Abstract
Background:
Progress in precision psychiatry is predicated on identifying reliable individual-level diagnostic biomarkers. For psychosis, measures of structural and functional connectivity could be promising biomarkers given consistent reports of dysconnectivity across psychotic disorders using magnetic resonance imaging.
Methods:
We leverage data from 4 independent cohorts of psychosis patients and controls with observations from approximately 800 individuals. We use group-level analyses and two supervised machine learning algorithms (support vector machines (SVM) and ridge regression) to test within, between, and across sample classification performance of white matter and resting-state connectivity metrics.
Results:
Although we replicate group-level differences in brain connectivity, individual-level classification was suboptimal. Classification performance within sample was variable across folds (highest AUC range= 0.30) and across datasets (average SVM AUC range= 0.50; average ridge regression AUC range= 0.18). Classification performance between samples was similarly variable or resulted in AUC values around 0.65, indicating a lack of model generalizability. Furthermore, collapsing across samples (rsfMRI N=888, DTI N=860) did not improve model performance (maximal AUC= 0.67). Ridge regression models generally outperformed SVM models, although classification performance was still suboptimal in terms of clinical relevance. Adjusting for demographic covariates did not greatly affect results.
Conclusions:
Connectivity measures were not suitable as diagnostic biomarkers for psychosis as assessed in the current study. Our results do not negate that other approaches may be more successful although it is clear that a systematic approach to individual-level classification with large independent validation samples is necessary to properly vet neuroimaging features as diagnostic biomarkers.
Keywords: Psychosis, biomarkers, machine learning, Magnetic Resonance Imaging (MRI), Connectivity
Introduction
Decisions related to the diagnosis and treatment of psychotic disorders like schizophrenia, bipolar disorder, and schizoaffective disorder, are currently dependent on patient report, behavioral observation, and clinical judgment rather than objective laboratory measures(1). Unfortunately, reliance on phenomenology limits the field’s attempts to join the precision medicine revolution(2) and likely contributes to the underinvestment in novel pharmacological treatments for psychotic illness(3). What is needed are biomarkers indexing core biological processes that more precisely predict clinical outcomes and provide novel insights into the pathophysiology of psychosis, psychosis risk, and psychosis treatment(4).
Neuroimaging measures acquired via magnetic resonance imaging (MRI) could serve as such biomarkers by acting as objective endpoints to evaluate treatment response or prognostic accuracy(5). Brain connectivity measures(6–8) are particularly promising given consistent reports of aberrant functional and structural connectivity across psychotic disorders(9–14) and at various stages of illness(15–19). Psychosis-related disruptions in functional connectivity (measured by resting state functional MRI-rsfMRI(20)) often include hypo-connectivity within and between large-scale cortical networks(6), especially those involving frontal and temporal cortex, whereas psychosis-related disruptions in structural connectivity (measured by Diffusion Tensor Imaging-DTI(21)) include brain-wide reductions in fractional anisotropy (FA), an indirect measure of white matter integrity(22). In fact, the two markers themselves are inter-related in both psychotic and healthy individuals(23, 24).
Although findings suggest that connectivity measures have great potential as diagnostic biomarkers for psychosis, conclusions related to their clinical utility remains unclear. Currently, psychotic dysconnectivity is primarily reported at the group level, however a successful biomarker should discriminate cases from controls at the individual level(25–27). Machine learning may bridge this disconnect(28, 29) by providing objective measures of individual-level classification while integrating large amounts of data that are characteristic of MRI connectivity analyses. While machine learning methods have been used to validate neuroimaging biomarkers in psychiatry, a recent review by Scheinost and colleagues(30) highlighted non-trivial shortcomings in their current implementation, including lack of consideration for covariate effects, failure to keep training and testing data independent, and lack of reporting metrics beyond accuracy to evaluate classification performance. Concerns about sample size and the absence of out-of-sample validation were also reported. Indeed, connectivity studies using machine learning seldom exceed 100 people per group, with larger sample sizes often failing to improve classification performance(28, 29, 31). Furthermore, reported classification accuracies are variable, reaching as high as 100%(28, 32, 33) without independent datasets for validation(34). DTI measures are also underrepresented in current studies(33, 35, 36), making their diagnostic utility unclear.
Our goal is to evaluate functional (rsfMRI) and structural (DTI) connectivity measures, separately and combined, as diagnostic biomarkers for psychosis(30) while systematically addressing the aforementioned concerns regarding machine learning analyses. To achieve this, we use four independent datasets with neuroimaging data. Initially, we perform group-level univariate analyses to confirm the presence of psychotic dysconnectivity in our samples. Next, we leverage our access to multiple independent datasets by implementing a multi-level data analysis strategy to evaluate individual-level classification performance. First, we perform within sample classification to determine whether functional and structural connectivity measures could consistently classify individuals regardless of variations in sample characteristics or data collection procedures(37). Second, we evaluate generalizability by building models in one dataset and testing in the remaining three, satisfying the requirement by the Psychiatric Neuroimaging Working Group of the American Psychiatric Association (APA) that biomarkers be validated in at least two independent samples(38). Third, we address key factors that could affect classification performance including demographic covariates, algorithm choice, and sample size. Finally, we share our feature sets and models so that others can implement their preferred machine learning technique or predict diagnostic status in their own samples.
Materials and Methods
Subjects
Participants were individuals with and without psychosis with rsfMRI and/or DTI data from 4 independent samples (Table 1): one from the Icahn School of Medicine at Mount Sinai (ISMMS), two from the Olin Center for Neuropsychiatric Research (Olin and a subset of BSNIP-2), and one collected at the Olin Center and the University of Maryland School of Medicine (BSNIP-1). Recruitment and scanning procedures for each sample are described in the Supplemental Materials and Supplementary Table 1. Most psychosis cases were mid-course and stably medicated.
Table 1.
Participant Demographics
| rsfMRI | DTI | rsfMRI + DTI | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ISMMS | Olin | BSNIP-1 | BSNIP-2 | ISMMS | Olin | BSNIP-1 | BSNIP-2 | ISMMS | Olin | BSNIP-1 | BSNIP-2 | |||||||||||||
| HC N=47 |
Psych N=96 |
HC N=167 |
Psych N=102 |
HC N=97 |
Psych N=237 |
HC N=51 |
Psych N=90 |
HC N=52 |
Psych N=127 |
HC N=180 |
Psych N=96 |
HC N=86 |
Psych N=213 |
HC N=35 |
Psych N=70 |
HC N=47 |
Psych N=94 |
HC N=147 |
Psych N=83 |
HC N=63 |
Psych N=173 |
HC N=31 |
Psych N=64 |
|
|
|
||||||||||||||||||||||||
| Demographics | ||||||||||||||||||||||||
| Age (years (sd.)) | 29.0 (7.6) |
27.2 (7.6) |
39.8 (14.2) |
42.0 (13.0) |
40.4 (12.2) |
35.0 (12.2) |
30.8 (12.9) |
37.6 (11.5) |
29.8 (8.3) |
27.2 (7.6) |
40.3 (14.0) |
41.6 (12.4) |
39.5 (12.4) |
34.4 (12.1) |
30.4 (12.5) |
38.8 (12.0) |
29.0 (7.6) |
27.1 (7.6) |
39.6 (14.0) |
41.7 (12.8) |
40.2 (12.3) |
34.9 (12.0) |
30.8 (12.9) |
39.0 (12.2) |
| Sex, male, n(%) | 27 (57%) |
69 (72%) |
86 (52%) |
54 (53%) |
36 (37%) |
136 (57%) |
22 (43%) |
37 (41%) |
31 (60%) |
93 (73%) |
93 (52%) |
56 (58%) |
38 (44%) |
125 (59%) |
14 (40%) |
28 (40%) |
27 (59%) |
68 (72%) |
76 (52%) |
47 (57%) |
26 (41%) |
99 (57%) |
11 (35%) |
26 (41%) |
| Educationa | 5.4 (1.0) |
3.9 (1.3) |
3.9 (1.4) |
3.4 (1.2) |
14.9 (2.4) |
13.3 (2.2) |
14.8 (2.2) |
13.0 (2.2) |
5.3 (1.0) |
3.9 (1.3) |
3.9 (1.5) |
3.5 (1.2) |
15.0 (2.4) | 13.2 (2.2) |
14.6 (2.3) |
13.1 (2.4) |
5.4 (1.0) |
3.9 (1.3) |
3.9 (1.4) |
3.5 (1.2) |
14.8 (2.3) |
13.3 (2.2) |
14.8 (2.2) |
12.9 (2.3) |
| IQ (sd.)b | 117.4 (16.2) |
96.3 (16.1) |
91.5 (13.2) |
87.4 (11.9) |
97.6 (15.4) |
97.2 (14.9) |
100.6 (11.0) |
95.2 (14.0) |
116.1 (16.7) |
96.3 (16.4) |
90.5 (13.2) |
88.3 (13.1) |
97.2 (15.6) |
98.4 (15.0) |
100.1 (12.3) |
97.3 (15.1) |
117.4 (16.2) |
96.5 (15.9) |
91.4 (13.3) |
87.6 (12.1) |
97.6 (14.6) |
98.2 (14.5) |
101.6 (11.8) |
96.0 (14.7) |
| Race n (%) | ||||||||||||||||||||||||
| Caucasian | 19 (40%) |
17 (17%) |
57 (58%) |
129 (54%) |
20 (40%) |
38 (42%) |
19 (37%) |
22 (17%) |
53 (61%) |
121 (57%) |
10 (29%) |
30 (43%) |
19 (41%) |
16 (17%) |
39 (62%) |
97 (56%) |
10 (33%) |
26 (41%) |
||||||
| African American | 12 (26%) |
40 (42%) |
167 (100%) |
102 (100%) |
28 (29%) |
75 (32%) |
15 (29%) |
18 (20%) |
17 (33%) |
50 (39%) |
180 (100%) |
96 (100%) |
23 (27%) |
63 (30%) |
15 (43%) |
16 (23%) |
12 (26%) |
40 (42%) |
147 (100%) |
83 (100%) |
16 (25%) |
52 (30% |
12 (39%) |
16 (25%) |
| Other | 12 (26%) | 36 (38%) | 12 (12%) | 32 (14%) | 12 (24%) | 34 (37%) | 12 (23%) | 49 (39%) | 10 (11%) | 28 (13%) | 9 (26%) | 24 (34%) | 12 (26%) | 35 (37%) | 8 (13%) | 23 (13%) | 8 (25%) | 22 (34%) | ||||||
| Diagnosis Breakdown | ||||||||||||||||||||||||
| BPP n(%) | 19 (20%) |
12 (12%) |
50 (21%) |
11 (12%) |
31 (24%) |
12 (13%) |
49 (23%) |
11 (16%) |
19 (20%) |
12 (15%) |
35 (20%) |
9 (14%) |
||||||||||||
| SAD n(%) | 27 (28%) |
26 (26%) |
64 (27%) |
43 (48%) |
32 (25%) |
27 (28%) |
61 (29%) |
30 (43%) |
25 (27%) |
21 (25%) |
48 (28%) |
28 (44%) |
||||||||||||
| SZ n(%) | 47 (49%) |
46 (45%) |
123 (52%) |
36 (40%) |
59 (46%) |
45 (47%) |
103 (48%) |
29 (41%) |
47 (51%) |
40 (48%) |
90 (52%) |
27 (42%) |
||||||||||||
| MDD n(%) | 3 (3%) |
8 (8%) |
5 (4%) |
5 (5%) |
3 (3%) |
5 (6%) |
||||||||||||||||||
| Other n(%) | 10 (9%) |
7 (7%) |
5 (6%) |
|||||||||||||||||||||
| Symptoms | ||||||||||||||||||||||||
| YMRS (sd.) | _ | 8.3 (7.6) |
4.9 (5.8) |
10.7 (6.5) |
_ | 7.8 (6.8) |
5.2 (5.9) |
10.3 (6.8) |
_ | 7.7 (6.8) | 4.8 (5.6) |
10.3 (6.9) |
||||||||||||
| BPRS (sd.) | 50.9 (19.3) |
45.6 (11.8) |
_ | _ | 49.6 (18.9) |
44.8 (11.3) |
_ | _ | 51.0 (18.6) | 45.1 (11.5) | _ | _ | ||||||||||||
| PANSS (sd.) | ||||||||||||||||||||||||
| Positive | _ | _ | 14.6 (5.5) |
_ | 14.8 (5.1) |
_ | _ | 14.5 (5.4) |
14.6 (5.4) |
_ | _ | 14.5 (5.3) |
14.7 (5.3) |
|||||||||||
| Negative | _ | _ | 14.3 (5.3) |
_ | 13.9 (4.9) |
_ | _ | 14.1 (5.3) |
13.8 (4.7) |
_ | _ | 14.1 (5.4) |
14.0 (4.7) |
|||||||||||
| General | _ | _ | 28.2 (7.9) |
_ | 29.2 (5.9) |
_ | _ | 28.5 (7.8) |
29.3 (5.5) |
_ | _ | 28.0 (7.6) |
29.5 (5.6) |
|||||||||||
| Total | _ | _ | 57.0 (15.3) |
_ | 58 (13.2) |
_ | _ | 57.1 (15.2) |
57.7( 13.2) |
_ | _ | 56.6 (14.8) |
58.2 (13.2) |
|||||||||||
| Medications | ||||||||||||||||||||||||
| Lithium use, n(%) | 0 (0%) |
13 (14%) |
0 (0%) |
2 (2%) |
0 (0%) |
26 (11%) |
0 (0%) |
14 (16%) |
0 (0%) |
22 (18%) |
0 (0%) |
2 (2%) |
0 (0%) |
28 (13%) |
0 (0%) |
12 (17%) |
0 (0%) |
13 (14%) |
0 (0%) |
2 (2%) |
0 (0%) |
20 (12%) |
0 (0%) |
12 (19%) |
| Antipsychotic use, n(%) | 0 (0%) |
86 (90%) |
2 (1%) |
62 (61%) |
0 (0%) |
211 (89%) |
0 (0%) |
68 (76%) |
0 (0%) |
112 (88%) |
2 (1%) |
59 (62%) |
0 (0%) |
187 (88%) |
0 (0%) |
54 (78%) |
0 (0%) |
85 (90%) |
2 (1%) |
5 2(63%) |
0 (0%) |
156 (90%) |
0 (0%) |
49 (78%) |
Dashes indicate unavailable data; blank cells indicate data not applicable. rsfMRI=resting state functional MRI, DTI=Diffusion Tensor Imaging, BPP=Bipolar Disorder with Psychosis, SAD=Schizoaffective Disorder, SZ=Schizophrenia, MDD=Major Depressive Disorder with Psychosis, BPRS=Brief Psychiatric Rating Scale, YMRS=Young Mania Rating Scale, PNASS=Positive and Negative Symptom Scale
An 8-point scale from the SCID screener was used for Mount Sinai and Olin samples; Total years was used for BSNIP-1 and BSNIP-2 samples.
Estimated with the WASI for Mt. Sinai and Olin samples; Estimated with the WRAT for BSNIP1 and BSNIP2 samples
Feature Generation
rsfMRI.
MRIQC v0.15.0(39) was used to generate visual reports and quality control (QC) metrics for rsfMRI data in each sample (Supplementary Table 2 and Supplemental Materials). Pre-processing was performed with fMRIPrep(40), version 1.3.2. Squares and derivatives of six motion parameters, frame-wise displacement, DVARS, and anatomical CompCor components(41) were regressed from the data. Residual mean time series were extracted from cortical and sub-cortical regions (N=250) using the Brainnetome atlas(42). Features for machine learning were unique values from the 250×250 matrix of correlation coefficients (converted to z-scores (z=.5×ln(1+r)/(1−r))) between the time courses for each pair of nodes for each subject. A second set of features was generated by residualizing for age, sex, and site (BSNIP-1 only). Further detail is noted in Supplemental Materials.
DTI.
Diffusion-weighted images were processed using FSL(43), version 5.0.10. Preprocessing included brain extraction(44), motion and eddy current correction(45), and tensor fitting, resulting in individual FA maps. A summary of QC metrics for each sample is listed in Supplementary Table 2. Preprocessed FA maps passing QC procedures (see Supplemental Methods) were fed into the Tract-Based Spatial Statistics (TBSS) pipeline(46); for each participant, average FA was calculated for 20 tracts from the John’s Hopkins University white matter tractography atlas(47). A second set of features was generated by residualizing for age, sex, and site (BSNIP-1 only). Further detail is noted in Supplemental Materials.
Group-level Univariate Analysis
We tested between-group differences in rsfMRI and DTI features for each sample using standard univariate null hypothesis testing methods. For rsfMRI, we performed two-sample t-tests comparing connectivity between each pair of nodes using the FSL Nets Analysis package, version 0.6.3 (http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FSLNets). Significance testing was performed using permutation tests via nets_glm (48). For DTI, we performed two-sample t-tests comparing FA values for each tract, considering FDR corrected p-values(49) < 0.05 as significant.
For each modality, we also quantified the aggregate effect across samples using random-effects inverse-variance weighted meta-analyses in R (metafor package, version 2.1–0 http://www.metafor-project.org/). For rsfMRI, we included connections showing significant differences between psychosis cases and healthy participants in one or more samples. To quantify how similar findings were in each sample, we performed Pearson correlations between case-control effect sizes in each pair of studies for both functional and structural connectivity measures.
Supervised Machine Learning
We used two machine learning algorithms, linear support vector machines (SVMs) and L2 logistic regression (ridge regression). For each algorithm, parameter optimization was performed using a grid search and nested cross validation (with 5 folds for both the outer and inner loops) over a range of values chosen to span several orders of magnitude, large enough to identify the optimal parameter space. SVMs classify individuals by projecting features into a multi-dimensional feature space and constructing a hyperplane that maximizes distances between data points of opposing groups by minimizing a cost function(50). Nested cross validation was performed with cost values [1,2,5,10,25,50,100,200,500,750,1000,5000]. Results were reported for models with a cost of 1 given that classification accuracies were not altered when optimizing the cost parameter. Ridge regression is a regularized form of logistic regression that mitigates multi-collinearity(51) by using a regularization parameter (λ). Optimization of λ was performed over 80 linearly spaced values between 1 and 5000. Regression model results are reported for lambda values that yielded the minimum mean cross-validated RMSE over training folds. In practice we found a high value of lambda was needed to sufficiently penalize the model. Training and testing datasets for all analyses were identical for each algorithm and covariate condition to ensure comparability.
For each covariate and algorithm condition, we considered features from each modality in separate and combined models and implemented three levels of analysis (outlined in Supplementary Figure 1). First, we evaluated within sample classification performance for each dataset using k-fold (k=5) cross-validation implemented with the “createDataPartition” function from the Caret package in R (52). Data was partitioned into training and testing folds using random selection with p = .8 (i.e. 80% of the data was used for training, 20% for testing), while preserving the proportion of psychosis cases to controls in the overall sample within each fold. Next, we tested model generalizability by systematically using each sample as a training dataset and testing classification performance on the remaining samples separately. Finally, we evaluated the effect of sample size on classification performance by combining all samples into one dataset and performing k-fold (k=5) cross-validation using the same procedures as within-sample classification. Note that in the combined sample case, we did not use sample as a confounding covariate due to concern that this could induce spurious associations (via condition on a collider (53)) since site is not randomly assigned and could be associated with psychological or biological factors (e.g. common effects of socioeconomic status). Classification performance for all models was evaluated using the area under the receiver operating curve (AUC of the ROC), a comprehensive measure of algorithm discriminability that is threshold-independent and results in a singular measure by which to compare classification success. We also report accuracy, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) from the midpoint of the ROC curve to compare our results to studies without ROC-based measures.
Results
Group-level Psychotic Dysconnectivity
Psychosis cases showed wide-spread reductions in resting state functional connectivity compared to heathy participants, but results were variable across samples (Figure 1 and Supplementary Figure 2 and 3). The ISMMS sample showed few significant case-control differences, with most disrupted connections linking the supplementary motor (SM) and default mode networks (DMN). In contrast, case-control differences in the Olin sample were dominated by connections within and between cognitive control networks [DMN, ventral attention network (VAN), and salience (SAL)] and between cognitive control networks and the auditory and SM networks. Case-control differences in BSNIP-1 were between visual networks and both lower-order sensory and higher-order association networks. BSNIP-1 psychosis cases also showed stronger connectivity than healthy participants in limbic and DMN networks, although only when residualizing features for covariates such as age, sex, and site. Lastly, case-control differences in BSNIP-2 were dominated by reductions in connectivity between sensory networks (visual and SM/auditory and SM) and between the DMN and the SAL and VAN networks. The aggregate effect across datasets included most connections that were weaker in psychosis in all four samples. In-scanner motion was significantly higher in cases than in healthy participants (ISMMS: t(106)= −2.31, p= 0.02; Olin: t(205.77)= −1.98, p= 0.05; BSNIP-1: t(250.96)= −4.43, p= 1.40 × 10−5; BSNIP-2: t(113.02)= −2.52, p= 0.01), although motion effects were regressed from time series before group-level analyses.
Figure 1. Group-wise Differences in Connectivity between Psychosis Cases and Healthy Participants: Non-Residualized Features.
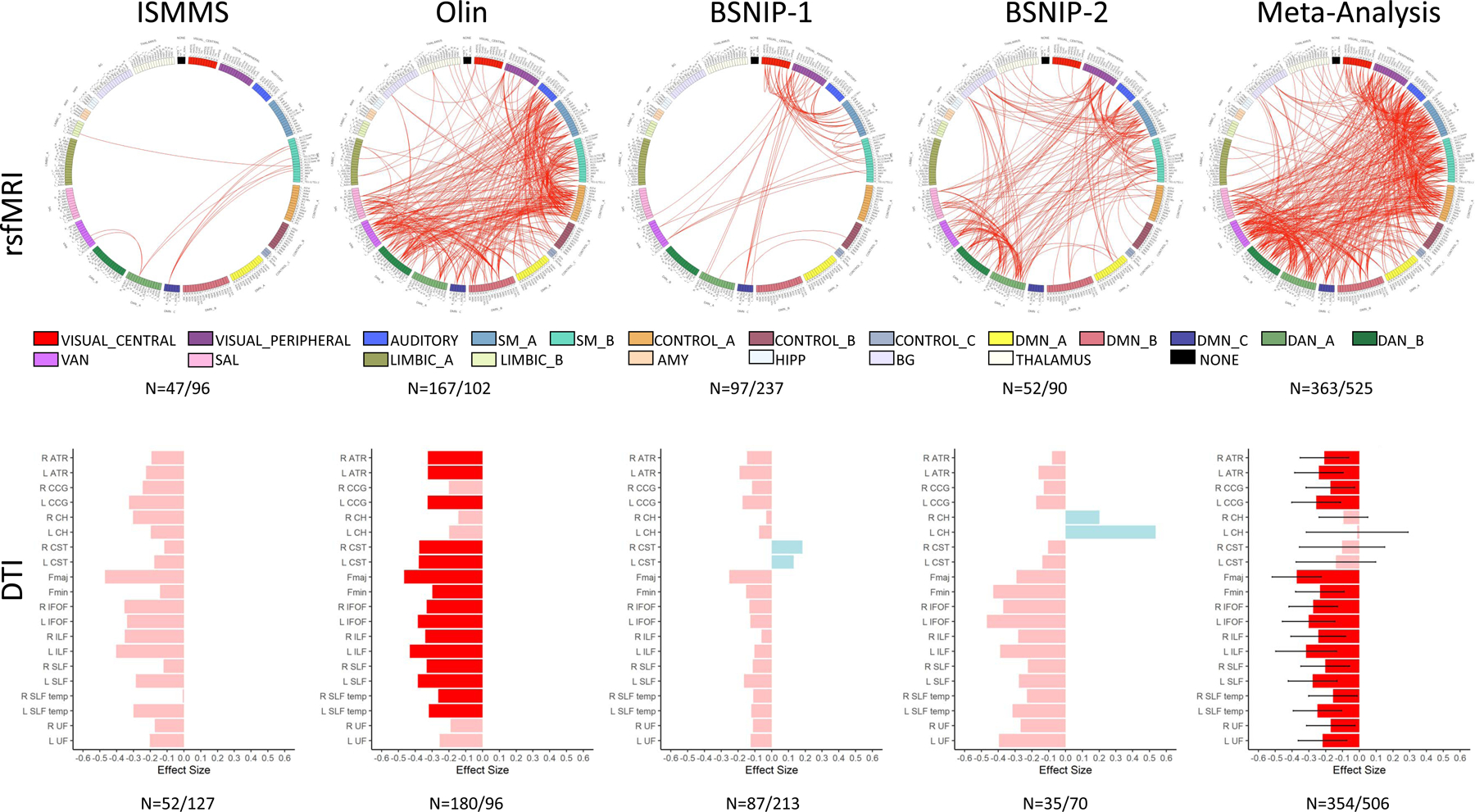
Reported N’s are the number of healthy participants and number of psychosis cases respectively. For rsfMRI (top), red lines show significant reductions in functional connectivity between nodes in psychosis cases compared to healthy participants for each sample (after permutation testing) and for the metanalysis across all samples (after FDR correction). To improve interpretability, Brainnentome nodes were assigned to one of the 17 networks defined in Yeo et al.(75) (https://github.com/ThomasYeoLab/CBIG/tree/master/stable_projects/brain_parcellation/Schaefer2018_LocalGlobal). See supplemental Table 7 for node order in circular plots. We further subdivided the “NONE” category into subcortical regions (AMY, HIPP, BG, and THALAMUS) for clarity. For DTI (bottom), bars show the Cohen’s d effect size for t-tests performed for the 20 white matter tracts in each sample and the metanalysis (error bars are confidence intervals). Negative effect sizes (pink/red) indicate psychosis cases FA < healthy participant FA. Darker shades indicate significant differences (FDR-corrected). Positive effect sizes (blue) indicate psychosis cases FA > healthy participant FA. rsfMRI= resting state MRI, DTI= Diffusion Tensor Imaging, SM=Supplementary Motor, DMN=Default Mode Network, DAN=Dorsal Attention Network, VAN=Ventral Attention Network, SAL=Salience Network, AMY=Amygdala, HIPP=Hippocampus, BG=Basal Ganglia, ATR=Anterior Thalamic Radiation, CgC=Cingulum Cortex, CgH=Cingulum Hippocampus, CST=Corticospinal Tract, Fmaj=Forceps major, Fmin=Forceps minor, IFOF=Inferior Fronto-Occipital Fasciculus, ILF=Inferior Longitudinal Fasciculus, SLF=Superior Longitudinal Fasciculus, tSLF=Superior Longitudinal Fasciculus Temporal Part, UF=Uncinate Fasciculus.
Psychosis cases also showed reductions in structural connectivity, with lower FA values than healthy participants in most white matter tracts (Figures 1 and Supplementary Figure 2). Effect sizes varied across samples (Supplementary Figure 3), although the aggregate effect included most white matter tracts. There were no significant group differences in in-scanner motion (average root mean square difference from the first volume)(ISMMS: t(177)= −0.51, p= 0.61; Olin: t(274)= −1.04, p= 0.30; BSNIP-2: t(103)= −0.29, p= 0.77); as we obtained preprocessed BSNIP-1 data that matched our standardized pipeline, the sample did not include the necessary information to obtain motion estimates).
Inclusion of demographic covariates (age, sex, and site (where appropriate)) did not significantly alter the pattern of group-wise results for either modality, except for BSNIP-1, where there was a large increase in the number of significantly weaker functional connections in the psychosis group, likely given the inclusion of site as a covariate.
Individual-level Classification Within-Sample Classification
SVM.
Classification performance was modality- and sample-specific (Figure 2A and Supplementary Table 3). rsfMRI model performance varied widely across samples (AUC range 0.26–0.73). DTI models were more consistent across samples but performed poorly (AUC values mostly < 0.50). Combining modalities did not improve either performance or variability. Model feature weights were also inconsistent across samples (Supplementary Figure 4A). For example, connections between the SM and limbic network, which had the largest positive weights in the rsfMRI model constructed in the ISMMS sample, were close to zero for models built in the Olin and BSNIP-2 samples, and negative in the BSNIP-1 sample. Within samples, fold-wise variability was also apparent with non-trivial standard deviations in AUC values and feature weights. Adjusting for covariates did not greatly affect classification performance (Supplementary Figure 5A and Supplementary Table 3) or feature weightings (Supplementary Figure 6A).
Figure 2. Within Sample Classification Results: Non-Residualized Features.
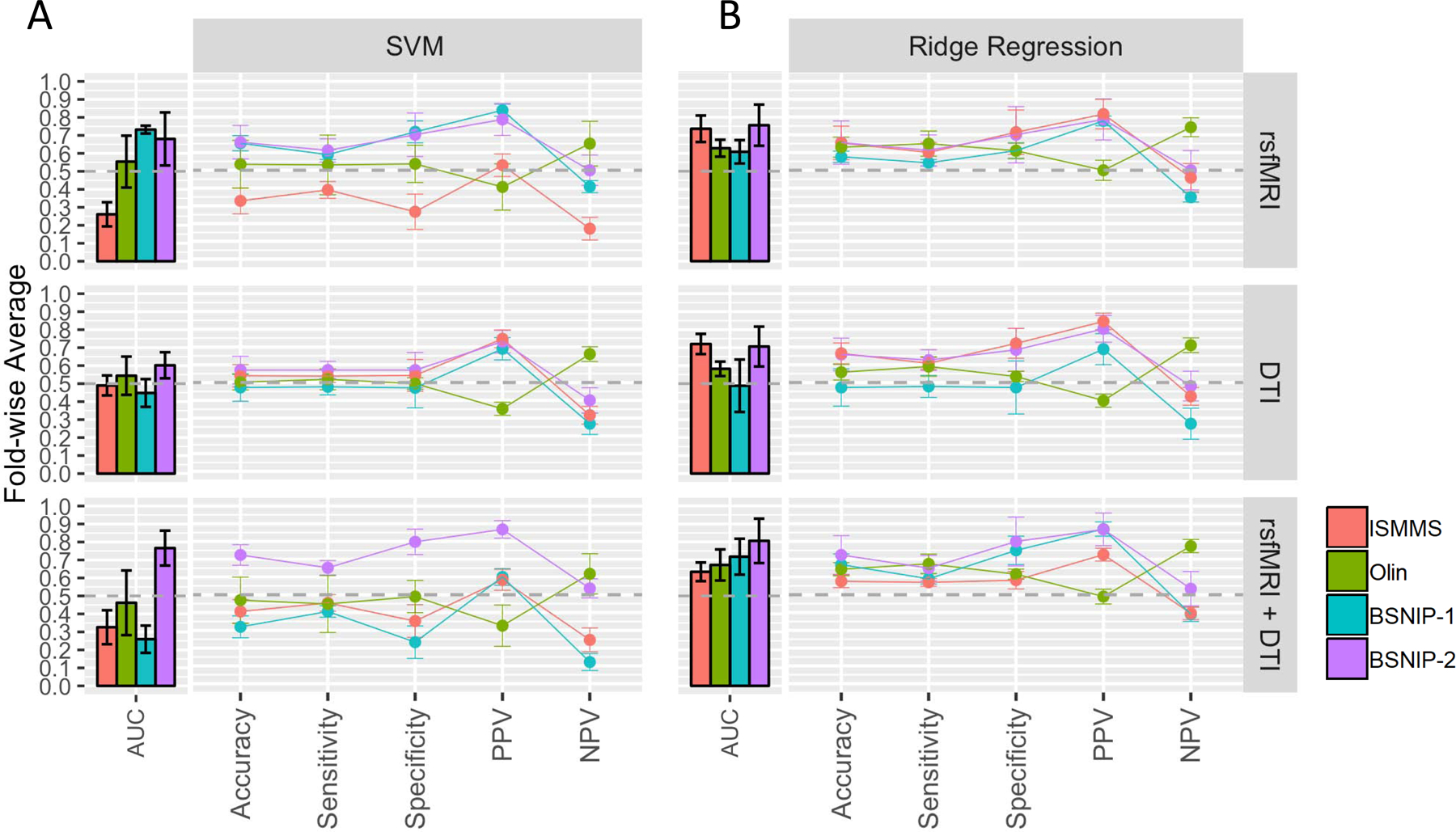
A. Performance profiles for the SVM algorithm and each feature modality. Colors correspond to samples. Bars show AUC (overall algorithm performance), while lines show point estimates from the midpoint of the ROC curve. Values on the y-axis are average values over 5 folds and error bars represent the standard deviation over folds. Values are for models using non-residualized features. Results for models using residualized features are shown in Supplementary Figure 5A and are largely similar. B. Same as A. but for ridge regression. Results for models using residualized features are shown in Supplementary Figure 5B. rsfMRI=resting state functional MRI, DTI=Diffusion Tensor Imaging, PPV=Positive Predictive Value, NPV=Negative Predictive Value, AUC=Area Under the Curve.
Ridge Regression.
Classification performance was similar to, or numerically better than, SVM performance (Figure 2B and Supplementary Table 3) (AUC values generally above 0.50). However, variability in performance across samples for each modality persisted. Inconsistency in feature weightings across datasets (Supplementary Figure 4B) and fold-wise variability in AUC also persisted. Adjusting for covariates did not globally affect results (Supplementary Figure 5B,6B, and Supplementary Table 3).
Between-Sample Classification
SVM.
Figure 3A shows classification performance for each sample as a training set (rows) and the remaining samples as independent test sets (colored bars/lines within panels) (also see Supplementary Table 4a). Performance was either inconsistent across testing datasets or poor regardless of the modality or dataset used for training. For example, rsfMRI models trained in BSNIP-2 had the most consistent performance across testing sets, although no AUC value exceeded 0.65. Classification success was also heavily dependent on which dataset was used to train the model. For example, BSNIP-1 AUCs for rsfMRI models were 0.41 when the model was trained in the ISMMS sample, 0.50 when trained in the Olin sample, and 0.65 when trained in the BSNIP-2 sample. Again, combining modalities did not result in superior model performance and results were unaffected by covariates (Supplementary Figure 7A and Supplementary Table 4a).
Figure 3. Between Sample Classification Results: Non-Residualized Features.
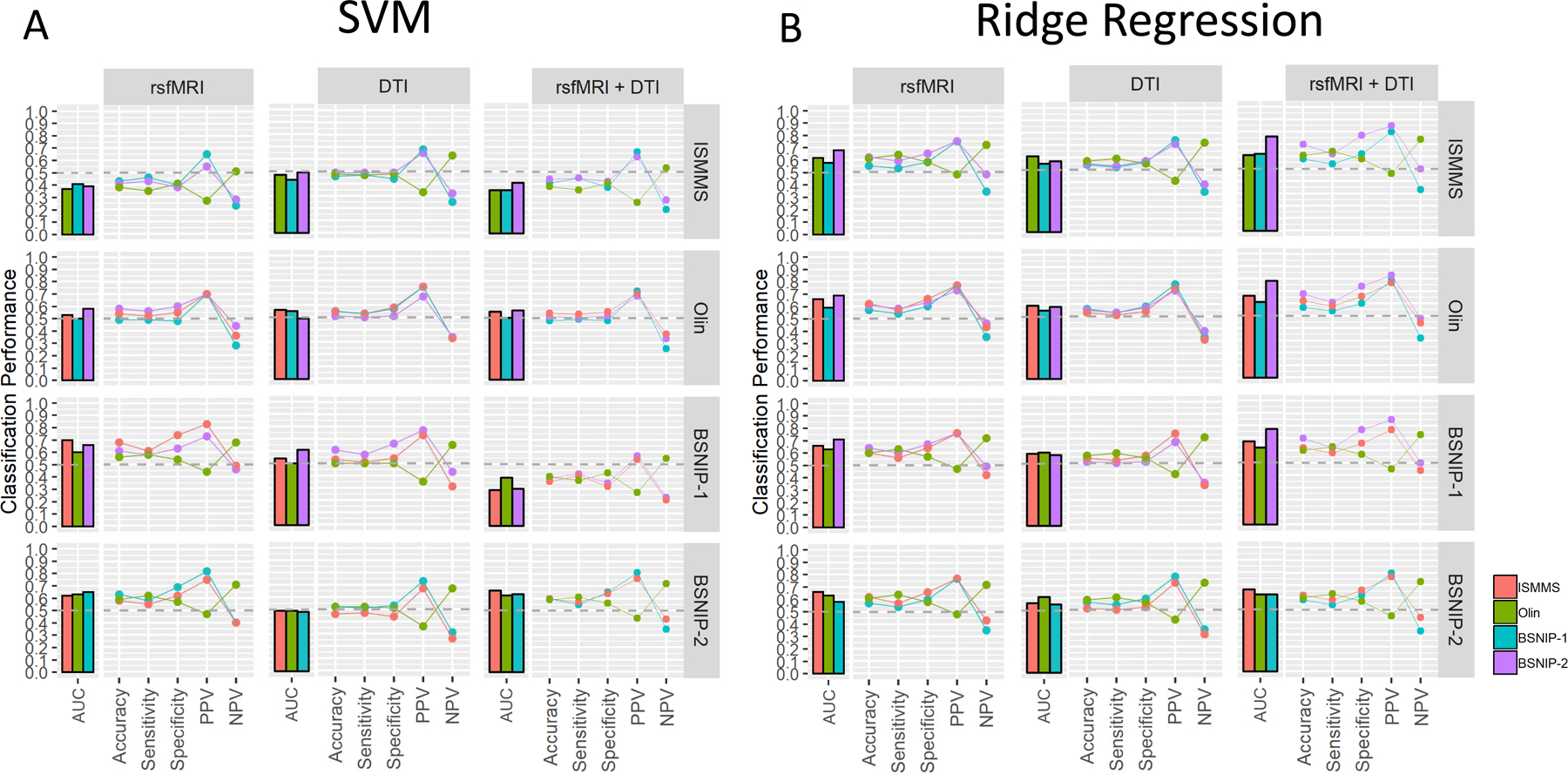
A. SVM performance profiles for between-sample sample classification for each feature modality (columns) when each sample is used as the training sample (rows). Each bar and line within a panel represents an independent testing sample for each training sample (n=3). Values are for models using non-residualized features; results for models using residualized features are shown in Supplementary Figure 7A and are largely similar B. Same as A but for the ridge regression algorithm. Again, models using residualized features are similar and shown in Supplementary Figure 7B. rsfMRI=resting state functional MRI, DTI=Diffusion Tensor Imaging, PPV=Positive Predictive Value, NPV=Negative Predictive Value, AUC=Area Under the Curve.
Ridge Regression.
Classification performance was similar to, or numerically better than that observed for SVM (Figure 3B and Supplementary Table 4b) with AUC values improving most for models using both feature modalities (compare right most panels in Figures 3B and 3A). While variability across datasets within a test set and variability within a test set across training sets was reduced, performance was still suboptimal with most AUC values below 0.70. Results were unaffected by adjusting for covariates (Supplementary Figure 7B Supplementary Table 4b).
Across-sample Classification
SVM.
Pooling data across samples resulted in ~500 psychosis cases and ~300 healthy participants (rsfMRI: HC=362, psychosis = 525; DTI: HCs = 353, Psychosis=506; rsfMRI+DTI: HC=287, Psychosis=413; see also Supplemental Table 5). Despite increases in sample size, there was no numerical advantage over models built within a dataset (Figure 4,Supplementary Figure 8, and Supplementary Table 6). Using features residualized for covariates reduced rsfMRI model performance (AUC=0.73 to AUC=0.53) and improved combined modality model performance (AUC=0.29 to AUC=0.50). DTI model performance was not dependent on adjusting for covariates.
Figure 4. Across Sample Classification.
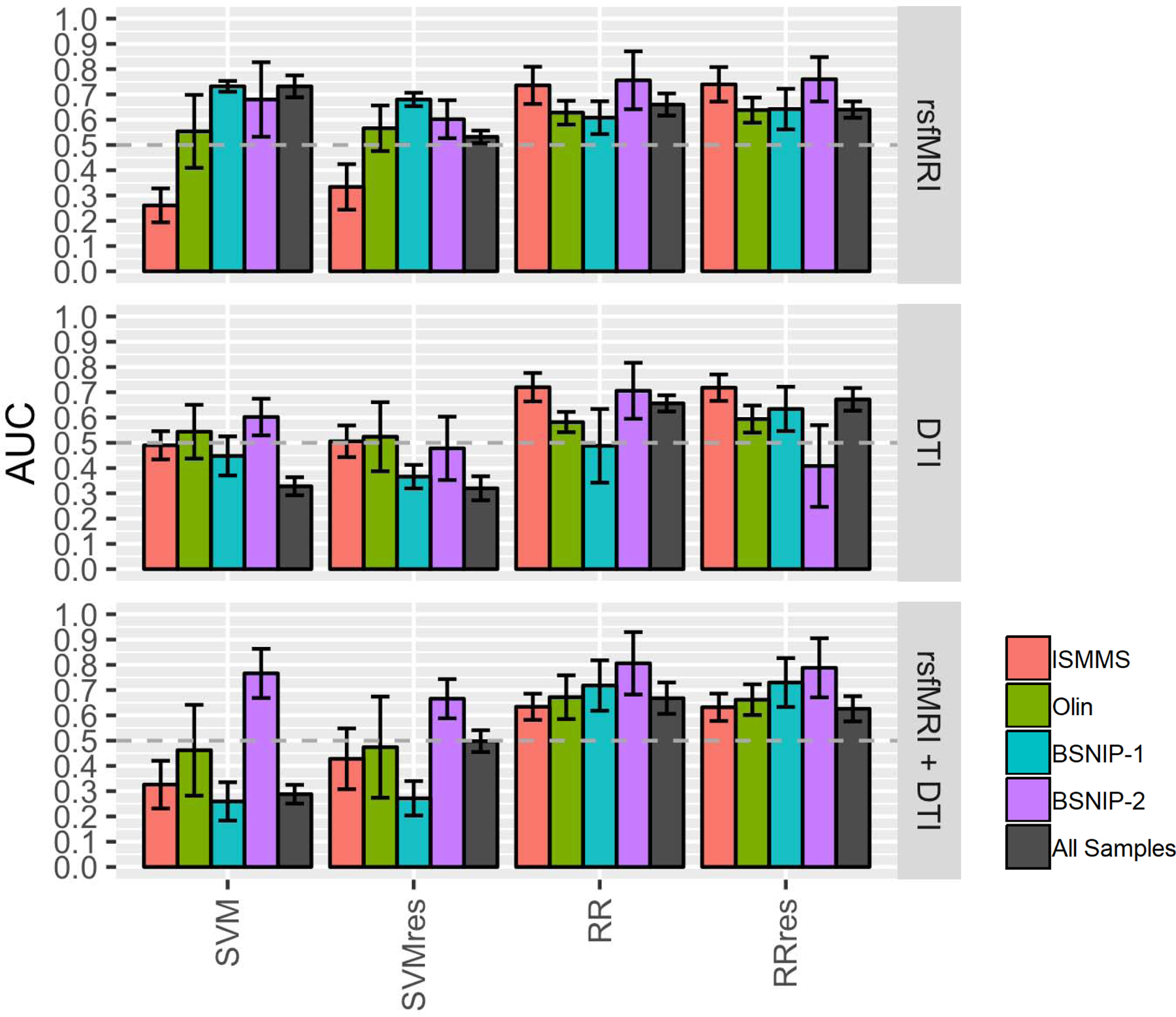
Bars are average AUC values over five folds with error bars representing the standard deviation over folds. Gray bars show across sample classification for each machine learning algorithm using residualized and non-residualized features compared to within sample classification for each dataset and each modality (colored bars). rsfMRI=resting state functional MRI, DTI=Diffusion Tensor Imaging, AUC=Area Under the Curve.
Ridge Regression.
Models using ridge regression performed equally across modalities with AUC values below 0.70 and were unaffected by covariate condition (gray bars in Figure 4, Supplementary Figure 8, and Supplementary Table 6).
Discussion
Successful biomarkers are reproducible, reliably identify case-control status at the individual level across samples(37), and generalize to independent samples(38). The MRI-based functional and structural connectivity measures examined here did not meet these standards, indicating that these measures are unsuitable for individual subject-level classification using SVM and ridge regression. As we describe below, it is possible that advances in acquisition or different classification algorithms (e.g. nonlinear) could improve the utility of connectivity measures as biomarkers for psychosis. Furthermore, given the myriad of machine learning approaches currently available and the rapid pace of algorithm development, we provide the feature sets utilized in these analyses to the research community (https://doi.org/10.5281/zenodo.4374644) in order to facilitate the direct comparison of classification methodologies. Our hope is that these data can facilitate new analytic approaches which may lead to more optimistic outcomes for connectivity-based psychosis biomarkers.
Despite confirming that functional and structural dysconnectivity are features of psychosis via group-level univariate analyses, we found substantial heterogeneity across samples (Figure 1; Supplementary Figure 2 and 3). Across-sample heterogeneity has major implications for biological models of psychosis based on neuroimaging data. For example, interpretation of group differences in functional connectivity would likely result in different conclusions about the nature of psychosis, from a disorder involving primarily sensory networks to one involving various higher order association networks. Yet, each of our findings is not unique and has been reported in other independent studies(6, 54–57) . Group differences in structural connectivity were more stable, although effect sizes were sample-dependent (Figure 1; Supplementary Figure 2 and 3). Effect size variability, however, was consistent with individual studies included in the recent ENIGMA meta-analysis(22). For both functional and structural connectivity, within-sample classification provided further evidence of sample heterogeneity as there was little overlap in the features utilized when building models in one sample vs. another. Variability across studies in either context could arise from multiple sources including methodological and/or biological variation. We attempted to minimize both by implementing nearly identical analytic pipelines and accounting for confounding factors such as age and sex. Additional factors, such as scanning protocols or other subject-specific factors (e.g. type of individuals included in the patient or control group) may have also contributed.
Results of within sample classification provided valuable information regarding the consistency of predicting an individual’s case-control status, even when capitalizing on sample-specific features. While we replicated within sample classification performance in other studies with AUC values as high as 0.80(58), this performance did not generalize across datasets with AUC values in some samples at or below 0.50. Classification performance was likewise inconsistent when building models within a dataset using 5-fold cross validation. For each dataset, AUC values over the five folds varied as little as 0.06 to as much as 0.38 (Supplementary Figure 9 and Supplementary Table 3), suggesting model performance was highly dependent on the exact participants included in the training and testing datasets.
Between sample classification performance is arguably the most important estimator of model validation and generalizability and is critical for future clinical applications(38). Our results did not wholly support the generalizability of the examined features. Some between sample models had modest performance (AUC approaching 0.80), but this was not observed across testing datasets for a given training dataset. Models using ridge regression and rsfMRI features were more consistent both across testing sets for a given training set and for a given testing set across training sets, although most AUC values were below .7. While these AUC values were above chance and their consistency is promising, this performance unfortunately falls short of requirements for practically useful and clinically relevant biomarkers. Furthermore, our between -sample classification results do not appear to be unique as we replicated two prior studies which reported similarly suboptimal classification performance when attempting to validate rsfMRI-derived models in independent datasets(59, 60).
Several methodological issues could have influenced the present findings, many of which we addressed directly. One was the effect of sample size. Both real and simulated data show that smaller sample sizes result in highly variable accuracy estimates with substantial errors (~10%)(61), which some suggest can be remedied by increasing the number of observations despite likely increases in heterogeneity(62). In our study, models utilizing all available participants (approximately 800 individuals) performed similar to, or slightly worse than, models built within datasets (Figure 4), although fold-wise stability was numerically better than that for within sample classification. Given the high level of inter-individual variation in our samples, it may be necessary to include vary large samples akin to those used in genetic studies for individual-level classification to be successful. This is particularly true if psychotic disorders are so heterogeneous that only small sub-samples share the same features.
Including a singular diagnosis like schizophrenia, as is common in most studies(63), may qualify as such a sub-sample. The present study was designed under the premise that a cross-diagnostic sample would improve the likelihood of discovering a marker for psychosis itself, given robust evidence that 1) traditional diagnoses do not reflect biologically distinct categories(64, 65) and 2) core psychotic features (cognitive-behavioral disorganization, hallucinations, delusions, etc.) may reflect a more homogeneous biological substrate than clinical diagnosis per se(65). This premise appears to be supported by a post-hoc analysis where we performed within-sample classification with our most promising metrics (ridge-regression and rsfMRI features) in our largest cohort (BSNIP-1) with only schizophrenia participants in the patient group. While performance was slightly better, it was still below .7 and fold-wise variability was increased (see supplemental Figure 10). We do not, however, negate the possibility that connectivity measures may be more suitable for predicting different criteria, be it continuous metrics (e.g. symptom severity, functional outcome, etc.) or other types of classification structures like the biotypes developed by the BSNIP consortium(65, 66). Changes in predicted targets may also ease projected sample size requirements mentioned previously. While we were unable to test these kinds of hypotheses given the variability and/or lack of data available for each sample, this should be a focus of future work.
An additional consideration was the inclusion of covariates. Adjusting for covariates did not largely alter results. The exception was for models built across samples using rsfMRI or combined features (a decrease and increase in AUC values respectively), although the difference was minimal and only apparent when using the SVM algorithm (Figure 4; Supplementary Figure 8). Algorithm choice appeared to influence performance in other classification contexts in that ridge regression performance metrics were often more stable across datasets and numerically higher than those achieved using SVM. However, using ridge regression did not resolve issues of variability or suboptimal performance to a sufficient degree to warrant that connectivity metrics are valid biomarkers for psychosis. We chose the current algorithms because they have shown promise in previous studies using machine learning and MRI features in schizophrenia and psychosis(67–69). We do not negate that more complex algorithms (e.g. non-linear models) or machine learning methods, like deep learning, may better classify individuals with psychosis. However, Schultz and colleagues(70) showed that simple linear models perform on par with complex techniques when using neuroimaging data for individual-level classification. Furthermore, increasingly complex models are more likely to overfit a singular dataset and subsequently less likely to translate to independent data.
Lastly, combining modalities did not consistently improve classification performance, regardless of the training/testing strategy, and despite evidence of different imaging data capturing unique and additive aspects of psychopathology(71, 72). This result was similar to those of Guo et al. (2018) which showed similar classification performance between rsfMRI features alone and a fusion of features from multiple modalities (73). Alternatively, it could be that different features altogether would perform better. We chose the features here because they were the most commonly used measures in the literature and there was consistent evidence for case-control differentiation at the group level with adequate sample sizes. For example, tractography measures may be better suited for classification using DTI data, however, we though it important to vet one of the most robustly demonstrated potential biomarkers provided via the ENIGMA meta-analysis(22). Given that those potential biomarkers were largely unsuccessful, it begs the question of what large differences at the group level tells us about individual-level prediction. Additionally for resting state, we used the Brainnetome atlas given reports that it may be superior to measures constructed using anatomical parcellations(74), whole-brain, or graph-based methods(58). As there were already a large number of model comparisons, we did not address the effect of feature selection. We do, however, recognize its importance and suggest that it and alternative feature types be the focus of future work.
In summary, leveraging data from over 800 individuals allowed us to confirm that measures of aberrant functional and structural connectivity are indeed present in psychosis at the group level(9, 15), but were not suitable for individual-level classification, at least with the algorithms and measures used here. Variability observed at both levels of analysis suggest that the field must focus on identifying measures that are more reproducible across sites and datasets. While it is possible that successful connectivity biomarkers may result from future innovation in technology, analytic pipelines, or perhaps other techniques that were not utilized in our analysis, neuroimaging researchers and clinicians awaiting useful biomarkers of the type they could employ to make consequential, person-level diagnoses should remain cautious.
Supplementary Material
Acknowledgements
This work was supported by funding from the National Instutiture of Mental Health. BSNIP-1 and BSNIP-2 Harford site: MH077945, BSNIP-1 Baltimore site: MH077852, Olin sample: MH106324, ISMMS sample: MH113619. Portions of this manuscript were presented at the 2018 American College of Neuropsychopharmacology and 2019 Society of Biological Psychiatry annual meetings.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
All authors report no biomedical financial interests or potential conflicts of interest.
References
- 1.Perez VB, Swerdlow NR, Braff DL, Näätänen R, Light GA (2014): Using biomarkers to inform diagnosis, guide treatments and track response to interventions in psychotic illnesses. Biomarkers in medicine. 8:9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins FS, Varmus H (2015): A new initiative on precision medicine. New England journal of medicine. 372:793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hyman SE (2013): Psychiatric drug development: diagnosing a crisis. Cerebrum: the Dana forum on brain science: Dana Foundation. [PMC free article] [PubMed]
- 4.Abi-Dargham A, Horga G (2016): The search for imaging biomarkers in psychiatric disorders. Nature medicine. 22:1248. [DOI] [PubMed] [Google Scholar]
- 5.Insel TR (2009): Translating scientific opportunity into public health impact: a strategic plan for research on mental illness. Archives of general psychiatry. 66:128–133. [DOI] [PubMed] [Google Scholar]
- 6.Baker JT, Dillon DG, Patrick LM, Roffman JL, Brady RO, Pizzagalli DA, et al. (2019): Functional connectomics of affective and psychotic pathology. Proceedings of the National Academy of Sciences. 116:9050–9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canu E, Agosta F, Filippi M (2015): A selective review of structural connectivity abnormalities of schizophrenic patients at different stages of the disease. Schizophr Res. 161:19–28. [DOI] [PubMed] [Google Scholar]
- 8.Satterthwaite TD, Baker JT (2015): How can studies of resting-state functional connectivity help us understand psychosis as a disorder of brain development? Curr Opin Neurobiol. 30:85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khadka S, Meda SA, Stevens MC, Glahn DC, Calhoun VD, Sweeney JA, et al. (2013): Is aberrant functional connectivity a psychosis endophenotype? A resting state functional magnetic resonance imaging study. Biological psychiatry. 74:458–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meda SA, Gill A, Stevens MC, Lorenzoni RP, Glahn DC, Calhoun VD, et al. (2012): Differences in resting-state functional magnetic resonance imaging functional network connectivity between schizophrenia and psychotic bipolar probands and their unaffected first-degree relatives. Biological psychiatry. 71:881–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du Y, Pearlson GD, Lin D, Sui J, Chen J, Salman M, et al. (2017): Identifying dynamic functional connectivity biomarkers using GIG-ICA: Application to schizophrenia, schizoaffective disorder, and psychotic bipolar disorder. Human brain mapping. 38:2683–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madre M, Canales-Rodríguez E, Ortiz-Gil J, Murru A, Torrent C, Bramon E, et al. (2016): Neuropsychological and neuroimaging underpinnings of schizoaffective disorder: a systematic review. Acta Psychiatrica Scandinavica. 134:16–30. [DOI] [PubMed] [Google Scholar]
- 13.Lu LH, Zhou XJ, Keedy SK, Reilly JL, Sweeney JA (2011): White matter microstructure in untreated first episode bipolar disorder with psychosis: comparison with schizophrenia. Bipolar disorders. 13:604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skudlarski P, Schretlen DJ, Thaker GK, Stevens MC, Keshavan MS, Sweeney JA, et al. (2013): Diffusion tensor imaging white matter endophenotypes in patients with schizophrenia or psychotic bipolar disorder and their relatives. American Journal of Psychiatry. 170:886–898. [DOI] [PubMed] [Google Scholar]
- 15.Wheeler AL, Voineskos AN (2014): A review of structural neuroimaging in schizophrenia: from connectivity to connectomics. Frontiers in human neuroscience. 8:653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woodward ND, Waldie B, Rogers B, Tibbo P, Seres P, Purdon SE (2009): Abnormal prefrontal cortical activity and connectivity during response selection in first episode psychosis, chronic schizophrenia, and unaffected siblings of individuals with schizophrenia. Schizophrenia Research. 109:182–190. [DOI] [PubMed] [Google Scholar]
- 17.Shim G, Oh JS, Jung WH, Jang JH, Choi C-H, Kim E, et al. (2010): Altered resting-state connectivity in subjects at ultra-high risk for psychosis: an fMRI study. Behavioral and Brain Functions. 6:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alonso-Solís A, Corripio I, de Castro-Manglano P, Duran-Sindreu S, Garcia-Garcia M, Proal E, et al. (2012): Altered default network resting state functional connectivity in patients with a first episode of psychosis. Schizophrenia research. 139:13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pérez-Iglesias R, Tordesillas-Gutiérrez D, Barker GJ, McGuire PK, Roiz-Santiañez R, Mata I, et al. (2010): White matter defects in first episode psychosis patients: A voxelwise analysis of diffusion tensor imaging. NeuroImage. 49:199–204. [DOI] [PubMed] [Google Scholar]
- 20.Biswal BB, Van Kylen J, Hyde JS (1997): Simultaneous assessment of flow and BOLD signals in resting-state functional connectivity maps. NMR Biomed. 10:165–170. [DOI] [PubMed] [Google Scholar]
- 21.Alexander AL, Lee JE, Lazar M, Field AS (2007): Diffusion Tensor Imaging of the Brain. Neurotherapeutics : the journal of the American Society for Experimental NeuroTherapeutics. 4:316–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly S, Jahanshad N, Zalesky A, Kochunov P, Agartz I, Alloza C, et al. (2017): Widespread white matter microstructural differences in schizophrenia across 4322 individuals: results from the ENIGMA Schizophrenia DTI Working Group. Molecular Psychiatry. 23:1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skudlarski P, Jagannathan K, Anderson K, Stevens MC, Calhoun VD, Skudlarska BA, et al. (2010): Brain connectivity is not only lower but different in schizophrenia: a combined anatomical and functional approach. Biological psychiatry. 68:61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skudlarski P, Jagannathan K, Calhoun VD, Hampson M, Skudlarska BA, Pearlson G (2008): Measuring brain connectivity: diffusion tensor imaging validates resting state temporal correlations. Neuroimage. 43:554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vignapiano A, DeLisi LE, Galderisi S (2019): Toward Clinical Translation of Neuroimaging Research in Schizophrenia and Other Primary Psychotic Disorders. Neuroimaging of Schizophrenia and Other Primary Psychotic Disorders: Springer, pp 327–345. [Google Scholar]
- 26.Orru G, Pettersson-Yeo W, Marquand AF, Sartori G, Mechelli A (2012): Using support vector machine to identify imaging biomarkers of neurological and psychiatric disease: a critical review. Neuroscience & Biobehavioral Reviews. 36:1140–1152. [DOI] [PubMed] [Google Scholar]
- 27.Singh I, Rose N (2009): Biomarkers in psychiatry. Nature. 460:202–207. [DOI] [PubMed] [Google Scholar]
- 28.Wolfers T, Buitelaar JK, Beckmann CF, Franke B, Marquand AF (2015): From estimating activation locality to predicting disorder: A review of pattern recognition for neuroimaging-based psychiatric diagnostics. Neuroscience & Biobehavioral Reviews. 57:328–349. [DOI] [PubMed] [Google Scholar]
- 29.Woo C-W, Chang LJ, Lindquist MA, Wager TD (2017): Building better biomarkers: brain models in translational neuroimaging. Nature neuroscience. 20:365–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheinost D, Noble S, Horien C, Greene AS, Lake EM, Salehi M, et al. (2019): Ten simple rules for predictive modeling of individual differences in neuroimaging. NeuroImage. [DOI] [PMC free article] [PubMed]
- 31.Schnack HG, Kahn RS (2016): Detecting Neuroimaging Biomarkers for Psychiatric Disorders: Sample Size Matters. Frontiers in Psychiatry. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du Y, Fu Z, Calhoun VD (2018): Classification and prediction of brain disorders using functional connectivity: Promising but challenging. Frontiers in neuroscience. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ardekani BA, Tabesh A, Sevy S, Robinson DG, Bilder RM, Szeszko PR (2011): Diffusion tensor imaging reliably differentiates patients with schizophrenia from healthy volunteers. Human brain mapping. 32:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janssen RJ, Mourão-Miranda J, Schnack HG (2018): Making individual prognoses in psychiatry using neuroimaging and machine learning. Biological Psychiatry: Cognitive Neuroscience and Neuroimaging. [DOI] [PubMed]
- 35.Ingalhalikar M, Kanterakis S, Gur R, Roberts TP, Verma R (2010): DTI based diagnostic prediction of a disease via pattern classification. Medical image computing and computer-assisted intervention : MICCAI International Conference on Medical Image Computing and Computer-Assisted Intervention. 13:558–565. [DOI] [PubMed] [Google Scholar]
- 36.Lee J, Chon M-W, Kim H, Rathi Y, Bouix S, Shenton ME, et al. (2018): Diagnostic value of structural and diffusion imaging measures in schizophrenia. NeuroImage: Clinical. 18:467–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woo C-W, Wager TD (2015): Neuroimaging-based biomarker discovery and validation. Pain. 156:1379–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Botteron K, Carter C, Castellanos FX, Dickstein DP, Drevets W, Kim KL, et al. (2012): Consensus report of the APA work group on neuroimaging markers of psychiatric disorders. Am Psychiatr Assoc.
- 39.Esteban O, Birman D, Schaer M, Koyejo OO, Poldrack RA, Gorgolewski KJ (2017): MRIQC: Advancing the automatic prediction of image quality in MRI from unseen sites. PloS one. 12:e0184661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esteban O, Markiewicz CJ, Blair RW, Moodie CA, Isik AI, Erramuzpe A, et al. (2019): fMRIPrep: a robust preprocessing pipeline for functional MRI. Nature methods. 16:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Behzadi Y, Restom K, Liau J, Liu TT (2007): A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. Neuroimage. 37:90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fan L, Li H, Zhuo J, Zhang Y, Wang J, Chen L, et al. (2016): The human brainnetome atlas: a new brain atlas based on connectional architecture. Cerebral cortex. 26:3508–3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith S, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-Berg H, et al. (2004): Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 23:S208–S219. [DOI] [PubMed] [Google Scholar]
- 44.Smith S (2002): Fast robust automated brain extraction. Human brain mapping. 17:143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andersson JLR, Sotiropoulos SN (2016): An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. NeuroImage. 125:1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith S, Jenkinson M, Johansen-Berg H, Rueckert D, Nichols T, Mackay CE, et al. (2006): Tract-based spatial statistics: Voxelwise analysis of multi-subject diffusion data. NeuroImage. 31:1487–1505. [DOI] [PubMed] [Google Scholar]
- 47.Hua K, Zhang J, Wakana S, Jiang H, Li X, Reich DS, et al. (2008): Tract Probability Maps in Stereotaxic Spaces: Analyses of White Matter Anatomy and Tract-Specific Quantification. NeuroImage. 39:336–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winkler AM, Ridgway GR, Webster MA, Smith SM, Nichols TE (2014): Permutation inference for the general linear model. Neuroimage. 92:381–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benjamini Y, Hochberg Y (1995): Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society Series B (Methodological). 289–300.
- 50.Cortes C, Vapnik V (1995): Support-vector networks. Machine Learning. 20:273–297. [Google Scholar]
- 51.Hoerl AE, Kennard RW (1970): Ridge regression: Biased estimation for nonorthogonal problems. Technometrics. 12:55–67. [Google Scholar]
- 52.Kuhn M (2020): caret: Classification and Regression Training. 6.0–86 ed: R package.
- 53.Cole SR, Platt RW, Schisterman EF, Chu H, Westreich D, Richardson D, et al. (2010): Illustrating bias due to conditioning on a collider. International journal of epidemiology. 39:417–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lang X, Wang L, Zhuo C-J, Jia F, Wang L-N, Wang C-L (2016): Reduction of Interhemispheric Functional Connectivity in Sensorimotor and Visual Information Processing Pathways in Schizophrenia. Chin Med J (Engl). 129:2422–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skåtun KC, Kaufmann T, Doan NT, Alnæs D, Córdova-Palomera A, Jönsson EG, et al. (2016): Consistent Functional Connectivity Alterations in Schizophrenia Spectrum Disorder: A Multisite Study. Schizophrenia Bulletin. 43:914–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaufmann T, Skatun KC, Alnaes D, Doan NT, Duff EP, Tonnesen S, et al. (2015): Disintegration of Sensorimotor Brain Networks in Schizophrenia. Schizophr Bull. 41:1326–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pettersson-Yeo W, Allen P, Benetti S, McGuire P, Mechelli A (2011): Dysconnectivity in schizophrenia: where are we now? Neurosci Biobehav Rev. 35:1110–1124. [DOI] [PubMed] [Google Scholar]
- 58.Lei D, Pinaya WH, van Amelsvoort T, Marcelis M, Donohoe G, Mothersill DO, et al. (2019): Detecting schizophrenia at the level of the individual: relative diagnostic value of whole-brain images, connectome-wide functional connectivity and graph-based metrics. Psychological Medicine. 1–10. [DOI] [PMC free article] [PubMed]
- 59.Cai XL, Xie DJ, Madsen KH, Wang YM, Bögemann SA, Cheung EF, et al. (2019): Generalizability of machine learning for classification of schizophrenia based on resting-state functional MRI data. Human brain mapping. [DOI] [PMC free article] [PubMed]
- 60.Ji GJ, Chen X, Bai T, Wang L, Wei Q, Gao Y, et al. (2019): Classification of schizophrenia by intersubject correlation in functional connectome. Human brain mapping. 40:2347–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Varoquaux G, Raamana PR, Engemann DA, Hoyos-Idrobo A, Schwartz Y, Thirion B (2017): Assessing and tuning brain decoders: cross-validation, caveats, and guidelines. NeuroImage. 145:166–179. [DOI] [PubMed] [Google Scholar]
- 62.Abraham A, Milham MP, Di Martino A, Craddock RC, Samaras D, Thirion B, et al. (2017): Deriving reproducible biomarkers from multi-site resting-state data: an autism-based example. NeuroImage. 147:736–745. [DOI] [PubMed] [Google Scholar]
- 63.Birur B, Kraguljac NV, Shelton RC, Lahti AC (2017): Brain structure, function, and neurochemistry in schizophrenia and bipolar disorder—a systematic review of the magnetic resonance neuroimaging literature. npj Schizophrenia. 3:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tamminga CA, Pearlson G, Keshavan M, Sweeney J, Clementz B, Thaker G (2014): Bipolar and Schizophrenia Network for Intermediate Phenotypes: Outcomes Across the Psychosis Continuum. Schizophrenia Bulletin. 40:S131–S137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clementz BA, Sweeney JA, Hamm JP, Ivleva EI, Ethridge LE, Pearlson GD, et al. (2015): Identification of distinct psychosis biotypes using brain-based biomarkers. American Journal of Psychiatry. 173:373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ji L, Meda SA, Tamminga CA, Clementz BA, Keshavan MS, Sweeney JA, et al. (2020): Characterizing functional regional homogeneity (ReHo) as a B-SNIP psychosis biomarker using traditional and machine learning approaches. Schizophrenia Research. 215:430–438. [DOI] [PubMed] [Google Scholar]
- 67.Salvador R, Canales-Rodríguez E, Guerrero-Pedraza A, Sarró S, Tordesillas-Gutiérrez D, Maristany T, et al. (2019): Multimodal Integration of Brain Images for MRI-Based Diagnosis in Schizophrenia. Frontiers in Neuroscience. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salvador R, Radua J, Canales-Rodríguez EJ, Solanes A, Sarró S, Goikolea JM, et al. (2017): Evaluation of machine learning algorithms and structural features for optimal MRI-based diagnostic prediction in psychosis. PloS one. 12:e0175683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Winterburn JL (2015): Comparing Techniques for Classifying Patients with Schizophrenia and Healthy Controls using Machine Learning and Magnetic Resonance Imaging.
- 70.Schulz M-A, Yeo T, Vogelstein J, Mourao-Miranada J, Kather J, Kording K, et al. (2019): Deep learning for brains?: Different linear and nonlinear scaling in UK Biobank brain images vs. machine-learning datasets. bioRxiv.757054.
- 71.Bora E, Fornito A, Radua J, Walterfang M, Seal M, Wood SJ, et al. (2011): Neuroanatomical abnormalities in schizophrenia: a multimodal voxelwise meta-analysis and meta-regression analysis. Schizophr Res. 127:46–57. [DOI] [PubMed] [Google Scholar]
- 72.Calhoun VD, Sui J (2016): Multimodal fusion of brain imaging data: A key to finding the missing link(s) in complex mental illness. Biological psychiatry Cognitive neuroscience and neuroimaging. 1:230–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo S, Huang C-C, Zhao W, Yang AC, Lin C-P, Nichols T, et al. (2018): Combining multi-modality data for searching biomarkers in schizophrenia. PloS one. 13:e0191202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dadi K, Rahim M, Abraham A, Chyzhyk D, Milham M, Thirion B, et al. (2019): Benchmarking functional connectome-based predictive models for resting-state fMRI. NeuroImage. 192:115–134. [DOI] [PubMed] [Google Scholar]
- 75.Yeo BTT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, et al. (2011): The organization of the human cerebral cortex estimated by intrinsic functional connectivity. Journal of neurophysiology. 106:1125–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


