Summary
Macular degeneration (MD) is characterized by the progressive deterioration of the macula and represents one of the most prevalent causes of blindness worldwide. Abnormal intracellular accumulation of lipid droplets and pericellular deposits of lipid-rich material in the retinal pigment epithelium (RPE) called drusen are clinical hallmarks of different forms of MD including Doyne honeycomb retinal dystrophy (DHRD) and age-related MD (AMD). However, the appropriate molecular therapeutic target underlying these disorder phenotypes remains elusive. Here, we address this knowledge gap by comparing the proteomic profiles of induced pluripotent stem cell (iPSC)-derived RPEs (iRPE) from individuals with DHRD and their isogenic controls. Our analysis and follow-up studies elucidated the mechanism of lipid accumulation in DHRD iRPE cells. Specifically, we detected significant downregulation of carboxylesterase 1 (CES1), an enzyme that converts cholesteryl ester to free cholesterol, an indispensable process in cholesterol export. CES1 knockdown or overexpression of EFEMP1R345W, a variant of EGF-containing fibulin extracellular matrix protein 1 that is associated with DHRD and attenuated cholesterol efflux and led to lipid droplet accumulation. In iRPE cells, we also found that EFEMP1R345W has a hyper-inhibitory effect on epidermal growth factor receptor (EGFR) signaling when compared to EFEMP1WT and may suppress CES1 expression via the downregulation of transcription factor SP1. Taken together, these results highlight the homeostatic role of cholesterol efflux in iRPE cells and identify CES1 as a mediator of cholesterol efflux in MD.
Keywords: age-related macular degeneration, Doyne honeycomb destrophy, DHRD, cholesterol efflux, drusen, RPE, CRISPR, isogenic, EGFR signaling, unfolded protein response, lipid accumulation
Introduction
Macular degeneration (MD) is a heterogenous group of severe neurodegenerative diseases characterized by retinal pigment epithelium (RPE) dysfunction, leading to progressive vision loss. Affecting more than 170 million individuals, MD is a leading cause of visual disabilities worldwide. Age is a risk factor for MD, and as the human lifespan increases, age-related macular degeneration (AMD [MIM: 610698]) will be a growing public health concern. By 2040, more than 288 million people are expected to be diagnosed with MD, outpacing all invasive cancers combined and more than double the prevalence of Alzheimer disease (MIM: 104300).1
Early stages of MD can be identified on fundus imaging by the presence of yellow spots called drusen, which correspond to deposits of excess lipids and proteins between the basal lamina of the RPE and Bruch’s membrane.2, 3, 4 Drusen are thought to cause vision loss through geographic atrophy and choroidal neovascularization. Studies exploring the mechanism of deposit formation have suggested the involvement of complement risk factors and the complement system.5, 6, 7, 8, 9, 10 Nonetheless, efforts to further elucidate the pathophysiology of disease and develop effective treatment strategies have fallen short as the vast majority of these studies have focused primarily on AMD. While by far the most prevalent and therefore devastating of all the different forms of MD, AMD as a disease model for MD is limited due to the variability in its genetic and environmental causes.
In this study, we aim to overcome the long-standing knowledge gap regarding drusen formation by focusing on a form of MD with a monogenic etiology: Doyne honeycomb retinal dystrophy (DHRD [MIM: 126600]). DHRD is a rare inherited macular dystrophy that causes irreversible central vision loss later in life due to geographic atrophy and choroidal neovascularization.11 Like other forms of MD, an early indication of DHRD is the development of drusen, which pattern the fundus in a honeycomb-like fashion. Although there is variability in disease progression,12 DHRD-affected individuals exhibit classical MD findings of RPE hypertrophy and abnormal subretinal fibrosis.
To date, the only identified causative gene for DHRD is EFEMP1 (MIM: 601548),11 which encodes epidermal growth factor (EGF)-containing fibulin-like extracellular matrix protein 1, also known as fibulin-3 (F3). EFEMP1 is one of eight glycoproteins in the fibulin family of extracellular matrix (ECM) glycoproteins. All proteins in this family contain a series of calcium-binding EGF domains followed by a C-terminal fibulin-type domain. These fibulin proteins are secreted and integrated into the ECM, where they play a critical role in basement membrane formation.13 Strikingly, mutations in three of the eight fibulin proteins—F3, F5, and F6—have been found or are suspected to contribute to the development of AMD or related retinal degeneration.14, 15, 16, 17, 18
DHRD is caused by a single heterozygous missense mutation (p.Arg345Trp [c.1033C>T]) in EFEMP1. The autosomal-dominant inheritance pattern suggests a toxic gain-of-function mechanism. Previous studies have generated different mouse models of DHRD with various levels of success. Both Efemp1R345W+ single dominant mice and knockout mice expressing no Efemp1 have no observable problems in the eye.19 On the other hand, Efemp1R345W/R345W double dominant mice develop pathological phenotypes in the retina including progressive development of drusen and RPE atrophy but do not genocopy individuals with DHRD.20,21
Cell culture studies have highlighted that both AMD and DHRD lead to significant EFEMP1 immunoreactivity around the RPE in the presence of drusen.22,23 To account for this observation, studies have suggested that the p.Arg345Trp mutation prevents the proper folding and secretion of the EFEMP1 protein, activating an unfolded protein response (UPR) that triggers MD. Subsequent studies have failed to validate this model, however, and have alternatively attributed the retinal degeneration to the complement pathway.24 As evident from this lack of consensus, the question of how the p.Arg345Trp mutation in EFEMP1 causes drusen formation in DHRD remains unanswered; as such, further investigation of the role of EFEMP1 in the retina is warranted.
In addition to EFEMP1R345W/R345W and EFEMP1−/− mouse models, previous studies have also characterized the EFEMP1 p.Arg345Trp variant by overexpressing EFEMP1R345W in ARPE-19 cells, a human retinal pigment epithelial cell line.25,26 These systems, however, are limited in their capacity as disease models for DHRD as they are not translatable to humans. At the same time, heterozygous knock-in of the R345W EFEMP1 mutation in mice results in insignificant phenotypes. To overcome these problems, we used human induced pluripotent stem cells (iPSC)-derived RPE (iRPE) cells to study the pathogenesis of drusen formation in DHRD.
Material and methods
iPSC culture and the differentiation of iRPE
Fibroblasts from three white individuals with DHRD (aged 37, 47, and 59 years) and three healthy donors (aged 14, 55, and 64 years) were reprogrammed into iPSCs using the CytoTune-iPS 2.0 Sendai Reprogramming Kit (Thermo Fisher Scientific). The generated iPSCs were tested for their quality and pluripotency by stem cell marker staining and karyotyping as previously described.27,28 All iPSC lines were passaged every 3–6 days while being maintained in mTeSR-1 medium (STEM CELL Technologies). The iPSC lines were then differentiated into RPE (iRPE) cells. To do this, they were cultured in 6-well culture plates precoated with 1:50 diluted matrigel (CORNING). For the first 2 weeks, the differentiation medium consisted of DMEM (Thermo Fisher Scientific), 15% serum replacement (Thermo Fisher Scientific), 2 mM glutamine (Thermo Fisher Scientific), 50 U/mL penicillin/streptomycin (Thermo Fisher Scientific), 1% non-essential amino acids (Thermo Fisher Scientific), and 10 mM nicotinamide (Sigma-Aldrich). For the following 2 weeks, the culture medium was supplemented with 100 ng/mL human Activin-A (PeproTech). Starting from week 5, the Activin-A was removed. The pigmented flat clusters formed in the plate were manually transferred to another matrigel-coated dish for further expansion.
CRISPR-meditated gene correction for isogenic line
To correct the p.Arg345Trp mutation in iPSCs from individuals with DHRD, cells were transfected by nucleofection using the P3 Primary Cell 4D-Nucleofector X Kit (Lonza) and the program DS150, according to the manufacturer’s instructions. Briefly, a 20 μL electroporation solution was prepared for each reaction, including 15.2 μL of the P3 nucleofector solution, 3.6 μL of the supplement, and 1.2 μL of ribonucleoprotein mixture, which consists of 1 μg Cas9 protein, 300 ng gRNA, and 200 pmol single-stranded oligo donor (ssODN). iPSCs with 60%–70% confluency were used for electroporation. Accutase (Stem Cell Technology) was used to dissociate iPSCs from the plate. Approximately 2 × 105 cells were pelleted and mixed with the 20 μL electroporation solution before being transferred into the cuvette. Nucleofection was conducted on an Amaxa Nucleofector 4D. After nucleofection the cuvette, the cells were immediately transferred to one 10 cm matrigel-coated Petri dish in mTeSR1 medium with 10 μM ROCK inhibitor (Selleck Chemical). Two days later, the iPSC culture was subcultured again and split into several 10 cm Petri dishes at a density of 200 cells/dish. After 1 week of culture, iPSC colonies of appropriate size were manually picked and transferred into matrigel-coated 96-well plate for colonial expansion. The cells in each well were sampled and extracted for genomic DNA. ScrFI (New England Biolabs) was used to carry out restriction fragment length polymorphism (RFLP) assay for the screening of gene-corrected clones. The quality of iPSC lines after gene correction was confirmed by karyotyping and immunostaining (Figure S1). Off-target site prediction was performed using Benchling webtool, and potential off-target loci in the gene-corrected clones were amplified by PCR and then analyzed by Sanger sequencing (Table S1).
Retinal organoid differentiation
Human iPSCs lines were maintained on Matrigel (BD)-coated plates in mTeSR medium (STEMCELL Technologies) and passaged with ReleSR (STEMCELL Technologies). Retinal organoid differentiation was carried out using the agarose microwell array seeding and scraping (AMASS) method.29 In brief, iPSCs at 90% confluence were detached with ReleSR (STEMCELL Technologies). After cell counting, cells were seeded at 2,000 cells per microwell (each microwell array mold contains 81 microwells) and incubated with (±)blebbistatin in mTeSR medium overnight and subsequently transitioned from mTeSR to Neural Induction Medium 1 (NIM)-1 over the next 3 days to form embryoid bodies (EBs). On differentiation day (DD)7 EBs were transferred to Matrigel-coated wells till DD28, with a transition from NIM-1 to NIM-2 medium at DD16. Using the checkerboard-scrapping method, neuroepithelia were lifted. Once lifted, retinal organoids were maintained with NIM-2 until DD41 in poly-HEMA (Sigma)-coated wells. Retinal lamination medium 1 (RLM-1) is used from DD42 to DD69, RLM-2 from DD70 to DD97, and RLM-3 from DD98 for long-term culture. NIM1 (50 mL): 48.95 mL DMEM/F12, 10 μL 10 mg/mL heparin (final concentration, 2 μg/mL), 0.5 mL Media-Non Essential Amino Acids (100×, MEM NEAA), 0.5 mL N2 supplement (100×). NIM2 (50 mL): 48 mL DMEM/ F12 (3:1), 0.5 mL MEM NEAA, 1 mL B27 Supplement (50×, minus vitamin A), 0.5 mL penicillin-streptomycin (P/S, 10,000 U/mL). RLM1 (50 mL): 42.9 mL DMEM/ F12 (3:1), 0.1 mL taurine (100 μM final concentration), 5 mL FBS, 1 mL B27, 0.5 mL MEM NEAA, 0.5 mL P/S. 15. RLM2: RLM1 supplemented with 0.1 μL per mL of 10 mM retinoic acid. RLM3: RLM1 without B27, replaced with N2 supplement and retinoic acid reduced to 0.05 μL per mL.
Comparative proteomic profiling
To prepare samples for mass spectrometer analysis, each cell pellet was homogenized with 1% NP-40 lysis buffer (Thermo Fisher Scientific) with protease & phosphatase cocktails (Thermo Fisher Scientific). Enhanced BCA Protein Quantification assay (Thermo Fisher Scientific) was used to determine the total amount of protein in each sample. Proteins were further purified by mini S-trap columns (Protifi) and digested on column by trypsin. The Thermo Quantitative Fluorometric Peptide Assay was used to quantify peptide concentrations prior to TMT labeling. 40 μg peptides were labeled with TMT 6plex isobaric reagent and mixed for high pH reverse phase peptide fractionation. Thermo Orbitrap Fusion Tribrid Mass Spectrometer was used for MS/MS analysis (MS3 data acquisition method). Two iRPE lines from each group were analyzed with three biological replicates. Proteome Discoverer software (v.2.1) was used to search the acquired MS/MS data against human protein database downloaded from the UniProt website and to generate TMT ratios. Positive identification was set at 5% peptide FDR, and at least 1 unique peptide needed to be identified per protein. Duplicated protein identifications from database were removed. A total of 366 human proteins were quantified and included in the final data. TMT ratios (each tag/common reference) were calculated by PD 2.1 and normalized by total peptide amount. Qlucore Omics Explorer and Prism 6 Software were used to perform correlation and statistical analysis. KNN imputation was used for missing values.
Differentially expressed proteins (DEPs) between groups were identified by 2 fold-change and p value < 0.001 using Wilcoxon signed-rank test. The DEPs were identified and visualized by volcano plots and Venn diagrams. The identified DEPs between the EFEMP1corrected and EFEMP1R345W were analyzed by gene ontology analysis via ShinyGO v.0.61, and the five most enriched groups in biological process were used to create the circus plot.
Immunofluorescence
Anti-BEST1 (NB300-164, Novus Biological), anti-RPE65(NB100-355, Novus Biological), and anti-ZO-1 (40-2300, Invitrogen) antibodies were used to detect mature RPE markers and verify differentiation by immunostaining. To detect UPR markers, anti-HSPA5 (GRP78/BIP) (Sigma) and anti-DDIT3 (CHOP) (Cell Signaling) antibodies were used. The iRPE culture was washed by PBS twice and fixed by 4% paraformaldehyde for 30 min. 5% bovine serum albumin (BSA, Thermo Fisher Scientific) was used for blocking for 2 h at room temperature. The cells were then incubated with the primary antibody diluted (1:500) with 2% BSA in PBS overnight. Alexa Fluor 488- or Alexa Fluor 555-conjugated secondary antibodies (1:500, Thermo Fisher Scientific) were used for the detection of primary antibody. Hochest staining was done at the end for counter staining.
For the detection of CES1 in human retina, human retinal paraffin sections (Biomax) were de-paraffinized with xylene for 3 min. The sections were further hydrated with 100%, 90%, 70%, and 50% alcohol for 3 min each. The sections were then incubated in running water for 10 min. To retrieve the antigen, the slides were incubated in Antigen Unmasking Solution (Thermo Fisher Scientific) for 20 min at 95°C. The slides were washed with running water for 10 min before the staining procedure.
Anti-CES1 (AF4920, R&D) and anti-EGFR (AB32077, Abcam) antibodies were diluted (1:200) with 5% BSA in PBS before being added to the slide for overnight incubation at 4°C. After 3 washes, the sections were stained with Alexa Fluor 555-conjugated secondary antibody (1:500, Thermo Fisher Scientific) for 1 h. Hochest staining was done at the end of antibody staining.
Nile red staining
To stain the intracellular neutral lipid, the iRPE cell cultures were treated with 2.0 μg/mL Nile red (Sigma Aldrich) for 30 min at 37°C. The cells were washed with PBS three times before microscopy observation.
Since mature iRPE culture is usually 100% confluent, the Nile red-positively stained lipid droplets were quantified using ImageJ in a fixed area of 90,000 square micrometers for the number and size of Nile red-positive signals. Fluorescent particles smaller than 0.0001 square micrometers was excluded as noise.
Cholesterol efflux assay
Cholesterol Efflux Assay Kit (Abcam) was used to determine the cholesterol efflux rate in iRPE according to the manufacturer’s protocol. In brief, fluorescence-labeled cholesterol was added to iRPE culture and incubated overnight. On the next day, the cells were washed twice with PBS, and the efflux of cholesterol was induced by 2% human serum. At designated time points, the culture supernatant and cell lysate were collected and measured by fluorescence at Ex/Em: 482/515 nm. The percentage of cholesterol efflux was calculated by fluorescence intensity of media fluorescence/ (fluorescence intensity of cell lysate + media) × 100.
Lentiviral transduction
For the knockdown of CES1 (MIM: 114835) in iRPE cells, the iRPE cells from healthy donor were cultured in 6-well plate. The lentiviral particles carrying CES1 shRNA (Locus ID 1066, Origene) were transduced into the iRPE cells at a MOI of 10 overnight. To establish fair comparison, the control iRPE cells were transduced with viral particles expressing scramble shRNA. The medium was refreshed the following day, and the cell were incubated for 7 days before analysis. To achieve overexpression of CES1 in iRPE cells, lentiviral particles (Origene) expressing CMV promoter-driven CES1 (GenBank: NM_001025194) were transduced following the same procedure.
To achieve overexpression of EFEMP1WT and EFEMP1R345W, the lentiviral particles carrying EFEMP1WT (GenBank: NM_001039348) and EFEMP1R345W cDNA (Origene) were transduced into the iRPE cells at a MOI of 10 overnight. The medium was refreshed the next day, and the cell were incubated for 90 days before analysis.
Immunoblot
To verify the differentiation of iRPE, CRALBP (Ab15051, Abcam) and RPE65 (401.8B11.3D9, Novas Biologicals) antibodies were used to detect mature RPE markers including CRALBP and RPE65 in iRPE cell lysates via western blot. To determine EFEMP1 expression in iRPE, both cell lysates and culture supernatant were assayed using anti-EFEMP1 antibody (MA5-25740, Thermo Fisher Scientific). To detect CES1 expression in iRPE, anti-CES1 antibody (AF4920, R&D) was used. Anti-alpha tubulin (T5168, Sigma Aldrich), anti-beta actin (A5316, Sigma), or anti-GAPDH (5174, Cell Signaling) antibodies were used to detect house-keeping proteins. The cell lysates were collected using RIPA buffer (Thermo Fisher Scientific).
To understand the upstream regulators of CES1 expression, the iRPE cells from healthy donor were treated with Gefitnib (5 μM, Sigma Aldrich), ML385 (10 μM, Sigma Aldrich), LY294002 (10 μM, Sigma Aldrich), GW4064 (5 μM, Sigma Aldrich), or GW3965 (3 μM, Sigma Aldrich) for 72 h. Anti-CES1(AF4920, R&D) and anti-alpha tubulin (T5168, Sigma Aldrich) antibodies were used to detect CES1 and tubulin expression, respectively. The signal was detected using Immobilon Western Chemiluminescent HRP Substrate (Millipore). The blot was quantified using ImageJ for signal intensity.
Phospho-array analysis
To understand the immediate response triggered by EFEMP1WT or EFEMP1R345W, the iRPE cells were treated with the lysates collected from HEK293 cells transfected with either EFEMP1WT cDNA, EFEMP1R345W cDNA, or empty pcDNA3.1 vector by Lipofectamine 2000 (Invitrogen). Each well in a 6-well plate received 5 μg of respective plasmid. The expression of EFEMP1 protein in the lysates was confirmed by western blot (Figure S5). The HEK293 lysate from one well was transferred into one iRPE cell culture well in a 12-well plate. After 12 h of incubation, the HEK293 lysates were washed away with PBS. The iRPE cell culture was extracted by Antibody Array Assay kit (Full Moon Biosystems). The extracts were further analyzed with EGF Pathway Phospho Antibody Array (Full Moon Biosystems) according to manufacturer’s instruction.
Real-time PCR
To extract mRNA from iRPE cells, the iRPE cells were harvested at the indicated time and lysed with TRIZOL reagent (Invitrogen). Total RNA was isolated according to the manufacturer’s instructions. DNase I (Invitrogen) treatment was then performed to prevent genomic DNA contamination. The reverse transcription reaction was conducted by Superscript III Reverse Transcription kit, and the oligo-dT (Invitrogen) was used to generate the cDNA. Real-time PCR method was performed using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) with CFX Connect Real-Time PCR Detection System (Bio-Rad) to quantify gene expression levels. The gene expression was normalized by ACTB (MIM: 102630). The primers used in this study are listed in Table S2.
To verify that SP1 transactivates CES1, the EFEMP1WT iRPE was treated with or without mithracin with a final concentration at 200 nM for 24 h before RNA extraction.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed to determine whether SP1 directly binds to the CES1 promoter. The EMSA was carried out using the EMSA kit (Signosis), according to manufacturer’s instruction. Briefly, the iRPE nuclear extracts were collected using the Nuclear Extraction Kit (Signosis). Biotinylated DNA binding consensus sequence was also purchased from Signosis as the hot probe. 2 μg of the nuclear extract was mixed with 0.5 μL of hot probe and 1 μL of poly(I:C) for 30 min. For the competition binding experiments, 5 μL of cold probe (unlabeled oligonucleotide) were added to reaction mixtures. Samples were then loaded onto a 6.5% non-denaturing polyacrylamide gel and separated at 100 V, and the proteins were transferred to a membrane at 60 V for 1 h at 4°C. The membrane was then fixed with UV Cross-Linker (Stratagene). Streptavidin-HRP Conjugate provided by the EMSA kit to detect the signal.
Transcription factor activity assay
The nuclear extracts of EFEMP1WT or EFEMP1R345W-treated iRPE cells were extracted using the Nuclear Extraction Kit (Signosis). TF Activation Profiling Plate Array I kit (Signosis) was used to determine the activity of transcription factors in the iRPE cells as per the manufacturer’s instructions. The signal detected in the EFEMP1R345W-treated iRPE cells was divided by the signal of the EFEMP1WT group to reflect the change in transcription factor activity induced by the R345W mutation.
Enzyme-linked immunosorbent assay
IL-1, IL-6, IL-18, TNF-alpha, TGF-beta, and IFN-alpha levels in culture supernatants were measured using IL-1 (eBioscience), IL-6 (Thermo Fisher Scientific), IL-18 kit (eBioscience), TNF-alpha (Thermo Fisher Scientific), TGF-beta (Thermo Fisher Scientific), and IFN-alpha (Origene) ELISA kits following the manufacturer’s instructions. The quantification was normalized to the volume of supernatant.
Chromatin immunoprecipitation (ChIP)-qPCR assay
The EFEMP1WT iRPE cells were washed with PBS buffer and incubated with 1% formaldehyde at room temperature for 15 min, followed by the addition of 1.25M glycine to a final concentration of 125 mM for another 5 min. The cells were further washed by PBS buffer and lysed using ChIP assay high sensitivity kit (Abcam) following manufacturer’s protocol. The SP1 antibody (Millipore, 07-645) was used for immunoprecipitation. The primers used to detect CES1 promoter sequencing are listed in Table S2.
Statistics
All statistical analysis was performed in Microsoft Excel or GraphPad Prism 8. Two-group comparisons were performed using two-tailed unpaired Student’s t test. Statistical analysis for multiple group comparisons was performed using one-way ANOVA. Significance was determined at p ≤ 0.05.
Study approval
All procedures were performed as described in Columbia University Irving Medical Center Institutional Review Board-approved protocol AAAF1849. Written informed consent was received from all participants prior to creating iRPE.
Result
Gene correction of EFEMP1 affects EFEMP1 secretion but not RPE differentiation
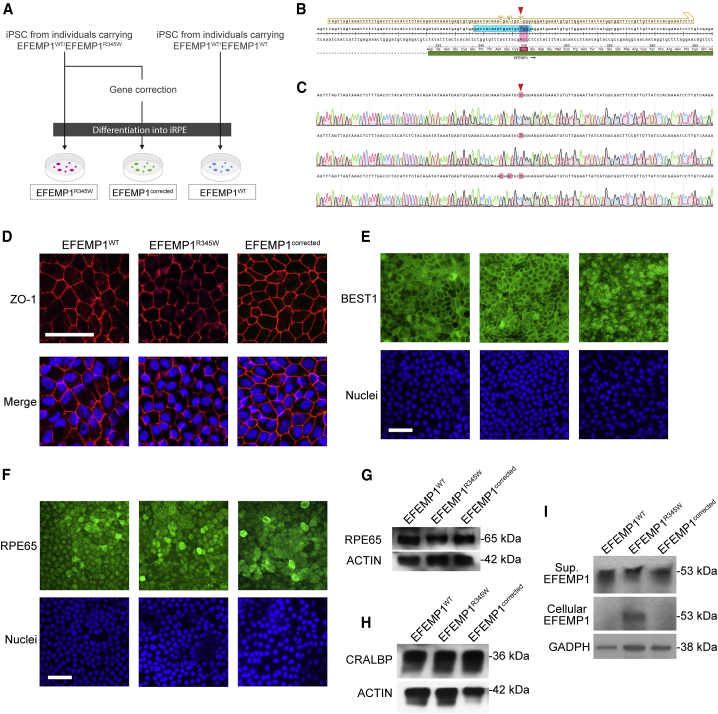
To model DHRD disease, we generated iRPE strains from three DHRD-affected individuals and healthy donor iPSCs (Figure 1A). To minimize noise from individual difference, we used the CRISPR-Cas9 gene editing strategy to correct the pathogenic mutation (c.1033C>T mutation in EFEMP1) in the DHRD iPSCs to serve as isogenic control. The SpCas9 protein, gRNA, and donor template were delivered together into DHRD iPSCs via nucleofection to create three lines of gene-corrected iPSCs (Figure 1B). In total, three different kinds of cell lines were created: EFEMP1WT (EFEMP1WT/EFEMP1WT), EFEMP1R345W (EFEMP1WT/EFEMP1R345W), and EFEMP1corrected (EFEMP1WT/EFEMP1corrected). The EFEMP1WT iRPE cells were created from iPSCs derived from healthy donor; the EFEMP1R345W iRPE cells were generated using iPSCs from DHRD-affected individuals, harboring a heterozygous p.Arg345Trp mutation; while EFEMP1corrected iRPE cells carry gene-corrected EFEMP1 with silent mutations. To confirm the genotypes, the wildtype, p.Arg345Trp mutant, and gene-corrected alleles were cloned into TOPO-TA vector and confirmed by sanger sequencing (Figure 1C). RT-PCR was also used to confirm that the silent mutations do not affect normal mRNA splicing (data not shown). The quality of these iPSCs were confirmed by karyotyping and immunostaining for stem cell markers (Figure S1). These iPSCs were then differentiated into iRPE cells using a previously established protocol.27 After differentiation, the iPRE cells from all three lines exhibited a hexagonal conformation and expressed ZO-1 in the tight junctions (Figure 1D). The p.Arg345Trp mutation and gene correction did not alter the expression pattern of RPE markers such as BEST1 (Figure 1E), RPE65 (Figures 1F and 1G), and CRALBP (Figure 1H). In our iRPE culture, we observed that EFEMP1WT protein is nearly undetectable in cell lysate but preserved in the culture supernatant (Figure 1I). In contrast, there was an intracellular accumulation of EFEMP1R345W mutant in iRPE. This disease phenotype is consistent with previous observation in DHRD-affected individual RPE staining and thus demonstrates that iRPE from DHRD-affected individuals are viable platforms for disease modeling.23 Notably, reduction of EFEMP1 secretion was remedied by the gene correction as evidenced by the disappearance of intracellular EFEMP1 in EFEMP1corrected iRPE cells. EFEMP1 can potentially generate different isoforms with the expected size ranging from 53 kDa to 5 kDa. However, only one species of 53 kDa was detected in our experiments.
Figure 1.
Gene correction of EFEMP1R345W in iPSC from individuals with DHRD
(A) Flowchart of the preparation of iRPEs used in this study.
(B) The design of gRNA and donor template used for the gene correction. The expected cutting site is indicated by the red arrow. The p.Arg345Trp mutation is marked by red box. The gRNA protospacer is marked in cyan while the donor template is outlined in yellow. The donor template contains two silent mutations (N342 and E343) in addition to the R345 mutation to enable screening by RFLP.
(C) The Sanger sequencing result of the WT (top), p.Arg345Trp (middle), and gene-corrected (bottom) alleles. The c.1033 nucleotide is indicated by the red arrow. The difference between the three alleles are marked with red boxes. (D) The ZO.1 (red) immunostaining of iRPE with DAPI counterstaining (blue).
(E) Immunostaining results of BEST1.
(F) Immunostaining results of RPE65.
(G) Representative western blot result of RPE65 expression in iRPE lysates.
(H) Representative western blot result of CRALBP expression in iRPE lysates.
(I) Representative western blot result of EFEMP1 expression in iRPE lysates and culture supernatants.
Scale bar = 100 μm.
Differentially expressed genes in iRPE cells derived from DHRD-affected individuals
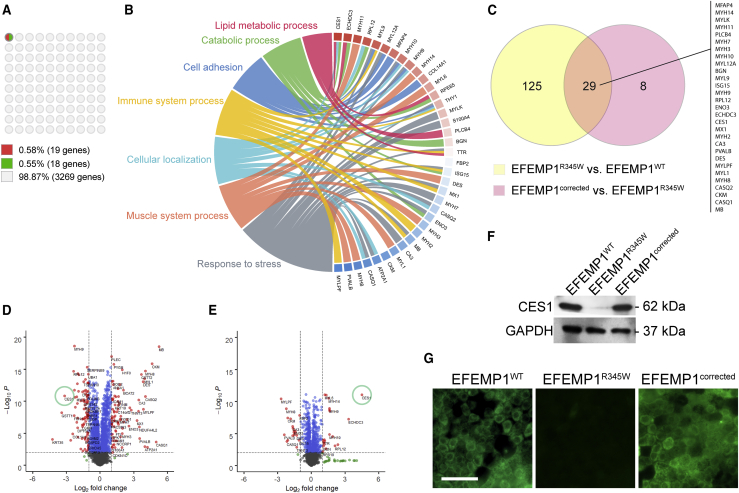
To characterize molecular changes induced by the EFEMP1R345W mutation at a global level, we compared the proteomic profiles of the EFEMP1WT, EFEMP1R345W, and EFEMP1corrected iRPE cells. All groups were analyzed in hexicate. A total of 3,269 proteins were analyzed. When EFEMP1WT was compared to EFEMP1R345W, 154 proteins (~5%) were differently expressed (DEPs, fold change > 2 and p value < 0.001). However, when we compared EFEMP1R345W iRPE clones to their isogenic controls, the number of DEP was reduced to 37 proteins (~1%) (Figure 2A), suggesting the validity of the isogenic controls. Of these 37 DEPs, 19 (0.58%) were upregulated and 18 (0.55%) were downregulated after gene correction.
Figure 2.
Proteomic analysis of iRPE cells
(A) 10 × 10 dot plot presenting the percentage of differentially expressed proteins (DEPs) after gene correction. The criteria defining DEP: >2-fold difference, p < 0.001.
(B) Circos plot presenting the biological process attributes of the DEPs shown in (A). The colored boxes next to the gene labels indicate the change in expression after gene correction (red: increase; blue: decrease).
(C) Venn diagram of the DEPs.
(D and E) Volcano plots presenting the DEPs in different comparisons: (D) EFEMP1R345W (n = 6) versus EFEMP1WT (n = 6); (E) EFEMP1corrected (n = 12) versus EFEMP1R345W (n = 6).
(F) Representative western blot result of CES1 expression of iRPE culture.
(G) Representative immunocytochemistry result of CES1 staining in iRPE culture. Scale bar = 100 μm.
To better understand the biological functions impacted by EFEMP1R345W, the DEPs were analyzed by gene ontology (GO) analysis to identify commonly altered functions between the EFEMP1R345W and EFEMP1corrected cells. The most significantly enriched biological process terms among all these DEPs were response to stress, muscle system process, cellular localization, immune system process, cell adhesion, catabolic process, and lipid metabolic process (Figure 2B). Interestingly, correction of the EFEMP1R345W mutation resulted in mostly downregulation of proteins involved in immune system processes but upregulation of those participating in lipid metabolism, implicating lipid metabolism and immune system response in DHRD phenotype development. Among the 37 DEPs, 29 were shared between the EFEMP1WT versus EFEMP1R345W and EFEMP1corrected versus EFEMP1R345W comparisons (Figure 2C).
We next used volcano plot (Figures 2D and 2E) analysis to explore the differential protein expression signatures based on correlation between different groups. We found that CES1 was remarkably reduced by 7- to 9-folds in EFEMP1R345W cells when compared to EFEMP1WT iRPE (Figures 2D and S2 and Table S3). Western blot was performed to confirm the expression of CES1 in these iRPE cells. CES1 protein can be detected in the cell lysates derived from EFEMP1WT cells, but its expression was nearly undetectable in EFEMP1R345W cells (Figure 2F). This reduction of CES1 expression in DHRD iPRE was further confirmed in two other individuals with DHRD and two wild-type donor iRPE cells (Figure S3). Strikingly, the expression level of CES1 was notably recovered after gene correction (Figures 2E and 2F) and comparable to the level found in EFEMP1WT iRPE clones (Figure S2). The downregulation of CES1 in EFEMP1R345W cells and its restoration in EFEMP1corrected cells were also confirmed by immunocytochemistry (Figure 2G).
EFEMP1R345W iRPE cells show no inflammatory cytokine release or UPR response
Inflammatory cytokine release has been reported in ex vivo RPE cultures of EFEMP1R345W mouse.24 Consistently, upregulation of immune response genes such as MX1 and ISG15 was detected in our proteomic profiling (Figures 2B and 2C). We performed ELISA assays to further investigate whether the EFEMP1R345W variant can elicit an immune response by releasing cytokines in iRPE cell culture. The medium from mature iRPE cell culture was used to detect interleukin-1 (IL-1), IL-6, IL-18, interferon alpha (IFN-α), tumor necrosis factor-alpha (TNF-α), and transforming growth factor (TGF)-beta. Interestingly, the cytokine level in all culture media was nearly undetectable, and there was no significant difference between groups (Figures S4A–S4F).
Alternatively, it has also been suggested that the misfolded mutant EFEMP1R345W protein can cause ER stress when overexpressed in a RPE cell line.26 To investigate this phenomenon, we carried out quantitative real-time PCR (Figure S4G) and immunocytochemistry (Figure S5) to detect the expression of UPR-related genes in EFEMP1R345W iRPE. However, no obvious change in UPR markers was detected in EFEMP1R345W iRPE clones and EFEMP1corrected cells.
Reduce CES1 expression results in lipid accumulation in iRPE cells
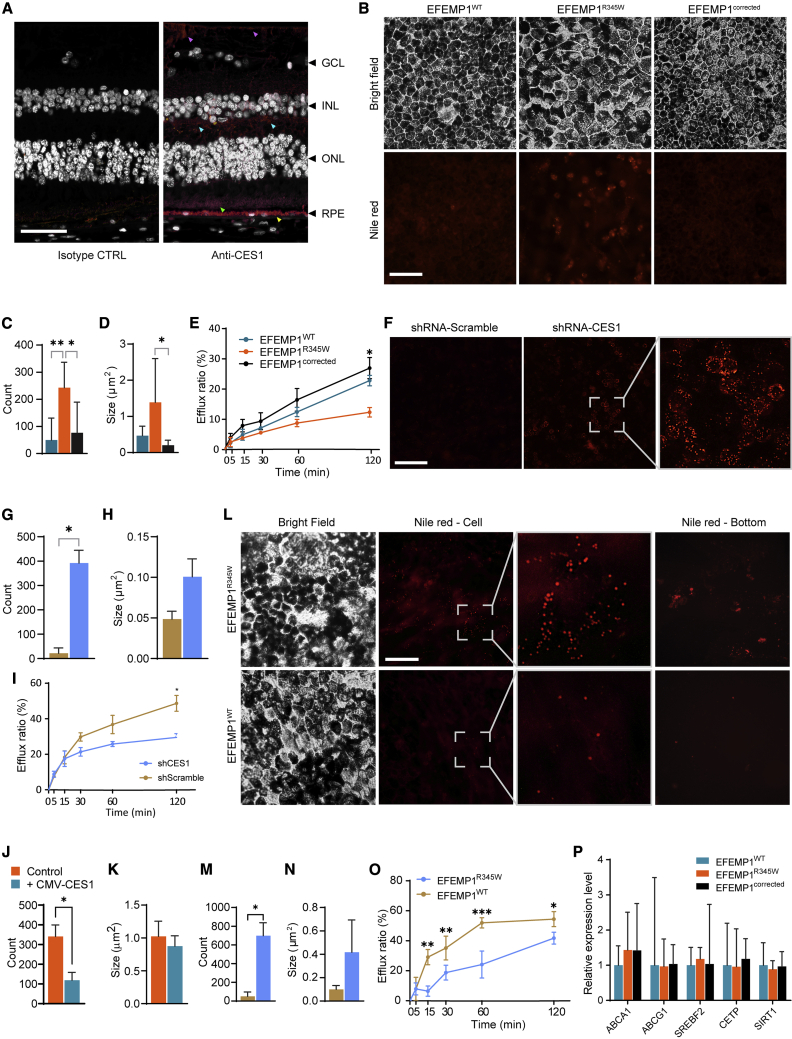
CES1 is an intracellular protein predominantly expressed in the lumen of the endoplasmic reticulum.30 CES1 has been reported to be expressed in hepatocytes and macrophages,31, 32, 33 but its expression in the eye remains unclear. To confirm its expression in the eye, we performed immunostaining on human eye sections. We found that CES1 was predominantly detected in the RPE layer, but weak signal was also observed in the outer segment of photoreceptor, outer plexiform layer, and inner limiting membrane (Figure 3A). We also used qPCR to quantify the expression pattern of CES1 in retina tissue from autopsy and iPSC-derived retinal organoid. We found that the expression of CES1 in autopsied RPE was 9.18 times higher than in neural retina (Figure S6). However, CES1 levels in organoid retina was 1.47 times higher than in organoid RPE. These results may be attributable to the fluctuation of CES1 level during retinal development. Given that CES1 participates in converting cholesteryl ester to free cholesterol,32 we hypothesized that CES1 deficiency may hamper secretion of cholesterol from RPE cells, as this hydrolytic reaction is the rate-limiting step in mobilizing stored cholesteryl ester.
Figure 3.
Intracellular lipid accumulation induced by EFEMP1R345W via reducing cholesterol efflux
(A) Representative CES1 staining of human retina section. A positive staining signal was detected in the RPE cells (yellow arrow), outer segment of photoreceptor (green arrow), outer plexiform layer (cyan arrows), and inner limiting membrane (purple arrows). Scale bar = 50 μm (n = 3).
(B) Nile red staining of iRPE cells. Scale bar = 100 μm.
(C and D) The amount (C) and size (D) of the lipid droplet detected in (B) (n = 7).
(E) The cholesterol efflux of iRPE cells shown in (B) (n = 2).
(F) Nile red staining of EFEMP1WT iRPE cells after CES1 knockdown. Scale bar = 100 μm.
(G and H) The amount (G) and size (H) of the lipid droplet detected in (F) (n = 2).
(I) The cholesterol efflux of iRPE cells shown in (F) (n = 2).
(J and K) The amount (J) and size (K) of the lipid droplets detected in the EFEMP1R345W iRPE with or without CMV-CES1 overexpression (n = 2).
(L) Nile red staining of EFEMP1WT iRPE cells overexpressing either EFEMP1WT or EFEMP1R345W. Scale bar = 100 μm.
(M and N) The amount (M) and size (N) of the lipid droplet detected in (L) (n = 2).
(O) The cholesterol efflux of iRPE cells shown in (L) (n = 2).
(P) Relative expression levels of cholesterol efflux-related genes in EFEMP1WT, EFEMP1R345W, and EFEMP1corrected iRPE cells.
Data in (C)–(E), (G)–(K), and (M)–(O) represented as mean ± SD. Data in (P) represented as geometric mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
To test this hypothesis, we first visualized the lipid droplets in iRPE via Nile red staining (Figure 3B). We found that the lipid droplets are 5 times more numerous in EFEMP1R345W than in EFEMP1WT iPRE (p = 0.0014) (Figure 3C) and 3 times larger in size (p = 0.0727) (Figure 3D), which indicates lipid accumulation. However, this accumulation of lipid droplets was not observed in EFEMP1corrected iRPE (amount: p = 0.0112; size: p = 0.024). We then determined cholesterol efflux rate of these iRPEs and found the EFEMP1R345W iPRE exhibits < 50% efflux rate when compared to either EFEMP1WT or EFEMP1corrected at 120 min (p = 0.0197) (Figure 3E).
To elucidate whether CES1 downregulation can result in accumulation of lipid droplet in iRPE, we used shRNA to knockdown the expression of CES1 in EFEMP1WT iRPE cells. Two weeks after transduction, the lipid content in the iRPE cells was examined. In the scrambled shRNA-treated group, the signal was very weak (Figure 3F) and comparable to that in untreated EFEMP1WT iRPE cells (data not shown). However, in the CES1 knockdown group, we observed a significant increase in the number (p = 0.0115) (Figure 3G) and slight increase in the size (p = 0.0928) (Figure 3H) of lipid droplets. Moreover, shRNA-treated iRPE cells exhibited a 30%–40% decrease in cholesterol efflux compared to the scramble-treated control at 120-min time point (p = 0.0318) (Figure 3I). These results suggest that the lipid accumulation in iRPE cells may be attributable to CES1 knockdown, due to a dysregulation of cholesterol efflux. To further verify the role of CES1 in lipid accumulation, we overexpressed CES1 in EFEMP1R345W iRPE using lentiviral vector expressing human CES1 cDNA. After 2 weeks of transduction, though the effect in size was not obvious, the amount of lipid droplets was found to be significantly decreased 66.5% when compared to the control (p = 0.0478) (Figures 3J and 3K). Given that CES1 is downregulated in EFEMP1R345W iRPE cells, we hypothesized that lipid accumulation can be induced by the mutant EFEMP1R345W protein. To observe long-term effects, we used lentiviral vectors to transduce either a WT or p.Arg345Trp mutant copy of EFEMP1 into EFEMP1WT iRPE cells. After 3 months of culture, the cells were examined by using Nile red staining. Compared to iRPE cells overexpressing WT EFEMP1, those expressing mutant EFEMP1 exhibited remarkable lipid accumulation (Figures 3L–3N). After removing the cells by trypsin, we observed stained debris in the dish of iRPE cells overexpressing EFEMP1R345W but not EFEMP1WT (Figure 3L, rightmost column). We tested the cholesterol efflux in these cells and found that overexpression of EFEMP1R345W reduced cholesterol efflux by 55% at 60 min (p < 0.001) and by 25% at 120 min (p = 0.033) (Figure 3O).
In addition to CES1, we wanted to determine whether there were any other cholesterol efflux-related proteins affected by the EFEMP1R345W mutation that were not identified by proteomic profiling. We used real-time PCR to analyze the expression of the transporter proteins ATP-binding cassette (ABC) subfamily A member 1 (A1 [MIM: 600046]) and ABC subfamily G member 1 (G1 [MIM: 603076]), the enzymes cholesterol ester transfer protein (CETP [MIM: 118470]) and sirtuin 1 (SIRT1), and the transcription factor sterol regulatory element-binding protein 1 (SREBF2 [MIM: 601510]) in iRPE cells (Figure 3P). No significant change in the expression of these proteins was detected in EFEMP1R345W iRPE cells when compared to either EFEMP1WT or EFEMP1corrected clones, further supporting the causative role of CES1. Taken together, these results suggest that EFEMP1R345W reduces CES1 expression and cholesterol efflux, causing lipid accumulation in iRPE cells.
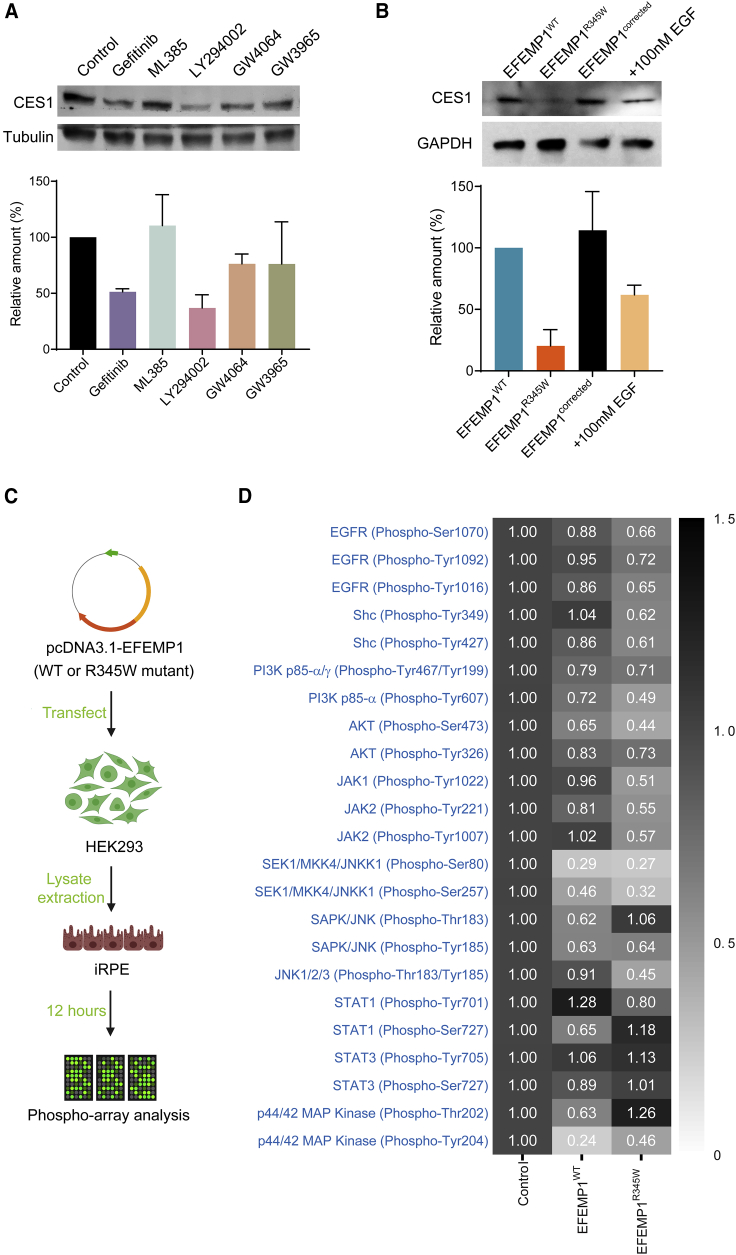
EFEMP1R345W regulates CES1 expression via EGFR-Akt signaling
Next, we sought to understand how EFEMP1R345W suppresses CES1 expression. Since EFEMP1 is an ECM protein, we suspected that EFEMP1 may affect CES1 expression through plasma membrane receptor signaling. It has been reported that EFEMP1 can bind to EGFR directly and inhibit EGFR signaling in glioma cells.34 Since RPE cells express EGFR,35 we hypothesized that EFEMP1 may modulate CES1 expression through EGFR signaling. We treated EFEMP1WT iRPE cells with the EGFR inhibitor gefitinib. After a 72 h incubation, we found that CES1 expression level in iRPE cells was remarkably reduced compared to untreated control, confirming that EGFR is involved in EFEMP1-induced regulation of CES1 (Figure 4A, lanes 1 and 2).
Figure 4.
EFEMP1R345W regulates CES1 expression through EGFR signaling
(A) Representative western blot result of CES1 expression levels in normal iRPE cells after 72 h of treatment with different agonists and antagonists (n = 2).
(B) Representative western blot result of CES1 expression of EFEMP1WT, EFEMP1R345W, and EFEMP1corrected iRPE cells (lane 1–3) and EFEMP1R345W cells pre-treated with 100 nM EGF for 12 h (lane 4) (n = 2).
(C) Sample preparation flowchart for phosphor-array analysis.
(D) Heatmap of phosphorylation status of the EGFR downstream targets measured by phosphor-array (n = 6). Data represented as mean ± SD.
Nuclear factor erythroid 2-related factor 2 (NRF2) and phosphatidylinositol 3-kinase (PI3K)/AKT are downstream targets of EGFR, and NRF2 has been reported to transactivate CES1 expression in the human hepatoma cell line HepG2.36 To determine whether either of these pathways regulate CES1 in iRPE cells, we used ML385 and LY294002 to inhibit NRF2 and PI3K/AKT, respectively. Unexpectedly, only LY294002 reduced CES1 expression (Figure 4A, lanes 3 and 4), suggesting that the EGFR-AKT pathway is necessary for controlling CES1 levels in iRPE cells while NRF2 is not essential. Since farnesoid X receptor (FXR) and liver X receptor (LXR) were also reported to regulate CES1,31 we treated EFEMP1WT iRPE cells with their respective agonists, GW4064 and GW3965, but observed no change in CES1 expression (Figure 4A, lanes 5 and 6). To further confirm EGFR signaling has positive impact on CES1 expression, we treated EFEMP1R345W iPRE with 100 nM EGF. After 72 h of incubation, the CES1 level in EGF-treated iRPE became remarkably higher than that in untreated cells (Figure 4B).
Given that the p.Arg345Trp mutation resides in the last EGF-like domain, we hypothesized that EFEMP1R345W may impact EGFR signaling. To focus on the direct effect of EFEMP1R345W protein on EGFR signaling and to avoid secondary effects, we treated EFEMP1WT iRPE cells with the lysates of HEK293 cells that were previously transfected with plasmids carrying either EFEMP1WT or EFEMP1R345W (Figure 4C and S5). After 12 h of incubation, the HEK293 lysate was removed. The iRPE cell lysate was extracted and analyzed using an EFGR signaling microarray. Notably, most of the downstream cascade, including PI3K/AKT1 and janus kinase (JAK)/c-Jun N-terminal kinase (JNK), were hyper-dephosphorylated in the EFEMP1R345W-treated group when compared to the EFEMP1WT-treated group (Figure 4D). These results suggest that EFEMP1R345W has a hyper-inhibitory effect on EGFR signaling.
Since EGFR signaling can positively modulate CES1 expression, we tested the treatment of 100 mM EGF on EFEMP1R345W iRPE. After 2 weeks of incubation, we noted a significant decrease in the amount of lipid droplets in the cells (p = 0.0478) (Figure S7A), but the effect was not obvious in the droplet size (Figure S7B). We also observed a 26% increase in cholesterol efflux in EGF-treated cells than in the untreated control (p = 0.0518) (Figure S7C).
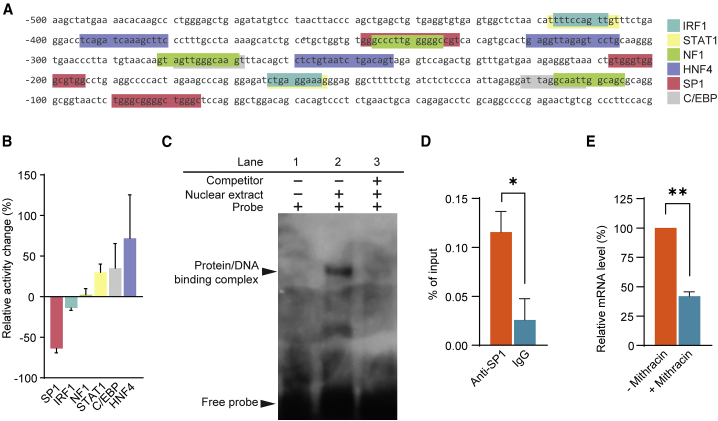
CES1 transcription is controlled by SP1
We next sought to understand how EFEMP1R345W-induced reduction of EGFR signaling impacts the transcription of CES1 in iRPE cells. We analyzed the proximal promoter (~500 bp) of CES1 and found potential binding sites for six transcription factors: interferon regulator factor 1 (IRF1), signal transducer and activator of transcription 1 (STAT1), neurofibromin 1 (NF1), hepatocyte nuclear factor 4 (HNF4), SP1, and CCAAT enhancer binding protein (C/EBP) (Figure 5A). To study the activity of these transcription factors, the nuclear extracts of EFEMP1WT or EFEMP1R345W-treated iRPE cells were used for analysis. We found that SP1 activity in the EFEMP1R345W group was reduced by 63.9% (Figure 5B). In contrast, the activity of STAT1, C/EBP, and HNF4 increased by 30.0%, 35.1% and 71.9%, respectively, and there was no change in IRF1 or NF1 activity. In light of these results, we hypothesized that EFEMP1R345W-induced CES1 downregulation may be mediated by SP1.
Figure 5.
EFEMP1R345W perturbs CES1 expression via SP1
(A) Predicted transcription factor binding sites located within 500 bp upstream of the CES1 transcription start site.
(B) Relative activity of transcription factors in iRPE cells. Data represented as EFEMP1R345W group/EFEMP1WT group. Data represented as mean ± SD (n = 2).
(C) Electrophoretic mobility shift assay of nuclear extract from EFEMP1WT iRPE cells.
(D) ChIP-qPCR was performed to detect the binding of SP1 to CES1 promoter. Data represented as geometric mean ± SD (n = 2).
(E) qPCR result of CES1 expression level in iRPE cells after treatment of SP1-specific inhibitor mithracin. Data represented as geometric mean ± SD (n = 2). ∗ = p<0.05; ∗∗ = p<0.01.
To determine whether SP1 can bind to the CES1 promoter, EMSA was conducted using a DNA probe containing the sequence of the predicted SP1 binding site in the CES1 promoter (5′-aactgtgggtgggcgtggcctgaggcccc-3′) (Figure 5C). Incubation of this probe with nuclear extracts of EFEMP1WT iRPE cells yielded a number of DNA-protein complexes on the gel (Figure 5C, lane 1 and 2). This complex formation was eliminated by the addition of excess unlabeled probe (Figure 5C, lane 3). Chromatin immunoprecipitation (ChIP)-qPCR was also carried out to test the binding activity of SP1 in EFEMP1WT iRPE. ChIP of SP1 showed a 5 times higher enrichment than the IgG control (p = 0.0387) (Figure 5D). To further validate whether SP1 directly controls CES1 expression, we treated EFEMP1WT iRPE cells with the SP1-specific inhibitor mithracin. After 24 h of incubation, expression of CES1 was reduced by 58.0% (p = 0.0019) (Figure 5E). Altogether, these results demonstrate that SP1 binds to CES1 promoter and contributes to its transactivation.
Discussion
Drusen are characterized as extracellular deposits of debris that accumulate between the basal lamina of the RPE layer and the inner layer of the Bruch’s membrane. On color fundus examinations, drusen manifest as small yellow-white deposits in the macular area and periphery of the retina. Drusen consist of a combination of lipids, polysaccharides, glycosaminoglycans, and proteins.37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49 Lipids—specifically, phosphatidylcholine and both esterified and unesterified cholesterol—are thought to comprise the bulk of drusen.39,50, 51, 52 Consistently, extracellular proteins responsible for cholesterol mobilization including apolipoprotein B and E have also been closely associated with drusen.53,54 Nonetheless, the specific mechanism underlying drusen formation remains unclear.
The presence of drusen is a clinical hallmark of various forms of MD including DHRD and AMD. The phenotypic similarity between these two diseases suggests that they share the same pathology. However, unlike AMD’s heterogenous etiology, DHRD is monogenic and thus offers a more approachable experimental model for drusen pathogenesis. Prior studies investigating DHRD have suggested two possible mechanisms for how mutant EFEMP1 contributes to drusen formation: the UPR and complement system activation. Roybal et al. found that overexpression of EFEMP1R345W in ARPE-19 elicited UPR activation and increased VEGF expression.26 In contrast, using EFEMP1R345W/R345W mice as an in vivo disease model, Fu et al. found no UPR activation as measured by Hspa5 (Grp78) expression, but instead detected activated complement component C3 in the RPE and Bruch’s membrane.24 More recently, Fernandez-Godino et al. suggested that increased production of C3a in EFEMP1R345W/R345W mice stimulated the release of cytokines IL-6 and IL-1B.20 These findings have yet to be reproduced and require further investigation.
Here, we used iRPE derived from three distinct individuals with DHRD as an in vitro disease model and leveraged isogenic comparisons to investigate how the EFEMP1R345W variant contributes to DHRD. Conventional study designs investigating DHRD pathology have used family members as control subjects. However, family members are not genetically identical, and different single-nucleotide polymorphisms (SNPs) can impact disease expressivity. Engineering an isogenic cell line from the same parental line removes noise from the genetic background and yields an opportunity to elucidate specific molecular mechanisms and pathways underlying disease.
Given that previous studies implicated UPR26 and the complement pathway20,24 in DHRD, we sought to reproduce and validate those claims. Interestingly, we did not detect any inflammatory cytokine release or UPR in our DHRD iRPE cell culture. One possible explanation for this discrepancy may lie in the limited translatability of the disease models used in the previous studies as overexpression of the EFEMP1R345W variant and homozygous p.Arg345Trp mutations do not mimic most cases of DHRD, which are caused by a single heterozygous mutation. Though we detected no inflammatory cytokines or chemokines in our in vitro iRPE model, immune response may still play a role in DHRD development in vivo. An EFEMP1-humanized model with p.Arg345Trp mutation in mouse or other animal models may offer additional insights and clarifications into this question.
We next turned to an unbiased genetic approach to determine DEPs in our iRPE model. We performed three iRPE cell comparisons: (1) EFEMP1R345W versus EFEMP1WT; (2) EFEMP1R345W versus EFEMP1corrected; and (3) EFEMP1R345W versus EFEMP1WT. Notably, the number of DEPs decreased significantly in the comparisons involving isogenic controls. Only 37 proteins (1.13% of the total number of screened proteins) exhibited differential expression between EFEMP1R345W and its isogenic control EFEMP1corrected. The number of DEPs was further reduced to eight proteins when comparing EFEMP1R345W to both EFEMP1corrected and EFEMP1WT iRPE cells. Overall, the isogenic control helped exclude around 80%–95% of the genetic noise. Importantly, the protein expression patterns of the 37 DEPs in EFEMP1corrected iRPE cells were more similar to those of EFEMP1WT iRPE cells than to those of either DHRD iRPE cells, indicating the involvement of these 37 proteins in the pathogenesis of DHRD.
We then used GO to analyze the 37 DEGs and grouped these genes into enriched terms, including lipid metabolic process, catabolic process, cell adhesion, immune system process, cellular localization, muscle system process, and response to stress. Based on current literature, most of these genes do not directly participate in cholesterol metabolism. Nonetheless, gene correction induced a significant increase in certain genes including CES1, phospholipase C beta 4 (PLCB4), and RPE65, all of which participate in lipid metabolic or catabolic processes. CES1 is responsible for the hydrolysis of ester- and amide-bond-containing xenobiotics as well as long-chain fatty acid esters and thioesters.33,55 CES1 acts as a rate-limiting enzyme in reverse cholesterol transport—specifically, the conversion of cholesteryl ester to free cholesterol—in macrophages during regression of atherosclerosis.32 PLCB4 catalyzes the formation of inositol 1,4,5-trisphosphate and diacylglycerol from phosphatidylinositol 4,5-bisphosphate, an important reaction in intracellular transduction of extracellular signals in the retina. RPE65 converts all-trans-retinyl esters to 11-cis-retinol, the rate limiting step for the retinoid cycle.
We focused on CES1 because CES1 exhibited the largest change between DHRD and healthy iRPE cells. Gene correction increased the expression level of CES1 by ~20-fold compared to that of diseased iRPE cells from either of the two DHRD-affected individuals. Interestingly, CES1 levels in EFEMP1Corrected was ~3-fold higher than in EFEMP1WT cells, indicating a nontrivial variation from endogenous CES1 expression. This might explain the variability and partial penetrance of DHRD.12,56,57 The role of CES1 has not been well studied in RPE. However, in macrophage/foam cells, CES1 has been reported to play a major role in reverse cholesterol transport during atherosclerosis regression. RPE cells resemble macrophages in their roles in active phagocytosis and cholesterol efflux.58 RPE cells are the most actively phagocytic cells in the human body. Within the retina, RPE cells are responsible for phagocytosing shed photoreceptor outer segments and preventing subsequent accumulation of excess lipid via lipid export. It is therefore possible that RPE cells and macrophages share the same pathways/mechanisms for controlling cholesterol transport.
Notably, we discovered that downregulation of CES1 leads to abnormal lipid accumulation in iRPE cells. Strikingly, EFEMP1R345W had a hyper-inhibitory effect on EGFR signaling, possibly by deactivating PI3K/AKT signaling. EGFR signaling was associated with increased expression of CES1 in iRPE cells. Taking these results together, we propose the following model: the p.Arg345Trp mutation enhances EFEMP1’s inhibitory activity on the EGFR-Akt pathway, downregulating CES1. Reduced expression of CES1 disrupts cholesterol efflux and leads to abnormal lipid accumulation in the RPE cells. This model is consistent with our current understanding of MD pathology. Cholesterol efflux genes such as apolipoprotein E (APOE) and ABCA1 have been identified as risk factors for AMD, and deficiency in cholesterol efflux is thought to accelerate AMD progression by promoting deposition of drusen and other extracellular lipids underneath the retina.59, 60, 61 Interestingly, the expression level of EFEMP1 was recently found to increase with age.62 Given that EFEMP1 inhibits the expression of CES1, aging may decrease cholesterol efflux in RPE cells by perturbing ECM homeostasis.
Lipid metabolism and immune responses are closely integrated through converging pathways in many tissues.63 Previous studies have demonstrated that the absence of infectious diseases and prolonged nutrient excess induce chronic low-level sterile metaflammation through immune pathway activation.64 Consistent with these observations, our GO analysis revealed that a number of downregulated proteins, including ISG15 and MX1, participates in immune response. While all of these proteins can be induced by interferons, few anti-viral cytokine secretions were detected in our iRPE cell culture. One possible explanation for this phenomenon is that the high expression of immune-related genes in DHRD iRPE may be an aftermath of the excessive accumulation of cytoplasmic lipids. Metabolic disorders involve a complex interplay of dysregulated pathways, leading to lipotoxicity, inflammation, and associated stress responses. This intricate dynamic makes it challenging to elucidate the exact mechanism of disease and identify interventional targets. One important finding is that SP1 binds to the CES1 promoter. As a zinc finger transcription factor, SP1 is involved in many different cellular processes including immune response. As such, one direction for future study is clarifying how the pathways associated with CES1 in our study are linked to SP1.
In this study we observed lipid deposits after we removed the iRPE from the polystyrene cell culture plate. This phenomenon was also observed in a previous study, where iRPE cultures were grown on Transwell membrane.65 Drusen has been previously shown to contain high levels of lipids such as cholesteryl ester and phosphatidylcholine.49 This finding suggests that iRPE may produce drusen-like sub-RPE deposit in a cell-autologous fashion. There was no sign of massive cell death in the EFEMP1R345W iRPE cell culture by the 3-month endpoint of our experiment (data not shown), and we did not further investigate the effect of aberrant lipid accumulation on the iRPE cells. However, lipid accumulation in RPE cells is believed to be harmful if persistent.66,67 For example, excessive lipids may perturb calcium homeostasis by increasing ER or mitochondrial stress.68, 69, 70 This intracellular lipotoxicity may eventually contribute to RPE atrophy which has been observed in DHRD- and AMD-affected individuals.13 If lipotoxicity kills cells, cholesteryl ester together with dead cell debris may remain precipitated around the RPE/BrM and become drusen.39,51,71,72 As such, the resultant formation of drusen may be partially attributed to the immobility of cholesteryl ester. Its clearance, in turn, must be achieved through ingestion by another cell (e.g., macrophage). This hypothesis might explain why esterified cholesterol, which usually exists intracellularly, constitutes a large part of drusen.39,51
In this study we showed that accumulation of intracellular cholesterol ester in DHRD-affected individual-derived RPE cells is a result of dysregulated cholesterol efflux. Cholesterol imbalance can also be exacerbated by increased uptake of lipids. Whether a low-fat diet as a supplement to gene editing-based therapy can postpone the onset of drusen or ameliorate drusen severity is an interesting direction for the development of treatments for not only DHRD but also other forms of MD such as AMD.
Declaration of interests
S.H.T. has received financial benefits from Spark Therapeutics and research support from Abeona Therapeutics, Inc and Emendo.
Acknowledgments
We thank the Columbia Stem Cell Initiative Core Facility for the use of Amaxa Nucleofector 4D and Proteomic Core Facility for mass spectrometry. The Jonas Children’s Vision Care and Bernard & Shirlee Brown Glaucoma Laboratory are supported by the National Institutes of Health (5P30CA013696, U01EY030580, U54OD020351, R24EY028758, R24EY027285, 5P30EY019007, R01EY018213, R01EY024698, R01EY024091, R01EY026682, R21AG050437), the Schneeweiss Stem Cell Fund, New York State (SDHDOH01-C32590GG-3450000), the Foundation Fighting Blindness New York Regional Research Center Grant (PPA-1218-0751-COLU), Nancy & Kobi Karp, the Crowley Family Funds, the Rosenbaum Family Foundation, Alcon Research Institute, the Gilbert Family Foundation, the Research to Prevent Blindness (RPB) Physician-Scientist Award, and unrestricted funds from RPB, New York, NY. The sponsors and funding organizations had no role in the design or conduct of this research.
Published: April 27, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.04.006.
Data and code availability
The MS/MS proteomic data supporting the current study have not been deposited in a public repository due to the sensitivity of patient information but will be made available by the corresponding author upon request.
Web resources
OMIM, https://www.omim.org/
Supplemental information
Reference
- 1.Pennington K.L., DeAngelis M.M. Epidemiology of age-related macular degeneration (AMD): associations with cardiovascular disease phenotypes and lipid factors. Eye Vis. (Lond.) 2016;3:34. doi: 10.1186/s40662-016-0063-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryan J.A., 3rd, Campochiaro P.A. A retinal pigment epithelial cell-derived growth factor(s) Arch. Ophthalmol. 1986;104:422–425. doi: 10.1001/archopht.1986.01050150124042. [DOI] [PubMed] [Google Scholar]
- 3.Karamali F., Esfahani M.N., Hajian M., Ejeian F., Satarian L., Baharvand H. Hepatocyte growth factor promotes the proliferation of human embryonic stem cell derived retinal pigment epithelial cells. J. Cell. Physiol. 2019;234:4256–4266. doi: 10.1002/jcp.27194. [DOI] [PubMed] [Google Scholar]
- 4.Sarks S., Cherepanoff S., Killingsworth M., Sarks J. Relationship of Basal laminar deposit and membranous debris to the clinical presentation of early age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2007;48:968–977. doi: 10.1167/iovs.06-0443. [DOI] [PubMed] [Google Scholar]
- 5.Anderson D.H., Mullins R.F., Hageman G.S., Johnson L.V. A role for local inflammation in the formation of drusen in the aging eye. Am. J. Ophthalmol. 2002;134:411–431. doi: 10.1016/s0002-9394(02)01624-0. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez-Godino R., Pierce E.A. C3a triggers formation of sub-retinal pigment epithelium deposits via the ubiquitin proteasome pathway. Sci. Rep. 2018;8:9679. doi: 10.1038/s41598-018-28143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seddon J.M., Yu Y., Miller E.C., Reynolds R., Tan P.L., Gowrisankar S., Goldstein J.I., Triebwasser M., Anderson H.E., Zerbib J. Rare variants in CFI, C3 and C9 are associated with high risk of advanced age-related macular degeneration. Nat. Genet. 2013;45:1366–1370. doi: 10.1038/ng.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bressler N.M., Bressler S.B., Fine S.L. Age-related macular degeneration. Surv. Ophthalmol. 1988;32:375–413. doi: 10.1016/0039-6257(88)90052-5. [DOI] [PubMed] [Google Scholar]
- 9.de Jong P.T. Age-related macular degeneration. N. Engl. J. Med. 2006;355:1474–1485. doi: 10.1056/NEJMra062326. [DOI] [PubMed] [Google Scholar]
- 10.Jager R.D., Mieler W.F., Miller J.W. Age-related macular degeneration. N. Engl. J. Med. 2008;358:2606–2617. doi: 10.1056/NEJMra0801537. [DOI] [PubMed] [Google Scholar]
- 11.Haimovici R., Wroblewski J., Piguet B., Fitzke F.W., Holder G.E., Arden G.B., Bird A.C. Symptomatic abnormalities of dark adaptation in patients with EFEMP1 retinal dystrophy (Malattia Leventinese/Doyne honeycomb retinal dystrophy) Eye (Lond.) 2002;16:7–15. doi: 10.1038/sj.eye.6700018. [DOI] [PubMed] [Google Scholar]
- 12.Michaelides M., Jenkins S.A., Brantley M.A., Jr., Andrews R.M., Waseem N., Luong V., Gregory-Evans K., Bhattacharya S.S., Fitzke F.W., Webster A.R. Maculopathy due to the R345W substitution in fibulin-3: distinct clinical features, disease variability, and extent of retinal dysfunction. Invest. Ophthalmol. Vis. Sci. 2006;47:3085–3097. doi: 10.1167/iovs.05-1600. [DOI] [PubMed] [Google Scholar]
- 13.Hulleman J.D. Malattia Leventinese/Doyne Honeycomb Retinal Dystrophy: Similarities to Age-Related Macular Degeneration and Potential Therapies. Adv. Exp. Med. Biol. 2016;854:153–158. doi: 10.1007/978-3-319-17121-0_21. [DOI] [PubMed] [Google Scholar]
- 14.Klein M.L., Schultz D.W., Edwards A., Matise T.C., Rust K., Berselli C.B., Trzupek K., Weleber R.G., Ott J., Wirtz M.K., Acott T.S. Age-related macular degeneration. Clinical features in a large family and linkage to chromosome 1q. Arch. Ophthalmol. 1998;116:1082–1088. doi: 10.1001/archopht.116.8.1082. [DOI] [PubMed] [Google Scholar]
- 15.Schultz D.W., Weleber R.G., Lawrence G., Barral S., Majewski J., Acott T.S., Klein M.L. HEMICENTIN-1 (FIBULIN-6) and the 1q31 AMD locus in the context of complex disease: review and perspective. Ophthalmic Genet. 2005;26:101–105. doi: 10.1080/13816810590968023. [DOI] [PubMed] [Google Scholar]
- 16.Stone E.M., Braun T.A., Russell S.R., Kuehn M.H., Lotery A.J., Moore P.A., Eastman C.G., Casavant T.L., Sheffield V.C. Missense variations in the fibulin 5 gene and age-related macular degeneration. N. Engl. J. Med. 2004;351:346–353. doi: 10.1056/NEJMoa040833. [DOI] [PubMed] [Google Scholar]
- 17.Stone E.M., Lotery A.J., Munier F.L., Héon E., Piguet B., Guymer R.H., Vandenburgh K., Cousin P., Nishimura D., Swiderski R.E. A single EFEMP1 mutation associated with both Malattia Leventinese and Doyne honeycomb retinal dystrophy. Nat. Genet. 1999;22:199–202. doi: 10.1038/9722. [DOI] [PubMed] [Google Scholar]
- 18.Thompson C.L., Klein B.E., Klein R., Xu Z., Capriotti J., Joshi T., Leontiev D., Lee K.E., Elston R.C., Iyengar S.K. Complement factor H and hemicentin-1 in age-related macular degeneration and renal phenotypes. Hum. Mol. Genet. 2007;16:2135–2148. doi: 10.1093/hmg/ddm164. [DOI] [PubMed] [Google Scholar]
- 19.McLaughlin P.J., Bakall B., Choi J., Liu Z., Sasaki T., Davis E.C., Marmorstein A.D., Marmorstein L.Y. Lack of fibulin-3 causes early aging and herniation, but not macular degeneration in mice. Hum. Mol. Genet. 2007;16:3059–3070. doi: 10.1093/hmg/ddm264. [DOI] [PubMed] [Google Scholar]
- 20.Fu L., Garland D., Yang Z., Shukla D., Rajendran A., Pearson E., Stone E.M., Zhang K., Pierce E.A. The R345W mutation in EFEMP1 is pathogenic and causes AMD-like deposits in mice. Hum. Mol. Genet. 2007;16:2411–2422. doi: 10.1093/hmg/ddm198. [DOI] [PubMed] [Google Scholar]
- 21.Marmorstein L.Y., McLaughlin P.J., Peachey N.S., Sasaki T., Marmorstein A.D. Formation and progression of sub-retinal pigment epithelium deposits in Efemp1 mutation knock-in mice: a model for the early pathogenic course of macular degeneration. Hum. Mol. Genet. 2007;16:2423–2432. doi: 10.1093/hmg/ddm199. [DOI] [PubMed] [Google Scholar]
- 22.Hulleman J.D., Kaushal S., Balch W.E., Kelly J.W. Compromised mutant EFEMP1 secretion associated with macular dystrophy remedied by proteostasis network alteration. Mol. Biol. Cell. 2011;22:4765–4775. doi: 10.1091/mbc.E11-08-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marmorstein L.Y., Munier F.L., Arsenijevic Y., Schorderet D.F., McLaughlin P.J., Chung D., Traboulsi E., Marmorstein A.D. Aberrant accumulation of EFEMP1 underlies drusen formation in Malattia Leventinese and age-related macular degeneration. Proc. Natl. Acad. Sci. USA. 2002;99:13067–13072. doi: 10.1073/pnas.202491599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez-Godino R., Garland D.L., Pierce E.A. A local complement response by RPE causes early-stage macular degeneration. Hum. Mol. Genet. 2015;24:5555–5569. doi: 10.1093/hmg/ddv287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez-Godino R., Bujakowska K.M., Pierce E.A. Changes in extracellular matrix cause RPE cells to make basal deposits and activate the alternative complement pathway. Hum. Mol. Genet. 2018;27:147–159. doi: 10.1093/hmg/ddx392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roybal C.N., Marmorstein L.Y., Vander Jagt D.L., Abcouwer S.F. Aberrant accumulation of fibulin-3 in the endoplasmic reticulum leads to activation of the unfolded protein response and VEGF expression. Invest. Ophthalmol. Vis. Sci. 2005;46:3973–3979. doi: 10.1167/iovs.05-0070. [DOI] [PubMed] [Google Scholar]
- 27.Li Y., Tsai Y.T., Hsu C.W., Erol D., Yang J., Wu W.H., Davis R.J., Egli D., Tsang S.H. Long-term safety and efficacy of human-induced pluripotent stem cell (iPS) grafts in a preclinical model of retinitis pigmentosa. Mol. Med. 2012;18:1312–1319. doi: 10.2119/molmed.2012.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maminishkis A., Chen S., Jalickee S., Banzon T., Shi G., Wang F.E., Ehalt T., Hammer J.A., Miller S.S. Confluent monolayers of cultured human fetal retinal pigment epithelium exhibit morphology and physiology of native tissue. Invest. Ophthalmol. Vis. Sci. 2006;47:3612–3624. doi: 10.1167/iovs.05-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cowan C.S., Renner M., De Gennaro M., Gross-Scherf B., Goldblum D., Hou Y., Munz M., Rodrigues T.M., Krol J., Szikra T. Cell Types of the Human Retina and Its Organoids at Single-Cell Resolution. Cell. 2020;182:1623–1640.e34. doi: 10.1016/j.cell.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lian J., Nelson R., Lehner R. Carboxylesterases in lipid metabolism: from mouse to human. Protein Cell. 2018;9:178–195. doi: 10.1007/s13238-017-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu J., Li Y., Chen W.D., Xu Y., Yin L., Ge X., Jadhav K., Adorini L., Zhang Y. Hepatic carboxylesterase 1 is essential for both normal and farnesoid X receptor-controlled lipid homeostasis. Hepatology. 2014;59:1761–1771. doi: 10.1002/hep.26714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao B., Song J., Ghosh S. Hepatic overexpression of cholesteryl ester hydrolase enhances cholesterol elimination and in vivo reverse cholesterol transport. J. Lipid Res. 2008;49:2212–2217. doi: 10.1194/jlr.M800277-JLR200. [DOI] [PubMed] [Google Scholar]
- 33.Ghosh S. Macrophage cholesterol homeostasis and metabolic diseases: critical role of cholesteryl ester mobilization. Expert Rev. Cardiovasc. Ther. 2011;9:329–340. doi: 10.1586/erc.11.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu Y., Gao H., Vo C., Ke C., Pan F., Yu L., Siegel E., Hess K.R., Linskey M.E., Zhou Y.H. Anti-EGFR function of EFEMP1 in glioma cells and patient prognosis. Oncoscience. 2014;1:205–215. doi: 10.18632/oncoscience.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan F., Hui Y.N., Li Y.J., Guo C.M., Meng H. Epidermal growth factor receptor in cultured human retinal pigment epithelial cells. Ophthalmologica. 2007;221:244–250. doi: 10.1159/000101926. [DOI] [PubMed] [Google Scholar]
- 36.Maruichi T., Fukami T., Nakajima M., Yokoi T. Transcriptional regulation of human carboxylesterase 1A1 by nuclear factor-erythroid 2 related factor 2 (Nrf2) Biochem. Pharmacol. 2010;79:288–295. doi: 10.1016/j.bcp.2009.08.019. [DOI] [PubMed] [Google Scholar]
- 37.Mullins R.F., Hageman G.S. Human ocular drusen possess novel core domains with a distinct carbohydrate composition. J. Histochem. Cytochem. 1999;47:1533–1540. doi: 10.1177/002215549904701205. [DOI] [PubMed] [Google Scholar]
- 38.Abdelsalam A., Del Priore L., Zarbin M.A. Drusen in age-related macular degeneration: pathogenesis, natural course, and laser photocoagulation-induced regression. Surv. Ophthalmol. 1999;44:1–29. doi: 10.1016/s0039-6257(99)00072-7. [DOI] [PubMed] [Google Scholar]
- 39.Curcio C.A., Presley J.B., Malek G., Medeiros N.E., Avery D.V., Kruth H.S. Esterified and unesterified cholesterol in drusen and basal deposits of eyes with age-related maculopathy. Exp. Eye Res. 2005;81:731–741. doi: 10.1016/j.exer.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 40.Curcio C.A., Presley J.B., Millican C.L., Medeiros N.E. Basal deposits and drusen in eyes with age-related maculopathy: evidence for solid lipid particles. Exp. Eye Res. 2005;80:761–775. doi: 10.1016/j.exer.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 41.Hageman G.S., Luthert P.J., Victor Chong N.H., Johnson L.V., Anderson D.H., Mullins R.F. An integrated hypothesis that considers drusen as biomarkers of immune-mediated processes at the RPE-Bruch’s membrane interface in aging and age-related macular degeneration. Prog. Retin. Eye Res. 2001;20:705–732. doi: 10.1016/s1350-9462(01)00010-6. [DOI] [PubMed] [Google Scholar]
- 42.Hageman G.S., Mullins R.F. Molecular composition of drusen as related to substructural phenotype. Mol. Vis. 1999;5:28. [PubMed] [Google Scholar]
- 43.Hageman G.S., Mullins R.F., Russell S.R., Johnson L.V., Anderson D.H. Vitronectin is a constituent of ocular drusen and the vitronectin gene is expressed in human retinal pigmented epithelial cells. FASEB J. 1999;13:477–484. doi: 10.1096/fasebj.13.3.477. [DOI] [PubMed] [Google Scholar]
- 44.Jiang K., To E., Cui J.Z., Cao S., Gao J., Matsubara J.A. Drusen and Pro-inflammatory Mediators in the Post-Mortem Human Eye. J. Clin. Exp. Ophthalmol. 2012;3:208. doi: 10.4172/2155-9570.1000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamei M., Hollyfield J.G. TIMP-3 in Bruch’s membrane: changes during aging and in age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 1999;40:2367–2375. [PubMed] [Google Scholar]
- 46.Luibl V., Isas J.M., Kayed R., Glabe C.G., Langen R., Chen J. Drusen deposits associated with aging and age-related macular degeneration contain nonfibrillar amyloid oligomers. J. Clin. Invest. 2006;116:378–385. doi: 10.1172/JCI25843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malek G., Li C.M., Guidry C., Medeiros N.E., Curcio C.A. Apolipoprotein B in cholesterol-containing drusen and basal deposits of human eyes with age-related maculopathy. Am. J. Pathol. 2003;162:413–425. doi: 10.1016/S0002-9440(10)63836-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mullins R.F., Russell S.R., Anderson D.H., Hageman G.S. Drusen associated with aging and age-related macular degeneration contain proteins common to extracellular deposits associated with atherosclerosis, elastosis, amyloidosis, and dense deposit disease. FASEB J. 2000;14:835–846. [PubMed] [Google Scholar]
- 49.Wang L., Clark M.E., Crossman D.K., Kojima K., Messinger J.D., Mobley J.A., Curcio C.A. Abundant lipid and protein components of drusen. PLoS ONE. 2010;5:e10329. doi: 10.1371/journal.pone.0010329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Curcio C.A., Millican C.L., Bailey T., Kruth H.S. Accumulation of cholesterol with age in human Bruch’s membrane. Invest. Ophthalmol. Vis. Sci. 2001;42:265–274. [PubMed] [Google Scholar]
- 51.Haimovici R., Gantz D.L., Rumelt S., Freddo T.F., Small D.M. The lipid composition of drusen, Bruch’s membrane, and sclera by hot stage polarizing light microscopy. Invest. Ophthalmol. Vis. Sci. 2001;42:1592–1599. [PubMed] [Google Scholar]
- 52.Pauleikhoff D., Zuels S., Sheraidah G.S., Marshall J., Wessing A., Bird A.C. Correlation between biochemical composition and fluorescein binding of deposits in Bruch’s membrane. Ophthalmology. 1992;99:1548–1553. doi: 10.1016/s0161-6420(92)31768-3. [DOI] [PubMed] [Google Scholar]
- 53.Anderson D.H., Ozaki S., Nealon M., Neitz J., Mullins R.F., Hageman G.S., Johnson L.V. Local cellular sources of apolipoprotein E in the human retina and retinal pigmented epithelium: implications for the process of drusen formation. Am. J. Ophthalmol. 2001;131:767–781. doi: 10.1016/s0002-9394(00)00961-2. [DOI] [PubMed] [Google Scholar]
- 54.Curcio C.A., Johnson M., Huang J.D., Rudolf M. Aging, age-related macular degeneration, and the response-to-retention of apolipoprotein B-containing lipoproteins. Prog. Retin. Eye Res. 2009;28:393–422. doi: 10.1016/j.preteyeres.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang D., Zou L., Jin Q., Hou J., Ge G., Yang L. Human carboxylesterases: a comprehensive review. Acta Pharm. Sin. B. 2018;8:699–712. doi: 10.1016/j.apsb.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edwards A.O., Klein M.L., Berselli C.B., Hejtmancik J.F., Rust K., Wirtz M.K., Weleber R.G., Acott T.S. Malattia leventinese: refinement of the genetic locus and phenotypic variability in autosomal dominant macular drusen. Am. J. Ophthalmol. 1998;126:417–424. doi: 10.1016/s0002-9394(98)00097-x. [DOI] [PubMed] [Google Scholar]
- 57.Evans K., Gregory C.Y., Wijesuriya S.D., Kermani S., Jay M.R., Plant C., Bird A.C. Assessment of the phenotypic range seen in Doyne honeycomb retinal dystrophy. Arch. Ophthalmol. 1997;115:904–910. doi: 10.1001/archopht.1997.01100160074012. [DOI] [PubMed] [Google Scholar]
- 58.Storti F., Grimm C. Active Cholesterol Efflux in the Retina and Retinal Pigment Epithelium. Adv. Exp. Med. Biol. 2019;1185:51–55. doi: 10.1007/978-3-030-27378-1_9. [DOI] [PubMed] [Google Scholar]
- 59.Apte R.S. Targeting Tissue Lipids in Age-related Macular Degeneration. EBioMedicine. 2016;5:26–27. doi: 10.1016/j.ebiom.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Biswas L., Zhou X., Dhillon B., Graham A., Shu X. Retinal pigment epithelium cholesterol efflux mediated by the 18ckDa translocator protein, TSPO, a potential target for treating age-related macular degeneration. Hum. Mol. Genet. 2017;26:4327–4339. doi: 10.1093/hmg/ddx319. [DOI] [PubMed] [Google Scholar]
- 61.Sene A., Khan A.A., Cox D., Nakamura R.E., Santeford A., Kim B.M., Sidhu R., Onken M.D., Harbour J.W., Hagbi-Levi S. Impaired cholesterol efflux in senescent macrophages promotes age-related macular degeneration. Cell Metab. 2013;17:549–561. doi: 10.1016/j.cmet.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pool F.M., Kiel C., Serrano L., Luthert P.J. Repository of proposed pathways and protein-protein interaction networks in age-related macular degeneration. NPJ Aging Mech. Dis. 2020;6:2. doi: 10.1038/s41514-019-0039-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hotamisligil G.S. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 64.Ertunc M.E., Hotamisligil G.S. Lipid signaling and lipotoxicity in metaflammation: indications for metabolic disease pathogenesis and treatment. J. Lipid Res. 2016;57:2099–2114. doi: 10.1194/jlr.R066514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Galloway C.A., Dalvi S., Hung S.S.C., MacDonald L.A., Latchney L.R., Wong R.C.B., Guymer R.H., Mackey D.A., Williams D.S., Chung M.M. Drusen in patient-derived hiPSC-RPE models of macular dystrophies. Proc. Natl. Acad. Sci. USA. 2017;114:E8214–E8223. doi: 10.1073/pnas.1710430114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lakkaraju A., Finnemann S.C., Rodriguez-Boulan E. The lipofuscin fluorophore A2E perturbs cholesterol metabolism in retinal pigment epithelial cells. Proc. Natl. Acad. Sci. USA. 2007;104:11026–11031. doi: 10.1073/pnas.0702504104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tabas I. Consequences of cellular cholesterol accumulation: basic concepts and physiological implications. J. Clin. Invest. 2002;110:905–911. doi: 10.1172/JCI16452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fu S., Fan J., Blanco J., Gimenez-Cassina A., Danial N.N., Watkins S.M., Hotamisligil G.S. Polysome profiling in liver identifies dynamic regulation of endoplasmic reticulum translatome by obesity and fasting. PLoS Genet. 2012;8:e1002902. doi: 10.1371/journal.pgen.1002902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fu S., Yang L., Li P., Hofmann O., Dicker L., Hide W., Lin X., Watkins S.M., Ivanov A.R., Hotamisligil G.S. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature. 2011;473:528–531. doi: 10.1038/nature09968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tumova J., Andel M., Trnka J. Excess of free fatty acids as a cause of metabolic dysfunction in skeletal muscle. Physiol. Res. 2016;65:193–207. doi: 10.33549/physiolres.932993. [DOI] [PubMed] [Google Scholar]
- 71.Ban N., Lee T.J., Sene A., Choudhary M., Lekwuwa M., Dong Z., Santeford A., Lin J.B., Malek G., Ory D.S., Apte R.S. Impaired monocyte cholesterol clearance initiates age-related retinal degeneration and vision loss. JCI Insight. 2018;3:3. doi: 10.1172/jci.insight.120824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li C.M., Chung B.H., Presley J.B., Malek G., Zhang X., Dashti N., Li L., Chen J., Bradley K., Kruth H.S., Curcio C.A. Lipoprotein-like particles and cholesteryl esters in human Bruch’s membrane: initial characterization. Invest. Ophthalmol. Vis. Sci. 2005;46:2576–2586. doi: 10.1167/iovs.05-0034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS/MS proteomic data supporting the current study have not been deposited in a public repository due to the sensitivity of patient information but will be made available by the corresponding author upon request.