Summary
ANKRD17 is an ankyrin repeat-containing protein thought to play a role in cell cycle progression, whose ortholog in Drosophila functions in the Hippo pathway as a co-factor of Yorkie. Here, we delineate a neurodevelopmental disorder caused by de novo heterozygous ANKRD17 variants. The mutational spectrum of this cohort of 34 individuals from 32 families is highly suggestive of haploinsufficiency as the underlying mechanism of disease, with 21 truncating or essential splice site variants, 9 missense variants, 1 in-frame insertion-deletion, and 1 microdeletion (1.16 Mb). Consequently, our data indicate that loss of ANKRD17 is likely the main cause of phenotypes previously associated with large multi-gene chromosomal aberrations of the 4q13.3 region. Protein modeling suggests that most of the missense variants disrupt the stability of the ankyrin repeats through alteration of core structural residues. The major phenotypic characteristic of our cohort is a variable degree of developmental delay/intellectual disability, particularly affecting speech, while additional features include growth failure, feeding difficulties, non-specific MRI abnormalities, epilepsy and/or abnormal EEG, predisposition to recurrent infections (mostly bacterial), ophthalmological abnormalities, gait/balance disturbance, and joint hypermobility. Moreover, many individuals shared similar dysmorphic facial features. Analysis of single-cell RNA-seq data from the developing human telencephalon indicated ANKRD17 expression at multiple stages of neurogenesis, adding further evidence to the assertion that damaging ANKRD17 variants cause a neurodevelopmental disorder.
Keywords: ANKRD17, ankyrin repeats, intellectual disability, neurodevelopmental syndrome, speech delay, dysmorphism, Hippo pathway, Yorkie, Mask
Main text
Ankyrin repeat domain 17 (ANKRD17) belongs to a protein family characterized by the presence of ankyrin repeats, one of the most widespread structural motifs in eukaryotes. Each Ankyrin repeat consists of approximately 33 amino acids, with multiple repeats organized into linear arrays that typically serve as protein-protein interaction surfaces. The motif is found in many proteins with a wide variety of functions including transcriptional regulation, cytoskeletal organization, and signal transduction.1 ANKRD17 (MIM: 615929) is widely expressed2,3 and encodes for a protein containing two distinct clusters of ankyrin repeats within its amino-terminal half, and a KH domain in its carboxy-terminal half (Figure 1). Homozygous Ankrd17 deficiency in mice results in abnormal vascular maturation, hemorrhage, and lethality by embryonic day (E) 11.5,3 so the role of this gene at subsequent developmental stages has not been studied. The Drosophila ortholog of ANKRD17, Mask, is required for tissue growth, acting as a co-factor to the Yorkie transcriptional coactivator, an effector of the Hippo pathway.4,5 In vitro studies suggest that ANKRD17 also interacts with cyclin E/CDK2 and stimulates cell cycle progression, with overexpression promoting S phase entry and depletion resulting in inhibition of DNA replication.2 In addition, a role for ANKRD17 in both anti-bacterial immunity, via the NOD1- and NOD2-mediated immune responses,6 and anti-viral immunity, via the RIG-1-like receptor-mediated signaling pathway,7 has been proposed. ANKRD17 is highly intolerant to loss of function in the human population, with a gnomAD pLI score (probability that the gene is intolerant to loss of function) of 1.0 and an observed/expected LoF score of 0.02 (90% CI 0.01–0.06). There is also an increased constraint against missense variation, with a positive z score (for deviation of observed from expected number of variants) of 5.36 in gnomAD (with a higher z score reflecting intolerance to missense variation).8 These gene-wide constraint metrics do not necessarily capture the variability in regional constraint since some regions within a gene are more highly conserved than others. However, ANKRD17 is one of a small subset of genes having multiple constrained coding regions in the 99th percentile or higher (measured as length of coding sequence without gnomAD variation).9
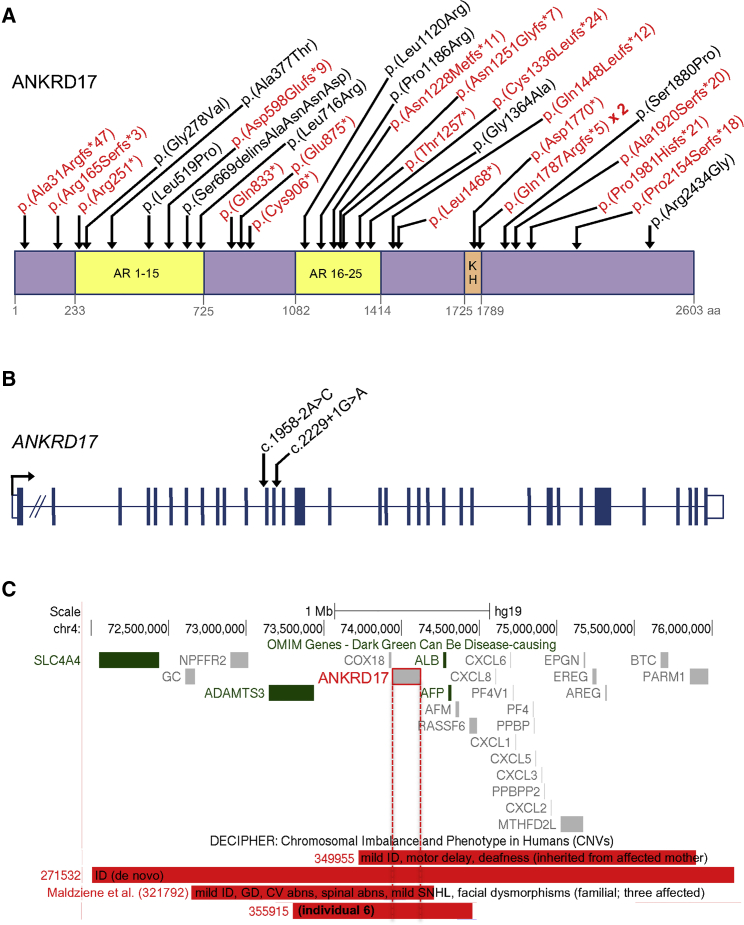
Figure 1.
Distribution of pathogenic ANKRD17 variants and 4q13.3 deletions
(A) Variants affecting coding sequence. Truncating variants are in red. Domain boundaries are based on Uniprot entry O75179. AR, ankyrin repeats; KH, K Homology domain.
(B) Variants affecting essential splice sites. Exon-intron structure drawn approximately to scale (apart from intron 1), according to GenBank: NM_032217.4.
(C) Deletions of the region containing ANKRD17 on chromosome 4q13.3. Among the disease-associated OMIM genes in the interval (those in green), the mode of inheritance or associated phenotype is not compatible with that of the deletions shown. Only deletions under 5 Mb from the DECIPHER database are shown. For DECIPHER individuals 349955 and 271532, publicly available clinical information is listed; individual 321792 is one of three affected individuals of the previously described familial case10 (the phenotypes listed on the figure are a summary of all three family members); and individual 355915 corresponds to individual 6 in the present report (see Table S1 for details). ID, intellectual disability; GD, growth delay; CV, cardiovascular; abns, abnormalities; SNHL, sensorineural hearing loss.
Here, we report 34 individuals with heterozygous pathogenic variants or a microdeletion of ANKRD17 (GenBank: NM_032217.4) and presenting with neurodevelopmental features, ascertained through an international collaborative effort utilizing GeneMatcher11 and DECIPHER.12 In total there were 19 females and 15 males, with an age range of 4 months to 34 years. Apart from one familial case (an affected mother and son) and a set of monozygotic female twins, all affected individuals were simplex cases. After obtaining written informed consent for either diagnostic or institutional review board-approved research sequencing, all individuals were enrolled for whole-exome or whole-genome sequencing (with the exception of individual 6, whose microdeletion was identified on array-CGH) according to standard protocols, details of which can be found in the supplemental information. Consent for publication of images was obtained from parents or legal guardians.
The mutational spectrum of our cohort is shown in Table 1 and Figure 1. Of the variants identified in the 32 probands, there were a total of 22 likely to result in loss of function, comprising 7 nonsense variants, 12 small deletions or duplications resulting in frameshifts, 2 essential splice site variants, and 1 large multi-gene deletion. The latter was a de novo 1.16 Mb microdeletion, identified by array-CGH, encompassing ANKRD17 and six other protein-coding genes in individual 6, who presented with intellectual disability (ID), absent speech, and dysmorphism (Figure 1C). The six other genes in the deletion comprised three for which bi-allelic loss of function is implicated in unrelated autosomal-recessive disease (ADAMTS3 [MIM: 605011], ALB [MIM: 103600], and AFP [MIM: 104150]) and three not known to be disease associated, each with a pLI of 0 (COX18 [MIM: 610428], AFM [MIM: 104150], and RASSF6 [MIM: 612620]). ANKRD17 was therefore determined to be the most likely candidate gene within the deleted region. None of the variants identified in our cohort have previously been described in gnomAD,8 with the exception of two cases: c.90dup (p.Ala31Argfs∗47) (individual 18), reported once in gnomAD v.2.1.1, although only as a filtered variant, i.e, it did not meet gnomAD’s quality control requirements, and c.2497C>T (p.Gln833∗) (individual 22), also reported once. The distribution of truncating ANKRD17 variants appeared random along the protein, although one de novo frameshift, c.5360–5363del (p.Gln1787Argfs∗5), was recurrent in two unrelated individuals. There were nine probands with de novo ANKRD17 missense variants, and one with an in-frame deletion-insertion, all of which affect highly conserved amino acids (see below).
Table 1.
ANKRD17 variants
| Individual No. | Technique | Chr4(GRCh37)a | NM_032217.4 (ANKRD17) | Amino acid | Exon | Inheritance | Predicted effect | gnomADballele count (frequency) | SIFT | Poly-Phen-2 | CADD scorec | Sanger confirmation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | trio WES | g.74008486T>G | c.1958−2A>C | N/A | intron 11 | de novo | splicing | 0 | N/A | N/A | N/A | + |
| 2 | trio WES | g.73984502C>G | c.4091G>C | p.Gly1364Ala | 22 | de novo | missense | 0 | tolerated | probably damaging | 27.7 | + |
| 3 | trio WES | g.73979568_73979571del | c.4341_4344del | p.Gln1448Leufs∗12 | 24 | de novo | frameshift | 0 | N/A | N/A | N/A | Q |
| 4 | trio WES | g.73990763A>C | c.3359T>G | p.Leu1120Arg | 18 | de novo | missense | 0 | deleterious | benign | 25.4 | Q |
| 5 | trio WES | g.74014541A>G | c.1556T>C | p.Leu519Pro | 8 | de novo | missense | 0 | deleterious | probably damaging | 26.2 | Q |
| 6 | array-CGH | 4q13.3 (73303180-74459331) | N/A | N/A | N/A | de novo | large deletion | – | N/A | N/A | N/A | N/A |
| 7 | trio WES | g.74005615G>T | c.2718C>A | p.Cys906∗ | 15 | de novo | nonsense | 0 | N/A | N/A | N/A | + |
| 8 | trio WES | g.73957982_73957985del | c.5360_5363del | p.Gln1787Argfs∗5 | 29 | de novo | frameshift | 0 | N/A | N/A | N/A | + |
| 9 | trio WES | g.73959817_73959823del | c.5304_5310del | p.Asp1770∗ | 28 | de novo | nonsense | 0 | N/A | N/A | N/A | Q |
| 10 | trio WES | g.73957707A>G | c.5638T>C | p.Ser1880Pro | 29 | de novo | missense | 0 | tolerated | probably damaging | 26.3 | Q |
| 11d | trio WGS | g.74005710C>A | c.2623G>T | p.Glu875∗ | 15 | inherited | nonsense | 0 | N/A | N/A | N/A | + |
| 12d | WGS | g.74005710C>A | c.2623G>T | p.Glu875∗ | 15 | ND | nonsense | 0 | N/A | N/A | N/A | + |
| 13 | trio WES | g.74007958C>T | c.2229+1G>A | N/A | intron 13 | de novo | splicing | 0 | N/A | N/A | N/A | Q |
| 14 | trio WES | g.73957590dup | c.5756dup | p.Ala1920Serfs∗20 | 29 | de novo | frameshift | 0 | N/A | N/A | N/A | + |
| 15 | trio WES | g.73956886_73956887del | c.6460_6461del | p.Pro2154Serfs∗18 | 29 | de novo | frameshift | 0 | N/A | N/A | N/A | + |
| 16 | trio WES | g.74021837G>A | c.751C>T | p.Arg251∗ | 4 | suspected paternal mosaicism | nonsense | 0 | N/A | N/A | N/A | + |
| 17 | trio WGS | g.73968263A>C | c.4403T>G | p.Leu1468∗ | 25 | de novo | nonsense | 0 | N/A | N/A | N/A | + |
| 18 | trio WES | g.74124302dup | c.90dup | p.Ala31Argfs∗47 | 1 | de novo | frameshift | 1 (0.000018)e | N/A | N/A | N/A | Q |
| 19 | trio WES | g.74008436_74008437delinsTCATTATTAGC | c.2005_2006delinsGCTAATAATGA | p.Ser669delinsAlaAsnAsnAsp | 12 | de novo | in-frame indel | 0 | N/A | N/A | N/A | + |
| 20 | duo WES | g.74043148_74043149delinsG | c.495_496delinsC | p.Arg165Serfs∗3 | 2 | absent in mother, father unavailable | frameshift | 0 | N/A | N/A | N/A | + |
| 21 | WES | g.73985897del | c.4007del | p.Cys1336Leufs∗24 | 21 | ND | frameshift | 0 | N/A | N/A | N/A | + |
| 22 | trio WES | g.74005836G>A | c.2497C>T | p.Gln833∗ | 15 | de novo | nonsense | 1 (0.0000040) | N/A | N/A | N/A | + |
| 23 | trio WES | g.73986689_73986696delinsGTCC | c.3751_3758delinsGGAC | p.Asn1251Glyfs∗7 | 20 | de novo | frameshift | 0 | N/A | N/A | N/A | Q |
| 24 | trio WES | g.73957982_73957985del | c.5360_5363del | p.Gln1787Argfs∗5 | 29 | de novo | frameshift | 0 | N/A | N/A | N/A | Q |
| 25 | trio WES | g.73957398_73957404del | c.5942_5948del | p.Pro1981Hisfs∗21 | 29 | de novo | frameshift | 0 | N/A | N/A | N/A | Q |
| 26 | trio WGS | g.74021755C>A | c.833G>T | p.Gly278Val | 4 | de novo | missense | 0 | deleterious | probably damaging | 31 | Q |
| 27 | trio WGS | g.73944467G>C | c.7300C>G | p.Arg2434Gly | 31 | de novo | missense | 0 | deleterious | probably damaging | 26 | Q |
| 28 | trio WGS | g.74012557dup | c.1793dup | p.Asp598Glufs∗9 | 10 | de novo | frameshift | 0 | N/A | N/A | N/A | Q |
| 29 | trio WES | g.73987412G>C | c.3557C>G | p.Pro1186Arg | 19 | de novo | missense | 0 | deleterious | probably damaging | 27.6 | Q |
| 30f | trio WES | g.74019702C>T | c.1129G>A | p.Ala377Thr | 6 | de novo | missense | 0 | tolerated | probably damaging | 26.8 | Q |
| 31f | trio WES | g.74019702C>T | c.1129G>A | p.Ala377Thr | 6 | de novo | missense | 0 | tolerated | probably damaging | 26.8 | Q |
| 32 | trio WES | g.74008041A>C | c.2147T>G | p.Leu716Arg | 13 | de novo | missense | 0 | deleterious | probably damaging | 28.6 | Q |
| 33 | trio WES | g.73986678_73986681del | c.3769_3772del | p.Thr1257∗ | 20 | de novo | nonsense | 0 | N/A | N/A | N/A | + |
| 34 | trio WES | g.73986765del | c.3683del | p.Asn1228Metfs∗11 | 20 | de novo | frameshift | 0 | N/A | N/A | N/A | + |
N/A, not applicable; Q quality criteria met, see Table S2 for details.
Mutation nomenclature was verified using Mutalyzer.
Refers to total allele counts and total frequencies in gnomAD dataset v2.1.1.
CADDv1.4 scores range from 1 to 99, with a higher score indicating greater deleteriousness.
Individual 11 is the offspring of individual 12.
Quality criteria not met for variant to be included in gnomAD (failed random forest filters).
Individuals 30 and 31 are monozygotic twins.
The ANKRD17 variants were shown to be de novo in 29 of the 34 individuals. On trio WGS, the truncating variant in individual 11 was found to be inherited from a parent (individual 12) with intellectual abilities in the borderline range (FSIQ 74). This individual had been schooled in the special education system and had a history of depression and anxiety. There was no suspicion of mosaicism in individual 12 (reference / alternate read ratio of 27/39). We were unable to determine whether her variant was de novo, due to lack of availability of parental DNA. The nonsense variant identified in individual 16 was present in paternal blood DNA at a frequency of five reads out of a total read count of 135 (3.7%), suggestive of low-level somatic mosaicism. This father was healthy, non-dysmorphic, and of normal intelligence. In individual 20, the variant was absent in maternal DNA but paternal DNA was unavailable for testing. Inheritance status could not be determined in individual 21 due to lack of parental DNA.
The frequencies of phenotypic characteristics are found in Table 2. The counts on which these frequencies were calculated are found in Table S1. More detailed phenotypic descriptions can be found in Table S2. Global developmental delay (DD)/ID was the most common feature, affecting 31 individuals. The severity of DD/ID was variable, with 19 individuals in the moderate to severe range and 12 in the mild or borderline range. The individuals with typical intellectual functioning were individual 22, an 11-year-old male with autism spectrum disorder (ASD) and behavioral difficulties, and individual 31, a 25-year-old female with renal agenesis, scoliosis, and a history of delayed speech but who is now independent and in the workforce. Individual 34 is only 4 months old and had severe feeding difficulties but reportedly normal development. Her age precludes assessment of her developmental and cognitive status. Speech development was reported as delayed in 29 individuals, including 6 with absent speech (no meaningful words) and 4 who used fewer than 10 words meaningfully (all over the age of 4 years). Absence of speech was observed in individuals with varying degrees of DD/ID, including one individual in the borderline and one in the mild range of ID. Detailed neuropsychiatric evaluation was available in five individuals (see Table S2) and confirmed a discrepancy between verbal IQ and performance IQ (verbal IQ < performance IQ) in three of these individuals. Less commonly reported neurodevelopmental phenotypes include ASD (n = 8) and ADHD (n = 4).
Table 2.
Frequencies of phenotypic characteristics of individuals with ANKRD17 variants
| Frequency | |
|---|---|
| Sex | F = 19, M = 15 |
| Growth | |
| Height < −2 SD | 12/31 |
| Weight < −2 SD | 9/30 |
| OFC < −2 SD | 7/31 |
| OFC > 2 SD | 4/31 |
| Development | |
| DD or ID | 31/34 |
| severe | 7 |
| moderate | 12 |
| mild | 5 |
| borderline | 7 |
| Motor delay | 20/29 |
| Speech delaya | 29/32 |
| Other | ASD, n = 8; ADHD, n = 4 |
| Neurology | |
| Epilepsy | 9/33 |
| Abnormal EEG | 10/23 |
| Brain MRI abnormalities | 11/23 |
| Gait or balance abnormalities | 9/25 |
| Spasticity or hypertonia | 4/26 |
| Other | |
| Recurrent infections | 11/33 |
| Feeding problems | 11/27 |
| Palate abnormalities | 3/34 |
| Hypermobility | 9/29 |
| Ophthalmological abnormalities | 13/23 |
| Miscellaneous | |
| Minor digital anomalies | 6 |
| Genitourinary abnormalities | 5 |
| Pigmentary abnormalities | 4 |
| Scoliosis | 3 |
| Abnormal bone mineralization | 2 |
| Prominent blood vessels | 2 |
ADHD, attention deficit hyperactivity disorder; ASD, autism spectrum disorder
For details see Table S1
Neonatal growth parameters were normal in the majority of individuals (Table S2) but postnatal growth failure was a feature of almost half of the individuals (height < −2 SD in n = 12 and weight < −2 SD in n = 9). One individual with marked growth failure (individual 3, height −3.8 SD) was under treatment with growth hormone (GH), although GH stimulation testing was normal. Feeding difficulties, especially reduced oral intake, were reported at some stage in 11 individuals, 5 of whom required G-tube nutritional supplementation. Postnatal microcephaly (OFC < −2SD) was noted in seven individuals, and macrocephaly in four (one of these individuals, however, also harbored a pathogenic de novo NSD1 variant (GenBank: NM_022455.4, c.2615T>G [p.Leu872∗]). Epilepsy was reported in nine individuals (individuals 1, 2, 16, 19, 21, 25, 27, 28, and 33), with an age of onset of under 2 years for five individuals (individuals 1, 2, 16, 19, and 25). Focal seizures with secondary generalization was the most common seizure subtype, present in five individuals (individuals 1, 2, 21, 25, and 27). One individual had Lennox-Gastaut epilepsy (individual 16), one had tonic seizures with head deviation (individual 19), one had mixed myoclonic and tonic-clonic epilepsy (individual 33), and another a mixture of tonic-clonic and absence seizures (individual 28). Seizures were well controlled (less frequent than every 2 years) in five individuals (individuals 2, 21, 25, 28, and 33), all of whom were on three or fewer anti-epileptic drugs (AEDs). Moderate control, with seizures every 2–3 months, was reported in individual 1, who was on Valproate monotherapy. Two individuals had refractory epilepsy during at least parts of their disease course—individual 19 who had frequent tonic seizures in infancy that resolved with topiramate monotherapy and individual 16 who had multiple seizures every day despite three AEDs. Further details of epilepsy phenotype, including previously trialled AEDs, are noted in Table S2. There were four individuals without epilepsy in whom an abnormal EEG was recorded. Other neurological features include poor balance and/or abnormal gait (9/25) and peripheral spasticity (4/26, one of whom one was microcephalic). Neuroimaging abnormalities were identified in 11 of the 23 individuals in whom an MRI was recorded. Abnormalities include decreased white matter volume (individuals 14, 16, and 18), thinning of the corpus callosum (individuals 14 and 19), optic nerve hypoplasia (individuals 18 and 19), a localized hyperintensity (individuals 7 and 31), right temporal sclerosis (individual 16), dilated Virchow-Robin spaces (individual 6), periventricular nodular heterotopia (individual 30), and an arachnoid (individual 24) and pineal cyst (individual 16). Ophthalmological abnormalities were reported in 13/23 individuals.
There were nine individuals with recurrent bacterial infections, one with recurrent viral infections, and one individual with recurrent infections that were both viral and bacterial. The source of bacterial infection was primarily the upper and lower respiratory system and the middle ear (nine individuals) and in some cases required hospitalization. Two individuals were on low-dose prophylactic antibiotics for recurrent otitis media or respiratory tract infections. Notably, individual 26 had a history of pseudomonas and methicillin-resistant staphylococcal aureus (MRSA) infection on his toes. Immunology assessments were recorded in five individuals, details of which can be found in Table S2, with no obvious immunodeficiency identified in these individuals. Generalized joint hypermobility was reported in 9/29 individuals. Notably, there were two individuals with cleft palate in the context of Pierre Robin sequence (PRS) and another with cleft lip and palate. Other infrequent features include minor digital anomalies (n = 6), genitourinary abnormalities (n = 5, of whom three had unilateral renal agenesis), abnormal skin pigmentation (n = 4), scoliosis (n = 3), abnormality of bone mineralization (n = 2), and cutaneous prominence of blood vessels (n = 2).
Figure 2 shows the facial features of individuals with the ANKRD17-related neurodevelopmental disorder. Key dysmorphic features include a triangular-shaped face found in 10 of the 24 individuals for whom photos were available with a high anterior hairline (19/24), eyes which are either deep-set (5/24) or almond shaped (8/24) with periorbital fullness (6/24), thick nasal alae and flared nostrils (9/24), full cheeks (7/24), and a thin upper lip (12/24). The degree of dysmorphism was variable, with several individuals (particularly individuals 8 and 10) presenting with only subtle dysmorphic characteristics. Persistence of the high anterior hairline, periorbital fullness, and full cheeks into adulthood is demonstrated in individual 12 (age 30 years) and individual 25 (age 34 years).
Figure 2.
Dysmorphic facial features of the ANKRD17-related disorder
Physical characteristics include a triangular face (I1, 4, 5, 6, 9, 15, 22, 30, 31, and 33), high anterior hairline (I1-10, 12, 15, 18, 25, 29, 30, 31, 32, and 33), deep set (I2, 3, 6, 7, 30) or almond-shaped (I1, 4, 5, 12, 15, 22, 29, and 33) eyes with periorbital fullness (I1, 3, 4, 5, 8, 12), full cheeks (I2, 6, 7, 12, 18, 26, and 29), thick alae nasi with flared nostrils (I2, 3, 5, 6, 8, 9, 12, 25, 31), and a thin upper lip (I1, 3, 4, 5, 9, 10, 11, 15, 22, 26, 30, and 31).
A number of diagnoses had been considered in several individuals prior to the identification of an ANKRD17 variant, including SATB2-associated syndrome (MIM: 612313) in individual 5 who presented with PRS, triangular facies and speech delay, and Floating-Harbour syndrome (MIM: 136140) in individual 9 who presented with marked short stature (height < −3 SD), microcephaly (head circumference < −2.5 SD), dysmorphic features, and borderline ID. This highlights the phenotypic overlap of the ANKRD17-related disorder with a number of other genetic syndromes, notably those with expressive language delay. In our cohort, significant speech delay was reported in most individuals (n = 29) even in those with IQ in the borderline range. The finding that verbal IQ was reduced relative to performance IQ in three of the five individuals for whom deep neuropsychological phenotyping was available adds further evidence to our observation that expressive language is particularly affected in this disorder.
All nine missense variants identified in our cohort occur at amino acids that are very highly conserved across vertebrate ANKRD17 orthologs (Figure S1). The human genome has one closely related ANKRD17 paralog, ANKHD1 (MIM: 610500), with 60% overall amino acid identity (GenBank: NP_115593.3 versus NP_060217.1; Needleman-Wunsch global alignment) and the same protein domain organization, i.e., two ankyrin repeat arrays made up of 15 and 10 repeats, followed by a KH domain. Indeed, the ankyrin repeat arrays have 93%–95% amino acid identity between ANKRD17 and ANKHD1. For the nine ANKRD17 missense variants, alignment with the corresponding region of ANKHD1 indicated conservation of the affected amino acid in each case (Figure S1). Furthermore, alignment of the regions containing the seven ANKRD17 missense variants that fall in the ankyrin repeats, with the corresponding regions of Mask, the Drosophila ortholog of vertebrate ANKRD17/ANKHD1, also indicated conservation of each (Figure S1). The presence of two missense variants, c.5638T>C (p.Ser1880Pro) and c.7300C>G (p.Arg2434Gly), outside of known functional domains, yet falling on residues conserved not only across ANKRD17 orthologs but also conserved at the equivalent position in ANKHD1, highlights the likely functional significance of these previously unexplored regions of each protein (Figure S1).
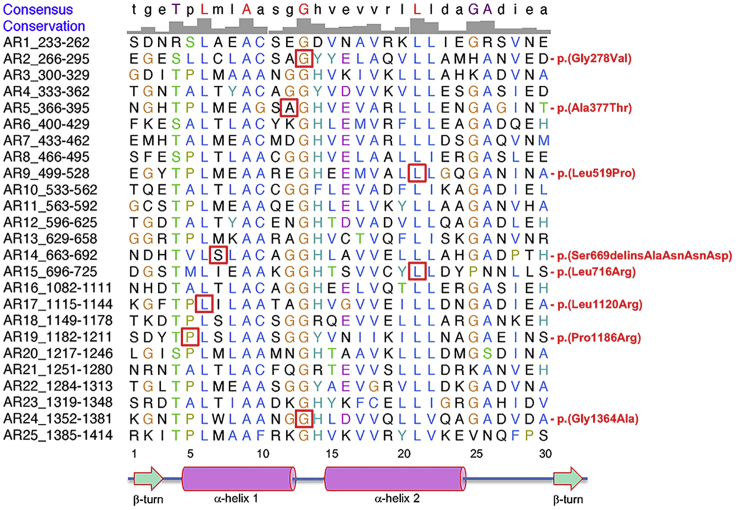
Beyond the high level of conservation of affected ANKRD17 residues across homologous proteins, we also analyzed conservation of these positions across the 25 ankyrin repeats of ANKRD17 itself. Remarkably, five of the seven ankyrin repeat missense variants affect amino acids that are invariant in all 25 repeats (Figure 3), with c.833G>T (p.Gly278Val) and c.4091G>C (p.Gly1364Ala) both affecting the 13th glycine of their respective repeats and c.1556T>C (p.Leu519Pro) and c.2147T>G (p.Leu716Arg) both affecting the 21st leucine in different repeats. The extreme inter- (across ANKRD17 orthologs and paralogs) and intra- (across ANKRD17 repeats) homology of amino acids affected by these missense variants in our cohort strongly supports their pathogenicity. The in-frame variant c.2005_2006delinsGCTAATAATGA (p.Ser669delinsAlaAsnAsnAsp) also occurs at a position that is conserved across homologous proteins (Figure S1), but its major effect is likely to involve disruption of the strict spacing of residues necessary for folding of ankyrin repeat 14 (Figure 3). The variant c.3557C>G (p.Pro1186Arg) affects a proline within a motif that is thought to be important for ankyrin repeat stability (the “TPLH tetrapeptide”13). Finally, the amino acid affected by the variant c.1129G>A (p.Ala377Thr) displays high inter-species conservation (Figure S1) but low inter-repeat conservation (Figure 3), suggesting this variant may disrupt a ligand interaction surface rather than the structural core of the ankyrin repeat it falls in.
Figure 3.
Conservation of amino acids affected by ANKRD17 non-truncating variants, across ankyrin repeats
Alignment of the 25 ankyrin repeats of human ANKRD17. The repeats, as defined by UniProtKB, were downloaded and aligned in UCSF Chimera. Amino acids affected by non-truncating variants are boxed in red. In the Consensus row, amino acids in red are 100% identical. Beneath the alignment, the numbering refers to the 30-amino acid repeating sequence of each ankyrin repeat, and the predicted positions of structural motifs that define ankyrin repeats are depicted14 (note that sequence corresponding to the canonical C-terminal beta-turn was not included in the UniProtKB-defined ANKRD17 ankyrin repeats).
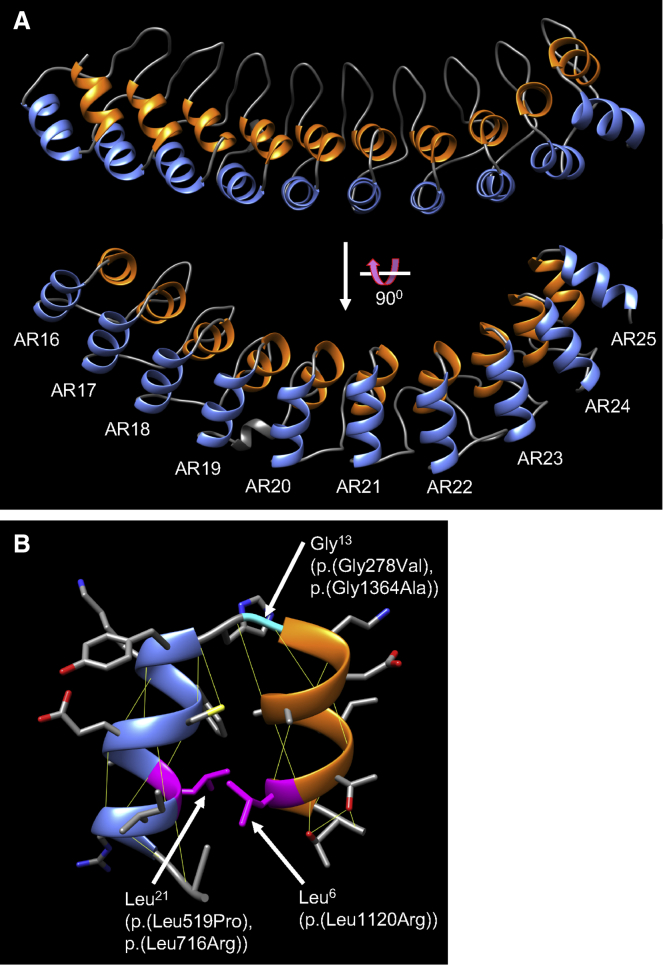
In order to further assess the effects of the ANKRD17 missense variants at the molecular level, we used homology modeling to generate a three-dimensional structure of the ANKRD17 ankyrin repeats. The model produced for ANKRD17 ankyrin repeats 16–25 conforms to several experimentally determined structures of other ankyrin domain-containing proteins1,14 with each ankyrin repeat containing two alpha helices connected by a short loop, and arranged in a slightly curved, linear array (Figure 4A). Examination of one representative repeat shows that the invariant glycine altered in individuals 2 and 26 is positioned at the beginning of the sharp turn between helices 1 and 2 (Figure 4B). The constant presence of a glycine at this position may be because of the high flexibility of glycine relative to other amino acids, a property that is likely important for maintenance of the turn. The leucines at the 6th and 21st positions of the repeat, altered in individuals 4, 5, and 32, participate in formation of the hydrophobic core of each ankyrin repeat (Figure 4B),14 a property that is perturbed upon mutation to the hydrophilic residue arginine in individuals 4 and 32, while in individual 5 the leucine is changed to the helix-disrupting residue proline. Indeed, leucines at the 6th and 21st positions and glycine at the 13th position are among the most conserved residues of ankyrin repeats across all three domains of life, suggesting their importance for basic structural integrity of the motif.1,15 The ANKRD17 missense variants affecting these residues are therefore more likely to disrupt the core structure of individual repeats, leading to protein destabilization, rather than disrupting specific ligand-interaction surfaces.
Figure 4.
Predicted three-dimensional structure of ANKRD17 ankyrin repeats and amino acids affected by missense variants
(A) Ankyrin repeats 16–25 of human ANKRD17 (GenBank: NP_115593) were modeled using the SWISS-MODEL homology-modeling server and visualized in UCSF Chimera. For each repeat, alpha-helix 1 is in orange and alpha-helix 2 is in blue.
(B) Detail of ANKRD17 ankyrin repeat 23, showing the positions of the invariant leucines (magenta) and glycine (cyan), which are altered at the equivalent positions in other repeats of ANKRD17 (indicated in brackets). Superscripts for Leu6, Gly13, and Leu21 refer to numbering within the 30-amino acid repeating sequence presented in Figure 3. Thin yellow lines indicate predicted hydrogen bonds.
Previously, three individuals with de novo ANKRD17 variants (two missense and one in-frame deletion of one amino acid) were listed in large-scale sequencing studies of autism, without further phenotypic information.16, 17, 18 The significance of these ANKRD17 variants was not explored in those reports, but we note that two of them fall outside of the ankyrin repeats, making their interpretation difficult, while one is within ankyrin repeat 25. Large deletions of a region harboring ANKRD17 and several other genes on chromosome 4q13.3 have previously been identified in individuals with ID, growth delay, and dysmorphic facial features10,19 (Figure 1C). Our work now strongly supports the possibility that haploinsufficiency of ANKRD17 is the major cause of these phenotypes in individuals with large alterations of 4q13.3. In turn, the phenotypic similarities between individuals with deletions containing ANKRD17, and individuals described here with ANKRD17 point mutations, support haploinsufficiency as the underlying disease basis in the latter.
Pathogenic variants in several other genes in the large ANKRD family have been implicated in Mendelian disease. Heterozygous loss-of-function variants in ANKRD11 (MIM: 611192) are causative of a syndromic ID, the variable but recognizable KBG syndrome (MIM: 148050) characterized by neurodevelopmental delay, macrodontia, short stature, and skeletal anomalies.20 Haploinsufficiency of SHANK3 (MIM: 606230) is responsible for Phelan-McDermid syndrome21 (MIM: 606232). Variants in the 5′ UTR of ANKRD26 (MIM: 610855) are causative of autosomal-dominant thrombocytopaenia-222 (MIM: 188000). Note that the above genes do not code for proteins similar to ANKRD17 in overall protein domain structure, aside from the presence of ankyrin repeats.
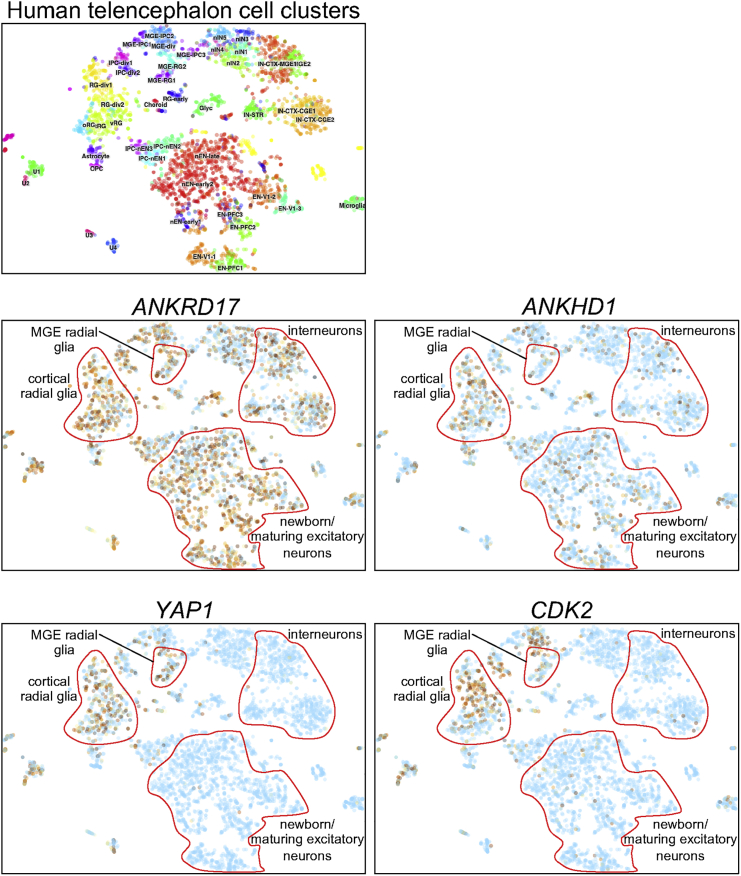
Interestingly, a de novo frameshift variant has been reported in the highly related ANKRD17 paralog ANKHD1, in an individual with a neurodevelopmental syndrome including speech and growth delay.23 As for ANKRD17, ANKHD1 has a pLI score of 1.0 in the gnomAD database, suggesting further heterozygous pathogenic variants in ANKHD1 may yet be discovered. Notably, knockdown of Ankhd1 in the developing mouse neocortex disrupts neurogenesis.24 Although ANKRD17 and ANKHD1 have very similar protein domain organization, have high sequence identity, and have both been reported to be widely expressed,2,3,24,25 the degree of redundancy between the two is unclear. Analysis of a publicly available single-cell RNA-seq (scRNA-seq) dataset of gene expression in the human telencephalon during key stages of neurogenesis26 indicates broad expression of ANKRD17, including in radial glial progenitors (the neural stem cells of the cortex), excitatory neurons, and interneurons, while ANKHD1 expression appears more limited (Figure 5). Mask, the Drosophila ortholog of ANKRD17/ANKHD1, is required for survival, proliferation, and differentiation of photoreceptor progenitors27 and for growth of eye and wing,4,5 while overexpression of Mask in the Drosophila brain reduces axonal outgrowth.28 The Hippo pathway is a key regulator of organ growth in animals, and its growth-promoting role is mediated by the transcriptional coactivator Yorkie in Drosophila or YAP in mammals. In Drosophila, Mask is required for expression of Yorkie target genes and for Yorkie’s growth-promoting activity.4,5 Yorkie/YAP can also interact with Mask/ANKHD1/ANKRD17.4,5 Predominantly nuclear localization of ANKRD17 has been reported in human cell lines,2 although nuclear versus cytoplasmic distribution was shown to be cell density dependent,5 and Mask/ANKHD1 have been shown to play a role in promoting nuclear import of Yorkie/YAP.29 Interestingly, important roles for the Hippo pathway in brain development have been described in several animal models,30 and analysis of human telencephalon scRNA-seq data suggested overlap of YAP1 (MIM: 606608) and ANKRD17 expression in radial glia (Figure 5), raising the possibility that the neurodevelopmental phenotypes associated with ANKRD17 variants may be due to disruption of this pathway.
Figure 5.
Single-cell RNA-seq analysis of ANKRD17, ANKHD1, YAP1, and CDK2 expression in the developing human telencephalon
Single-cell RNA-seq data26 were visualized at the UCSC Cell Browser (dataset: Cortex development). The dataset was generated from 4,261 cells obtained from the telencephalon (cortex and/or ganglionic eminences) of 48 fetuses, ranging in age from 6 to 37 post-conception weeks (pcw) (with the majority of cells from 9–16.5 pcw samples). Data are plotted using the “t-SNE on WGCNA” layout. Upper left panel indicates clusters of cell types in detail (see https://cells.ucsc.edu/?ds=cortex-dev for definitions). In the gene panels, the degree of brown shading in each cell represents transcript abundance, light blue shading indicates absence of detected expression, and broad categories of cell types of interest are outlined in red. MGE, medial ganglionic eminence.
While heterozygous Ankrd17-deficient mice are viable and fertile, homozygous Ankrd17 deficiency results in lethality by E11.5 due to catastrophic hemorrhages, with incomplete vascular maturation likely due to a reduction in the number of smooth muscle cells associating with endothelial tubes.3 There were no clinical features suggestive of vascular abnormalities in our cohort. Given the early embryonic death of the knockout mice, central nervous system development was not studied. The development of conditional loss-of-function mouse models may shed further light on the central nervous system features of the ANKRD17 disorder.
Several other molecular functions have been reported for ANKRD17 and its homologs. In human cell lines, ANKRD17 interacts with the cyclin E/CDK2 complex, promotes cell cycle progression, and associates with DNA replication factors and chromatin.2 Interestingly, scRNA-seq data suggest that CDK2 expression overlaps with that of ANKRD17 in neuronal progenitors of the developing brain (Figure 5). Interaction of ANKHD1 with microRNAs via its KH domain leads to increased proliferation of renal carcinoma cells.31 Previous demonstration of ANKRD17 as a binding partner of NOD2 in the NOD1/NOD2 innate immunity pathway,6 and as a positive regulator of the RIG-1-like receptor-mediated signaling pathway,7 suggests a plausible mechanism for the predisposition to bacterial or viral infections, respectively, identified in some individuals in our cohort. Of further relevance to immune system defects, a functional screen in Drosophila identified Mask as a positive regulator of cytokine receptor stability.32 Also, a membrane-deformation ability has been identified for the ANKHD1 ankyrin repeats (and is also present in ANKRD17) and plays a role in promoting vesiculation during early endosome enlargement.33 How the apparently diverse molecular activities of ANKRD17 family proteins are coordinated needs further exploration.
With this report, we delineate a neurodevelopmental disorder in 34 individuals caused by pathogenic variants in ANKRD17, most of which are de novo. The high frequency of truncating variants in our cohort, the structural predictions for the damaging effects of the identified missense and in-frame variants, and the known intolerance of haploinsufficiency for this gene in the general population support a strong gene-disease association. The true prevalence of this disorder in the neurodevelopmental disorder population remains to be seen. Although our cohort is relatively large, it was assembled over 3 years from multiple sources, making determination of the denominator difficult. It is hoped that with this report, more individuals with this previously unrecognized condition will be diagnosed and better prevalence estimates can be made. Although recurrent bacterial infections, cleft palate with Pierre Robin sequence, and unilateral renal agenesis are useful discriminating features, they occur in only a minority of patients. We suggest that individuals in whom a rare ANKRD17 variant of uncertain significance is identified should be evaluated for these features, the presence of which may add weight to the interpretation of the variant as causal.
Although roles for ANKRD17 in cell cycle progression and tissue growth have been proposed, the exact mechanism by which ANKRD17 disruption leads to the clinical phenotype, particularly with respect to neuronal development and function, is yet to be understood. Given the previous evidence that the Mask/ANKHD1/ANKRD17 family regulates the activity of Yorkie/YAP, it is plausible that dysregulation of the Hippo pathway contributes to the developmental features of the ANKRD17-related disorder. In the future, loss-of-function studies in an appropriate animal model may shed light on the precise cell types and transcriptional outputs regulated by ANKRD17.
Consortia
The investigators of the CAUSES Study include Shelin Adam, Christele Du Souich, Alison Elliott, Anna Lehman, Jill Mwenifumbo, Tanya Nelson, Clara Van Karnebeek, and Jan Friedman.
Declaration of interests
S.Y., R.W., J.J., and J. Semotok are employees of GeneDx, Inc., a wholly owned subsidiary of OPKO Health, Inc.
Acknowledgments
The authors acknowledge the individuals and families for their participation in this study and the Genomic and Bioinformatic facilities of the Institut Imagine. This work was supported by grants from the Agence Nationale de la Recherche (CranioRespiro project and “Investissements d’Avenir” program [ANR-10-IAHU-01]); MSDAvenir (Devo-Decode project); The Ministry of Health and Family Welfare, Government of India (project entitled “Clinical and Molecular Characterization of Leukodystrophies in Indian Children” [V.25011/379/2015-GIA/HR]); Alabama Genomic Health Initiative (an Alabama-State earmarked project F170303004) through the University of Alabama in Birmingham; the Netherlands Organization for Health Research and Development (ZonMw grant 91718310 to T.K.); the National Human Genome Research Institute of the National Institutes of Health (award number U01HG009599); AXA (“Tete et Coeur” project); Genomics Aotearoa; and Curekids NZ.
The content of the manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The CAUSES study is funded by Mining for Miracles, British Columbia Children’s Hospital Foundation (grant number F15-01355), and Genome British Columbia (grant number F16-02276). This work was in part generated within the European Reference Network ITHACA.
Sequencing and analysis provided by the Broad Institute of MIT and Harvard Center for Mendelian Genomics (Broad CMG) were funded by the National Human Genome Research Institute, the National Eye Institute, and the National Lung and Blood Institute grant UM1 HG008900, and in part by National Human Genome Research Institute grant R01 HG009141.
See supplemental information for DDD and 100,000 Genomes acknowledgment statements.
Published: April 27, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.04.007.
Contributor Information
Maya Chopra, Email: maya.chopra@childrens.harvard.edu.
Christopher T. Gordon, Email: chris.gordon@inserm.fr.
Data and code availability
This study did not generate datasets or code.
Web resources
Clustal Omega, http://www.ebi.ac.uk/Tools/msa/clustalo/
DECIPHER, https://decipher.sanger.ac.uk/
GeneMatcher, https://genematcher.org/
gnomAD Browser, https://gnomad.broadinstitute.org/
Needleman-Wunsch global alignment, https://blast.ncbi.nlm.nih.gov/Blast.cgi
OMIM, https://www.omim.org/
PolyPhen-2 v.2.2.2, http://genetics.bwh.harvard.edu/pph2/
SIFT v.1.03, https://sift.bii.a-star.edu.sg/
SWISS-MODEL, https://swissmodel.expasy.org/
UCSC Cell browser, https://cells.ucsc.edu/?ds=cortex-dev
UCSC Genome Browser, https://genome.ucsc.edu
UCSF Chimera, https://www.cgl.ucsf.edu/chimera/
UniProtKB, https://www.uniprot.org
Supplemental information
References
- 1.Li J., Mahajan A., Tsai M.-D. Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry. 2006;45:15168–15178. doi: 10.1021/bi062188q. [DOI] [PubMed] [Google Scholar]
- 2.Deng M., Li F., Ballif B.A., Li S., Chen X., Guo L., Ye X. Identification and functional analysis of a novel cyclin e/cdk2 substrate ankrd17. J. Biol. Chem. 2009;284:7875–7888. doi: 10.1074/jbc.M807827200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hou S.-C., Chan L.-W., Chou Y.-C., Su C.-Y., Chen X., Shih Y.-L., Tsai P.-C., Shen C.-K.J., Yan Y.-T. Ankrd17, an ubiquitously expressed ankyrin factor, is essential for the vascular integrity during embryogenesis. FEBS Lett. 2009;583:2765–2771. doi: 10.1016/j.febslet.2009.07.025. [DOI] [PubMed] [Google Scholar]
- 4.Sansores-Garcia L., Atkins M., Moya I.M., Shahmoradgoli M., Tao C., Mills G.B., Halder G. Mask is required for the activity of the Hippo pathway effector Yki/YAP. Curr. Biol. 2013;23:229–235. doi: 10.1016/j.cub.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sidor C.M., Brain R., Thompson B.J. Mask proteins are cofactors of Yorkie/YAP in the Hippo pathway. Curr. Biol. 2013;23:223–228. doi: 10.1016/j.cub.2012.11.061. [DOI] [PubMed] [Google Scholar]
- 6.Menning M., Kufer T.A. A role for the Ankyrin repeat containing protein Ankrd17 in Nod1- and Nod2-mediated inflammatory responses. FEBS Lett. 2013;587:2137–2142. doi: 10.1016/j.febslet.2013.05.037. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y., Tong X., Li G., Li J., Deng M., Ye X. Ankrd17 positively regulates RIG-I-like receptor (RLR)-mediated immune signaling. Eur. J. Immunol. 2012;42:1304–1315. doi: 10.1002/eji.201142125. [DOI] [PubMed] [Google Scholar]
- 8.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., Genome Aggregation Database Consortium The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Havrilla J.M., Pedersen B.S., Layer R.M., Quinlan A.R. A map of constrained coding regions in the human genome. Nat. Genet. 2019;51:88–95. doi: 10.1038/s41588-018-0294-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maldžienė Ž., Vaitėnienė E.M., Aleksiūnienė B., Utkus A., Preikšaitienė E. A case report of familial 4q13.3 microdeletion in three individuals with syndromic intellectual disability. BMC Med. Genomics. 2020;13:63. doi: 10.1186/s12920-020-0711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Y., Yuan C., Tian F., Huang K., Weghorst C.M., Tsai M.-D., Li J. Contributions of conserved TPLH tetrapeptides to the conformational stability of ankyrin repeat proteins. J. Mol. Biol. 2010;399:168–181. doi: 10.1016/j.jmb.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mosavi L.K., Cammett T.J., Desrosiers D.C., Peng Z.Y. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jernigan K.K., Bordenstein S.R. Ankyrin domains across the tree of life. PeerJ. 2014;2:e264. doi: 10.7717/peerj.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krumm N., Turner T.N., Baker C., Vives L., Mohajeri K., Witherspoon K., Raja A., Coe B.P., Stessman H.A., He Z.-X. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 2015;47:582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iossifov I., O’Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S., DDD Study. Homozygosity Mapping Collaborative for Autism. UK10K Consortium Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quintela I., Barros F., Fernandez-Prieto M., Martinez-Regueiro R., Castro-Gago M., Carracedo A., Gomez-Lado C., Eiris J. Interstitial microdeletions including the chromosome band 4q13.2 and the UBA6 gene as possible causes of intellectual disability and behavior disorder. Am. J. Med. Genet. A. 2015;167A:3113–3120. doi: 10.1002/ajmg.a.37291. [DOI] [PubMed] [Google Scholar]
- 20.Sirmaci A., Spiliopoulos M., Brancati F., Powell E., Duman D., Abrams A., Bademci G., Agolini E., Guo S., Konuk B. Mutations in ANKRD11 cause KBG syndrome, characterized by intellectual disability, skeletal malformations, and macrodontia. Am. J. Hum. Genet. 2011;89:289–294. doi: 10.1016/j.ajhg.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durand C.M., Betancur C., Boeckers T.M., Bockmann J., Chaste P., Fauchereau F., Nygren G., Rastam M., Gillberg I.C., Anckarsäter H. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat. Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pippucci T., Savoia A., Perrotta S., Pujol-Moix N., Noris P., Castegnaro G., Pecci A., Gnan C., Punzo F., Marconi C. Mutations in the 5¢ UTR of ANKRD26, the ankirin repeat domain 26 gene, cause an autosomal-dominant form of inherited thrombocytopenia, THC2. Am. J. Hum. Genet. 2011;88:115–120. doi: 10.1016/j.ajhg.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anazi S., Maddirevula S., Salpietro V., Asi Y.T., Alsahli S., Alhashem A., Shamseldin H.E., AlZahrani F., Patel N., Ibrahim N. Expanding the genetic heterogeneity of intellectual disability. Hum. Genet. 2017;136:1419–1429. doi: 10.1007/s00439-017-1843-2. [DOI] [PubMed] [Google Scholar]
- 24.Hermann R. 2014. Regulation of Neural Progenitor Proliferation by ANKHD1.https://archiv.ub.uni-heidelberg.de/volltextserver/16467/ [Google Scholar]
- 25.Poulin F., Brueschke A., Sonenberg N. Gene fusion and overlapping reading frames in the mammalian genes for 4E-BP3 and MASK. J. Biol. Chem. 2003;278:52290–52297. doi: 10.1074/jbc.M310761200. [DOI] [PubMed] [Google Scholar]
- 26.Nowakowski T.J., Bhaduri A., Pollen A.A., Alvarado B., Mostajo-Radji M.A., Di Lullo E., Haeussler M., Sandoval-Espinosa C., Liu S.J., Velmeshev D. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science. 2017;358:1318–1323. doi: 10.1126/science.aap8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith R.K., Carroll P.M., Allard J.D., Simon M.A. MASK, a large ankyrin repeat and KH domain-containing protein involved in Drosophila receptor tyrosine kinase signaling. Development. 2002;129:71–82. doi: 10.1242/dev.129.1.71. [DOI] [PubMed] [Google Scholar]
- 28.Lou W.P.-K., Mateos A., Koch M., Klussman S., Yang C., Lu N., Kumar S., Limpert S., Göpferich M., Zschaetzsch M. Regulation of Adult CNS Axonal Regeneration by the Post-transcriptional Regulator Cpeb1. Front. Mol. Neurosci. 2018;10:445. doi: 10.3389/fnmol.2017.00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sidor C., Borreguero-Munoz N., Fletcher G.C., Elbediwy A., Guillermin O., Thompson B.J. Mask family proteins ANKHD1 and ANKRD17 regulate YAP nuclear import and stability. eLife. 2019;8:8. doi: 10.7554/eLife.48601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu V., Plouffe S.W., Guan K.-L. The Hippo pathway in organ development, homeostasis, and regeneration. Curr. Opin. Cell Biol. 2017;49:99–107. doi: 10.1016/j.ceb.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fragiadaki M., Zeidler M.P. Ankyrin repeat and single KH domain 1 (ANKHD1) drives renal cancer cell proliferation via binding to and altering a subset of miRNAs. J. Biol. Chem. 2018;293:9570–9579. doi: 10.1074/jbc.RA117.000975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fisher K.H., Fragiadaki M., Pugazhendhi D., Bausek N., Arredondo M.A., Thomas S.J., Brown S., Zeidler M.P. A genome-wide RNAi screen identifies MASK as a positive regulator of cytokine receptor stability. J. Cell Sci. 2018;131:jcs209551. doi: 10.1242/jcs.209551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitamata M., Hanawa-Suetsugu K., Maruyama K., Suetsugu S. Membrane-Deformation Ability of ANKHD1 Is Involved in the Early Endosome Enlargement. iScience. 2019;17:101–118. doi: 10.1016/j.isci.2019.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate datasets or code.