Summary
Dysregulated transforming growth factor TGF-β signaling underlies the pathogenesis of genetic disorders affecting the connective tissue such as Loeys-Dietz syndrome. Here, we report 12 individuals with bi-allelic loss-of-function variants in IPO8 who presented with a syndromic association characterized by cardio-vascular anomalies, joint hyperlaxity, and various degree of dysmorphic features and developmental delay as well as immune dysregulation; the individuals were from nine unrelated families. Importin 8 belongs to the karyopherin family of nuclear transport receptors and was previously shown to mediate TGF-β-dependent SMADs trafficking to the nucleus in vitro. The important in vivo role of IPO8 in pSMAD nuclear translocation was demonstrated by CRISPR/Cas9-mediated inactivation in zebrafish. Consistent with IPO8’s role in BMP/TGF-β signaling, ipo8−/− zebrafish presented mild to severe dorso-ventral patterning defects during early embryonic development. Moreover, ipo8−/− zebrafish displayed severe cardiovascular and skeletal defects that mirrored the human phenotype. Our work thus provides evidence that IPO8 plays a critical and non-redundant role in TGF-β signaling during development and reinforces the existing link between TGF-β signaling and connective tissue defects.
Keywords: IPO8, connective tissue disorder, TGF-β signaling, Loeys-Dietz syndrome, joint hyperlaxity, arterial dilatation
Main text
First isolated as a RanGTP-binding protein, importin 8 encoded by IPO8 (MIM: 605600) was then identified as a member of the β-karyopherin family, the largest group of nuclear transport receptors.1,2 Accordingly, in vitro studies have implicated importin 8 in cytoplasm-to-nucleus shuttling of a broad spectrum of cargos, including the signal recognition particle protein SRP19,3 Argonaute-microRNAs complexes,4,5 the c-Jun protein,6 the NF-κB p65 subunit,7 the eukaryotic translation initiation factor eIF4E,8 and a set of receptor-activated mothers against decapentaplegic homolog (SMAD) transcription factors9 that play a critical role downstream of the large family of tumor growth factor β (TGF-β) and bone morphogenetic protein (BMP) cytokines. In contrast, in vivo data on IPO8 functions are lacking except for a report in Drosophila showing that Msk, the ortholog of mammalian importins 7 and 8 (which share 60% homology), mediates nuclear accumulation of phosphorylated MAD downstream of decapentaplegic, the homolog of mammalian BMP.10 Msk inactivation was embryonically lethal, suggesting its key role in Drosophila development, but no link was clearly established with impairment of BMP signaling.10
Strikingly, the TGF-β/BMP cytokine family exerts pleiotropic functions during embryonic development, tissue homeostasis, and tissue repair as well as within the immune system.11 Accordingly, dysregulation of TGF-β signaling is the cause of severe congenital disorders characterized by developmental defects with or without impaired immune regulation.12 This is notably the case of Loeys-Dietz syndrome (LDS), which is caused by heterozygous loss-of-function (LOF) variants in TGFBR1 (MIM: 190181), TGFBR2 (MIM: 190182), TGFB2 (MIM: 190220) or TGFB3 (MIM: 190230), and SMAD2 (MIM: 601366) or SMAD3 (MIM: 603109);13 of Shprintzen-Goldberg syndrome (SGS), which results from heterozygous variants in SKI (MIM: 182212), a negative regulator of the TGF-β signaling pathway;14 and of Marfan syndrome (MFS) (MIM: 154700) caused by variants in FBN1 (MIM: 134797), encoding fibrillin-1, the main component of extracellular matrix microfibrils that, in turn, scaffolds latent TGF-β. Thus, individuals with LDS, SGS, and MFS display systemic connective tissue disorders of variable expressivity, which notably affect the vascular tree and the skeleton, causing life-threatening arterial aneurysms, dysmorphic features, pectus deformity, scoliosis, and joint hyperlaxity.14,15 In keeping with the immunoregulatory functions of TGF-β,16,17 developmental defects of LDS have been associated with increased frequency of allergic manifestations18 and a 10-fold increase in the risk of inflammatory bowel diseases (IBDs).19 More recently, bi-allelic LOF variants in TGFB1 (MIM: 190180) were identified as the cause of a severe syndrome combining very early-onset IBD and encephalopathy.20
Here, we report 12 individuals with bi-allelic pathogenic IPO8 variants who displayed a complex syndrome reminiscent of LDS and SGS that variably combined cardiovascular, neurologic, skeletal and immunologic abnormalities along with dysmorphic features; the individuals are from nine unrelated families (Table 1, Figures 1 and S1). Affected individuals were recruited through an international collaborative effort facilitated by Genematcher.21 All procedures were performed in accordance with the Helsinki Declaration and were approved by the ethics committees and the institutional review boards from each center. Sequencing (targeted/whole-exome sequencing and Sanger sequencing) was performed after obtaining informed written consent from all affected individuals or their legal guardians. Table 1 summarizes the main phenotypic features in the cohort. Seven individuals showed early-onset (before the age of 1 year for the youngest) dilatation of the ascending aorta. Individuals 1 and 2 (I-1 and I-2), the two oldest siblings (59 and 53 years old at the time of the first assessment, respectively) had diffuse arterial frailty with multiple aneurysms affecting the abdominal aorta, iliac, coronary, and renal arteries and, in I-1, the thoracic aorta (Figure 1A). Arterial tortuosity similar to that observed in LDS22 was noted in four individuals. Recurrent spontaneous pneumothorax was noted in I-1 and I-2. Pulmonary emphysema was present in I-1 (Figure 1B) as well as in two younger individuals (I-5 and I-6). Facial dysmorphism was observed in ten individuals (Figure 1C, Table 1), while 11 had joint hyperlaxity complicated by multiple joint dislocations. All displayed skeletal features, including scoliosis (n = 9), pectus excavatum or carinatum (n = 6), arachnodactyly (n = 6), or pes planus or talipes equinovarus (n = 6) (Figures 1D–1F and S1). Eight individuals developed myopia complicated by retinal detachment for two of them and by early-onset cataract for two others, but none of them had lens dislocation. In line with a generalized connective tissue disorder, skin hyperextensibility and/or hernia were observed in 11 individuals (Figure 1G). Delayed motor milestone presumably consecutive to joint instability was observed in seven individuals; four of them also had mild (I-5, I-6, and I-8) or severe (I-7) intellectual disability. Predisposition to allergic or inflammatory diseases, which was previously documented in LDS,18,19 was evidenced in six individuals (I-5, I-6, I-7, I-8, I-10, and I-12), some of whom also displayed immunological parameters consistent with impaired TGF-β signaling, including hyperIgE, hyperIgG, hypoIgA, and hypereosinophilia.
Table 1.
Clinical features of individuals carrying IPO8 variants
| Family 1 | Family 2 | Family 3 | Family 4 | Family 5 | Family 6 | Family 7 | Family 8 | Family 9 | Total | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Individual | I-1 | I-2 | I-3 | I-4 | I-5 | I-6 | I-7 | I-8 | I-9 | I-10 | I-11 | I-12 | – |
| Sex | M | F | M | M | F | M | M | M | F | F | M | F | – |
| Age at last examination | 62 years | 62 years | 1 year 8 months | 2 years 2 months | 16 years | 15 years | 9 years 6 months | 13 years | 22 years | 33 years | 7 years 6 months | 26 years | – |
| Vascular abnormalities | |||||||||||||
| Dilatation of the ascending aorta | yes | yes | yes | no | yes | yes | yes | yes | yes | yes | yes | yes | 11/12 |
| Other abnormal great vessels | dilated and calcified iliac arteries, AAA | calcified coronary arteries, AAA, dilated iliac arteries | no | no | carotid artery tortuosity | carotid artery tortuosity | no | no | carotid artery tortuosity | no | no | internal carotid tortuosity and ectasia | 6/12 |
| Heart malformation | MVP | no | ASD, left atrium and ventricle mild dilatation | no | ASD, VSD | no | N/A | N/A | ASD | ASD, VSD | MVP | no | 6/10 |
| Ocular abnormalities | |||||||||||||
| Myopia | severe | severe | no | no | severe | no | no | severe | severe | mild | severe | severe | 8/12 |
| Retinal detachment | yes | yes | no | no | no | no | no | no | no | no | no | no | 2/12 |
| Dysmorphic features | |||||||||||||
| Proptosis | no | no | yes | no | no | yes | yes | no | yes | yes | yes | yes | 7/12 |
| Micrognathia | no | no | yes | yes | yes | yes | yes | no | yes | no | no | yes | 7/12 |
| Hypertelorism | no | no | yes | yes | yes | yes | yes | no | no | no | yes | no | 6/12 |
| Frontal bossing | no | no | yes | no | yes | yes | no | no | no | no | no | no | 3/12 |
| Ptosis | no | no | yes | no | no | no | yes | no | no | no | yes | no | 3/12 |
| Abnormal palate | no | no | cleft uvula | no | no | no | no | no | no | no | cleft uvula | no | 2/12 |
| Skeletal abnormalities | |||||||||||||
| Hyperlaxity | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes | no | yes | 11/12 |
| Recurrent joint dislocations | yes | yes | yes | no | no | no | yes | no | yes | yes | no | yes | 7/12 |
| Pectus | no | no | carinatum | carinatum | excavatum | excavatum | no | no | excavatum | no | carinatum | no | 6/12 |
| Scoliosis | yes | yes | no | no | yes | yes | yes | yes | yes | yes | yes | no | 9/12 |
| Arachnodactyly | no | no | yes | yes | yes | yes | no | no | yes | no | yes | no | 6/12 |
| Feet malposition | no | no | yes | yes | no | no | no | yes | yes | yes | yes | no | 6/12 |
| Connective tissue abnormalities | |||||||||||||
| Skin hyperextensibility | no | no | yes | yes | yes | yes | no | yes | no | yes | yes | yes | 8/12 |
| Hernia | umbilical hernia | spigelian and umbilical hernia | inguinal, umbilical, diaphragmatic hernia | umbilical hernia | umbilical and inguinal hernia | umbilical and inguinal hernia | umbilical and inguinal hernia | umbilical hernia | no | umbilical and abdominal hernia | umbilical hernia | umbilical and abdominal hernia | 11/12 |
| Developmental delay/intellectual disability | no | no | yes | yes | yes | yes | yes | yes | no | no | yes | no | 7/12 |
| Immunological abnormalities | |||||||||||||
| HyperIgE | N/A | N/A | N/A | N/A | yes | yes | N/A | yes | N/A | N/A | N/A | N/A | 3/3 |
| HyperIgG | N/A | N/A | N/A | N/A | yes | yes | N/A | yes | N/A | N/A | N/A | N/A | 3/3 |
| HypoIgA | N/A | N/A | N/A | N/A | yes | yes | N/A | yes | N/A | N/A | no | N/A | 3/4 |
| Hypereosinophilia | mild | N/A | N/A | N/A | yes | yes | N/A | yes | N/A | N/A | no | N/A | 4/5 |
| Intestinal inflammation | no | no | no | no | severe colitis | celiac disease/ gastritis | no | dysimmune gastroenterocolitis | no | no | chronic gastritis and duodenitis | celiac disease | 5/12 |
| Allergic symptoms | no | no | no | no | asthma, eczema | asthma, eczema | asthma | eczema, rhinoconjunctivitis |
no | asthma | no | drug allergies | 6/12 |
| Urogenital/kidney anomalies | large cortical cyst | ischemic nephropathy | ureterohydronephrosis | no | pyelo-ureteral duplication | no | N/A | ureterohydronephrosis | no | no | hydronephrosis | no | 6/11 |
AAA, ascending aortic aneurysm; ASD, atrial septal defect; MVP, mitral valve prolapse; VSD, ventricular septal defect; N/A, not available. Severe myopia was defined as of −6.00 diopters or greater.
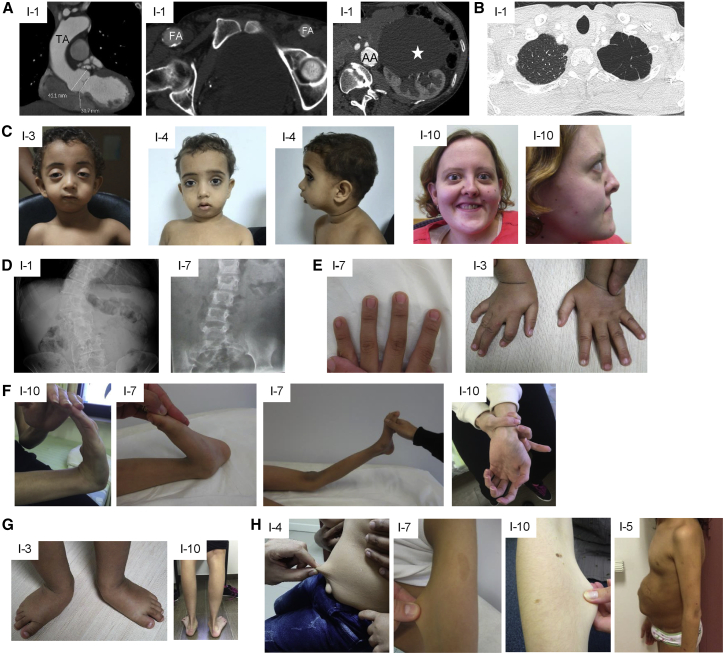
Figure 1.
Clinical features of affected individuals
(A) CT scan showing dilated aortic root and thoracic aorta (TA), calcified asymmetric large femoral arteries (FA), calcified and dilated abdominal aorta (AA), and large renal cyst (white star) in individual 1.
(B) CT scan showing emphysema of the apex of the left lung in individual 1.
(C) Proptosis, micrognathia, and hypertelorism in individuals 3, 4, and 10.
(D) X-ray showing scoliosis in individuals 1 and 7.
(E) Arachnodactyly in individuals 3 and 7.
(F) Hyperlaxity of small and large joints in individuals 10 and 7.
(G) Pes planus and talipes equinovarus in individuals 10 and 3, respectively.
(H) Skin hyperextensibility in individuals 4, 7, and 10 and umbilical hernia in individual 5.
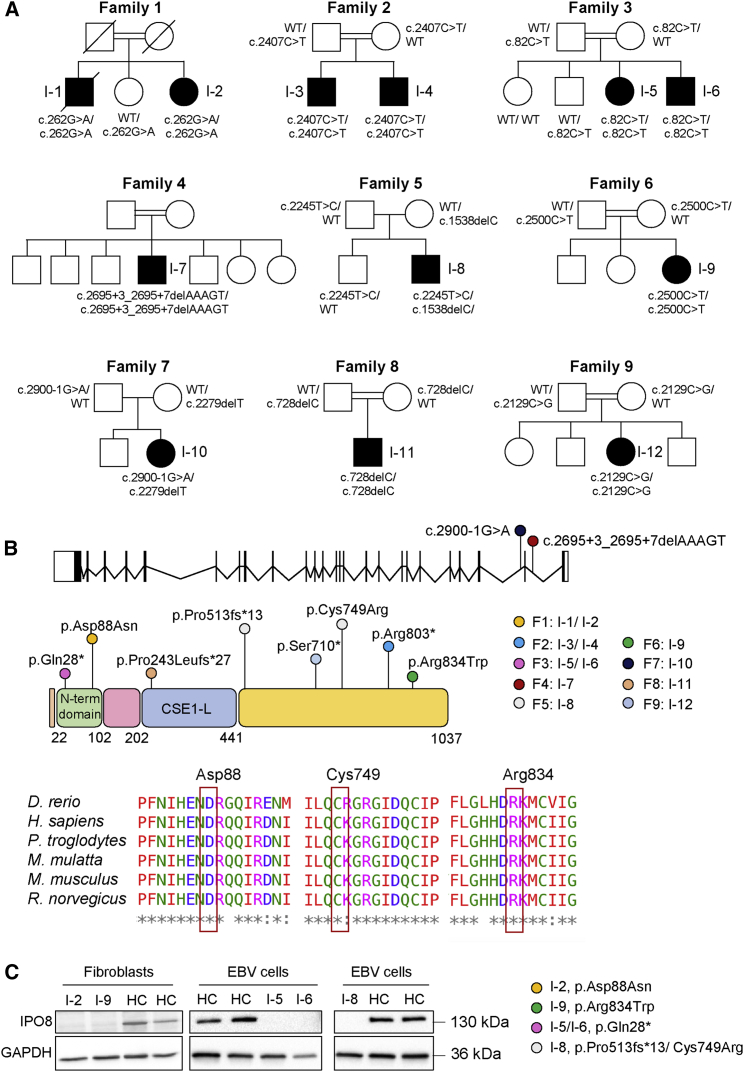
As detailed in Table 2, 11 variants were identified in the 12 individuals: seven as homozygous and four as compound heterozygous in IPO8 (GenBank: NM_006390.3). This gene (25 exons, 24 introns) encodes a 1,037 amino acid protein with the β-importin N-terminal domain (22–102 aa) and a CSE1-like domain (202–441 aa) containing a RanGTPase-binding motif characteristic of β-importins (Figures 2A and 2B). Segregation of the variant within each family was analyzed by Sanger sequencing (Figure S2) whenever DNA was available (n = 8). They were all exceedingly rare (MAF < 0.01%), and only three were found in the gnomAD at a heterozygous state (Table 2).
Table 2.
Characteristics of IPO8 variants identified by next-generation sequencing in affected individuals
| Individual | Chromosome position (Hg19-GRCh37) chr12 | cDNA change (GenBank: NM_006390.3) | Amino acid change | SIFT | PolyPhen | DANN | CADD | Mutation Taster | Allele frequency gnomAD |
|---|---|---|---|---|---|---|---|---|---|
| 1–2 | g.30837296C>T | c.262G>A | p.Asp88Asn | damaging | probably damaging | 0.9989 | 25.3 | disease causing | not observed |
| 3–4 | g.30792531G>A | c.2407C>T | p.Arg803∗ | N/A | N/A | 0.9979 | 36 | disease causing | 3.98E−6, no homozygotes |
| 5–6 | g.30848500G>A | c.82C>T | p.Gln28∗ | N/A | N/A | 0.9984 | 39 | disease causing | not observed |
| 7 | g.30789909_30789913del | c.2695+3_2695+7delAAAGT | N/A | N/A | N/A | N/A | 22.8 | disease causing | not observed |
| 8 | g.30816479del | c.1538delC | p.Pro513Leufs∗13 | N/A | N/A | N/A | 33 | disease causing | not observed |
| g.30802094A>G | c.2245T>C | p.Cys749Arg | damaging | probably damaging | 0.9978 | 28.4 | disease causing | not observed | |
| 9 | g.30790111G>A | c.2500C>T | p.Arg834Trp | damaging | probably damaging | 0.9993 | 32 | disease causing | 4.03E−6, no homozygotes |
| 10 | g.30784946C>T | c.2900−1G>A | N/A | N/A | N/A | 0.9935 | 34 | disease causing | not observed |
| g.30792659del | c.2279delT | p.Leu760Profs∗10 | N/A | N/A | N/A | 33 | disease causing | 1.06E−5, no homozygotes | |
| 11 | g. 30829433del | c.728del | p.Pro243Leufs∗27 | N/A | N/A | N/A | 33 | disease causing | not observed |
| 12 | g.30805169G>C | c.2129C>G | p.Ser710∗ | N/A | N/A | 0.9962 | 38 | disease causing | not observed |
Figure 2.
Molecular features of identified variants
(A) Pedigrees of families 1–9.
(B) Schematic representation of IPO8 gene (25 exons, 24 introns) and importin 8 protein (1,037 amino acids), which features the importin N-terminal domain and the CSE1-like domain, containing tandem HEAT repeats and a RanGTP-binding motif. IPO8 variants identified in this study as well as their location are shown. Protein sequence alignments of IPO8 orthologs are provided for each affected residue.
(C) Importin 8 expression in fibroblasts or EBV cells derived from individuals 2, 5, 6, 8, and 9. GAPDH served as loading control.
Out of the seven homozygous variants, four were likely LOF variants (three nonsense, c.2407C>T [p.Arg803∗], c.82C>T [p.Gln28∗], and c.2129C>G [p.Ser710∗], and one frameshift c.728delC [p.Pro243Leufs∗27]), one was a splicing variant (c.2695+3_2695+7delAAAGT), and two were missense (c.262G>A [p.Asp88Asn] and c.2500C>T [p.Arg834Trp]). The compound heterozygous variants were frameshift variants leading to a premature stop codon (c.2279delT [p.Leu760ProfsTer10] and c.1538delC [p.Pro513Leufs∗13]) in trans with a splicing (c.2900−1G>A) and a missense variant (c.2245T>C [p.Cys749Arg]), respectively. Both, c.2695+3_2695+7delAAAGT and c.2900−1G>A variants were predicted to impact the splicing according to SpliceAI.23 Although the lack of available biospecimens or DNA from I-10 (family 7) prevented assessment of the splicing variant C.2900−1 G>A, analysis of the c.2695+3_2695+7delAAAGT variant by minigene assay demonstrated that it resulted in exon 22 skipping and activation of a cryptic splicing site (Figure S3). The IPO8 missense variants, namely p.Asp88Asn, p.Cys749Arg, and p.Arg834Trp, affected highly conserved residues among IPO8 orthologs and were predicted to be damaging by several in silico tools, including CADD score (Figure 2B).
The impact of variants on protein level was evaluated by immunoblot analysis of protein lysates from primary cells, either fibroblasts or Epstein-Barr virus-immortalized B cell lines (EBV-B cells). A striking reduction of IPO8 level was observed in the four individuals who could be tested, including I-2 and I-9, who carried homozygous missense variants (Figure 2C). The overlap of clinical features between all the affected individuals and the rarity or absence of the identified IPO8 variants in population databases, including LOF variants and three missense variants impairing protein level and/or with predicted damaging effect on protein function, strongly supported IPO8 deficiency as disease causing in all the 12 individuals.
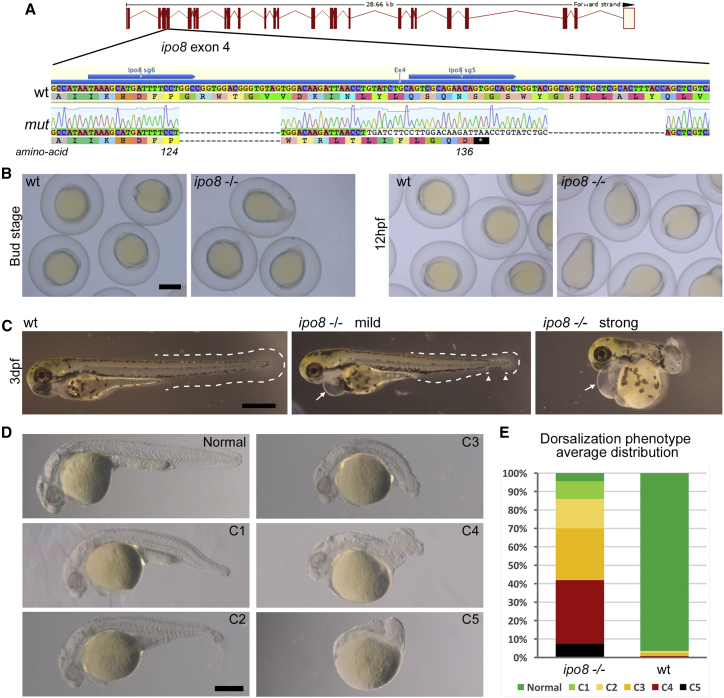
We next assessed how ipo8 disruption might affect development by using a zebrafish model and focused attention on early dorso-ventral patterning defects that are a telltale sign of altered TGF-β/BMP signaling, as well as on skeletal and cardiovascular defects that were the main clinical hallmarks in the cohort. The zebrafish genome encodes one single ipo8 ortholog with 72% identity and 85% similarity with human IPO8. IPO8 mutants were generated via CRISPR/Cas9 genome editing and two RNA guides simultaneously. The selected fish line carried both one insertion and two deletions (indels) in exon 4 that overall resulted in alternative translation from amino acid 125 and introduction of an early stop codon after amino acid 136 (Figure 3A). Zygotic mutants derived from the incross of two heterozygous parents (ipo8idv1/+ or simply ipo8+/−) did not develop any obvious phenotype and could be grown to adulthood, a result in keeping with the fact that maternal factors stored as mRNAs and proteins in the egg can compensate for zygotic loss of function during embryonic stages of zebrafish.24,25 In order to obtain maternal-zygotic mutants (MZ ipo8−/−, here referred to simply as ipo8−/−) that lacked wild-type (WT) ipo8 provided in the egg by the mother, we incrossed homozygous ipo8 zygotic mutant adults. Their homozygous WT siblings were incrossed as controls. At 10 h post fertilization (hpf) (bud stage), ipo8−/− MZ embryos appeared ovoid rather than round (Figure 3B). Moreover, during the following early somite stages, their tail bud failed to extend around the yolk but extended off prematurely, generating elongated, pear-shaped embryos (86% of 36 embryos from 3 clutches). These early morphological changes correlated with increased death rate that varied between 20% to 100% of the embryos per clutch. As development proceeded, a range of tail elongation defects became apparent, from embryos displaying an entirely normal or only partially absent ventral tail fin to embryos with a strongly twisted body axis resembling a snail shell-like trunk (Figure 3C). In addition, at 3 days post fertilization (dpf), most embryos (81% of 83 larvae from 6 clutches) developed heart edemas (as highlighted by arrows in Figure 3C). Morphological changes were strongly pronounced in the tail region, while anterior parts appeared comparatively normal with well-developed eyes and visible midbrain-hindbrain boundaries. These features are typical of dorsalized zebrafish mutants that result from mutations affecting the specification of ventral regions.26 To quantify the dorsalization phenotypic spectrum of the ipo8−/− mutants, we evaluated each analyzed clutch at 24 hpf and divided the phenotypes into five severity classes as previously described.26 A normal phenotype was observed in 96.5% of WT embryos but only 4.5% of ipo8−/− embryos, which displayed a whole spectrum of dorsalization phenotypes (Figures 3D and 3E).
Figure 3.
Zebrafish ipo8 mutants show a range of dorsalization phenotypes
(A) Schematics of CRISPR/Cas9-mediated gene disruption at the ipo8 genomic locus. The sgRNAs (sg5 and sg6, blue arrows) targeted exon 4. Compared to the wild-type (WT) sequence, the mutated allele from the founder fish (mut) displayed two deletions of 18 and 22 bases and an insertion of 34 bp generating a frameshift from amino acid 124 and a premature STOP codon after 136 amino acids.
(B) Bright field pictures of bud (10 hpf) and early somite stages (12 hpf) embryos showing the elongating shape of the ipo8−/− embryos compared to WT controls. Scale bar represents 500 μm.
(C) Bright field pictures of 3 dpf larvae presenting the variable penetrance of the ipo8−/− phenotype (mild and strong) compared to WT controls. The tail fin is circled with dashed lines when visible. Gaps are highlighted by arrowheads. Arrows point at heart edema. Scale bar represents 200 μm.
(D) Nomarski pictures depicting the different classes of the dorsalization phenotype at 24 hpf as described in Mullins et al.26 (from severe C5 to mild C1) in the ipo8−/− mutants compared to the normal phenotype of a WT embryo. Scale bar represents 200 μm.
(E) Quantification of the distribution of the dorsalization classes in ipo8−/− mutants (5 clutches, n = 903) compared to WT clutches (4 WT clutches, n = 503). Average values in ipo8−/− mutants were 7.3% C5, 34.6% C4, 28.0% C3, 16.0% C2, 9.6% C1, and 4.5% normal, and average values in WT controls were 0% C5, 0.8% C4, 1.6% C3, 1.1% C2, 0% C1, and 96.5% normal.
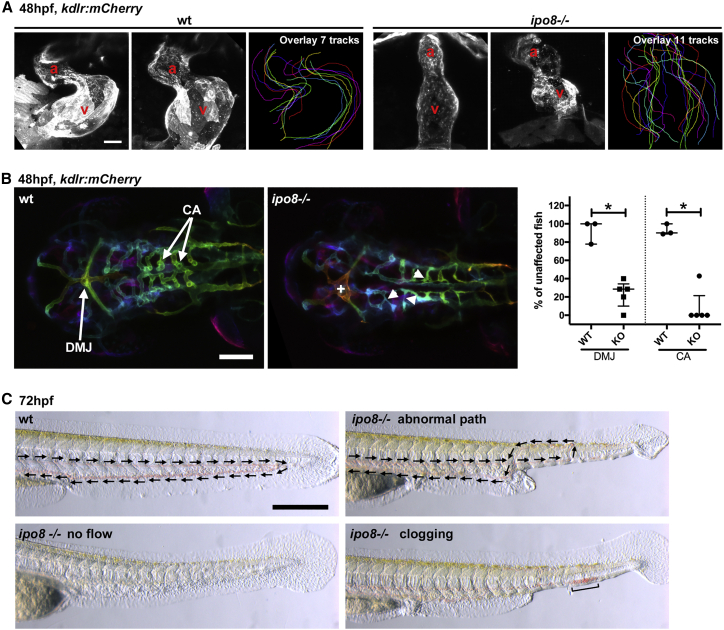
To dissect these morphological defects at the molecular level, we performed whole-mount in situ hybridization analysis to reveal mRNA expression of early patterning genes in embryos (Figure S4). Ntl showed a shorter and broader expression pattern indicating defects in axial mesoderm elongation (Figure S4A). Gata1 showed reduced expression limited to the posterior side, indicating ventral mesoderm specification defects (Figure S4B). In addition, the paraxial mesoderm marker myoD showed a shorter and broader pattern, underlying defects in convergent-extension gastrulation movements (Figure S4C). Overall, these defects in ipo8 mutants appeared comparable to those reported in zebrafish dorsalized mutants, which affect members of BMP and TGF-β signaling (e.g., bmp2, bmp7, alk8, and smad5).27,28 To investigate skeletal defects in the ipo8−/− zebrafish model, we analyzed expression of the transcription factor sox9a, a marker of chondrogenic precursor development (Figure S4D). Shorter trabeculae of the neurocranium were observed in mutant embryos by 2 dpf. In the tail region, sox9a expression was strongly reduced or appeared in irregular patches along the spinal cord axis, consistent with defects in vertebrae formation. Heart and vessel morphologies were analyzed after crossing the ipo8 mutant line with a transgenic line expressing a reporter fluorescently labeling blood vessels in living embryos (Tg(kdrl:Hsa.HRAS-mCherry)). Analysis at 2 dpf revealed severe defects in heart chamber formation and atrial and ventricular chambers were less or not delimited in ipo8−/− mutants (Figure 4A). Ipo8−/− mutants also exhibited arterio-venous malformations in the head and over 70% of mutant embryos in 5 independent clutches showed abnormal arterio-venous connections in the dorsal midline junction and poorly differentiated central arteries that appeared irregular, thin, and poorly lumenized (Figure 4B). By 3 dpf, various severe cardiovascular defects were observed in mutant embryos, including heart edema and, in the subset of embryos with mild tail morphological defects (normal or C1 class in Figure 3D), various blood vessel patterning abnormalities that resulted in the absence of blood circulation or abnormal blood flow in the tail region (e.g., looping or clogging, Figure 4C).
Figure 4.
Ipo8 deficiency causes cardiovascular defects in zebrafish
(A) Defects in heart chamber formation exemplified by two maximal projections of confocal Z stacks of the hearts of WT and ipo8−/− 48 hpf embryos carrying the kdrl:mCherry transgene to label endothelial cells. For each line, the third panel shows the overlays of the outlines of the 7 (WT) and 11 (ipo8−/−) analyzed hearts. a, atrium; v, ventricle. Scale bar represents 30 μm.
(B) Maximal projections of depth color-coded confocal Z stacks of the head vessels of WT and ipo8−/− embryos at 48 hpf, and quantifications of the percentage of fish showing dorsal midline junction (DMJ) defects and central arteries (CA) differentiation defects. Twenty-three WT and 30 ipo8−/− embryos from 3 and 5 clutches, respectively, were analyzed. Median and IQR are shown. p values were calculated by Mann Whitney test (∗p < 0.05). +, abnormal DMJ; arrowheads, abnormal CA. Scale bar represents 100 μm.
(C) Maximal projections of time-lapses of Nomarsky imaging of the tails of 3 dpf larvae highlighting absence of blood circulation (no flow) or defects in blood vessel patterning (abnormal path and clogging) in ipo8−/− mutants. Arrows indicate flow direction. Scale bar represents 200 μm.
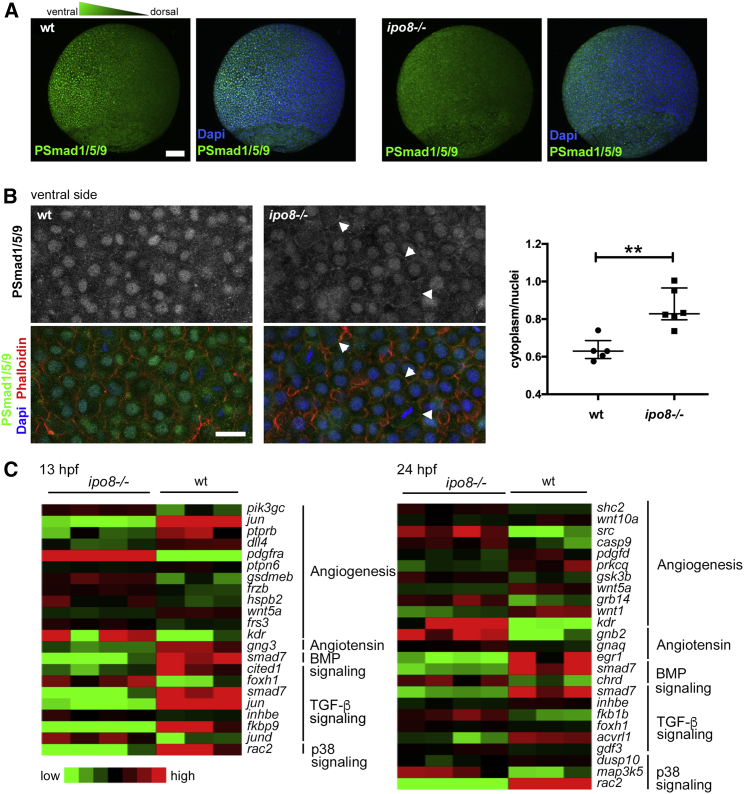
Putative defects in TGF-β/BMP signaling in the ipo8−/− mutants were next investigated with an antibody specific for pSmad1/5/9. This antibody revealed the expected ventral to dorsal gradient of pSmad1/5/9 detected in WT gastrulating embryos with nuclear localization in the ventral part. In ipo8−/− embryos, nuclear localization of pSmad was significantly reduced (Figures 5A and 5B). In contrast and in keeping with a defect in pSMAD translocation in ipo8−/− embryos, the pSMAD signal was detected in the cytoplasm and at membranes, as shown by its colocalization with phalloidin at cell outlines.
Figure 5.
pSMAD nuclear translocation defects in ipo8−/− embryos highlight impaired TGF-β/BMP signaling
(A) Maximal projections of confocal Z stacks of WT and ipo8−/− embryos labeled for pSmad1/5/9 (green) and DAPI (blue) showing well-defined ventral to dorsal gradient of pSmad5 in WT gastrulating zebrafish embryos (left panel) but not in the ipo8−/− embryos (right panel). Scale bar represents 100 μm.
(B) Confocal images of the ventral sides of WT and ipo8−/− embryos labeled for pSmad1/5/9 (green), phalloidin (red), and DAPI (blue). Arrowheads highlight the membrane localization of the pSmad staining in the ipo8−/− mutants. Scale bar represents 20 μm. Dot plots representative of three experiments show quantifications of the ratio of pSmad1/5/9 cytoplasmic signal over nuclear signal; 5 WT and 6 ipo8−/− embryos were analyzed. Median and IQR are shown. p values were calculated by Mann Whitney test (∗∗p < 0.01).
(C) Heatmaps of differentially expressed genes between WT and ipo8−/− embryos at 13 and 24 hpf. Three to four biological replicates are shown per group. The top enriched gene ontology term for biological process is highlighted for each cluster.
To further demonstrate the role of ipo8 in TGF-β/BMP signaling during development, we compared the transcriptomes of WT and ipo8−/− embryos at two different time points during early embryogenesis (13 and 24 hpf) (Figure 5C). We chose these early developmental stages to avoid secondary effects deriving from abnormalities developing later on. Ipo8 itself, as expected, was significantly downregulated in ipo8−/− fish (Figure S5A). Principal-component analysis (PCA) showed close clustering of biological replicates and a clear segregation of the ipo8−/− samples from WT samples both at 13 and 24 hpf (Figure S5B). Genes differentially regulated between the ipo8−/− and WT embryos (adjusted p value < 0.01) were analyzed by pathway enrichment analysis (Figure 5C). Strikingly, genes differentially regulated upon ipo8 depletion encoded multiple components of the TGF-β/BMP pathway as well as genes involved in angiotensin/angiogenesis pathways. Notably, expression of smad7, one of the direct transcriptional targets of the TGF-β/BMP pathway, was strongly decreased in ipo8−/− embryos compared to WT at both 13 and 24 hpf, compatible with lack of translocation to the nucleus and, as a consequence, impaired downstream activation of SMAD-dependent transcription.
Overall, our zebrafish model demonstrates that importin 8 plays a critical role during the early stage of development by controlling pSmad nuclear translocation and downstream TGF-β/BMP-dependent transcription. Because homozygous ipo8−/− embryos born from heterozygous parents were normal, and because the abnormal phenotype was observed only in MZ mutant embryos, it is likely that Ipo8 is not essential after the initial embryogenesis occurs. This may be due to genetic compensation29 or simply to Ipo8 function’s being redundant with that of a paralog. We cannot, however, exclude subtler cellular/organ phenotypes that may occur later in development or in adult animals and have not been characterized in this study. Importantly, data in the zebrafish model support a causative role of IPO8 deficiency in the vascular and skeleton defects observed in all affected individuals. While our results indicate a role for Ipo8 via the nuclear translocation of phosphorylated SMAD proteins, future investigation remains necessary to define whether and how the role of IPO8 in the translocation of other putative cargoes3, 4, 5, 6, 7 (see above) may participate in the phenotypic features of IPO8 deficiency.
In summary, we have identified IPO8 deficiency as the cause of a previously uncharacterized syndrome that is inherited in an autosomal-recessive pattern. This syndrome is characterized by deregulation of TGF-β signaling pathway and overlaps clinically with other TGF-β signalopathies: MFS, LDS, and SGS as depicted in Figure S6 and Table S1. Common features observed in IPO8 individuals and shared with LDS, MFS, and SGS include cardiovascular anomalies with notably strong predisposition for ascending aorta aneurysm and facial and skeletal anomalies (Table S1). Other less recurrent manifestations displayed by IPO8-deficient individuals were immune dysregulation and allergic diseases, which have also been reported in a subset of LDS-affected individuals,30 and developmental delay that was observed in SGS.14 The accompanying article by Van Gucht et al.31 in this issue of The American Journal of Human Genetics reports largely overlapping developmental abnormalities in seven additional IPO8-deficient individuals.31 Their data showing that the loss of ipo8 causes severe early-onset thoracic aortic aneurysm in a mouse model supports the causative role of the gene defect in the most severe manifestation of the disease and complements our demonstration that IPO8 plays a crucial role in TGF-β-dependent organogenesis and in cardio-vascular development in the zebrafish model. Overall, the identification of IPO8 deficiency as cause of TGF-β signalopathy stresses the essential functions of this pathway in development, patterning, and homeostasis of the affected tissues.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
We are grateful to the families who participated in this work. We thank Nicole Van Bergen, MCRI, for performing the confirmatory Sanger sequencing in individual 10. This work was supported by the Agence Nationale de la Recherche under “Investissements d’avenir” program (ANR-10-IAHU-01), institutional grants from INSERM, the European grant ERC-2013-AdG-339407-IMMUNOBIOTA, and grants from Fondation Princesse Grace and “Fondation Maladies Rares” (to N.C.B.); by institutional grants from the CNRS and the University of Angers (to D.B.); by intramural funding (fortüne) at the University of Tübingen (2545-1-0) and the Ministry of Science, Research and Art Baden-Württemberg (to B.V.); and by the Programme Investissements d’Avenir IHU FOReSIGHT (ANR-18-IAHU-01) (to F.D.B.). C.R. was supported by an EU Horizon 2020 Marie Skłodowska-Curie Action fellowship (H2020-MSCA-IF-2014 #661527). K.M.G. and K.K. are jointly funded by the Indian Council of Medical Research (file no. 5/7/1508/2016 to K.M.G.) and the Federal Ministry of Education and Research (01DQ17003 to K.K.). Sequencing and analysis for individual 10 were provided by the Broad Institute of MIT and Harvard Center for Mendelian Genomics (Broad CMG) and was funded by the National Human Genome Research Institute, the National Eye Institute, and the National Heart, Lung, and Blood Institute grant UM1 HG008900 and in part by National Human Genome Research Institute grant R01 HG009141. Funding for the UDP-Vic was provided by philanthropic donation and the Murdoch Children’s Research Institute. The research conducted at the Murdoch Children’s Research Institute was supported by the Victorian Government’s Operational Infrastructure Support Program, the Harbig Family Foundation, and The Royal Children's Hospital Foundation.
Published: May 18, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.04.020.
Contributor Information
Filippo Del Bene, Email: filippo.del-bene@inserm.fr.
Marianna Parlato, Email: marianna.parlato@inserm.fr.
Data and code availability
The IPO8 variants were submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) (GenBank: NM_006390.3; accession numbers SCV001571677, SCV001571678, SCV001571679, SCV001571680, SCV001571681, SCV001571682, SCV001571683, SCV001571684, SCV001571685, SCV001571686, and SCV001571687). The WES datasets supporting this study have not been deposited in a public repository because of ethical restriction but are available from the corresponding author on request.
Web resources
GeneMatcher, https://genematcher.org/
Mutation Taster, http://www.mutationtaster.org/
OMIM, https://omim.org
PolyPhen2, http://genetics.bwh.harvard.edu/pph2/
Supplemental information
References
- 1.Görlich D., Dabrowski M., Bischoff F.R., Kutay U., Bork P., Hartmann E., Prehn S., Izaurralde E. A novel class of RanGTP binding proteins. J. Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 3.Dean K.A., von Ahsen O., Görlich D., Fried H.M. Signal recognition particle protein 19 is imported into the nucleus by importin 8 (RanBP8) and transportin. J. Cell Sci. 2001;114:3479–3485. doi: 10.1242/jcs.114.19.3479. [DOI] [PubMed] [Google Scholar]
- 4.Wei Y., Li L., Wang D., Zhang C.-Y., Zen K. Importin 8 regulates the transport of mature microRNAs into the cell nucleus. J. Biol. Chem. 2014;289:10270–10275. doi: 10.1074/jbc.C113.541417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinmann L., Höck J., Ivacevic T., Ohrt T., Mütze J., Schwille P., Kremmer E., Benes V., Urlaub H., Meister G. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell. 2009;136:496–507. doi: 10.1016/j.cell.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 6.Schreck I., Al-Rawi M., Mingot J.-M., Scholl C., Diefenbacher M.E., O’Donnell P., Bohmann D., Weiss C. c-Jun localizes to the nucleus independent of its phosphorylation by and interaction with JNK and vice versa promotes nuclear accumulation of JNK. Biochem. Biophys. Res. Commun. 2011;407:735–740. doi: 10.1016/j.bbrc.2011.03.092. [DOI] [PubMed] [Google Scholar]
- 7.Liang P., Zhang H., Wang G., Li S., Cong S., Luo Y., Zhang B. KPNB1, XPO7 and IPO8 mediate the translocation ofNF-κB/p65 into the nucleus. Traffic. 2013;14:1132–1143. doi: 10.1111/tra.12097. [DOI] [PubMed] [Google Scholar]
- 8.Volpon L., Culjkovic-Kraljacic B., Osborne M.J., Ramteke A., Sun Q., Niesman A., Chook Y.M., Borden K.L.B. Importin 8 mediates m7G cap-sensitive nuclear import of the eukaryotic translation initiation factor eIF4E. Proc. Natl. Acad. Sci. USA. 2016;113:5263–5268. doi: 10.1073/pnas.1524291113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao X., Chen X., Cottonham C., Xu L. Preferential utilization of Imp7/8 in nuclear import of Smads. J. Biol. Chem. 2008;283:22867–22874. doi: 10.1074/jbc.M801320200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu L., Yao X., Chen X., Lu P., Zhang B., Ip Y.T. Msk is required for nuclear import of TGF-β/BMP-activated Smads. J. Cell Biol. 2007;178:981–994. doi: 10.1083/jcb.200703106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.David C.J., Massagué J. Contextual determinants of TGFβ action in development, immunity and cancer. Nat. Rev. Mol. Cell Biol. 2018;19:419–435. doi: 10.1038/s41580-018-0007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacFarlane E.G., Haupt J., Dietz H.C., Shore E.M. TGF-β Family Signaling in Connective Tissue and Skeletal Diseases. Cold Spring Harb. Perspect. Biol. 2017;9:a022269. doi: 10.1101/cshperspect.a022269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schepers D., Tortora G., Morisaki H., MacCarrick G., Lindsay M., Liang D., Mehta S.G., Hague J., Verhagen J., van de Laar I. A mutation update on the LDS-associated genes TGFB2/3 and SMAD2/3. Hum. Mutat. 2018;39:621–634. doi: 10.1002/humu.23407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doyle A.J., Doyle J.J., Bessling S.L., Maragh S., Lindsay M.E., Schepers D., Gillis E., Mortier G., Homfray T., Sauls K. Mutations in the TGF-β repressor SKI cause Shprintzen-Goldberg syndrome with aortic aneurysm. Nat. Genet. 2012;44:1249–1254. doi: 10.1038/ng.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loeys B.L., Chen J., Neptune E.R., Judge D.P., Podowski M., Holm T., Meyers J., Leitch C.C., Katsanis N., Sharifi N. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat. Genet. 2005;37:275–281. doi: 10.1038/ng1511. [DOI] [PubMed] [Google Scholar]
- 16.Chen W., Ten Dijke P. Immunoregulation by members of the TGFβ superfamily. Nat. Rev. Immunol. 2016;16:723–740. doi: 10.1038/nri.2016.112. [DOI] [PubMed] [Google Scholar]
- 17.Kelly A., Houston S.A., Sherwood E., Casulli J., Travis M.A. Regulation of Innate and Adaptive Immunity by TGFβ. Adv. Immunol. 2017;134:137–233. doi: 10.1016/bs.ai.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 18.Frischmeyer-Guerrerio P.A., Guerrerio A.L., Oswald G., Chichester K., Myers L., Halushka M.K., Oliva-Hemker M., Wood R.A., Dietz H.C. TGFβ receptor mutations impose a strong predisposition for human allergic disease. Sci. Transl. Med. 2013;5:195ra94. doi: 10.1126/scitranslmed.3006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guerrerio A.L., Frischmeyer-Guerrerio P.A., Huang C., Wu Y., Haritunians T., McGovern D.P.B., MacCarrick G.L., Brant S.R., Dietz H.C. Increased Prevalence of Inflammatory Bowel Disease in Patients with Mutations in Genes Encoding the Receptor Subunits for TGFβ. Inflamm. Bowel Dis. 2016;22:2058–2062. doi: 10.1097/MIB.0000000000000872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotlarz D., Marquardt B., Barøy T., Lee W.S., Konnikova L., Hollizeck S., Magg T., Lehle A.S., Walz C., Borggraefe I. Human TGF-β1 deficiency causes severe inflammatory bowel disease and encephalopathy. Nat. Genet. 2018;50:344–348. doi: 10.1038/s41588-018-0063-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loeys B.L., Schwarze U., Holm T., Callewaert B.L., Thomas G.H., Pannu H., De Backer J.F., Oswald G.L., Symoens S., Manouvrier S. Aneurysm syndromes caused by mutations in the TGF-beta receptor. N. Engl. J. Med. 2006;355:788–798. doi: 10.1056/NEJMoa055695. [DOI] [PubMed] [Google Scholar]
- 23.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B. Predicting Splicing from Primary Sequence with Deep Learning. Cell. 2019;176:535–548.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 24.Abrams E.W., Mullins M.C. Early zebrafish development: it’s in the maternal genes. Curr. Opin. Genet. Dev. 2009;19:396–403. doi: 10.1016/j.gde.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stainier D.Y.R., Raz E., Lawson N.D., Ekker S.C., Burdine R.D., Eisen J.S., Ingham P.W., Schulte-Merker S., Yelon D., Weinstein B.M. Guidelines for morpholino use in zebrafish. PLoS Genet. 2017;13:e1007000. doi: 10.1371/journal.pgen.1007000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mullins M.C., Hammerschmidt M., Kane D.A., Odenthal J., Brand M., van Eeden F.J., Furutani-Seiki M., Granato M., Haffter P., Heisenberg C.P. Genes establishing dorsoventral pattern formation in the zebrafish embryo: the ventral specifying genes. Development. 1996;123:81–93. doi: 10.1242/dev.123.1.81. [DOI] [PubMed] [Google Scholar]
- 27.Tucker J.A., Mintzer K.A., Mullins M.C. The BMP signaling gradient patterns dorsoventral tissues in a temporally progressive manner along the anteroposterior axis. Dev. Cell. 2008;14:108–119. doi: 10.1016/j.devcel.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammerschmidt M., Mullins M.C. Dorsoventral patterning in the zebrafish: bone morphogenetic proteins and beyond. Results Probl. Cell Differ. 2002;40:72–95. doi: 10.1007/978-3-540-46041-1_5. [DOI] [PubMed] [Google Scholar]
- 29.Rossi A., Kontarakis Z., Gerri C., Nolte H., Hölper S., Krüger M., Stainier D.Y.R. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–233. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- 30.Cannaerts E., van de Beek G., Verstraeten A., Van Laer L., Loeys B. TGF-β signalopathies as a paradigm for translational medicine. Eur. J. Med. Genet. 2015;58:695–703. doi: 10.1016/j.ejmg.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 31.Van Gucht I., Meester J.A.N., Bento J.R., Bastiaansen M., Bastianen J., Luyckx I., Van Den Heuvel L., Neutel C.H.G., Guns P.J., Vermont M. A human importin-β-related disorder: Syndromic thoracic aortic aneurysm caused by bi-allelic loss-of-function variants in IPO8. Am. J. Hum. Genet. 2021 doi: 10.1016/j.ajhg.2021.04.019. Published online May 18, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The IPO8 variants were submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) (GenBank: NM_006390.3; accession numbers SCV001571677, SCV001571678, SCV001571679, SCV001571680, SCV001571681, SCV001571682, SCV001571683, SCV001571684, SCV001571685, SCV001571686, and SCV001571687). The WES datasets supporting this study have not been deposited in a public repository because of ethical restriction but are available from the corresponding author on request.