Abstract
Alternative pathways of energy transfer guarantee the functionality and productivity in marine food webs that experience strong seasonality. Nevertheless, the complexity of zooplankton interactions is rarely considered in trophic studies because of the lack of detailed information about feeding interactions in nature. In this study, we used DNA metabarcoding to highlight the diversity of trophic niches in a wide range of micro- and mesozooplankton, including ciliates, rotifers, cladocerans, copepods and their prey, by sequencing 16- and 18S rRNA genes. Our study demonstrates that the zooplankton trophic niche partitioning goes beyond both phylogeny and size and reinforces the importance of diversity in resource use for stabilizing food web efficiency by allowing for several different pathways of energy transfer. We further highlight that small, rarely studied zooplankton (rotifers and ciliates) fill an important role in the Baltic Sea pelagic primary production pathways and the potential of ciliates, rotifers and crustaceans in the utilization of filamentous and picocyanobacteria within the pelagic food web. The approach used in this study is a suitable entry point to ecosystem-wide food web modelling considering species-specific resource use of key consumers.
Keywords: zooplankton, food web, trophic niche diversity, metabarcoding, rotifer
1. Introduction
The ability for ecosystems to maintain functionality and productivity under annual and seasonal variation in primary production relies on energy transfer pathways sustained by a network of diverse primary consumers [1,2]. In marine food webs, functionally diverse assemblages of planktonic bacteria, protists and metazoans regulate the flow of energy from primary producers to higher trophic levels [3–5]. While crustacean zooplankton (e.g. copepods and cladocerans) constitute the primary link between phytoplankton and planktivorous fish [6], microzooplankton are main grazers of primary production at times when the biomass of phytoplankton is low or inedible [3,7]. In order to estimate the resilience of marine ecosystems, a mechanistic understanding of resource use by the primary consumers is needed [8]. However, in most food web studies, the trophic niche is based on size or phylogeny due to a lack of detailed information about feeding interactions in nature. Consequently, the entire niche diversity of the zooplankton community is not accurately considered [9,10].
Variation in temporal abundance, feeding traits, size, phenotypic plasticity, growth rate and predation resistance contribute to the total diversity of zooplankton functional groups in marine food webs [11]. While most trophic studies have clustered zooplankton into broad phylogenetic groups [9], recent studies show that incorporating traits, particularly size, has consequences for interpreting food web dynamics and productivity [12–14]. For example, the rotifer phylum contains members of different size classes [15], as well as organisms with various feeding behaviours including filter feeders [16], selective feeders [17–19] and in some cases even carnivores [20]. Similarly, copepods and cladocerans can perform different feeding strategies including, among others, feeding-current and ambush feeding [21], thereby using a wide spectrum of resources. Consequently, trophic niche partitioning at a high phylogenetic level, such as class or family, will underestimate the trophic niche diversity in zooplankton guilds.
Traditionally, method limitations have made it difficult to assess trophic niche differences between zooplankton species. Experiments to estimate the grazing impact of zooplankton are time-consuming and elaborate, and the amount of biological material required for biogeochemical tracer studies on plankton communities [22] often exceeds what is feasible to sort out from diverse zooplankton samples. Consequently, few species are often included in these studies, limiting the ability to describe key niche differences between them. Similarly, the diet spectrum in plankton experimental studies is often limited to a small number of a priori hypothetical prey species and may not display the food web's full complexity with enough resolution. The challenges and limitations of studying feeding traits of zooplankton species have further created a biased knowledge towards larger organisms in the food web that are more frequently studied [23]. Since most of the zooplankton feeding studies are conducted at different sites and times of the year, often using different methods, the comparison of existing information on zooplankton trophic niches is laborious. DNA metabarcoding of selected organisms has proven to be a useful tool for resolving trophic interactions [24,25] and is increasingly being used for studying trophic interactions of zooplankton [26–28]. With the high sensitivity of the polymerase chain reaction, metabarcoding requires very little biological material and is a non-a priori method with high taxonomic resolution. Metabarcoding allows for a food web-oriented approach as several zooplankton species can be investigated simultaneously [29], thereby providing detailed insights on trophic interactions and better linking the trophic niche diversity with energy flow.
Detailed knowledge about trophic interactions can be of particular importance for coastal ecosystems that experience a shift in phytoplankton community with an increase of cyanobacteria due to climate warming and eutrophication [30–32]. The Baltic Sea is a prime example, illustrating that nitrogen fixed by cyanobacteria supports the productivity of upper trophic levels [33,34]. Yet, as filamentous cyanobacteria are often considered unpalatable for copepods [35], the mechanism of trophic incorporation is not fully understood. While rotifers and microzooplankton are abundant in the Baltic Sea (figure 1), their potential trophic link with cyanobacteria is seldom considered. Without knowledge about trophic partitioning between micro and mesozooplankton, the possible fate and sinks of cyanobacterial production in the plankton food web remain largely unknown.
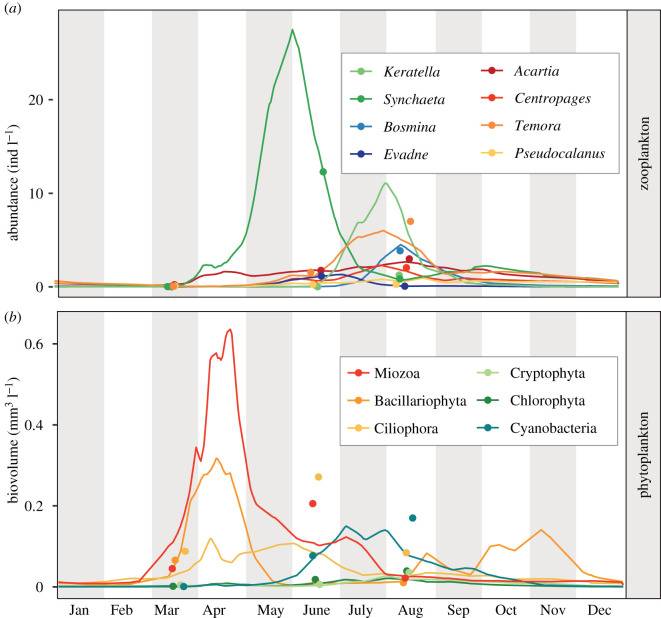
Figure 1.
(a) Abundance of zooplankton and (b) biovolume of phytoplankton at Landsort Deep in the Baltic Sea. Interpolated daily means over the years 2006–2018. The data are available at the Swedish national archive for oceanographic data: https://sharkweb.smhi.se/. Samples are taken weekly to bi-weekly during the spring and summer period and monthly during winter. The points indicate (a) taxa abundance and (b) biovolume at the date of sampling during this study. (Online version in colour.)
In this study, we aimed to investigate the trophic niches of functionally diverse groups of zooplankton spanning both size and phylum. By sequencing 18S rRNA and 16S rRNA genes, we analysed zooplankton-associated prey of selected individuals of different size classes, including a ciliate, rotifers, copepods and cladocerans. The study was done at an offshore station in the Baltic Sea, where ciliates, rotifers and crustaceans at times dominate the zooplankton community. Our results show that the trophic niche diversity extends beyond broad phylogenetic groups and size classes and that small, rarely studied zooplankton fill an important role in the pathways of the coastal pelagic primary production.
2. Methods
(a) . Sampling
Zooplankton and water samples were collected at Landsort Deep monitoring station BY31 (58′35 N18′14 E) located in the eastern Baltic Sea proper, an offshore station at the deepest location of the Baltic Sea with 495 m depth. This frequently monitored offshore station experiences strong seasonal changes in biotic and abiotic properties (see electronic supplementary material, figures S1 and S2). To capture the seasonality of zooplankton (figure 1), samples were collected on 19 June and 15 August 2017, and on 16 March 2018, synchronized with the Swedish national pelagic monitoring programme [36].
Water samples, used for validation and to describe the potential zooplankton prey present in the water, were collected with 10 l Niskin bottles with 5 m depth intervals above the thermocline (0–30 m depth). The depths were mixed by adding an equal volume of water from the Niskin bottles; 1–3 l was sequentially filtered onto 25 mm diameter filters with 20 µm (nylon), 2 µm and 0.2 µm (polycarbonate) pore size. Filters were stored frozen at −80°C until further analysis. Zooplankton samples were collected with three vertical hauls from 0–30 m, 30–60 m and 60–100 m using a 90 µm WP2 closing plankton net (Hydrobios, Kiel, Germany). Ciliates were sampled with a 55 µm hand-towed plankton net in the upper 10 m layer (Hydrobios, Kiel, Germany). The zooplankton and ciliate samples were immediately preserved in 95% ethanol.
(b) . Zooplankton sorting and DNA metabarcoding
Individuals of abundant zooplankton species were identified under a stereomicroscope and selected from depth layers where they were most abundant (electronic supplementary material, table S1 and figure S2). This includes the rotifers Synchaeta baltica, Synchaeta monopus and Keratella spp., the cladocerans Evadne nordmanni and Bosmina spp., and the copepods Temora longicornis, Acartia spp., Pseudocalanus spp. and Centropages hamatus. All individual rotifers were rinsed five times in ethanol; crustaceans were rinsed five times in miliQ water and after that soaked for 30 s in a 1% bleach solution to remove contamination of external DNA. Five to 12 individuals from each species were randomly pooled into one sample tube and stored in 180 µl ALT lysis buffer (Qiagen, Hilden, Germany). A representative of the protozooplankton community, the ciliate Helicostomella, was transferred from the zooplankton samples onto a PET membrane-coated glass slide (Zeiss, Oberkochen, Germany) and covered with resin-based liquid cover glass (Zeiss). Single cells of Helicostomella were collected using a laser capture microdissection microscope (Zeiss) and 10–15 individuals per sample pooled into 10 µl ALT lysis buffer (Qiagen). All of the sorted zooplankton samples were prepared in at least five replicates that were treated separately in all downstream analyses.
In the DNA metabarcoding analysis, we amplified a 500 bp long fragment of the V3–V4 region of the 16S rRNA gene (16S) using universal primers 341F and 805R targeting both bacterial and plastidial 16S of phototrophic eukaryotes [37,38], and a 400 bp long fragment of the V4 region of the 18S rRNA gene (18S) using the primers 528F and 706R [39]. The amplicon libraries were sequenced on MiSeq (MSC 2.5.0.5/RTA 1.18.54) pair-end set-up (2 × 300 bp, v. 3, Illumina, San Diego, California). DNA sequences and associated metadata were uploaded to the European Nucleotide Archive (ENA) under accession no. PRJEB39191.
16S sequences were assigned to a custom-made database combining the SILVA 16S reference database [40] with the PhytoREF database [41] to achieve an adequate taxonomic resolution for both prokaryotes and photoautotrophic eukaryotes. 18S sequences were assigned to the Protist Ribosomal Reference database [42]. Details of sample processing, sequencing and bioinformatic analysis can be found in the electronic supplementary material.
(c) . Data analysis and visualization
Data filtering and statistical analysis were facilitated by the Phyloseq R package [43]. All sequences originating from the respective zooplankton consumer species in each sample were removed before data visualization. Heterogeneous sequencing depth was controlled for using subsampling (rarefaction) and subsequent conversion to relative abundance. Non-metric multidimensional scaling plots were based on Bray–Curtis distances and calculated with the ‘metaMDS’ function in the Vegan R package [44]. We used Bray–Curtis similarity index to assess diet overlap between samples (1-Bray–Curtis distance). Differences in the proportion of the specific diet of consumers were modelled with β regression using the ‘betareg’ function in R. We used the ‘simper’ function in the Vegan package to decide which prey species contributed most to the differences in diet overlap.
Figures were made using the ggplot2 R package [45]. The most important prevalent taxa (determined as taxa occupying at least 0.1 per cent of the sequences in at least 70 per cent of the samples in each sample group) were visualized in bipartite networks made in the Circlize R package [46]. All data used for the statistical analysis and plotting together with the R scripts to generate the figures were uploaded to the Dryad Digital Repository [47].
3. Results
(a) . Diversity of biotic associations
The Illumina sequencing effort produced over 37 million sequence reads that passed quality control. The 16S rRNA gene (16S) that targets bacteria and photoautotrophic eukaryotes (plastids), generated 1492 unique ribosomal sequence variants (RSVs) of which 988 were found in the bulk water samples and 996 found in the selective zooplankton samples. The 18S rRNA gene (18S) that targets all eukaryotes generated 3267 RSVs, of which 2258 were in the bulk water samples and 1394 found in the zooplankton samples. We found a broad range of organisms associated with the zooplankton organisms, including heterotrophic and autotrophic bacteria, phytoplankton, protozoans and metazoans.
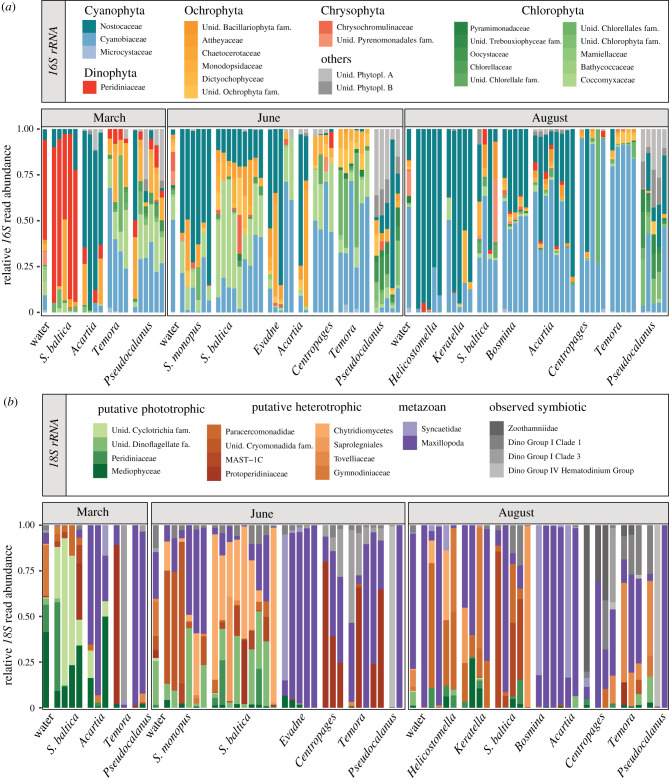
We found that, on average, 85% of the 16S sequence reads associated with the zooplankton samples were proteobacteria, which varied between zooplankton species and season (electronic supplementary material, appendix S1 and figure S4). Among photoautotrophic taxa (cyanobacteria and plastic-containing eukaryotes), associations of zooplankton consumer samples were dominated by cyanobacteria, green algae (Chlorophyta), diatoms (Bacillariophyta) and dinoflagellates (Dinophyceae) (figure 2a). Based on the 18S reads, the zooplankton species were associated with a diversity of eukaryotic organisms, comprising both photoautotrophic and heterotrophic plankton and a diversity of potential symbiotic or parasitic organisms with oomycetes and dinoflagellates (figure 2b).
Figure 2.
Relative abundance of sequence counts per family of (a) 16S rRNA gene reads (photoautotrophic organisms only) and (b) 18S rRNA, for different zooplankton consumer species and months in the Baltic Sea. The bars represent unique biological replicates. (Online version in colour.)
(b) . Trophic niche diversity in spring
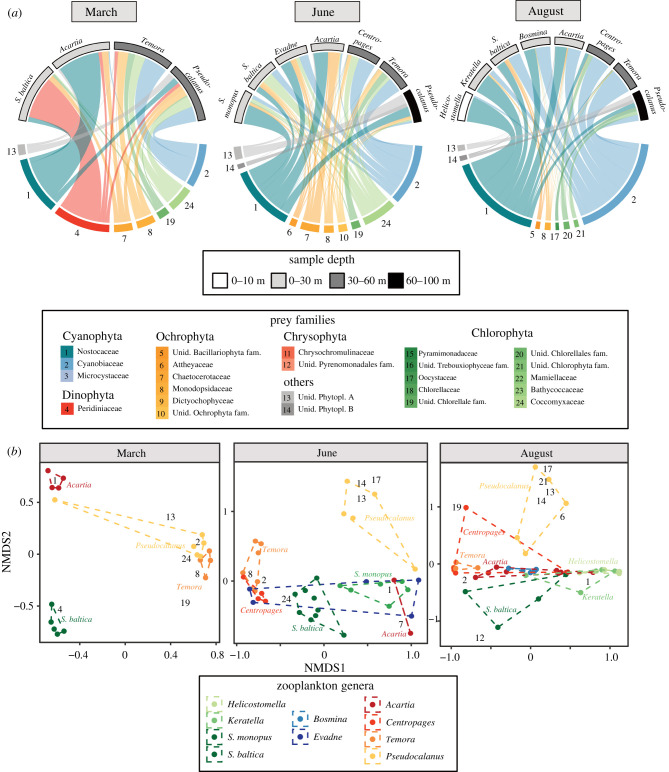
During the spring months, from March to June, the rotifer Synchaeta baltica was the dominating zooplankton species in the Baltic Sea proper, accompanied by less abundant copepod species (figure 1a). The main primary producers were bloom-forming dinoflagellates and diatoms, but also the mixotrophic ciliate Myrionecta (figure 1b). In March, at the beginning of spring bloom, diet overlap between the zooplankton species was relatively low, according to the 16S reads. The rotifer S. baltica had a diet overlap between 0.1 and 0.17 with the copepod groups, while the highest overlap in diet was found between the copepods Temora and Pseudocalanus (0.53) (figure 3b; electronic supplementary material, figure S5). The rotifer S. baltica was mainly associated with the bloom-forming dinoflagellate Peridiniella (occupying on average 76% of the 16S reads) (figure 3a). The copepods Temora and Pseudocalanus were associated with fewer sequences of Peridiniella compared to the rotifer (on average 6% of 16S reads, d.f. = 3, z = 16, p < 0.001), but instead associated with various groups of small phytoplankton and picocyanobacteria. In March, Acartia was almost exclusively associated with filamentous cyanobacteria (figure 3a). The 18S sequences supported the association between S. baltica and Peridiniella but also revealed associations with the ciliate Myrionecta. The 18S sequences further revealed associations between all zooplankton species and diatoms (figure 2b).
Figure 3.
(a) Zooplankton consumer species (upper) with their most prevalent prey families (lower) based on 16S rRNA gene reads. The thickness of the bars is proportional to relative rRNA read abundance. (b) Non-metric multidimensional scaling plot of Bray–Curtis distances between zooplankton samples (represented by coloured points) based on their prey (16S rRNA reads). The prey families responsible for the largest percentage of dissimilarity between any pair of zooplankton species are represented as numbers.
Synchaeta baltica reached its peak abundance in the Baltic Sea towards the end of the spring, in June, coordinated with the decline of dinoflagellates (Miozoa) (figure 1). Diet overlap between zooplankton species became more apparent but did not cluster according to phylogenetic affiliation. In June, S. baltica had an equally high diet overlap with the copepod Centropages (0.48) and the cladoceran Evadne (0.39), compared to the sister species S. monopus (0.40) (figure 3b; electronic supplementary material, appendix S1 and figure S3). Similarly, the copepod Acartia had a higher diet overlap with S. monopus (0.53) than with the other copepods (overlap of 0.11 with Temora) (electronic supplementary material, figure S5). At the end of spring, cyanobacteria became more apparent in the diet of the rotifers, indicating a transition from a spring to a summer prey community (figure 3a).
(c) . Trophic niche diversity in summer
In August, the abundance and diversity of crustacean zooplankton increased in the Baltic Sea, and Keratella was the most abundant rotifer. The rotifer Synchaeta baltica was still present but with low abundance (figure 1a). The primary production was characterized by extensive blooms of filamentous cyanobacteria (figure 1b). In August, a large part of the variation in zooplankton diet read abundance could be explained by the abundance of filamentous cyanobacteria (Nostocaceae) and picocyanobacteria (Cyanobiaceae) (figure 3a).
The highest diet overlap was found between the heterotrophic ciliate Helicostomella and the rotifer Keratella in August (0.75), as they were mostly associated with filamentous cyanobacteria (occupying 93% and 74% of 16S reads, respectively) (figure 3; electronic supplementary material, figure S5). Synchaeta baltica together with the cladoceran Bosmina and the copepod Acartia were associated with a lower proportion of filamentous cyanobacteria than the Keratella and Helicostomella (on average 39%, d.f. = 4, z = 5.7, p < 0.001), but with a larger proportion of picocyanobacteria (50%, d.f. = 5, z = 5.7, p < 0.001) as well as diverse small phytoplankton. Thus, the diet overlap between the two rotifer species Keratella and S. baltica was lower (0.42) than the overlap both between Keratella and the heterotrophic ciliate Helicostomella (0.75) and between S. baltica and the copepod Acartia (0.54). The copepods Temora and Centropages were associated with a low proportion of filamentous cyanobacteria (8%) and were almost exclusively associated with a higher relative proportion of picocyanobacteria (80%) compared with Acartia, Bosmina and S. baltica (d.f. = 5, z = 4.6, p < 0.001). Consequently, the copepod Acartia had a higher diet overlap with the cladoceran Bosmina (0.70) than with the other copepods (e.g. Temora, 0.52). Finally, Pseudocalanus, clustering alone, was associated with a significant proportion of unclassified organisms (up to 31% of 16S reads) (figure 3a).
The 18S sequences revealed various groups of heterotrophic flagellates associated with S. baltica, Keratella and Helicostomella. Small phytoplankton (chlorophytes and eustigmatophytes), heterotrophic protozoans of different phyla, as well as metazoans dominated the 18S sequences of the cladocerans and copepods in summer (figure 2b).
4. Discussion
(a) . Niche diversity and overlap
In order to resolve the trophic niche diversity of zooplankton, we analysed the physical associations of several micro and mesozooplankton species using 18S and 16S rRNA gene sequencing of selected zooplankton. The trophic position and role of zooplankton in food webs are often derived from taxonomic groupings and size estimation [12,48]. However, our results highlight that clustering zooplankton by size or phylogeny does not capture the true differences in diet niche and leads to an underestimation of the trophic niche diversity of primary consumers in the pelagic food web. Rotifers in the Baltic Sea are often grouped with microzooplankton and are referred to as obligate filter feeders [49,50]. Despite this, we can not find support for a high diet overlap or clustering between rotifer species (figure 3b). Instead, our study indicates that Synchaeta baltica (approx. 350 µm) has a trophic niche more similar to cladocerans and copepods than to the other rotifers, S. monopus, Keratella and the ciliate Helicostomella. The diet of S. baltica in spring included bloom-forming phytoplankton taxa, including the dinoflagellate Peridiniella (Peridiniaceae, 20–35 µm) and the mixotrophic ciliate Myrionecta (Cyclotrichia, 45–55 µm) (figure 3), findings that are in line with previous studies that have observed predation on large phytoplankton and protozoa up to 50 µm by Synchaeta [16–19].
In contrast with Synchaeta, the smaller sized rotifer Keratella peaks in abundance during the summer (figure 1) and was mainly associated with larger filamentous cyanobacteria (Nostocaceae). Keratella revealed a higher diet overlap with the tintinnid ciliate Helicostomella than with S. baltica (figure 3). The size of the Keratella (150 µm) and Helicostomella (100 µm) compared to cyanobacteria filaments that often exceed 1 mm suggests that these consumers do not feed directly on filamentous cyanobacteria, but rather on degraded filaments. This is supported by experiments indicating a filter-feeding behaviour of Keratella [15] that prefers partially degraded food (detritus) over living cells [51]. The same can be expected for Helicostomella, although few studies have investigated the selectivity of this ciliate [52–54]. The detritivorous feeding niche of Keratella and Helicostomella suggested here is further supported by a relatively high proportion of associated crustacean DNA (figure 2b), which for similar reasons is unlikely to be preyed upon directly and is likely ingested in the form of particulate organic matter. We suggest that filamentous cyanobacteria likely contribute to a pool of organic matter that is both available and attractive for detritivorous rotifers and ciliates. The results point to the importance of grouping zooplankton according to function rather than taxa, with some rotifers occupying the function of selective feeders similar to copepods, a distinction already proposed in a study by Arndt in 1993 [15].
Similar to rotifers, there was no clear clustering within copepod and cladoceran species based on their prey composition. While Temora and Centropages shared high diet overlap, mostly associated with picocyanobacteria, Acartia had a diet more similar to the cladocerans and the rotifer Synchaeta baltica during the summer months, relying on a large diversity of resources, including both filamentous and picocyanobacteria (figure 3). On the other side, the copepod Pseudocalanus occupies its own niche, feeding in all seasons on various unclassified organisms. The different feeding niches of copepods may reflect their vertical distribution (electronic supplementary material, figure S2), as Temora and Centropages are more abundant at 30–60 m depth compared to Acartia that dominates in the upper 30 m. By contrast, Pseudocalanus extends deeper in the water column compared to the other zooplankton [55,56]. The association of Pseudocalanus with unidentified taxa might indicate that the components of its natural diet are not well represented in the taxonomic databases. The differences in trophic niches of copepods are supported by a more intense seasonal sample analysis of crustacean zooplankton described in the previous study [29] and more extensive sampling several locations in the Baltic Sea proper (Baptiste Serandour 2021, pers. comm).
(b) . Food web implications
The trophic niche partitioning of zooplankton has implications in the food web dynamics in spring and summer. As the decline of dinoflagellates in spring coincides with the peak of Synchaeta (figure 1), grazing by rotifers should be considered a potential cause of the phytoplankton spring bloom decline in addition to nutrient limitation in the upper water column [57]. This is further supported by a study from the Mediterranean Sea, where Synchaeta was estimated to consume up to 80% of the daily production of a dinoflagellate bloom [58]. Given that cladocerans and copepods are temporally decoupled from the spring bloom (figure 1), Synchaeta is likely a major pathway of energy transfer to higher trophic levels in the Baltic Sea and possibly in other ecosystems where this species is abundant [58].
Pico and filamentous cyanobacteria are favoured under climate warming and eutrophication and are increasing in both marine and freshwater systems [30,59]. Due to their significance as food resources in the summer community of the Baltic Sea [33,49,60], understanding the pathways of cyanobacteria incorporation into food webs is important. The trophic role of filamentous cyanobacteria is widely debated as they are the main source of nitrogen fixation in the Baltic Sea, which is suggested to be nitrogen limited in summer [61]. Several studies suggest little or no grazing on filamentous cyanobacteria by zooplankton [34,62,63], but our study shows that the microzooplankton Helicostomella and Keratella, but also to some extent Bosmina and Acartia grazed on filamentous cyanobacteria, either living or degraded.
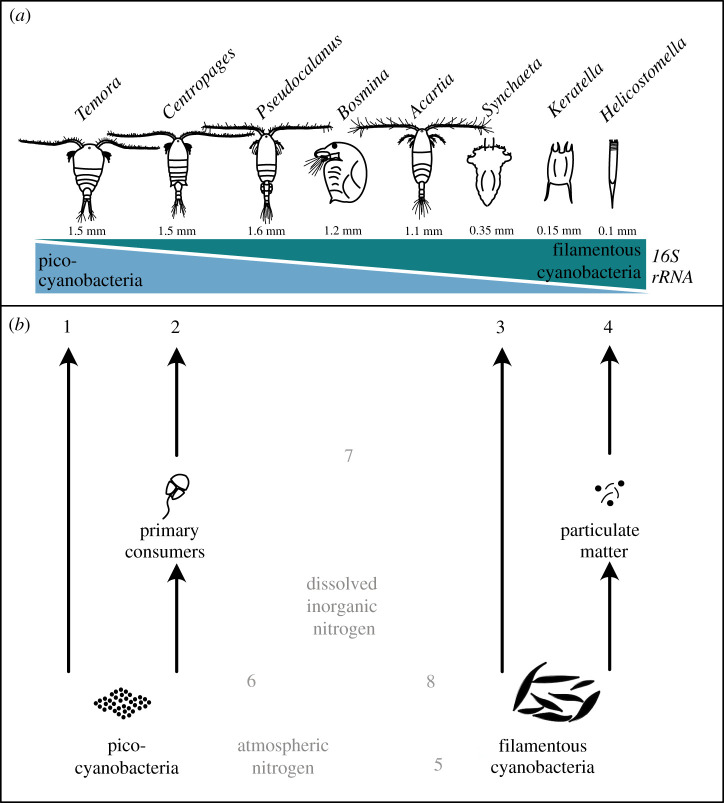
Zooplankton feeding on filamentous cyanobacteria may also act as important vectors of diazotroph nitrogen availability to upper trophic levels by stimulating the microbial food web. This is supported by an experiment by Arndt [15] showing that the presence of filter-feeding Keratella stimulates the growth of both heterotrophic flagellates and bacteria. Arndt proposed that Keratella, through its feeding, enhances leaking of dissolved matter from the algae [4], thereby supporting the increased biomass of both bacteria and protozoa. Tracer studies suggest that diazotroph nitrogen is mainly incorporated in the Baltic Sea food web by passive leaking of nitrogen compounds by filamentous cyanobacteria [64,65] that stimulate the production of heterotrophic bacteria and picocyanobacteria [33,34,66]. Our study confirms that picocyanobacteria are a key resource for the larger zooplankton species, primarily Temora and Centropages, but also, to some extent, Acartia, Bosmina and Synchaeta (figure 4a), and are as such indirectly supported by filamentous cyanobacteria. In addition to passive leaking, feeding by copepods, cladocerans, rotifers and ciliates on filamentous cyanobacteria actively enhances leaking of diazotroph nitrogen. This constitutes an alternative pathway of cyanobacteria incorporation into the pelagic food web and enhances the support for copepods that rely on the microbial food web (figure 4b).
Figure 4.
Illustration of dominant zooplankton consumers and alternative energy transfer pathways in the pelagic food web. (a) Size range of selected zooplankton species aligned with the relative read abundance of associated pico- and filamentous cyanobacteria. (b) Black arrows illustrate alternative pathways of energy transfer from primary producers to zooplankton consumers. Ingestion of picocyanobacteria can be directly (1) or via primary consumers (2). Filamentous cyanobacteria can be ingested alive (3) or in the state of decay (4). Shaded arrows denote putative pathways of nitrogen fixed by filamentous cyanobacteria (5) that may enter the food web either via consumption (3, 4) but may also stimulate the production of picocyanobacteria (6). Grazers of filamentous cyanobacteria may enhance the release of dissolved inorganic nitrogen (7), in contrast with previously suggested passive leaking (8). (Online version in colour.)
While DNA metabarcoding has become more frequently used over the last decade, few studies have until now exploited the potential of investigating the feeding niche diversity of the entire zooplankton community spanning several phyla and size classes. We show that the method used here is applicable for several metazoan taxa and potentially protozooplankton, using the ciliate Helicostomella as an example. DNA metabarcoding could be relevant in future investigations to unveil the role of rarer species and better comprehend the ecosystem function. Our approach shows the advantage of investigating the prey composition of diverse species in natural systems with DNA metabarcoding that reveals the entire food spectrum. By putting weight on the relative comparison between zooplankton species, we could capture key differences in zooplankton resource. While we can discuss possible ecosystem effects of diverse zooplankton feeding, the metabarcoding data is inevitably proportional. Thus, the data do not reveal information about feeding rates or biomasses. Despite this, metabarcoding has the potential to serve as an important complement to food web models that implement population biomasses and metabolic energy demands [67], by bringing details of species-specific feeding interactions to the model.
Our results highlight a large variation in resource use between groups of zooplankton that may stabilize energy transfer in food webs by pathways of energy flow that are rarely described, particularly during seasons when primary producers include pico and filamentous cyanobacteria. The presence of multitrophic species with the ability to prey on different food web components may contribute to ecosystem resilience. We emphasize the importance of understanding the trophic niche diversity of key zooplankton taxa to generate an accurate understanding of ecosystem functioning. Food web models based on size or phylogeny may not capture the important role of individual species and may not be detailed enough to predict energy pathways of plankton food webs and thus the vulnerability of ecosystems to environmental change. Combined with estimates of prey biomass and predator feeding rates, the approach used in this study is a suitable entry point to food web modelling and ecosystem network analysis.
Supplementary Material
Acknowledgements
We would like to direct our gratitude to the pelagic monitoring group at Stockholm University, especially Stefan Svensson and Jakob Walve for hosting us during the sampling effort. We also acknowledge support from the National Genomics infrastructure in Stockholm and Uppsala, funded by Science for Life Laboratory, the Knut and Alice Wallenberg Foundation and the Swedish Research Council, and SNIC/Uppsala Multidisciplinary Centre for Advanced Computational Science for assistance with massively parallel sequencing and access to the UPPMAX computational infrastructure.
Data accessibility
DNA sequences and associated metadata were uploaded to the ENA under accession no. PRJEB39191 (https://www.ebi.ac.uk/ena/browser/view/PRJEB39191). RSV data files used for statistical analysis, together with the R code to generate the figures are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.gb5mkkwpw [47].
Authors' contributions
A.N.: conceptualization, data curation, formal analysis, investigation, methodology, project administration, software, validation, visualization, writing-original draft, writing-review and editing; S.Z.-T: conceptualization, data curation, investigation, methodology, project administration, validation, writing-review and editing; M.W.: conceptualization, funding acquisition, project administration, resources, supervision, writing-review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Competing interests
We have no competing interests.
Funding
The research was funded by the Swedish Research Council project no. 2016-04685.
References
- 1.Cadotte MW, Carscadden K, Mirotchnick N, Cadotte MW, Carscadden K, Mirotchnick N. 2011. Beyond species: functional diversity and the maintenance of ecological processes and services. J. Appl. Ecol. 48, 1079-1087. ( 10.1111/j.1365-2664.2011.02048.x) [DOI] [Google Scholar]
- 2.Naeem S, Duffy JE, Zavaleta E. 2012. The functions of biological diversity in an age of extinction. Science 336, 1401-1406. ( 10.1126/science.1215855) [DOI] [PubMed] [Google Scholar]
- 3.Calbet A, Landry MR. 2004. Phytoplankton growth, microzooplankton grazing, and carbon cycling in marine systems. Limnol. Oceanogr. 49, 51-57. ( 10.4319/lo.2004.49.1.0051) [DOI] [Google Scholar]
- 4.Steinberg DK, Landry MR. 2017. Zooplankton and the ocean carbon cycle. Ann. Rev. Mar. Sci. 9, 413-444. ( 10.1146/annurev-marine-010814-015924) [DOI] [PubMed] [Google Scholar]
- 5.Mitra A, Davis C. 2010. Defining the ‘to’ in end-to-end models. Prog. Oceanogr. 84, 39-42. ( 10.1016/j.pocean.2009.09.004) [DOI] [Google Scholar]
- 6.Cushing DH. 1990. Plankton production and year-class strength in fish populations: an update of the match/mismatch hypothesis. Adv. Mar. Biol. 26, 249-293. ( 10.1016/S0065-2881(08)60202-3) [DOI] [Google Scholar]
- 7.Sherr EB, Sherr BF. 2002. Significance of predation by protists in aquatic microbial food webs. Antonie Van Leeuwenhoek 81, 293-308. [DOI] [PubMed] [Google Scholar]
- 8.Bindoff NL, et al. 2019. Changing ocean, marine ecosystems, and dependent communities. In Special report on the ocean and cryosphere in a changing climate (eds Pörtner H-O, et al.), pp. 447-588. New York, NY: IPCC. [Google Scholar]
- 9.Mitra A, et al. 2014. Bridging the gap between marine biogeochemical and fisheries sciences; configuring the zooplankton link. Prog. Oceanogr. 129, 176-199. ( 10.1016/j.pocean.2014.04.025) [DOI] [Google Scholar]
- 10.Heneghan RF, Everett JD, Blanchard JL, Richardson AJ. 2016. Zooplankton are not fish: improving zooplankton realism in size-spectrum models mediates energy transfer in food webs. Front. Mar. Sci. 3, 1-15. ( 10.3389/fmars.2016.00201) [DOI] [Google Scholar]
- 11.Petchey OL, Gaston KJ. 2006. Functional diversity: back to basics and looking forward. Ecol. Lett. 9, 741-758. ( 10.1111/j.1461-0248.2006.00924.x) [DOI] [PubMed] [Google Scholar]
- 12.Boyce DG, Frank KT, Leggett WC. 2015. From mice to elephants: overturning the ‘one size fits all’ paradigm in marine plankton food chains. Ecol. Lett. 18, 504-515. ( 10.1111/ele.12434) [DOI] [PubMed] [Google Scholar]
- 13.Sommer U, Stibor H. 2002. Copepoda – Cladocera – Tunicata: the role of three major mesozooplankton groups in pelagic food webs. Ecol. Res. 17, 161-174. ( 10.1046/j.1440-1703.2002.00476.x) [DOI] [Google Scholar]
- 14.Stibor H, et al. 2004. Copepods act as a switch between alternative trophic cascades in marine pelagic food webs. Ecol. Lett. 7, 321-328. ( 10.1111/j.1461-0248.2004.00580.x) [DOI] [Google Scholar]
- 15.Arndt H. 1993. Rotifers as predators on components of the microbial web (bacteria, heterotrophic flagellates, ciliates) – a review. In Hydrobiologia, pp. 231-246. Dordrecht, The Netherlands: Springer Netherlands. ( 10.1007/BF00025844) [DOI] [Google Scholar]
- 16.Pourriot R. 1977. Food and feeding habits of Rotifera. Hydrobiol. Beih. Ergebn. Limnol. 8, 243-260. [Google Scholar]
- 17.Bogdan KG, Gilbert JJ, Starkweather PL. 1980. In situ clearance rates of planktonic rotifers. In Hydrobiologica, pp. 73-77. Dordrecht, The Netherlands: Springer Netherlands. ( 10.1007/978-94-009-9209-2_14) [DOI] [Google Scholar]
- 18.Bogdan KG, Gilbert JJ. 1982. Seasonal patterns of feeding by natural populations of Keratella, Polyarthra, and Bosmina: clearance rates, selectivities, and contributions to community grazing1. Limnol. Oceanogr. 27, 918-934. ( 10.4319/lo.1982.27.5.0918) [DOI] [Google Scholar]
- 19.Gilbert JJ, Jack JD. 1993. Rotifers as predators on small ciliates. In Rotifer symposium VI, pp. 247-253. Dordrecht, The Netherlands: Springer Netherlands. ( 10.1007/978-94-011-1606-0_32) [DOI] [Google Scholar]
- 20.Gilbert JJ. 1980. Observations on the susceptibility of some protists and rotifers to predation by asplanchna girodi. Hydrobiologia 73, 87-91. ( 10.1007/BF00019431) [DOI] [Google Scholar]
- 21.Kiørboe T. 2011. How zooplankton feed: mechanisms, traits and trade-offs. Biol. Rev. 86, 311-339. ( 10.1111/j.1469-185X.2010.00148.x) [DOI] [PubMed] [Google Scholar]
- 22.Post DM. 2002. Using stable isotopes to estimate trophic position: models, methods, and assumptions. Ecology 83, 703. ( 10.2307/3071875) [DOI] [Google Scholar]
- 23.Gutiérrez-Rodríguez A, Décima M, Popp BN, Landry MR. 2014. Isotopic invisibility of protozoan trophic steps in marine food webs. Limnol. Oceanogr. 59, 1590-1598. ( 10.4319/lo.2014.59.5.1590) [DOI] [Google Scholar]
- 24.Pompanon F, Deagle BE, Symondson WOCC, Brown DS, Jarman SN, Taberlet P. 2012. Who is eating what: diet assessment using next generation sequencing. Mol. Ecol. 21, 1931-1950. ( 10.1111/j.1365-294X.2011.05403.x) [DOI] [PubMed] [Google Scholar]
- 25.Roslin T, Majaneva S. 2016. The use of DNA barcodes in food web construction—terrestrial and aquatic ecologists unite!. Genome 59, 603-628. ( 10.1139/gen-2015-0229) [DOI] [PubMed] [Google Scholar]
- 26.Ray JL, et al. 2016. Metabarcoding and metabolome analyses of copepod grazing reveal feeding preference and linkage to metabolite classes in dynamic microbial plankton communities. Mol. Ecol. 25, 5585-5602. ( 10.1111/mec.13844) [DOI] [PubMed] [Google Scholar]
- 27.Yi X, Huang Y, Zhuang Y, Chen H, Yang F, Wang W, Xu D, Liu G, Zhang H. 2017. In situ diet of the copepod Calanus sinicus in coastal waters of the South Yellow Sea and the Bohai Sea. Acta Oceanol. Sin. 36, 68-79. ( 10.1007/s13131-017-0974-6) [DOI] [Google Scholar]
- 28.Craig C, Kimmerer WJ, Cohen CS. 2014. A DNA-based method for investigating feeding by copepod nauplii. J. Plankton Res. 36, 271-275. ( 10.1093/plankt/fbt104) [DOI] [Google Scholar]
- 29.Zamora-Terol S, Novotny A, Winder M. 2020. Reconstructing marine plankton food web interactions using DNA metabarcoding. Mol. Ecol. 29, 3380-3395. ( 10.1111/mec.15555) [DOI] [PubMed] [Google Scholar]
- 30.Paerl HW, Huisman J. 2008. Blooms like it hot. Am. Assoc. Adv. Sci. 320, 57-58. ( 10.1210/jcem-10-10-1361) [DOI] [PubMed] [Google Scholar]
- 31.Cloern JE, et al. 2016. Human activities and climate variability drive fast-paced change across the world's estuarine-coastal ecosystems. Glob. Chang. Biol. 22, 513-529. ( 10.1111/gcb.13059) [DOI] [PubMed] [Google Scholar]
- 32.Schmidt K, et al. 2020. Increasing picocyanobacteria success in shelf waters contributes to long-term food web degradation. Glob. Chang. Biol. 26, 5574-5587. ( 10.1111/gcb.15161) [DOI] [PubMed] [Google Scholar]
- 33.Karlson AML, et al. 2015. Nitrogen fixation by cyanobacteria stimulates production in Baltic food webs. Ambio 44, 413-426. ( 10.1007/s13280-015-0660-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loick-Wilde N, Fernández-Urruzola I, Eglite E, Liskow I, Nausch M, Schulz-Bull D, Wodarg D, Wasmund N, Mohrholz V. 2019. Stratification, nitrogen fixation, and cyanobacterial bloom stage regulate the planktonic food web structure. Glob. Chang. Biol. 25, 794-810. ( 10.1111/gcb.14546) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engstrom J. 2000. Feeding interactions of the copepods Eurytemora affinis and Acartia bifilosa with the cyanobacteria Nodularia sp. J. Plankton Res. 22, 1403-1409. ( 10.1093/plankt/22.7.1403) [DOI] [Google Scholar]
- 36.Naturvårdsverket. 2009. Beskrivning av delprogram Fria vattenmassan. See https://www.havochvatten.se/download/18.64f5b3211343cffddb280005451/1348912810782/beskrivning-delprogram-fria-vattenmassan.pdf (accessed on 3 April 2018).
- 37.Herlemann DPR, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, Andersson AF. 2011. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5, 1571-1579. ( 10.1038/ismej.2011.41) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu YOO, Karlson B, Charvet S, Andersson AF. 2016. Diversity of pico- to mesoplankton along the 2000 km salinity gradient of the Baltic Sea. Front. Microbiol. 7, 679. ( 10.3389/fmicb.2016.00679) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho TW, Hwang J-S, Cheung MK, Kwan HS, Wong CK. 2017. DNA-based study of the diet of the marine calanoid copepod Calanus sinicus. J. Exp. Mar. Bio. Ecol. 494, 1-9. ( 10.1016/j.jembe.2017.04.004) [DOI] [Google Scholar]
- 40.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188-7196. ( 10.1093/nar/gkm864) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Decelle J, et al. 2015. PhytoREF: a reference database of the plastidial 16S rRNA gene of photosynthetic eukaryotes with curated taxonomy. Mol. Ecol. Resour. 15, 1435-1445. ( 10.1111/1755-0998.12401) [DOI] [PubMed] [Google Scholar]
- 42.Guillou L, et al. 2013. The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 41, D597-D604. ( 10.1093/nar/gks1160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217. ( 10.1371/journal.pone.0061217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oksanen J, Kindt R, Legendre P, O'Hara B, Simpson GL, Solymos PM, Stevens MHH, Wagner H. 2007. The vegan package. Commun. Ecol. Packag. 10, 613-637. [Google Scholar]
- 45.Wickham H. 2016. Ggplot2: elegant graphics for data analysis. Berlin, Germany: Springer. [Google Scholar]
- 46.Gu Z, Gu L, Eils R, Schlesner M, Brors B. 2014. circlize implements and enhances circular visualization in R. Bioinformatics 30, 2811-2812. ( 10.1093/bioinformatics/btu393) [DOI] [PubMed] [Google Scholar]
- 47.Novotny A, Zamora-Terol S, Winder M. 2021. Data from: DNA metabarcoding reveals trophic niche diversity of micro and mesozooplankton species. Dryad Digital Repository. ( 10.5061/dryad.gb5mkkwpw) [DOI] [PMC free article] [PubMed]
- 48.Sommer F, Stibor H, Sommer U, Velimirov B. 2000. Grazing by mesozooplankton from Kiel Bight, Baltic Sea, on different sized algae and natural seston size fractions. Mar. Ecol. Prog. Ser. 199, 43-53. ( 10.3354/meps199043) [DOI] [Google Scholar]
- 49.Motwani NH, Gorokhova E. 2013. Mesozooplankton grazing on picocyanobacteria in the baltic sea as inferred from molecular diet analysis. PLoS ONE 8, e79230. ( 10.1371/journal.pone.0079230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grinienė E, Šulčius S, Kuosa H. 2016. Size-selective microzooplankton grazing on the phytoplankton in the Curonian Lagoon (SE Baltic Sea). Oceanologia 58, 292-301. ( 10.1016/j.oceano.2016.05.002) [DOI] [Google Scholar]
- 51.Starkweather PL, Bogdan KG. 1980. Detrital feeding in natural zooplankton communities: discrimination between live and dead algal foods. Hydrobiologia 73, 83-85. ( 10.1007/BF00019430) [DOI] [Google Scholar]
- 52.Mironova E, Telesh I, Skarlato S. 2012. Diversity and seasonality in structure of ciliate communities in the Neva Estuary (Baltic Sea). J. Plankton Res. 34, 208-220. ( 10.1093/plankt/fbr095) [DOI] [Google Scholar]
- 53.Aberle N, Lengfellner K, Sommer U. 2007. Spring bloom succession, grazing impact and herbivore selectivity of ciliate communities in response to winter warming. Oecologia 150, 668-681. ( 10.1007/s00442-006-0540-y) [DOI] [PubMed] [Google Scholar]
- 54.Capriulo GM. 1982. Feeding of field collected tintinnid micro-zooplankton on natural food. Mar. Biol. 71, 73-86. ( 10.1007/BF00396994) [DOI] [Google Scholar]
- 55.Renz J, Hirche HJ. 2006. Life cycle of Pseudocalanus acuspes Giesbrecht (Copepoda, Calanoida) in the Central Baltic Sea: I. Seasonal and spatial distribution. Mar. Biol. 148, 567-580. ( 10.1007/s00227-005-0103-5) [DOI] [Google Scholar]
- 56.Schulz J, et al. 2012. Spatial and temporal habitat partitioning by zooplankton in the Bornholm Basin (central Baltic Sea). Prog. Oceanogr. 107, 3-30. ( 10.1016/j.pocean.2012.07.002) [DOI] [Google Scholar]
- 57.Tamminen T, Andersen T. 2007. Seasonal phytoplankton nutrient limitation patterns as revealed by bioassays over Baltic Sea gradients of salinity and eutrophication. Mar. Ecol. Prog. Ser. 340, 121-138. ( 10.3354/meps340121) [DOI] [Google Scholar]
- 58.Calbet A, Vaqué D, Felipe J, Vila M, Alcaraz M, Estrada M. 2003. Relative grazing impact of microzooplankton and mesozooplankton. Mar. Ecol. Prog. Ser. 259, 303-309. ( 10.3354/meps259303) [DOI] [Google Scholar]
- 59.Winder M, Sommer U. 2012. Phytoplankton response to a changing climate. Hydrobiologia 698, 5-16. ( 10.1007/s10750-012-1149-2) [DOI] [Google Scholar]
- 60.Motwani NH, Duberg J, Svedén JB, Gorokhova E. 2018. Grazing on cyanobacteria and transfer of diazotrophic nitrogen to zooplankton in the Baltic Sea. Limnol. Oceanogr. 63, 672-686. ( 10.1002/lno.10659) [DOI] [Google Scholar]
- 61.Granéli E, Wallström K, Larsson U, Granéli W, Elmgren R. 1990. Nutrient limitation of primary production in the Baltic Sea area. Ambio 19, 142-151. [Google Scholar]
- 62.Loick-Wilde N, Dutz J, Miltner A, Gehre M, Montoya JP, Voss M. 2012. Incorporation of nitrogen from N2 fixation into amino acids of zooplankton. Limnol. Oceanogr. 57, 199-210. ( 10.4319/lo.2012.57.1.0199) [DOI] [Google Scholar]
- 63.Wannicke N, Korth F, Liskow I, Voss M. 2013. Incorporation of diazotrophic fixed N2 by mesozooplankton: case studies in the southern Baltic Sea. J. Mar. Syst. 117–118, 1-13. ( 10.1016/j.jmarsys.2013.03.005) [DOI] [Google Scholar]
- 64.Ploug H, Musat N, Adam B, Moraru CL, Lavik G, Vagner T, Bergman B, Kuypers MMM. 2010. Carbon and nitrogen fluxes associated with the cyanobacterium Aphanizomenon sp. in the Baltic Sea. ISME J. 4, 1215-1223. ( 10.1038/ismej.2010.53) [DOI] [PubMed] [Google Scholar]
- 65.Ploug H, Adam B, Musat N, Kalvelage T, Lavik G, Wolf-Gladrow D, Kuypers MMM. 2011. Carbon, nitrogen and O 2 fluxes associated with the cyanobacterium Nodularia spumigena in the Baltic Sea. ISME J. 5, 1549-1558. ( 10.1038/ismej.2011.20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eglite E, Graeve M, Dutz J, Wodarg D, Liskow I, Schulz-Bull D, Loick-Wilde N. 2019. Metabolism and foraging strategies of mid-latitude mesozooplankton during cyanobacterial blooms as revealed by fatty acids, amino acids, and their stable carbon isotopes. Ecol. Evol. 9, 9916-9934. ( 10.1002/ece3.5533) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kortsch S, et al. 2021. Disentangling temporal food web dynamics facilitates understanding of ecosystem functioning. J. Anim. Ecol. 90, 1-12. ( 10.1111/1365-2656.13447) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Novotny A, Zamora-Terol S, Winder M. 2021. Data from: DNA metabarcoding reveals trophic niche diversity of micro and mesozooplankton species. Dryad Digital Repository. ( 10.5061/dryad.gb5mkkwpw) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
DNA sequences and associated metadata were uploaded to the ENA under accession no. PRJEB39191 (https://www.ebi.ac.uk/ena/browser/view/PRJEB39191). RSV data files used for statistical analysis, together with the R code to generate the figures are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.gb5mkkwpw [47].