Summary
Proteins involved in transcriptional regulation harbor a demonstrated enrichment of mutations in neurodevelopmental disorders. The Sin3 (Swi-independent 3)/histone deacetylase (HDAC) complex plays a central role in histone deacetylation and transcriptional repression. Among the two vertebrate paralogs encoding the Sin3 complex, SIN3A variants cause syndromic intellectual disability, but the clinical consequences of SIN3B haploinsufficiency in humans are uncharacterized. Here, we describe a syndrome hallmarked by intellectual disability, developmental delay, and dysmorphic facial features with variably penetrant autism spectrum disorder, congenital malformations, corpus callosum defects, and impaired growth caused by disruptive SIN3B variants. Using chromosomal microarray or exome sequencing, and through international data sharing efforts, we identified nine individuals with heterozygous SIN3B deletion or single-nucleotide variants. Five individuals harbor heterozygous deletions encompassing SIN3B that reside within a ∼230 kb minimal region of overlap on 19p13.11, two individuals have a rare nonsynonymous substitution, and two individuals have a single-nucleotide deletion that results in a frameshift and predicted premature termination codon. To test the relevance of SIN3B impairment to measurable aspects of the human phenotype, we disrupted the orthologous zebrafish locus by genome editing and transient suppression. The mutant and morphant larvae display altered craniofacial patterning, commissural axon defects, and reduced body length supportive of an essential role for Sin3 function in growth and patterning of anterior structures. To investigate further the molecular consequences of SIN3B variants, we quantified genome-wide enhancer and promoter activity states by using H3K27ac ChIP-seq. We show that, similar to SIN3A mutations, SIN3B disruption causes hyperacetylation of a subset of enhancers and promoters in peripheral blood mononuclear cells. Together, these data demonstrate that SIN3B haploinsufficiency leads to a hitherto unknown intellectual disability/autism syndrome, uncover a crucial role of SIN3B in the central nervous system, and define the epigenetic landscape associated with Sin3 complex impairment.
Keywords: SINB, SIN3A, epigenetics, autism, intellectual disability, acetylation, mutation, HDAC, transcription, zebrafish
Main text
Impairment of transcriptional regulation has been linked closely to the molecular etiology of intellectual disability (ID) and autism spectrum disorders (ASDs).1, 2, 3 Specifically, multiple genes mutated in Mendelian disorders with a neurodevelopmental or neuroanatomical aspect, and notably ID/ASD genes, have been found to encode chromatin regulators.4, 5, 6 Eukaryotic chromatin structure is organized in histone octamers of the four core histones, H2A, H2B, H3, and H4, forming nucleosomes to ensure DNA compaction. Post-translational modifications of histone N termini, particularly acetylation, play decisive roles in the dynamic regulation of gene transcription. Histone deacetylation is executed by the histone deacetylase (HDAC) enzymes HDAC1 and HDAC2 and is dependent upon complexes containing Sin3, MecP2,7 NuRD, and CoREST.8, 9, 10 Recruitment of the Sin3 corepressor module leads to deacetylation of histones H3 and H4.11 The core SIN3-HDAC complex acts as a scaffold for the assembly of multiple cofactors.12, 13, 14 In mammals, two genes encode master factors of the Sin3 complex, SIN3A (MIM: 607776) and SIN3B (MIM: 607777). SIN3A disruption in humans causes syndromic ID with craniofacial defects (Witteveen-Kolk syndrome [MIM: 613406]).15 Consequences of SIN3B disruption in humans are hitherto poorly understood.
In this study, we report the phenotypic features associated with haploinsufficiency at the SIN3B locus through combined analysis of clinical and cerebral MRI data collected for nine individuals harboring rare SIN3B variants (seven de novo and two of undetermined origin due to an inaccessible parental sample). We identified five copy number variant (CNV) deletions at 19q13.11, which encompass SIN3B (0.427 to 1.5 Mb; smallest region of overlap, SRO, ∼230 kb, hg19), and four single nucleotide variants (SNV) in the coding regions of SIN3B (Table 1). To investigate the consequences of SIN3B loss in vivo, we ablated sin3b in zebrafish (Danio rerio) larvae with a transgenic reporter of cartilage formation in the pharyngeal skeleton by CRISPR/Cas9 genome editing and transient morpholino (MO)-based suppression. We recapitulate multiple aspects of the human phenotype in zebrafish and demonstrate pathogenicity for the two missense variants. Furthermore, we investigated the epigenetic consequences of SIN3B variation by epigenomic profiling the peripheral blood mononuclear cells (PBMCs) from an affected CNV deletion-bearing individual compared with his family members and compared with data generated from PBMCs from a SIN3A CNV deletion pedigree. The results suggest that SIN3B disruption leads to increased histone acetylation in this individual, which is consistent with SIN3A ablation and the repressor function of the Sin3 complex.
Table 1.
Clinical features of five individuals with de novo 19p13.11 microdeletions encompassing SIN3B and four individuals with point mutations in SIN3B
| Individual identifier | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | Total | Aten et al., 200916 | Bens et al., 201117 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DECIPHER identifier (v.9.21) | 332280 | 262142 | NA | NA | 308455 | 325602 | NA | NA | NA | NA | 4101 | NA |
| Genotypea,b | ||||||||||||
| Variant type | CNV, deletion | CNV, deletion | CNV, deletion | CNV, deletion | CNV, deletion | SNV, frameshift | SNV, frameshift | SNV, nonsynonymous | SNV, nonsynonymous | 5 CNV del, 4 SNV | CNV, deletion | CNV, deletion |
| Inheritance | de novo | de novo | de novo | de novo | de novo | de novo | parents unavailable for testing | de novo | father unavailable for testing | 7 confirmed de novo | de novo | de novo |
| Variant details | 19: 16848440–17871985 | 19: 15978604–17500427 | 19: 16599950–17469382 | 19: 16456955–17333482 | 19: 16652215–17079033 | c.31delA (p.Ser11Alafs∗11) | c.1579delC (p.Arg527Glyfs∗12) | c.249C>G (p.Ile83Met) | c.58G>A (p.Gly20Arg) | NA | 19: 16548375–17547292 | 19: 16611808–17733344 |
| Size | 1.02 Mb | 1.52 Mb | 869 kb | 877 kb | 427 kb | NA | NA | NA | NA | NA | 0.99 Mb | 1.12 Mb |
| Clinical phenotypes | ||||||||||||
| Sex | M | F | M | F | M | M | M | M | M | NA | M | F |
| Age at last clinical examination | 20 years | 11 years | 2 years 6 months | 8 years 10 months | 3 years 10 months | 15 years | 50 years 4 months | 3 years | 5 years | NA | 6 years | 4 years |
| Growth parameters | W: −4 SD, H: −2 SD, OFC: 0 SD | W: +1 SD, H: −0.5 SD, OFC: −0.5 SD | W: −0.5 SD, H: N, OFC: −3 SD | W: −1 SD, H: +0.5 SD, OFC: −1.5 SD | W: −0.75 SD, H: −1.04 SD, OFC: −0.97 SD | W: +3 SD, H: +2.5 SD, OFC: +2.5 SD | W: −1 SD, H: −2 SD, OFC: N | W: +1.2 SD, H: −0.5 SD, OFC: −0.5 SD | W: +2 SD, H: +2 SD, OFC: +2 SD | NA | W: ND, H: −0.3 SD, OFC: −1 SD | W: −1.4 SD, H: −1.9 SD, OFC: −3.5 SD |
| DD/ID | − (IQ 96) | + (IQ 41) | + (IQ ND) | + (WISC-IV) | + (IQ ND) | + (IQ ND) | + (IQ 56) | + (IQ ND) | + (IQ ND) | 8/9 | + (IQ ND) | + (IQ ND) |
| DD/ID severity | NA | moderate | moderate | mild | mild | mild | mild | moderate | mild | 5 mild, 3 moderate | severe | severe |
| Age at independent walking | 18 months | 2 years 8 months | 2 years 4 months | 13 months | 24 months | 24 months | ND | 22 months | 18 months | NA | ND | ND |
| Speech delay | − | + | + | − (articulation problems) | − | + | + | + | + | 6/9 | + | + |
| ASD | + | − | − | − | − | + | − | − | + | 3/9 | − | − |
| Other behavioral disorders | ADHD | ADHD | − | hyperactivity, impulsivity, low frustration tolerance, anxiety | − | aggressive behavior | echolalia | − | ADHD | 6/9 | − | − |
| Epilepsy | − | − | − | − | − | − | + | − | − | 1/9 | − | − |
| Brain MRI | N | N | short CC, olfactory bulb agenesis | mild tonsillar ectopia | ND | defects in CC and subependymal nodular heterotopia | N | ND | ND | 3/6 | ND | ND |
| Dysmorphic featuresc | + | + | + | + | + | + | + | + | + | 9/9 | + | + |
| Limb abnormalities | short hands | bilateral fifth fingers clinodactyly, genu recurvatum | − | clinodactyly of the 2nd and 5th digits, tapered distal phalanges | − | − | − | − | bilateral radio-ulnar synostosis | 4/9 | SHFM | − |
| MCA | − | VSD, bifid uvula, strabismus | VSD, left iris coloboma, ocular anterior segment dysgenesis, left cleft lip and palate, micropenis | bifid uvula, strabismus | tetralogy of Fallot, preauricular pit | − | − | intestinal malrotation | ND | 5/8 | tetralogy of Fallot | − |
| Additional phenotypic features | myopia, daytime hypersomnia, decreased melatonin urinary excretion | ataxia | − | umbilical hernia, clumsiness, myopia, hypotonia | pectus carinatum | conductive hearing loss | hand tremor | − | ND | NA | strabismus | initial poor feeding, premature pubarche, strabismus, ataxia |
| Additional genetic findings | − | − | CNV deletion (1.6 Mb) de novo, 18: 3192682–4854252 | − | − | − | − | − | CNOT1 c.4861A>G (p.Ile1621Val) (unknown inheritance) | NA | ND | ND |
Abbreviations are as follows: NA, not applicable; M, male; F, female; W, weight; H, height; OFC, occipitofrontal head circumference; SD, standard deviation; ND, no data; N, normal; DD, developmental delay; ID, intellectual disability; WISC-IV, Wechsler Intelligence Scale for Children; ASD, autism spectrum disorder; MRI, magnetic resonance imaging; MCA, multiple congenital anomalies; ADHD, attention deficit hyperactivity disorder; CC, corpus callosum; VSD, ventricular septal defect; SHFM, split hand and foot malformation; p, percentile; +, affected; −, not affected; CNV, copy number variant; del, deletion; SNV, single-nucleotide variant.
The reference genome used for annotations is GRCh37/hg19.
Coding DNA/protein variant described according to the nomenclature HGVS v.2.0 established by the Human Genome Variation Society; GenBank: NM_015260.4 and NP_056075.1.
For dysmorphic features, see supplemental notes.
Nine affected individuals were referred independently for genetic counseling and clinical genetic testing because of unexplained ID or ASD (Table 1). We obtained written informed consent from each affected individual and available family members prior to inclusion in genetics research in accordance with each respective institution’s human subjects ethics committee. All participants were assessed by at least one expert clinical geneticist from each respective participating center (Table 1; supplemental notes).
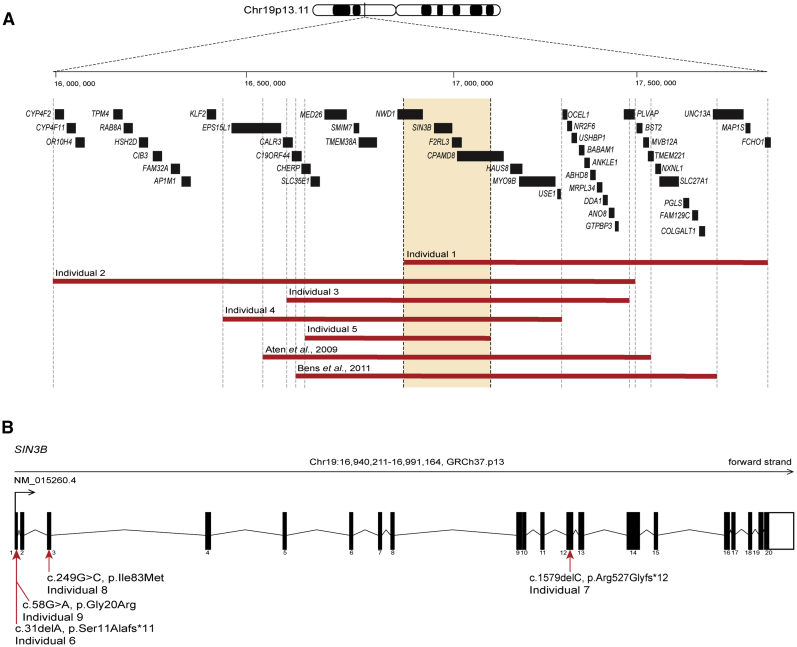
To investigate the possibility that CNVs could contribute to the ID/ASD features, we performed chromosomal microarray studies (Table S1). Individuals 1, 2, 3, 4, and 5 carry microdeletions of 1.02, 1.5, 0.87, 0.88, and 0.43 Mb, respectively, at the 19p13.11 locus, all of which encompass SIN3B (Figure 1A; Table S2). Notably, de novo deletions of this chromosomal region have been implicated previously in multi-system pathologies in humans, including neurodevelopmental disorders. Aten and colleagues reported an affected male who harbored a 0.99 Mb de novo deletion that contains 28 genes (Table 1; Table S2),16 and Bens et al. described a female pediatric affected individual with an overlapping 1.12 Mb microdeletion at 19p13.11 (Table 1; Table S2).17 Comparison of the microdeletions reported in these two affected individuals with the five individuals in our study yielded a smallest region of overlap (SRO) of ∼230 kb, which contains four genes (NWD1 [MIM: 616250], SIN3B, F2RL3 [MIM: 602779], and CPAMD8 [MIM: 608841]; Figure 1A; Table S2). In silico predictions suggested SIN3B to be the most likely candidate disease driver gene because of its high probability of loss of function (LoF) intolerance (pLI) score18 (SIN3B pLI > 0.9; NWD1, F2RL3, CPAMD8 pLI = 0; Table S2). Individual 3 carried an additional de novo deletion of 1.6 Mb at 18p11.31 (chr18: 3,192,682–4,854,252, GRCh37/hg19), encompassing four genes (MYOM1 [MIM: 603508], MYL12B [MIM: 609211], TGIF1 [MIM: 602630], and DLGAP1 [MIM: 605445]). TGIF1 loss-of-function variants have been described in holoprosencephaly type 4 (MIM: 142946).19 MYOM1, MYL12B, and DLGAP1 have not been linked to a human disorder. However, this second mutational event most likely plays a role in the phenotype of individual 3. Additionally, the individual described by Aten et al. presents with split hand and foot malformation (SHFM) (Table 1).16 EPS15L1 (MIM: 616826) was considered to be responsible for the limb phenotype and is not included in the SRO reported in our study (Figure 1; Table S2).
Figure 1.
Copy number variants and single nucleotide variants altering SIN3B cause intellectual disability with autistic features
(A) Schematic depicting SIN3B (GRCh37.p13; chr19: 16,940,211–16,991,164) at the 19p13.11 locus. CNV deletions (red bars) encompassing SIN3B in individuals 1, 2, 3, 4, and 5, as well as two previously reported individuals,16,17 are shown. Gene positions are indicated by black boxes, and genes located in smallest region of overlap are included in orange box between vertical dashed lines.
(B) Exon structure of longest SIN3B transcript, containing 20 exons (GenBank: NM_015260.4). Variants c.31delA (p.Ser11Alafs∗11) (individual 6); c.1579delC (p.Arg527Glyfs∗12) (individual 7); c.249C>G (p.Ile83Met) (individual 8); and c.58G>A (p.Gly20Arg) (individual 9) are indicated with red arrows.
Next, we asked whether pathogenic variants in any of these genes in the SRO had been implicated in phenotypes overlapping with 19p13.11 deletion carriers. NWD1 and F2RL3 have not been implicated previously in human genetic disorders. Bi-allelic mutations in CPAMD8 cause anterior segment dysgenesis20 (MIM: 617319), but carriers of heterozygous loss-of-function variants are not known to have neurodevelopmental symptoms. Individual 3 presents with ocular anterior segment dysgenesis (Table 1), and we cannot exclude the possibility of a variant in CPAMD8 on the non-deleted allele that could explain a part of this individual’s ocular phenotype. However, a likely disruptive variant was identified in SIN3B in a previous study within a large syndromic ID cohort21 but was only reported as a candidate because of a lack of genetic evidence in support of causality. Here, we report the same individual with the frameshift de novo variant located in exon 1 of SIN3B (c.31delA [p.Ser11Alafs∗11] [GenBank: NM_015260.4]) as individual 6 (Figure 1B; Table 1). This SNV is absent from >240,000 alleles in the Genome Aggregation Database (gnomAD; accessed September 22, 2019), and the variant transcript is predicted to be degraded by nonsense-mediated decay, resulting in SIN3B haploinsufficiency. However, cell lines derived from the affected individual were not available to test this possibility experimentally.
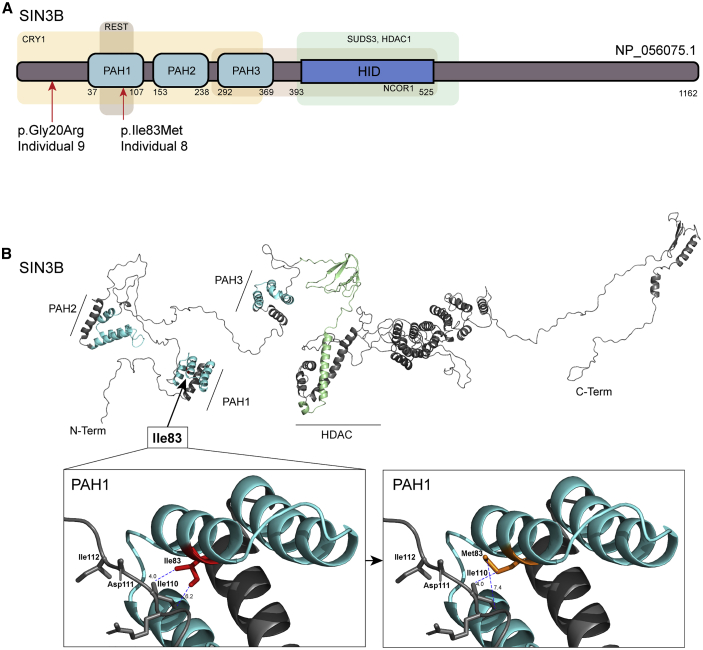
We turned to community data-sharing platforms22 to ask whether additional affected individuals might be present with deleterious mutations and overlapping clinical features at this locus. We identified individual 7, who carries a c.1579delC (p.Arg527Glyfs∗12) frameshift variant in exon 12 (Figure 1B; Table 1). Parental samples were not available for segregation analysis. Exome sequencing (ES) of individual 8 identified a de novo c.249C>G (p.Ile83Met) change that is predicted to be damaging by in silico classifiers and is absent from gnomAD (Figures 1B, 2, and S1; Tables 1, S1, and S3). This change affects a conserved residue within the first paired amphipathic helix (PAH) domain of SIN3B, which is responsible for interaction with members of Sin3/HDAC complex (Figures 2 and S1). Individual 9 harbors a heterozygous c.58G>A (p.Gly20Arg) variant (Figures 1B, 2, and S1; Tables 1 and S1). This variant is absent from gnomAD and is predicted to be deleterious in silico (Table S3). This change was not detected in genomic DNA from individual 9’s mother, but the paternal sample was not available to confirm the mode of inheritance. Individual 9 also harbors a variant of uncertain significance in CNOT1 (MIM: 604917), a locus known to cause either dominant holoprosencephaly23,24 (MIM: 618500) or Vissers-Bodmer syndrome25 (MIM:619033; c.4861A>G [p.Ile1621Val] [GenBank: NM_001265612.2]), for which contribution to the phenotype cannot be excluded (Table 1). Even so, we considered this SIN3B variant, in combination with individual 9’s neurodevelopmental phenotypes, to be potentially relevant to our case series. Together, these two ultra-rare missense variants were supportive, but not conclusive, in implicating SIN3B involvement in the ID/developmental delay (DD) phenotypes observed in individuals 1–5, who harbor CNVs on 19p13.11, and in individuals 6 and 7, who harbor frameshift variants.
Figure 2.
In silico protein modeling of SIN3B nonsynonymous variants indicate likely disruption of protein function
(A) SIN3B protein structure (GenBank: NP_056075.1). Three paired amphipathic helix (PAH domains) predicted on the N-terminal region of SIN3B are indicated by light blue boxes. Regions that interact (predicted by similarity with murine Sin3b) with CRY1 (cryptochrome circadian clock 1), REST (RE1-silencing transcription factor), SUDS (Sin3 histone deacetylase corepressor complex component SDS), HDAC1 (histone deacetylase 1), NCOR1 (nuclear receptor corepressor 1), and HID (HDAC-interacting domain) are indicated by light-colored boxes.
(B) In silico three-dimensional view of SIN3B. Variant c.249C>G encodes p.Ile83Met, which affects a residue located in the PAH1 domain nearest to the N terminus, which is predicted to mediate protein-protein interaction with transcriptional corepressors REST and CRY1. Protein Data Bank (PDB) file was generated by RaptorX and analyzed in Pymol 2.0. PAH domains are colored in turquoise and HID is colored in light green. Distance (Å) between side chains of wild-type (Ile83; left box) and mutant (Met83; right box) SIN3B and the closest residue on the opposing side of the protein are measured as indicated (blue dashed lines).
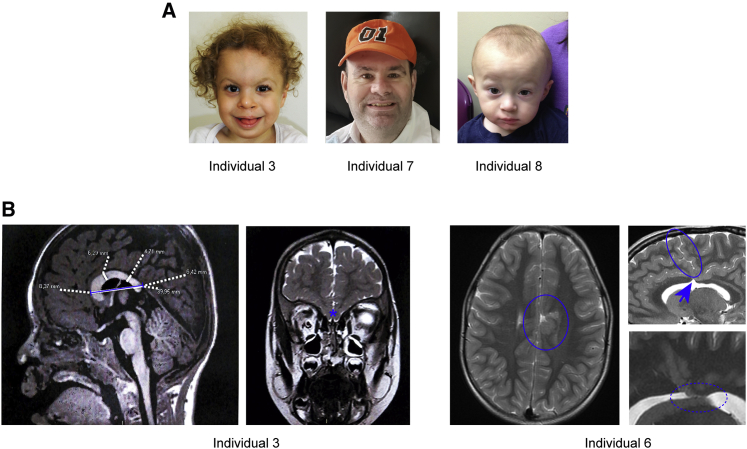
All nine individuals presented with a constellation of cognitive and neuroanatomical phenotypes. The most frequently reported features included ID and DD (in 8/9), classified as mild (individuals 4, 5, 6, 7, and 9) and moderate (individuals 2, 3, and 8; Table 1). In addition to ID/DD, three individuals fulfilled clinical diagnostic criteria for ASD (individuals 1, 6, and 9). Moreover, three individuals had attention deficit hyperactivity disorder (ADHD; individuals 1, 2, and 9). Cerebral MRI showed variable features; these included corpus callosum defects and subependymal nodular heterotopia (individual 6, Figure 3B), hypoplastic corpus callosum (individual 3, who also has a TGIF1 deletion), and mild tonsillar ectopia (individual 4); the other half of the cohort presented no detectable MRI abnormalities at last assessment or did not have brain imaging (Table 1). Individual 7 had generalized tonic-clonic seizures starting at age 3. Some individuals presented with short stature for age (individuals 1 and 7). Additionally, cardiac defects were present in three individuals, specifically ventricular septal defect (VSD; individuals 2 and 3) and tetralogy of Fallot (individual 5). Three individuals presented with labiopalatine cleft or bifid uvula (individuals 2, 3, and 4). Although several affected individuals displayed dysmorphic facial features, including broad nasal root, arched and full eyebrows, synophrys, or epicanthus, no unifying facial gestalt was evident across all affected individuals (Figure 3A).
Figure 3.
Facial photographs and magnetic resonance imaging (MRI) of individuals who harbor CNVs or SNVs impacting SIN3B
(A) Front view of individual 3, who has an 869 kb deletion CNV; note the bulbous nose, arched eyebrows, epicanthus, broad nasal root, and prominent forehead. Front view of individual 7, who harbors the c.1579delC (p.Arg527Glyfs∗12) variant; note large ears and full and arched eyebrows. Front view of individual 8, carrying the c.249C>G (p.Ile83Met) variant; note prominent coronal suture, arched eyebrows, and small palpebral fissures.
(B) Brain MRI from individuals 3 and 6. Individual 3: note short corpus callosum (blue segment) and olfactory bulb agenesis (asterisk). Individual 6: note polymicrogyria (blue circle) with corpus callosum defects (blue arrow) and subependymal nodular heterotopia (dotted blue circle).
Zebrafish mutants harboring truncating mutations in sin3b display locomotion defects, delayed ossification, and shortened body length.26 With the exception of variable growth phenotypes in our cohort (Table 1), these aberrant zebrafish mutant phenotypes did not have discrete anatomical correlates with features of our human cohort. To test whether we could link anatomical phenotypes observed in individuals with SIN3B haploinsufficiency, we disrupted the SIN3B ortholog in zebrafish (Ensembl: ENSDARG00000062472, GRCz10; 63% identity, 74% similarity versus human [GenBank: NP_056075.1]; Figure S2A). All experiments involving zebrafish were approved by the Duke University Institutional Animal Care and Use Committee. These experiments were performed at least twice with similar results, with the investigator masked to experimental condition, and all statistical comparisons were performed with a non-parametric Kruskal-Wallis test. We generated F0 mutant models by targeting exon 13 of sin3b by using CRISPR/Cas9 genome editing; we confirmed a high level of mosaicism for frameshifting events through heteroduplex analysis and sequencing of PCR amplicons flanking the target sites (>90%; Figures S2A and S2B).
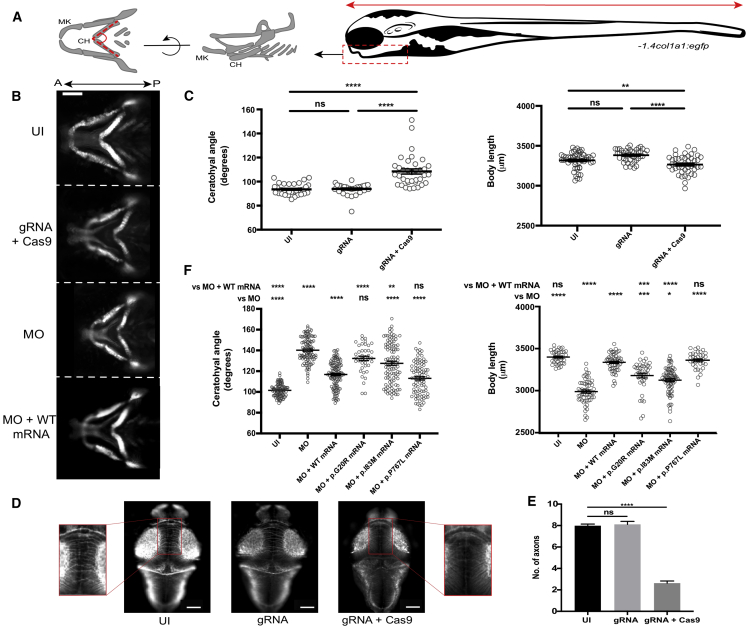
Given that several individuals in our study displayed dysmorphic facial features (Figure 3A), we first focused on cartilage patterning as a readout for sin3b effects on development. We and others have shown previously that morphometric assessment of the pharyngeal skeleton in zebrafish is directly relevant to craniofacial features associated with neurodevelopmental defects in humans.27, 28, 29, 30, 31, 32, 33 We injected guide RNA (gRNA) and Cas9 protein into the cell of single cell-staged embryos carrying a 1.4col1a1:egfp transgene,34 performed live bright-field and fluorescent imaging at 3 days post fertilization (dpf), and analyzed cartilaginous ventral structures of the developing larval head by measuring the ceratohyal (CH) angle. sin3b F0 mutants display a significantly increased CH angle compared with control larvae or larvae injected with gRNA alone, supporting a role for the Sin3 complex in patterning of anterior facial structures (p < 0.0001, gRNA + Cas9 versus gRNA alone or controls; n = 21–34 larvae/batch; repeated, masked scoring; Figures 4A–4C). Additionally, and consistent with stable sin3b mutants,26 sin3b F0 larvae display significantly shorter body length compared with control larvae or larvae injected with gRNA alone (p < 0.0001, gRNA + Cas9 versus gRNA alone; p < 0.005, gRNA + Cas9 versus controls; n = 21–27 larvae/batch; repeated, masked scoring; Figure 4C). Further, to detect neuroanatomical defects, we investigated the integrity of commissural axon tracts in the zebrafish brain at 3 dpf as a proxy for the corpus callosum defects observed in two affected individuals with SIN3B variants. We fixed larvae, immunostained with acetylated tubulin antibody, and quantified the number of intertectal neurons crossing the dorsal midline between the optic tecta as described.35 We observed a significant depletion of commissural axons in sin3b F0 mutants compared with both gRNA alone batches or uninjected controls (Figures 4D and 4E; p < 0.0001 for gRNA + Cas9 versus gRNA alone or controls; n = 49–50 larvae/batch; repeated).
Figure 4.
Modeling of sin3b disruption in zebrafish shows abnormal craniofacial patterning, reduced body length, and pathogenicity of case-associated missense variants
(A) Schematic of structures of interest used for in vivo measurements in developing zebrafish larvae at 3 days post fertilization (dpf); cartilaginous craniofacial structures (left, angle between dashed lines); body length (right, red horizontal arrow); CH, ceratohyal cartilage; MK, Meckel cartilage. −1.4col1a1:egfp transgenic zebrafish larvae were positioned with the Vertebrate Automated Screening Technology (VAST) BioImager and ventral fluorescent images were captured as indicated (right panel, red vertical arrow).
(B) Representative ventral images of in vivo models of sin3b disruption in zebrafish. CH angle was measured as indicated in (A). UI, uninjected; MO, morpholino; WT, human SIN3B mRNA. Scale bar, 100 μm.
(C) Quantification of craniofacial defects and body length in CRISPR/Cas9 F0 mutants. Small insertions and deletions were introduced into sin3b exon 13 via a high-efficiency guide RNA (gRNA). Targeting with gRNA leads to an increase of CH angle (left) and decrease in body length (right), indicating a crucial role for the Sin3 complex in the patterning of anterior structures and growth (n = 21–34 and 21–27 larvae/batch, repeated, for craniofacial and body length, respectively).
(D) Representative images showing fluorescent signal detected from the dorsal aspect of fixed and acetylated tubulin antibody-stained 3dpf larvae. Zoomed insets (left and right) show the region assessed for commissural neurons that cross the midline.
(E) Quantification of intertectal neurons in sin3b F0 mutants as a proxy for corpus callosum defects in SIN3B mutation-bearing humans. n = 49–50 larvae/batch, repeated.
(F) Transient suppression models mimic sin3b CRISPR F0 mutants. sin3b morphants (injected with 9 ng of e2i2 splice-blocking [sb] MO) display a broadened CH angle (left) and shortened body length (right) compared with uninjected controls. Coinjection of sin3b e2i2 splice-blocking MO with 100 pg of SIN3B WT human mRNA rescues this phenotype significantly. Coinjection of MO with case-specific variant mRNA (encoding p.Gly20Arg and p.Ile83Met) resulted in significantly reduced ability to rescue CH and body length compared with SIN3B WT mRNA (n = 41–61 larvae/batch, repeated). p.Pro767Leu is a negative control variant (rs117307745; six homozygotes in gnomAD). Statistical analyses were performed via a non-parametric Kruskal-Wallis test. ∗∗∗∗p < 0.0001; ∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05; ns, not significant; error bars represent standard error of the mean.
We then confirmed the specificity of the craniofacial and body length phenotypes in F0 mutants by performing MO-based knockdown of sin3b. We obtained a splice-blocking (sb) MO targeting the splice donor site of sin3b exon 2 (e2i2), injected it into embryo batches at the one-to-four cell stage, and generated cDNA from whole larvae at 3 dpf to monitor mRNA splicing. We performed RT-PCR to amplify and sequence regions of interest encompassing exon 2 and confirmed a frameshifting event induced by exclusion of sin3b exon 2 (Figures S3A–S3C). Next, we injected increasing concentrations of e2i2 sb MO (3, 6, and 9 ng) into −1.4col1a1:egfp embryos and subjected larvae to the same craniofacial phenotyping paradigm described for F0 mutants. We observed a dose-dependent increase in CH angle in sin3b morphants (p < 0.0001; n = 35–58 larvae/condition; Figure S3D). Moreover, coinjection of e2i2 sb MO with wild-type (WT) human SIN3B mRNA (GenBank: NM_015260.4; generated with a commercially obtained ORF clone; Genecopoeia, Z4616) as described36 rescued these craniofacial defects, indicating MO specificity (p < 0.0001 for MO versus MO+WT mRNA; n = 34–72 larvae/batch; repeated; Figure S3E). To test the pathogenicity of the p.Gly20Arg and p.Ile83Met variants, we compared the efficiency of variant SIN3B mRNA to rescue the morphant phenotype versus WT mRNA for both craniofacial patterning and body length. We used a common variant from gnomAD, p.Pro767Leu, as a negative control (dbSNP: rs117307745, six homozygotes in 282,500 individuals). In replicate experiments, the case-associated SNVs scored as hypomorphic (significantly less rescue than WT mRNA but also different from MO alone). However, the negative control variant scored consistently as benign (not significantly different from WT mRNA rescue; Figure 4F). Finally, to rule out a gain-of-function hypothesis for the nonsynonymous variants and exclude an effect of WT mRNA alone on the detected phenotypes, we confirmed that expression of WT or case-associated variant human SIN3B mRNA does not induce craniofacial defects or body length variations in injected larvae at 3 dpf (n = 36–46 larvae/batch; repeated; Figures S4A and S4B). Together, our in vivo complementation data in larval zebrafish with mutation or suppression of sin3b reinforce the deleterious effects of SIN3B haploinsufficiency resulting in the clinical features noted in our human cohort.
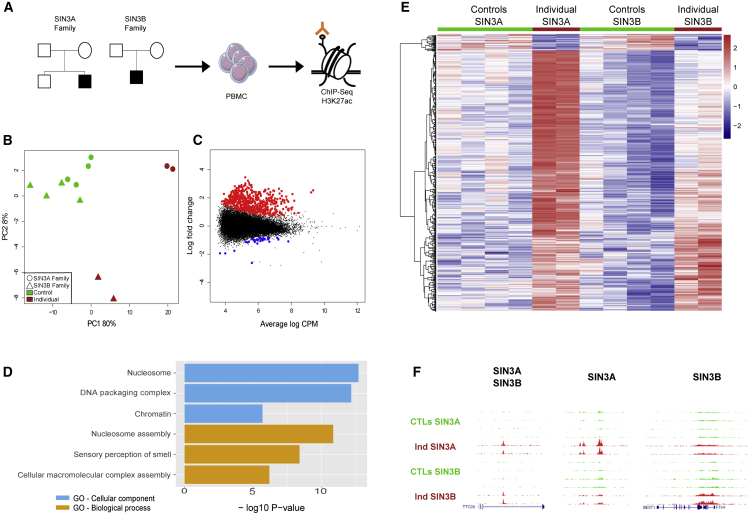
To investigate the consequences of SIN3B alteration on gene regulation, we used histone H3 K27 acetylation (H3K27ac) ChIP-seq to map genome-wide enhancer and promoter states in cells derived from an individual carrying a de novo SIN3B deletion and his healthy family members (individual 1; Figure 5A). H3K27ac is a histone mark found at active gene regulatory elements (enhancers and promoters) and is thought to be highly cell type specific.37,38 This mark is therefore used to define active gene regulatory elements, and because of its robustness, this mark has been used to identify disease-specific mechanisms, especially for neurodevelopmental and degenerative disorders.39,40 Here, we used the genome-wide distribution of this mark to test whether the regulatory landscape is altered in PBMCs, which would suggest that the putative repressor gene function of SIN3B is abrogated. Because SIN3A is thought to have a similar molecular function,41 we also profiled an individual with a SIN3A deletion (individual 4 from Witteveen et al.15 with a ∼350 kb microdeletion) to test whether SIN3A and SIN3B gene disruption leads to similar epigenetic alterations. For comparison with healthy individuals, we also profiled PBMCs from the healthy parents of the SIN3A and SIN3B deletion-bearing individuals as well as the unaffected brother of the individual with the SIN3A (Figure 5A).
Figure 5.
Histone acetylation (H3K27ac) analysis in cells derived from affected individuals with SIN3A or SIN3B deletion CNVs compared with within-pedigree control individuals reveals hyperacetylation in affected individuals
(A) Diagram showing the experimental approach of the ChIP-seq H3K27ac analysis performed on peripheral blood mononuclear cells (PBMCs) from affected individuals compared with their healthy family members. We characterized genome-wide histone acetylation changes in one individual with a SIN3A deletion (individual 4 in Witteveen et al. 15 with a ∼350 kb microdeletion) and individual 1 from this study with a SIN3B deletion (see Figure 1).
(B) PCA (principal-component analysis) plot of peak heights from the 532 differentially acetylated peaks between all control individuals compared with affected individuals.
(C) MA (log ratio-mean average) plot showing hyperacetylated (red) and hypoacetylated (blue) peaks for both SIN3A/B-affected individuals versus all control individuals.
(D) Top three most significant functional groups corresponding to hyperacetylated peaks. GO, gene ontology.
(E) Heatmap of unsupervised clustering of differentially acetylated peaks; Z scores were calculated from normalized counts per million reads.
(F) Representative ChIP-seq genome browser tracks highlighting commonly hyperacetylated peaks in SIN3A and SIN3B deletion-bearing individuals (left) and exemplar hyperacetylated peaks distinct to either SIN3A (center)- or SIN3B (right)-affected individuals. CTL, control; Ind, affected individual.
For each individual, we isolated PBMCs and performed two replicate H3K27ac ChIP-seq experiments. We sequenced ChIP-seq libraries and performed quality control analysis; two samples were removed because of a low fraction of reads in peaks (FrIP) or low non-redundant fraction (NRF) (see supplemental methods and Table S5). For each of the remaining samples, we called peaks by using dfilter42 (average N of peaks = 23,043), merged all the peaks, and counted reads for each sample within the merged peak set. We used these peaks to calculate differential acetylation peaks between the individuals with SIN3A and SIN3B CNVs and their within-pedigree control individuals. In total, we found 500 upregulated and 32 downregulated peaks at an FDR cut-off of 5%. Principal-component analysis (PCA) of differential peaks reveals that while the healthy control individuals cluster together, the profiles from SIN3A and SIN3B CNV carriers are distinct, suggesting that their epigenomic state is altered (Figure 5B). Notably, more peaks were upregulated than downregulated, suggesting that SIN3A and SIN3B contribute to repressor function of enhancers and promoters in PBMCs (Figure 5C). To categorize the biological functions of genes with altered H3K27ac profiles, we performed Gene Ontology analysis. Among the gene regulatory elements upregulated in either mutant context, we found that various gene pathways are altered, and nucleosome assembly was the most significantly enriched (Figure 5D).
To investigate potential differences in regulatory elements influenced by either SIN3A or SIN3B independently, we analyzed differential peaks identified in each sample. These were detected when each individual was compared with his respective family members (Figure S5A), suggesting that haploinsufficiency of either SIN3A or SIN3B leads to hyperacetylation at several loci. Importantly, the epigenetic alterations observed in affected individuals with SIN3A and SIN3B CNVs were distinct in their intensity, suggesting that although they may be part of the same complex, their regulatory function appears to be non-redundant (Figure 5E). For example, analysis of specific loci shows that some peaks are altered similarly (e.g., TTC28 [MIM: 615098]), while other peaks are SIN3A or SIN3B specific (Figure 5F). Overall, fold changes between affected individuals and control individuals were similar for 16% of hyperacetylated peaks (logFC > 1.5; Figure S5B). These data are supported further by the observation that peaks significantly upregulated in the context of SIN3A haploinsufficiency are also significantly upregulated in the SIN3B mutant cells as compared with in-family controls (p < 0.001); there was a similar observation in the reciprocal context (peaks significantly upregulated in SIN3B mutant cells are also significantly upregulated in SIN3A CNV-bearing cells [p < 0.001]; Figure S5C). Taken together, these results suggest that, as is the case for heterozygous deletion of SIN3A, heterozygous SIN3B deletion leads to epigenetic alterations at specific loci in circulating immune cells, a cell type shown previously as a reasonable proxy for neurodevelopmental traits.43 However, we are cautious in the interpretation of our data and recognize the need to analyze cells from additional SIN3A and SIN3B pedigrees, particularly those with single-gene deletions or pathogenic SNVs.
Here, we show genetic and in vivo modeling evidence that de novo SIN3B pathogenic variants cause a neurodevelopmental syndrome characterized by syndromic ID/DD with variable ASD, growth defects, congenital anomalies, and dysmorphic craniofacial features. Variants in epigenetic regulators, and particularly in genes modulating histone acetylation, have been associated previously with several neurodevelopmental disorders, reinforcing the importance for tightly regulated histone post-translational modification events. For example, disruptive variants in HDAC4 (MIM: 605314) likely resulting in haploinsufficiency have been described in individuals with brachydactyly-mental retardation syndrome44,45 (MIM: 600430). Additionally, HDAC8 (MIM: 300269) loss-of-function variants cause an X-linked form of Cornelia de Lange syndrome46,47 (MIM: 300882), although the likely pathogenic mechanism is impairment of cohesin subunit SMC3 deacetylation by HDAC8. These rare disorders exemplify how histone deacetylation acts as a key molecular process during human development. Moreover, although no Mendelian disorder has been associated with disrupted HDAC7 (MIM: 606542) in humans, Hdac7−/− mice are embryonic lethal.48 Hdac1 and Hdac2 disruption in mice leads to developmental defects.48 Homozygous mouse Sin3b mutants display embryonic development defects at embryonic day (E) 13.5 with increased mortality rate and a growth retardation phenotype.49
The closely related SIN3A paralog (48% similarity to SIN3B protein sequence, CLUSTALW v.1.81, multiple sequence alignment, GenBank: NP_056075; Figure S6), located on chromosome 15q24.2, has been associated with syndromic ID and ASD.15 Clinical features of SIN3A-affected individuals include mild ID, recognizable facial gestalt, abnormalities in brain MRIs including ventricular dilatation, corpus callosum dysgenesis, and aberrant cortical development. Some affected individuals were also described with ASD, seizures, microcephaly, and short stature. In vivo knockdown of Sin3a in mice impairs cortical neurogenesis, leading to a decreased number of cortical progenitors and altered cortical projections in the developing brain. Clinical features described in our SIN3B cohort overlap with previously described phenotypes associated with the SIN3A-related disorder. Our H3K27ac ChIP-seq experiments on cells from SIN3A and SIN3B CNV deletion pedigrees offer initial molecular insight to explain these similarities as well as subtle differences.
In summary, our data expand the phenotypic spectrum associated with Sin3 complex haploinsufficiency to syndromic ID/ASD, arguing against the functional redundancy of SIN3A and SIN3B. Our findings confirm the importance of Sin3 complex integrity for central nervous system development, anterior cartilage patterning, and growth and highlight a major role for SIN3B in this process.
Declaration of interests
S.S. and K.M. are employees of GeneDx, Inc., a wholly owned subsidiary of OPKO Health, Inc. N.K. is a paid consultant for and holds significant stock of Rescindo Therapeutics, Inc. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from clinical genetic testing conducted at Baylor Genetics Laboratory.
Acknowledgments
We are grateful to individuals who participated in the study. We also thank Mr. Z. Kupchinsky and Mr. I. Pediaditakis for zebrafish husbandry; Mr. D. Morrow for assisting with reagents for in vivo modeling studies; and members of the Center for Human Disease Modeling for helpful discussions. This work was supported by funds from US NIH grant R01 HD096326 to N.K. and R01 MH106826 to E.E.D.
Published: April 2, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.03.017.
Contributor Information
Jeremie Poschmann, Email: jeremie.poschmann@univ-nantes.fr.
Erica E. Davis, Email: eridavis@luriechildrens.org.
Bertrand Isidor, Email: bertrand.isidor@chu-nantes.fr.
Data and code availability
The NCBI reference sequence numbers for human SIN3B transcript and zebrafish sin3b transcript are GenBank NM_015260.4 and NP_056075.1 (hg19) and GenBank: NM_001044945 and NP_001038410 (GRCz10), respectively.
Web resources
1000 Genomes, https://www.internationalgenome.org/
CHOPCHOP, http://chopchop.cbu.uib.no/
Database of Genomic Variants (DGV), http://dgv.tcag.ca/dgv/app/home
DECIPHER, https://decipher.sanger.ac.uk/
ExAC browser, http://exac.broadinstitute.org/
Exome Variant Server, https://evs.gs.washington.edu/EVS/
GeneMatcher, https://genematcher.org/
gnomAD browser, https://gnomad.broadinstitute.org/
OMIM, https://omim.org/
Primer3, https://bioinfo.ut.ee/primer3
Supplemental information
References
- 1.Izumi K. Disorders of Transcriptional Regulation: An Emerging Category of Multiple Malformation Syndromes. Mol. Syndromol. 2016;7:262–273. doi: 10.1159/000448747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tebbenkamp A.T.N., Willsey A.J., State M.W., Sestan N. The developmental transcriptome of the human brain: implications for neurodevelopmental disorders. Curr. Opin. Neurol. 2014;27:149–156. doi: 10.1097/WCO.0000000000000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen L.-F., Zhou A.S., West A.E. Transcribing the connectome: roles for transcription factors and chromatin regulators in activity-dependent synapse development. J. Neurophysiol. 2017;118:755–770. doi: 10.1152/jn.00067.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larizza L., Finelli P. Developmental disorders with intellectual disability driven by chromatin dysregulation: Clinical overlaps and molecular mechanisms. Clin. Genet. 2019;95:231–240. doi: 10.1111/cge.13365. [DOI] [PubMed] [Google Scholar]
- 5.Lopez-Atalaya J.P., Valor L.M., Barco A. Epigenetic factors in intellectual disability: the Rubinstein-Taybi syndrome as a paradigm of neurodevelopmental disorder with epigenetic origin. Prog. Mol. Biol. Transl. Sci. 2014;128:139–176. doi: 10.1016/B978-0-12-800977-2.00006-1. [DOI] [PubMed] [Google Scholar]
- 6.Bjornsson H.T. The Mendelian disorders of the epigenetic machinery. Genome Res. 2015;25:1473–1481. doi: 10.1101/gr.190629.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nan X., Ng H.H., Johnson C.A., Laherty C.D., Turner B.M., Eisenman R.N., Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y., Iratni R., Erdjument-Bromage H., Tempst P., Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 9.Knoepfler P.S., Eisenman R.N. Sin meets NuRD and other tails of repression. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- 10.Grimes J.A., Nielsen S.J., Battaglioli E., Miska E.A., Speh J.C., Berry D.L., Atouf F., Holdener B.C., Mandel G., Kouzarides T. The co-repressor mSin3A is a functional component of the REST-CoREST repressor complex. J. Biol. Chem. 2000;275:9461–9467. doi: 10.1074/jbc.275.13.9461. [DOI] [PubMed] [Google Scholar]
- 11.Kadamb R., Mittal S., Bansal N., Batra H., Saluja D. Sin3: insight into its transcription regulatory functions. Eur. J. Cell Biol. 2013;92:237–246. doi: 10.1016/j.ejcb.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 12.van Oevelen C., Wang J., Asp P., Yan Q., Kaelin W.G., Jr., Kluger Y., Dynlacht B.D. A role for mammalian Sin3 in permanent gene silencing. Mol. Cell. 2008;32:359–370. doi: 10.1016/j.molcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alland L., David G., Shen-Li H., Potes J., Muhle R., Lee H.-C., Hou H., Jr., Chen K., DePinho R.A. Identification of mammalian Sds3 as an integral component of the Sin3/histone deacetylase corepressor complex. Mol. Cell. Biol. 2002;22:2743–2750. doi: 10.1128/MCB.22.8.2743-2750.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laherty C.D., Yang W.M., Sun J.M., Davie J.R., Seto E., Eisenman R.N. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 15.Witteveen J.S., Willemsen M.H., Dombroski T.C.D., van Bakel N.H.M., Nillesen W.M., van Hulten J.A., Jansen E.J.R., Verkaik D., Veenstra-Knol H.E., van Ravenswaaij-Arts C.M.A. Haploinsufficiency of MeCP2-interacting transcriptional co-repressor SIN3A causes mild intellectual disability by affecting the development of cortical integrity. Nat. Genet. 2016;48:877–887. doi: 10.1038/ng.3619. [DOI] [PubMed] [Google Scholar]
- 16.Aten E., den Hollander N., Ruivenkamp C., Knijnenburg J., van Bokhoven H., den Dunnen J., Breuning M. Split hand-foot malformation, tetralogy of Fallot, mental retardation and a 1 Mb 19p deletion-evidence for further heterogeneity? Am. J. Med. Genet. A. 2009;149A:975–981. doi: 10.1002/ajmg.a.32748. [DOI] [PubMed] [Google Scholar]
- 17.Bens S., Haake A., Tönnies H., Vater I., Stephani U., Holterhus P.-M., Siebert R., Caliebe A. A de novo 1.1Mb microdeletion of chromosome 19p13.11 provides indirect evidence for EPS15L1 to be a strong candidate for split hand split foot malformation. Eur. J. Med. Genet. 2011;54:e501–e504. doi: 10.1016/j.ejmg.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gripp K.W., Wotton D., Edwards M.C., Roessler E., Ades L., Meinecke P., Richieri-Costa A., Zackai E.H., Massagué J., Muenke M., Elledge S.J. Mutations in TGIF cause holoprosencephaly and link NODAL signalling to human neural axis determination. Nat. Genet. 2000;25:205–208. doi: 10.1038/76074. [DOI] [PubMed] [Google Scholar]
- 20.Cheong S.-S., Hentschel L., Davidson A.E., Gerrelli D., Davie R., Rizzo R., Pontikos N., Plagnol V., Moore A.T., Sowden J.C. Mutations in CPAMD8 Cause a Unique Form of Autosomal-Recessive Anterior Segment Dysgenesis. Am. J. Hum. Genet. 2016;99:1338–1352. doi: 10.1016/j.ajhg.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martínez F., Caro-Llopis A., Roselló M., Oltra S., Mayo S., Monfort S., Orellana C. High diagnostic yield of syndromic intellectual disability by targeted next-generation sequencing. J. Med. Genet. 2017;54:87–92. doi: 10.1136/jmedgenet-2016-103964. [DOI] [PubMed] [Google Scholar]
- 22.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Franco E., Watson R.A., Weninger W.J., Wong C.C., Flanagan S.E., Caswell R., Green A., Tudor C., Lelliott C.J., Geyer S.H. A Specific CNOT1 Mutation Results in a Novel Syndrome of Pancreatic Agenesis and Holoprosencephaly through Impaired Pancreatic and Neurological Development. Am. J. Hum. Genet. 2019;104:985–989. doi: 10.1016/j.ajhg.2019.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kruszka P., Berger S.I., Weiss K., Everson J.L., Martinez A.F., Hong S., Anyane-Yeboa K., Lipinski R.J., Muenke M. A CCR4-NOT Transcription Complex, Subunit 1, CNOT1, Variant Associated with Holoprosencephaly. Am. J. Hum. Genet. 2019;104:990–993. doi: 10.1016/j.ajhg.2019.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vissers L.E.L.M., Kalvakuri S., de Boer E., Geuer S., Oud M., van Outersterp I., Kwint M., Witmond M., Kersten S., Polla D.L., DDD Study De Novo Variants in CNOT1, a Central Component of the CCR4-NOT Complex Involved in Gene Expression and RNA and Protein Stability, Cause Neurodevelopmental Delay. Am. J. Hum. Genet. 2020;107:164–172. doi: 10.1016/j.ajhg.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moravec C.E., Yousef H., Kinney B.A., Salerno-Eichenholz R., Monestime C.M., Martin B.L., Sirotkin H.I. Zebrafish sin3b mutants are viable but have size, skeletal, and locomotor defects. Dev. Dyn. 2017;246:946–955. doi: 10.1002/dvdy.24581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaw N.D., Brand H., Kupchinsky Z.A., Bengani H., Plummer L., Jones T.I., Erdin S., Williamson K.A., Rainger J., Stortchevoi A. SMCHD1 mutations associated with a rare muscular dystrophy can also cause isolated arhinia and Bosma arhinia microphthalmia syndrome. Nat. Genet. 2017;49:238–248. doi: 10.1038/ng.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rooryck C., Diaz-Font A., Osborn D.P.S., Chabchoub E., Hernandez-Hernandez V., Shamseldin H., Kenny J., Waters A., Jenkins D., Kaissi A.A. Mutations in lectin complement pathway genes COLEC11 and MASP1 cause 3MC syndrome. Nat. Genet. 2011;43:197–203. doi: 10.1038/ng.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beunders G., Voorhoeve E., Golzio C., Pardo L.M., Rosenfeld J.A., Talkowski M.E., Simonic I., Lionel A.C., Vergult S., Pyatt R.E. Exonic deletions in AUTS2 cause a syndromic form of intellectual disability and suggest a critical role for the C terminus. Am. J. Hum. Genet. 2013;92:210–220. doi: 10.1016/j.ajhg.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stankiewicz P., Khan T.N., Szafranski P., Slattery L., Streff H., Vetrini F., Bernstein J.A., Brown C.W., Rosenfeld J.A., Rednam S., Deciphering Developmental Disorders Study Haploinsufficiency of the Chromatin Remodeler BPTF Causes Syndromic Developmental and Speech Delay, Postnatal Microcephaly, and Dysmorphic Features. Am. J. Hum. Genet. 2017;101:503–515. doi: 10.1016/j.ajhg.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Küry S., Besnard T., Ebstein F., Khan T.N., Gambin T., Douglas J., Bacino C.A., Craigen W.J., Sanders S.J., Lehmann A. De Novo Disruption of the Proteasome Regulatory Subunit PSMD12 Causes a Syndromic Neurodevelopmental Disorder. Am. J. Hum. Genet. 2017;100:352–363. doi: 10.1016/j.ajhg.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frosk P., Arts H.H., Philippe J., Gunn C.S., Brown E.L., Chodirker B., Simard L., Majewski J., Fahiminiya S., Russell C., FORGE Canada Consortium. Canadian Rare Diseases: Models & Mechanisms Network A truncating mutation in CEP55 is the likely cause of MARCH, a novel syndrome affecting neuronal mitosis. J. Med. Genet. 2017;54:490–501. doi: 10.1136/jmedgenet-2016-104296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Isrie M., Breuss M., Tian G., Hansen A.H., Cristofoli F., Morandell J., Kupchinsky Z.A., Sifrim A., Rodriguez-Rodriguez C.M., Dapena E.P. Mutations in Either TUBB or MAPRE2 Cause Circumferential Skin Creases Kunze Type. Am. J. Hum. Genet. 2015;97:790–800. doi: 10.1016/j.ajhg.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kague E., Gallagher M., Burke S., Parsons M., Franz-Odendaal T., Fisher S. Skeletogenic fate of zebrafish cranial and trunk neural crest. PLoS ONE. 2012;7:e47394. doi: 10.1371/journal.pone.0047394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ansar M., Ullah F., Paracha S.A., Adams D.J., Lai A., Pais L., Iwaszkiewicz J., Millan F., Sarwar M.T., Agha Z. Bi-allelic Variants in DYNC1I2 Cause Syndromic Microcephaly with Intellectual Disability, Cerebral Malformations, and Dysmorphic Facial Features. Am. J. Hum. Genet. 2019;104:1073–1087. doi: 10.1016/j.ajhg.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niederriter A.R., Davis E.E., Golzio C., Oh E.C., Tsai I.-C., Katsanis N. In vivo modeling of the morbid human genome using Danio rerio. J. Vis. Exp. 2013;78:e50338. doi: 10.3791/50338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marzi S.J., Leung S.K., Ribarska T., Hannon E., Smith A.R., Pishva E., Poschmann J., Moore K., Troakes C., Al-Sarraj S. A histone acetylome-wide association study of Alzheimer’s disease identifies disease-associated H3K27ac differences in the entorhinal cortex. Nat. Neurosci. 2018;21:1618–1627. doi: 10.1038/s41593-018-0253-7. [DOI] [PubMed] [Google Scholar]
- 40.Sun W., Poschmann J., Cruz-Herrera Del Rosario R., Parikshak N.N., Hajan H.S., Kumar V., Ramasamy R., Belgard T.G., Elanggovan B., Wong C.C.Y. Histone Acetylome-wide Association Study of Autism Spectrum Disorder. Cell. 2016;167:1385–1397.e11. doi: 10.1016/j.cell.2016.10.031. [DOI] [PubMed] [Google Scholar]
- 41.Bainor A.J., Saini S., Calderon A., Casado-Polanco R., Giner-Ramirez B., Moncada C., Cantor D.J., Ernlund A., Litovchick L., David G. The HDAC-Associated Sin3B Protein Represses DREAM Complex Targets and Cooperates with APC/C to Promote Quiescence. Cell Rep. 2018;25:2797–2807.e8. doi: 10.1016/j.celrep.2018.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar V., Muratani M., Rayan N.A., Kraus P., Lufkin T., Ng H.H., Prabhakar S. Uniform, optimal signal processing of mapped deep-sequencing data. Nat. Biotechnol. 2013;31:615–622. doi: 10.1038/nbt.2596. [DOI] [PubMed] [Google Scholar]
- 43.Migliavacca E., Golzio C., Männik K., Blumenthal I., Oh E.C., Harewood L., Kosmicki J.A., Loviglio M.N., Giannuzzi G., Hippolyte L., 16p11.2 European Consortium A Potential Contributory Role for Ciliary Dysfunction in the 16p11.2 600 kb BP4-BP5 Pathology. Am. J. Hum. Genet. 2015;96:784–796. doi: 10.1016/j.ajhg.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Villavicencio-Lorini P., Klopocki E., Trimborn M., Koll R., Mundlos S., Horn D. Phenotypic variant of Brachydactyly-mental retardation syndrome in a family with an inherited interstitial 2q37.3 microdeletion including HDAC4. Eur. J. Hum. Genet. 2013;21:743–748. doi: 10.1038/ejhg.2012.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wheeler P.G., Huang D., Dai Z. Haploinsufficiency of HDAC4 does not cause intellectual disability in all affected individuals. Am. J. Med. Genet. A. 2014;164A:1826–1829. doi: 10.1002/ajmg.a.36542. [DOI] [PubMed] [Google Scholar]
- 46.Harakalova M., van den Boogaard M.-J., Sinke R., van Lieshout S., van Tuil M.C., Duran K., Renkens I., Terhal P.A., de Kovel C., Nijman I.J. X-exome sequencing identifies a HDAC8 variant in a large pedigree with X-linked intellectual disability, truncal obesity, gynaecomastia, hypogonadism and unusual face. J. Med. Genet. 2012;49:539–543. doi: 10.1136/jmedgenet-2012-100921. [DOI] [PubMed] [Google Scholar]
- 47.Deardorff M.A., Bando M., Nakato R., Watrin E., Itoh T., Minamino M., Saitoh K., Komata M., Katou Y., Clark D. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature. 2012;489:313–317. doi: 10.1038/nature11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Montgomery R.L., Hsieh J., Barbosa A.C., Richardson J.A., Olson E.N. Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc. Natl. Acad. Sci. USA. 2009;106:7876–7881. doi: 10.1073/pnas.0902750106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.David G., Grandinetti K.B., Finnerty P.M., Simpson N., Chu G.C., Depinho R.A. Specific requirement of the chromatin modifier mSin3B in cell cycle exit and cellular differentiation. Proc. Natl. Acad. Sci. USA. 2008;105:4168–4172. doi: 10.1073/pnas.0710285105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The NCBI reference sequence numbers for human SIN3B transcript and zebrafish sin3b transcript are GenBank NM_015260.4 and NP_056075.1 (hg19) and GenBank: NM_001044945 and NP_001038410 (GRCz10), respectively.