Abstract
By feeding tryptophan to the marine-derived fungus Aspergillus sp. HNMF114 from the bivalve mollusk Sanguinolaria chinensis, 3 new quinazoline-containing indole alkaloids, named aspertoryadins H–J (1–3), along with 16 known ones (4–19), were obtained. The structures of the new compounds were elucidated by the analysis of spectroscopic data combined with quantum chemical calculations of nuclear magnetic resonance (NMR) chemical shifts and electron capture detector (ECD) spectra. Structurally, compound 3 represents the first example of this type of compound, bearing an amide group at C-3. Compounds 10 and 16 showed potent α-glucosidase inhibitory activity with IC50 values of 7.18 and 5.29 μM, and compounds 13 and 14 showed a clear activation effect on the ryanodine receptor from Spodoptera frugiperda (sfRyR), which reduced the [Ca2+]ER by 37.1 and 36.2%, respectively.
Keywords: marine-derived fungus, Aspergillus sp., indole alkaloids, α-glucosidase inhibitory activity, ryanodine receptor
Introduction
While plants and terrestrial microorganisms have been in the limelight of natural product research for several decades, the marine environment is one of the current hotspots for the bio-prospection of new bioactive molecules, and it is considered to be a new reservoir for drug discovery (Montaser and Luesch, 2011). Marine microorganisms are some of the most prolific sources of structurally novel and biologically active compounds (Bugni and Ireland, 2004; Shen et al., 2009; Arasu et al., 2013; Manimegalai et al., 2013; Xiong et al., 2013).
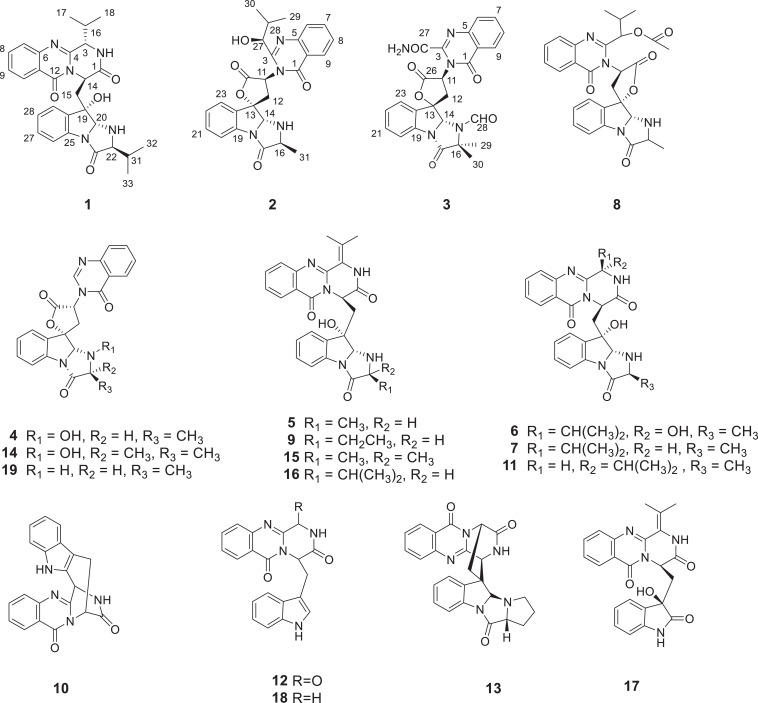
Among the marine-derived fungi, the genus Aspergillus is a prolific source of bioactive secondary metabolites (Blunt et al., 2017). In our search for marine active natural products, the fungus Aspergillus sp. HNMF114 was isolated and identified from a bivalve mollusk, Sanguinolaria chinensis, collected in Haikou Bay, China. Our previous research (Kong et al., 2019) on the secondary metabolites of this fungus reported a series of quinazoline-containing indole alkaloids. Biogenetically, these compounds are all derived from tryptophan and anthranilic acid, along with other amino acids. It has been reported that feeding tryptophan is beneficial to the production of quinazoline-containing indole alkaloids (Guo et al., 2020). Thus, in order to tap the metabolic potential of this fungus, the continuous chemical investigation on the fermentation broth of the fungus Aspergillus sp. HNMF114 supplemented with L-tryptophan was carried out, which led to the isolation of 3 new quinazoline-containing indole alkaloids, aspertoryadins H–J (1–3), along with 16 known ones, tryptoquivaline H (4) (Buttachon et al., 2012), norquinadoline A (5) (Peng et al., 2013), epi-fiscalin D (6) (Qian et al., 2018), epi-fiscalin A (7) (Buttachon et al., 2012), deoxynortryptoquivaline (8) (Lin et al., 2013), scequinadolines D (9) (Huang et al., 2017), lapatin A (10) (Wu and Ma, 2013), fiscalin A (11) (Buttachon et al., 2012), oxoglyantrypine (12) (Peng et al., 2013), quinadoline B (13) (Koyama et al., 2008), tryptoquivalines L (14) (Buttachon et al., 2012), quinadoline A (15) (Koyama et al., 2008), scequinadolines E (16) (Huang et al., 2017), scequinadoline G (17) (Huang et al., 2017), glyantrypine (18) (Penn et al., 1992), and tryptoquivalines F (19) (Buttachon et al., 2012) (Figure 1). Interestingly, these compounds were all different from those of our previous report (Kong et al., 2019). Herein, the isolation, structure elucidation, and bioactivities of these compounds are reported.
FIGURE 1.
The structures of compounds 1–19.
Materials and Methods
General Experimental Procedure
Nuclear magnetic resonance (NMR) spectra were obtained on a Bruker AV-600 spectrometer (Bruker, Bremen, Germany) with tetramethylsilane (TMS) as an internal standard. HRESIMS data were determined on a mass spectrometer API QSTAR Pulsar (Bruker, Bremen, Germany). Optical rotations were achieved on a JASCO P-1020 digital polarimeter and IR spectra were recorded on a Shimadzu UV2550 spectrophotometer. UV spectra were measured on a Beckman DU 640 spectrophotometer. Electron capture detector (ECD) data were collected using a JASCO J-715 spectropolarimeter. Silica gel (200–300 mesh, Qingdao Marine Chemical Inc., Qingdao, China) and Sephadex LH-20 (40–70 μm, Merck, Darmstadt, Germany) were used for column chromatography. Semipreparative high-performance liquid chromatography (HPLC) equipped with octadecyl silane column (YMC-pack ODS-A, 10 × 250 mm, 5 μm, 4 mL/min). Spots were visualized by heating silica gel plates sprayed with 10% H2SO4 in EtOH.
Fungal Material
The fungal information of Aspergillus sp. HNMF114 has been described in our previous report (Kong et al., 2019).
Fermentation, Extraction, and Isolation
The fungus was cultured was cultured for 30 days in 100 × 1,000 mL erlenmeyer flasks each containing 100 g of rice, 100 mL H2O (33 g sea salt, and 2 g tryptophan per liter of tap water) were autoclaved at 121°C for 25 min. The fermented material was extracted three times with EtOAc (20 L for each time) to give 230 g of crude extract. The extract was extracted between petroleum ether and 90% methanol (1:1) to remove the oil. The methanolic extract (119 g) was subjected to a silica gel vacuum-liquid chromatographed (VLC) column, eluting with a stepwise gradient of petroleum ether/EtOAc (10:1, 8:1, 6:1, 4:1, 2:1, 1:1, 1:2, v/v) to afford 10 fractions (Fr.1–Fr.10). Fr.5 (3.0 g) was applied to octadecylsilane (ODS) silica gel with a gradient elution of MeOH-H2O (1:4, 1:3, 2:3, 1:1, 3:2, 4:1, 9:1, and 1:0) to get eight subfractions (Fr.5-1-Fr.5-8). Fr. 5-3 (368 mg) was separated by ODS silica gel MeCN-H2O (1:3, 2:3, 1:1, 3:2, 4:1, 9:1, and 1:0) to afford two subfractions. Subfraction Fr.5-3-1 was applied to semipreparative HPLC (60% MeOH-H2O; 4 mL/min) to give compounds 3 (tR = 5.8 min; 2.3 mg), 4 (tR = 7.3 min; 3.3 mg), and 6 (tR = 8.3 min; 6.1 mg). Fr.5-3-2 was applied to ODS silica gel with gradient elution of MeCN-H2O (1:3 to 4:1) semipreparative HPLC (C 60% MeOH-H2O; 4 mL/min) to obtain compounds 5 (tR = 8.2 min; 5.0 mg), 9 (tR = 10.4 min; 5.3 mg), and 10 (tR = 10.7 min; 3.4 mg). Fr. 5-5 (56.0 mg) was further chromatographed by silica gel CC eluted with gradient Petroleum ether-EtOAc (10:1, 8:1, 6:1, 4:1, 2:1, 1:1, 1:2, v/v) to afford seven subfractions (Fr.5-5-1–Fr.5-5-7). Fr.5-5-3 were purified by semipreparative HPLC (55% MeOH-H2O; 4 mL/min) to yield compounds 11 (tR = 13.3 min; 1.8 mg), 12 (tR = 13.8 min; 3.4 mg), 14 (tR = 15.2 min; 4.7 mg), and 16 (tR = 17.8 min; 2.7 mg). Fr.6 (2.7 g) was applied to an open ODS column chromatography eluted with stepwise gradient of MeOH-H2O (1:5, 1:4, 1:3, 2:3, 1:1, 3:2, 4:1, 9:1, 1:0) to get nine subfractions (Fr.6-1 to Fr.6-9). Fr.6-4 (158 mg) was further separated by Sephadex LH-20 column chromatography, followed by semipreparative HPLC (50% MeCN-H2O; containing 0.1% TFA; 4 mL/min) to afford compounds 2 (tR = 10.3 min; 1.9 mg), 8 (tR = 17.8 min; 3.8 mg), and 1 (tR = 27.7 min; 3.4 mg). Fr. 6-5 was subjected to an ODS column chromatography by stepwise gradient elution of MeOH-H2O (1:5, 1:4, 1:3, 2:3, 1:1, 3:2, 4:1, 9:1, and 1:0) to get nine subfraction (Fr.6-5-1 to Fr.6-5-9). The subfraction 6-5-4 (53.2 mg) was purified by semipreparative HPLC (45% MeCN-H2O; 4 mL/min) to obtain compounds 7 (tR 8.8 min; 2.3 mg), 13 (tR 11.4 min; 37.7 mg), 15 (tR 15.2 min; 4.3 mg), 19 (tR 15.6 min; 2.8 mg). Compounds 17 (36.2 mg) and 18 (15.3 mg) were combined and crystallized from the Fr.6-5-5 and Fr.6-5-6, respectively.
Aspertoryadin H (1): yellow powder solid; [α]25 D -131 (c 0.1, MeOH); UV (MeOH) λmax (log ε): 200 (3.6), 229 (3.4), 279 (2.9, and 308 (2.6) nm; ECD (MeOH) λmax (Δε): 197 (−5.78), 213 (14.45), 232 (−21.54), 330 (−0.17) nm; IR (KBr) υmax (cm–1): 3,345 (−OH), 2,961 (−CH), 2,925 (−CH), 1,684 (−C=O), 1,602 (−NH), 1,475(−C−N), and 1,031 (−C-C−); 1H NMR and13C NMR data, see Table 1; HRESIMS m/z [M − H]– 500.2310 (calcd for C28H30N5O4, 500.2303).
TABLE 1.
1H (600 MHz) and 13C NMR (150 MHz) data of compounds 1–3.
| 1 (in DMSO-d6) |
2 (in DMSO-d6) |
3 (in DMSO-d6) |
||||
| δC | δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | |
| 1 | 160.3 | 161.2 | 160.9 | |||
| 3 | 57.7 | 4.79 (1H, d, 1.8) | 156.6 | 150.2 | ||
| 4 | 151.3 | |||||
| 5 | 146.2 | 146.4 | ||||
| 6 | 146.8 | 127.3 | 7.72 (1H, d, 7.8) | 128.0 | 7.82 (1H, d, 9.0) | |
| 7 | 127.2 | 7.66 (1H, d, 8.4) | 135.3 | 7.89 (1H, t, 7.8) | 136.1 | 7.97 (1H, t, 9.0) |
| 8 | 134.6 | 7.83 (1H, dd, 7.2, 8.4) | 127.5 | 7.60 (1H, t, 7.8) | 128.9 | 7.69 (1H, t, 9.0) |
| 9 | 126.9 | 7.53 (1H, dd, 7.2, 8.4) | 126.2 | 8.19 (1H, d, 7.8) | 126.9 | 8.27 (1H, d, 9.0) |
| 10 | 126.3 | 8.15 (1H, d, 8.4) | 120.1 | 121.4 | ||
| 11 | 120.1 | 54.8 | 6.07 (1H, t, 9.6) | 56.8 | 5.73 (1H, dd, 10.2, 12.0) | |
| 12 | 160.4 | 30.8 | 2.75 (1H, dd, 9.6, 12.6) | 27.0 | 3.31 (1H, dd, 10.2, 16.8) | |
| 3.33 (1H, overlap) | 3.40 (1H, dd, 13.2, 16.8) | |||||
| 13 | 86.1 | 84.4 | ||||
| 14 | 52.7 | 5.35 (1H, dd, 3.6, 7.8) | 85.1 | 5.44 (1H, d, 7.2) | 81.5 | 6.09 (1H, s) |
| 15 | 37.6 | 2.80 (1H, dd, 3.6, 14.4) | ||||
| 2.67 (1H, dd, 7.8, 14.4) | ||||||
| 16 | 28.3 | 2.98 (1H, m) | 59.5 | 3.61 (1H, m) | 64.6 | |
| 17 | 19.0 | 1.16 (3H, d, 7.2) | 177.2 | 172.9 | ||
| 18 | 15.4 | 0.91 (3H, d, 7.2) | ||||
| 19 | 74.9 | 140.9 | 138.8 | |||
| 20 | 83.0 | 5.01 (1H, d, 7.2) | 117.1 | 7.45 (1H, d, 7.8) | 116.8 | 7.56 (1H, d, 9.0) |
| 21 | 131.5 | 7.55 (1H, td, 1.2, 7.8) | 132.2 | 7.61 (1H, t, 9.0) | ||
| 22 | 69.9 | 3.47 (1H, dd, 4.2, 4.2) | 126.0 | 7.38 (1H, td, 1.2, 7.8) | 127.2 | 7.45 (1H, t, 9.0) |
| 23 | 173.4 | 125.5 | 7.67 (1H, d, 7.8) | 127.3 | 8.01 (1H, d, 9.0) | |
| 24 | 132.3 | 133.9 | ||||
| 25 | 137.7 | |||||
| 26 | 114.9 | 7.36 (1H, d, 7.8) | 171.1 | 171.5 | ||
| 27 | 129.6 | 7.33 (1H, t, 7.8) | 80.2 | 4.35 (1H, dd, 4.8, 9.6) | 163.5 | |
| 28 | 124.8 | 7.13 (1H, t, 7.8) | 32.6 | 2.14 (1H, m) | 162.5 | 8.64 (1H, s) |
| 29 | 124.9 | 7.43 (1H, d, 7.8) | 19.7 | 1.14 (3H, d, 6.0) | 25.9 | 1.53 (3H, s) |
| 30 | 138.3 | 19.2 | 0.83 (3H, d, 6.6) | 27.0 | 1.70 (3H, s) | |
| 31 | 31.2 | 1.81 (1H, m) | 17.6 | 1.36 (3H, d, 7.2) | ||
| 32 | 18.6 | 0.74 (3H, d, 6.6) | ||||
| 33 | 17.5 | 0.76 (3H, d, 6.6) | ||||
| 2-NH | 8.45 (1H, br s) | |||||
| 19-OH | 5.66 (1H, s) | |||||
| 15-NH | 4.01 (1H, dd, 7.2, 7.2) | |||||
| 21-NH | 3.13 (1H, overlap) | |||||
| 27-OH | 6.56 (1H, d, 4.8) | |||||
| 27-CONH2 | 8.36 (1H, br s) | |||||
| 8.66 (1H, br s) | ||||||
Aspertoryadin I (2): yellow powder solid; [α]25 D −144 (c 0.1, MeOH); UV (MeOH) λmax (log ε): 205 (3.6), 226 (3.4), 279 (2.8), and 309 (2.5) nm; ECD (MeOH) λmax (Δε): 200 (2.59), 210 (27.82), 231 (−22.02), 316 (−0.95) nm; IR (KBr) υmax (cm–1): 3,354 (−OH), 2,965 (−CH), 2,926 (−CH), 1,677(−C=O), 1,602 (−NH), 1,475(-C-N), and 1,066 (−C-C-); 1H NMR and 13C NMR data, see Table 1; HRESIMS m/z [M − H]– 473.1833 (calcd for C26H25N4O5, 473.1830).
Aspertoryadin J (3): yellow powder solid; [α]25 D +32 (c 0.1, MeOH); UV (MeOH) λmax (log ε): 206 (3.8), 228 (3.7), 276 (3.1), and 304 (3.9) nm; ECD (MeOH) λmax (Δε): 219 (−5.83), 230 (−47.81), 300 (−4.07), 320 (−2.66), 360 (−0.14); IR (KBr) υmax (cm–1): 3,412 (−OH), 2,925 (−CH), 1,678 (−C=O), 1,474 (−C−N), and 1,027(−C-C-); 1H NMR and 13C NMR data, see Table 1; HRESIMS m/z [M − H]– 486.1421 (calcd for C25H20N5O6, 486.1419).
Computational Section
The initial conformational search was carried out in Confab (O’Boyle et al., 2011) using the MMFF94 molecular mechanics force field. Density functional theory calculations were performed using the Gaussian 16 package (Frisch et al., 2019). These conformers were optimized at B3LYP/6-31G (d) in the gas phase, and the conformers with a population over 1% were kept. Then, these conformers were further subjected to geometry optimizations at B3LYP/6-311G (d) in the gas phase, and frequency analysis of all optimized conformations was also performed at the same level of theory to exclude the imaginary frequencies. NMR shielding tensors were calculated with the gauge-independent atomic orbital (GIAO) method at mPW1PW91/6-311G (d,p) level with the IEFPCM solvent model in DMSO. The shielding constants obtained were converted into chemical shifts by referencing to TMS at 0 ppm (δcal = σTMS – σcal), where the σTMS was the shielding constant of TMS calculated at the same level. For each candidate, the parameters a and b of the linear regression δcal = aδexp + b; the correlation coefficient, R2; the mean absolute error (MAE) defined as Σn |δcal – δexp| /n; the corrected mean absolute error (CMAE), defined as Σn |δcorr – δexp|/n, where δcorr = (δcal – b)/a, were calculated. DP4+ probability analysis was performed using the calculated NMR shielding tensors (Grimblat et al., 2015). The ECD spectra were calculated by the TDDFT methodology at the B3LYP/TZVP utilizing IEFPCM in methanol. ECD spectra were simulated using SpecDis 1.71 (Bruhn et al., 2013) with σ = 0.30 eV.
α-Glucosidase Inhibition Assay
Yeast α-glucosidase (0.2 U/mL) in 0.1 M phosphate buffer (pH 6.8), was used as an enzyme source. The substrate solution of p-nitrophenyl-α-D-glucopyranoside (PNPG, 2.5 mM), was prepared in 0.1 M phosphate buffer (pH 6.8) solution. The tested compounds were prepared at different concentrations (250, 500, 1,000, 2,000, and 4,000 μg/mL) in DMSO. The tested compound solution (10 μL) was pre-incubated with 100 μL α-glucosidase solution for 15 min. After pre-incubation, 45 μL of the substrate was added and further incubated for another 15 min at room temperature. The absorbance of each well was measured in Microplate Reader (Thermo Fisher Scientific, United States) reader at 405 nm. All experiments were carried out in triplicate (Dan et al., 2019).
Homology Modeling of α-Glucosidase
Since there is no structural information of α-glucosidase, we model the homology of it by modeler 9.20 (Webb and Sali, 2016). The amino acid sequence of α-glucosidase AL12 from Baker’s yeast was retrieved from UniProt protein knowledgebase (accession number P53341). The crystal structure of isomaltase (PDBID:3A4A), with 75% sequence identity with the target sequence as the template for homology modeling, was selected after a search in the Protein Data Bank (PDB) at National Center or Biotechnology and Information (NCBI) using BLAST. The constructed model was validated by Procheck, ERRATE, and verify3D programs (Laskowski et al., 1993; Eisenberg et al., 1997).
Molecular Docking
The 3D structures of the active compounds 10 and 16 were constructed and minimized by MM2 force field using Chemoffice 14.0 software. All hydrogen atoms and gasteiger charges were added to modeled receptor by AutoDock Tools. Docking was performed centered at the active pocket of the α-glucosidase with Autodock vina software (Trott and Olson, 2010). The poses were ranked by their binding affinities and the lowest one was selected as the predicted protein–ligand complexes. The results were presented by Pymol1.
Time-Lapse [Ca2+]ER Measurements
The biological activity of compounds against the insect ryanodine receptor (RyR) was tested using HEK cells stably expressing RyR from Spodoptera frugiperda (sfRyR) or RyR1 from rabbit (rRyR1), and R-CEPIA1er, an engineered endoplasmic reticulum (ER)-targeting fluorescent protein used to measure ER luminal Ca2+ concentration ([Ca2+]ER) (Suzuki et al., 2014). Cells were cultured in Dulbecco’s modified Eagle medium containing 10% fetal bovine serum, 15 mg/mL blasticidin, 100 mg/mL hygromycin, and 400 mg/mL G418 first in Petri dishes, and later seeded into 96-well plates at a density of 104 cells/well. After seeding for 24 h, 2 mg/mL doxycycline was added to induce the expression of RyR. After 48 h of induction, the medium was replaced by HEPES-buffered Krebs solution, and [Ca2+]ER was measured using FlexStation 3 fluorometer (Molecular Devices) by monitoring the fluorescence signal changes. The R-CEPIAer signal, which is excited at 560 nm and emitted at 610 nm, was captured every 10s for 300s. The compounds for screening were added 100s after the recording started. The ratio of the average fluorescence for the last 100s (F) and first 100s (F0), F/F0, was used to report the fluorescence change caused by the compounds. All experiments were carried out in triplicate and repeated twice.
Results and Discussion
Structure Elucidation
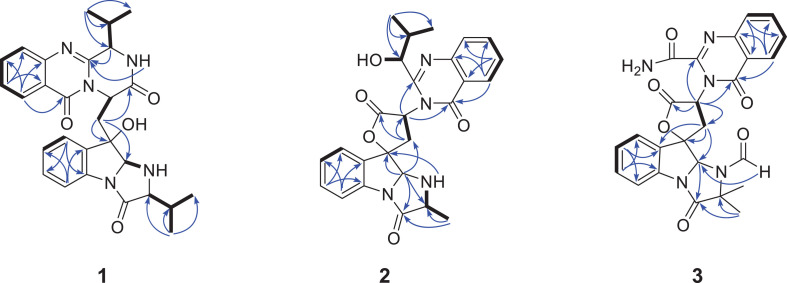
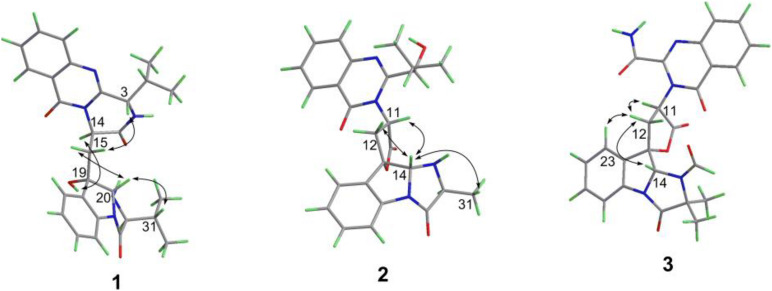
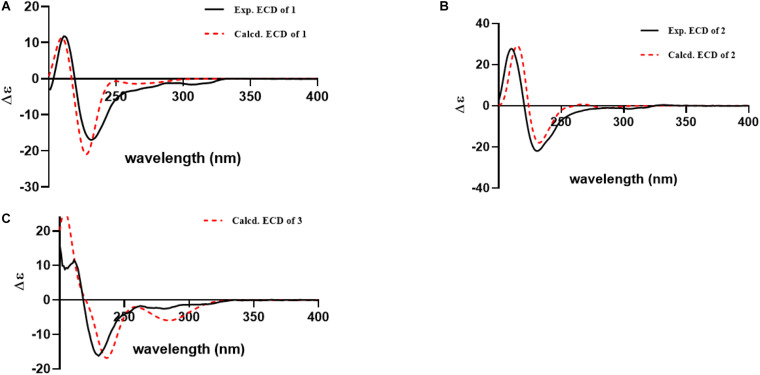
Compound 1 was obtained as yellow powder solid, and its molecular formula was determined to be C28H31N5O4 on the basis of HRESIMS, indicating 16 degrees of unsaturation. The 1H NMR spectrum (Table 1) showed the obvious resonances for eight aromatic protons attributed to two disubstituted benzene rings and four methyl protons. Its 13C, DEPT, and HSQC NMR data (Table 1) revealed a total of 28 carbons, including four methyls, one methylene, 14 methines (six sp3 and eight olefinic), and nine quaternary carbons (including three carbonyl groups and five olefinic). A detailed comparison of the NMR data of 1 with those of epi-fiscalin C (Buttachon et al., 2012) indicated that they shared the same skeleton with the only difference being the presence of an isopropyl at C-22 in 1 instead of two methyls in epi-fiscalin C. The key 1H-1H COSY cross-peaks of H-22/H-31/H-32 (H-33) and the HMBC correlations from H3-32 and H3-33 to C-31 and C-22 confirmed the assignment of the isopropyl at C-22. Thus, the planar structure of 1 was established as shown in Figure 2. The relative configuration of the quinazoline and 5/5/6 tri-cyclic rings (Figure 3) were determined by the rotating frame overhauser effect spectroscopy (ROESY) spectrum, in which the correlations of H-20 with H-15a (δH 2.67) and H-31 revealed that H-20, H-31, and CH2-15 were on the same face of the 5/5/6 tri-cyclic ring system. The ROESY correlation between H-3 and H-15b indicated that H-3 and C-15 should be placed on the same face of the quinazoline ring. Due to the flexibility of the single bonds C14-C15 and C15-C19, the relative configuration of 1 could not be clearly determined by ROESY spectrum. Therefore, a theoretical NMR calculation with DP4+ analysis was applied to clarify the relative configuration of 1. The chemical shifts of two isomers (3S,14R,19S,20R,22S)-1 and (3R,14S,19S,20R,22S)-1, were predicted using the GIAO method. DP4+ probability analysis showed that (3S,14R,19S,20R,22S)-1 was the most likely candidate structure, with a 100% DP4+ (all data). As for the absolute configuration of 1, the ECD spectrum of (3S,14R,19S,20R,22S)-1 was calculated, the result of which matched well with an experimental curve (Figure 4A), establishing the absolute configuration of 1 as presented in Figure 1.
FIGURE 2.
The key HMBC (arrows) and COSY (bold) correlations for compounds 1–3.
FIGURE 3.
Key ROESY correlations (double arrows) of compounds 1–3.
FIGURE 4.
Experimental and calculated ECD curves for compound 1 (A); compound 2 (B); compound 3 (C).
Compound 2 was isolated as yellow powder solid and had the molecular formula C26H26N4O5 as established by HRESIMS data with 16 degrees of unsaturation. Its 13C (Table 1), DEPT, and HSQC NMR spectra revealed a total of 26 carbons including 13 aromatic carbons (eight protonated), one sp3 methylene, three carbonyl carbons, five sp3 methine, three methyls, and one oxygenated sp3 quaternary carbon. The above data were similar to those of aspertoryadin A (Kong et al., 2019), indicating that they had a similar skeleton. The obvious structural difference between them is that a hydrogen on N-15 in 2 replaced the methyl sulfonyl group in aspertoryadin A. In addition, there are two methyls at C-16 in aspertoryadin A, but one methyl in 2. The above deduction was supported by the contiguous COSY cross-peaks (Figure 2) of H-14/NH-15/H-16/H-31 and the key HMBC correlations (Figure 2) from H3-31 and H-14 to C-16 and C-17. The relative configuration of 2 was established by ROESY spectrum. The ROESY cross-peaks (Figure 3) of H-11/H-14/H-12a and H-14/H3-31 led to the assignment of the relative configurations for the stereocenters C-13, C-14, C-11, and C-16. The stereocenter C-27 was far away from other stereocenters and no ROE correlation was available for the assignment of its relative configuration. Thus, the chemical shifts of two isomers (11S,13S,14R,16S,27S)-2 and (11S,13S,14R,16S,27R)-2, were calculated and the former showed 100% DP4+ (all data) probability, enable assignment of 27S∗ configuration for 2. The absolute configuration of 2 was confirmed as 11S,13S,14R,16S,27S by comparison of its experimental ECD spectrum with the calculated ECD curves of 2 (Figure 4B).
Compound 3, a yellow powder solid, was assigned the molecular formula C25H21N5O6 by its HRESIMS, requiring 18 degrees of unsaturation. The 13C NMR and DEPT spectra displayed five carbonyl carbons (including an aldehyde carbon and four amide or ester carbonyls), 13 aromatic carbons (eight protonated), one sp3 methylene, two sp3 methines, and two sp3 quaternary carbons (one oxygenated). These data were also indicative of a quinazoline-containing indole alkaloid skeleton as that of 2. A direct comparison of the NMR data of 3 with those of 2 revealed that the differences between them are the presence of an additional aldehyde group and one additional methyl group at the N-15 and C-16 in 3, respectively, as well as the attachment of an amide moiety in 3 instead of an isobutyl unit at C-3 as in 2. The additional groups were supported by the HMBC correlations (Figure 2) from H-28 (δH 8.64) to C-14 (δC 81.5) and from H3-29 and H3-30 to C-16, and C-17. The ROESY cross-peaks of H-12a/H-14/H-11 and H-23/H-12b indicated that 3 possessed the same relative configurations at C-11, C-13, and C-14 as aspertoryadin A (Figure 3). The absolute configuration was determined to be 11S, 13S, and 14R by ECD calculation (Figure 4C).
α-Glucosidase Inhibition Assay
All of the compounds isolated were evaluated for α-glucosidase inhibitory activity (Apostolidis et al., 2007). Compounds 10 and 16 exhibited potent α-glucosidase inhibitory activities with the IC50 values of 7.18 and 5.29 μM, respectively (Acarbose as a positive control, IC50: 213.0 μM).
Molecular Docking
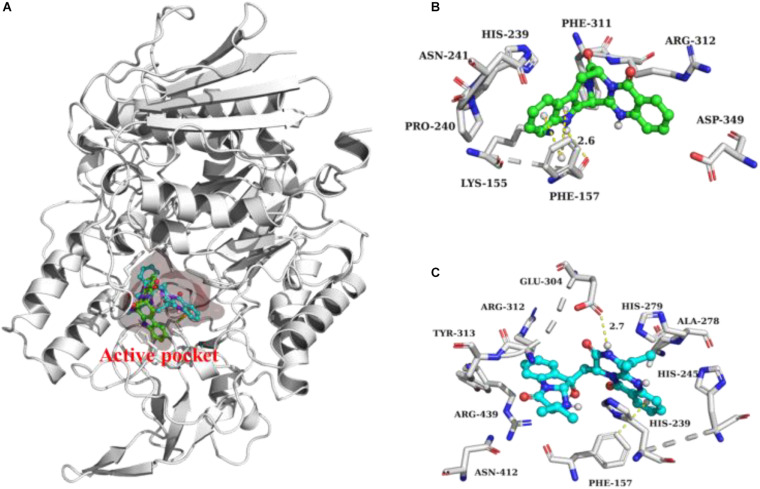
The intermolecular interaction and potential binding sites between compounds 10, 16, and α-glucosidase were investigated via molecular docking simulations. The docking simulation results (Figure 5) demonstrated that compound 10 could interact with α-glucosidase by forming one hydrogen bond and π–π interactions with residues PHE 157 (Figure 5B). While compound 16 could generate one hydrogen bond with residues GLU 340 and also form π–π interactions with residues PHE 157 in α-glucosidase (Figure 5C).
FIGURE 5.
Docking analysis of 10 (green ball and stick) and 16 (cyan ball and stick) with α-glucosidase (A). 3D cartoon diagram of the interactions of 10 (B) and 16 (C) with α-glucosidase.
Biological Activity Against RyR
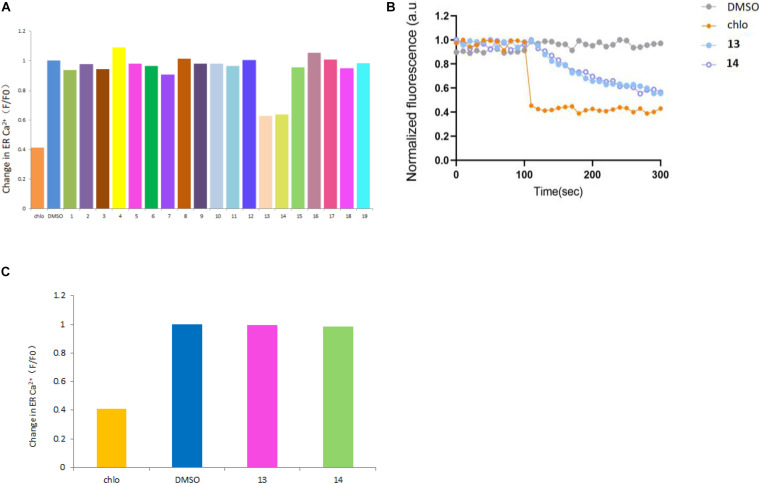
Ryanodine receptor, an intracellular calcium channel located on ER membrane, is a well-known insecticide target. The top-selling diamide insecticides, such as flubendiamide, chlorantraniliprole, and cyanotraniliprole, all target insect RyRs. The insecticidal activities of all compounds were tested against RyRs from an agricultural pest, S. frugiperda, using time-lapse [Ca2+]ER measurements. At 100 μM concentration, compounds 13 and 14, showed a clear activation effect against sfRyR, which reduced the [Ca2+]ER by 37.1 and 36.2%, respectively (Figure 6A). While their effect in intracellular Ca2 release is similar to that of the positive control chlo, the release rate is much slower (Figure 6B) suggesting different binding sites and different mechanisms of action on the sfRyR. Compounds 1, 2, and 3 also showed some weak activation effects on sfRyR (Figure 6A). The species selectivity of 13 and 14 was further characterized by comparing their activity against rabbit RyR1 (rRyR1). Interestingly, both compounds showed no clear activation activity on rRyR1, suggesting that they can selectively act on insect RyRs and have good potential to be developed into insecticidal molecules (Figure 6C).
FIGURE 6.
Summary of against activities of compounds 1–19 on sfRyR (A) and compounds 13 and 14 on rRyR1 (C) at 100 μM; Time-lapse R-CEPIA1er fluorescence measurement curves of 13 and 14 (B) with chlorantraniliprole (chlo) and DMSO as positive and negative controls, respectively.
Conclusion
In summary, 19 quinazoline-containing indole alkaloids (1–19), including 3 new ones, were isolated from the marine-derived fungus Aspergillus sp. HNMF114 by supplemented L-tryptophan to its fermentation broth. Among them, compounds 10 and 16 showed α-glucosidase inhibitory activity, that significant activity with potential for further development. Compounds 1, 2, and 3 showed weak activities against sfRyR, and 13 and 14 showed moderate activities against sfRyR. Compounds 13 and 14 also have no clear activation activity on rRyR1, which means 13 and 14 could selectively act on insect RyRs and have good potential for the development of insecticidal drugs.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
Y-XZ and D-QL contributed to the conception and design of the study. LYan and F-DK determined the plane structure and absolute configuration. S-SL wrote the first draft of the manuscript and performed all of the experimental work. Q-YM and Q-YX contributed to the isolation of compounds. J-HZ, LYao, and L-MZ contributed to the bioactivity assay. Z-GYC and H-FD improved the manuscript. All authors contributed to manuscript revision as well as read and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This research was supported by the National Key R&D Program of China (2017YFD0201401), the Natural Science Foundation of Hainan Province (2019CXTD411), the Financial Fund of the Ministry of Agriculture and Rural Affairs, China (NFZX2018), the China Agriculture Research System (CARS-21), the Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (1630052017002 and 1630052021019), and the Specific research project of Guangxi for research bases and talents (AD18126005).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.680879/full#supplementary-material
References
- Apostolidis E., Kwon Y. I., Shetty K. (2007). Inhibitory potential of herb, fruit, and fungal-enriched cheese against key enzymes linked to type 2 diabetes and hypertension. Innov. Food Sci. Emerg. 1 46–54. 10.1016/j.ifset.2006.06.001 [DOI] [Google Scholar]
- Arasu M. V., Duraipandiyan V., Ignacimuthu S. (2013). Antibacterial and antifungal activities of polyketide metabolite from marine Streptomyces sp. AP-123 and its cytotoxic effect. Chemosphere 90 479–487. 10.1016/j.chemosphere.2012.08.006 [DOI] [PubMed] [Google Scholar]
- Blunt J. W., Copp B. R., Keyzers R. A., Munro M. H., Prinsep M. R. (2017). Marine natural products. Nat. Prod. Rep. 34 235–294. 10.1039/C7NP00052A [DOI] [PubMed] [Google Scholar]
- Bruhn T., Schaumloffel A., Hemberger Y., Bringmann G. (2013). SpecDis: quantifying the comparison of calculated and experimental electronic circular dichroism spectra. Chirality 25 243–249. 10.1002/chir.22138 [DOI] [PubMed] [Google Scholar]
- Bugni T. S., Ireland C. M. (2004). Marine-derived fungi: a chemically and biologically diverse group of microorganisms. Nat. Prod. Rep. 21 143–163. 10.1039/b301926h [DOI] [PubMed] [Google Scholar]
- Buttachon S., Chandrapatya A., Manoch L., Silva A., Gales L., Bruyére C., et al. (2012). Sartorymensin, a new indole alkaloid, and new analogues of tryptoquivaline and fiscalins produced by Neosartorya siamensis (KUFC 6349). Tetrahedron 68 3253–3262. 10.1016/j.tet.2012.02.024 [DOI] [Google Scholar]
- Dan W. J., Zhang Q., Zhang F., Wang W. W., Gao J. M. (2019). Benzonate derivatives of acetophenone as potent α-glucosidase inhibitors: synthesis, structure-activity relationship and mechanism. J. Enzyme Inhib. Med. Chem. 34 937–945. 10.1080/14756366.2019.1604519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D., Luthy R., Bowie J. U. (1997). VERIFY3D: assessment of protein models with three-dimensional profiles. Method. Enzymol. 277 396–404. 10.1016/s0076-6879(97)77022-8 [DOI] [PubMed] [Google Scholar]
- Frisch M. J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., et al. (2019). Gaussian 16, Revision C.01. Wallingford CT: Gaussian, Inc. [Google Scholar]
- Grimblat N., Zanardi M. M., Sarotti A. M. (2015). Beyond DP4: an improved probability for the stereochemical assignment of isomeric compounds using quantum chemical calculations of NMR shifts. J. Org. Chem. 80 12526–12534. 10.1021/acs.joc.5b02396 [DOI] [PubMed] [Google Scholar]
- Guo Y. W., Liu X. J., Yuan J., Li H. J., Mahmud T., Hong M. J., et al. (2020). L-tryptophan induces a marine-derived Fusarium sp. to produce indole alkaloids with activity against the Zika Virus. J. Nat. Prod. 11 3372–3380. 10.1021/acs.jnatprod.0c00717 [DOI] [PubMed] [Google Scholar]
- Huang L. H., Xu M. Y., Li H. J., Li J. Q., Chen Y. X., Ma W. Z., et al. (2017). Amino acid-directed strategy for inducing the marine-derived fungus Scedosporium apiospermum F41-1 to maximize alkaloid diversity. Org. Lett. 19 4888–4891. 10.1021/acs.orglett.7b02238 [DOI] [PubMed] [Google Scholar]
- Kong F. D., Zhang S. L., Zhou S. Q., Ma Q. Y., Xie Q. Yi, Chen J. P., et al. (2019). Quinazoline-containing indole alkaloids from the marine-derived fungus Aspergillus sp. HNMF114. J. Nat. Prod. 82 3456–3463. 10.1021/acs.jnatprod.9b00845 [DOI] [PubMed] [Google Scholar]
- Koyama N., Inoue Y., Sekine M., Hayakawa Y., Homma H., Omura S., et al. (2008). Relative and absolute stereochemistry of quinadoline B, an inhibitor of lipid droplet synthesis in macrophages. Org. Lett. 22 5273–5276. 10.1021/ol802089p [DOI] [PubMed] [Google Scholar]
- Laskowski R. A., Macarthur M. W., Moss D. S., Thornton J. M. (1993). PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26 283–291. 10.1107/S0021889892009944 [DOI] [Google Scholar]
- Lin T., Tan T., Liu T. X. (2013). A study on secondary metabolites PJX-41 mangrove-derived fungi and their antitumor activity. J. Nanjing Agric. Univ. 3 117–123. 10.7685/j.issn.1000-2030.2013.03.020 [DOI] [Google Scholar]
- Manimegalai K., Devi N. K. A., Padmavathy S. (2013). Marine fungi as a source of secondary metabolites of antibiotics. Int. J. Biotechnol. Bioeng. Res. 4 275–282. [Google Scholar]
- Montaser R., Luesch H. (2011). Marine natural products: a new wave of drugs. Future Med. Chem. 3 1475–1489. 10.4155/fmc.11.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle N. M., Vandermeersch T., Flynn C. J., Maguire A. R., Hutchison G. R. (2011). Confab–systematic generation of diverse low-energy conformers. J. Cheminform. 3:8. 10.1186/1758-2946-3-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J. X., Lin T., Wang W., Xin Z. H., Zhu T. J., Gu Q. Q., et al. (2013). Antiviral alkaloids produced by the mangrove-derived fungus Cladosporium sp. PJX-41. J. Nat. Prod. 76 1133–1140. 10.1021/np400200k [DOI] [PubMed] [Google Scholar]
- Penn J., Mantle P. G., Bilton J. N., Sheppard R. N. (1992). Glyantrypine, a novel anthranilic acid-containing metabolite of Aspergillus clavatus. J. Chem. Soc. Pak. 1 1495–1496. 10.1039/P19920001495 [DOI] [Google Scholar]
- Qian S. Y., Yang C. L., Khan A., Chen R. X., Wu M. S., Tuo L., et al. (2018). New pyrazinoquinazoline alkaloids isolated from a culture of Stenotrophomonas maltophilia QB-77. Nat. Prod. Res. 9 1387–1391. 10.1080/14786419.2018.1475381 [DOI] [PubMed] [Google Scholar]
- Shen S., Li W., Ouyang M. A., Wu Z. J., Lin Q. Y., Xie L. H. (2009). Identification of two marine fungi and evaluation of their antivirus and antitumor activities. Acta. Microbiol. Sin. 49 1240–1246. 10.13343/j.cnki.wsxb.2009.09.008 [DOI] [PubMed] [Google Scholar]
- Suzuki J., Kanemaru K., Ishii K., Ohkura M., Okubo Y., Iino M., et al. (2014). Imaging intraorganellar Ca2+ at subcellular resolution using CEPIA. Nat. Commun. 5 4153–4165. 10.1038/ncomms5153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A. J. (2010). AutoDock vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B., Sali A. (2016). Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 54 5.6.1–5.6.37. 10.1002/cpbi.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Ma D. (2013). Total syntheses of (±)-spiroquinazoline, (-)-alantryphenone, (+)-lapatin A, and (-)-quinadoline B. Angew. Chem. Int. Ed. 52 9759–9762. 10.1002/anie.201303893 [DOI] [PubMed] [Google Scholar]
- Xiong Z. Q., Wang J. F., Hao Y. Y., Wang Y. (2013). Recent advances in the discovery and development of marine microbial natural products. Mar. Drugs 11 700–717. 10.3390/md11030700 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.