Abstract
Severe acute respiratory syndrome coronavirus 2 is responsible for coronavirus disease 2019 (COVID‐19). While COVID‐19 is often benign, a subset of patients develops severe multilobar pneumonia that can progress to an acute respiratory distress syndrome. There is no cure for severe COVID‐19 and few treatments significantly improved clinical outcome. Dexamethasone and possibly aspirin, which directly/indirectly target the biosynthesis/effects of numerous lipid mediators are among those options. Our objective was to define if severe COVID‐19 patients were characterized by increased bioactive lipids modulating lung inflammation. A targeted lipidomic analysis of bronchoalveolar lavages (BALs) by tandem mass spectrometry was done on 25 healthy controls and 33 COVID‐19 patients requiring mechanical ventilation. BALs from severe COVID‐19 patients were characterized by increased fatty acids and inflammatory lipid mediators. There was a predominance of thromboxane and prostaglandins. Leukotrienes were also increased, notably LTB4, LTE4, and eoxin E4. Monohydroxylated 15‐lipoxygenase metabolites derived from linoleate, arachidonate, eicosapentaenoate, and docosahexaenoate were also increased. Finally yet importantly, specialized pro‐resolving mediators, notably lipoxin A4 and the D‐series resolvins, were also increased, underscoring that the lipid mediator storm occurring in severe COVID‐19 involves pro‐ and anti‐inflammatory lipids. Our data unmask the lipid mediator storm occurring in the lungs of patients afflicted with severe COVID‐19. We discuss which clinically available drugs could be helpful at modulating the lipidome we observed in the hope of minimizing the deleterious effects of pro‐inflammatory lipids and enhancing the effects of anti‐inflammatory and/or pro‐resolving lipid mediators.
Keywords: COVID‐19, docosanoids, eicosanoids, eoxins, specialized pro‐resolving mediators, thromboxane
Abbreviations
- AA
arachidonic acid
- ALA
linolenic acid
- BAL
bronchoalveolar lavage
- DGLA
dihomo‐γ‐linolenic acid
- DHA
docosahexaenoic acid
- DPA
docosapentaenoic acid
- EPA
eicosapentaenoic acid
- EX
eoxin
- HDHA
hydroxy‐docosahexaenoic acid
- HETE
hydroxyeicosatetraenoic acid
- HEPE
hydroxyeicosapentaenoic acid
- 12‐HHTrE
12‐hydroxy‐heptadecatrienoic acid
- HODE
hydroxyoctadecadienoic acid
- KETE
oxo‐eicosatetraenoic acid
- KODE
oxo‐octadecadienoic acid
- LA
linoleic acid
- LO
lipoxygenase
- LT
leukotriene
- LX
lipoxin
- PDX
protectin D1
- PG
prostaglandin
- Rv
resolvin
- SPM
specialized pro‐resolving mediators
- TX
thromboxane
1. INTRODUCTION
Coronavirus disease 2019 (COVID‐19) is the infectious disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). 1 SARS‐CoV‐2 spreads predominantly from respiratory droplets of infected individuals to epithelial cells in the upper airways and oral cavity. 2 COVID‐19 manifestations are broad, from asymptomatic carriers to severe viral pneumonia leading to respiratory failure and ultimately acute respiratory distress syndrome (ARDS) requiring mechanical ventilation. 3 , 4 In some patients, there is a yet to be explained hyperinflammatory response.
Autopsies from severe COVID‐19 patients unmasked a heterogeneous disease that can be characterized by diffuse alveolar and/or epithelial damage, thrombosis, pulmonary embolism, and lymphocytic and/or granulocytic inflammatory cell infiltration. 4 , 5 , 6 An uncontrolled systemic inflammatory response known as the cytokine storm is suggested as an important contributor of SARS‐CoV‐2 lethality and is the consequence of an important release of cytokines such as tumor necrosis factor‐α, interleukin (IL)‐1β, and IL‐6, 4 , 7 although this remains unclear. 8 The disease heterogeneity suggests that, aside cytokines/chemokines, other immunological effectors contribute to the sustained inflammatory responses observed in severe COVID‐19 cases.
Bioactive lipids, notably eicosanoids, likely promote their fair share of deleterious effects by enhancing leukocyte recruitment/activation, promoting the exudate formation and by stimulating platelet aggregation and thrombus formation. In contrast, specialized pro‐resolving mediators (SPMs), mainly consisting of docosanoids, could dampen inflammation and promote its resolution. 9 Of note, some of these lipids are elevated in the blood of COVID‐19 patients. 10 , 11 While therapeutic approaches targeting the biosynthesis/effects of inflammatory lipids are readily available, the levels of those lipids in the lungs during severe COVID‐19 were unknown.
Herein, we performed a targeted lipidomic analysis of bronchoalveolar lavages fluids (BALs) from healthy volunteers and severe COVID‐19 patients. Numerous eicosanoids, notably thromboxane, and SPMs were found in severe COVID‐19 patients’ BALs. Our study further highlights that few associations exist between clinical parameters and the bioactive lipids present in the primary site of disease, the lungs.
2. METHODS
2.1. Materials
All lipids were obtained from Cayman Chemical (Ann Arbor, MI, USA) and all LC‐MS grade solvents were purchased from Fisher Scientific (Ottawa, Canada).
2.2. Ethics
This study was approved by the local ethics committee (Cheikh Zaid Hospital in Rabat Morocco (Project: CEFCZ/PR/2020‐ PR04); Québec City Heart & Lung Institute; CHU de Québec‐Université Laval). The study complies with the Declaration of Helsinki and all subjects signed a consent form.
2.3. Subject selection
Non‐smoking healthy subjects taking no other medication than anovulants, without any documented acute or chronic inflammatory disease and without recent (8 weeks) airway infection were recruited for bronchoalveolar lavages at the Québec City Heart & Lung Institute (Canada). Severe COVID‐19 patients were enrolled based on the inclusion criteria for intubation and the need of mechanical ventilation at Cheikh Zaid Hospital (Rabat, Morocco) between May and June 2020, before COVID‐19 therapies such as remdesevir and dexamethasone were greenlit for treating COVID‐19. Within 2 hours after intubation and the initiation of mechanical ventilation, blood sampling was done to assess the clinical variables (Table 1, Table S1 for each individual subjects). Patients were ventilated in the prone decubitus position with a tidal volume of 6 mL/kg and a PEEP varying between 8 and 14 cm H2O. The FiO2 was adjusted between 0.6 and 1.0 to obtain a SpO2 ≥ 92%.
TABLE 1.
Clinical characteristics of BAL donors
| COVID‐19 | Healthy controls | |
|---|---|---|
| N | 33 | 25 |
| Women/Men | 16/17 | 16/9 |
| Smoker (Y/N) | 5/28 | 0/25 |
| Death (Y/N) | 2/31 | N/A |
| SARS‐CoV‐2 detection in BAL (Ct) | 25 ± 1 | N/A |
| Age | 58 ± 3 | 26 ± 1 |
| Weight (kg) | 73 ± 3 | N/A |
| Hospital stay (days) | 21 ± 2 | N/A |
| Days before BAL | 3 ± 1 | N/A |
| D‐Dimers (mg/mL) | 1.21 ± 0.13 | N/A |
| CRP (mg/l) | 20.65 ± 2.53 | N/A |
| ALT (U/l) | 32.6 ± 2.1 | N/A |
| AST (U/l) | 35.1 ± 3.0 | N/A |
| LDH (U/l) | 616 ± 41 | N/A |
| Hemoglobin (g/l) | 123 ± 2 | N/A |
| Blood Monocytes (million/mL) | 0.3 ± 0.03 | N/A |
| Blood Neutrophils (million/mL) | 2.9 ± 0.19 | N/A |
| Blood Eosinophils (million/mL) | 0.03 ± 0.003 | N/A |
| Blood Platelets (million/mL) | 182 ± 13 | N/A |
| Blood Lymphocytes (million/mL) | 1.13 ± 0.13 | N/A |
| Platelet to Lymphocyte ratio (PLR) | 223.89 ± 34.93 | N/A |
| BAL Neutrophils (million/mL) | 24.7 ± 2.2 | 0.001 ± 0.0002 |
| BAL Eosinophils (million/mL) | 0.2 ± 0.08 | 0.000 ± 0.000 |
| BAL Lymphocytes (million/mL) | 21.8 ± 2.4 | 0.004 ± 0.001 |
| BAL macrophages (million/mL) | N/A | 0.073 ± 0.007 |
Data are presented as the mean ±SEM.
Abbreviations: ALT, alanine transaminase; AST, aspartate transaminase; CRP, C‐reactive protein; LDH, Lactate dehydrogenase; N/A, not available.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
2.4. Bronchoalveolar lavages
BAL fluids from healthy volunteers were obtained as follows: One 50 mL bolus of sterile 0.9% saline was injected in a sub‐segmental bronchi of the right middle lobe. The obtained lavages were centrifuged (350 g, 10 minutes) to pellet cells and the supernatants were immediately frozen (−80° Celsius) until further processing. A 5 mL aliquot of the samples was thawed then reduced to ~500 µL using a stream of nitrogen. BAL fluids from severe COVID‐19 patients were obtained less than 2 hours following intubation as follows: a total of 100 mL of sterile 0.9% saline (2 boli of 50 mL) was injected in a sub‐segmental bronchi of the right middle lobe. The obtained lavages were pooled and centrifuged (320 g, 15 minutes) to pellet cells. Supernatants were next concentrated as for the BALs of healthy donors and immediately frozen (−80° Celsius) until further processing.
2.5. LC‐MS/MS analyses
Concentrated samples (500 µL) were denatured with 500 µL of LC‐MS–grade MeOH containing the internal standards then warmed at 60º Celsius for 30 minutes to inactivate SARS‐CoV‐2, denatured overnight (−20° Celsius), and then centrifuged (10,000 g) to remove the denaturated proteins. Under these conditions, no significant degradation of internal standards and selected lipids and lipid mediators was observed (Figure S1). Supernatants were diluted with water containing 0.01% acetic acid and lipids were extracted by solid phase extraction using Strata‐X cartridges (Polymeric Reversed Phase, Phenomenex, USA) as described before. 12 Lipids were separated using the same column and LC program as in, 12 quantification was done using a Shimadzu 8050 triple quadrupole mass spectrometer and analyses were performed using multiple reaction monitoring for the specific mass transitions of each lipid as well as the deuterated internal standard used (Table S2). Due to the large number of analytes being quantitated and/or commercial availability, some lipid mediators were analyzed with a surrogate, deuterated internal standard.
2.6. Statistical analysis
Statistical analysis was made using the GraphPad Prism 9 software. The normality assumption was verified using the Shapiro‐Wilk test and p‐values were calculated using the Mann‐Whitney test. Spearman correlations were calculated using corr.test function from pshych CRAN package and the correlation matrices were generated using the corrplot package. Values of P < .05 were considered significant.
3. RESULTS
While the cytokine profile observed in severe COVID‐19 patients is relatively well‐defined, 4 , 13 , 14 , 15 , 16 no study investigated the lipid content of BALs. We thus performed a targeted lipidomic analysis to define whether lipid mediators were present/increased in the lungs of severe COVID‐19 patients.
In contrast to the BALs of healthy controls, numerous eicosanoids were detected in those of severe COVID‐19 patients. As such, BALs from COVID‐19 patients had increased AA levels as well as all measured COX metabolites (Figure 1B‐D). The most increased COX metabolites were TXB2 >> PGE2 ~ 12‐HHTrE > PGD2. Leukotrienes also regulate the inflammatory response in the airway/lung, notably during ARDS, 17 , 18 and their involvement in COVID‐19 has been postulated. 19 Leukotrienes were not detected in the BALs of healthy volunteers, with the exception of LTB4 (4/25 subjects). In contrast, COVID‐19 patients had increased levels of LTB4 and its CYP4F3A metabolite 20‐COOH‐LTB4 (Figure 1F) as well as minor metabolites (20‐OH‐LTB4 and 12‐oxo‐LTB4). Noteworthy, LTB4 levels in the BALs of COVID‐19 patients are comparable to those found in BALs of ARDS patients. 20 Moreover, the ratio between the sum of 20‐OH‐ and 20‐COOH‐LTB4 to LTB4 ((20‐OH‐ + 20‐COOH‐LTB4)/LTB4) was 2.843 ± 0.532 (mean ± SEM), reflecting an increased omega oxidation of LTB4. The cysteinyl‐leukotrienes LTE4 and the 15‐LO‐derived eoxin E4 (EXE4) were also found in most COVID‐19 patients (Figure 1G). Surprisingly, EXE4 levels trended to be higher than those of LTE4. In line with the increase in both 5‐LO‐ and 15‐LO‐derived leukotrienes, other 5‐, 12‐, and 15‐LO metabolites, were increased in the BALs of COVID‐19 patients, notably 5‐HETE, 12‐HETE, 15‐HETE, and 15‐KETE (Figure 1I,J). Dual LO metabolites from AA were also significantly increased. Of note, 12‐ and 15‐HEPE derived from EPA metabolism, were also increased in the BAL of COVID‐19 patients despite comparable levels of EPA between healthy controls and COVID‐19 patients (Figure 1K,L), while the EPA‐derived SPM RvE1 was not detected. Overall, the eicosanoid profile of intubated COVID‐19 patients mainly consisted of TXB2 and prostaglandins (62%), followed by the different HETEs and LTs (Figure 1L).
FIGURE 1.
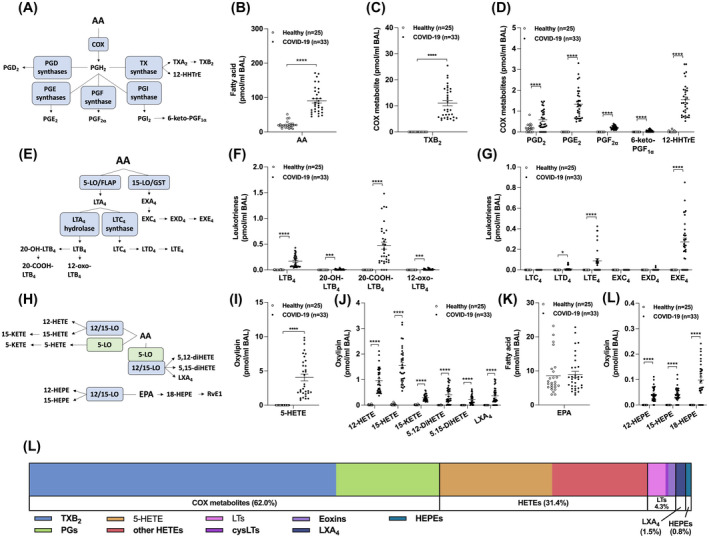
Levels of eicosanoids in the BAL fluids of healthy and COVID‐19 patients. BALs were obtained and processed as described in Methods. Lipids then were extracted and quantitated by LC‐MS/MS. A, Metabolism of arachidonic acid (AA) by the cyclooxygenase (COX) pathway. B, AA levels. C,D, Levels of COX‐derived metabolites. E, Leukotriene (LT) biosynthetic pathways. F,G, Levels of LTs and eoxins (EX). H, Metabolism of AA and eicosapentaenoic acid (EPA) by 5‐, 12‐, and 15‐lipoxygenase (LO). I,J, Levels of LO‐derived metabolites of AA. K, EPA levels. L, Levels of LO‐derived EPA metabolites. L, Proportions of eicosanoids from panel C, D, F, G, I, and K. Results are from the BALs of 25 healthy subjects and 33 COVID‐19 patients. P values were obtained by performing a Mann‐Whitney test using the GraphPad Prism 9 software: *** = P < .001, **** = P < .0001
Many SPMs arise from the metabolism of docosanoids 9 and given their documented pro‐resolving actions and antimicrobial activities, SPMs were recently postulated as being a potential approach for diminishing the inflammatory burden of COVID‐19. 21 Both DPA(n‐3) and DHA levels were increased in the BALs of COVID‐19 patients (Figure 2B). We also observed significant increases in 17‐HDPA(n‐3), 14‐HDHA and 17‐HDHA (Figure 2C). In line with these findings, D‐serie resolvins (aside from RvD3) and Protectin D1 (PDX) were also increased in the BALs of COVID‐19 patients (Figure 2D). Despite our good sensitivity (Table S1), Maresin‐1 and −2 were not detected.
FIGURE 2.
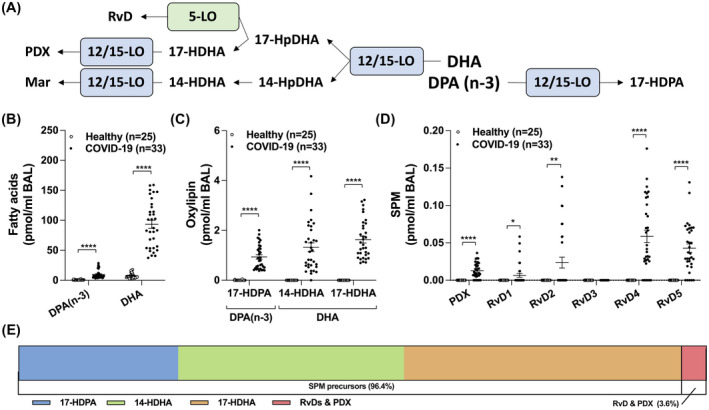
Levels of docosanoids in the BAL fluids of healthy and COVID‐19 patients. BALs were obtained and processed as described in Methods. Lipids then were extracted and quantitated by LC‐MS/MS. A, Docosanoid biosynthetic pathways. B, Levels of docosapentaenoic acid n‐3 (DPAn‐3) and docosahexaenoic acid (DHA). C,D, Docosanoid levels. E, Proportions of the different docosanoids from panel C and D. Results are from the BALs from 25 healthy subjects and 33 COVID‐19 patients. P values were obtained by performing a Mann‐Whitney test using the GraphPad Prism 9 software: * = P < .05, ** = P < .01, *** = P < .001, **** = P < .0001
Lastly, we quantitated the levels of 15‐LO metabolites derived from linoleic acid (LA), linolenic acid (ALA) and dihomo‐γ‐linolenic acid (DGLA). LA levels were increased in the BAL of COVID‐19 patients (Figure 3B), as well as its 15‐LO‐derived metabolites 13‐HODE and 13‐KODE (Figure 3C,D). Furthermore, 13‐HOTrE (derived from ALA) and 15‐HETrE (derived from DGLA) were both increased in COVID‐19 patients (Figure 3D3C). We also observed an increase in the non‐enzymatic metabolite 9‐HODE (derived from LA) (Figure 3C). We also found that other auto‐oxidative (or minor enzymatic) products were increased in the BAL of COVID‐19 patients, namely 8‐HETE and 11‐HETE (both from AA) as well as 18‐HEPE (from EPA), suggesting to an increased oxidative burst in the lungs of intubated COVID‐19 patients (not shown).
FIGURE 3.
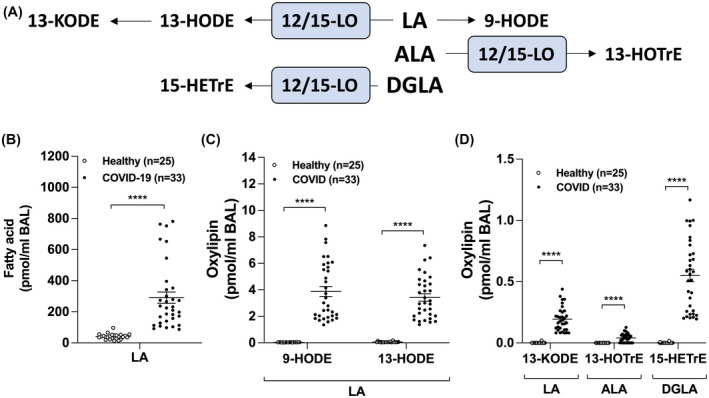
Levels of lipid mediators derived from linoleic acid (LA), α‐linolenic acid or dihomo‐γ‐linolenic acid (DGLA) in the BAL fluids of healthy and COVID‐19 patients. BALs were obtained and processed as described in Methods. Lipids then were extracted and quantitated by LC‐MS/MS. A, Metabolism of LA, ALA, and DGLA by the 12/15‐lipoxygenase (LO) pathways. B, Levels of LA. C,D, Levels of oxidized mediators from LA, ALA, and DGLA. Results are from the BALs from 25 healthy subjects and 33 COVID‐19 patients. P values were obtained by performing a Mann‐Whitney test using the GraphPad Prism 9 software: **** = P < .0001
Given that the increased in most lipids mediators we quantitated, we next addressed if correlations between lipids existed in the BAL of intubated COVID‐19 patients. Most lipids consistently correlated with each other with few exceptions (Figure 4). Indeed, fatty acids levels were not a key determinant for the levels of their metabolites, indicating that downstream enzymes (cyclooxygenases, lipoxygenases) are likely more important in that respect. Interestingly, all fatty acids correlated with each other. As expected, lipids within the same metabolic pathway correlated with each other. Moreover, all 12‐ and 15‐LO‐derived mediators (HETEs, diHETEs, RvDs, and PDX) correlated with each other, regardless of their fatty acid of origin. Unsurprisingly, the levels of LXA4 correlated with those of 5‐HETE and 15‐HETE, given that LXA4 requires both the 5‐ and the 15‐LO. Interestingly, LTE4 and EXE4 had a trend toward a negative correlation (r = −0.322, P = .0676) and they correlated with different bioactive lipid subsets, LTE4 positively correlating with fatty acid and 15‐HETrE levels while EXE4 mostly correlating with other 15‐lipoxygenase metabolites and LTB4. This suggests that some COVID‐19 patients are more prone to biosynthesize LTC4 while others are more prone to biosynthesize EXC4.
FIGURE 4.

Correlation between bioactive lipid mediators in the BALs of COVID‐19 patients. A Spearman correlation matrix between the different lipid mediators is shown. Positive correlations are shown using shades of blue. P < .05 was the threshold of significance and non‐significant correlations (P > .05) are marked by an X
We also investigated whether lipids from the BAL of COVID‐19 patients correlated with the clinical parameters shown in Table 1 and found a very limited number of correlations (Figure 5). Fatty acids negatively correlated with AST, ALT and body weight (not significant for DPAn‐3 and DHA), and positively correlated with the circulating platelet‐to‐lymphocyte ratio. LTE4 negatively correlated with blood eosinophil counts and ALT, while positively correlating with hemoglobin. In contrast, EXE4 positively correlated with blood eosinophils and we found a trend toward a positive correlation with ALT and toward an almost significant negative correlation with hemoglobin (r = −0.339, P‐value = 0.054). Interestingly, LXA4, PDX and, RvD4 positively correlated with blood eosinophil counts.
FIGURE 5.
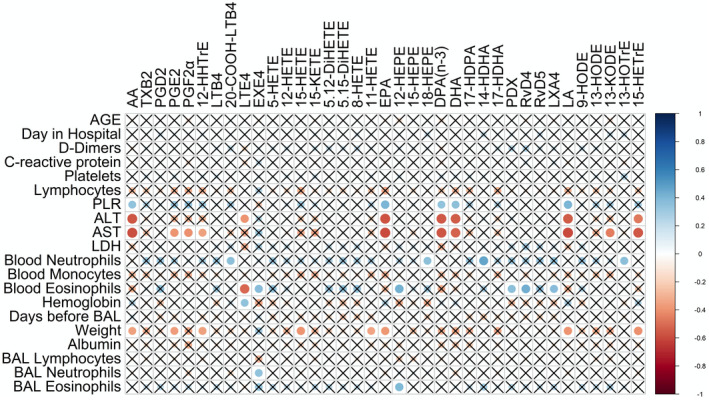
Correlation between bioactive lipids in the BALs of COVID‐19 patients and clinical parameters. A non‐parametric Spearman correlation matrix between Lipid mediators and clinical parameters was done using the corrplot package. Positive correlations are shown using shades of blue and negative correlations are shown using shades of red. P < .05 was the threshold of significance and non‐significant correlations (P > .05) are marked by an X
4. DISCUSSION
Bioactive lipids are recognized pharmaceutical targets in numerous inflammatory diseases and lipid modulators have been utilized over the last fifty years with great success. Defining whether bioactive lipid levels were increased in severe COVID‐19 patients was thus imperative. The lipidome arising from the metabolism of polyunsaturated fatty acids containing 20 carbons (eicosanoids) or 22 carbons (docosanoids) is continually expanding. While we performed a targeted lipidomic analysis on a significant fraction of them, we could not quantitate all of them, notably SPMs, due to our inability to secure appropriate internal standards. Thus, we cannot speculate on the levels and/or the potential roles of eicosanoids and docosanoids that we did not analyze in SARS‐CoV‐2‐infected people.
Herein, we provide evidence that BALs of intubated COVID‐19 patients contain extensive levels of bioactive lipids, which likely participate in the deleterious inflammatory response. More specifically, we highlight (1) an increase in COX metabolites, notably thromboxane; (2) an increase in leukotrienes, notably LTB4 LTE4, and EXE4; (3) an increase in 15‐LO metabolites from LA, AA, EPA, DPA, and DHA; (4) an increase in SPMs; (5) correlations between most lipid mediators, (6) a limited number of correlations between BAL lipids vs blood markers and/or clinical parameters.
The important increase in COX metabolites (Figure 1C) was expected, notably because of the previously documented increase in IL‐1β, 20 the latter being a good COX‐2 inducer. 22 TXA2 promotes bronchoconstriction and activate leukocytes and platelets, which is consistent with the enhanced platelet activation and thrombosis in COVID‐19 and the strong link between TXB2 and ARDS. 10 , 23 , 24 , 25 , 26 PGD2 acting via the DP2 receptor promotes the recruitment/activation of eosinophils, basophils, mastocytes, and innate lymphoid cells. 27 , 28 In contrast, PGE2 is mostly anti‐inflammatory via the EP2 and EP4 receptors 29 and PGI2 is a recognized microvasculature dilator. In that respect, a retrospective observational study showed that the administration of PGI2 (epoprostenol) was effective at improving the PaO2/FiO2 ratio in 50% of COVID‐19 patients. 30 Therapies targeting the prostaglandin pathway are numerous and usually effective at controlling inflammation. Dexamethasone limits COX‐2 expression 22 , 31 and might diminish COVID‐19 severity and mortality, at least in part, by diminishing COX metabolites. 32 Aspirin was shown to significantly diminish ICU admissions and the need of mechanical ventilation, 33 and to diminish disease duration and mortality, 34 which is in line with a possible clinical benefit of early aspirin for the prevention of ARDS. 26 Altogether, our data support the concept that dexamethasone might improve severe COVID‐19 by diminishing the prostaglandins/thromboxane storm in the lungs and that aspirin is also probably safe and beneficial for patients severely affected by COVID‐19 although this needs to be investigated. To that end, aspirin would be a cheap and easily accessible alternative to limit the lipid storm and its putative burden during severe COVID‐19 episodes. Finally, testing whether blocking the deleterious effects of PGD2 and TXA2 with the dual DP2/TP antagonist Ramatroban might be beneficial in COVID‐19 is of interest, as proposed recently. 35
Leukotrienes (LTB4, LTE4, and EXE4) were also increased in the BALs of COVID‐19 patients (Figure 1F,G). This indicates that LTB4 and cysteinyl‐leukotrienes might also participate in the inflammatory burden of severe COVID‐19 patients. 19 While LTB4 is known to stimulate host defense during viral infections, 35 the observed ω‐LTB4/LTB4 ratio of 2.84 is comparable to those found in cystic fibrosis and severely burned patients, two conditions in which infections are poorly controlled. 36 , 37 Conceptually, this might diminish LTB4 effectiveness at promoting its host defense‐related functions. 38 The trend toward an inverse correlation between LTE4 and EXE4, which is supported by their distinct correlation pattern with other lipids (Figure 4) and clinical parameters (Figure 5), suggests that these cysteinyl‐LTs are coming from different cellular sources. To that end, EXE4 levels in the BALs positively and significantly correlated with blood eosinophils counts while there was a strong trend between EXE4 and BAL eosinophils counts. This suggests that eoxin presence/biosynthesis is partly linked to eosinophils although the involvement of other 15‐LO expressing cells, notably epithelial cells cannot be excluded. 39 Furthermore, the detection of EXE4 in the BAL is of importance, since this mediator has rarely been detected in biological samples. Our data also suggest that the use of phosphodiesterase 4 inhibitors (eg, Roflumilast) in COVID‐19 could be of interest as they might diminish inflammation by increasing intracellular cAMP concentrations, thereby decreasing AA release and eicosanoid biosynthesis. 40 , 41 , 42 , 43 , 44
The metabolism of the omega‐3 fatty acids EPA, DPA, and DHA notably leads to SPM biosynthesis, which are anti‐inflammatory and pro‐resolving lipids. Their high levels and correlations with eicosanoids were unexpected as they generally appear during the resolution of inflammation, after prostaglandin/leukotriene biosynthesis occurred, according to the class‐switch model. 45 However, our data indicate that prostaglandins, leukotrienes, and SPMs co‐exist and participate in the inflammatory cascade during the acute phase of inflammation/infection in which resolution has not fully been engaged. Moreover, given their documented anti‐inflammatory effects and their host‐defense boosting functions, they could be viewed as potential mediators to enhance to limit the infection/inflammation. To that end, it was proposed that dexamethasone might promote some of its beneficial effects by upregulating SPM biosynthesis/effects in moderate to severe COVID‐19 patients. 46 As of today, no resolution pharmacology treatment has been approved by governmental health agencies with the exception of EPA‐ethyl ester (Vascepa). Nonetheless, EPA, DHA as well as some SPM precursors (18‐HEPE, 17‐HDHA) are available as dietary supplements. Finally, we must point out that only a fraction of docosanoids belonging to SPMs was analyzed and that other SPMs might also play a role during SARS‐CoV‐2 infections.
While it is largely accepted that the cytosolic phospholipase A2 releases most of AA in activated human leukocytes (eg, in 47), the increased levels of LA, DPA, and DHA indicate that other phospholipases are also involved in the lipidome we found. The correlation between all fatty acids might also indicate that some fatty acids are released by the same phospholipases. Those phospholipases are likely secreted phospholipases A2, which can release fatty acids other than AA 48 , 49 and are contributing to lung inflammation 50 , 51 but our findings do not allow to pinpoint which additional phospholipases are involved.
One of the striking finding of this study is the very poor correlations between the lipid mediators from the BALs and the blood markers available. The significant negative correlations between liver enzymes and BAL fatty acids are puzzling and difficult to interpret as it is counterintuitive. While a liver‐lung axis may promote damages in ARDS, 52 it is unclear to which extent such an axis participates in the pathogenesis and lipid mediator biosynthesis/effects in COVID‐19. 53 , 54 Finally, even if blood TXB2 levels are increased in COVID‐19 patients, 10 those found in the plasma of our intubated COVID‐19 patients did not correlate with those found in their BALs (data not shown), supporting the idea that the lipid mediator profile found in the lungs is not reflected in the circulation.
Our study has, however, some limitations. (1) BALs from healthy controls are from younger individuals than those from severe COVID‐19 patients. This could have an impact on some mediators, notably PGD2, which participate in the worsening of infections mediated by SARS‐CoV and are increased in aged mice 55 ; (2) The procedure to obtain BAL fluids is slightly different between healthy controls and severe COVID‐19 patients. Indeed, while lipid mediator analyses were done on the BAL return from one 50 mL saline bolus in healthy people, that of severe COVID‐19 patients were done on two boli. Although the possible outcome of this discrepancy could be and underestimation of lipid levels in one group as opposed to the other, these comparisons have permitted to estimate whether candidate lipids were to be expected in BAL in normal conditions. While some might argue that the first saline bolus contains more lipids than the following one(s), others might argue that lipid levels are consistent in each bolus. Given that, in our hands, the number of BAL cells is usually greater in the first bolus than in the subsequent ones (when multiple boli are utilized), we are confident that the BAL fluid from the first bolus likely contains more lipids than the second one. This would thus lead to a slight underestimation of lipids in the COVID‐19 group. (3) Unfortunately, BAL fluid leukocyte counts of severe COVID‐19 patients did not include the number of alveolar macrophages and it is thus impossible to determine a clear differential count of leukocytes in those samples. Furthermore, the eosinophil counts were somewhat limited by the apparatus as it could estimate the counts to ±105/mL cells, which likely impacts the different correlations involving eosinophils, notably with EXE4; (4) We could not compare the levels of all lipids in the BALs of severe COVID‐19 patients to those found in the blood. This appears important given the limited number of correlations found between BAL lipids and most recognized COVID‐19‐related biomarkers such as CRP and D‐dimers. To that end, a recent study indicated differences in some of the herein investigated lipids in the circulation. 11 It will thus be important to pursue our investigations and determine whether there is a transposition of the BAL lipidome to the circulation although our preliminary data indicate this is not the case.
In conclusion, our data indicate that lung fluids of severe COVID‐19 patients, in addition to the cytokine storm previously documented, a lipid mediator storm also occurs. The correlations of most lipid mediators with each other regardless of their origin or biosynthetic pathways suggests an increase production of lipid mediators in general in the lungs of COVID‐19 patients, as we observe for cytokine production. The rich library of clinically approved and low‐cost lipid modulators might provide beneficial and important add‐ons to the current therapeutic arsenal utilized to diminish COVID‐19 severity and possibly that of other coronavirus‐mediated infections.
CONFLICT OF INTEREST
The authors have stated explicitly that there are no conflicts of interest in connection with this article.
AUTHOR CONTRIBUTIONS
ASA, YZ, YA, AC, HF, AEH, YT, AC, ML, EB, LF, and NF designed the research. ASA, YZ, VR, CT, ED, ID, CM, performed the research. ASA, YZ, OF, LF, and NF analyzed the data. All authors wrote the paper.
Supporting information
Fig S1
Fig S2
ACKNOWLEDGMENTS
ASA, AC, VR, ML, and NF are members of the Quebec Respiratory Health research Network. This work was supported by the Cheikh Zaid Foundation awarded to YZ and grants from the Canadian New Frontier Research Fund (NFRN‐2019‐00004) awarded to LF (PI), EB (co‐PI) and NF (co‐PI) and the Natural Sciences and Engineering Research Council of Canada awarded to NF. ASA received a doctoral award from the Canadian Institutes of Health Research. ED and EB are respectfully recipient of doctoral and senior awards from the Fonds de Recherche du Québec en Santé (FRQS).
Archambault A‐S, Zaid Y, Rakotoarivelo V, et al. High levels of eicosanoids and docosanoids in the lungs of intubated COVID‐19 patients. The FASEB Journal. 2021;35:e21666. 10.1096/fj.202100540R
Anne‐Sophie Archambault and Younes Zaid are co‐first authors.
Contributor Information
Louis Flamand, Email: Louis.Flamand@crchudequebec.ulaval.ca.
Nicolas Flamand, Email: Nicolas.Flamand@criucpq.ulaval.ca.
REFERENCES
- 1. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang XS, Duchaine C. SARS‐CoV‐2 and health care worker protection in low‐risk settings: a review of modes of transmission and a novel airborne model involving inhalable particles. Clin Microbiol Rev. 2020;34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Verdoni L, Mazza A, Gervasoni A, et al. An outbreak of severe Kawasaki‐like disease at the Italian epicentre of the SARS‐CoV‐2 epidemic: an observational cohort study. Lancet. 2020;395:1771‐1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Damiani S, Fiorentino M, De Palma A, et al. Pathological post mortem findings in lungs infected with Sars‐Cov 2. J Pathol. 2020. [DOI] [PubMed] [Google Scholar]
- 6. Barton LM, Duval EJ, Stroberg E, Ghosh S, Mukhopadhyay S. COVID‐19 autopsies, Oklahoma, USA. Am J Clin Pathol. 2020;153:725‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Amigues I, Pearlman AH, Patel A, et al. Coronavirus disease 2019: investigational therapies in the prevention and treatment of hyperinflammation. Expert Rev Clin Immunol. 2020;16(12):1185‐1204. 10.1080/1744666X.2021.1847084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rubin EJ, Longo DL, Baden LR. Interleukin‐6 receptor inhibition in Covid‐19 ‐ cooling the inflammatory soup. N Engl J Med. 2021;384:1564‐1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Serhan CN, Levy BD. Resolvins in inflammation: emergence of the pro‐resolving superfamily of mediators. J Clin Invest. 2018;128:2657‐2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zaid Y, Guessous F, Puhm F, et al. Platelet reactivity to thrombin differs between patients with COVID‐19 and those with ARDS unrelated to COVID‐19. Blood Adv. 2021;5:635‐639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schwarz B, Sharma L, Roberts L, et al. Cutting edge: Severe SARS‐CoV‐2 infection in humans is defined by a shift in the serum lipidome, resulting in dysregulation of eicosanoid immune mediators. J Immunol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Turcotte C, Dumais E, Archambault AS, et al. Human leukocytes differentially express endocannabinoid‐glycerol lipases and hydrolyze 2‐arachidonoyl‐glycerol and its metabolites from the 15‐lipoxygenase and cyclooxygenase pathways. J Leukoc Biol. 2019;106(6):1337‐1347. 10.1002/JLB.3A0919-049RRR [DOI] [PubMed] [Google Scholar]
- 13. Del Valle DM, Kim‐Schulze S, Huang HH, et al. An inflammatory cytokine signature predicts COVID‐19 severity and survival. Nat Med. 2020;26(10):1636‐1643. 10.1038/s41591-020-1051-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tang Y, Liu J, Zhang D, Xu Z, Ji J, Wen C. Cytokine storm in COVID‐19: The current evidence and treatment strategies. Front Immunol. 2020;11:1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xiong Y, Liu Y, Cao L, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID‐19 patients. Emerg Microbes Infect. 2020;9:761‐770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hu B, Huang S, Yin L. The cytokine storm and COVID‐19. J Med Virol. 2021;93:250‐256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stephenson AH, Lonigro AJ, Hyers TM, Webster RO, Fowler AA. Increased concentrations of leukotrienes in bronchoalveolar lavage fluid of patients with ARDS or at risk for ARDS. Am Rev Respir Dis. 1988;138:714‐719. [DOI] [PubMed] [Google Scholar]
- 18. Ratnoff WD, Matthay MA, Wong MY, et al. Sulfidopeptide‐leukotriene peptidases in pulmonary edema fluid from patients with the adult respiratory distress syndrome. J Clin Immunol. 1988;8:250‐258. [DOI] [PubMed] [Google Scholar]
- 19. Funk CD, Ardakani A. A novel strategy to mitigate the hyperinflammatory response to COVID‐19 by targeting leukotrienes. Front Pharmacol. 2020;11:1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liao M, Liu Y, Yuan J, et al. Single‐cell landscape of bronchoalveolar immune cells in patients with COVID‐19. Nat Med. 2020;26:842‐844. [DOI] [PubMed] [Google Scholar]
- 21. Panigrahy D, Gilligan MM, Huang S, et al. Inflammation resolution: a dual‐pronged approach to averting cytokine storms in COVID‐19? Cancer Metastasis Rev. 2020;39:337‐340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mitchell JA, Belvisi MG, Akarasereenont P, et al. Induction of cyclo‐oxygenase‐2 by cytokines in human pulmonary epithelial cells: regulation by dexamethasone. Br J Pharmacol. 1994;113:1008‐1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zaid Y, Puhm F, Allaeys I, et al. Platelets can associate with SARS‐Cov‐2 RNA and are hyperactivated in COVID‐19. Circ Res. 2020;127:1404‐1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Manne BK, Denorme F, Middleton EA, et al. Platelet gene expression and function in patients with COVID‐19. Blood. 2020;136:1317‐1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hottz ED, Azevedo‐Quintanilha IG, Palhinha L, et al. Platelet activation and platelet‐monocyte aggregate formation trigger tissue factor expression in patients with severe COVID‐19. Blood. 2020;136:1330‐1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abdulnour RE, Gunderson T, Barkas I, et al. Early intravascular events are associated with development of acute respiratory distress syndrome. a substudy of the LIPS‐A clinical trial. Am J Respir Crit Care Med. 2018;197:1575‐1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hirai H, Tanaka K, Yoshie O, et al. Prostaglandin D2 selectively induces chemotaxis in T helper type 2 cells, eosinophils, and basophils via seven‐transmembrane receptor CRTH2. J Exp Med. 2001;193:255‐261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chang JE, Doherty TA, Baum R, Broide D. Prostaglandin D2 regulates human type 2 innate lymphoid cell chemotaxis. J Allergy Clin Immunol. 2014;133(899–901):e893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Medeiros A, Peres‐Buzalaf C, Fortino Verdan F, Serezani CH. Prostaglandin E2 and the suppression of phagocyte innate immune responses in different organs. Mediators Inflamm. 2012;2012:327568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sonti R, Pike CW, Cobb N. Responsiveness of inhaled epoprostenol in respiratory failure due to COVID‐19. J Intensive Care Med. 2020;885066620976525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Crofford LJ, Wilder RL, Ristimaki AP, et al. Cyclooxygenase‐1 and ‐2 expression in rheumatoid synovial tissues. Effects of interleukin‐1b, phorbol ester, and corticosteroids. J Clin Invest. 1994;93:1095‐1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tomazini BM, Maia IS, Cavalcanti AB, et al. Effect of dexamethasone on days alive and ventilator‐free in patients with moderate or severe acute respiratory distress syndrome and COVID‐19: the CoDEX randomized clinical trial. JAMA. 2020;324:1307‐1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chow JH, Khanna AK, Kethireddy S, et al. Aspirin use is associated with decreased mechanical ventilation, ICU admission, and in‐hospital mortality in hospitalized patients with COVID‐19. Anesth Analg. 2020;132(4):930‐941. [DOI] [PubMed] [Google Scholar]
- 34. Merzon E, Green I, Vinker S, et al. The use of aspirin for primary prevention of cardiovascular disease is associated with a lower likelihood of COVID‐19 infection. FEBS J. 2021. 10.1111/febs.15784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gupta A, Kalantar‐Zadeh K, Reddy ST. Ramatroban as a novel immunotherapy for COVID‐19. J Mol Genet Med. 2020;14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang J, Eiserich JP, Cross CE, Morrissey BM, Hammock BD. Metabolomic profiling of regulatory lipid mediators in sputum from adult cystic fibrosis patients. Free Radic Biol Med. 2012;53:160‐171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brom J, Konig W, Koller M, Gross‐Weege W, Erbs G, Muller F. Metabolism of leukotriene B4 by polymorphonuclear granulocytes of severely burned patients. Prostaglandins, Leukot Med. 1987;27:209‐225. [DOI] [PubMed] [Google Scholar]
- 38. Archambault AS, Poirier S, Lefebvre JS, et al. 20‐Hydroxy‐ and 20‐carboxy‐leukotriene (LT) B4 downregulate LTB4‐mediated responses of human neutrophils and eosinophils. J Leukoc Biol. 2019;105:1131‐1142. [DOI] [PubMed] [Google Scholar]
- 39. Brunnstrom A, Tryselius Y, Feltenmark S, et al. On the biosynthesis of 15‐HETE and eoxin C4 by human airway epithelial cells. Prostaglandins Other Lipid Mediat. 2015;121:83‐90. [DOI] [PubMed] [Google Scholar]
- 40. Ham EA, Soderman DD, Zanetti ME, Dougherty HW, McCauley E, Kuehl FA Jr. Inhibition by prostaglandins of leukotriene B4 release from activated neutrophils. Proc Natl Acad Sci USA. 1983;80:4349‐4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fonteh AN, Winkler JD, Torphy TJ, Heravi J, Undem BJ, Chilton FH. Influence of isoproterenol and phosphodiesterase inhibitors on platelet‐activating factor biosynthesis in the human neutrophil. J Immunol. 1993;151:339‐350. [PubMed] [Google Scholar]
- 42. Denis D, Riendeau D. Phosphodiesterase 4‐dependent regulation of cyclic AMP levels and leukotriene B4 biosynthesis in human polymorphonuclear leukocytes. Eur J Pharmacol. 1999;367:343‐350. [DOI] [PubMed] [Google Scholar]
- 43. Flamand N, Boudreault S, Picard S, et al. Adenosine, a potent natural suppressor of arachidonic acid release and leukotriene biosynthesis in human neutrophils. Am J Respir Crit Care Med. 2000;161:S88‐94. [DOI] [PubMed] [Google Scholar]
- 44. Flamand N, Plante H, Picard S, Laviolette M, Borgeat P. Histamine‐induced inhibition of leukotriene biosynthesis in human neutrophils: involvement of the H2 receptor and cAMP. Br J Pharmacol. 2004;141:552‐561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Buckley CD, Gilroy DW, Serhan CN. Proresolving lipid mediators and mechanisms in the resolution of acute inflammation. Immunity. 2014;40:315‐327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Andreakos E, Papadaki M, Serhan CN. Dexamethasone, pro‐resolving lipid mediators and resolution of inflammation in COVID‐19. Allergy. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Flamand N, Picard S, Lemieux L, Pouliot M, Bourgoin SG, Borgeat P. Effects of pyrrophenone, an inhibitor of group IVA phospholipase A2, on eicosanoid and PAF biosynthesis in human neutrophils. Br J Pharmacol. 2006;149:385‐392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hanasaki K, Ono T, Saiga A, et al. Purified group X secretory phospholipase A2 induced prominent release of arachidonic acid from human myeloid leukemia cells. J Biol Chem. 1999;274:34203‐34211. [DOI] [PubMed] [Google Scholar]
- 49. Murakami M, Masuda S, Shimbara S, et al. Cellular arachidonate‐releasing function of novel classes of secretory phospholipase A2s (groups III and XII). J Biol Chem. 2003;278:10657‐10667. [DOI] [PubMed] [Google Scholar]
- 50. Hallstrand TS, Lai Y, Altemeier WA, et al. Regulation and function of epithelial secreted phospholipase A2 group X in asthma. Am J Respir Crit Care Med. 2013;188:42‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Arbibe L, Koumanov K, Vial D, et al. Generation of lyso‐phospholipids from surfactant in acute lung injury is mediated by type‐II phospholipase A2 and inhibited by a direct surfactant protein A‐phospholipase A2 protein interaction. J Clin Invest. 1998;102:1152‐1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Herrero R, Sanchez G, Asensio I, et al. Liver‐lung interactions in acute respiratory distress syndrome. Intensive Care Med Exp. 2020;8:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Weber S, Hellmuth JC, Scherer C, Muenchhoff M, Mayerle J, Gerbes AL. Liver function test abnormalities at hospital admission are associated with severe course of SARS‐CoV‐2 infection: a prospective cohort study. Gut. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Portincasa P, Krawczyk M, Machill A, Lammert F, Di Ciaula A. Hepatic consequences of COVID‐19 infection. Lapping or biting? Eur J Intern Med. 2020;77:18‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhao J, Zhao J, Legge K, Perlman S. Age‐related increases in PGD2 expression impair respiratory DC migration, resulting in diminished T cell responses upon respiratory virus infection in mice. J Clin Invest. 2011;121:4921‐4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2


