Summary
Truncating variants in exons 33 and 34 of the SNF2-related CREBBP activator protein (SRCAP) gene cause the neurodevelopmental disorder (NDD) Floating-Harbor syndrome (FLHS), characterized by short stature, speech delay, and facial dysmorphism. Here, we present a cohort of 33 individuals with clinical features distinct from FLHS and truncating (mostly de novo) SRCAP variants either proximal (n = 28) or distal (n = 5) to the FLHS locus. Detailed clinical characterization of the proximal SRCAP individuals identified shared characteristics: developmental delay with or without intellectual disability, behavioral and psychiatric problems, non-specific facial features, musculoskeletal issues, and hypotonia. Because FLHS is known to be associated with a unique set of DNA methylation (DNAm) changes in blood, a DNAm signature, we investigated whether there was a distinct signature associated with our affected individuals. A machine-learning model, based on the FLHS DNAm signature, negatively classified all our tested subjects. Comparing proximal variants with typically developing controls, we identified a DNAm signature distinct from the FLHS signature. Based on the DNAm and clinical data, we refer to the condition as “non-FLHS SRCAP-related NDD.” All five distal variants classified negatively using the FLHS DNAm model while two classified positively using the proximal model. This suggests divergent pathogenicity of these variants, though clinically the distal group presented with NDD, similar to the proximal SRCAP group. In summary, for SRCAP, there is a clear relationship between variant location, DNAm profile, and clinical phenotype. These results highlight the power of combined epigenetic, molecular, and clinical studies to identify and characterize genotype-epigenotype-phenotype correlations.
Keywords: SRCAP, Floating-Harbor syndrome, speech delay, intellectual disability, DNA methylation signature, neurodevelopmental disorders, non-FLHS SRCAP-related NDD, genotype-phenotype correlation, nonsense-mediated decay, epigenomics
Introduction
Chromatin remodelers and other epigenetic regulators play a central role in several neurodevelopmental processes.1 Therefore, pathogenic variants in their encoding genes commonly result in specific neurodevelopmental disorders (NDDs).2 SRCAP encodes the SNF2-related CREBBP activator protein (SRCAP [MIM: 611421]), which is an important component of the SRCAP chromatin remodeling complex which regulates transcription of various target genes by incorporating H2A.Z-H2B dimers into nucleosomes.3 Truncating variants in SRCAP cause Floating-Harbor syndrome (FLHS [MIM: 136140]).4 To date, all FLHS-causing variants have been mapped to the last two exons (exons 33 and 34) of SRCAP, upstream of the AT-hook region, in a locus further referred as the “FLHS locus.”4, 5, 6, 7 FLHS is a well-recognizable syndrome characterized by a clinical triad consisting of (1) typical craniofacial features (including triangular face, deep-set eyes, broad nose with bulbous tip, low-hanging columella, short philtrum, and thin lips), (2) expressive and receptive speech and language delay, and (3) proportionate short stature with delayed bone age. In addition, most individuals show some degree of developmental delay (DD) or intellectual disability (ID).8 Although SRCAP is widely studied in the context of FLHS,3 the consequences of SRCAP variants located outside of the FLHS-causing locus are poorly understood, despite their increasing identification through next generation sequencing
Different pathogenic variant types and locations within the same gene can be associated with distinct neurodevelopmental disorders.9, 10, 11, 12, 13 When these occur in epigenetic regualtory genes, DNA methylation (DNAm) is an emerging functional tool to identify and characterize such disorders using specific patterns of genome-wide DNAm in peripheral blood which we call DNAm signatures.14 To date, we and others have described >50 DNAm signatures associated with epigenetic regulatory genes.14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24 These signatures are particularly useful for classifying variants of uncertain significance (VUSs) in these genes as pathogenic or benign as they provide a functional readout of pathogenicity. DNAm signatures can also help to discriminate between related disorders in a differential diagnosis.18,21,23 Truncating variants associated with FLHS are known to be associated with a DNAm signature in blood;24 however, DNAm patterns associated with truncating variants in other regions of SRCAP are not well understood.
In this study, we investigate the effect of variants located in SRCAP proximal or distal to the FLHS locus on DNAm and clinical phenotype. We report that proximal variants are distinguished by an overlapping but distinct DNAm signature from FLHS. CpGs in both signatures map to genes relevant to SRCAP molecular function. We also show that individuals with proximal or distal truncating SRCAP variants are clinically distinct from FLHS, showing DD/ID, normal stature, hypotonia, behavioral and psychiatric problems, non-specific dysmorphic features, and musculoskeletal issues but lacking the classic FLHS triad.
Subjects and methods
Cohort recruitment
We collected 33 unrelated individuals with truncating SRCAP variants outside of the FLHS locus (GenBank: NP_006653.2; p.2329–2748). We describe the location of the variants based on their position relative to the FLHS locus: 28 individuals had truncating variants upstream of the 33rd exon (further referred as proximal SRCAP group) and five individuals had truncating variants downstream of the locus and the first AT-hook domain (further referred as distal SRCAP group). We focused on affected individuals with truncating variants because SRCAP is intolerant only to loss-of-function variants (pLI = 1) but not to missense variants (Z = 2.1). The individuals with SRCAP truncating variants outside the FLHS locus were identified and recruited through a collaborative network of research and diagnostic centers, the Dutch Genome Diagnostic Laboratories (VKGL) variant sharing database,25 and by using MSSNG,26 GeneMatcher,27 DECIPHER,28 and the Simons Simplex Collection (SSC).29 Clinical and molecular data were provided by the individuals’ clinicians, which were compared to data previously described in the literature from cohorts of individuals with FLHS.6, 7, 8 This study was approved by the institutional review board “Commissie Mensgebonden Onderzoek Regio Arnhem-Nijmegen” under number 2011/188.
Variant identification
The SRCAP variants were identified by large gene panel sequencing or whole-exome sequencing in diagnostic settings in clinical laboratories or in research settings. The sequencing data analysis was performed as described previously.30, 31, 32, 33, 34, 35, 36, 37, 38, 39 Variants in SRCAP were annotated using the GRCh37 reference genome and GenBank: NM_006662.3 transcript. For all individuals, a truncating SRCAP variant (25/33 de novo) was considered to be the most likely cause of the individuals’ phenotype. A summary of other molecular findings are provided (Table S1). Variants are reported in ClinVar (accession SCV001477310–SCV001477340).
DNAm research participants
Informed consent for DNAm analysis was obtained from all research participants according to the protocol approved by the Hospital for Sick Children Research Ethics Board (REB# 1000038847). DNAm analysis was performed using a subset of case samples and age- and sex-matched typically developing control subjects (Table S2). The non-FLHS affected individuals were recruited as described above. Additional samples (individuals with FLHS, Rubinstein-Taybi syndrome, and Menke-Hennekam syndrome) used to compare the signature, were obtained from the Hospital for SickChildren Diagnostic Lab. DNA samples obtained from individuals with ID or multiple congenital anomalies with missense variants in SRCAP (n = 4) were also included (Table S2). Control samples were obtained from the Province of Ontario Neurodevelopmental Disorders (POND) Network, The Hospital for Sick Children, and The University of Michigan (Dr. Gregory Hanna).40 “Typically developing” was defined as healthy and developmentally normal by using formal cognitive/behavioral assessments (samples from POND and The University of Michigan) or via physician/parental screening questionnaires (SickKids). Blood DNA samples were available for eight individuals with FLHS, nine with proximal SRCAP variants, and all five individuals with distal SRCAP variants. Case samples harboring SRCAP variants were split into discovery (n = 4/8 FLHS group and n = 5/9 proximal SRCAP group) and validation cohorts (Table S2). Due to low sample numbers, a DNAm signature was not defined for the distal SRCAP samples.
DNAm microarray data processing
DNAm microarray data processing was performed as previously described.19,41 Briefly, whole blood DNA samples were bisulfite converted using the EpiTect Bisulfite Kit (EpiTect PLUS Bisulfite Kit, QIAGEN). Sodium bisulfite converted DNA was then hybridized to the Illumina Infinium Human MethylationEPIC BeadChip to interrogate more than 850,000 CpG sites in the human genome at The Center for Applied Genomics (TCAG), Hospital for Sick Children Research Institute, Toronto, Ontario, Canada. Sample groups were run in five batches, with balanced cases and controls in each batch and on each chip, randomly assigned a chip position. The minfi Bioconductor package in R was used to preprocess data including quality control, Illumina normalization and background subtraction, followed by extraction of β values. Standard quality control metrics in minfi were used, including median intensity QC plots, density plots, and control probe plots; one FLHS sample (EX0537) and one distal SRCAP sample (distal SRCAP #5) had lower median channel intensity values than recommended by minfi standards, so neither was used for signature discovery. Probes with detection flaws (n = 644), probes near SNPs with minor allele frequencies above 1% (n = 29,958), cross-reactive probes20 (n = 41,975), probes with raw beta = 0 or 1 in >0.25% of samples (n = 16), non-CpG probes (n = 2,925), and X and Y chromosome probes (n = 57,969) were removed, so a total of n = 774,580 probes remained for differential methylation analysis.
DNA methylation signatures
To assess DNAm patterns, we identified differentially methylated sites in whole-blood DNA: one comparison for FLHS variants and one for the proximal SRCAP variants by comparing samples from affected individuals to typically developing control subjects. We were not able to generate a signature for the distal SRCAP cases due to the low sample number, so we opted to classify these samples using the FLHS and proximal SRCAP DNAm signatures. For all samples, we applied the blood cell-type proportion estimation tool in minfi based on Illumina EPIC array data from FACS-sorted blood cells.42 As there is a substantial effect of age on DNAm,43 we used only DNA samples from affected individuals and control subjects older than 18 months of age to generate each signature. To match the age and sex distributions of each case group, a distinct (but overlapping) control cohort was used for each comparison (Table S2). We identified differentially methylated sites using limma regression covaried by age, sex, batch, and five of the six predicted cell types (i.e. all but neutrophils). For the FLHS DNAm signature, we compared affected individuals with genetically and clinically confirmed FLHS (n = 4) with matched control samples (n = 35). This identified 464 probes with a Benjamini-Hochberg adjusted p value < 0.05 and a |Δβ| > 0.20. For the proximal SRCAP signature, we compared cases with a proximal truncating variant (n = 5) with matched control samples (n = 32). This identified 347 probes with a Benjamini-Hochberg adjusted p value < 0.05 and a |Δβ| > 0.20 (20% methylation difference).
Machine learning classification models
We developed two machine learning models, one using each DNAm signature. Using the R package caret, CpG sites with correlations equal to or greater than 90% to other signature CpGs were removed as previously described.18 This led to a set of n = 175 non-redundant CpGs from the proximal SRCAP signature and n = 255 from the FLHS signature. Next, we developed two support vector machine (SVM) models with linear kernel trained on the non-redundant CpG sites. Each model was trained using the methylation values for the discovery cases compared to their matched discovery controls, i.e., for the proximal SRCAP SVM model, proximal SRCAP cases (n = 5) versus controls (n = 32); for the FLHS model, FLHS cases (n = 4) versus controls (n = 35). The models were set to “probability” mode to generate SVM scores ranging between 0 and 1 (0%–100%), classifying samples as “positive” (score > 0.5) or “negative” (score < 0.5). To test model specificity, EPIC array data from additional typically developing controls (n = 97) were scored. To test model sensitivity, validation samples (FLHS n = 4; proximal SRCAP n = 4) were classified. We also classified distal SRCAP cases (n = 5) as well as pathogenic CREB-binding protein (CREBBP [MIM: 600140]) and E1A-Binding Protein, 300-KD (EP300 [MIM: 602700]) variants from individuals clinically diagnosed with Rubinstein-Taybi syndrome (n = 10 [MIM: 180849]) or Menke-Hennekam syndrome 1 (n = 1 [MIM: 618332]) to assess the specificity of the model because of their clinical similarity to FLHS and known interactions between the SRCAP and CREBBP/EP300 proteins.
Gene ontology analysis
The lists of CpG positions comprising each DNAm signature were submitted to GREAT (Genomic Regions Enrichment of Annotations Tool) for gene ontology (GO) enrichment analysis.44 Enrichment of each GO term within the gene list was calculated using a foreground/background hypergeometric test over genomic regions, using the set of CpG sites after minfi probe quality control (n = 774,580) as a background set. Overlapping genes were mapped using default GREAT settings with the following exceptions: the cut-off to annotate a CpG as overlapping with a gene (“distal gene mapping” setting) was set to 10 kb, and only enriched terms with two or more gene hits were reported.
Quantitative facial phenotyping
Individuals’ faces were analyzed from provided frontal photos using the hybrid model reported previously.45,46 This model combines two algorithms used for facial recognition (OpenFace47 and Clinical Face Phenotype Space48) to create a 468-dimensional vector of an individuals’ facial features. These vectors are used to calculate the clustering impact factor (CIF) of an analyzed group, which is a measurement of how a group of individuals cluster within a group of control subjects. The control subjects used for the analysis are age-, sex-, and ethnicity-matched individuals with ID, as reported previously.45 A Mann-Whitney U test is used to determine whether the CIF is statistically significantly higher than expected from a random chance. This was performed for all the individuals with FLHS and proximal SRCAP variants, as the number of available individuals with distal SRCAP variants was not sufficient for the analysis. A p value < 0.05 was considered significant. Further, we tested whether the facial photos of individuals with proximal and distal SRCAP variants clustered with the FLHS-affected individuals or control subjects.
Results
Study cohort
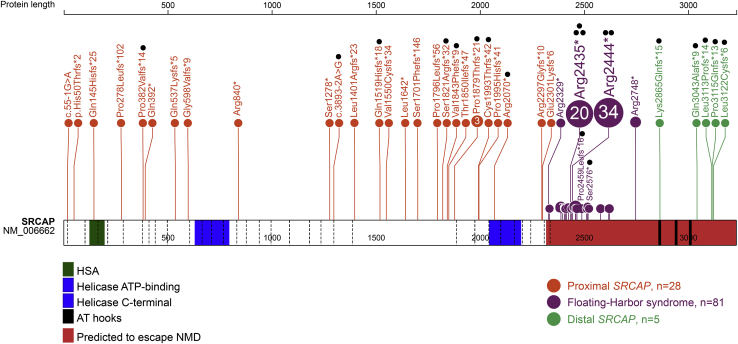
We identified and collected clinical data from 33 unrelated individuals with truncating SRCAP variants outside of the FLHS locus. We defined the boundaries of this locus based on reported FLHS-causing variants, from the most proximal variant located in the 33rd exon (GenBank: NM_006662.3 (SRCAP); c.6985C>T [p.Arg2329∗]) to the most distal variant, located in the 34th exon upstream to the AT-hooks (GenBank: NM_006662.3 (SRCAP); c.8242C>T [p.Arg2748∗]; Figure 1). Genetic investigations were performed for these individuals based on a presentation of neurodevelopmental and/or musculoskeletal issues. None of these individuals had a clinical diagnosis of FLHS. We identified 28 individuals with truncating variants upstream of the 33rd exon (proximal SRCAP group) and five with truncating variants downstream of the FLHS locus and the first AT-hook domain (distal SRCAP group; Figure 1). Most of the variants were de novo (23/28 proximal and 2/5 distal SRCAP). In two individuals, a proximal SRCAP variant was inherited from a healthy parent who was mosaic for the variant and for three individuals in each group, inheritance could not be established because one or both parental samples were unavailable. Unlike FLHS, for which two recurrent variants have been identified in the majority of cases (GenBank: NM_006662.3 (SRCAP); c.7303C>T [p.Arg2435∗] or GenBank: NM_006662.3 (SRCAP); c.7330C>T [p.Arg2444∗]), almost all of the variants in our cohort (31/33) were unique to each individual. The only recurrent variant was GenBank: NM_006662.3 (SRCAP); c.5633dup (p.Pro1879Thrfs∗21). Of the 31 unique truncating variants, 24 are frameshift (including all five distal SRCAP variants), five are nonsense, and two are splice acceptor-site variants (Table S1).
Figure 1.
Spectrum and location of the SRCAP truncating variants
Schematic representation of the SRCAP protein (GenBank: NP_006653.2), its functional domains, and variants used in this study. Exon structure, based on GenBank: NM_006662.3, is provided by dashed lines. Green, HSA-domain (124–196); blue, helicase ATP-binding (630–795) and C-terminal (2,044–2,197) domain; black, AT-hooks (2,857–2,869; 2,936–2,948; 3,004–3,016). Locus, predicted to escape NMD (<55 bp from the last exon/intron junction), is shown in red. Proximal and distal truncating SRCAP variants identified in this study are shown in orange and green, respectively. Floating-Harbor syndrome-causing variants are depicted in purple (recurrent and the most distant variants are specified). Black dots indicate samples used for DNA methylation analysis.
Two distinct DNAm signatures associated with SRCAP
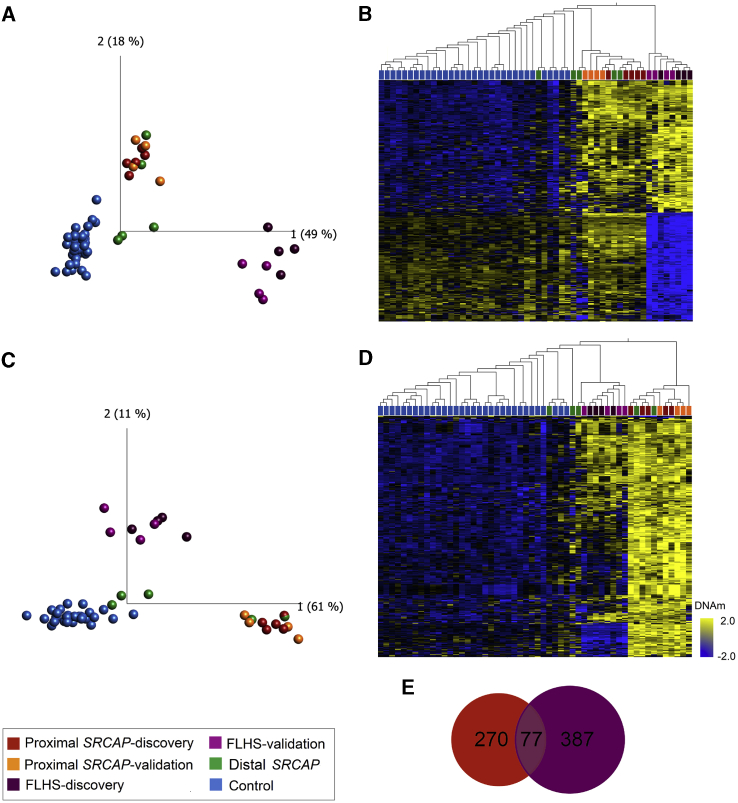
DNAm analysis was performed using a subset of case samples (Table S2). Blood DNA samples were available for eight individuals with FLHS, nine with proximal SRCAP variants, and all five individuals with distal SRCAP variants. Case samples harboring SRCAP variants were split into discovery cohorts to define the signatures (n = 4 FLHS and n = 5 proximal SRCAP) and the remaining samples were used as validation cohorts (n = 4 FLHS and n = 4 proximal SRCAP). We first sought to characterize the proximal and distal variants using the FLHS DNAm signature to determine whether the individuals with different position of truncating SRCAP variants had a comparable DNAm profile. To do this, we generated genome-wide DNAm profiles on FLHS-affected individuals and analyzed them against matched control subjects using limma regression (Table S3). We identified an FLHS DNAm signature of 464 differentially methylated CpG sites (q < 0.05, |Δβ| > 0.20). We then characterized the DNAm profile of our proximal and distal SRCAP cohorts at the FLHS signature sites using principal components analysis (PCA). None of our non-FLHS SRCAP cohort clustered with FLHS-affected individuals using DNAm values at these FLHS signature sites (Figure 2A). Notably, all tested proximal SRCAP subjects (n = 9) and two of the five distal SRCAP subjects clustered together, intermediately between the FLHS-affected subjects and control subjects, while the remaining distal SRCAP subjects clustered closer to the control subjects (Figure 2A). The intermediate profile of the two subjects was also evident using hierarchical clustering (Figure 2B). This intermediate profile was recapitulated when we clustered all samples using CpG sites made available from a previously published FLHS signature24 (Figure S1). In other disorders of the epigenetic machinery, a case group clustering out from control subjects using one signature indicates the possible existence of a second overlapping but distinct signature for these subjects.18,22,49 We hypothesized that the intermediate clustering of the proximal SRCAP subjects was indicative of a unique DNAm signature associated with these cases, overlapping that of FLHS.
Figure 2.
Loss-of-function variants in SRCAP are associated with two distinct but overlapping DNAm signatures based on variant position
(A and B) FLHS DNAm signature: (A) principal components analysis (PCA) and (B) heatmap showing clustering of FLHS discovery subjects (n = 4; dark purple), FLHS validation subjects (n = 4; light purple), proximal SRCAP discovery subjects (n = 5; dark orange), proximal SRCAP validation subjects (n = 4; light orange), distal SRCAP subjects (green) and discovery control subjects (n = 35; blue) using DNAm values at 464 CpG sites in the FLHS DNAm signatures. FLHS subjects clearly segregate from all other samples, while all proximal and some distal SRCAP cases cluster intermediately.
(C and D) Proximal SRCAP DNAm signature: (C) PCA and (D) heatmap showing clustering of the same subjects from (A) and (B) and matched control subjects (n = 32; blue) using DNAm values at 347 CpG sites in the proximal SRCAP DNAm signature. Proximal SRCAP discovery and validation subjects and some distal subjects clearly separate from control subjects, with FLHS subjects clustering intermediately. The heatmap color gradient indicates the normalized DNAm value ranging from −2.0 (blue) to 2.0 (yellow). Euclidean distance metric is used in the heatmap clustering dendrograms.
(E) Venn diagram showing the CpG sites are shared and distinct between the proximal SRCAP and FLHS signatures.
To test this hypothesis, we then compared a discovery cohort of proximal SRCAP subjects (n = 5) with age- and sex-matched typically developing control subjects (n = 32) to identify whether proximal SRCAP variants are indeed associated with a distinct DNAm signature. We identified a proximal SRCAP DNAm signature of 347 differentially methylated CpG sites (q < 0.05, |Δβ| > 0.20; Table S4). Clustering of all samples at the proximal SRCAP DNAm signature sites showed that all proximal SRCAP validation subjects (n = 4) clustered clearly with proximal SRCAP discovery subjects, using PCA (Figure 2C) and hierarchical clustering (Figure 2D). Given the limited sample size and divergent/disparate clustering of the distal SRCAP subjects, we were not able to generate a signature for this group.
Shared and distinct genomic features of SRCAP signatures
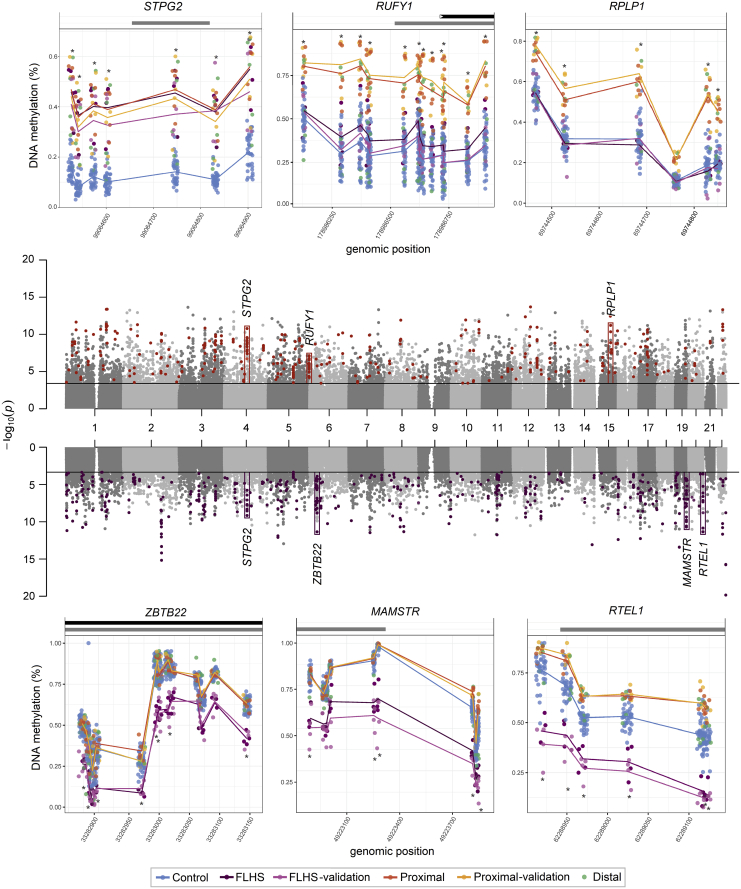
We identified a subset of 77 differentially methylated CpG sites shared by the FLHS and proximal SRCAP DNAm signatures (Table S5). The FLHS signature is composed of both hypo- and hypermethylated sites, with 255/464 (55%) of sites being hypermethylated compared with mean control methylation. In contrast, the proximal SRCAP DNAm signature is predominately composed of hypermethylated sites, with 344/347 (99%) of sites having increased DNAm in affected individuals compared with control subjects. Both DNAm signatures are characterized by clusters of differentially methylated CpGs overlapping the same genomic regions in the same direction of methylation change. Correlated differentially methylated CpGs which are co-localized in regulatory regions have been shown to have direct impacts on gene expression.50 In the proximal SRCAP DNAm signature, 106/347 (30%) signature CpGs are within 100 bp of another signature CpG and 159/347 (46%) are within 1 kb of another signature CpG. In the FLHS signature, 125/464 (27%) signature CpGs are within 100 bp of another signature CpG and 220/464 (47%) are within 1 kb of another signature CpG. All these neighboring differentially methylated CpGs display the same direction of methylation change between affected individuals and control subjects (Table S4). Furthermore, intervening CpG sites that are not in the signature did meet statistical significance (q < 0.05 cutoff) but not the stringent effect size cut-off (|Δβ| > 0.20) used in signature derivation. This is illustrated by plotting the β values for six selected examples of these contiguous regions (Figure 3). This plot also illustrates the different DNAm patterns in each group. For example, seven probes in a CpG island/enhancer region upstream of STPG2 show a similar level of increased DNAm in both groups (Figure 3). In contrast, differentially methylated CpG sites in the proximal SRCAP signature at RUFY1 (MIM: 610327) and RPLP1 (MIM: 180520) have DNAm levels overlapping controls in individuals with FLHS. Finally, hypomethylated RTEL1 (MIM:608833) CpG sites in the FLHS signature have increased DNAm in proximal SRCAP subjects, but these changes do not meet statistical cutoffs for the proximal SRCAP signature. In summary, these findings show that there is a more complex relationship between the two signatures than is captured by simply comparing the number of overlapping CpG sites with stringent cut-off criteria.
Figure 3.
Distribution of differentially methylated regions in SRCAP DNAm signatures
The Manhattan plots (center) show the CpG site p values for each DNAm signature discovery comparison; sites that meet the |Δβ| > 0.20 cut off in the proximal SRCAP signature (upper) are colored orange, those that meet the cut off in the FLHS signature (lower) are colored purple. Six differentially methylated regions are boxed on the Manhattan, with the full plots for sample groups shown above and below. These six illustrate different patterns of DNAm between the proximal SRCAP and FLHS groups. Each is named for the gene body/promoter to which they map. Lines connect the average beta values for each corresponding group. Grey bars above the plots indicate CpG islands, black represent gene bodies. β values for each sample are indicated with mean lines for groups (except the distal group, since two classified positively and three negatively). The CpGs mapping to STGP2 are present in both signatures, RUFY1 and RPLP1 are in the proximal SRCAP signature, and ZBTB2, MAMSTR, and RTEL1 are in the FLHS signature. There is increased DNAm at the RTEL1 sites in the Proximal group, though not a large enough Δβ to be in the proximal SRCAP DNAm signature. In all cases, β values in the discovery and validation cohorts are concordant. RPLP1, RUFY1, and RTEL1 are encoded on the plus strand, STPG2, ZBTB22, and MAMSTR are on the minus strand.
Machine learning classification of samples using SRCAP DNAm signatures
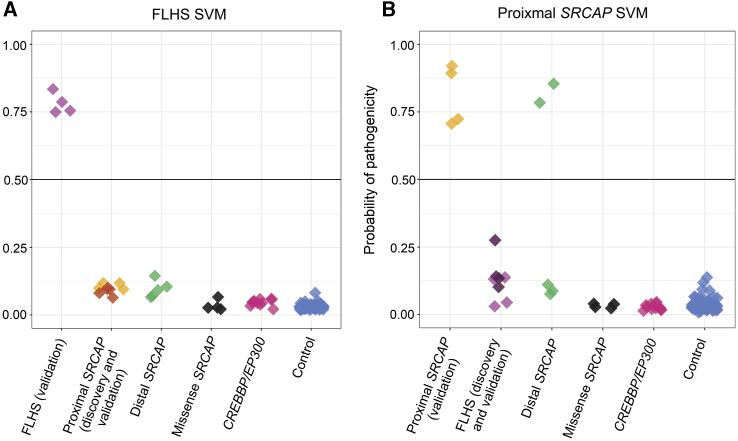
In order to robustly classify each sample, we trained two support vector machine (SVM) models on the DNAm data from the proximal SRCAP and FLHS DNAm signatures, respectively (Figure 4). We then used these models to classify the remaining samples: FLHS validation (n = 4), proximal SRCAP validation (n = 4), distal SRCAP validation (n = 5), and control validation (n = 97). We also obtained samples from individuals with pathogenic variants in CREBBP and EP300 to use for classification. Each model generated a probability of pathogenicity score from 0 to 1 for each sample to which it is applied, with 0.5 being the boundary between a positive and a negative classification (Tables S6 and S7). We validated the FLHS SVM model using FLHS validation subjects (n = 4) which classified positively demonstrating 100% model sensitivity, and additional control subjects (n = 97), all of which classified negatively demonstrating 100% model specificity (Figure 4A). The FLHS model also clearly negatively classified all proximal SRCAP subjects (n = 9). To assess the proximal SRCAP SVM model sensitivity and specificity, we tested the model using proximal SRCAP validation subjects (n = 4), all of which classified positively, demonstrating 100% model sensitivity, and additional control subjects (n = 97) all of which classified negatively, demonstrating 100% specificity. The proximal SRCAP model also clearly classified all FLHS samples negatively (n = 8), despite the intermediate clustering described above (Figure 4B).
Figure 4.
Classification of samples using SVM machine learning models based on each DNAm signature
Sample groups were scored using (A) the FLHS support vector machine (SVM) model and (B) the proximal SRCAP SVM model. FLHS validation subjects (n = 4) classified positively using the FLHS model; similarly proximal SRCAP validation subjects (n = 4) classified positively using the proximal SRCAP model, demonstrating 100% sensitivity of both models. Using the FLHS model, proximal SRCAP subjects (n = 9) and validation control subjects (n = 97) classified negatively, demonstrating 100% specificity of the model. Using the proximal model, FLHS subjects (n = 8) and validation control subjects (n = 97) classified negatively demonstrating 100% specificity of the model. SRCAP missense variants (n = 4) classified negatively using both models, suggesting them to be benign. Distal SRCAP subjects (n = 5) all classified negatively on the FLHS signature, suggesting these subjects do not have FLHS. Two distal SRCAP subjects classified positively on the proximal SRCAP model (distal SRCAP individual #1 and #2) demonstrating concordant DNAm profiles of these subjects with the proximal SRCAP subjects, while three classified negatively (distal SRCAP individual #3, #4, and #5). Subjects with a pathogenic variant in CREBBP (n = 10) or EP300 (n = 1) all classified negatively using both models.
Next, we classified all distal SRCAP subjects (n = 5) using both signature models. Using the FLHS SVM model, all five distal SRCAP subjects classified negatively, demonstrating that these subjects do not have FLHS (Figure 4). Using the proximal SRCAP SVM model, two distal SRCAP subjects classified positively and three classified negatively (Figure 4). This demonstrates that these two distal SRCAP subjects have the same DNAm profile as the proximal SRCAP subjects at the proximal SRCAP DNAm signature sites, possibly indicating that these variants result in the same disorder as the proximal SRCAP variants. Clinical features of the distal SRCAP group are non-specific and do not differentiate the positive from negative classifying case subjects (see below clinical features section). A negative classification typically indicates a benign variant for the condition tested, although it is possible that these variants have another signature distinct from the individuals with proximal SRCAP variants and FLHS. Additionally, all four (Table S2) obtained samples from individuals with different SRCAP missense variants with intellectual disability or multiple congenital anomalies classified negatively using both models (Figure 4). Finally, we classified pathogenic CREBBP and EP300 variants from individuals clinically diagnosed with Rubinstein-Taybi syndrome (n = 10) or Menke-Hennekam syndrome 1 (n = 1), given the similar clinical features to FLHS and the known interactions of the SRCAP complex and CREBBP/EP300; all samples classified negatively using both models (Figure 4).
Gene ontology of SRCAP DNAm signatures
Finally, we assessed the ontology of the genes overlapping the CpGs in each signature using GREAT.44 There were 124 genes overlapping the proximal SRCAP DNAm signature and 148 for the FLHS signature. Both gene lists were characterized by multiple enriched GO terms related to regulation of chromosome structure and DNA repair (Tables S8–S13) which are related to SRCAP molecular function. The top biological process for the proximal SRCAP DNAm signature sites is “DNA recombination” (Table S8). Of the 22 CpGs in this term, 11 are within 400 bp of the transcriptional start site of EID3 (EP300 Interacting Inhibitor of Differentiation 3 [MIM: 612986]). EID3 encodes a transcriptional repressor that is predicted to function by interfering with CREBBP-dependent transcription factors.51 Several of the top terms were related to other DNA metabolism processes like translation (Table S8). For the FLHS signature, several similar terms were identified but with different genes implicated. The top biological processes for the FLHS group were related to regulation of telomeres and neural development (Table S11), largely due to six CpGs in the promoter of RTEL1 (Regulator of Telomere Elongation Helicase 1 [MIM: 608833]). In summary, both signatures map to distinct genes related to SRCAP molecular function.
SRCAP truncating variants proximal to the FLHS locus result in a neurodevelopmental disorder with clinical features distinct from FLHS
We undertook a detailed clinical characterization of our cohort (n = 33; Table S1). We considered the individuals with proximal SRCAP variants (n = 28) to be a distinct group, based on the DNAm results, which we refer to as non-FLHS SRCAP-related NDD. We compared features of the individuals with the non-FLHS SRCAP-related NDD with those with FLHS diagnosis reported in literature and individuals with the distal SRCAP variants (Table 1). Some of the features were not assessed, or not possible to assess in all patients, so the frequency of the features is reported among the number of individuals in whom a feature was evaluated. Most of the individuals presented with neurodevelopmental and behavioral issues. Speech and motor delay were reported in 24/25 individuals, and ID was reported in 13/24 individuals. Among individuals with ID, mostly mild ID was reported although there were a few individuals with moderate or severe ID. Additionally, learning difficulties were reported in four individuals with normal IQ. Autism spectrum disorder (ASD) was present in nearly half of the individuals with the non-FLHS SRCAP-related NDD (10/24). Although DD/ID are also common among individuals with FLHS, ASD is not typically reported as a feature of FLHS (Table 1). Additionally, in the non-FLHS SRCAP-related NDD group, 16/25 individuals had various behavioral problems other than ASD including challenging behavior, anger, anxiety, and attention deficit and hyperactivity disorder (ADHD). Tics, including Tourette syndrome, and psychoses/schizophrenia were each reported in four non-FLHS SRCAP-related NDD subjects.
Table 1.
Phenotype comparison between the non-FLHS SRCAP-related NDD (proximal SRCAP) and distal SRCAP groups with the Floating-Harbor syndrome reported in the literature
|
Features |
Literature |
This study |
||||
|---|---|---|---|---|---|---|
| References | FLHS features reported by Le Goff et al.7(reported/observed) | FLHS features reported by Nikkel et al.8(reported/observed) | FLHS features reported by Seifert et al.6(reported/observed) | Total frequency of FLHS features (reported/observed) | Non-FLHS SRCAP-related NDD (reported/observed) | Distal SRCAP individuals (reported/observed) |
| Individuals included | 6 | 52 | 5 | 63 | 28 | 5 |
| FLHS facial gestalt | 6/6 | 52/52 | 5/5 | 63/63 (100%) | 0/28 (0%) | 1/5 (20%) |
| Delayed bone age | 6/6 | 23/25 | 4/4 | 33/35 (94%) | 0/25 (0%) | 0/5 (0%) |
| Short stature (< − 2SD) | 6/6 | 41/52 | 5/5 | 52/63 (83%) | 0/25 (0%) | 0/5 (0%) |
| OFC < −2SD | 3/6 | 9/43 | 0/5 | 12/54 (22%) | 1/24 (4%) | 0/5 (0%) |
| Macrocephaly | 0/6 | 0/43 | 0/5 | 0/54 (0%) | 2/24 (8%) | 0/5 (0%) |
| Speech delay | 6/6 | 52/52 | 5/5 | 63/63 (100%) | 24/25 (96%) | 4/5 (80%) |
| ID/borderline IQ/special education | 1/6 | 37/41 | 4/5 | 42/52 (81%) | 17/24 (71%) | 5/5 (100%) |
| Schizophrenia/psychoses | NR | NR | NR | not typical | 4/25 (16%) | 0/5 (0%) |
| Behavioral problems | NR | 5/25 (9/32 with ADHD) | 3/5 | 8/30 (27%) | 16/25 (64%) | 2/5 (40%) |
| ASD | NR | NR | NR | not typical | 10/24 (42%) | 2/5 (40%) |
| Seizures | 1/6 | 6/52 | 0/5 | 7/63 (11%) | 3/27 (11%) | 0/5 (0%) |
| Joint hypermobility/musculosceletal problems | NR | NR (4/52 with hip dysplasia) | NR | not typical | 13/27 (48%) | 4/5 (80%) |
| Hypotonia | NR | NR | NR | not typical | 16/25 (64%) | 2/5 (40%) |
| Broad thumbs, broad fingertips, brachydactyly | 5/6 | 10/17 | 5/5 | 20/28 (72%) | 0/28 (0%) | 0/5 (0%) |
| Hearing loss | NR | 9/52 | 2/5 | 9/52 (17%) | 3/26 (12%) | 0/5 (0%) |
| Myopia/hypermetropia | NR | 5/43 | NR | 5/43 (12%) | 11/26 (42%) | 2/5 (40%) |
| Strabismus | NR | 7/43 | NR | 7/43 (16%) | 3/25 (12%) | 0/5 (0%) |
| Cryptorchidism | 0/2 | 5/24 | NR | 5/26 (19%) | 0/13 (0%) | 1/4 (25%) |
| Genitourinary malformations | 1/6 | 7/52 | 0/5 | 8/63 (13%) | 4/28 (14%) | 2/5 (40%) |
| High pitched voice | NR | 8/11 | NR | 8/11 (73%) | 0/26 (0%) | 0/5 (0%) |
NR, not reported; ASD, autism spectrum disorder; ADHD, attention deficit and hyperactivity disorder.
Unlike individuals with FLHS, the individuals with non-FLHS SRCAP-related NDD have normal or tall stature and do not have brachydactyly, broad thumbs, or fingertips. A range of other non-specific skeletal and connective tissue features not typical for FLHS-affected individuals were commonly present. Joint hypermobility is reported in 7/27 individuals while chronic musculoskeletal pain was reported in three adult individuals (proximal SRCAP individuals #10, #11, and #16). In fact, two individuals (proximal SRCAP individuals #10 and #16) were evaluated by rheumatologists regarding the chronic pain without a conclusive diagnosis. As pain is present only in adults, it may develop gradually over time. Additionally, features such as pectus excavatum or carinatum were reported in five individuals and scoliosis in three. Notably, these issues were one of the main reasons for the genetic investigation of three individuals.
In general, the affected organ systems and severity of the non-FLHS SRCAP group phenotype is variable. For example, seizures and genitourinary anomalies were each reported in 3/27 and 3/28 individuals, respectively. Noticeably, three individuals were diagnosed at the neonatal or infant age. At infant age, hypotonia, gastro-esophageal reflux disease, and tracheo/laryngomalacia were reported.
Although the number of individuals with distal SRCAP truncating variants is small (n = 5), they seem to have a similar phenotype to the individuals with the non-FLHS SRCAP-related NDD harboring proximal SRCAP truncating variants. They have developmental delay with mild ID reported in 4/5 individuals. ASD and other behavioral problems were reported in 3/5 and 2/5 individuals, respectively. Additionally, musculoskeletal issues (scoliosis, pectus anomalies, joint hypermobility, and pain) were reported in 4/5 individuals. Importantly, none of the individuals with distal SRCAP variant have short stature and delayed bone age and other typical FLHS features. It is impossible to clinically distinguish the two individuals with distal SRCAP variants who classified positively on the proximal SRCAP DNAm signature (distal SRCAP individual #1 and #2) from the three that classified negatively since the phenotype is non-specific.
Craniofacial dysmorphic features
Facial photos of eight individuals with proximal SRCAP variants and three individuals with distal SRCAP variants (one proximal SRCAP DNAm signature positive and two negative) are shown (Figure 5). Most individuals presented with a long face and long philtrum, prominent forehead, and thin vermilion upper lip with everted lower vermilion and wide mouth. Other typical features were narrow palpebral fissures, epicanthal folds, periorbital fullness, wide nasal bridge, prominent ears, as well as retro- or prognathia. Despite the fact that most individuals presented with some dysmorphic features, these are nonspecific and variable. Notably, none of the individuals that were positive on the proximal SRCAP DNAm signature have a facial gestalt characteristic of FLHS.
Figure 5.
Facial features of individuals with proximal and distal truncating SRCAP variants
Phenotype of nine individuals with the proximal and three individuals with the distal truncating SRCAP gene variants. Photos at age of 2, 8, and 25 years are available for the proximal SRCAP individual #9. Shared facial (non-specific) phenotypic features of the proximal SRCAP group individuals are seen: long face and long, wide philtrum, prominent forehead, thin upper lip vermilion and everted lower vermilion, wide mouth, typical (narrow) palpebral fissures, epicanthal folds, periorbital fullness, wide nasal bridge, prominent ears, and retro- or prognathia. Distal SRCAP individual #4 has some FLHS facial features although he does not have short stature or other typical FLHS features and has Marfanoid habitus with pectus excavatum (similarly to distal SRCAP individual #1).
To objectively evaluate the facial phenotype similarities and differences, we compared clinical photos of 16 individuals with FLHS from the literature and 14 proximal individuals with age-, sex-, and ethnicity-matched control individuals with various non-specific NDDs (one matched individual per tested individual) by utilizing a facial feature recognition algorithm as described previously.46 First, we validated that individuals with FLHS cluster together (p = 0.001) and have a significantly different gestalt than the general NDD cohort, thus confirming that FLHS has a specific and recognizable gestalt. Next, we compared individuals with proximal (n = 14) SRCAP variants with the NDD control subjects. The analysis was not able to identify specific facial features for the group and did not discriminate case subjects from control subjects (p = 0.698). This supports our observation that the individuals with proximal SRCAP variants do not have a typical gestalt. As the number of the distal SRCAP individuals was insufficient (n = 3), we were not able to test whether these individuals have different facial gestalt from the control subjects. Lastly, we compared individuals with proximal and distal SRCAP variants with the individuals with FLHS diagnosis. One individual with distal SRCAP variant (distal SRCAP individual #4) was classified as FLHS. Indeed, this individual has facial features suggestive of FLHS, such as a triangular face with prominent forehead and prominent nose, but he also presented with a Marfanoid habitus and does not have other typical FLHS features. Therefore, we did not classify this individual as FLHS. Surprisingly, one individual with the proximal SRCAP variant (proximal SRCAP individual #18) was also classified by the tool as FLHS, though our clinical observation, both regarding facial and other features, was not in agreement with this. Although facial feature recognition tools can be useful, these data show that they are not always well suited as a diagnostic tool in isolation and should be used in conjunction with other information.
Since the clinical and dysmorphic features of subjects with proximal SRCAP truncating variants are rather non-specific, it is also not possible to phenotypically distinguish proximal SRCAP from distal SRCAP individuals (regardless of proximal SRCAP DNAm signature status).
Discussion
Truncating variants clustering within the last two exons of the SRCAP gene cause FLHS, but the molecular and phenotypic consequences of variants outside this locus have been poorly understood. In this study, we show that truncating variants proximal to the FLHS locus result in a non-FLHS SRCAP-related NDD with behavioral and psychiatric problems, non-specific dysmorphic features, musculoskeletal problems, and hypotonia. In addition, these individuals demonstrate a specific DNAm signature that can be used to positively identify affected individuals and distinguish them from FLHS. These findings support the non-FLHS SRCAP-related NDD as representing a distinct disorder from FLHS. We also show that truncating variants located distally to the known FLHS locus do not cause Floating-Harbor syndrome, and they seem to result in an NDD different from FLHS, but further collection of additional affected individuals is needed to confirm this.
FLHS is characterized clinically by a typical facial gestalt, short stature with delayed bone age, and developmental delay (especially expressive speech delay) with or without mild to moderate ID,8 and is characterized molecularly by a specific DNAm signature.24 We found that none of the 14 tested individuals with a truncating variant outside of the FLHS locus were positive using the FLHS DNAm signature. This led us to identify a distinct DNAm signature in our subjects with proximal SRCAP truncating variants, which demonstrates some overlap with the FLHS signature. The SVM model derived from this signature provided clear positive or negative classifications for all samples (Figure 3). This model positively classified all the proximal SRCAP subjects, and 2/5 distal SRCAP subjects, while none of them were positively classified using our FLHS SVM model. These clear binary categories (positive or negative) demonstrate very different DNAm profiles for each condition and strongly suggest they are distinct disorders. Importantly, a sample from the SSC cohort (13857.p1) with a proximal SRCAP truncating variant (GenBank: NM_006662.3 (SRCAP); c.6409_6419del [p.Asp2137Glufs∗25]) but limited clinical information, classified positively using the proximal SRCAP SVM model and negatively on the FLHS model. Without detailed clinical examination, it is not possible to confirm the diagnosis, but given the recruitment criteria for SSC (children 4–18 years old with confirmed ASD and without severe neurological deficit and with negative ASD family history29) and DNAm classification, we expect that this individual has the non-FLHS SRCAP-related NDD. We also found individuals with pathogenic CREBBP and EP300 variants classified negatively using both (the proximal SRCAP and FLHS) models, demonstrating the ability of these models to discriminate the non-FLHS SRCAP-related NDD and FLHS from clinically and molecularly related disorders.
We found that none of the individuals in our cohort have typical FLHS clinical features. Although DD/ID are seen in both FLHS and our study cohort, the individuals from our cohort (both with proximal SRCAP and distal SRCAP truncating variants) do not have a recognizable facial gestalt nor short stature with delayed bone age and therefore are clinically distinguishable from individuals with FLHS. Moreover, while uncommon among individuals with FLHS, the non-FLHS SRCAP-related NDD individuals commonly have mild to severe psychiatric and behavioral issues, reported as one of the main challenges for these individuals and their families. Additionally, a high proportion of these individuals have various musculoskeletal problems (e.g., pectus anomalies and scoliosis) as well as joint hypermobility/instability reported at younger age and joint pain presenting in adulthood. Coupled with the DNAm data, these clinical differences support the view that the non-FLHS SRCAP-related NDD and FLHS are distinct conditions.
SRCAP is a chromatin remodeler, activating transcription of various genes by depositing the H2A.Z-HSB dimers in promoter regions, targeting ∼10% of promoters.52 Deposition of H2A.Z is an important step for the DNA-end resection required for DNA repair.53 Both sets (proximal SRCAP and FLHS) of the identified signature CpGs map to genes relevant to SRCAP function. Both were enriched for GO terms related to chromosome structure and DNA repair. “DNA recombination” was the top biological processes hit for the proximal SRCAP signature, in part due to EID3, a transcriptional repressor expected to interfere with CREBBP-dependent transcription factors, acting in opposition to SRCAP.51 Hypermethylation and, therefore, possible silencing of this gene in the context of SRCAP haploinsufficiency may further impair CREB-mediated transcription. The FLHS signature was enriched for different genes regulating chromosome structure including RTEL1, which encodes a telomeric DNA helicase that appears to be important during early brain development.54 Some DNA-replication-regulating genes were present in both signatures (e.g., BRCA1). Clusters of signature CpGs have been observed in other signatures as well, but the number and length in the two SRCAP-associated signatures is notable. The CpGs exclusive to each signature can demonstrate DNAm values overlapping controls in the other signature (e.g., RPLP1, MAMSTR) while some can demonstrate values distinct from controls (e.g., RTEL1; Figure 4).
We show that the position of truncating variants in SRCAP determines the phenotype, which likely occurs via different molecular mechanisms. Based on the available evidence, we hypothesize that the phenotype of the individuals with proximal SRCAP variants could be explained by haploinsufficiency. First, all reported variants do not cluster at any specific domain and spread from intron 3 to exon 32 and, therefore, they are expected to undergo nonsense-mediate decay (NMD) or to result in a loss of a significant functional part of the protein if escaping NMD. Second, the gene is intolerant to the loss-of-function variants (pLI = 1), which is suggestive of the gene being haploinsufficient.55 In fact, there are 11 rare loss-of-function variants in gnomAD v.2.1.1., but 7/11 variants have skewed allele balance (20%–35%), which suggests somatic origin of the variants, and two additional frameshift variants seem to be a single complex indel. Therefore, this confirms that individuals with truncating SRCAP variants are depleted from the population database of adults without severe pediatric disorders. Third, Gerundino et al. reported a subject with overlapping features with our cohort with a de novo 186 kb 16p11.2 microdeletion that encompasses SRCAP and eight other genes.56 This individual demonstrates a partial clinical overlap with our proximal SRCAP cohort including facial features, global developmental delay, normal IQ, and behavioral problems (ADHD, inadequate social skills) although the individual’s features of microcephaly and short stature do not overlap;56 however, these additional features could be the result of the other gene deletions in the region. There is currently no evidence that SRCAP missense variants can be pathogenic (based on the low Z score, as well as different phenotype and negative DNAm data of the 4 tested individuals); however, more variants should be tested to reach a definitive conclusion. Based on all these data, we hypothesize that truncating variants upstream of the FLHS locus cause a distinct NDD via SRCAP haploinsufficiency. This would make FLHS and the non-FLHS SRCAP-related NDD analogous to Marshall-Smith syndrome (MIM: 602535) caused by variants in NFIX (MIM: 164005) that escape NMD, and Sotos syndrome 2 (MIM: 614753) caused by variants leading to NFIX haploinsufficiency.57 These conditions have distinct clinical features and are not considered part of a single disorder spectrum.
Most of the FLHS-causing variants are recurrent stop-gain variants which cluster at the 3′ end of the gene predicted to result in a truncated protein that lacks AT-hooks by escaping nonsense-mediated mRNA decay.8 The AT-hooks are necessary for the direct DNA binding by epigenetic regulators.58 It has been shown that the AT-hooks can also serve as nuclear localization signals.59 Therefore, it is hypothesized that FLHS is caused by the (trans) dominant negative effect (i.e., antimorph) of the truncated SRCAP protein which competes with the wild-type protein to form the SRCAP complex, which can result in a mislocalization of the complex.3,4 However, there are currently no functional data available supporting this hypothesis. For MECP2, truncating variants resulting in a complete loss of the second AT-hook do not cause mislocalization of the protein but rather an impaired chromatin binding and altered chromatin conformation, while only mild reduction of activity was shown for a truncating variant downstream to the AT-hook.60 Therefore, we propose that a gain-of-function (i.e., neomorph) mechanism should be investigated as the FLHS causal mechanism.
Various mechanisms of distal truncating variant pathogenesis are possible. Our DNAm data suggest that different distal SRCAP variants have differing effects on the protein function and resulting phenotype. At this time, the DNAm data cannot definitively determine whether the two positive distal SRCAP individuals have the same disorder as the individuals with proximal SRCAP variants. The phenotype of the individuals with the distal truncating SRCAP variants (both positive and negative on the proximal SRCAP DNAm signature) is similar to the individuals with the proximal SRCAP variants; however, the overlapping clinical features cannot be used to conclude whether individuals with proximal and distal truncating SRCAP variants are affected with the same disorder because the phenotype of the non-FLHS SRCAP-related NDD is non-specific. Identification of a distal SRCAP DNAm signature that positively classifies the proximal SRCAP variants will be necessary to determine whether they truly share the same signature and are the same disorder. The three individuals with distal SRCAP variants who classified negatively on the proximal SRCAP DNAm signature are not phenotypically distinct from other non-FLHS subjects. Genetically, there is a notable pattern: the three negative subjects are the most distal, i.e., nearest the end of the gene, while the two positive subjects are closer to the FLHS region and AT-hook domains. It may be that the three negative distal SRCAP frameshift variants escape NMD and, therefore, with the AT-hooks intact, result in a functional protein.12 Unfortunately, no suitable three-dimensional SRCAP complex structure is currently available to evaluate the role of the distal part of the protein. If a functional protein is produced, the clinical features seen in these individuals might be caused by another yet unknown variant. For all these reasons, we currently classify these variants as VUSs.
In addition to the utility of the DNAm data described here to discriminate between SRCAP-related conditions, they demonstrate the robustness and complexities of DNAm signatures. Previous work has suggested that there may be genotype-epigenotype-phenotype correlations for disorders of the epigenetic machinery, i.e., differences in variant location, DNAm signature, and clinical phenotype are mirrored in each other. The degree of overlap between signatures can reflect the degree of clinical overlap between conditions, demonstrating epigenotype-phenotype correlations.14,18,19,23 Genotype-epigenotype correlations have also been reported in ADNP syndrome,61 with variant location reflecting changes in DNAm signatures; however, this study did not find any correlation with differences in clinical features. Recent work in SMARCA2 shows correlations between variant location, DNAm signature, and clinical phenotype.12 For SRCAP, it is clear there is a genotype-epigenotype-phenotype correlation, with different truncating variant locations within the gene associated with distinct clinical presentations and DNAm signatures.
Similar to other recently described novel NDDs, the phenotype of the individuals with non-FLHS SRCAP pathogenic variants is non-specific and clinically not recognizable.12,62 Thus, shared typical phenotypic features, historically used as the basis of the novel syndrome discoveries, cannot be used as the main evidence that individuals are indeed affected by the same disorder. DNA methylation signatures can provide clarity in these cases, demonstrating, as we did here for SRCAP, that individuals with proximal SRCAP truncating variants have the same condition, which is distinct from FLHS. The DNAm data also raise questions for further study, namely the pathogenicity of the distal SRCAP variants. This work illustrates that the partnership of comprehensive clinical assessment and DNAm signature research have great power both to identify conditions and to utilize molecular data to support syndrome delineation and stratification.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
We are grateful to all the study participants and their families and to the many clinicians who recruited them into this study. This work was supported by Canadian Institutes of Health Research (CIHR) grants (IGH-155182 and MOP-126054), the Province of Ontario Neurodevelopmental Disorders (POND) network (IDS-11-02), and McLaughlin Center (MC 2015-16) grants to R.W., by the Dutch Research Council grants to T.K. (015.014.036) and L.E.L.M.V. (015014066), Netherlands Organisation for Health Research and Development grant to T.K. (91718310) and L.E.L.M.V. (843002608, 846002003), by Donders Junior Researcher Grant 2019 to L.E.L.M.V. and B.B.A.d.V., by Estonian Research Council grants to K.Õ. and K.R. (PRG471) and S.P. (PUTJD827, MOBTP175), by a grant of the Raregenomics network, financed by the Dirección de General de Universidades e Investigación de la Comunidad de Madrid (S2017 / BMD-3721) to M.P.-M., and by Deutsche Forschungsgemeinschaft to D.L. (LE4223/1-1). This work contributes toward the goals of the Solve-RD project that has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No 779257 (to H.B. and L.E.L.M.V.). Several authors of this publication are members of the European Reference Network on congenital malformations and rare intellectual disability (ITHACA). We also acknowledge the technical assistance of Khadine Wiltshire, Youliang Lou, and Chunhua Zhao. Thank you as well to Dr. Greg Hanna for contributing blood DNA samples from typically developing control individuals who had undergone cognitive/behavioral assessments. We also acknowledge Dr. Marleen E.H. Simon, Dr. Alanna Strong, Dr. Femke Tammer, and Dr. Bregje van Bon for providing DNA samples of individuals with FLHS and SRCAP missense variants.
Published: April 27, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.04.008.
Contributor Information
David A. Koolen, Email: david.koolen@radboudumc.nl.
Rosanna Weksberg, Email: rweksb@sickkids.ca.
Data and code availability
All genotype and phenotype data supporting findings of this study are available within the manuscript and supplement files. The reported variants are available at the ClinVar database with accession numbers SCV001477310–SCV001477340. The DNAm datasets are not publicly available due to institutional ethics restrictions. R code used is publicly available and cited in the methods.
Web resources
DECIPHER, https://www.deciphergenomics.org
GeneMatcher, https://genematcher.org/
MSSNG, https://research.mss.ng/
OMIM, https://www.omim.org/
ProteinPaint, https://proteinpaint.stjude.org
Simons Simplex Collection, https://www.sfari.org/resource/simons-simplex-collection/
Supplemental information
References
- 1.Kleefstra T., Schenck A., Kramer J.M., van Bokhoven H. The genetics of cognitive epigenetics. Neuropharmacology. 2014;80:83–94. doi: 10.1016/j.neuropharm.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 2.Fahrner J.A., Bjornsson H.T. Mendelian disorders of the epigenetic machinery: postnatal malleability and therapeutic prospects. Hum. Mol. Genet. 2019;28(R2):R254–R264. doi: 10.1093/hmg/ddz174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Messina G., Atterrato M.T., Dimitri P. When chromatin organisation floats astray: the Srcap gene and Floating-Harbor syndrome. J. Med. Genet. 2016;53:793–797. doi: 10.1136/jmedgenet-2016-103842. [DOI] [PubMed] [Google Scholar]
- 4.Hood R.L., Lines M.A., Nikkel S.M., Schwartzentruber J., Beaulieu C., Nowaczyk M.J., Allanson J., Kim C.A., Wieczorek D., Moilanen J.S., FORGE Canada Consortium Mutations in SRCAP, encoding SNF2-related CREBBP activator protein, cause Floating-Harbor syndrome. Am. J. Hum. Genet. 2012;90:308–313. doi: 10.1016/j.ajhg.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kehrer M., Beckmann A., Wyduba J., Finckh U., Dufke A., Gaiser U., Tzschach A. Floating-Harbor syndrome: SRCAP mutations are not restricted to exon 34. Clin. Genet. 2014;85:498–499. doi: 10.1111/cge.12199. [DOI] [PubMed] [Google Scholar]
- 6.Seifert W., Meinecke P., Krüger G., Rossier E., Heinritz W., Wüsthof A., Horn D. Expanded spectrum of exon 33 and 34 mutations in SRCAP and follow-up in patients with Floating-Harbor syndrome. BMC Med. Genet. 2014;15:127. doi: 10.1186/s12881-014-0127-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Goff C., Mahaut C., Bottani A., Doray B., Goldenberg A., Moncla A., Odent S., Nitschke P., Munnich A., Faivre L., Cormier-Daire V. Not all floating-harbor syndrome cases are due to mutations in exon 34 of SRCAP. Hum. Mutat. 2013;34:88–92. doi: 10.1002/humu.22216. [DOI] [PubMed] [Google Scholar]
- 8.Nikkel S.M., Dauber A., de Munnik S., Connolly M., Hood R.L., Caluseriu O., Hurst J., Kini U., Nowaczyk M.J., Afenjar A., FORGE Canada Consortium The phenotype of Floating-Harbor syndrome: clinical characterization of 52 individuals with mutations in exon 34 of SRCAP. Orphanet J. Rare Dis. 2013;8:63. doi: 10.1186/1750-1172-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuvertino S., Stuart H.M., Chandler K.E., Roberts N.A., Armstrong R., Bernardini L., Bhaskar S., Callewaert B., Clayton-Smith J., Davalillo C.H., DDD Study ACTB Loss-of-Function Mutations Result in a Pleiotropic Developmental Disorder. Am. J. Hum. Genet. 2017;101:1021–1033. doi: 10.1016/j.ajhg.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menke L.A., van Belzen M.J., Alders M., Cristofoli F., Ehmke N., Fergelot P., Foster A., Gerkes E.H., Hoffer M.J., Horn D., DDD Study CREBBP mutations in individuals without Rubinstein-Taybi syndrome phenotype. Am. J. Med. Genet. A. 2016;170:2681–2693. doi: 10.1002/ajmg.a.37800. [DOI] [PubMed] [Google Scholar]
- 11.Martinez F., Marín-Reina P., Sanchis-Calvo A., Perez-Aytés A., Oltra S., Roselló M., Mayo S., Monfort S., Pantoja J., Orellana C. Novel mutations of NFIX gene causing Marshall-Smith syndrome or Sotos-like syndrome: one gene, two phenotypes. Pediatr. Res. 2015;78:533–539. doi: 10.1038/pr.2015.135. [DOI] [PubMed] [Google Scholar]
- 12.Cappuccio G., Sayou C., Tanno P.L., Tisserant E., Bruel A.L., Kennani S.E., Sá J., Low K.J., Dias C., Havlovicová M., Telethon Undiagnosed Diseases Program De novo SMARCA2 variants clustered outside the helicase domain cause a new recognizable syndrome with intellectual disability and blepharophimosis distinct from Nicolaides-Baraitser syndrome. Genet. Med. 2020;22:1838–1850. doi: 10.1038/s41436-020-0898-y. [DOI] [PubMed] [Google Scholar]
- 13.Banka S., Sayer R., Breen C., Barton S., Pavaine J., Sheppard S.E., Bedoukian E., Skraban C., Cuddapah V.A., Clayton-Smith J. Genotype-phenotype specificity in Menke-Hennekam syndrome caused by missense variants in exon 30 or 31 of CREBBP. Am. J. Med. Genet. A. 2019;179:1058–1062. doi: 10.1002/ajmg.a.61131. [DOI] [PubMed] [Google Scholar]
- 14.Choufani S., Cytrynbaum C., Chung B.H., Turinsky A.L., Grafodatskaya D., Chen Y.A., Cohen A.S., Dupuis L., Butcher D.T., Siu M.T. NSD1 mutations generate a genome-wide DNA methylation signature. Nat. Commun. 2015;6:10207. doi: 10.1038/ncomms10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aref-Eshghi E., Bend E.G., Colaiacovo S., Caudle M., Chakrabarti R., Napier M., Brick L., Brady L., Carere D.A., Levy M.A. Diagnostic Utility of Genome-wide DNA Methylation Testing in Genetically Unsolved Individuals with Suspected Hereditary Conditions. Am. J. Hum. Genet. 2019;104:685–700. doi: 10.1016/j.ajhg.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aref-Eshghi E., Bend E.G., Hood R.L., Schenkel L.C., Carere D.A., Chakrabarti R., Nagamani S.C.S., Cheung S.W., Campeau P.M., Prasad C. BAFopathies’ DNA methylation epi-signatures demonstrate diagnostic utility and functional continuum of Coffin-Siris and Nicolaides-Baraitser syndromes. Nat. Commun. 2018;9:4885. doi: 10.1038/s41467-018-07193-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aref-Eshghi E., Rodenhiser D.I., Schenkel L.C., Lin H., Skinner C., Ainsworth P., Paré G., Hood R.L., Bulman D.E., Kernohan K.D., Care4Rare Canada Consortium Genomic DNA Methylation Signatures Enable Concurrent Diagnosis and Clinical Genetic Variant Classification in Neurodevelopmental Syndromes. Am. J. Hum. Genet. 2018;102:156–174. doi: 10.1016/j.ajhg.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butcher D.T., Cytrynbaum C., Turinsky A.L., Siu M.T., Inbar-Feigenberg M., Mendoza-Londono R., Chitayat D., Walker S., Machado J., Caluseriu O. CHARGE and Kabuki Syndromes: Gene-Specific DNA Methylation Signatures Identify Epigenetic Mechanisms Linking These Clinically Overlapping Conditions. Am. J. Hum. Genet. 2017;100:773–788. doi: 10.1016/j.ajhg.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chater-Diehl E., Ejaz R., Cytrynbaum C., Siu M.T., Turinsky A., Choufani S., Goodman S.J., Abdul-Rahman O., Bedford M., Dorrani N. New insights into DNA methylation signatures: SMARCA2 variants in Nicolaides-Baraitser syndrome. BMC Med. Genomics. 2019;12:105. doi: 10.1186/s12920-019-0555-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y.A., Choufani S., Grafodatskaya D., Butcher D.T., Ferreira J.C., Weksberg R. Cross-reactive DNA microarray probes lead to false discovery of autosomal sex-associated DNA methylation. Am. J. Hum. Genet. 2012;91:762–764. doi: 10.1016/j.ajhg.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siu M.T., Butcher D.T., Turinsky A.L., Cytrynbaum C., Stavropoulos D.J., Walker S., Caluseriu O., Carter M., Lou Y., Nicolson R. Functional DNA methylation signatures for autism spectrum disorder genomic risk loci: 16p11.2 deletions and CHD8 variants. Clin. Epigenetics. 2019;11:103. doi: 10.1186/s13148-019-0684-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choufani S., Gibson W.T., Turinsky A.L., Chung B.H.Y., Wang T., Garg K., Vitriolo A., Cohen A.S.A., Cyrus S., Goodman S. DNA Methylation Signature for EZH2 Functionally Classifies Sequence Variants in Three PRC2 Complex Genes. Am. J. Hum. Genet. 2020;106:596–610. doi: 10.1016/j.ajhg.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aref-Eshghi E., Kerkhof J., Pedro V.P., Barat-Houari M., Ruiz-Pallares N., Andrau J.-C., Lacombe D., Van-Gils J., Fergelot P., Dubourg C., Groupe DI France Evaluation of DNA Methylation Episignatures for Diagnosis and Phenotype Correlations in 42 Mendelian Neurodevelopmental Disorders. Am. J. Hum. Genet. 2020;106:356–370. doi: 10.1016/j.ajhg.2020.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hood R.L., Schenkel L.C., Nikkel S.M., Ainsworth P.J., Pare G., Boycott K.M., Bulman D.E., Sadikovic B. The defining DNA methylation signature of Floating-Harbor Syndrome. Sci. Rep. 2016;6:38803. doi: 10.1038/srep38803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fokkema I.F.A.C., van der Velde K.J., Slofstra M.K., Ruivenkamp C.A.L., Vogel M.J., Pfundt R., Blok M.J., Lekanne Deprez R.H., Waisfisz Q., Abbott K.M. Dutch genome diagnostic laboratories accelerated and improved variant interpretation and increased accuracy by sharing data. Hum. Mutat. 2019;40:2230–2238. doi: 10.1002/humu.23896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.C Yuen R.K., Merico D., Bookman M., L Howe J., Thiruvahindrapuram B., Patel R.V., Whitney J., Deflaux N., Bingham J., Wang Z. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci. 2017;20:602–611. doi: 10.1038/nn.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischbach G.D., Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–195. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 30.Neveling K., Feenstra I., Gilissen C., Hoefsloot L.H., Kamsteeg E.J., Mensenkamp A.R., Rodenburg R.J., Yntema H.G., Spruijt L., Vermeer S. A post-hoc comparison of the utility of sanger sequencing and exome sequencing for the diagnosis of heterogeneous diseases. Hum. Mutat. 2013;34:1721–1726. doi: 10.1002/humu.22450. [DOI] [PubMed] [Google Scholar]
- 31.Guillen Sacoto M.J., Tchasovnikarova I.A., Torti E., Forster C., Andrew E.H., Anselm I., Baranano K.W., Briere L.C., Cohen J.S., Craigen W.J., Undiagnosed Diseases Network De Novo Variants in the ATPase Module of MORC2 Cause a Neurodevelopmental Disorder with Growth Retardation and Variable Craniofacial Dysmorphism. Am. J. Hum. Genet. 2020;107:352–363. doi: 10.1016/j.ajhg.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frederiksen A.L., Larsen M.J., Brusgaard K., Novack D.V., Knudsen P.J., Schrøder H.D., Qiu W., Eckhardt C., McAlister W.H., Kassem M. Neonatal High Bone Mass With First Mutation of the NF-κB Complex: Heterozygous De Novo Missense (p.Asp512Ser) RELA (Rela/p65) J. Bone Miner. Res. 2016;31:163–172. doi: 10.1002/jbmr.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holla Ø.L., Busk Ø.L., Tveten K., Hilmarsen H.T., Strand L., Høyer H., Bakken A., Skjelbred C.F., Braathen G.J. Clinical exome sequencing – Norwegian findings. Tidsskr. Nor. Laegeforen. 2015;135:1833–1837. doi: 10.4045/tidsskr.14.1442. [DOI] [PubMed] [Google Scholar]
- 34.Haer-Wigman L., van Zelst-Stams W.A., Pfundt R., van den Born L.I., Klaver C.C., Verheij J.B., Hoyng C.B., Breuning M.H., Boon C.J., Kievit A.J. Diagnostic exome sequencing in 266 Dutch patients with visual impairment. Eur. J. Hum. Genet. 2017;25:591–599. doi: 10.1038/ejhg.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cappuccio G., Pinelli M., Torella A., Alagia M., Auricchio R., Staiano A., Nigro V., Brunetti-Pierri N., TUDP Expanding the phenotype of DST-related disorder: A case report suggesting a genotype/phenotype correlation. Am. J. Med. Genet. A. 2017;173:2743–2746. doi: 10.1002/ajmg.a.38367. [DOI] [PubMed] [Google Scholar]
- 36.Hempel M., Cremer K., Ockeloen C.W., Lichtenbelt K.D., Herkert J.C., Denecke J., Haack T.B., Zink A.M., Becker J., Wohlleber E. De Novo Mutations in CHAMP1 Cause Intellectual Disability with Severe Speech Impairment. Am. J. Hum. Genet. 2015;97:493–500. doi: 10.1016/j.ajhg.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Sluijs P.J., Aten E., Barge-Schaapveld D.Q.C.M., Bijlsma E.K., Bökenkamp-Gramann R., Donker Kaat L., van Doorn R., van de Putte D.F., van Haeringen A., Ten Harkel A.D.J. Putting genome-wide sequencing in neonates into perspective. Genet. Med. 2019;21:1074–1082. doi: 10.1038/s41436-018-0293-0. [DOI] [PubMed] [Google Scholar]
- 38.Terhal P.A., Vlaar J.M., Middelkamp S., Nievelstein R.A.J., Nikkels P.G.J., Ross J., Créton M., Bos J.W., Voskuil-Kerkhof E.S.M., Cuppen E. Biallelic variants in POLR3GL cause endosteal hyperostosis and oligodontia. Eur. J. Hum. Genet. 2020;28:31–39. doi: 10.1038/s41431-019-0427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pajusalu S., Kahre T., Roomere H., Murumets Ü., Roht L., Simenson K., Reimand T., Õunap K. Large gene panel sequencing in clinical diagnostics-results from 501 consecutive cases. Clin. Genet. 2018;93:78–83. doi: 10.1111/cge.13031. [DOI] [PubMed] [Google Scholar]
- 40.Hanna G.L., Liu Y., Isaacs Y.E., Ayoub A.M., Torres J.J., O’Hara N.B., Gehring W.J. Withdrawn/Depressed Behaviors and Error-Related Brain Activity in Youth With Obsessive-Compulsive Disorder. J. Am. Acad. Child Adolesc. Psychiatry. 2016;55:906–913.e2. doi: 10.1016/j.jaac.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodman S.J., Cytrynbaum C., Chung B.H.-Y., Chater-Diehl E., Aziz C., Turinsky A.L., Kellam B., Keller M., Ko J.M., Caluseriu O. EHMT1 pathogenic variants and 9q34.3 microdeletions share altered DNA methylation patterns in patients with Kleefstra syndrome. J. Transl. Genet. Genom. 2020;4:144–158. [Google Scholar]
- 42.Salas L.A., Koestler D.C., Butler R.A., Hansen H.M., Wiencke J.K., Kelsey K.T., Christensen B.C. An optimized library for reference-based deconvolution of whole-blood biospecimens assayed using the Illumina HumanMethylationEPIC BeadArray. Genome Biol. 2018;19:64. doi: 10.1186/s13059-018-1448-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Donk R., Jansen S., Schuurs-Hoeijmakers J.H.M., Koolen D.A., Goltstein L.C.M.J., Hoischen A., Brunner H.G., Kemmeren P., Nellåker C., Vissers L.E.L.M. Next-generation phenotyping using computer vision algorithms in rare genomic neurodevelopmental disorders. Genet. Med. 2019;21:1719–1725. doi: 10.1038/s41436-018-0404-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diets I.J., van der Donk R., Baltrunaite K., Waanders E., Reijnders M.R.F., Dingemans A.J.M., Pfundt R., Vulto-van Silfhout A.T., Wiel L., Gilissen C. De Novo and Inherited Pathogenic Variants in KDM3B Cause Intellectual Disability, Short Stature, and Facial Dysmorphism. Am. J. Hum. Genet. 2019;104:758–766. doi: 10.1016/j.ajhg.2019.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baltrusaitis T., Zadeh A., Lim Y., Morency L.-P. 2018. OpenFace 2.0: Facial Behavior Analysis Toolkit. [Google Scholar]
- 48.Ferry Q., Steinberg J., Webber C., FitzPatrick D.R., Ponting C.P., Zisserman A., Nellåker C. Diagnostically relevant facial gestalt information from ordinary photos. eLife. 2014;3:e02020. doi: 10.7554/eLife.02020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turinsky A.L., Choufani S., Lu K., Liu D., Mashouri P., Min D., Weksberg R., Brudno M. EpigenCentral: Portal for DNA methylation data analysis and classification in rare diseases. Hum. Mutat. 2020 doi: 10.1002/humu.24076. Published online July 5, 2020. [DOI] [PubMed] [Google Scholar]
- 50.Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 51.Båvner A., Matthews J., Sanyal S., Gustafsson J.A., Treuter E. EID3 is a novel EID family member and an inhibitor of CBP-dependent co-activation. Nucleic Acids Res. 2005;33:3561–3569. doi: 10.1093/nar/gki667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong M.M., Cox L.K., Chrivia J.C. The chromatin remodeling protein, SRCAP, is critical for deposition of the histone variant H2A.Z at promoters. J. Biol. Chem. 2007;282:26132–26139. doi: 10.1074/jbc.M703418200. [DOI] [PubMed] [Google Scholar]
- 53.Dong S., Han J., Chen H., Liu T., Huen M.S.Y., Yang Y., Guo C., Huang J. The human SRCAP chromatin remodeling complex promotes DNA-end resection. Curr. Biol. 2014;24:2097–2110. doi: 10.1016/j.cub.2014.07.081. [DOI] [PubMed] [Google Scholar]
- 54.Le Guen T., Jullien L., Touzot F., Schertzer M., Gaillard L., Perderiset M., Carpentier W., Nitschke P., Picard C., Couillault G. Human RTEL1 deficiency causes Hoyeraal-Hreidarsson syndrome with short telomeres and genome instability. Hum. Mol. Genet. 2013;22:3239–3249. doi: 10.1093/hmg/ddt178. [DOI] [PubMed] [Google Scholar]
- 55.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., Genome Aggregation Database Consortium The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gerundino F., Marseglia G., Pescucci C., Pelo E., Benelli M., Giachini C., Federighi B., Antonelli C., Torricelli F. 16p11.2 de novo microdeletion encompassing SRCAP gene in a patient with speech impairment, global developmental delay and behavioural problems. Eur. J. Med. Genet. 2014;57:649–653. doi: 10.1016/j.ejmg.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 57.Malan V., Rajan D., Thomas S., Shaw A.C., Louis Dit Picard H., Layet V., Till M., van Haeringen A., Mortier G., Nampoothiri S. Distinct effects of allelic NFIX mutations on nonsense-mediated mRNA decay engender either a Sotos-like or a Marshall-Smith syndrome. Am. J. Hum. Genet. 2010;87:189–198. doi: 10.1016/j.ajhg.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodríguez J., Mosquera J., Couceiro J.R., Vázquez M.E., Mascareñas J.L. The AT-Hook motif as a versatile minor groove anchor for promoting DNA binding of transcription factor fragments. Chem. Sci. (Camb.) 2015;6:4767–4771. doi: 10.1039/c5sc01415h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cattaruzzi G., Altamura S., Tessari M.A., Rustighi A., Giancotti V., Pucillo C., Manfioletti G. The second AT-hook of the architectural transcription factor HMGA2 is determinant for nuclear localization and function. Nucleic Acids Res. 2007;35:1751–1760. doi: 10.1093/nar/gkl1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baker S.A., Chen L., Wilkins A.D., Yu P., Lichtarge O., Zoghbi H.Y. An AT-hook domain in MeCP2 determines the clinical course of Rett syndrome and related disorders. Cell. 2013;152:984–996. doi: 10.1016/j.cell.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bend E.G., Aref-Eshghi E., Everman D.B., Rogers R.C., Cathey S.S., Prijoles E.J., Lyons M.J., Davis H., Clarkson K., Gripp K.W. Gene domain-specific DNA methylation episignatures highlight distinct molecular entities of ADNP syndrome. Clin. Epigenetics. 2019;11:64. doi: 10.1186/s13148-019-0658-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vissers L.E.L.M., Kalvakuri S., de Boer E., Geuer S., Oud M., van Outersterp I., Kwint M., Witmond M., Kersten S., Polla D.L., DDD Study De Novo Variants in CNOT1, a Central Component of the CCR4-NOT Complex Involved in Gene Expression and RNA and Protein Stability, Cause Neurodevelopmental Delay. Am. J. Hum. Genet. 2020;107:164–172. doi: 10.1016/j.ajhg.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genotype and phenotype data supporting findings of this study are available within the manuscript and supplement files. The reported variants are available at the ClinVar database with accession numbers SCV001477310–SCV001477340. The DNAm datasets are not publicly available due to institutional ethics restrictions. R code used is publicly available and cited in the methods.