduced Smad6 RNA levels failed to align under homeostatic laminar flow, as indicated by cell axis ratio and nuclear displacement angle measured parallel to the direction of flow (Fig. 1a, b, Supp. Fig. 1C–F). Endothelial cell misalignment in response to homeostatic laminar flow occurred with multiple siRNAs targeting Smad6 (Smad6-1 and Smad6-2), including a Smad6 siRNA pool (Smad6-3) (Supp. Fig. 1G, H). Both venous and arterial endothelial cells depleted for Smad6 also had mis-positioned Golgi and centrosomes under flow compared to controls (Fig. 1c, d, Supp. Fig. I–K, not shown), indicating that SMAD6 is important for endothelial cell polarization in response to homeostatic laminar flow. These results show that endothelial cell SMAD6 expression is flow-regulated and induced by homeostatic laminar flow, and that SMAD6 is functionally required for homeostatic flow-mediated endothelial cell alignment and polarization.
Fig. 1.
SMAD6 is required for homeostatic endothelial cell flow-mediated alignment and polarization. a Representative panels of HUVEC stained with VE-cadherin (green, junctions) and DRAQ7 (white, nucleus) under indicated conditions with indicated treatments. White arrow, flow vector. Scale bar, 20 μM. b Quantification of cell axis ratio. Statistical analysis, One-way ANOVA; ****p ≤ 0.0001. N = 3, representative experiment shown. c Representative panels of HUVEC stained with GM130 (red, Golgi) and DAPI (white, nucleus) under indicated conditions with indicated treatments. White arrow, flow vector. Scale bar, 50 μM. d Quantification of Golgi localization relative to nucleus in indicated conditions. ≥ 30 cells/condition were analyzed. Statistical analysis, One-way ANOVA; **p ≤ 0.01; ***p ≤ 0.001. NT non-targeting (control) siRNA
SMAD6 is required downstream of Notch for flow-mediated alignment of endothelial cells
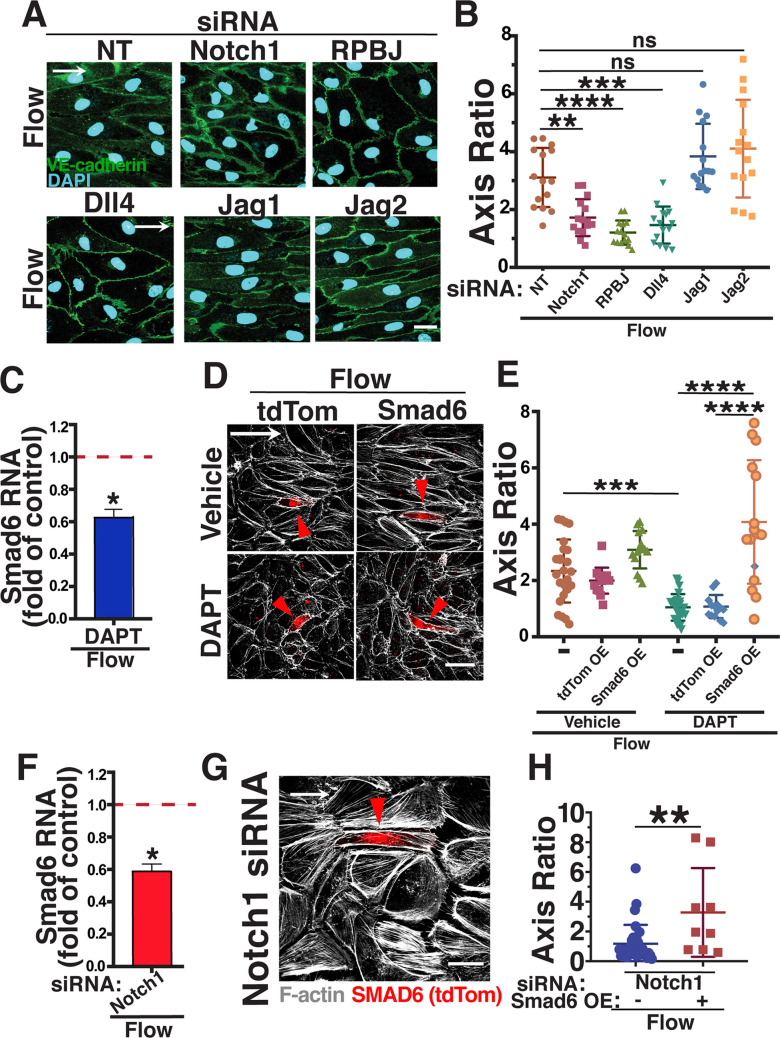
SMAD6 is both a negative regulator of BMP signaling and a transcriptional target of the pathway [38, 44–46]. Consistent with the importance of canonical BMP signaling for endothelial cell flow alignment [47], endothelial cells were misaligned under homeostatic flow when incubated with the BMP inhibitor Crossveinless-2 (CV2) (Supp. Fig. 2A, B). However, since BMP receptors are not identified as direct mechanotransducers of flow-mediated signals, we searched for another pathway more directly linked to mechanotransduction that regulates SMAD6. Notch1 is a direct mechanotransducer of flow-mediated signaling [21, 22], and Notch regulates SMAD6 expression under static (non-flow) conditions [40], leading us to hypothesize that SMAD6 functions downstream of Notch in endothelial cell responses to homeostatic laminar flow. Reduced Notch signaling, via siRNA depletion of Notch1 or by treatment with the γ-secretase inhibitor DAPT which blocks Notch signaling, prevented endothelial cell alignment under homeostatic laminar flow (Fig. 2a, b, Supp. Fig. 2C, D). The misalignment induced by reduced Notch was accompanied by reduced expression of Smad6 RNA relative to controls under flow (Fig. 2c, f), indicating that SMAD6 expression levels are regulated downstream of Notch in endothelial cells and are important for homeostatic flow alignment. Consistent with this idea, ectopic SMAD6 expression rescued the flow-mediated misalignment of endothelial cells downstream of reduced Notch. Upon Notch blockade via DAPT, control endothelial cells transfected with empty vector remained misaligned when exposed to homeostatic laminar flow and did not differ from nearby untransfected cells; in contrast, similarly treated endothelial cells expressing SMAD6 aligned in response to homeostatic laminar flow while nearby untransfected cells remained misaligned (Fig. 2d, e). Endothelial cells over-expressing SMAD6 after Notch1 KD also aligned to homeostatic laminar flow while nearby cells remained misaligned (Fig. 2g, h). Thus SMAD6 over-expression rescued Notch loss-induced endothelial cell misalignment, indicating that SMAD6 is a functional effector of Notch-mediated homeostatic flow alignment in endothelial cells.
Fig. 2.
SMAD6 is downstream of Notch in homeostatic endothelial cell flow-mediated alignment. a Representative panels of HUVEC stained with VE-cadherin (green, junctions) and DAPI (blue, nucleus) under indicated conditions with indicated treatments. White arrow, flow vector. Scale bar, 20 μM. b Quantification of cell axis ratio. Statistical analysis, One-way ANOVA; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001. N = 3, representative experiment shown. c qPCR RNA levels (normalized to vehicle control under flow) of SMAD6 in HUVEC treated with DAPT under flow. Statistical analysis, Student’s t-test; *p ≤ 0.05. d Representative panels of HUVEC stained with Phalloidin (F-actin, white) with indicated treatments and expression constructs (Empty Vector (EV) or SMAD6) under flow conditions. White arrow, flow vector. Red arrowhead, positive HUVEC. Scale bar, 50 μM. e Quantification of cell axis ratio. Statistical analysis, One-way ANOVA; ***p ≤ 0.001; ****p ≤ 0.0001. N = 3, representative experiment shown. f qPCR RNA levels (normalized to vehicle control under flow) in HUVEC treated with Notch1 siRNA under flow. Statistical analysis, Student’s t-test; *p ≤ 0.05. g Representative panels of HUVEC stained with Phalloidin (F-actin, white) with indicated treatments and over-expression construct (SMAD6) under flow conditions. White arrow, flow vector. Red arrowhead, positive EC. Scale bar, 20 μM. h Quantification of cell axis ratio. Statistical analysis, One-way ANOVA; **p ≤ 0.01. N = 3, representative experiment shown
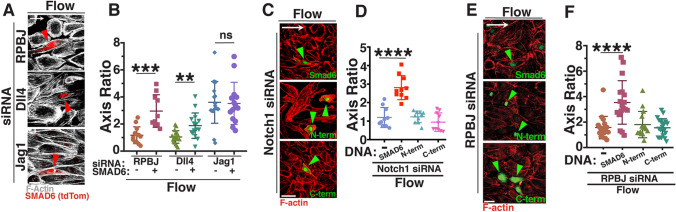
We next asked which Notch signaling components are required for homeostatic endothelial cell flow alignment. RPBJ is a transcriptional co-activator required for canonical downstream Notch signaling, and endothelial cells with reduced levels of RPBJ had reduced expression of Smad6 RNA (Supp. Fig. 2E). Moreover, RPBJ knockdown led to misalignment of both arterial and venous endothelial cells in response to homeostatic laminar flow (Fig. 2a, b; Supp. Figure 2F–I). Several Notch ligands are expressed in endothelial cells and implicated in Notch responses to flow. Reduced levels of Dll4, but not Jagged1 or Jagged2, resulted in endothelial cells misaligned in response to homeostatic laminar flow (Fig. 2a, b; Supp. Fig. 2F, G), indicating that Dll4-mediated activation of Notch1 signaling is important for homeostatic endothelial cell flow alignment via canonical Notch signaling.
The misalignment induced by RPBJ or Dll4 knockdown was rescued by expression of SMAD6 in HUVEC (Fig. 3a, b), and SMAD6 expression also rescued alignment of arterial endothelial cells with homeostatic laminar flow after RPBJ KD (Supp. Fig. 2H, I), suggesting that Notch regulation of SMAD6 expression is an important component of endothelial cell responses to homeostatic flow. The SMAD6 protein consists of two major domains connected by a linker (Supp Fig. 2J); the N-terminal portion includes several arginine residues that are methylated to regulate SMAD6 activity [48], while the C-terminal portion contains the MH2 protein-interacting domain [49]. Since both major domains are required for the regulatory role of SMAD6 [50], we hypothesized that SMAD6 rescue of homeostatic endothelial cell alignment downstream of Notch required full-length SMAD6. Reduced Notch signaling, either via Notch1 or RPBJ depletion, led to endothelial cell misalignment in response to homeostatic flow that was not rescued by expression of constructs encoding only either the N-terminal or C-terminal portion of SMAD6 (Fig. 3c–f). These data indicate that full-length SMAD6 is required to mediate the effects of Notch signaling on homeostatic endothelial cell flow alignment.
Fig. 3.
Full-length SMAD6 rescues Notch loss-induced endothelial cell homeostatic flow mis-alignment. a Representative panels of HUVEC stained with Phalloidin (F-actin, white) with indicated siRNA treatments and SMAD6 expression construct under flow conditions. White arrow, flow vector. Red arrowheads, positive EC. Scale bar, 50 μM. b Quantification of cell axis ratio. Statistical analysis, One-way ANOVA; **p ≤ 0.01; ***p ≤ 0.001; ns not significant. N = 3, representative experiment shown. c Representative panels of HUVEC stained with Phalloidin (F-actin, red) with Notch1 siRNA treatments and expression constructs (full-length SMAD6, N-terminal SMAD6, or C-terminal SMAD6; green) under flow conditions. White arrow, flow vector. Green arrowhead, positive EC. Scale bar, 50 μM. d Quantification of cell axis ratio. Statistical analysis, One-way ANOVA; ****p ≤ 0.0001. N = 3, representative experiment shown. e Representative panels of HUVEC stained with Phalloidin (F-actin, red) with RPBJ siRNA treatment and expression constructs (full-length SMAD6, N-terminal SMAD6, or C-terminal SMAD6; green) under flow conditions. White arrow, flow vector. Green arrowhead, positive EC. Scale bar, 50 μM. f Quantification of cell axis ratio. Statistical analysis, One-way ANOVA; ****p ≤ 0.0001. N = 3, representative experiment shown
SMAD6 regulates endothelial cell proliferation
To better understand the effects of reduced SMAD6 function on flow-mediated endothelial cell responses, we examined the transcriptome of endothelial cells under homeostatic laminar flow relative to non-flow conditions, and with depleted Smad6 levels, via RNA-seq analysis. Pearson Correlation Analysis revealed good correlation between experimental replicates of each condition (Supp. Fig. 3A). Principle Component Analysis (PCA) distinguished transcriptomes of control (non-targeting siRNA) and SMAD6 depleted endothelial cells, and transcriptomes also clustered by flow status (Supp. Fig. 3B). Overall comparisons (33,694 genes) showed that, when binned by flow status, only 1.2% of transcripts were significantly up- or down-regulated with reduced Smad6 levels under non-flow (static) conditions, while 6.9% of transcripts changed with reduced Smad6 levels under homeostatic flow conditions (Supp. Table 1). When binned by depletion condition, control endothelial cells under static vs. flow conditions had 6.7% of transcripts showing expression level changes, while Smad6 KD cells had 8.9% of transcripts changed in static vs. flow conditions. These numbers suggest that the magnitude of flow-mediated changes is greatest in endothelial cells with reduced Smad6 levels, consistent with a role for SMAD6 in endothelial cell flow responses.
Despite the fact that flowed EC with Smad6 KD morphologically resembled non-flowed EC [NT (static) vs. Smad6 KD (flow)], 7.9% of analyzed genes (2655/33,694) differed in relative expression between these gene sets, even more than the 6.7% that differed between static and flow without Smad6 manipulation (Supp. Table 1). Finally, the greatest change in gene expression (11.2%) was seen when comparing flowed normal EC to non-flowed EC with Smad6 KD (Supp. Table 1). We next looked at individual gene sets, and found that a significant number of genes normally flow-responsive (up or down) became non-responsive with Smad6 KD (1041/2262, 46%), and that an even larger group of genes that were normally non-responsive to flow became flow-responsive (up or down) with Smad 6 KD (1782) (Supp. Fig. 3C–F shows top 50 genes/category; Supp Fig. 3G shows overlap). Finally, many genes that appeared concordant in direction of flow-responsiveness between control and Smad6 KD had baseline (non-flow) changes (up or down) that led to expression changes under flow in Smad6 KD relative to control (data not shown). We conclude that reduced SMAD6 levels have multiple impacts on the EC transcriptome under homeostatic laminar flow: (1) dampened expression changes for some flow-responsive genes; (2) about half of flow-responsive genes becoming non-responsive; and (3) aberrant up- and down-regulation of genes usually not responsive to flow. This analysis supports that SMAD6 is a key regulator of endothelial cell flow responses.
Smad6 expression is upregulated in lung EC of adult mice relative to infants [51], suggesting a role in vascular quiescence. Consistent with this idea, Gene Ontogeny (GO) Analysis to identify cellular processes significantly affected by Smad6 depletion revealed that transcripts associated with the cell cycle were enriched in endothelial cells with reduced Smad6 levels, independent of flow status (Supp Fig. 3H). We hypothesized that endothelial cell proliferation is negatively regulated by SMAD6, and we found that expression of the proliferation marker Ki67 was increased with Smad6 depletion under both static and homeostatic flow conditions (Supp Fig. 3I, J). BrdU incorporation, which labels S-phase cells, was increased upon Smad6 depletion in both static conditions and trending upward under flow conditions (Supp Fig. 3K, L). Thus, repression of endothelial cell proliferation that is a prerequisite of vascular quiescence requires SMAD6.
SMAD6 regulates endothelial cell barrier function and junctions
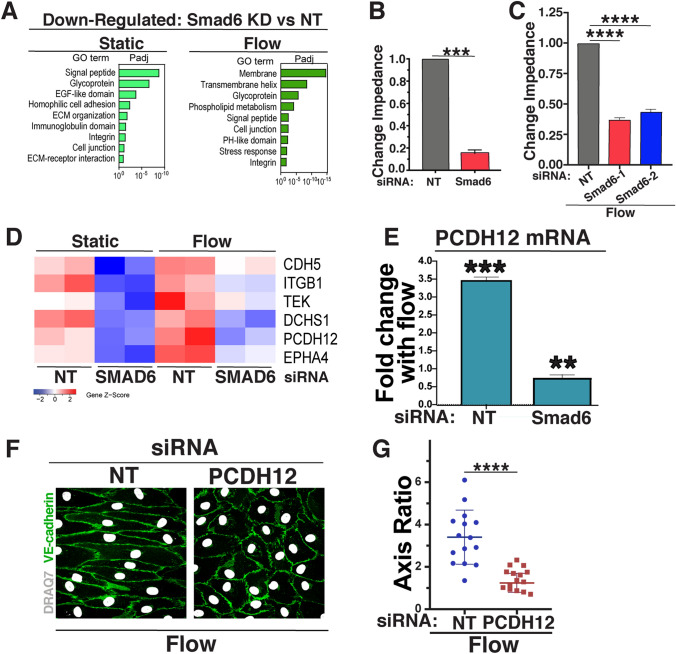
GO Analysis also indicated that expression of genes associated with cell–cell junctions was down-regulated in endothelial cells with reduced Smad6 levels (Fig. 4a). Since SMAD6 regulates junction morphology in the absence of flow [42], we hypothesized that the barrier formed by endothelial cell–cell junctions and important for proper vascular function was compromised by SMAD6 depletion. Endothelial cells with depleted Smad6 levels had reduced barrier function relative to controls, as measured by trans-endothelial electrical impedance (Fig. 4b), and the defective resistance downstream of reduced Smad6 levels was also significant under flow conditions (Fig. 4c).
Fig. 4.
SMAD6 regulates endothelial cell barrier function and cell–cell junction genes under homeostatic flow. a Gene ontology (GO) analysis performed on differentially expressed genes from bulk RNA-seq data sets using DAVID. GO terms significantly enriched (p adjusted < 0.1) in down-regulated are shown. b Change in impedence after 24 h of EC with indicated siRNAs under static (control) conditions, normalized to control. Statistical analysis, One-way ANOVA; ***p ≤ 0.001. c Change in impedence after 72 h of EC with indicated siRNAs under flow, normalized to control. Statistical analysis, One-way ANOVA; ****p ≤ 0.0001. d Heat map showing subset of cell–cell-adhesion genes (see text for details) down-regulated with reduced Smad6 levels under indicated conditions. e qPCR of relative PCDH12 RNA levels under flow relative to static (control) with indicated siRNAs. f Representative panels of HUVEC stained with VE-cadherin (green, junctions) and DAPI (white, nucleus) under flow conditions with indicated treatments. White arrow, flow vector. Scale bar, 50 μM. g Quantification of cell axis ratio. Statistical analysis, student’s t-test; ****p ≤ 0.0001. N = 3, representative experiment shown
To further examine how SMAD6 manipulations affect endothelial cell–cell junctions, we focused on expression differences in cell–cell adhesion and junction genes between control and Smad6 KD endothelial cells under both static and flow conditions. Overlapping genes from these two genelists were identified, and a small subset with significant overall expression levels was examined further. Most junction genes were upregulated with homeostatic laminar flow in controls, and relative expression of a subset of these genes was reduced in endothelial cells depleted for Smad6, regardless of flow status. The net effect was that a group of cell junction genes had reduced expression in Smad6-depleted endothelial cells relative to contols when both were exposed to homeostatic laminar flow (Fig. 4d, compare Flow-NT to Flow-Smad6 KD). Of these, the protocadherin PCDH12 was chosen for further analysis, since PCDH12 is selectively expressed in arterial endothelial cells, and its deletion leads to changes in murine arterial blood pressure, while human mutations are associated with brain arterial calcification [52–54]. Validation of PCDH12 expression changes via qRT-PCR showed significant upregulation of PCDH12 expression with homeostatic laminar flow, and this increase was blunted to 60% of static control levels in flowed endothelial cells with reduced Smad6 levels (Fig. 4e). We next asked whether PCDH12 functions in flow alignment responses, by subjecting HUVEC treated with PCDH12 siRNA to homeostatic laminar flow, and found that PCDH12 depleted endothelial cells failed to align (Fig. 4f, g; Supp Fig. 3M). These results show that SMAD6 is required for full PCDH12 expression in response to flow, and that PCDH12 expression is necessary for proper endothelial cell alignment under homeostatic flow conditions. These findings suggest that SMAD6 regulation of PCDH12 contributes to endothelial cell homeostatic flow-mediated responses downstream of Notch-induced mechanotransduction.
Discussion
This work reveals a functional requirement for SMAD6 in endothelial cell responses to homeostatic laminar flow, which is the shear stress experienced by arterial endothelial cells and is considered atheroprotective. SMAD6 functions downstream of Notch1; since Notch1 is a mechanotransducer, it is likely that shear stress signals transduced by Notch1 signaling mediate flow responses that maintain vascular homeostasis in part through SMAD6. Expression profiling reveals numerous SMAD6-dependent changes in endothelial gene expression under flow, including downregulation of cell cycle/proliferation genes and upregulation of cell-cell adhesion genes, consistent with a role for SMAD6 in regulating the maintenance of endothelial cell quiescence and morphogenesis in response to laminar flow. Thus SMAD6 is a critical transducer of Notch-dependent endothelial cell flow responses that promote the maintenance of vascular homeostasis.
Notch regulation of homeostatic flow responses is linked to its regulation of SMAD6 expression levels. Loss of Notch1 signaling significantly reduced Smad6 RNA levels under homeostatic flow conditions, and restored expression of SMAD6 was sufficient to rescue flow-mediated alignment downstream of loss of Notch signaling. Interestingly, Notch-dependent endothelial cell alignment under homeostatic flow also utilizes the ligand Dll4 and requires Notch-mediated transcription (this study), while initial flow-induced endothelial cell responses require Notch but not its transcriptional activity [22], although subsequent flow responses may depend on Notch transcriptional activity [21]. These findings suggest that equilibration to laminar flow involves a switch from a non-transcriptional to a transcriptional program. Consistent with a the idea of a switch, analysis of endothelial cell junction changes under flow showed immediate effects on VE-cadherin clustering, followed by later junction and cell shape changes [11]. A switch model is also consistent with SMAD6 expression regulation being critical for vascular homeostasis in response to flow, and suggests that endothelial cells may have evolved different mechanisms for an acute response to changes in mechanotransduction vs. equilibration to ongoing mechanotransduction inputs.
SMAD6 functions as a negative regulator of BMP signaling, and canonical BMP signaling also affects flow responses in complex ways, so SMAD6 may regulate flow responses downstream of both Notch and BMP inputs. Alternatively, SMAD6 also has BMP-independent functions in innate immunity and other cellular processes [55–57], so SMAD6 may also affect endothelial cell flow responses in BMP-independent ways. Moreover, it is likely that other pathways involved in endothelial cell flow responses, such as KLF2 and/or KLF4, function upstream or downstream of Notch and SMAD6 [58]. In any case, expression profiling showed that SMAD6 manipulations change transcriptional endothelial cell flow responses in both directions, and more genes change expression when SMAD6 levels are depleted under homeostatic flow conditions (6.9%) vs. non-flow conditions (1.2%), indicating that homeostatic flow amplifies SMAD6-dependent transcriptional differences in endothelial cells. While some of the genes whose expression profiles change with Smad6 manipulations have Smad-binding motifs in their promoters, we hypothesize that most gene expression changes are downstream of initial Smad-binding and indirect, as the readout is homeostatic flow after 72 h of equilibration.
Loss of SMAD6 led to a more “activated” endothelial cell phenotype, with cell cycle/proliferation pathways upregulated and cell–cell junction pathways down-regulated, and these changes were accompanied by significant loss of barrier function. These findings are consistent with SMAD6 regulating the atheroprotective endothelial cell quiescence phenotype that accompanies flow-mediated alignment under homeostatic flow and important for barrier function, and it is also consistent with the hemorrhage phenotype of Smad6−/− mutant embryos [42] and the increase in Smad6 expression in lungs of adult mice relative to infants [51]. Interestingly, cell cycle pathways were also upregulated in endothelial cells with reduced levels of Notch1 [21], consistent with SMAD6 being downstream of Notch activation. Although expression profiling suggests that endothelial cell–cell adhesion changes with reduced SMAD6 are likely to be complex and involve multiple adhesion receptors, the arterial-expressed protocadherin PCDH12 was significantly upregulated by homeostatic laminar flow, and this upregulation was dramatically blunted by Smad6 depletion. Since independent reduction of PCDH12 levels prevents endothelial cell flow-mediated alignment, it is likely that PCDH12 is one SMAD6 target normally upregulated downstream of SMAD6 under homeostatic flow that contributes to endothelial cell barrier function and quiescence. Thus, our data support a model whereby homeostatic flow-mediated mechanotransduction from Notch1 regulates expression of the effector SMAD6, and SMAD6 levels affect endothelial cell alignment, proliferation and barrier function to maintain vascular homeostasis. These findings provide new pathway intersections, and potential new therapeutic targets for diseases such as atherosclerosis that are linked to loss of vascular homeostasis.
Methods
Cell culture
HUVEC (Lonza, #C2519A) and HAEC (Lonza, #C2535) were maintained according to manufacturer’s recommendations and used at passage 2–4. For HUVEC and HAEC culture, EBM-2 (Endothelial Cell Growth Basal Medium-2, Lonza, #CC-3156) was supplemented with EGM-2 SingleQuots Supplements (Lonza, #CC-4176) (called EBM-2+). Main experiments were independently replicated with a different lot of HUVEC, and key experiments were replicated in HAEC at least two independent times. HUVEC and HAEC were certified mycoplasma-free by the UNC Tissue Culture Facility.
Endothelial cell flow experiments
HUVEC and HAEC were plated at 100% confluency in each lane of a µ-Slide VI0.4 (Ibidi, #80601) coated with fibronectin (Millipore Sigma, F2006, 5μg/mL) 4 h prior to the experiment in EGM-2+ medium. After 2 h for cell attachment and spread, slides were washed 3× in flow medium [EBM-2 with 10% FBS (Gibco, #26140-079) and 1× Antibiotic-Antimycotic (Gibco, #15240-062)], and incubated in flow medium for 2 h prior to flow onset. Uniform laminar shear stress was generated by attaching slide chambers to a pump system (Ibidi, #10902) with both the slide and the pump apparatus kept at 37°C and 5% CO2. To reduce cell shearing, flow was applied for 30 min at 5 d/cm2, then 30 min at 10 d/cm2, followed by 72 h at 15 d/cm2. For siRNA knockdown, siRNA incubation was for 30 h prior to plating and flow initiation.
Endothelial cell shear stress measurements
All data presented under “flow” conditions is at 15 d/cm2 laminar flow for 72 h (Ibidi system) and considered to be homeostatic laminar flow. At least 10 cells/condition were measured, and each cell is represented as an individual data point. Experiments were replicated at least three times.
Cell axis ratio
Cell shape and alignment were measured by staining for VE-cadherin or PECAM1 as described below. Each cell size measurements were recorded by measuring total cell length and width using ImageJ. Cell axis ratio was length divided by width, with length the direction of flow.
Nuclear displacement angle
Nuclear displacement angle was measured using the angle measurement tool in Image J. Measured angles were degrees separating a line perpendicular to the flow vector and a line through the nucleus along its long axis (see Supp. Fig. 1C).
Cell polarization
We determined the Golgi or centrosome location relative to the nucleus. HUVEC and HAEC stained for the Golgi (GM130) or the centrosome (TUBGCP2) and nuclei (DAPI) were analyzed by dividing the nucleus into equal thirds and and binning the organelle relative to the flow vector: upstream, middle, or downstream. At least five fields/experiment were quantified per condition.
Endothelial cell transfection
HUVEC (Lonza) were transfected with non-targeting siRNA (NT, Life Technologies, #4390847) or experimental siRNAs [single siRNAs or siRNA pools—Supp. Table 2 (Key Resources)] using the standard Lipofectamine 2000 (Invitrogen, 11668027) manufacturer's protocol. Briefly, for each siRNA to be tested: 24 μl of 10uM siRNA was diluted in 476 μl of opti-MEM media and separately, 24 μl of Lipofectamine 2000 reagent was diluted in 476 μl of opti-MEM medium. Each mixture was incubated separately 5 min at RT, then mixed 15 min at RT. This mixture was added to a 10cm plate of ~70% confluent HUVEC or HAEC in EGM-2+ medium without antibiotics (EBM-2 media, added BulletKit without gentamicin supplement) and incubated 24 h at 37 °C, then incubated for 6 h in fresh non-antibiotic EGM-2+ prior to plating in flow channels to start the experiment. Smad6-tdTomato was transfected into HUVEC as described [40] and provided approximately three to eightfold increase in baseline expression levels.
Immunofluorescence
HUVEC and HAEC were fixed for 10 min in 4% PFA, washed 3× with PBS, then permeabilized for 10 min in 0.1% Triton X-100. Cells were blocked for 30 min at RT in 1% BSA (Millipore Sigma, #A-4503) in PBS, then incubated with primary antibodies (1:100—see Supp. Table 2) in 1% BSA for 45 min at RT. After washing 5× with 1% BSA, samples were incubated with Alexa-fluor-conjugated anti-species secondary antibodies (1:250—see Supp. Table 2) plus Alexa-fluor-conjugated phalloidin (1:50—Invitrogen, #A12379 or #A12381) and DAPI or DRAQ7 (1:300—Sigma #10236276001 or Abcam #ab109202, respectively) in 1% BSA for 30 min at RT. Samples were washed 5× in 1% BSA and mounted by washing 3× in 80% glycerol (Millipore Sigma, #G5516) in PBS. Immunofluorescent imaging was done either immediately after mounting or slides were wrapped in foil at 4°C for up to 2 weeks.
BrdU incorporation was an adaptation [59]. Briefly, HUVEC were incubated with 10 μM BrdU (Millipore Sigma, #B5002) in flow media for the last 90 min of flow and fixed in ice-cold 100% methanol. Samples were blocked for 30 min at RT in 1% BSA, then acid treated as follows: 10 min in cold (4°C) 1N HCl on ice, rinse in 2N HCl and 10 min at RT, then incubated in 10mM citric acid solution (pH 7.4, in 0.2M Na+2HPO4) for 10 min at RT, rinsed with 1% BSA 3× and processed for antibody staining as detailed above with a sheep polyclonal antibody to BrdU (1:100—Abcam, #ab1893), then Alexa Fluor Donkey anti-sheep 594 secondary (1:250—Life Technologies #A-11016), along with DAPI and phalloidin as described above.
All fluorescent imaging was done using an Olympus FV3000 Laser Scanning Confocal Microscope and Flow View software. Olympus OIB file formats were imported into ImageJ using Bio-Formats Importer20 for analysis and quantification.
Quantitative RT-PCR
Primers are listed in Supp. Table 2. cDNA was generated from 1 μg mRNA using iScript reverse transcription kit (Bio-Rad, #1708891) and diluted 1:3 in water. qRT–PCR was performed using iTaq Universal SYBR Green SuperMix (Bio-Rad, #1725121). SYBR Green real-time PCR was performed in triplicate on the Applied Biosystems QuantStudio 6 Flex Real-Time PCR System. For quantification, relative expression of each gene to GAPDH in each sample was calculated by 2^(CT of gene−CT of GAPDH). Statistical significance was determined by one-sample T-test compared to a reference value of onefold change.
RNA sequencing and analysis
RNA was extracted using TRIzol (Invitrogen) from two biological replicates (independent experiments), and TruSeq Stranded mRNA Library Prep Kit (Illumina) was used to prepare cDNA and Illumina libraries for sequencing (HiSeq4000). 2–5 × 107 50-bp paired-end reads per sample were obtained and mapped to human genome GRCh38-1.2.0 downloaded from https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/advanced/references with TopHat/2.1.1 using default settings. Mapping rate was >92% for all samples, and gene expression was determined with Htseq-count/0.6.1 using the union mode (https://htseq.readthedocs.io/en/master/). PCA analysis was performed with the top 400 genes selected by largest weight (loading) contribution to PCs 1, 2 or 3 using the R package SINGuLAR. Differential expression analysis was performed with DESeq2 in R, and lists of differentially expressed genes were obtained (FDR < 0.05). Heat maps were generated using the heatmap.2 function in the ‘gplots’ package in R. Gene ontology analysis was performed using the DAVID functional annotation tool version 6.8 (https://david.ncifcrf.gov/). All gene ontology terms shown in this study have a corrected P value (the “Benjamini” value from DAVID) < 0.1.
Barrier function analysis
Real-time cell analysis (RTCA) experiments
Barrier properties were measured using a commercially available system [xCELLigence Real-Time Cell Analyzer (RTCA)]; Acea Biosciences/Roche Applied Science, Basel, Switzerland). RTCA measures electrical impedance as a readout for the barrier status of cells grown on top of microelectrode coated surfaces. HUVEC were pre-treated with siRNAs or drug for 24 h prior to plating an equal cell number onto the microelectrode surface of the E-plate (E-plate 16, Roche Applied Science). Impedance readings were taken automatically every 5 min for 24 h.
Electric cell-substrate impedance sensing system (ECIS) experiments
Endothelial barrier function analysis under static and flow conditions was performed using impedance-based cell monitoring using ECIS zeta theta (Applied Biophysics) in conjunction with the Ibidi pump system. HUVEC were seeded onto an ibidi flow chamber with 8 microelectrodes on the bottom (ECIS Flow Array 1E). Experiments proceeded exactly as ibidi flow experimental setup, with the addition of impedance readings taken every five min across each electrode for 72 h.
Statistical analysis
All statistical analyses were performed using Prism v8.01 (www.graphpad.com), with an α of 0.05. For two-sample data sets with equal variances (control -v- a single experimental condition) unpaired, two-tailed Student’s t-test was used as reported in figure legends. For data sets with greater than two conditions and equal variances, one-way analysis of variance (ANOVA) with Tukey’s post-hoc test was used as reported in figure legends. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. ns not significant.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Bautch lab members for input and constructive discussion, Dr. Keith Burridge for the use of TEER apparatus, and the UNC High Throughput Sequencing Facility (HTSF) for support. We thank Dr. Suk-Won Jin for constructive comments on the manuscript. The authors declare no competing financial interests.
Author contributions
Dana L Ruter (DLR) and Victoria L Bautch (VLB) conceptualized the work; DLR, Ziqing Liu (ZL), Kimlynn M Ngo (KMN), Shaka X (SX), and Elise J Kidder (EJK) performed and analyzed experiments; DLR and VLB wrote the manuscript; DLR, ZL, KMN, SX, Allison Marvin (AM), Danielle B Buglak (DBB) and VLB reviewed and edited the manuscript; VLB provided study supervision and oversight.
Funding
NIH R01 HL43174, HL116719, HL117256, R35 HL139950 (VLB); NIH-NCI (T32 CA009156), AHA Postdoctoral Fellowship (19POST34380916) (DLR); Integrated Vascular Biology TG (T32HL069768-17), AHA Predoctoral Fellowship (PRE34380887) (DBB); NIH Postbaccalaureate Research Education Program (PREP) (R25GM089569) (SX).
Data availability
The RNA-seq data that support the findings of this study are available in the Gene Expression Omnibus (GEO) under the accession number GSE147036.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Augustin HG, Koh GY. Organotypic vasculature: from descriptive heterogeneity to functional pathophysiology. Science. 2017;357(6353):eaal2379. doi: 10.1126/science.aal2379. [DOI] [PubMed] [Google Scholar]
- 2.Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473(7347):298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chappell JC, Wiley DM, Bautch VL. How blood vessel networks are made and measured. Cell Tissue Organs. 2012;195:94–107. doi: 10.1159/000331398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baeyens N, et al. Endothelial fluid shear stress sensing in vascular health and disease. J Clin Invest. 2016;126(3):821–828. doi: 10.1172/JCI83083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies PF, Zilberberg J, Helmke BP. Spatial microstimuli in endothelial mechanosignaling. Circ Res. 2003;92(4):359–370. doi: 10.1161/01.RES.0000060201.41923.88. [DOI] [PubMed] [Google Scholar]
- 6.Gimbrone MA, et al. Endothelial dysfunction, hemodynamic forces, and atherogenesisa. Annal NY Acad Sci. 2000;902(1):230–240. doi: 10.1111/j.1749-6632.2000.tb06318.x. [DOI] [PubMed] [Google Scholar]
- 7.Hoffman BD, Grashoff C, Schwartz MA. Dynamic molecular processes mediate cellular mechanotransduction. Nature. 2011;475(7356):316–323. doi: 10.1038/nature10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claesson-Welsh L. Vascular permeability—the essentials. Upsala J Med Sci. 2015;120(3):135–143. doi: 10.3109/03009734.2015.1064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Küppers V, et al. Phosphatases and kinases as regulators of the endothelial barrier function. Cell Tissue Res. 2014;355(3):577–586. doi: 10.1007/s00441-014-1812-1. [DOI] [PubMed] [Google Scholar]
- 10.Lampugnani MG, Dejana E, Giampietro C. Vascular endothelial (VE)-cadherin, endothelial adherens junctions, and vascular disease. Cold Spring Harb Perspect Biol. 2018 doi: 10.1101/cshperspect.a029322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seebach J, et al. Regulation of endothelial barrier function during flow-induced conversion to an arterial phenotype. Cardiovasc Res. 2007;75(3):598–607. doi: 10.1016/j.cardiores.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 12.Dieterich P, et al. Quantitative morphodynamics of endothelial cells within confluent cultures in response to fluid shear stress. Biophys J. 2000;79(3):1285–1297. doi: 10.1016/S0006-3495(00)76382-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCue S, et al. Shear stress regulates forward and reverse planar cell polarity of vascular endothelium in vivo and in vitro. Circ Res. 2006;98(7):939–946. doi: 10.1161/01.RES.0000216595.15868.55. [DOI] [PubMed] [Google Scholar]
- 14.Noria S, et al. Transient and steady-state effects of shear stress on endothelial cell adherens junctions. Circ Res. 1999;85(6):504–514. doi: 10.1161/01.res.85.6.504. [DOI] [PubMed] [Google Scholar]
- 15.Schnittler H-J, Püschel B, Drenckhahn D. Role of cadherins and plakoglobin in interendothelial adhesion under resting conditions and shear stress. Am J Physiol Heart Circ Physiol. 1997;273(5):H2396–H2405. doi: 10.1152/ajpheart.1997.273.5.H2396. [DOI] [PubMed] [Google Scholar]
- 16.Shikata Y, et al. Differential effects of shear stress and cyclic stretch on focal adhesion remodeling, site-specific FAK phosphorylation, and small GTPases in human lung endothelial cells. Exp Cell Res. 2005;304(1):40–49. doi: 10.1016/j.yexcr.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Conway DE, Schwartz MA. Flow-dependent cellular mechanotransduction in atherosclerosis. J Cell Sci. 2013;126(22):5101–5109. doi: 10.1242/jcs.138313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies PF, et al. Endothelial heterogeneity associated with regional athero-susceptibility and adaptation to disturbed blood flow in vivo. Semin Thromb Hemost. 2010;36(03):265–275. doi: 10.1055/s-0030-1253449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahn C, Schwartz MA. Mechanotransduction in vascular physiology and atherogenesis. Nat Rev Mol Cell Biol. 2009;10(1):53–62. doi: 10.1038/nrm2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mack JJ, Iruela-Arispe ML. NOTCH regulation of the endothelial cell phenotype. Curr Opin Hematol. 2018;25(3):212–218. doi: 10.1097/MOH.0000000000000425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mack JJ, et al. NOTCH1 is a mechanosensor in adult arteries. Nat Commun. 2017;8(1):1620. doi: 10.1038/s41467-017-01741-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polacheck WJ, et al. A non-canonical Notch complex regulates adherens junctions and vascular barrier function. Nature. 2017;552(7684):258–262. doi: 10.1038/nature24998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang JS, et al. Shear-induced Notch-Cx37-p27 axis arrests endothelial cell cycle to enable arterial specification. Nat Commun. 2017;8(1):2149. doi: 10.1038/s41467-017-01742-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawson ND, et al. Notch signaling is required for arterial-venous differentiation during embryonic vascular development. Development. 2001;128(19):3675–3683. doi: 10.1242/dev.128.19.3675. [DOI] [PubMed] [Google Scholar]
- 25.Pitulescu ME, et al. Dll4 and Notch signalling couples sprouting angiogenesis and artery formation. Nat Cell Biol. 2017;19(8):915–927. doi: 10.1038/ncb3555. [DOI] [PubMed] [Google Scholar]
- 26.Cai J, et al. BMP signaling in vascular diseases. FEBS Lett. 2012;586(14):1993–2002. doi: 10.1016/j.febslet.2012.04.030. [DOI] [PubMed] [Google Scholar]
- 27.Chappell JC, Wiley DM, Bautch VL. Regulation of blood vessel sprouting. Semin Cell Dev Biol. 2011;22(9):1005–1011. doi: 10.1016/j.semcdb.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.David L, Feige J-J, Bailly S. Emerging role of bone morphogenetic proteins in angiogenesis. Cytokine Growth Factor Rev. 2009;20(3):203–212. doi: 10.1016/j.cytogfr.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 29.García de Vinuesa A, et al. BMP signaling in vascular biology and dysfunction. Cytokine Growth Factor Rev. 2016;27:65–79. doi: 10.1016/j.cytogfr.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 30.Gallione C, et al. Overlapping spectra of SMAD4 mutations in juvenile polyposis (JP) and JP–HHT syndrome. Am J Med Genet A. 2010;152A(2):333–339. doi: 10.1002/ajmg.a.33206. [DOI] [PubMed] [Google Scholar]
- 31.Johnson DW, et al. Mutations in the activin receptor–like kinase 1 gene in hereditary haemorrhagic telangiectasia type 2. Nat Genet. 1996;13(2):189–195. doi: 10.1038/ng0696-189. [DOI] [PubMed] [Google Scholar]
- 32.McAllister KA, et al. Endoglin, a TGF-β binding protein of endothelial cells, is the gene for hereditary haemorrhagic telangiectasia type 1. Nat Genet. 1994;8(4):345–351. doi: 10.1038/ng1294-345. [DOI] [PubMed] [Google Scholar]
- 33.Garrido-Martin Eva M, et al. Common and distinctive pathogenetic features of arteriovenous malformations in hereditary hemorrhagic telangiectasia 1 and hereditary hemorrhagic telangiectasia 2 animal models—brief report. Arterioscler Thromb Vasc Biol. 2014;34(10):2232–2236. doi: 10.1161/ATVBAHA.114.303984. [DOI] [PubMed] [Google Scholar]
- 34.Roman BL, Hinck AP. ALK1 signaling in development and disease: new paradigms. Cell Mol Life Sci. 2017;74(24):4539–4560. doi: 10.1007/s00018-017-2636-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tual-Chalot S, Oh P, Arthur H. Mouse models of hereditary haemorrhagic telangiectasia: recent advances and future challenges. Front Genet. 2015 doi: 10.3389/fgene.2015.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Urness LD, Sorensen LK, Li DY. Arteriovenous malformations in mice lacking activin receptor-like kinase-1. Nat Genet. 2000;26:328–331. doi: 10.1038/81634. [DOI] [PubMed] [Google Scholar]
- 37.Itoh F, et al. Synergy and antagonism between Notch and BMP receptor signaling pathways in endothelial cells. EMBO J. 2004;23(3):541–551. doi: 10.1038/sj.emboj.7600065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larrivee B, et al. ALK1 signaling inhibits angiogenesis by cooperating with the Notch pathway. Dev Cell. 2012;22(3):489–500. doi: 10.1016/j.devcel.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rostama B, et al. DLL4/Notch1 and BMP9 interdependent signaling induces human endothelial cell quiescence via P27KIP1 and thrombospondin-1. Arterioscler Thromb Vasc Biol. 2015;35(12):2626–2637. doi: 10.1161/ATVBAHA.115.306541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mouillesseaux KP, et al. Notch regulates BMP responsiveness and lateral branching in vessel networks via SMAD6. Nat Commun. 2016;7:13247. doi: 10.1038/ncomms13247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Topper JN, et al. Vascular MADs: two novel MAD-related genes selectively inducible by flow in human vascular endothelium. Proc Natl Acad Sci. 1997;94(17):9314–9319. doi: 10.1073/pnas.94.17.9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wylie LA, et al. Developmental SMAD6 loss leads to blood vessel hemorrhage and disrupted endothelial cell junctions. Dev Biol. 2018;442(2):199–209. doi: 10.1016/j.ydbio.2018.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galvin KM, et al. A role for Smad6 in development and homeostasis of the cardiovascular system. Nat Genet. 2000;24:171–174. doi: 10.1038/72835. [DOI] [PubMed] [Google Scholar]
- 44.Afrakhte M, et al. Induction of inhibitory Smad6 and Smad7 mRNA by TGF-β family members. Biochem Biophys Res Commun. 1998;249(2):505–511. doi: 10.1006/bbrc.1998.9170. [DOI] [PubMed] [Google Scholar]
- 45.Imamura T, et al. Smad6 inhibits signalling by the TGF-β superfamily. Nature. 1997;389:622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- 46.Ishida W, et al. Smad6 Is a Smad1/5-induced Smad Inhibitor: characterization of bone morphogenetic protein-responsive element in the mouse Smad6 promoter. J Biol Chem. 2000;275(9):6075–6079. doi: 10.1074/jbc.275.9.6075. [DOI] [PubMed] [Google Scholar]
- 47.Poduri A, et al. Endothelial cells respond to the direction of mechanical stimuli through SMAD signaling to regulate coronary artery size. Development. 2017;144(18):3241–3252. doi: 10.1242/dev.150904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu J, et al. Arginine methylation initiates BMP-induced Smad signaling. Mol Cell. 2013;51(1):5–19. doi: 10.1016/j.molcel.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hanyu A, et al. The N domain of Smad7 is essential for specific inhibition of transforming growth factor-beta signaling. J Cell Biol. 2001;155(6):1017–1027. doi: 10.1083/jcb.200106023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakayama T, Berg LK, Christian JL. Dissection of inhibitory Smad proteins: both N- and C-terminal domains are necessary for full activities of xenopus Smad6 and Smad7. Mech Dev. 2001;100(2):251–262. doi: 10.1016/s0925-4773(00)00533-5. [DOI] [PubMed] [Google Scholar]
- 51.Schlereth K, et al. The transcriptomic and epigenetic map of vascular quiescence in the continuous lung endothelium. eLife. 2018;7:e34423. doi: 10.7554/eLife.34423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nicolas G, et al. Brain calcifications and PCDH12 variants. Neurol Genet. 2017;3(4):e166–e166. doi: 10.1212/NXG.0000000000000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Philibert C, et al. Protocadherin-12 deficiency leads to modifications in the structure and function of arteries in mice. Pathol Biol. 2012;60(1):34–40. doi: 10.1016/j.patbio.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 54.Rampon C, et al. Protocadherin 12 (VE-cadherin 2) is expressed in endothelial, trophoblast, and mesangial cells. Exp Cell Res. 2005;302(1):48–60. doi: 10.1016/j.yexcr.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 55.Choi K-C, et al. Smad6 negatively regulates interleukin 1-receptor–Toll-like receptor signaling through direct interaction with the adaptor Pellino-1. Nat Immunol. 2006;7:1057–1065. doi: 10.1038/ni1383. [DOI] [PubMed] [Google Scholar]
- 56.Lee YS, et al. Inhibition of lethal inflammatory responses through the targeting of membrane-associated Toll-like receptor 4 signaling complexes with a Smad6-derived peptide. EMBO Mol Med. 2015;7(5):577–592. doi: 10.15252/emmm.201404653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang T, et al. Smad6 methylation represses NFκB activation and periodontal inflammation. J Dent Res. 2018;97(7):810–819. doi: 10.1177/0022034518755688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.García-Cardeña G, Slegtenhorst BR. Hemodynamic control of endothelial cell fates in development. Ann Rev Cell Dev Biol. 2016;32(1):633–648. doi: 10.1146/annurev-cellbio-100814-125610. [DOI] [PubMed] [Google Scholar]
- 59.Yu Z, et al. Excess centrosomes induce p53-dependent senescence without DNA damage in endothelial cells. FASEB J. 2017;31(10):4295–4304. doi: 10.1096/fj.201601320R. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data that support the findings of this study are available in the Gene Expression Omnibus (GEO) under the accession number GSE147036.