Abstract
Antimicrobial resistance has become a worldwide medical challenge [1], so impactful that vancomycin-resistant Enterococcus (VRE) and methicillin-resistant Staphylococcus aureus (MRSA) have entered the common vernacular. We have attempted to reduce the selective pressure through antimicrobial stewardship, curtail the spread by identifying and isolating carriers and individuals with symptomatic infection, and treat antibiotic-resistant organisms (AROs) by developing novel antimicrobials. Despite these extraordinary measures, the challenge of AROs continues to grow. The gut microbiome, the ecosystem of microbes (ie, the microbiota) and metabolites present upon and within all humans, is an emerging target for both the risk for colonization and defense against infection with AROs. Here, informed from experiences and successes with understanding the role of the microbiome in mediating risk of Clostridioides difficile infection (CDI), we (1) review our understanding of the risk from ARO acquisition; (2) review our current understanding of the gut microbiome’s ability to resist colonization with AROs; (3) describe how experimental model systems can test these initial, global insights to arrive at more granular, mechanistic ones; and (4) suggest a path forward to make further progress in the field.
Keywords: antimicrobial resistance, microbiome, fecal microbiota transplantation, microbial ecology, next-generation sequencing, metagenomics
EVIDENCE FOR THE RISK FROM ARO COLONIZATION
The gut can be colonized with AROs, serving as a reservoir and potential transmission source for the AROs [2], while not making the host overtly ill; this has prompted many hospitals to employ screening and isolation efforts for AROs, with MRSA or VRE as examples [3]. Such efforts have become increasingly controversial as we have learned more about the downsides for care teams and patients with isolation [4], complexities for patient flow within a hospital, and with posthospital care. Ideally, we would reduce ARO colonization rates.
It is also clear and not surprising that being colonized with AROs in the gut, especially at high levels, raises the risk of infections such as bacteremia from the same AROs [5, 6]. More intriguingly, however, is the evidence that domination of an intestinal community from one such ARO can raise the risk of clinical infection from a separate, unrelated ARO [7, 8]. This hints at a shared, underlying mechanism of the healthy gut microbiome that protects against colonization by different AROs, which in turn suggests therapeutics that can work across different ARO mitigation goals.
EVIDENCE THAT THE GUT MICROBIOME MEDIATES RISK OF ARO ACQUISITION AND INFECTION
Colonization with AROs appears more likely after disruption of the gut microbiome [9], and resolves in some patients when the gut microbiome is restored with fecal microbiota transplantation (FMT). One study of a microbiota-based drug to treat recurrent CDI noted that of the 11 patients with preexisting colonization with VRE, 8 (72%) converted to negative, with some remaining VRE-free upon 6 months of follow-up [10]. Another study of more traditional FMT noted that it could successfully clear colonization with Enterobacteriaceae that produce extended-spectrum β-lactamase in 20% of subjects after one FMT, rising to 40% after a second FMT [11]. The effects of FMT on antimicrobial resistance may extend beyond what we observe for colonization with clinically relevant AROs, as studies have observed a general reduction of antibiotic resistance genes in the microbiome as a whole following FMT [12, 13]. In individual case reports, FMT has also been shown to successfully treat patients with persistent pneumonia and recurrent urinary tract infections with AROs [14, 15]; in a prospective cohort trial, FMT recipients had a reduced incidence of subsequent bacteremia [16]. Finally, FMT is a risk for ARO acquisition as well as infection with acquired ARO from FMT [17].
Together, these mixed results speak to the magnitude of the challenge of translating the microbiome science around AROs to novel therapies. As an example of our limited understanding of this complex phenomenon, while VRE colonization is detrimental to colonization resistance against other AROs, one study found that VRE colonization may be protective against typical, non-ARO enteric infections [16].
FMT successfully treats recurrent CDI across a wide variety of permutations in the details of the products and procedures used [18, 19] and despite significant variation among donors and the transplanted microbiota. It is notable that the efficacy for FMT-mediated ARO decolonization is both limited and inconsistent, as not all patients in the studies above benefited. We were, in a sense, lucky that C. difficile is not “picky” about donor stool community composition, but are not so lucky when attempting to use FMT to treat other conditions such as inflammatory bowel disease or to decolonize AROs [20, 21]. Why were we lucky with CDI? How can we better handle circumstances in which success depends on the details of the permutations above?
Motivated by these questions, we next turn to how emerging research tools can help develop this mechanistic understanding of the underlying complexity of the microbiome.
FOR CDI, THE CAUSAL CHAIN WAS THE FIRST OBSERVATION: AN ANTIBIOTIC-ASSOCIATED DIARRHEA
What followed was the microbiology, the identification of C. difficile, and finally next-generation sequencing (NGS) revealing the broad disruption of the gut microbial community after antibiotics and associated with the development of CDI. We would argue that the picture with C. difficile differs from AROs in that through both human studies and animal models, we had a better understanding of the causal chain leading to CDI, specifically regarding the role of antibiotic-induced microbiome disruption in attenuating colonization resistance to CDI [22]. This showed how patients and animals with very different gut microbial community structures and compositions could still have a shared function, namely colonization resistance, due at least in part to shared metabolic environments such as the presence of secondary bile acids [23].
Viewed this way, there is no real reason to think that this untargeted FMT approach would work for other conditions, including ARO colonization and infection. Indeed, even for CDI the necessary and sufficient components of feces in mediating the desired therapeutic effect is unknown, and it is not even fully established that transplanting microbes is essential, as a case series of sterile fecal filtrates for recurrent CDI showed efficacy [24].
THE ROLE OF MODEL SYSTEMS
Gnotobiotic animals have been the most fruitful experimental strategy for microbiome science to establish causality [25, 26]. Microbes, metabolites, or entire microbial communities can be transferred into germ-free animals to study the physiological effects. Emerging in vitro organoid-based models offer new opportunities and allow for controlled experiments not possible previously. These systems are currently being deployed and tested in part through a National Institutes of Health (NIH) program titled the Novel, Alternative Model Systems for Enteric Disease (NAMSED). These systems can test hypotheses from observational studies to arrive at mechanistic insights, which can again be taken back to human subjects for further testing and analysis. This cycle of moving from human subjects, to analysis of multidimensional datasets, to experiments in model systems, and then back to humans constitutes the systems biology approach. This systems biology approach is increasingly being recognized as a fruitful avenue to arrive at actionable insights, including multiple funding mechanisms through NIH to support such work.
An example of how a holistic, systems biology approach could lead to insights that can directly impact patient care can be gleaned from the seminal work showing how the therapeutic efficacy of checkpoint inhibitors (eg, PD-1 inhibitors) is modulated by the gut microbiome. This trio of articles published in the same issue of Science effectively hints at the power of a true systems biology approach, though conducted by separate teams, starting by showing that antibiotic consumption is associated with poor response to immunotherapeutic PD-1 blockade and that nonresponding lung and kidney cancer patients had low gut levels of Akkermansia muciniphila [27]. This was followed by experiments showing that oral supplementation with A. muciniphila to antibiotic-treated mice restored the response to PD-1 inhibitors. Two additional studies on melanoma patients receiving PD-1 blockade [28, 29] found a lower abundance of “good” bacteria in the guts of nonresponding patients, and this correlated with impaired immune cell activity, suggesting that the modulation by the microbiome of PD-1 inhibitor efficacy was not limited to lung and kidney cancers. In one of these studies, Matson and colleagues took these insights from analysis of the human microbiome of responders and tested them in a mouse model [28]. They did this by reconstituting germ-free mice with fecal material from responders and found this led to improved tumor control, augmented T-cell responses, and greater efficacy of anti–PD-1 therapy.
These findings have already led to a phase 1 clinical trial testing FMT for nonresponders to PD-1 inhibitor treatment, with some (but not all) previously nonresponsive patients having a treatment response following FMT [30], and with FMT being associated with favorable changes in immune cell infiltrates and gene expression profiles in the gut lamina propria and the tumor microenvironment. In a separate but concurrent phase 1 trial [31], responders had increased abundance of taxa previously shown to be associated with response to anti–PD-1, distinct proteomic and metabolomic signatures with transkingdom network analyses supporting the gut microbiome as a regulator of these changes, increased CD8+ T-cell activation, and decreased frequency of interleukin-8–expressing myeloid cells. Despite these advances, we still do not have a way of precisely and reliably manipulating the gut microbiome in patients treated with PD-1 inhibitors.
Future studies around how the microbiome affects ARO colonization should make use of these novel and emerging model systems of host–microbe interactions in the context of a systems biology approach. In particular, these in vivo and in vitro model systems offer a pathway to establish which aspects of the gut microbiome are responsible for ARO colonization resistance—such as live microbes, specific metabolites, or microbe–microbe interactions. To support such studies, novel model-driven analytic approaches of human observational data and these model systems are needed.
ANALYTIC APPROACHES
Due to its complexity, multidimensionality, and extreme variability even in an individual over time, analyzing microbiome data to arrive at generalizable and mechanistic insights is challenging. Further compounding the challenge are the complex ways by which antibiotic resistance develops in microbes. While there are identifiable strains or species of bacteria with vertically inherited antimicrobial resistance, resistance can arise by horizontal gene transfer, shifts in the allelic frequency of specific microbial genes, mutations in protein coding genes, and/or mutations in intergenic transcriptional regulating sequences. Most of these mechanisms are opaque to NGS analysis techniques in wide use. As with our successes with CDI, we suggest starting with a testable conceptual framework, and employing NGS as a tool to test and refine the framework over time.
For example, butyrate production by the gut microbiome has defined mechanisms by which it can affect host inflammation and immunity [32, 33]. Butyrate production is a ubiquitous feature of the healthy human gut microbiome [34]. The causal framework that the loss of butyrate production by the gut microbiome after hematopoietic cell transplantation increases the risk for graft-vs-host disease has been successfully tested with a study that combined NGS and targeted metabolomics [35]. This opens the door to microbiome-oriented interventions to preserve and boost butyrate production [36] and improve outcomes after hematopoietic cell transplantation. Similar associations to specific species of bacteria have led to probiotic trials to test the causal framework that specific species are protective in irritable bowel syndrome (NCT03721107).
A comprehensive framework for the microbiome and ARO should incorporate what we have learned about the effects of antimicrobials on the microbiome, how specific disruptions of the gut microbiome affect colonization resistance, and in turn how the state of the host epithelium (eg, age, inflammation, cellular respiration) affects both the gut microbiome and AROs.
For example, in people living in skilled nursing facilities, broad spectrum antibiotic exposure is associated with domination with aerobic bacilli such as Enterococcus species and eventually AROs organisms [8]. Other nonantibiotic medications can affect the composition and structure of the gut microbiome [37], as can age and inflammatory diseases of the gut itself.
Metabolic interactions between microbes and between the microbial community and the host are clearly implicated in colonization resistance to C. difficile and are likely involved in resiliency against ARO colonization as well. For example, agonism of the PPAR-family of intracellular receptors by microbially produced short-chain fatty acids is critical for the maintenance of the anaerobic environment within the gut, and in turn the strict anaerobic microbes typically dominant in a healthy community [38]. Bile catabolism, resistant starch degradation, and metabolism of riboflavin, indole, and sphingolipids are other well-defined metabolic interactions between microbes and the host that can be tested by NGS and targeted metabolomics to see if they impact ARO colonization or infection.
We have begun to appreciate that the state of the host colonic epithelium can affect both the microbial community and the nature of host–microbe interactions. For example, butyrate has radically different effects on undifferentiated colonic stem cells compared to mature and fully differentiated colonic epithelium [39], which in turn corresponds to treatment refractory colitis when butyrate-generating microbes are present [40]. In a similar manner, we can expect colonic epithelium from older individuals to respond in distinct ways to the same microbes and metabolites.
NEXT-GENERATION SEQUENCING ANALYSIS OF ARO–MICROBIOME INTERACTIONS
Microbe–microbe interactions are a possible contributor to both susceptibility for and resiliency against colonization with AROs. Detecting such microbe–microbe interactions with microbiome data (whether from 16S ribosomal RNA [rRNA] gene amplicon sequencing or shotgun metagenomics) is a surprisingly challenging task [41, 42] and an area of active development. As new statistical methods and tools become available, there will be an opportunity to revisit extant human observational datasets and find critical microbe–microbe interactions that could prevent ARO colonization.
Antimicrobial resistance can arise from multiple mechanisms, many of which require less common analytic techniques of shotgun metagenomic data. The acquisition of a resistant allele, or the development of one in situ via mutation and selection by antimicrobials, is both a well-described mechanism of resistance, and only detectable with gene- and allele-oriented metagenomic techniques. Even the task of assembling an accurate and comprehensive catalog of protein-coding alleles present in a shotgun metagenome is a complex task, prone to both false positives and false negatives [43]. Dysregulation of genes is another very well described mechanism of antimicrobial resistance, with the most classic example being the dysregulation of chromosomal ampC genes under selective pressure from β-lactam antibiotics, and subsequent failure of treatments [44]. Gene-oriented metagenomics is a promising and emerging technique that could allow for the detection of some of these sorts of changes in a metagenome [43, 45, 46].
The detection and accurate cataloging of mutations in regulatory domains of microbial genes or operons from shotgun metagenomes is even more nascent at this time, and likely critical to detect and track this means of ARO acquisition. Databases of antibiotic resistance genes and alleles exist, such as the Comprehensive Antibiotic Resistance Database (CARD) [47], with significant effort made to categorize the antimicrobial resistance alleles into models that reflect our best understanding of how resistance can develop: overexpression, sequence variants, knockout, and acquisition of a gene that confers resistance (by homolog). The generation and maintenance of such databases is difficult to get supported by funders and academic institutions, and as such the reference databases for the interpretation of sequencing-based microbiome studies often fall into disrepair. One lesson we can take from the coronavirus disease 2019 pandemic is the immense utility of rapid, open, worldwide sharing of near real-time sequence data. A similar effort for antimicrobial resistance (the goal of sequencing and sharing in a cohesive and open database of the genome of every antibiotic-resistant clinical isolate) would require ongoing investment by scientific institutions, but in turn be an invaluable resource in the fight against the spread of AROs.
ESTABLISHING CAUSATION WITH STUDIES IN HUMANS: OVERVIEW
In contrast to inference of association, causal inference in statistics is the process of determining the independent effect of an exposure, and it analyzes the response of the effect variable when the putative cause (exposure) changes. The fundamental problem of causal inference is that directly observing causal effects is impossible, since an individual subject cannot be both exposed and unexposed. This does not make causal inference impossible, just difficult as assumptions must be made in order to estimate the missing counterfactuals. Three assumptions are both necessary and sufficient [48]: (1) exchangeability that the treated and untreated individuals are exchangeable such that the assignment to the exposure does not depend on the outcome (ie, there are no unmeasured confounders that cause both the exposure and the outcome); (2) positivity: every individual has a greater than zero chance of being exposed at every level of exposure and of having the outcome; and (3) consistency: the exposure assigned to one individual is unambiguous, could be hypothetically assigned to another individual, and the different routes by which the exposure could occur would produce the same effect.
So how does one design human subject studies with causal inference as a goal? A full discussion is out of scope and it is recommended that study teams include personnel with such expertise, but some common approaches are discussed here. The most logically straightforward solution is the randomized controlled trial (RCT), which through randomized treatment assignment for all subjects addresses our 3 necessary assumptions of exchangeability (equalizes the distribution of confounders), positivity (everyone is eligible to receive the treatment), and consistency (the treatment is unambiguous, could be assigned to any individual, and the method of exposure is consistent). However, RCTs do not come first, because they generally follow basic science and observational human subject research; meeting the assumptions for causal inference in the latter is possible but more challenging.
Addressing exchangeability in observational studies includes controlling for confounders through study design and statistical modeling with covariates, and positivity is generally evident in the design (eg, it is possible that control subjects could have been exposed to an antibiotic). Additionally, the likelihood of exposure, termed a propensity score, can be calculated [49], and this in turn can be used in several different ways to meet the exchangeability and positivity assumptions through techniques such as matching, inverse probability treatment weighting, and standardization, all of which aim to account for the likelihood of exposure among exposed and unexposed subjects, reducing bias despite the lack of randomization. In mediation analysis the effect of an exposure is partitioned into direct and indirect components, the latter working through mediators of interest that causally sit between the effect and outcome. These effects are estimated along with the proportion mediated, which quantifies how much of the total exposure effect works through a particular mediator [50]. Finally, though more frequently used in epidemiologic research, instrumental variables can be a useful tool in causal inference [51]. Instrumental variables are associated with the exposure but not the outcome, and they can be helpful when measuring the exposure directly is difficult or to account for unmeasured confounders. A common example is estimating exposure to smoking in a population, which is difficult to measure directly, using data on taxes collected from tobacco sales. Understanding if a variable should be treated as a confounder, mediator, or instrumental variable is study-dependent and relies on an underlying causal framework.
CAUSAL INFERENCE FOR THE MICROBIOME AND AROs: AN EXAMPLE OF DEPLOYING A CONCEPTUAL MODEL OF ARO COLONIZATION AND INFECTION
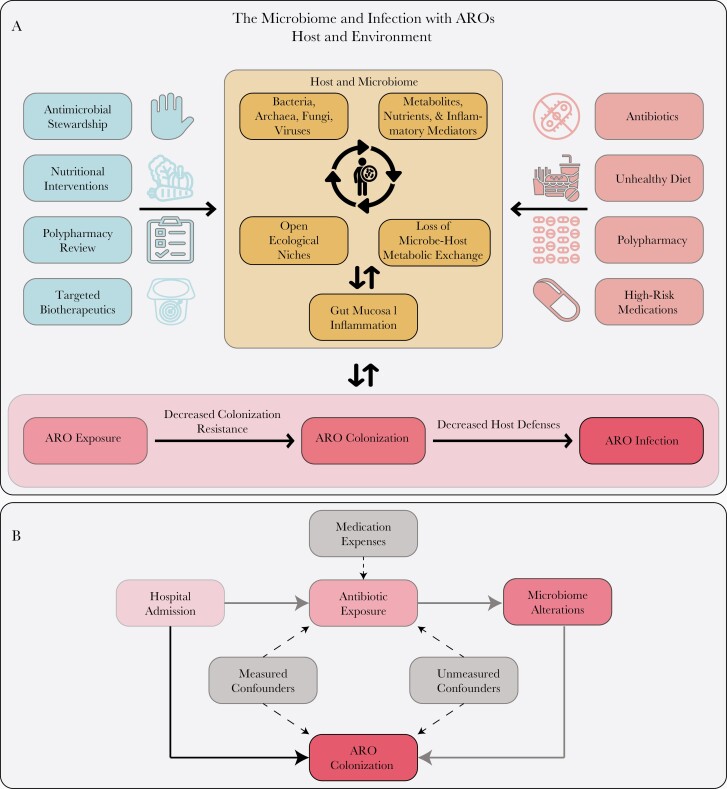
From the observations presented above, we can build a conceptual model for how the microbiome is involved in the colonization with and infection from AROs (Figure 1A). A resulting causal inference diagram (Figure 1B) can guide multivariable modeling to adjust for true confounders (ie, associates with both the predictor and the outcome), while also identifying variables that are mediators, since they are in the causal pathway to the outcome (eg, microbiome disruption). The overall model can also identify areas where cyclic or bidirectional influences can be present and, thus, typical linear modeling may not be sufficient to describe the phenomena studied without additional assumptions [43]. For example, if studying ARO colonization as a risk factor for subsequent invasive infection, it would be appropriate to adjust for confounders such as antibiotic/device exposure and comorbid conditions, as we have previously performed in an analysis of intestinal domination with members of the order Enterobacterales [52]. However, if the primary aim is to study antibiotic association and subsequent risk of infection, including microbiome-derived variables as covariates may only dilute the association needlessly, as microbiome disruption is an effect of antibiotic exposure and in the putative causal pathway from antibiotic exposure to infection, not a confounder. Instead, a formal mediation analysis conducted within the context of this causal inference diagram could not only identify the microbiome as a mediator but also quantify its contribution to risk of infection (the indirect pathway) vs antibiotic exposure (the direct pathway). Finally, this could be done in the context of a study design addressing exchangeability through calculation of a propensity score for antibiotic exposure followed by matching of cases to controls. The sum of these approaches can begin to meet the requirements of causal inference and enable/support subsequent RCTs and other experimental studies.
Figure 1.
A, Example conceptual framework for testing hypotheses about the microbiome and subsequent risk of infection with antibiotic-resistant organisms (AROs). Beneficial and detrimental host/environmental factors are shown on the left and right, respectively. The microbiome is modeled as a system of microbes and local environmental factors that interact to fill ecological niches and maintain metabolic exchange with the host, disruption of which can influence gut mucosal inflammation. The main hypothesis being tested should progress from a risk factor/predictor through the various components leading to ARO infection. Alternate pathways distinct from the main hypothesis that also lead from host factors to ARO infection should prompt consideration of these as putative confounders or mediators. Modeling the microbiome itself may require special techniques due to cyclic or nonlinear dynamics. ARO colonization here is modeled to be in the causal pathway between loss of colonization resistance and infection and, thus, it should be modeled as a mediator in multivariable analyses. However, colonization itself can lead to microbiome alterations, so this simplified framework may not apply in all scenarios. B, Here we see a specific putative causal inference diagram from hospital admission to ARO colonization. The direct pathway is shown in solid black and the indirect in solid gray. Antibiotic exposure and microbiome alterations are mediators, while medication expenses could serve as instrumental variables. Here confounders are depicted between antibiotic exposure and ARO colonization, but confounding can exist for each variable in the causal pathway and must be accounted for before formal mediation analysis can proceed.
CONCLUSIONS AND RECOMMENDATIONS FOR STUDYING AROs AND THE MICROBIOME
In summary, although there is compelling evidence linking colonization and infection with AROs to the structure and function of the gut microbiome, a more fundamental mechanistic understanding of the involved processes is needed to make further progress in developing therapeutic strategies to manipulate the microbiome for the benefit of the patient. These new approaches can benefit from a focus on careful study design set in a causal inference framework—having such expertise on the study team is important—and from leveraging existing associative/epidemiological insights from prior studies and public databases. Additionally, 16S rRNA–based analyses remain useful, and other -omics modalities should augment, rather than replace, 16S approaches. Analysis strategies such as dimensionality reduction, characterization of longitudinal microbiome data, and novel machine learning approaches can help wrangle the complexity of microbiome data, but they are not a replacement for proper study design. The systems biology and NAMSED-derived approaches leveraging new in vitro models can carry us further toward our goal. Funders and academic institutions should value and support efforts to maintain an open, accessible, and continuously updated catalog of the genomes of antimicrobial-resistant microbes. These approaches allow us to test specific hypotheses in model systems and then circle back with these additional insights to apply them in human cohorts. While we got lucky with FMT for recurrent CDI, we feel that developing successful microbiome-based treatments for ARO colonization and infection will require a more holistic approach to tease apart the signal from the noise.
Notes
Financial support. K. R. is supported by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health (grant number U01AI124255) and from the Agency for Healthcare Research and Quality (grant number R01HS027431).
Potential conflicts of interest. K. R. is supported in part from an investigator-initiated grant from Merck & Co, and has consulted for Bio-K Plus International, Roche Molecular Systems, and Seres Therapeutics. J. L. G. reports no potential conflicts of interest.
Both authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Centers for Disease Control and Prevention. Antibiotic-resistant germs: new threats. 2020. https://www.cdc.gov/drugresistance/biggest-threats.html. Accessed 9 July 2020.
- 2. Donskey CJ. The role of the intestinal tract as a reservoir and source for transmission of nosocomial pathogens. Clin Infect Dis 2004; 39:219–26. [DOI] [PubMed] [Google Scholar]
- 3. Banach DB, Bearman G, Barnden M, et al. Duration of contact precautions for acute-care settings. Infect Control Hosp Epidemiol 2018; 39:127–44. [DOI] [PubMed] [Google Scholar]
- 4. Abad C, Fearday A, Safdar N. Adverse effects of isolation in hospitalised patients: a systematic review. J Hosp Infect 2010; 76:97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Taur Y, Xavier JB, Lipuma L, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis 2012; 55:905–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shimasaki T, Seekatz A, Bassis C, et al. ; Centers for Disease Control and Prevention Epicenters Program . Increased relative abundance of Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae within the gut microbiota is associated with risk of bloodstream infection in long-term acute care hospital patients. Clin Infect Dis 2019; 68:2053–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Collingwood A, Blostein F, Seekatz AM, et al. Epidemiological and microbiome associations between Klebsiella pneumoniae and vancomycin-resistant Enterococcus colonization in intensive care unit patients. Open Forum Infect Dis 2020; 7:ofaa012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang J, Cassone M, Gibson K, et al. Gut microbiota features on nursing home admission are associated with subsequent acquisition of antibiotic-resistant organism colonization. Clin Infect Dis 2020; 71:3244–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ananthakrishnan AN, Luo C, Yajnik V, et al. Gut Microbiome Function Predicts Response to Anti-integrin Biologic Therapy in Inflammatory Bowel Diseases. Cell Host Microbe. 2017; 21:603–10.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dubberke ER, Mullane KM, Gerding DN, et al. Clearance of vancomycin-resistant Enterococcus concomitant with administration of a microbiota-based drug targeted at recurrent Clostridium difficile infection. Open Forum Infect Dis 2016; 3:ofw133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Singh R, de Groot PF, Geerlings SE, et al. Fecal microbiota transplantation against intestinal colonization by extended spectrum beta-lactamase producing Enterobacteriaceae: a proof of principle study. BMC Res Notes 2018; 11:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Millan B, Park H, Hotte N, et al. Fecal microbial transplants reduce antibiotic-resistant genes in patients with recurrent Clostridium difficile infection. Clin Infect Dis 2016; 62:1479–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jouhten H, Mattila E, Arkkila P, Satokari R. Reduction of antibiotic resistance genes in intestinal microbiota of patients with recurrent Clostridium difficile infection after fecal microbiota transplantation. Clin Infect Dis 2016; 63:710–1. [DOI] [PubMed] [Google Scholar]
- 14. Ueckermann V, Hoosien E, De Villiers N, Geldenhuys J. Fecal microbial transplantation for the treatment of persistent multidrug-resistant Klebsiella pneumoniae infection in a critically ill patient. Case Rep Infect Dis 2020; 2020:8462659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grosen AK, Povlsen JV, Lemming LE, Jørgensen SMD, Dahlerup JF, Hvas CL. Faecal microbiota transplantation eradicated extended-spectrum beta-lactamase-producing Klebsiella pneumoniae from a renal transplant recipient with recurrent urinary tract infections. Case Rep Nephrol Dial 2019; 9:102–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ianiro G, Murri R, Sciumè GD, et al. Incidence of bloodstream infections, length of hospital stay, and survival in patients with recurrent Clostridioides difficile infection treated with fecal microbiota transplantation or antibiotics: a prospective cohort study. Ann Intern Med 2019; 171:695–702. [DOI] [PubMed] [Google Scholar]
- 17. DeFilipp Z, Bloom PP, Torres Soto M, et al. Drug-resistant E. coli bacteremia transmitted by fecal microbiota transplant. N Engl J Med 2019; 381:2043–50. [DOI] [PubMed] [Google Scholar]
- 18. Rao K, Young VB, Malani PN. Capsules for fecal microbiota transplantation in recurrent Clostridium difficile infection: the new way forward or a tough pill to swallow? JAMA 2017; 318:1979–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Malani PN, Rao K. Expanded evidence for frozen fecal microbiota transplantation for Clostridium difficile infection: a fresh take. JAMA 2016; 315:137–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moayyedi P, Surette MG, Kim PT, et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology 2015; 149:102–9.e6. [DOI] [PubMed] [Google Scholar]
- 21. Kump PK, Gröchenig HP, Lackner S, et al. Alteration of intestinal dysbiosis by fecal microbiota transplantation does not induce remission in patients with chronic active ulcerative colitis. Inflamm Bowel Dis 2013; 19:2155–65. [DOI] [PubMed] [Google Scholar]
- 22. Britton RA, Young VB. Role of the intestinal microbiota in resistance to colonization by Clostridium difficile. Gastroenterology 2014; 146:1547–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Winston JA, Theriot CM. Impact of microbial derived secondary bile acids on colonization resistance against Clostridium difficile in the gastrointestinal tract. Anaerobe 2016; 41:44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ott SJ, Waetzig GH, Rehman A, et al. Efficacy of sterile fecal filtrate transfer for treating patients with Clostridium difficile infection. Gastroenterology 2017; 152:799–811.e7. [DOI] [PubMed] [Google Scholar]
- 25. Abrams GD, Bishop JE. Effect of the normal microbial flora on gastrointestinal motility. Proc Soc Exp Biol Med 1967; 126:301–4. [DOI] [PubMed] [Google Scholar]
- 26. Bhattarai Y, Kashyap PC. Germ-free mice model for studying host–microbial interactions. In: Proetzel G, Wiles MV, eds. Mouse models for drug discovery: methods and protocols. New York: Springer New York, 2016:123–35. [DOI] [PubMed] [Google Scholar]
- 27. Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018; 359:91–7. [DOI] [PubMed] [Google Scholar]
- 28. Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018; 359:104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018; 359:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baruch EN, Youngster I, Ben-Betzalel G, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 2021; 371:602–9. [DOI] [PubMed] [Google Scholar]
- 31. Davar D, Dzutsev AK, McCulloch JA, et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 2021; 371:595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu H, Wang J, He T, et al. Butyrate: A Double-Edged Sword for Health? Adv Nutr Bethesda Md 2018; 9:21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Donohoe DR, Garge N, Zhang X, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab 2011; 13:517–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 2017; 19:29–41. [DOI] [PubMed] [Google Scholar]
- 35. Romick-Rosendale LE, Haslam DB, Lane A, et al. Antibiotic Exposure and Reduced Short Chain Fatty Acid Production after Hematopoietic Stem Cell Transplant. Biol Blood Marrow Transplant J Am Soc Blood Marrow Transplant 2018; 24:2418–24. [DOI] [PubMed] [Google Scholar]
- 36. Baxter NT, Schmidt AW, Venkataraman A, et al. Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. mBio 2019; 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maier L, Pruteanu M, Kuhn M, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 2018; 555:623–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Byndloss MX, Olsan EE, Rivera-Chávez F, et al. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 2017; 357:570–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kaiko GE, Ryu SH, Koues OI, et al. The colonic crypt protects stem cells from microbiota-derived metabolites. Cell 2016; 165:1708–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Golob JL, DeMeules MM, Loeffelholz T, et al. Butyrogenic bacteria after acute graft-versus-host disease (GVHD) are associated with the development of steroid-refractory GVHD. Blood Adv 2019; 3:2866–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Weiss S, Van Treuren W, Lozupone C, et al. Correlation detection strategies in microbial data sets vary widely in sensitivity and precision. ISME J 2016; 10:1669–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sugihara G, May R, Ye H, et al. Detecting causality in complex ecosystems. Science 2012; 338:496–500. [DOI] [PubMed] [Google Scholar]
- 43. Golob JL, Minot SS. In silico benchmarking of metagenomic tools for coding sequence detection reveals the limits of sensitivity and precision. BMC Bioinformatics 2020; 21:459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tamma PD, Doi Y, Bonomo RA, Johnson JK, Simner PJ, Antibacterial resistance leadership group. A primer on ampC β-Lactamases: necessary knowledge for an increasingly multidrug-resistant World. Clin Infect Dis Off Publ Infect Dis Soc Am. 2019; 69:1446–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Minot SS, Willis AD. Clustering co-abundant genes identifies components of the gut microbiome that are reproducibly associated with colorectal cancer and inflammatory bowel disease. Microbiome 2019; 7:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Minot SS, Barry KC, Kasman C, Golob JL, Willis AD. Gene-level metagenomics identifies genome islands associated with immunotherapy response [Internet]. Bioinformatics; 2020. Oct. Available from: http://biorxiv.org/lookup/doi/10.1101/2020.10.09.333971 [DOI] [PMC free article] [PubMed]
- 47. Alcock BP, Raphenya AR, Lau TTY, et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 2020; 48:D517–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cole SR, Frangakis CE. Commentary: the Consistency statement in causal inference: a definition or an assumption? Epidemiology. Lippincott Williams & Wilkins; 2009; 20:3–5. [DOI] [PubMed] [Google Scholar]
- 49. Haukoos JS, Lewis RJ. The propensity score. JAMA 2015; 314:1637–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee H, Herbert RD, McAuley JH. Mediation analysis. JAMA 2019; 321:697–8. [DOI] [PubMed] [Google Scholar]
- 51. Maciejewski ML, Brookhart MA. Using instrumental variables to address bias from unobserved confounders. JAMA 2019; 321:2124–5. [DOI] [PubMed] [Google Scholar]
- 52. Rao K, Seekatz A, Bassis C, Sun Y, Mantlo E, Bachman MA. Enterobacterales infection after intestinal dominance in hospitalized patients. mSphere 2020; 5:e00450-20. [DOI] [PMC free article] [PubMed] [Google Scholar]