Summary
Latent transforming growth factor β (TGFβ)-binding proteins (LTBPs) are microfibril-associated proteins essential for anchoring TGFβ in the extracellular matrix (ECM) as well as for correct assembly of ECM components. Variants in LTBP2, LTBP3, and LTBP4 have been identified in several autosomal recessive Mendelian disorders with skeletal abnormalities with or without impaired development of elastin-rich tissues. Thus far, the human phenotype associated with LTBP1 deficiency has remained enigmatic. In this study, we report homozygous premature truncating LTBP1 variants in eight affected individuals from four unrelated consanguineous families. Affected individuals present with connective tissue features (cutis laxa and inguinal hernia), craniofacial dysmorphology, variable heart defects, and prominent skeletal features (craniosynostosis, short stature, brachydactyly, and syndactyly). In vitro studies on proband-derived dermal fibroblasts indicate distinct molecular mechanisms depending on the position of the variant in LTBP1. C-terminal variants lead to an altered LTBP1 loosely anchored in the microfibrillar network and cause increased ECM deposition in cultured fibroblasts associated with excessive TGFβ growth factor activation and signaling. In contrast, N-terminal truncation results in a loss of LTBP1 that does not alter TGFβ levels or ECM assembly. In vivo validation with two independent zebrafish lines carrying mutations in ltbp1 induce abnormal collagen fibrillogenesis in skin and intervertebral ligaments and ectopic bone formation on the vertebrae. In addition, one of the mutant zebrafish lines shows voluminous and hypo-mineralized vertebrae. Overall, our findings in humans and zebrafish show that LTBP1 function is crucial for skin and bone ECM assembly and homeostasis.
Keywords: LTBP1, transforming growth factor beta; cutis laxa syndrome; craniosynostosis; Danio rerio; extracellular matrix; short stature; collagen fibrillogenesis; tissue mineral density
Introduction
Latent transforming growth factor β (TGFβ)-binding proteins (LTBPs) are microfibril-associated multidomain proteins essential for the sequestration of TGFβ in the extracellular matrix (ECM). Mature TGFβ growth factor dimers associate non-covalently with the latency-associated peptide (LAP) in order to form the small latent complex (SLC), which is covalently tethered via two disulfide-bridges to LTBPs.1 SLCs of the three human TGFβ isoforms were shown to bind to LTBP1 and LTBP3, while TGFβ1 SLC exclusively interacts with LTBP4.2 Most LTBPs are targeted to the ECM via their amino- and carboxy-terminal regions. LTBP1, LTBP2, and LTBP4 interact through their C-terminal region with fibrillin-1 (FBN1),3, 4, 5 while the N-terminal region of LTBP1 and LTBP4 interact with fibronectin (FN).6,7 Moreover, LTBP4 facilitates the deposition of tropoelastin onto the microfibrillar scaffold through interaction with fibulin-4 (EGF-containing fibulin extracellular matrix protein 2: EFEMP2) and fibulin-5 (FBLN5).8, 9, 10, 11, 12, 13 Similar to LTBP4, LTBP2 facilitates the deposition of tropoelastin onto the microfibrillar scaffold through interaction with fibulin-5.14 Dysfunction of any member of the LTBP superfamily has multiple consequences on the TGFβ bioavailability and elastic fiber assembly in various tissues both in vitro and in vivo.9, 10, 11,15, 16, 17, 18, 19
Pathogenic variants in LTBP genes have been identified in several autosomal recessive (AR) Mendelian disorders presenting with impaired development of the skeleton and/or elastin-rich tissues. Pathogenic variants in LTBP2 cause AR primary congenital glaucoma (MIM: 613086), AR microspherophakia and/or megalocornea, with ectopia lentis and with or without secondary glaucoma (MIM: 251750), and AR Weill-Marchesani syndrome (MIM: 614819).20, 21, 22 Pathogenic variants in LTBP3 cause AR platyspondyly with amelogenesis imperfecta (MIM: 601216) and geleophysic dysplasia 3 (MIM: 617809).23,24 In addition, homozygous loss-of-function (LOF) variants in LTBP3 were reported in syndromic forms of thoracic aortic aneurysm and dissection (TAAD).25 Finally, pathogenic variants in LTBP4 cause AR cutis laxa (CL) type 1C, characterized by loose redundant skin folds, emphysema, and diverticula of the gastrointestinal and urinary tract.26, 27, 28 Thus far, the human phenotype associated with LTBP1 (MIM: 150390) deficiency has remained enigmatic.
Nevertheless, the molecular consequences of Ltbp1 deficiency have been studied in mice. In most vertebrates, LTBP1 encodes two alternatively spliced isoforms: a long (LTBP1L) and a short (LTBP1S) isoform. Mice lacking Ltbp1L only or both Ltbp1S and Ltbp1L show a persistent truncus arteriosus and an interrupted aortic arch that associates with perinatal lethality.15,29 At the embryonal stage, the outflow tract of Ltbp1L−/− mouse hearts show decreased TGFβ activity.15 Mice lacking Ltbp1S while still retaining expression of an alternatively spliced form of Ltbp1L (Δ55 variant) are viable, show mild craniofacial and skeletal abnormalities and impaired ovarian function, and are less prone to hepatic fibrosis after bile duct ligation, which was attributed to decreased bio-availability of TGFβ.29, 30, 31 Together, data from animal studies indicate that Ltbp1L, and hence intact TGFβ signaling, is required for proper embryonal cardiovascular development, while Ltbp1S could play a role in craniofacial development.15,29,30
Here, we report homozygous premature truncating LTBP1 variants in eight affected individuals from four unrelated consanguineous families. Affected individuals present with cutis laxa, craniofacial dysmorphism, mild variable heart defects, and altered skeletal development, including short stature, craniosynostosis, brachydactyly, clinodactyly, and syndactyly, which we propose to coin as LTBP1-related CL syndrome.32 In vitro studies on proband dermal fibroblasts indicate distinct molecular consequences and effects on TGFβ signaling depending on the position of the variant in LTBP1. For in vivo validation, we generated and characterized ltbp1−/− Δ29 and ltbp1−/− Δ35 zebrafish. We found abnormal collagen fibrillogenesis in the skin and in the intervertebral ligaments. In addition, ltbp1−/− Δ29 zebrafish show hypo-mineralized vertebrae with ectopic bone formation and increased vertebral volume. These observations were not investigated in the majority of the affected individuals. Our data indicate that LTBP1 has dual functions in humans and zebrafish affecting cutaneous and skeletal development.
Subjects and methods
Clinical assessment
Informed consents were obtained from all individuals or from their parents in the case of minor individuals, including specific consent to publish the clinical pictures in Figure 1. All individuals were evaluated at one of the collaborating referral centers and clinical data were recorded with a clinical checklist (Table S1). Skin biopsies were obtained from several probands for dermal fibroblast culture (F1:IV-2 and F4:II-1) and transmission electron microscopy (TEM) (F1:IV-2). This study was conducted in accordance with the Declaration of Helsinki and approved by the Ghent University Hospital ethical committees (registration number B6702020000194). Family 2, family 3, and family 4 were identified through GeneMatcher.33
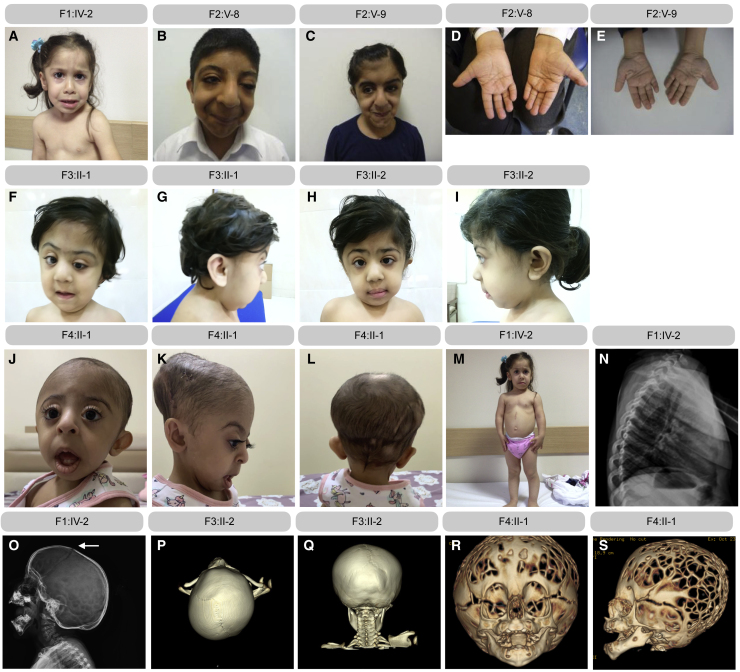
Figure 1.
Clinical characteristics
Clinical pictures of F1:IV-2 (at age 3 years) (A), F2:V-8 (at age 17 years) (B), F2:V-9 (at age 9 years) (C), F3:II-1 (at age 1.6 years) (F and G), F3:II-2 (at age 4 years) (H and I), and F4:II-1 (at age 2 years) (J–L). Brachydactyly is observed in multiple families, but clinical pictures (D and E) are only available from family 2. Short stature and ovoid-shaped vertebral bodies are observed in F1:IV-2 (at age 3 years) (M and N). A copper beaten calvarium and a coronal suture (arrow) are present in F1:IV-2 (at age 3 years) (O). Craniosynostosis involving the right coronal and sagittal suture is observed in F3:II-2 (at age 6 months) (P and Q). A copper beaten calvarium due to high intracranial pressure is present in F4:II-1 (R and S). Pedigrees of all affected families can be found in Figure S1.
Exome sequencing
Exome sequencing (ES) was performed on genomic DNA (gDNA) extracted from blood leukocytes of each person. gDNA was enriched with the SureselectXT Human All Exon v.6 kit (Agilent Technologies, Santa Clara, CA, USA) followed by sequencing on a HiSeq 3000 platform (family 1) (Illumina, San Diego, CA, USA). LTBP1 (GenBank: NM_206943.4) nucleotides were numbered according to the Human Genome Variation Society guidelines with nucleotide “A” of the ATG start codon of the long isoform of LTBP1 = c.1. The following algorithms were used to predict the consequences of variants identified with ES: PolyPhen-2, PhD-SNP, SIFT, SNAP, MAPP, and REVEL. Allele frequencies were evaluated via the gnomAD population database. Homozygosity mapping was performed prior to ES in family 2 via an Affymetrix Genome-Wide Human SNP Array 6.0 (Thermo Fisher, Waltham, MA, USA). Segregation analysis was performed in parents of affected individuals via Sanger sequencing. ES (family 3 and family 4) was done as previously described.34,35
Transmission electron microscopy
For human dermal biopsies, 3 mm skin fragments from an individual (F1:IV-2) and an age- and sex-matched control were initially immersed in a fixative solution of 4% glutaraldehyde for transport. Subsequently, samples were placed in 2.5% glutaraldehyde and 4% formaldehyde in 0.1 M Na-Cacodylate buffer in a vacuum oven for 30 min followed by further fixation for 3 h at room temperature on a sample rotator. This solution was then replaced with fresh fixative and samples were incubated overnight at 4°C on a sample rotator. After washing in double-distilled H2O, samples were post-fixed in 1% OsO4 with K3Fe(CN)6 in 0.1 M Na-Cacodylate buffer (pH 7.2). After washing in double-distilled H2O, samples were subsequently dehydrated through a graded ethanol series, including bulk staining with 2% uranyl acetate at the 50% ethanol step followed by embedding in Spurr’s resin. To select the area of interest on the block and in order to have an overview of the phenotype, we first cut semi-thin sections at 0.5 μm and stained them with toluidine blue. Ultrathin sections were cut with an ultramicrotome (Leica EM UC6, Wetzlar, Germany) followed by post-staining in a Leica EM AC20 for 40 min in uranyl acetate at 20°C and for 10 min in lead stain at 20°C. Sections were collected on formvar-coated copper slot grids. Grids were viewed with a JEM 1400plus transmission electron microscope (JEOL, Tokyo, Japan) operating at 80 kV. For zebrafish, skin biopsies and vertebrae of 4- to 6-month-old male zebrafish were fixed and processed for ultrastructural analysis as previously described.36 Sections were viewed with Jeol JEM 1010 TEM (Jeol, Tokyo, Japan) equipped with a charge-coupled device (CCD) side-mounted Veleta camera operating at 60 kV. Experiments were performed in collaboration with the TEM facility of the Nematology Research Unit at Ghent University. Results are representative of three independent experiments.
Cell culture
Dermal fibroblasts obtained from a skin biopsy from individuals F1:IV-2 and F4:II-1 and four healthy individuals (two control subjects, age- and sex-matched, for each individual, see Table S2) were cultured in Dulbecco’s modified Eagle’s medium (GIBCO; Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (PAN-Biotech, Aidenbach, Germany), 1% non-essential amino acids (GIBCO), 1% penicillin/streptomycin (GIBCO), and 0.1% fungizone (GIBCO) and incubated at 37°C with 5% CO2. Cells were tested for mycoplasma contamination by biochemical analysis of mycoplasmal enzymes (Lonza, Basel, Switzerland) and confirmed to be mycoplasma free.
Antibodies
The following primary antibodies and dilutions were used for immunoblot analysis: anti-Phospho-Smad2 (Ser465/467) (#3108, Cell Signaling Technologies [CST], Danvers, MA, USA, 1/500), anti-Smad2 (#5339, CST, 1/1,000), anti-Vinculin (#13901, CST, 1/1,000), and anti-fibronectin (ab23750, Abcam, Cambridge, UK, 1/1,500). Anti-rabbit IgG HRP-linked Antibody (#7074 CST, 1/2,500–1/4,000) was used as secondary antibody. Polyclonal rabbit anti-FBN1 antiserum (1/1,000 for immunofluorescence [IF] and 1/2,000 for western blot [WB]) was raised against the recombinantly produced N-terminal half of human fibrillin-1 (F90).37,38 Polyclonal rabbit anti-LTBP-1 antiserum (1/1,000 for IF) was raised against the last 214 C-terminal residues of human LTBP-1 L1K.3,38 Polyclonal rabbit anti-Fbn2 antibody was raised against the C-terminally double-strep-tagged N-terminal recombinant human FBN2 polypeptide rF86 (Gln29-Asp535).39 Polyclonal rabbit anti-LTBP-2 antiserum (1/1,000 for IF) was raised against the last 254 C-terminal residues of human LTBP-2 (Asp1568-Glut1821), similar to as described for L1K.38 Anti-collagen I antibody (#ab34710, Abcam, 1/1,000) and polyclonal goat anti-collagen III (#1330-01, Southern Biotech, 1/1,000) were used for IF. Polyclonal rabbit antibody recognizing human fibulin-4 (1/1,000 for IF) was a kind gift from Dr. Takako Sasaki (Oita University). Goat anti-Rabbit IgG Alexa Fluor 555 (A32732, Life Technologies, 1/800) was used as secondary antibody.
Recombinantly produced proteins
For recombinant protein production of LTBP1, cDNA encoding the wild-type (WT) human LTBP1 fragment of 541 C-terminal amino acid residues and corresponding fragments carrying c.4431T>A and c.4844del variants were overexpressed in HEK293 cells together with the unaffected control sequence. Encoding cDNAs were cloned into a variant of the pCEP-Pu vector, and stably transfected overexpressing cells were established after puromycin selection as previously described.9 Proteins were expressed with a C-terminally placed double-strep-tag and purified via affinity chromatography from collected serum-free culture medium. Fresh medium was filtered with a suitable membrane filter and then subjected to Strep-Tactin XT gravity flow column (2 mL beads; IBA GmbH, Germany) at 4°C overnight. LTBP1 proteins were eluted with elution buffer (100 mM Tris/HCl [pH 8.0] 150 mM NaCl, 1 mM EDTA, 2.5 mM desthiobiotin). The collected fractions were concentrated and exchanged to PBS by Amicon Ultra Centrifugal Filter, 3 kDa (Merck Millipore, MA, USA). Using the same protocol, we recombinantly produced the N-terminal region of human fibrillin-1 (after signal peptide cleavage site, up to the amino acids coding for the fourth epidermal growth factor [EGF4] domain, encompassing the binding site for LTBP1). Production and purification of the N-terminal region of human FBN2 (rF86) was as previously described.40
RT-qPCR
Total RNA was extracted from dermal fibroblast cultures from control subjects and individuals (F1:IV-2 and F4:II-1) via the RNeasy Kit (QIAGEN, Hilden, Germany) with DNase digestion of genomic DNA followed by cDNA synthesis with the iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA, USA). Gene expression of EFEMP2, FBLN5, LTBP3, LTBP4, FBN1, FBN2, FN, POSTN, and CTGF was investigated between control subjects’ and affected individuals’ (F1:IV-2 and F4:II-1) dermal fibroblast cultures. Gene expression of COL1A1, COL1A2, and COL3A1 was investigated between control subjects’ and affected individuals’ (F1:IV-2 and F4:II-1) dermal fibroblast cultures stimulated with 25 μg/mL ascorbate (Sigma-Aldrich, St. Louis, MO, USA) for 3 days. All measurements were obtained from three separate dermal fibroblast culture samples originating from individuals F1:IV-2 and F4:II-1 and from two control subjects. Average values of the two control subjects were plotted as “control” for each experiment. Total RNA was extracted from juvenile zebrafish in quintuplicate in which 10 zebrafish larvae were pooled per sample. Gene expression of ltbp1 was investigated between ltbp1−/− Δ29, ltbp1−/− Δ35, and WT zebrafish controls. Assays were prepared with the addition of SsoAdvanced SYBR Green supermix (Bio-Rad Laboratories) and were subsequently run on a LightCycler 480 Instrument II (Roche, Basel, Switzerland). Primers were designed via Primer-BLAST (Table S3). We used Biogazelle qBase+3.0 software for data analysis by using YWAHZ, HPRT1, and RLP13A for normalization of human dermal fibroblasts and loopern, hatn10, and tdr7 for normalization of zebrafish samples.41
Nonsense-mediated decay analysis
Dermal fibroblasts from individuals (F1:IV-2 and F4:II-1) and control subjects were incubated with 5 mg/mL cycloheximide for 17 h or vehicle followed by reverse transcription quantitative PCR (RT-qPCR). All measurements were obtained from three separate dermal fibroblast culture samples originating from individuals F1:IV-2 and F4:II-1 and from two control subjects. Average values of the two control subjects were plotted as “control” for each experiment.
Immunoblot analysis and determination of TGFβ levels
For the investigation of extracellular proteins, conditioned serum-free medium of dermal fibroblast cultures from control subjects and affected individuals (F1:IV-2 and F4:II-1) was collected at day 14 as previously described.42 Protein samples were subjected to 3%–8% Tris-acetate sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) before blotting, either by wet or dry blotting onto a polyvinylidene difluoride (PVDF) or nitrocellulose (NC) membrane. We used imperial protein staining (Life Technologies, Carlsbad, CA, USA) to visualize the total protein amount. For the investigation of intracellular proteins, cell lysate of confluent dermal fibroblast cultures from control subjects and affected individuals (F1:IV-2 and F4:II-1) was collected. Protein samples were subjected to 4%–12% Bis-Tris SDS-PAGE before dry blotting onto a PVDF membrane. Imaging was performed on an Amersham Imager 680 (GE Healthcare Life Sciences, Chicago, IL, USA). Resulting images were processed with Fiji software.43 We measured total TGFβ protein levels in conditioned serum-free medium of dermal fibroblast cultures from control subjects and affected individuals (F1:IV-2 and F4:II-1) collected at day 9 by using the Quantikine ELISA (#MB100B, R&D Systems, Minneapolis, MN, USA) according to manufacturer’s instructions. All measurements were obtained from three separate dermal fibroblast culture samples originating from individuals F1:IV-2 and F4:II-1 and from two control subjects. Average values of the two control subjects were plotted as “control” for each experiment. Recombinant human TGF-beta 1 (Bio-Techne Corporation, Minneapolis, MN, USA) was added at 2.5 ng/mL to one confluent control dermal fibroblast culture acting as positive control for Smad2 phosphorylation.
Immunofluorescence
For analysis of ECM network formation, cells were seeded on uncoated glass coverslips at a density of 8 × 104 cells/well in a 24-well plate. After culture, cells were washed with PBS, fixed at –20°C in methanol/acetone, blocked in a phosphate-buffered saline/1% bovine serum albumin solution, and subsequently incubated with primary and secondary antibodies diluted in the blocking solution. Images were obtained from three independent experiments.
Solid-phase binding assay
Multiwell plates were coated with purified LTBP1 (100 nM) in 50 mM carbonate/bicarbonate buffer (pH 9.6) at 4 °C overnight. Coated wells were blocked with 5% nonfat dry milk in TBS at room temperature for 1 h. Recombinant fibrillin-1 and -2 were serially diluted 1:2 in 2% milk, TBS, and incubated in the wells for 2 h followed by a 1 h incubation with anti-fibrillin-1 or -2 antibody (1/5,000). Color reaction of the enzyme immunoassay was achieved with the TMB (3,3′,5,5′-tetramefhyl-benzidine) Substrate Kit (Thermo Fisher Scientific, Waltham, MA, USA) and stopped with 0.1 M HCl after streptavidin-HRP (biotinylated) antibody incubation. Absorbance was read at 450 nm with a Microplate Reader Sunrise (Tecan, Maennedorf, Switzerland). We achieved curve fits to obtain affinity constants by employing Graphpad Prism 9 (La Jolla, CA, USA) and selecting the nonlinear one-site model.
Surface plasmon resonance
We performed surface plasmon resonance (SPR) experiments as described previously44 by using a Biacore 2000 system (Biacore AB, Uppsala, Sweden). We covalently coupled recombinant human fibrillin-1 covering the N-terminal region including EGF4 to CM5 sensor chips at 3,600 resonance units (RUs) by using the amine coupling kit following the manufacturer’s instructions (Cytiva, Uppsala, Sweden), and 0–320 nM of recombinant LTBP1 was flown over in HBS-P buffer (0.01 M HEPES [pH 7.4], 0.15 M NaCl, 10 mM CaCI2, and 0.005% [v/v] surfactant P20). Affinity constants (KDs) were calculated with nonlinear fitting (1:1 interaction model with mass transfer) to the association and dissociation curves according to the manufacturer’s instructions (BIAevaluation v.3.0 software). Apparent equilibrium dissociation constants (KD values) were then calculated as the ratio of kd/ka.
Zebrafish lines and maintenance
Zebrafish lines were housed in a Zebtec semi-closed recirculation housing system at a constant temperature (27°C–28°C), pH (~7.5), conductivity (~550 μS), and light/dark cycle (14/10). Fish were fed twice a day with dry food (Gemma Micro, Skretting) and once with artemia (Ocean Nutrition, Essen, Belgium). Ltbp1−/− Δ29 and ltbp1−/− Δ35 zebrafish were generated via CRISPR-Cas9 mutagenesis according to the workflow previously described.45 Zebrafish were genotyped with primers listed in Table S4. We adhered to the general guidelines, in agreement with EU Directive 2010/63/EU for laboratory animals, for zebrafish handling, mating, embryo collection, and maintenance.46,47 Approval for this study was provided by the local committee on the Ethics of Animal Experiments (Ghent University Hospital, Ghent, Belgium; permit number: ECD 17/63K and ECD 18/05).
Echocardiography
We performed ultrasound imaging on 10- to 11-month-old male zebrafish by using a dedicated ultrasound apparatus Vevo 2100 (Visualsonics, Toronto, Canada) equipped with a high-frequency linear array transducer (MS 700, frequency 30–70 MHz). Zebrafish were placed in an anesthetic chamber containing 200 mg/L tricaine (Sigma-Aldrich). Zebrafish were transferred to a 3D printed imaging chamber where the zebrafish was positioned ventral side up containing 100 mg/L tricaine to minimize movements. Water temperature was maintained at 28°C throughout the whole procedure. Image acquisition was conducted within 5 min after the induction of anesthesia. Echocardiographic images were obtained in two planes: long axis (LAX), enabling normalized 2D ventricular dimension parameters using the body surface area (BSA) normalization factor, and abdominal-cranial axis (ACX), for color Doppler and pulse-wave Doppler image acquisition, enabling cardiac function measurements.48,49 Measurements and functional calculations were performed in Vevo LAB 1.7.0. Volumes of systole and diastole are calculated in Vevo LAB 1.7.0. on the basis of the geometry of mammalian heart. Therefore, we reported the area of the systole and diastole normalized to the body surface area.50 Measurements were performed by a researcher blinded to the genotype.
Whole-mount staining with alizarin red S
Alizarin red staining for mineralized bone of 4-month-old adult zebrafish was performed as previously described.51 Stained specimens were analyzed for the presence of ectopic bone with a Leica M165 FC Fluorescent Stereo Microscope (Leica Microsystems, GmbH, Wetzlar, Germany). Ectopic bone counts started from the second caudal vertebral body (VB) (with complete neural and haemal arches and complete neural and haemal spines) (VB16–VB27). The vertebral columns were scored by two observers blinded to the genotype of the samples.
μCT analysis
For μCT-based phenotyping and quantification, 4-month-old adult zebrafish were euthanized via an overdose of tricaine, fixed in 4% PFA for 48 h, and transferred to a 70% ethanol solution for scanning. Whole-body μCT scans of ltbp1−/− Δ29 (n = 5) and ltbp1−/− Δ35 (n = 5) zebrafish and corresponding controls (n = 4–5) were acquired on a SkyScan 1275 (Bruker, Kontich, Belgium) with the following scan parameters: 0.25 mm aluminum filter, 50 kV, 160 μA, 65 ms integration time, 0.5° rotation step, 721 projections/360°, and 21 μm voxel size. We generated DICOM files of individual zebrafish, which we segmented in MATLAB with custom FishCuT software, by using NRecon v.1.7.3.2 (Bruker) software followed by data analysis in the R statistical environment as previously described.52,53
Statistical analysis
Statistical calculations, including multiple testing corrections, were performed with GraphPad Prism 9. p values < 0.05 were considered significant.
Results
Bi-allelic premature truncating variants in LTBP1 cause cutis laxa with impaired craniofacial, skeletal, and cardiac development
Table 1, Figure 1, and Table S1 summarize and illustrate the clinical findings in all eight affected individuals. Detailed case reports and pedigrees are available in the supplemental notes. Core clinical features include cutis laxa, craniosynostosis, a copper beaten calvarium, short stature, and discernible craniofacial characteristics. Affected individuals show facial asymmetry, coarse facial features, arched eyebrows, proptosis, downslanting palpebral fissures, long eyelashes, a prominent nose with convex nasal ridge, wide nasal bridge and broad nasal tip, sagging cheeks with prominent nasolabial folds, long philtrum, thick lower lip vermillion, and a highly arched palate. Skin features include mild to moderate cutis laxa, deep palmar creases, and inguinal hernia (F1:IV-2, F2:V-3, F2:V-8, F2:V-9, and F3:II-1). Individual F1:IV-2 further presents with a congenital diaphragmatic hernia, but this is also present in her carrier mother. All affected individuals, with the exception of F1:IV-2 and F2:V-4, present with craniosynostosis, involving the coronal suture (F2:V-3 and F2:V-9), coronal, sagittal, and lambdoid suture (F2:V-8), or right coronal and sagittal suture (F3:II-1 and F3:II-2). Individual F4:II-1 shows pansynostosis. In addition to short stature, most individuals show other skeletal abnormalities, including brachydactyly (F1:IV-2, F2:V-3, F2:V-4, F2:V-8, F2:V-9, F3:II-1, and F3:II-2), clinodactyly of the fifth finger (F1:IV-2, F2:V-3, F2:V-8, F2:V-9, F3:II-1, F3:II-2, and F4:II-1), syndactyly of the 2nd, 3rd, and 4th toe (F2:V-8 and F2:V-9), syndactyly of the 2nd and 3rd toe (F2:V-3), syndactyly of the 4th and 5th toe (F3:II-1 and F3:II-2), genua vara (F1:IV-2, F3:II-1, and F4:II-1), and joint hypermobility (F1:IV-2, F3:II-1, F3:II-2, and F4:II-1). Less frequent skeletal findings include scoliosis (F1:IV-2 and F3:II-2), lumbar hyperlordosis (F1:IV-2), hip dislocation (F2:V-3), and a short thorax with pectus excavatum (F4:II-1). X-ray images of the spine from F1:IV-2 showed “ovoid-”shaped vertebral bodies at the age of 3 years. No evidence for exostoses could be observed on the X-ray images, but no CT scan was made to exclude this with more certainty. Variable heart defects were found in three individuals. A moderate secundum atrial septum defect of congenital origin with mild right ventricular volume overload was observed in F3:II-1. F1:IV-2 shows mitral and tricuspid insufficiency, and mild concentric left ventricular hypertrophy is present in F2:V-3. Neurodevelopment was normal in most individuals, but F2:V-3 and F2:V-4 experience learning difficulties. Severe intellectual disability of unknown cause has been recorded in individual F2:V-4. Cranial nerve dysfunction occurred in families 2 and 3. In family 2, optic nerve hypoplasia and associated visual impairment is present in individuals F2:V-4, F2:V-8, and F2:V-9, while F2:V-3, F2:V-8, and F2:V-9 have hearing loss. Both affected individuals from family 3 display ophthalmoplegia due to a 3rd and 4th cranial nerve palsy. Feeding problems, attributable to gastresophageal reflux or poor appetite, were recorded in F2:V-4, F3:II-1, F3:II-2, and F4:II-1. Finally, urological abnormalities, including a low and small right kidney in F1:IV-2 and left hydroureter in F2:V-8, are observed in two families.
Table 1.
Overview of homozygous genotypes and clinical characteristics
| Proband 1, family F1:IV-2 | Proband 2, family F2:V-3 | Proband 3, family F2:V-4 | Proband 4, family F2:V-8 | Proband 5, family F2:V-9 | Proband 6, family F3:II-1 | Proband 7, family F3:II-2 | Proband 8, family F4:II-1 | Number and % affected individuals | |
|---|---|---|---|---|---|---|---|---|---|
| Demographic features | |||||||||
| Age at last evaluation | 3 years 4 months | 17 years 5 months | 16 years | 17 years | 9 years | 1 year 6 months | 4 years | 1 year 9 months | N/A |
| Sex | female | female | female | male | female | male | female | female | N/A |
| Parental consanguinity | + | + | + | + | + | + | + | + | N/A |
| Ethnicity | Turkish | Pakistani | Pakistani | Pakistani | Pakistani | Pakistani | Pakistani | Saudi Arabic | N/A |
| Craniofacial dysmorphism | |||||||||
| Coarse face | + | + | − | + | + | + | + | + | 7/8 (87.5%) |
| Arched eyebrows | − | + | + | + | + | + | + | − | 6/8 (75%) |
| Proptosis | − | + | − | + | + | + | + | + | 6/8 (75%) |
| Downslanted palpebral fissures | + | + | − | + | + | − | − | + | 5/8 (62.5%) |
| Long eyelashes | + | + | + | + | + | + | + | unknown | 7/8 (87.5%) |
| Convex nasal ridge | − | + | + | + | + | + | + | + | 7/8 (87.5%) |
| Wide nasal bridge and broad tip | + | + | + | + | + | + | + | + | 8/8 (100%) |
| Sagging cheeks | + | + | − | + | + | + | + | + | 7/8 (87.5%) |
| Prominent nasolabial folds | + | + | − | + | + | + | − | + | 6/8 (75%) |
| Long philtrum | + | + | − | + | + | + | + | + | 7/8 (87.5%) |
| Thick lower lip vermillion | + | + | − | + | + | + | + | + | 7/8 (87.5%) |
| Highly arched palate | − | − | + | + | + | + | + | − | 5/8 (62.5%) |
| Connective tissue features | |||||||||
| Cutis laxa | + | + | + | + | + | + | + | + | 8/8 (100%) |
| Deep palmar creases | + | + | − | + | + | + | + | + | 7/8 (87.5%) |
| Inguinal hernia | + | + | − | + | + | + | − | − | 5/8 (62.5%) |
| Skeletal features | |||||||||
| Craniosynostosis | − | + | − | + | + | + | + | + | 6/8 (75%) |
| Short stature | + | + | + | + | + | + | + | + | 8/8 (100%) |
| Brachydactyly | + | + | + | + | + | + | + | − | 7/8 (87.5%) |
| Clinodactyly | + | + | − | + | + | + | + | + | 7/8 (87.5%) |
| Syndactyly | − | + | − | + | + | + | + | − | 5/8 (62.5%) |
| Joint hyperlaxity | + | − | − | − | − | + | + | + | 4/8 (50%) |
| Genua vara | + | − | − | − | − | + | − | + | 3/8 (37.5%) |
| Additional features | |||||||||
| Learning difficulties | − | + | + | − | + | − | − | unknown | 3/8 (37.5%) |
| Cardiac abnormalities | + | + | − | − | − | + | − | − | 3/8 (37.5%) |
| Hearing loss | − | + | − | + | + | − | − | − | 3/8 (37.5%) |
| Feeding problems/GER | − | − | − | − | − | + | + | + | 3/8 (37.5%) |
| Urological abnormalities | + | − | − | − | + | − | − | − | 2/8 (25%) |
| Molecular characteristics | |||||||||
| Gene | LTBP1 | LTBP1 | LTBP1 | LTBP1 | LTBP1 | LTBP1 | LTBP1 | LTBP1 | N/A |
| cDNA change | c.4844del | c.4431T>A | c.4431T>A | c.4431T>A | c.4431T>A | c.3991_3995del | c.3991_3995del | c.1342C>T | N/A |
| Protein change | p.Asn1615Ilefs∗23 | p.Cys1477∗ | p.Cys1477∗ | p.Cys1477∗ | p.Cys1477∗ | p.Thr1331Asnfs∗20 | p.Thr1331Asnfs∗20 | p.Gln448∗ | N/A |
Exome sequencing (ES) identified homozygous premature truncating variants in LTBP1 (GenBank: NM_206943.4) in eight affected children from four different families. Prior to ES, homozygosity mapping in family 2 showed one shared 22.9 Mb homozygous region on chromosome 2 (20,605,248–43,530,418), containing LTBP1, between the three affected individuals tested and absent in unaffected family members. Families 1 and 3 harbor homozygous frameshift variants in LTBP1 consisting of a 1 base pair (bp) (c.4844del [p.Asn1615Ilefs∗23]) and 5 bp deletion (c.3991_3995del [p.Thr1331Asnfs∗20]), respectively. Families 2 and 4 harbor homozygous nonsense variants (c.4431T>A [p.Cys1477∗] and c.1342C>T [p.Gln448∗], respectively). All variants segregate in family members according to disease—and carrier status. According to different in silico algorithms, the identified variants are predicted to be disease causing and all variants are absent from the population databases. Schematic presentation of the corresponding alterations in LTBP1 and their amino acid homology in other species are shown in Figure 2. TEM analysis of the dermis from a skin biopsy was done in individual F1:IV-2 (Figure 3). The elastic fiber shows microfibril infiltration in its periphery with mild fragmentation of elastin that still formed a central core (Figures 3C and 3D). Collagen fibrils appear similar to the control subject with regular fiber diameters (Figures 3E–3H).
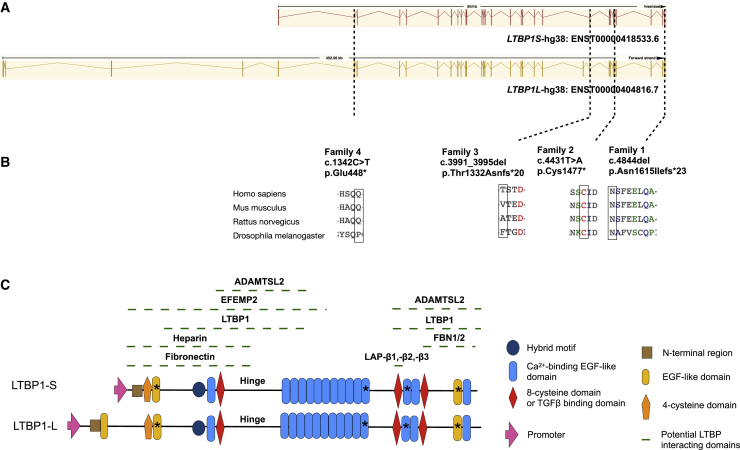
Figure 2.
Schematic representation of the premature truncating variants in LTBP1 and corresponding protein in four unrelated consanguineous families
(A) Schematic representation of the location of the four distinct LTBP1 variants identified in the affected families. The genomic position of each variant is indicated on the exon structure of both the short (LTBP1S) and long (LTBP1L) isoforms of LTBP1.
(B) Sequence alignment shows conservation of the mutated residues among different species.
(C) Schematic representation of the LTBP1 domains. LTBP1 consists of fifteen calcium-binding (cb) EGF-like domains, three EGF-like domains, two TGFβ-binding domains, a hybrid motif, and a 4-cysteine domain. The position of the corresponding alterations on the protein level are indicated by an asterisk.
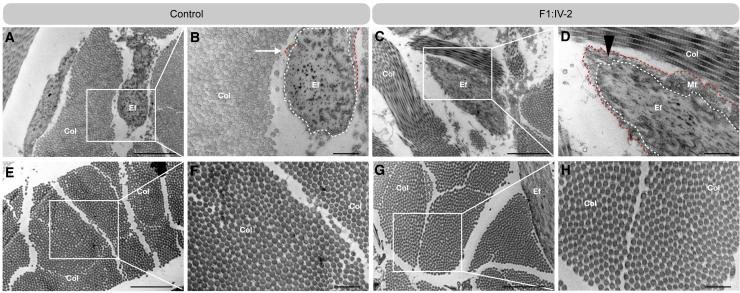
Figure 3.
Ultrastructural analysis of the ECM in dermal biopsies
(A–D) Ultrastructural analysis of the elastic fibers in a skin biopsy of affected individual F1:IV-2 (C and D) and a control subject (A and B).
(E–H) Ultrastructural analysis of collagen fibrils in a skin biopsy of affected individual F1:IV-2 (G and H) and a control subject (E and F). Scale bar represents 2 μm (A, C, E, and G) and scale bar represents 500 nm (B, D, F, and H). The elastin core (dotted while line) is surrounded by a spare mantle of microfibrils in the control subject (red dotted line, white arrow). Note that microfibrils infiltrate into the periphery of the elastic fiber in a skin biopsy of affected individual F1:IV-2 (red dotted line, black triangle). Col, collagen; Ef, elastic fiber; Mf, microfibrils.
LTBP1-deficient ECM responses are variant specific
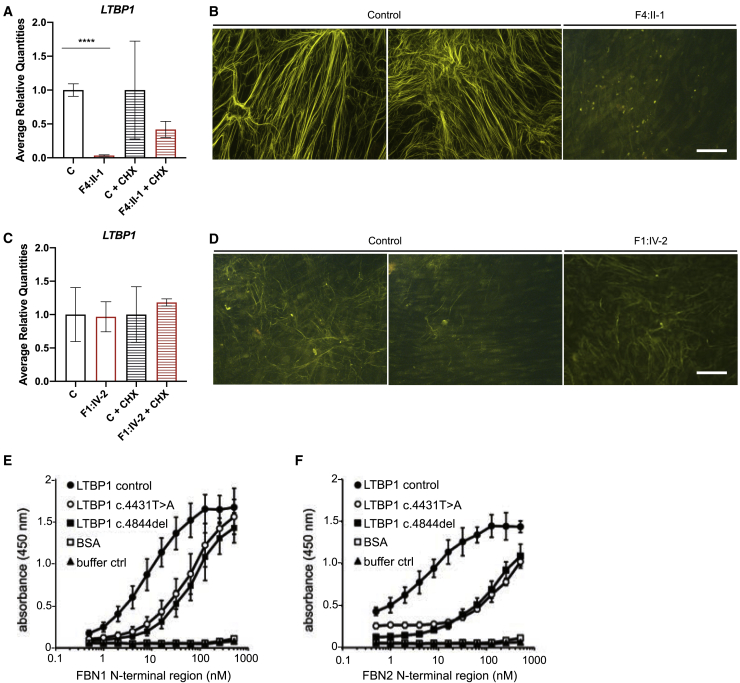
We used dermal fibroblasts cultured from skin biopsies of F1:IV-2 and F4:II-1 in this study. Skin biopsies of F2:V-3, F2:V-4, F2:V-8, F2:V-9, F3:II-1, and F3:II-2 are not available. We characterized LTBP1 transcript and protein levels in dermal fibroblasts derived from individuals F1:IV-2 and F4:II-1. Both variants (c.4844del and c.1342C>T, respectively) are predicted to be susceptible to nonsense-mediated decay (NMD).54 RT-qPCR indicates that LTBP1 mRNA expression is completely abolished in dermal fibroblast cultures of F4:II-1 (c.1342C>T) compared to control fibroblasts but is partly rescued upon cycloheximide treatment, indicative of NMD (Figure 4A). In contrast, LTBP1 mRNA expression in dermal fibroblasts of F1:IV-2 (c.4844del) is present at equal levels as in fibroblasts of control subjects (Figure 4B). In line with the mRNA expression data, immunofluorescent analysis at 9 days post confluency (dpc) shows complete absence of LTBP1 in dermal fibroblasts of F4:II-1 (Figure 4B) but rudimentary LTBP1 fibers in dermal fibroblasts of F1:IV-2 (Figure 4D). The C terminus of LTBP1 interacts with the N terminus of fibrillin-1 and fibrillin-2 (Figure 2). To evaluate the interaction of truncated LTBP1 with fibrillin-1, we used recombinantly expressed C-terminal LTBP1 fragments containing the c.4844del (p.Asn1615Ilefs∗23) and c.4431T>A (p.Cys1477∗) variants in solid-phase binding studies with the N-terminal region of fibrillin-1 and fibrillin-2. Binding studies using surface plasmon resonance (SPR) showed that both mutant LTBP1 fragments show negligible binding to the immobilized N-terminal region of fibrillin-1 when compared to the control fragment (KD = 12 ± 2) (Figures S2A–S2D). Solid-phase binding studies in the opposite direction (LTBP1 immobilized; fibrillin-1 and -2 incubated in solution) also show a significant reduction of binding affinity of the N-terminal regions of fibrillin-1 (8- to 12-fold) and fibrillin-2 (16- to 40-fold) to either mutant LTBP1 fragment (Figures 4E, 4F, andS2E). Taken together, these results suggest that LTBP1 is loosely anchored to the fibrillin microfibril network assembled by dermal fibroblasts derived from F1:IV-2.
Figure 4.
Effect of the LTBP1 variants on the assembly of LTBP1 in the ECM
(A and C) Quantification of LTBP1 expression with and without CHX treatment by RT-qPCR.
(B and D) Representative images of immunofluorescent analysis of LTBP1 in 9 dpc fibroblast cultures derived from affected individuals and control subjects. Scale bar represents 50 μm.
(E and F) Solid-phase binding assay of soluble LTBP1 fragments to immobilized N-terminal region of FBN1. The negative control was incubated with buffer only. Results are representative of three independently conducted experiments. Data are expressed as mean ± standard deviation (SD). ∗∗∗∗p value < 0.0001. Two-tailed unpaired t test with Welch’s correction was used for statistical analysis.
We next analyzed the mRNA expression and protein levels of fibronectin and fibrillin-1, the most important binding partners of LTBP1. FN mRNA (Figures S3A and S3D) and fibronectin protein level in the conditioned media (Figures S3G–S3J) and in the ECM fraction (Figures S3K and S3L) are unaltered in cultured dermal fibroblasts from both affected individuals (F1:IV-2 [c.4844del] and F4:II-1 [c.1342C>T]) compared to control fibroblasts. In cultured dermal fibroblasts of F1:IV-2, FBN1 mRNA expression (Figure S3B) and fibrillin-1 protein level in the conditioned media is equal to control fibroblasts (Figures S3M and S3N), but fibrillin-1 immunofluorescent analysis shows increased fibrillin-1 deposition in the ECM fraction (Figure S3Q). Of note, FBN2 mRNA expression was significantly increased in cultured dermal fibroblasts of F1:IV-2 (Figure S3C). In contrast, cultured dermal fibroblasts of F4:II-1 show significantly reduced FBN1 mRNA but normal FBN2 mRNA levels (Figures S3E and S3F) and, accordingly, significantly decreased fibrillin-1 protein present in the conditioned media compared to control fibroblasts (Figures S3O and S3P). Fibrillin-1 immunofluorescent analysis of the ECM fraction in cultured dermal fibroblasts of F4:II-1 is comparable to control fibroblasts, although the fibers appear more patchy (Figure S3R).
In cultured dermal fibroblasts of F1:IV-2, EFEMP2 (FBLN4) mRNA levels are normal, but EFEMP2 fibers are completely abolished in the ECM fraction (Figures S4A and S4E), suggesting that the presence of the c.4844del variant interferes with EFEMP2 ECM incorporation. In contrast, cultured dermal fibroblasts of F4:II-1 show normal abundance of EFEMP2 fibers in the ECM fraction but significantly decreased EFEMP2 mRNA levels (Figures S4B and S4I). We addressed gene expression and protein levels of other members of the LTBP protein family. Immunofluorescent analysis shows a remarkable increase in LTBP2 fibers in cultured dermal fibroblasts of F1:IV-2 at 9 dpc, while no change is detected for F4:II-1 fibroblasts compared to control fibroblasts (Figures S4C and S4D). LTBP3 expression was significantly increased in cultured dermal fibroblasts of F1:IV-2 (Figure S4G) but significantly decreased in cultured dermal fibroblasts of F4:II-1 (Figure S4K). LTBP4 expression remained equal between cultured dermal fibroblasts of F4:II-1, cultured dermal fibroblasts of F1:IV-2, and control fibroblasts (Figures S4H and S4L). Finally, FBLN5 expression is unchanged in both affected individuals (Figures S4F and S4J). Together, these data indicate that complete loss (c.1342C>T) of LTBP1 and the presence of C-terminally aberrant LTBP1 (c.4844del) each have a different effect on ECM assembly.
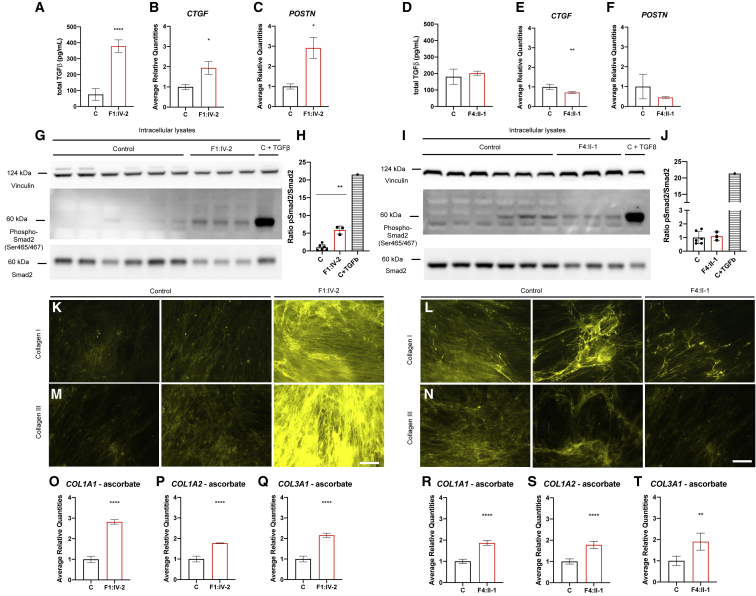
TGFβ signaling response to LTBP1 deficiency is variant specific
LTBP1 interacts with the SLC and plays an important role in regulating the bioavailability of TGFβ in the ECM. Therefore, we investigated the canonical TGFβ pathway in cultured fibroblasts derived from affected individuals and control subjects. Total TGFβ protein levels are significantly increased in conditioned media of F1:IV-2, but not of F4:II-1, compared to control subjects (Figures 5A and 5D). Also, expression levels of the canonical (SMAD2/3-dependent) TGFβ-target genes,55,56 CTGF and POSTN, are significantly upregulated in cultured dermal fibroblasts of F1:IV-2 at 1 dpc compared to control fibroblasts (Figures 5B and 5C), while expression of POSTN remains equal and CTGF expression is significantly decreased in cultured dermal fibroblasts of F4:II-1 at 1 dpc compared to control fibroblasts (Figures 5E and 5F). Accordingly, the pSmad2/Smad2 ratio is significantly increased in cultured dermal fibroblasts of F1:IV-2 at 1 dpc (Figures 5G, 5H, S5A, and S5B) but unaltered in cultured dermal fibroblasts of F4:II-1 compared to controls (Figures 5I and 5J). COL1A1, COL1A2, and COL3A1 are significantly upregulated in cultured dermal fibroblasts of both F1:IV-2 and F4:II-1 at 1 dpc (Figures 5O–5T). Immunofluorescent analysis shows a marked increase in collagen I and collagen III fibers in cultured dermal fibroblasts of F1:IV-2 at 9 dpc (Figures 5K and 5M), while collagen I and collagen III fibers are equal in F4:II-1 fibroblasts compared to control fibroblasts (Figures 4L and 4N). Together, these data indicate that complete loss (c.1342C>T [p.Gln448∗]) of LTBP1 and the presence of C-terminally aberrant LTBP1 (c.4844del [p.Asn1615Ilefs∗23]) each impact TGFβ signaling differently.
Figure 5.
LTBP1 variant-specific canonical TGFβ signaling responses
(A and D) Measurement of total TGFβ in 9 dpc conditioned media obtained from fibroblast cultures derived from individuals F1:IV-2 and F4:II-1 and respective sex- and age-matched control subjects.
(B, C, E, and F) Quantification of CTGF and POSTN expression by RT-qPCR.
(G–J) Immunoblot of intracellular lysates at 1 dpc obtained from fibroblast cultures derived from individuals F1:IV-2 and F4:II-1 and respective sex- and age-matched control subjects. One of the control subjects was stimulated with TGFβ as positive control.
(K–N) Representative images demonstrating immunofluorescent analysis of collagen I and collagen III fibers in 9 dpc fibroblast cultures stimulated with ascorbate. Scale bar represents 50 μm.
(O–T) Quantification of COL1A1, COL1A2, and COL3A1 expression by RT-qPCR after ascorbate stimulation. Data are expressed as mean ± SD. ∗p value < 0.05, ∗∗p value < 0.01, ∗∗∗∗p value < 0.0001. Two-tailed unpaired t test with Welch’s correction was used for statistical analysis.
Ltbp1 deficiency causes ectopic bone and reduced tissue mineral density in the zebrafish skeleton but does not affect cardiac function
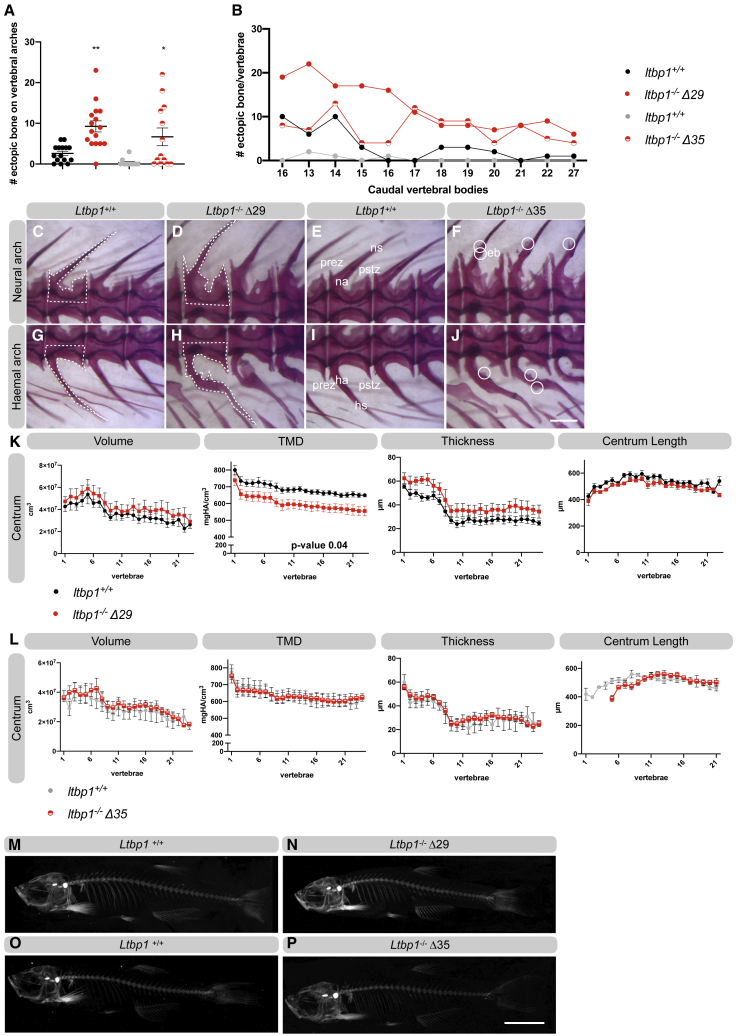
In order to further investigate the impact of ltbp1 deficiency in an in vivo setting, we generated zebrafish models. LTBP1 is well-conserved between humans and zebrafish, and zebrafish Ltbp1 shows a (predicted) domain homology similar to human LTBP1 (Figure S6). However, in contrast to humans, zebrafish only express a long form of the ltbp1, and no other isoforms are present in the genome. Using CRISPR-Cas9 technology, we generated two ltbp1−/− zebrafish models, one harboring a 1 bp deletion, c.3526del, in exon 29 and one harboring a 10 bp deletion, c.4294_4303del, in exon 35 (Figure S7). Both deletions result in a premature stop codon and cause reduced ltbp1 expression at the juvenile stage (Figure S8A). Ltbp1−/−Δ29 zebrafish lack two TGFβ-binding domains, three calcium-binding EGF-like domains, and one EGF-like domain at the Ltbp1 C terminus. Ltbp1−/−Δ35 zebrafish lack the last calcium-binding EGF-like domains and the last EGF-like domain.
Ltbp1−/−Δ29 and ltbp1−/−Δ35 zebrafish have similar weight and length compared to WT siblings, show Mendelian inheritance, and do not show premature mortality (Figures S8B and S8C). Investigation of the skeletal phenotype demonstrated that the neural and haemal arches of the vertebrae of ltbp1−/−Δ29 and ltbp1−/−Δ35 zebrafish have ectopic bone formation (of intramembranous origin) (Figures 6A–6J). In addition, the arch bases that sit on the vertebrae clearly show more intramembranous bone (white dotted lines in Figures 6C, 6D, 6G, and 6H). Quantitative μCT analysis of 4-month-old zebrafish reveals a significant decrease in tissue mineral density (TMD) of the vertebral centrum and a significant decrease in TMD of the neural- and haemal-associated elements of the skeleton in ltbp1−/−Δ29 zebrafish compared to WT siblings (Figures 6K, 6M, 6N, and S9). The volume of these skeletal elements tends to be increased in ltbp1−/−Δ29 zebrafish, although statistical significance is not reached. This finding is further supported by an increased volume of the vertebrae observed in alizarin red-stained ltbp1−/−Δ29 zebrafish vertebral columns (Figures 6D and 6H). Bone thickness also tends to be increased (p value < 0.07) in ltbp1−/−Δ29 zebrafish. Interestingly, quantitative μCT parameters were not different between ltbp1−/−Δ35 zebrafish and WT siblings (Figures 6L, 6O, 6P, and S9). Ltbp1−/−Δ29 and ltbp1−/−Δ35 zebrafish have normal interfrontal, coronal, sagittal, and lambdoid sutures (Figures S8D–S8K) and do not show alterations in cranial morphological structures, including the hyomandibula, premaxilla, and basioccipital bone (Figures S8D–S8K). Because the complete knockout of Ltbp1 in mice causes a severe cardiovascular phenotype, we investigated the cardiac parameters. Assessment of cardiovascular function in adult zebrafish by ultrasound imaging, however, revealed no significant differences between ltbp1−/−Δ29 zebrafish, ltbp1−/−Δ35 zebrafish, and corresponding WT siblings at 10 months of age (Figure S10). Taken together, ltbp1−/−Δ29 zebrafish reveal vertebral hypo-mineralization, voluminous vertebrae, and ectopic bone formation but normal heart function.
Figure 6.
Ltbp1−/−Δ29 zebrafish show hypo-mineralization and voluminous vertebrae with ectopic bone
(A) Quantification of the amount of ectopic bone present on the neural and haemal arches of the caudal vertebrae of ltbp1−/−Δ29 and ltbp1−/−Δ35 zebrafish and corresponding WT siblings. The amount of ectopic bone is significantly increased in ltbp1−/−Δ29 and ltbp1−/−Δ35 zebrafish.
(B) Graphical representation of the amount of ectopic bone on the individually scored VBs of ltbp1−/−Δ29 and ltbp1−/−Δ35 zebrafish and corresponding WT siblings.
(C–J) Representative images of the neural and haemal arches on the vertebrae of ltbp1−/−Δ29 and ltbp1−/−Δ35 zebrafish and their respective WT siblings. Note that the shape of the neural and haemal part of the vertebrae is indicated with dashed lines. Ltbp1−/−Δ29 zebrafish have more erratic and voluminous vertebral shapes than their respective WT siblings. Ectopic bone is indicated with a circle.
(K–L) Quantitative μCT analysis of the vertebral column in five ltbp1−/−Δ29 zebrafish versus five WT siblings and in five ltbp1−/−Δ35 zebrafish versus four WT siblings at the age of 4 months. The bone volume, tissue mineral density (TMD), and bone thickness were calculated from the vertebral centrum. The x axis represents individual abdominal and caudal VB along the anterior-posterior axis. The TMD is significantly reduced in the vertebral centrum of ltbp1−/−Δ29 zebrafish compared to WT siblings. In contrast, equal TMD is observed in the vertebral centrum of ltbp1−/−Δ35 zebrafish compared to WT siblings. Data were analyzed in the R statistical environment.
(M–P) Representative 2D μCT images of the skeleton of ltbp1−/−Δ29 and ltbp1−/−Δ35 zebrafish and corresponding WT siblings.
Data are expressed as mean ± standard error of the mean (SEM) and analyzed in the R statistical environment in (K) and (L). Data are expressed as mean ± SD in (A). ∗p value < 0.05, ∗∗p value < 0.01. Two-tailed unpaired t test with Welch’s correction was used for statistical analysis in (A). Eb, ectopic bone; ha, haemal arch; hs, haemal spine; HA, hydroxyapatite; na, neural arch; ns, neural spine; prez, prezygapophysis; pstz, postzygapophysis; vc, vertebral column.
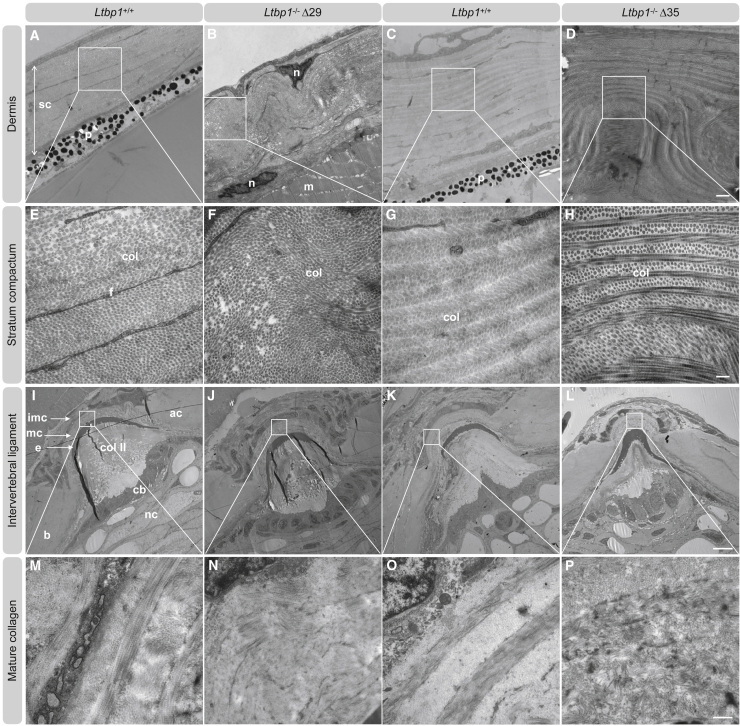
Ltbp1 deficiency causes abnormal collagen fibrillogenesis in zebrafish skin and intervertebral ligaments
TEM analysis of skin biopsies of ltbp1−/−Δ29 and ltbp1−/−Δ35 zebrafish demonstrated an abnormal dermal collagen architecture showing a folded appearance of the typical plywood-like organization. (Figures 7A–7H). In contrast, TEM of the notochord sheet, a part of the intervertebral disc, shows normal diameters and structural organization of collagen type II (Figures S11E–S11H). Also, the immature collagen deposited by osteoblasts in the outer edges of the intervertebral ligament appears normally structured (Figures S11A–S11D). However, the mature collagen structure (Figures 7I–7P) consistently shows a lack of the plywood-like organization with a chaotic assembly of the collagen fibrils in intervertebral ligament samples from ltbp1−/−Δ29 and ltbp1−/−Δ35 zebrafish. Taken together, our experiments highlight a role for ltbp1 in collagen architecture in vivo in zebrafish.
Figure 7.
LTBP1 deficiency causes abnormal collagen fibrillogenesis in skin and intervertebral ligaments
(A–H) Representative images of ultrathin sections taken from the dermis of 4-months old adult ltbp1−/− Δ29 zebrafish, ltbp1−/− Δ35 zebrafish, and corresponding WT siblings. Increased interfibrillar spaces and disorganized collagen architecture are noted in ltbp1−/−Δ29 and ltbp1−/−Δ35 zebrafish samples. Col, collagen; f, fibroblast; m, muscle; n, nucleus; p, pigmentation; sc, stratum compactum. Scale represents 1 μm in (A)–(D) and scale represents 200 nm in (E)–(H).
(I–P) Representative images of ultrathin parasagittal sections showing internal structures of zebrafish vertebral centra and intervertebral ligament of 4-months-old adult ltbp1−/−Δ29 and ltbp1−/−Δ35 zebrafish and corresponding WT siblings. Note that the notochord sheet is composed of collagen type II. Collagen type II is secreted by the chordoblasts lining the notochord sheet on the inside and in between the chordocytes and the notochord sheet. Abnormal mature collagen architecture is noted in adult ltbp1−/−Δ29 and ltbp1−/−Δ35 zebrafish compared to corresponding WT siblings. Ac, autocenter; b, bone; cb, chordoblasts; colII, collagen type II (notochord sheet); e, outer elastin layer; imc, immature collagen; mc, mature collagen; nc, vacuolated notochord cells (chordocytes). Scale represents 200 μm in (I)–(L) and scale represents 500 nm in (M)–(P).
Discussion
We describe an AR CL syndrome caused by bi-allelic truncating variants in LTBP1. The craniofacial features, short stature, brachydactyly, variable craniosynostosis, and variable mild heart defects clearly distinguish this AR CL syndrome from other subtypes of CL syndrome. Because of the pleiotropic manifestations, we propose the name LTBP1-related CL syndrome. The identified premature truncating variants are distributed across LTBP1 and correspond to protein alterations in the second (family 4, c.1342C>T) and third EGF-like domains (family 1, c.4844del) and the 12th (family 3, c.3991_3995del) and 13th calcium-binding EGF-like domains (family 2, c.4431T>A) of the long isoform of LTBP1.1 We demonstrate distinct molecular consequences of truncating variants in LTBP1 depending on their position within the gene. No NMD is observed for the c.4844del variant, allowing for rudimentary (altered) LTBP1 fiber formation in the ECM. In contrast, NMD is observed in the c.1342C>T variant, resulting in an absence of LTBP1 in the ECM layer. Mutant LTBP1 protein produced by c.4844del or c.4431T>A LTBP1 variants shows reduced binding affinity for the N-terminal regions of fibrillin-1 and fibrillin-2 causing loss of function.
Reduced LTBP1 binding to fibrillin-containing microfibrils would yield in large latency complexes (LLCs) that fail to be targeted correctly to the ECM, resulting in their inappropriate activation. Therefore, we hypothesize that the increased TGFβ levels observed in cultured dermal fibroblasts of F1:IV-2 is the result of an unstable anchorage of LTBP1 to fibrillin microfibrils. Our finding of activated TGFβ signaling in cultured dermal fibroblasts of F1:IV-2 that may still express a C-terminally truncated form of LTBP1 is consistent with a previous study in murine skin. Transgenic protein production of a truncated LTBP1 variant that is still capable to bind TGFβ but fails to interact with the ECM because of lack of the known N- and C-terminal ECM-binding regions resulted in an excess of active TGFβ.57,58 Moreover, strongly increased ECM production (as evidenced by mRNA expression and/or protein level of collagens, FBN1 and LTBP2) in cultured dermal fibroblasts expressing the c.4844del variant may be secondary to aberrant canonical TGFβ growth factor activation.55,56 In contrast, absence of LTBP1 in the ECM layer (c.1342>T variant) does not alter canonical TGFβ signaling and does not induce strong alterations of collagen I and III fiber incorporation in the ECM of cultured fibroblasts. Hence, functional redundancy of other LTBP family members may be sufficient for TGFβ transport and sequestering in absence of LTBP1.2 Nevertheless, newly produced collagen might be degraded by other specific factors such as matrix metalloproteinases and trigger other pathological cascades.59
In addition, absence of LTBP1 does not alter fibulin-4 deposition in the ECM layer, while the presence of altered LTBP1 impedes fibulin-4 incorporation into the ECM. Fibulin-4 acts as an adaptor molecule to guide tropoelastin and lysyl oxidase to fibrillin-containing microfibrils.60,61 Efemp2R/R mice have mild elastic fiber alterations62 in line with the observation of mild elastic fiber defects upon ultrastructural analysis of a skin biopsy of the individual harboring the LTBP1 c.4844del variant. Concomitantly, we observed increased deposition of LTBP2 in the ECM. LTBP2 is a known interaction partner of fibulin-5 and facilitates tropoelastin deposition in human dermal fibroblasts.14 It is tempting to hypothesize that LTBP2 and fibulin-5 might compensate for the loss of fibulin-4 incorporation in the ECM in human dermal fibroblasts. Further studies are needed to confirm the distinct molecular consequences related to a loss of LTBP1 or altered LTBP1 level in cultured dermal fibroblast samples derived from other diagnosed individuals with LTBP1 variants in similar regions.
However, some differences in clinical features between F1:IV-2 and F4:II-1 may be at least partly TGFβ related. For instance, F1:IV-2 shows mitral valve prolapse (MVP), which was suggested to be caused by increased TGFβ activity in a mouse model of Marfan syndrome.63 A homozygous premature truncation variant after 171 amino acids in LTBP3 also causes MVP,64 while a heterozygous missense mutation in LTBP3 resulted only in a mildly thickened mitral valve with mild mitral regurgitation.23 Skin fibroblast from individuals who harbor this less severe LTBP3 missense mutation also did not show any signs of increased total or activated levels of TGFβ,23 suggesting that only the LTBP3 truncation variant leads to activated TGFβ and MVP similar to our findings in F1:IV-2. In addition, the occurrence of hernias was reported to be a feature of neonates with Marfan syndrome,65, 66, 67 a disorder suggested to be generally driven by aberrant TGFβ activation. F4:II-1 did not present with mitral valve prolapse or hernias but was initially presented with deformities of the skull. Craniosynostosis, a pathology that is closely linked to dysregulated TGFβ signaling,68 was also reported to be caused by a reduced bioavailability of TGFβ within the bone matrix because of the genetic ablation of Ltbp3 in mice.17,69 Deformities of the skull were also reported in Ltbp1 null mice.30 These reports in mice are consistent with the idea of reduced TGFβ bioavailability in bone of F4:II-1. However, the mechanisms controlling the tissue bioavailability of TGFβ are most likely tissue specific. Depending on the tissue-specific ECM composition and biomechanical properties, LTBP deficiency may have different effects on TGFβ bioavailability. In addition, LTBP1 might have other, yet unknown functions that cannot be compensated by other LTBPs and are causative for the clinical features in the reported probands. These could include unknown roles in modulating the deposition of ECM components or cell-matrix interactions.
Little is known about the function of LTBP1 in chondrogenesis. LTBP1, fibrillin-1, and FN are localized in developing long bones of R. Novergicus. LTBP1 and fibrillin-1 are present in the longitudinal fibrillar structures in the outer periosteum and in the perichondrium and in the layer of osteoblasts adjacent to the surface of newly forming osteoid.70,71 Many microfibrillar genes, including LTBP2, LTBP3, ADAMTS10, ADAMTS17, ADAMTSL2, FBN1, and FBN2, have been associated with short stature.72 FBN1 and FBN2 variants might even cause opposite phenotypes depending on the domain harboring the pathogenic variant.73, 74, 75, 76 How these defects affect ECM interactions, microenvironment, and growth factor signaling pathways in chondrocytes is poorly understood.77 Genes involved in isolated and syndromic forms of craniosynostosis suggest a link between fibroblast growth factor and TGFβ signaling dysregulation,68 which suggests a delicate cellular and molecular interplay between osteoblastogenic and osteoclastogenic pathways.78 Of note, most craniosynostosis syndromes do not present with clear cutaneous manifestation or short stature. In this context, growth factor signaling in fibroblasts may not be fully representative for the molecular consequences in osteogenic pathways. Our study, however, adds an additional contributor to the short stature and craniosynostosis phenotypes.
Our in vivo experiments furthermore provide evidence that Ltbp1 is required for proper cutaneous and skeletal homeostasis in adult zebrafish. Both homozygous mutant zebrafish models have an abnormal dermal collagen architecture showing a folded plywood-like organization, indicating skin redundancy, the hallmark phenotype of CL syndrome, as well as abnormal fibrillogenesis in the intervertebral ligament. Ltbp1−/−Δ29 zebrafish have vertebral hypo-mineralization, voluminous vertebrae, and ectopic bone formation. Ltbp1−/−Δ35 zebrafish show normal mineralization but still display ectopic bone formed by intramembranous ossification. Increased vertebral volume in zebrafish associates with ECM defects.52 PLOD2 deficiency in zebrafish, phenotypically concordant with clinical findings in individuals with Bruck Syndrome, causes loss of the typical hourglass-shape morphology in zebrafish due to excessive bone formation and therefore increases vertebral body thickness and disrupts type 1 collagen fibrillar organization in the bone.52 Collagen maturation defects, which are clearly observed in ltbp1−/−Δ29 and ltbp1−/−Δ35 zebrafish, could potentially contribute to the observed ectopic bone formation. However, neither craniofacial abnormalities nor craniosynostosis were observed in both homozygous mutant zebrafish models. A possible explanation for the lack of these features could be the induction of genetic compensation mechanisms, which could partly rescue the craniosynostosis phenotype.79 A recent study showed that knockdown of ltbp1 leads to abnormal craniofacial cartilage structures in zebrafish embryos,80 suggesting a role in cartilage development, which we did not observe in our models (data not shown). Considering the reduction of mutant ltbp1 mRNA levels in ltbp1−/−Δ29 and ltbp1−/−Δ35 zebrafish (30%–10% of WT Ltbp1 levels, respectively), an RNA-less ltbp1 allele model, which would preclude activation of the genetic compensation mechanisms,81 might be informative in this context. However, we cannot exclude the possibility that mutant Ltbp1 might still retain some level of activity, mitigating the severity of the observed phenotype. The C-terminal TGFβ-binding domains, absent in ltbp1−/−Δ29 zebrafish but present in ltbp1−/−Δ35 zebrafish, may play a role in the observed differences in the bone mineral density. Indeed, excessive TGFβ signaling has been implicated in the pathogenesis of osteogenesis imperfecta (MIM: 259420).82 Unfortunately, this hypothesis could not be confirmed nor rejected because of lack of suitable zebrafish TGFβ antibodies. Further studies should delineate whether aberrant TGFβ signaling exists in ltbp1−/−Δ29 zebrafish using a luciferase reporter assay driven by a TGFβ responsive promotor as feasible readout. Remarkably, Ltbp1L−/− and Ltbp1−/− mice present with truncus arteriosus, interrupted aortic arch, and perinatal lethality. Our observations imply differences in functional redundancy of LTBP family members or other compensatory mechanisms for Ltbp1 deficiency in mice versus humans and teleosts.
In conclusion, we identified bi-allelic truncating variants in LTBP1 as disease causing for CL syndrome with altered skeletal development. Data from in vitro experiments on cultured fibroblasts show that different LTBP1-truncating variants have distinct molecular signatures on ECM development and TGFβ signaling, depending on the absence or presence of mutated protein. Moreover, ltbp1 deficiency in zebrafish confirms a prominent role for Ltbp1 in skeletal morphogenesis in vivo. Taken together, our data underscore the importance of the LTBP1 LLC in matrix assembly and bone homeostasis.
Declaration of interests
P.S. is an employee of Bruker-microCT. The remaining authors declare no competing interests.
Acknowledgments
The authors wish to indicate our deep gratitude to the probands and their families for their contribution to the study. We specially want to thank Jan Willem Bek for implementing the μCT scanning technique in our zebrafish phenotypic studies. Moreover, we want to thank Velislava Zoteva, Petra Vermassen, Hanna De Saffel, Lisa Caboor, and Myriam Claes for their technical support. B.C. is a senior clinical investigator of the Research Foundation Flanders. This work was supported by a starting grant of the Special Research Fund of Ghent University (01N04516C to B.C.), a Methusalem grant (BOFMET2015000401) from the Special Research Fund of Ghent University, a junior fundamental research project grant of the Fund for Scientific Research (G035620N), grants from Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) project numbers 73111208 (SFB 829/ B12), 397484323 (TRR 259/ B09), and FOR2722/ C2 to G.S., and the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement no. 794365 to P.S. F3 was collected as part of the SYNaPS Study Group collaboration funded by The Wellcome Trust and strategic award (Synaptopathies) funding (WT093205 MA and WT104033AIA), and research was conducted as part of the Queen Square Genomics group at University College London, supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre. H.H. is funded by the MRC (MR/S01165X/1, MR/S005021/1, and G0601943), the National Institute for Health Research University College London Hospitals Biomedical Research Centre, Rosetree Trust, Ataxia UK, MSA Trust, Brain Research UK, Sparks GOSH Charity, Muscular Dystrophy UK (MDUK), and Muscular Dystrophy Association (MDA USA). W.G.N. is supported by the Manchester NIHR BRC (IS-BRC-1215-20007).
Published: May 14, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.04.016.
Data and code availability
The published article includes all data and code generated or analyzed during this study.
Web resources
Supplemental information
References
- 1.Rifkin D.B., Rifkin W.J., Zilberberg L. LTBPs in biology and medicine: LTBP diseases. Matrix Biol. 2018;71–72:90–99. doi: 10.1016/j.matbio.2017.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saharinen J., Keski-Oja J. Specific sequence motif of 8-Cys repeats of TGF-beta binding proteins, LTBPs, creates a hydrophobic interaction surface for binding of small latent TGF-beta. Mol. Biol. Cell. 2000;11:2691–2704. doi: 10.1091/mbc.11.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Isogai Z., Ono R.N., Ushiro S., Keene D.R., Chen Y., Mazzieri R., Charbonneau N.L., Reinhardt D.P., Rifkin D.B., Sakai L.Y. Latent transforming growth factor beta-binding protein 1 interacts with fibrillin and is a microfibril-associated protein. J. Biol. Chem. 2003;278:2750–2757. doi: 10.1074/jbc.M209256200. [DOI] [PubMed] [Google Scholar]
- 4.Ono R.N., Sengle G., Charbonneau N.L., Carlberg V., Bächinger H.P., Sasaki T., Lee-Arteaga S., Zilberberg L., Rifkin D.B., Ramirez F. Latent transforming growth factor beta-binding proteins and fibulins compete for fibrillin-1 and exhibit exquisite specificities in binding sites. J. Biol. Chem. 2009;284:16872–16881. doi: 10.1074/jbc.M809348200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirani R., Hanssen E., Gibson M.A. LTBP-2 specifically interacts with the amino-terminal region of fibrillin-1 and competes with LTBP-1 for binding to this microfibrillar protein. Matrix Biol. 2007;26:213–223. doi: 10.1016/j.matbio.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Dallas S.L., Sivakumar P., Jones C.J., Chen Q., Peters D.M., Mosher D.F., Humphries M.J., Kielty C.M. Fibronectin regulates latent transforming growth factor-beta (TGF beta) by controlling matrix assembly of latent TGF beta-binding protein-1. J. Biol. Chem. 2005;280:18871–18880. doi: 10.1074/jbc.M410762200. [DOI] [PubMed] [Google Scholar]
- 7.Massam-Wu T., Chiu M., Choudhury R., Chaudhry S.S., Baldwin A.K., McGovern A., Baldock C., Shuttleworth C.A., Kielty C.M. Assembly of fibrillin microfibrils governs extracellular deposition of latent TGF beta. J. Cell Sci. 2010;123:3006–3018. doi: 10.1242/jcs.073437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noda K., Dabovic B., Takagi K., Inoue T., Horiguchi M., Hirai M., Fujikawa Y., Akama T.O., Kusumoto K., Zilberberg L. Latent TGF-β binding protein 4 promotes elastic fiber assembly by interacting with fibulin-5. Proc. Natl. Acad. Sci. USA. 2013;110:2852–2857. doi: 10.1073/pnas.1215779110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bultmann-Mellin I., Conradi A., Maul A.C., Dinger K., Wempe F., Wohl A.P., Imhof T., Wunderlich F.T., Bunck A.C., Nakamura T. Modeling autosomal recessive cutis laxa type 1C in mice reveals distinct functions for Ltbp-4 isoforms. Dis. Model. Mech. 2015;8:403–415. doi: 10.1242/dmm.018960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bultmann-Mellin I., Essers J., van Heijingen P.M., von Melchner H., Sengle G., Sterner-Kock A. Function of Ltbp-4L and fibulin-4 in survival and elastogenesis in mice. Dis. Model. Mech. 2016;9:1367–1374. doi: 10.1242/dmm.026005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dabovic B., Robertson I.B., Zilberberg L., Vassallo M., Davis E.C., Rifkin D.B. Function of latent TGFβ binding protein 4 and fibulin 5 in elastogenesis and lung development. J. Cell. Physiol. 2015;230:226–236. doi: 10.1002/jcp.24704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumra H., Nelea V., Hakami H., Pagliuzza A., Djokic J., Xu J., Yanagisawa H., Reinhardt D.P. Fibulin-4 exerts a dual role in LTBP-4L-mediated matrix assembly and function. Proc. Natl. Acad. Sci. USA. 2019;116:20428–20437. doi: 10.1073/pnas.1901048116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin S.J., Yanagisawa H. Recent updates on the molecular network of elastic fiber formation. Essays Biochem. 2019;63:365–376. doi: 10.1042/EBC20180052. [DOI] [PubMed] [Google Scholar]
- 14.Hirai M., Horiguchi M., Ohbayashi T., Kita T., Chien K.R., Nakamura T. Latent TGF-beta-binding protein 2 binds to DANCE/fibulin-5 and regulates elastic fiber assembly. EMBO J. 2007;26:3283–3295. doi: 10.1038/sj.emboj.7601768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Todorovic V., Frendewey D., Gutstein D.E., Chen Y., Freyer L., Finnegan E., Liu F., Murphy A., Valenzuela D., Yancopoulos G., Rifkin D.B. Long form of latent TGF-beta binding protein 1 (Ltbp1L) is essential for cardiac outflow tract septation and remodeling. Development. 2007;134:3723–3732. doi: 10.1242/dev.008599. [DOI] [PubMed] [Google Scholar]
- 16.Dabovic B., Levasseur R., Zambuto L., Chen Y., Karsenty G., Rifkin D.B. Osteopetrosis-like phenotype in latent TGF-beta binding protein 3 deficient mice. Bone. 2005;37:25–31. doi: 10.1016/j.bone.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 17.Dabovic B., Chen Y., Colarossi C., Zambuto L., Obata H., Rifkin D.B. Bone defects in latent TGF-beta binding protein (Ltbp)-3 null mice; a role for Ltbp in TGF-beta presentation. J. Endocrinol. 2002;175:129–141. doi: 10.1677/joe.0.1750129. [DOI] [PubMed] [Google Scholar]
- 18.Shipley J.M., Mecham R.P., Maus E., Bonadio J., Rosenbloom J., McCarthy R.T., Baumann M.L., Frankfater C., Segade F., Shapiro S.D. Developmental expression of latent transforming growth factor beta binding protein 2 and its requirement early in mouse development. Mol. Cell. Biol. 2000;20:4879–4887. doi: 10.1128/mcb.20.13.4879-4887.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoue T., Ohbayashi T., Fujikawa Y., Yoshida H., Akama T.O., Noda K., Horiguchi M., Kameyama K., Hata Y., Takahashi K. Latent TGF-β binding protein-2 is essential for the development of ciliary zonule microfibrils. Hum. Mol. Genet. 2014;23:5672–5682. doi: 10.1093/hmg/ddu283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ali M., McKibbin M., Booth A., Parry D.A., Jain P., Riazuddin S.A., Hejtmancik J.F., Khan S.N., Firasat S., Shires M. Null mutations in LTBP2 cause primary congenital glaucoma. Am. J. Hum. Genet. 2009;84:664–671. doi: 10.1016/j.ajhg.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Désir J., Sznajer Y., Depasse F., Roulez F., Schrooyen M., Meire F., Abramowicz M. LTBP2 null mutations in an autosomal recessive ocular syndrome with megalocornea, spherophakia, and secondary glaucoma. Eur. J. Hum. Genet. 2010;18:761–767. doi: 10.1038/ejhg.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haji-Seyed-Javadi R., Jelodari-Mamaghani S., Paylakhi S.H., Yazdani S., Nilforushan N., Fan J.B., Klotzle B., Mahmoudi M.J., Ebrahimian M.J., Chelich N. LTBP2 mutations cause Weill-Marchesani and Weill-Marchesani-like syndrome and affect disruptions in the extracellular matrix. Hum. Mutat. 2012;33:1182–1187. doi: 10.1002/humu.22105. [DOI] [PubMed] [Google Scholar]
- 23.McInerney-Leo A.M., Le Goff C., Leo P.J., Kenna T.J., Keith P., Harris J.E., Steer R., Bole-Feysot C., Nitschke P., Kielty C. Mutations in LTBP3 cause acromicric dysplasia and geleophysic dysplasia. J. Med. Genet. 2016;53:457–464. doi: 10.1136/jmedgenet-2015-103647. [DOI] [PubMed] [Google Scholar]
- 24.Noor A., Windpassinger C., Vitcu I., Orlic M., Rafiq M.A., Khalid M., Malik M.N., Ayub M., Alman B., Vincent J.B. Oligodontia is caused by mutation in LTBP3, the gene encoding latent TGF-beta binding protein 3. Am. J. Hum. Genet. 2009;84:519–523. doi: 10.1016/j.ajhg.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo D.C., Regalado E.S., Pinard A., Chen J., Lee K., Rigelsky C., Zilberberg L., Hostetler E.M., Aldred M., Wallace S.E., University of Washington Center for Mendelian Genomics LTBP3 Pathogenic Variants Predispose Individuals to Thoracic Aortic Aneurysms and Dissections. Am. J. Hum. Genet. 2018;102:706–712. doi: 10.1016/j.ajhg.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ritelli M., Cammarata-Scalisi F., Cinquina V., Colombi M. Clinical and molecular characterization of an 18-month-old infant with autosomal recessive cutis laxa type 1C due to a novel LTBP4 pathogenic variant, and literature review. Mol. Genet. Genomic Med. 2019;7:e00735. doi: 10.1002/mgg3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Callewaert B., Su C.T., Van Damme T., Vlummens P., Malfait F., Vanakker O., Schulz B., Mac Neal M., Davis E.C., Lee J.G. Comprehensive clinical and molecular analysis of 12 families with type 1 recessive cutis laxa. Hum. Mutat. 2013;34:111–121. doi: 10.1002/humu.22165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su C.T., Huang J.W., Chiang C.K., Lawrence E.C., Levine K.L., Dabovic B., Jung C., Davis E.C., Madan-Khetarpal S., Urban Z. Latent transforming growth factor binding protein 4 regulates transforming growth factor beta receptor stability. Hum. Mol. Genet. 2015;24:4024–4036. doi: 10.1093/hmg/ddv139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horiguchi M., Todorovic V., Hadjiolova K., Weiskirchen R., Rifkin D.B. Abrogation of both short and long forms of latent transforming growth factor-beta binding protein-1 causes defective cardiovascular development and is perinatally lethal. Matrix Biol. 2015;43:61–70. doi: 10.1016/j.matbio.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drews F., Knöbel S., Moser M., Muhlack K.G., Mohren S., Stoll C., Bosio A., Gressner A.M., Weiskirchen R. Disruption of the latent transforming growth factor-beta binding protein-1 gene causes alteration in facial structure and influences TGF-beta bioavailability. Biochim. Biophys. Acta. 2008;1783:34–48. doi: 10.1016/j.bbamcr.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 31.Dietzel E., Weiskirchen S., Floehr J., Horiguchi M., Todorovic V., Rifkin D.B., Jahnen-Dechent W., Weiskirchen R. Latent TGF-β binding protein-1 deficiency decreases female fertility. Biochem. Biophys. Res. Commun. 2017;482:1387–1392. doi: 10.1016/j.bbrc.2016.12.046. [DOI] [PubMed] [Google Scholar]
- 32.Biesecker L.G., Adam M.P., Alkuraya F.S., Amemiya A.R., Bamshad M.J., Beck A.E., Bennett J.T., Bird L.M., Carey J.C., Chung B. A dyadic approach to the delineation of diagnostic entities in clinical genomics. Am. J. Hum. Genet. 2021;108:8–15. doi: 10.1016/j.ajhg.2020.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mencacci N.E., Kamsteeg E.J., Nakashima K., R’Bibo L., Lynch D.S., Balint B., Willemsen M.A., Adams M.E., Wiethoff S., Suzuki K. De Novo Mutations in PDE10A Cause Childhood-Onset Chorea with Bilateral Striatal Lesions. Am. J. Hum. Genet. 2016;98:763–771. doi: 10.1016/j.ajhg.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monies D., Abouelhoda M., Assoum M., Moghrabi N., Rafiullah R., Almontashiri N., Alowain M., Alzaidan H., Alsayed M., Subhani S. Lessons Learned from Large-Scale, First-Tier Clinical Exome Sequencing in a Highly Consanguineous Population. Am. J. Hum. Genet. 2019;104:1182–1201. doi: 10.1016/j.ajhg.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huysseune A., Sire J.Y. Bone and cartilage resorption in relation to tooth development in the anterior part of the mandible in cichlid fish: a light and TEM study. Anat. Rec. 1992;234:1–14. doi: 10.1002/ar.1092340102. [DOI] [PubMed] [Google Scholar]
- 37.Sengle G., Charbonneau N.L., Ono R.N., Sasaki T., Alvarez J., Keene D.R., Bächinger H.P., Sakai L.Y. Targeting of bone morphogenetic protein growth factor complexes to fibrillin. J. Biol. Chem. 2008;283:13874–13888. doi: 10.1074/jbc.M707820200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hiepen C., Jatzlau J., Hildebrandt S., Kampfrath B., Goktas M., Murgai A., Cuellar Camacho J.L., Haag R., Ruppert C., Sengle G. BMPR2 acts as a gatekeeper to protect endothelial cells from increased TGFβ responses and altered cell mechanics. PLoS Biol. 2019;17:e3000557. doi: 10.1371/journal.pbio.3000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mularczyk E.J., Singh M., Godwin A.R.F., Galli F., Humphreys N., Adamson A.D., Mironov A., Cain S.A., Sengle G., Boot-Handford R.P. ADAMTS10-mediated tissue disruption in Weill-Marchesani syndrome. Hum. Mol. Genet. 2018;27:3675–3687. doi: 10.1093/hmg/ddy276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pilecki B., Holm A.T., Schlosser A., Moeller J.B., Wohl A.P., Zuk A.V., Heumüller S.E., Wallis R., Moestrup S.K., Sengle G. Characterization of Microfibrillar-associated Protein 4 (MFAP4) as a Tropoelastin- and Fibrillin-binding Protein Involved in Elastic Fiber Formation. J. Biol. Chem. 2016;291:1103–1114. doi: 10.1074/jbc.M115.681775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vanhauwaert S., Van Peer G., Rihani A., Janssens E., Rondou P., Lefever S., De Paepe A., Coucke P.J., Speleman F., Vandesompele J., Willaert A. Expressed repeat elements improve RT-qPCR normalization across a wide range of zebrafish gene expression studies. PLoS ONE. 2014;9:e109091. doi: 10.1371/journal.pone.0109091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Syx D., Van Damme T., Symoens S., Maiburg M.C., van de Laar I., Morton J., Suri M., Del Campo M., Hausser I., Hermanns-Lê T. Genetic heterogeneity and clinical variability in musculocontractural Ehlers-Danlos syndrome caused by impaired dermatan sulfate biosynthesis. Hum. Mutat. 2015;36:535–547. doi: 10.1002/humu.22774. [DOI] [PubMed] [Google Scholar]
- 43.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wohl A.P., Troilo H., Collins R.F., Baldock C., Sengle G. Extracellular Regulation of Bone Morphogenetic Protein Activity by the Microfibril Component Fibrillin-1. J. Biol. Chem. 2016;291:12732–12746. doi: 10.1074/jbc.M115.704734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boel A., Steyaert W., De Rocker N., Menten B., Callewaert B., De Paepe A., Coucke P., Willaert A. BATCH-GE: Batch analysis of Next-Generation Sequencing data for genome editing assessment. Sci. Rep. 2016;6:30330. doi: 10.1038/srep30330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beyens A., Albuisson J., Boel A., Al-Essa M., Al-Manea W., Bonnet D., Bostan O., Boute O., Busa T., Canham N. Arterial tortuosity syndrome: 40 new families and literature review. Genet. Med. 2018;20:1236–1245. doi: 10.1038/gim.2017.253. [DOI] [PubMed] [Google Scholar]
- 47.Westerfield M., Doerry E., Kirkpatrick A.E., Douglas S.A. Zebrafish informatics and the ZFIN database. Methods Cell Biol. 1999;60:339–355. doi: 10.1016/s0091-679x(08)61909-3. [DOI] [PubMed] [Google Scholar]
- 48.Hein S.J., Lehmann L.H., Kossack M., Juergensen L., Fuchs D., Katus H.A., Hassel D. Advanced echocardiography in adult zebrafish reveals delayed recovery of heart function after myocardial cryoinjury. PLoS ONE. 2015;10:e0122665. doi: 10.1371/journal.pone.0122665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang L.W., Huttner I.G., Santiago C.F., Kesteven S.H., Yu Z.Y., Feneley M.P., Fatkin D. Standardized echocardiographic assessment of cardiac function in normal adult zebrafish and heart disease models. Dis. Model. Mech. 2017;10:63–76. doi: 10.1242/dmm.026989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang H., Dvornikov A.V., Huttner I.G., Ma X., Santiago C.F., Fatkin D., Xu X. A Langendorff-like system to quantify cardiac pump function in adult zebrafish. Dis. Model. Mech. 2018;11:dmm034819. doi: 10.1242/dmm.034819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakata-Haga H., Uchishiba M., Shimada H., Tsukada T., Mitani M., Arikawa T., Shoji H., Hatta T. A rapid and nondestructive protocol for whole-mount bone staining of small fish and Xenopus. Sci. Rep. 2018;8:7453. doi: 10.1038/s41598-018-25836-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gistelinck C., Witten P.E., Huysseune A., Symoens S., Malfait F., Larionova D., Simoens P., Dierick M., Van Hoorebeke L., De Paepe A. Loss of Type I Collagen Telopeptide Lysyl Hydroxylation Causes Musculoskeletal Abnormalities in a Zebrafish Model of Bruck Syndrome. J. Bone Miner. Res. 2016;31:1930–1942. doi: 10.1002/jbmr.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hur M., Gistelinck C.A., Huber P., Lee J., Thompson M.H., Monstad-Rios A.T., Watson C.J., McMenamin S.K., Willaert A., Parichy D.M. MicroCT-based phenomics in the zebrafish skeleton reveals virtues of deep phenotyping in a distributed organ system. eLife. 2017;6:e26014. doi: 10.7554/eLife.26014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Popp M.W., Maquat L.E. Leveraging Rules of Nonsense-Mediated mRNA Decay for Genome Engineering and Personalized Medicine. Cell. 2016;165:1319–1322. doi: 10.1016/j.cell.2016.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verrecchia F., Chu M.L., Mauviel A. Identification of novel TGF-beta /Smad gene targets in dermal fibroblasts using a combined cDNA microarray/promoter transactivation approach. J. Biol. Chem. 2001;276:17058–17062. doi: 10.1074/jbc.M100754200. [DOI] [PubMed] [Google Scholar]
- 56.Gressner O.A., Lahme B., Siluschek M., Rehbein K., Weiskirchen R., Gressner A.M. Connective tissue growth factor is a Smad2 regulated amplifier of transforming growth factor beta actions in hepatocytes--but without modulating bone morphogenetic protein 7 signaling. Hepatology. 2009;49:2021–2030. doi: 10.1002/hep.22850. [DOI] [PubMed] [Google Scholar]
- 57.Mazzieri R., Jurukovski V., Obata H., Sung J., Platt A., Annes E., Karaman-Jurukovska N., Gleizes P.E., Rifkin D.B. Expression of truncated latent TGF-beta-binding protein modulates TGF-beta signaling. J. Cell Sci. 2005;118:2177–2187. doi: 10.1242/jcs.02352. [DOI] [PubMed] [Google Scholar]
- 58.Zeyer K.A., Reinhardt D.P. Fibrillin-containing microfibrils are key signal relay stations for cell function. J. Cell Commun. Signal. 2015;9:309–325. doi: 10.1007/s12079-015-0307-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Doren S.R. Matrix metalloproteinase interactions with collagen and elastin. Matrix Biol. 2015;44–46:224–231. doi: 10.1016/j.matbio.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choudhury R., McGovern A., Ridley C., Cain S.A., Baldwin A., Wang M.C., Guo C., Mironov A., Jr., Drymoussi Z., Trump D. Differential regulation of elastic fiber formation by fibulin-4 and -5. J. Biol. Chem. 2009;284:24553–24567. doi: 10.1074/jbc.M109.019364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Horiguchi M., Inoue T., Ohbayashi T., Hirai M., Noda K., Marmorstein L.Y., Yabe D., Takagi K., Akama T.O., Kita T. Fibulin-4 conducts proper elastogenesis via interaction with cross-linking enzyme lysyl oxidase. Proc. Natl. Acad. Sci. USA. 2009;106:19029–19034. doi: 10.1073/pnas.0908268106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hanada K., Vermeij M., Garinis G.A., de Waard M.C., Kunen M.G., Myers L., Maas A., Duncker D.J., Meijers C., Dietz H.C. Perturbations of vascular homeostasis and aortic valve abnormalities in fibulin-4 deficient mice. Circ. Res. 2007;100:738–746. doi: 10.1161/01.RES.0000260181.19449.95. [DOI] [PubMed] [Google Scholar]
- 63.Ng C.M., Cheng A., Myers L.A., Martinez-Murillo F., Jie C., Bedja D., Gabrielson K.L., Hausladen J.M., Mecham R.P., Judge D.P., Dietz H.C. TGF-beta-dependent pathogenesis of mitral valve prolapse in a mouse model of Marfan syndrome. J. Clin. Invest. 2004;114:1586–1592. doi: 10.1172/JCI22715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dugan S.L., Temme R.T., Olson R.A., Mikhailov A., Law R., Mahmood H., Noor A., Vincent J.B. New recessive truncating mutation in LTBP3 in a family with oligodontia, short stature, and mitral valve prolapse. Am. J. Med. Genet. A. 2015;167:1396–1399. doi: 10.1002/ajmg.a.37049. [DOI] [PubMed] [Google Scholar]
- 65.Parida S.K., Kriss V.M., Hall B.D. Hiatus/paraesophageal hernias in neonatal Marfan syndrome. Am. J. Med. Genet. 1997;72:156–158. doi: 10.1002/(sici)1096-8628(19971017)72:2<156::aid-ajmg6>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 66.Ming J.E., McDonald-McGinn D.M., Megerian T.E., Driscoll D.A., Elias E.R., Russell B.M., Irons M., Emanuel B.S., Markowitz R.I., Zackai E.H. Skeletal anomalies and deformities in patients with deletions of 22q11. Am. J. Med. Genet. 1997;72:210–215. doi: 10.1002/(sici)1096-8628(19971017)72:2<210::aid-ajmg16>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 67.Herman T.E., Siegel M.J., Mathur A., Vachharajani A. Neonatal marfan syndrome with hiatus hernia and intrathoracic stomach. J. Perinatol. 2013;33:652–653. doi: 10.1038/jp.2013.15. [DOI] [PubMed] [Google Scholar]
- 68.Chim H., Manjila S., Cohen A.R., Gosain A.K. Molecular signaling in pathogenesis of craniosynostosis: the role of fibroblast growth factor and transforming growth factor-β. Neurosurg. Focus. 2011;31:E7. doi: 10.3171/2011.5.FOCUS1197. [DOI] [PubMed] [Google Scholar]
- 69.Dabovic B., Chen Y., Colarossi C., Obata H., Zambuto L., Perle M.A., Rifkin D.B. Bone abnormalities in latent TGF-[beta] binding protein (Ltbp)-3-null mice indicate a role for Ltbp-3 in modulating TGF-[beta] bioavailability. J. Cell Biol. 2002;156:227–232. doi: 10.1083/jcb.200111080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dallas S.L. Measuring interactions between ECM and TGF beta-like proteins. Methods Mol. Biol. 2000;139:231–243. doi: 10.1385/1-59259-063-2:231. [DOI] [PubMed] [Google Scholar]
- 71.Dallas S.L., Keene D.R., Bruder S.P., Saharinen J., Sakai L.Y., Mundy G.R., Bonewald L.F. Role of the latent transforming growth factor beta binding protein 1 in fibrillin-containing microfibrils in bone cells in vitro and in vivo. J. Bone Miner. Res. 2000;15:68–81. doi: 10.1359/jbmr.2000.15.1.68. [DOI] [PubMed] [Google Scholar]
- 72.Stanley S., Balic Z., Hubmacher D. Acromelic dysplasias: how rare musculoskeletal disorders reveal biological functions of extracellular matrix proteins. Ann. N Y Acad. Sci. 2021;1490:57–76. doi: 10.1111/nyas.14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Le Goff C., Mahaut C., Wang L.W., Allali S., Abhyankar A., Jensen S., Zylberberg L., Collod-Beroud G., Bonnet D., Alanay Y. Mutations in the TGFβ binding-protein-like domain 5 of FBN1 are responsible for acromicric and geleophysic dysplasias. Am. J. Hum. Genet. 2011;89:7–14. doi: 10.1016/j.ajhg.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dietz H.C., Cutting G.R., Pyeritz R.E., Maslen C.L., Sakai L.Y., Corson G.M., Puffenberger E.G., Hamosh A., Nanthakumar E.J., Curristin S.M. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1991;352:337–339. doi: 10.1038/352337a0. [DOI] [PubMed] [Google Scholar]
- 75.Putnam E.A., Zhang H., Ramirez F., Milewicz D.M. Fibrillin-2 (FBN2) mutations result in the Marfan-like disorder, congenital contractural arachnodactyly. Nat. Genet. 1995;11:456–458. doi: 10.1038/ng1295-456. [DOI] [PubMed] [Google Scholar]
- 76.Peeters S., Decramer A., Cain S.A., Houpt P., Verstreken F., Noyez J., Hermans C., Jacobs W., Lammens M., Fransen E. Delineation of a new fibrillino-2-pathy with evidence for a role of FBN2 in the pathogenesis of carpal tunnel syndrome. J. Med. Genet. 2020 doi: 10.1136/jmedgenet-2020-107085. jmedgenet-2020-107085. [DOI] [PubMed] [Google Scholar]
- 77.Delhon L., Mahaut C., Goudin N., Gaudas E., Piquand K., Le Goff W., Cormier-Daire V., Le Goff C. Impairment of chondrogenesis and microfibrillar network in Adamtsl2 deficiency. FASEB J. 2019;33:2707–2718. doi: 10.1096/fj.201800753RR. [DOI] [PubMed] [Google Scholar]
- 78.Beederman M., Farina E.M., Reid R.R. Molecular basis of cranial suture biology and disease: Osteoblastic and osteoclastic perspectives. Genes Dis. 2014;1:120–125. doi: 10.1016/j.gendis.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.El-Brolosy M.A., Stainier D.Y.R. Genetic compensation: A phenomenon in search of mechanisms. PLoS Genet. 2017;13:e1006780. doi: 10.1371/journal.pgen.1006780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xiong Y.T., Sun R.R., Li J.Y., Wu Y., Zhang J.J. Latent TGF-beta binding protein-1 plays an important role in craniofacial development. J. Appl. Oral Sci. 2020 doi: 10.1590/1678-7757-2020-0262. Published online November 4, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tessadori F., de Bakker D.E.M., Barske L., Nelson N., Algra H.A., Willekers S., Nichols J.T., Crump J.G., Bakkers J. Zebrafish prrx1a mutants have normal hearts. Nature. 2020;585:E14–E16. doi: 10.1038/s41586-020-2674-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grafe I., Yang T., Alexander S., Homan E.P., Lietman C., Jiang M.M., Bertin T., Munivez E., Chen Y., Dawson B. Excessive transforming growth factor-β signaling is a common mechanism in osteogenesis imperfecta. Nat. Med. 2014;20:670–675. doi: 10.1038/nm.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all data and code generated or analyzed during this study.