Abstract
The bacterial, fungal, and helminthic species that comprise the microbiome of the mammalian host have profound effects on health and disease. Pathogenic viruses must contend with the microbiome during infection and likely have evolved to exploit or evade the microbiome. Both direct interactions between the virions and the microbiota and immunomodulation and tissue remodeling caused by the microbiome alter viral pathogenesis in either host- or virus-beneficial ways. Recent insights from in vitro and murine models of viral pathogenesis have highlighted synergistic and antagonistic, direct and indirect interactions between the microbiome and pathogenic viruses. This review will focus on the transkingdom interactions between human gastrointestinal and respiratory viruses and the constituent microbiome of those tissues.
Keywords: immune response, influenza, A virus, microbiome, respiratory syncytial virus, transkingdom
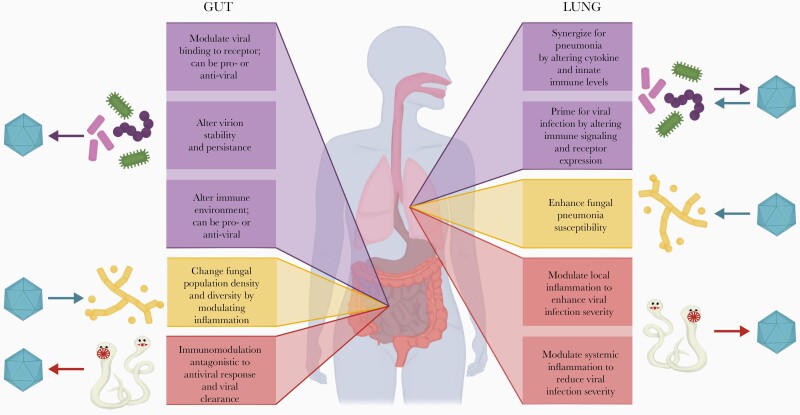
The recent advances in metagenomic sequencing have revealed the complex interactions between host mucosal sites and their associated microbiota (viruses, bacteria, and fungi) and macrobiota (helminths). Microbiota are crucial for mucosal homeostasis and immunity [1], and perturbations of this balance between host and microbes are associated with dysfunction and disease [2]. The extent to which antibiotics can disrupt this balance and affect the ability of the host to combat pathogenic viral infections remains an active area of research. Although the interaction between bacteria and bacteriophage and also aspects of the human virome in preventing and promoting disease are of great interest, this review summarizes our current knowledge of how direct and indirect transkingdom interactions (Table 1) between the mucosal micro- and macrobiota and pathogenic viruses can shift the balance between factors that protect the host and those that promote the virus, with a particular focus on interactions in the gut and lung. Figure 1 summarizes the current state of knowledge on transkingdom interactions in the lung and gut.
Table 1.
Direct and Indirect Transkingdom Interactions Important to Viral Pathogenesis and Representative Examples
| Mechanism | Virus | Organism(s) | |
|---|---|---|---|
| Direct | Virion stability | IAV | GI bacteria, respiratory bacteria [43, 58] |
| Innate immune response | Respiratory viruses | Helminths [45] | |
| Adaptive immune response | Respiratory viruses | Helminths [35, 63] | |
| Receptor availability | Herpesviruses | Bacteriophages [73] | |
| Indirect | Cytokine production | Respiratory viruses | Respiratory bacteria [51] |
| Microenvironment modulation | IAV | Aspergillus [64] | |
| Innate immune response | RSV | GI bacteria [54] | |
| Adaptive immune response | IAV | GI bacteria [57] | |
| Receptor availability | Rhinovirus | Haemophilus influenza [50] |
Abbreviations: GI, gastrointestinal; IAV, influenza A virus; RSV, respiratory syncytial virus.
Figure 1.
Summary of transkingdom interactions in the lung and gut. Interaction between viruses and bacterial, fungal, and helminthic components (top to bottom) of the microbiome. Arrows indicate primary direction of interaction between the virus and the component of the microbiome.
ENTERIC BACTERIAL-VIRAL INTERACTIONS
Most of what is understood about transkingdom interactions has been derived from mouse models, which have yielded strong evidence of direct and indirect interactions between the gut microbiome and enteric viruses. In the early 1990s, it was observed that a nonpathogenic murine picornavirus could be rendered pathogenic if coadministered with lipopolysaccharides [3]. Since then, numerous landmark studies of enteric viruses have demonstrated that bacteria and/or bacterial products can promote virus stability (eg, that of reovirus [4] or poliovirus [4]), entry and/or attachment to target cells (eg, by norovirus [5] or poliovirus [6]), disease (eg, that caused by norovirus [7]), and persistence (eg, of retrovirus [8] or norovirus [5]). Murine norovirus, in an indirect interaction, takes advantage of bacterial suppression of interferon lambda (IFN-λ) to prevent viral clearance [9, 10]. It was recently shown that this interaction is region specific: virus in antibiotic-treated animals is reduced in the proximal small intestine but increased in distal sites [11]. It is interesting to note that the spatial effect was associated with bile acid priming of IFN-λ [11]. In another example of the intertwined host-bacteria-virus complexity, antibiotic treatment was shown to indirectly suppress murine astrovirus infection of goblet cells in vivo by decreasing mucus secretion, which is a process that is partially regulated by microbiota sensing and is also critical for astrovirus replication [12]. Adding to these complexities, closely related viruses such as poliovirus and coxsackievirus were shown to differ in their interactions with distinct microbiota based on varying antibiotic treatments, demonstrating even greater nuances in what are likely bacterial species-specific interactions with enteric viruses [13].
Given the numerous proviral effects afforded by direct and indirect interactions with commensal bacteria, antibiotics could be seen as a means to inhibit enteric viruses; however, this approach is likely to prove ineffective in clinical practice because the homeostatic deficits associated with microbiome disruption [14] could outweigh the antiviral effects. It is important to note that antibiotics can have bacteria-independent effects on virus infection. It was recently shown that antibiotic treatment in vitro can alter the plaque size of coxsackievirus and reovirus [15]. Although the mechanism of this alteration is unclear, these data indicate that the effects of using antibiotics in experimental models and, most likely, in clinical practice can extend beyond direct antimicrobial effects.
Commensal gut bacteria can protect against pathogenic bacteria and pathogenic viruses. Clinical trials of probiotic Lactobacillus spp and Bifidobacterium spp demonstrated efficacy against rotavirus [16]. The significance of these findings likely extends beyond rotavirus, because Lactobacillus spp were also found to inhibit transmissible gastroenteritis virus, a porcine coronavirus [17]. Although the precise mechanisms have yet to be elucidated, Lactobacillus casei and Bacteroides thetaiotaomicron can alter host glycoproteins, which results in decreased binding of rotavirus to target cells [18]. Bacterial flagellin can also play a role in rotavirus clearance via Toll-like receptor signaling that induces proinflammatory cytokines [19]. More recently, segmented filamentous bacteria were shown to block rotavirus infection by directly neutralizing the virus and promoting epithelial cell turnover [20]. Commensal organisms coevolve with the development of the intestinal immunity [21], particularly T-cell immunity that is critically important for the clearance of numerous enteric viruses [22–25]. Although the indirect actions of this immunity on enteric virus pathogenesis remain to be explored, a common feature across these studies is that promoting innate and adaptive immunity via commensal bacteria and probiotics may be a viable strategy for reducing viral pathogenesis in the gut. Additional studies to explore these possibilities are needed.
ENTERIC FUNGAL-VIRAL INTERACTIONS
Despite the estimated presence of more than 180 fungal species in the intestines of healthy individuals, there is a paucity of information detailing nonbacterial transkingdom interactions with the human gut mycobiome [26]. To date, only a handful of studies have examined the mycobiome during the progression of viral diseases. Decreased mycobiome diversity and increased prevalence of Candida spp were observed in the gut of patients with human immunodeficiency virus (HIV) [27]. In contrast, a correlative increase in gut fungal diversity was observed during hepatitis B progression [28]. However, these studies did not establish causative or directional relations between fungi and viral infection; thus, substantial evidence of a fungal role in viral pathogenesis in the human gut is lacking. The known interactions between fungi and enteric viruses, whether direct or indirect, are poorly characterized. The probiotic yeast Saccharomyces boulardii has demonstrated some efficacy as a therapeutic intervention against rotavirus-induced acute diarrhea, although the specific antiviral interactions have not been well defined [29]. Therefore, the nature of the interplay between viral pathogens and the enteric mycobiome remains unclear and requires further investigation. The rise of multidrug-resistant fungal pathogens lends urgency to this topic.
ENTERIC HELMINTHIC-VIRAL INTERACTIONS
Helminths that reside in the human gut, most commonly nematodes, cestodes, and trematodes, are frequently found in populations in low-income countries [30]. Helminths provoke complex immunomodulatory effects in the mammalian gut in response to infections [31] that are typically characterized by Th2-mediated responses effected via the interleukin (IL)-4Rα and STAT6 pathways and includes release of cytokines IL-4, IL-5, IL-9, and IL-13 [32, 33]. Consequently, helminth infection can induce immune responses that may not be appropriate for or effective at controlling viral infections. For example, diminished Th1 cytokine and CD8+ cytotoxic T lymphocyte response during Schistosoma mansoni infection has been suggested to play a role in a decreased ability to fight viral infection during concurrent helminth infection [34]. Coinfection in mice with the nematodes Trichinella spiralis or Heligmosomoides polygyrus bakeri and murine norovirus resulted in a dampened CD8+ T-cell antiviral response and higher viral loads in the murine intestine, compared with monoinfected control animals [35]. Furthermore, modulation of antiviral immunity could be observed in germ-free mice, indicating that these changes were independent of the resident bacterial microbiota. Infecting mice with H polygyrus or administering S mansoni eggs was found to reactivate latent murine gammaherpesvirus 68, which is similar to the human gammaherpesviruses (Epstein-Barr virus and Kaposi’s sarcoma-associated herpesvirus) [36]. Here, IL-4-induced activation of the Stat6 transcription factor promoted viral replication in conjunction with antagonism of IFN-γ-mediated viral suppression in response to helminthic infection [36].
In contrast, helminth infections may also provide a protective antiviral response due to the dynamic changes in immune response over the course of the infection. For example, the Th1-mediated parasite antigen response to migrating schistosomes in mice elicits an increase in IFN-γ, which suppressed hepatitis B virus replication [37]. In addition, Th2-mediated IL-4 release in response to S mansoni eggs augmented CD8+ T-cell responses to murine herpesvirus 4, resulting in reduced viral disease severity [38]. The severe acute respiratory syndrome coronavirus 2 (SARS-CoV2) pandemic has raised discussion on the possible impact of helminth infection on coronavirus 2019 (COVID-19) disease severity. Some groups speculate that a concurrent helminth infection could negatively impact COVID-19 patients due to elevated Th2-associated cytokine levels. Others argue that a chronic helminth infection could benefit COVID-19 patients by reducing Th1-associated proinflammatory cytokines [39, 40]. Taken together, the results of these studies highlight that helminthic infections impacts viral pathogenesis. However, further studies are needed to evaluate the clinical significance of the contributions of helminths to viral infection.
PULMONARY BACTERIAL-VIRAL INTERACTIONS
Although the human upper and lower respiratory tract microbiomes are less well characterized than the gut, especially that of the healthy lung, transkingdom interactions are well described in both clinical practice and animal models. The best characterized of these interactions is the synergy between influenza A virus (IAV) and bacterial pathogens of the respiratory tract. Influenza A virus directly interacts with pathogenic constituents of the upper respiratory tract microbiome, including Streptococcus pneumoniae, Staphylococcus aureus, Haemophilus influenzae, and Moraxella catarrhalis [41, 42]. These interactions enhance bacterial pathogenesis [42] and alter immune responses to both S pneumoniae and IAV [41]. They can also alter IAV pathogenesis by promoting environmental survival and influencing transmissibility [43]. Infection with IAV causes lung cells to increase their expression of bacterial receptors [44], thereby enhancing bacterial adherence, and depletes alveolar macrophages [45], which are key to clearing bacterial infections in the lungs. Cytokines induced by IAV infection can exacerbate bacterial pneumonia caused by S aureus or S pneumoniae [46]. Likewise, respiratory syncytial virus (RSV) infection predisposes vulnerable populations to secondary bacterial pneumonia [47]. Direct interactions have been observed between RSV and both S pneumoniae and H influenzae [48], which can enhance bacterial and possibly viral pathogenesis [48].
The enhancement of respiratory viral pathogenesis by bacteria and bacterial products is due to direct and indirect mechanisms. Proteases released by bacteria or exposed on the bacterial surface enhance IAV pathogenesis by cleaving the viral hemagglutinin to enable receptor binding [49]. Preincubation of epithelial cells with H influenzae promotes the adherence of rhinovirus to the cells through increased expression of intercellular adhesion molecule (ICAM)-1 and enhances the pathogenesis of rhinovirus infection through upregulation of TLR3 and subsequent proinflammatory cytokine production [50]. These interactions do not require viable bacteria because heat-killed H influenzae also increased ICAM-1 expression, viral replication, and proinflammatory cytokine production when used to coinfect cells along with rhinovirus or RSV [51]. The enhancement of respiratory viral pathogenesis by bacterial products extends to heat-killed Pseudomonas aeruginosa, adenovirus, and influenza B virus, but not to heat-killed S pneumoniae [51], suggesting that the respiratory bacterial community exerts species-specific effects on viral pathogenesis. Insights into which bacterial communities in the upper and lower respiratory tract are proviral or antiviral could help stratify populations according to their risk of pneumonia and inform antibiotic treatments to spare antiviral commensal bacteria.
Finally, in the “gut-lung axis,” the bacterial gut microbiome can alter the response to respiratory infection [52] and viral upper respiratory infection can alter the bacterial gut microbiome [53]. Gut microbiome-derived compounds alter IFN signaling in the lungs, thereby conferring protection against RSV [54] and IAV [52] in murine models. In human transplant patients, the presence of butyrate-producing bacteria in the gut reduced the incidence of lower respiratory tract viral infection after bone marrow or kidney transplant [55, 56]. Together, these data support the immunomodulatory roles of the gut microbial community in regulating respiratory immune response and susceptibility to respiratory viral infection, even in the context of immunosuppressive posttransplant medication. The enteric community alters both cell-based and humoral responses to IAV [57]. Bacterial products found and made in the gut community can also destabilize respiratory viruses, including IAV [58] and coronaviruses [59], which suggests that there is a mechanism for tissue tropism and modulation of environmental stability. These findings, taken together, suggest the diverse roles played by the microbiota in the enteric and respiratory tracts in terms of modulating both the risk of acquisition and the severity of viral infection in healthy and high-risk hosts.
PULMONARY VIRAL-HELMINTH INTERACTIONS
When viruses and parasites coinfect a host, one infectious agent can skew the immune response away from the natural response to the other invader, even if the 2 agents are infecting distinct sites within the host, as in the “gut-lung axis.” In the case of viruses that cause immunopathologic disease, such as IAV and RSV, such interactions can be beneficial to the host. Coinfection with Trichinella spiralis and IAV causes a reduction in both tumor necrosis factor-α production and immune-cell infiltration into the lungs after IAV infection, leading to a quicker recovery [60]. Heligmosomoides polygyrus coinfection with IAV results in decreased viral loads and correspondingly decreased hemagglutination inhibition titers [61]. Likewise, H polygyrus coinfection with RSV results in decreased viral loads and reduced pulmonary inflammation [62]. More importantly, neither of these eukaryotic parasites have pulmonary stages in their life cycles; therefore, they apparently act remotely to modulate the immune microenvironment in the lung during viral infection.
More complex interactions occur when viruses and parasites both directly infect the lungs. Nippostrongylus brasiliensis infection leads to increased IAV lung titers, but only when larvae are actively migrating through the lungs. In contrast, quiescent parasites can induce proinflammatory states that are protective against secondary viral infection. Schistosoma mansoni oviposition results in granulatomous inflammation in the lung that can protect against secondary infection with IAV or pneumovirus [63]. These interactions, taken together, demonstrate that the multivariate nature of virus-parasite interactions in the lung is dependent on the species of virus and parasite, the life stage of the parasite, and the site of the parasite infection. As with enteric helminth infection, a better understanding of helminth-virus interactions will have particular benefit for global health and especially in low-income countries, where upper respiratory viruses are a leading cause of pediatric mortality.
PULMONARY VIRAL-FUNGAL INTERACTIONS
Emerging evidence suggests that fungi play important roles in viral pathogenesis in the lungs. Multiple case reports suggest that after influenza virus infection, immunocompetent patients can become infected with Aspergillus [64]. It is hypothesized that the lung microenvironment during and after influenza virus infection predisposes the lungs to Aspergillus infection. Treating influenza with steroids and/or neuraminidase inhibitors can further increase the risk of secondary fungal infections [64, 65]. In humans, pulmonary fungal infections are predominately associated with immunosuppressive disorders such as HIV infection. Fungal infections in patients with HIV infection are primarily caused by members of the Aspergillus, Cryptococcus, and Pneumocystis genera [66] and are one marker of progression to acquired immune deficiency syndrome. The immunosuppressive state induced by HIV infection enables the colonization of the lung by these opportunistic and environmental fungi. Human immunodeficiency virus and another immunosuppressive virus, measles virus, both trigger “vomocytosis,” which causes the expulsion of Cryptococcus cells from virally infected macrophages, potentially exacerbating disease severity and spread [67].
PATHOGENIC VIRUS-BACTERIOPHAGE INTERACTIONS
Bacteriophages are important components of the microbiome. They are primarily known for their role in intestinal homeostasis, immune tolerance, and mucosal immunity [68]. Bacteriophages that parasitize the bacterial population of the lung have been described in (1) healthy lungs [69], (2) lungs with chronic obstructive pulmonary disease [69], and (3) lungs with cystic fibrosis [70]. It is interesting to note that these bacteriophage communities can be diverse even within the same patient [70]. Bacteriophages encode virulence and antibiotic resistance genes [69–71]; therefore, antibiotic therapies that activate lysogenic bacteriophages risk causing the spread of bacterial virulence factors and, more importantly, contribute to the rise of antimicrobial resistance. Although bacteriophages primarily exert indirect effects on viral pathogenesis through their regulation of the microbiome, they have been shown to have more direct interactions with viral pathogens. Bacteriophages contain many of the pathogen-associated molecular patterns (PAMPs) that are common to pathogenic viruses and that can stimulate host immune responses. These PAMPs include bacteriophage nucleic acids, which can stimulate IFN production and predispose the host to mount an antiviral immune response [72]. In addition, bacteriophages can directly inhibit pathogenic viruses by competing with them in binding to receptors. This has been demonstrated for several viruses, particular herpesviruses, which can be outcompeted by bacteriophages in binding to their receptor [73]. Bacteriophage therapy, the use of bacteriophages to therapeutically kill bacteria, has been explored since the early 1900s and shows promise against the superbugs of today [74]. Future studies should consider the role of bacteriophages and bacteriophage therapy in viral pathogenesis.
CONCLUSIONS
The direct and indirect interactions between viruses and the micro-, myco-, and macrobiomes of the lung and the gut represent an important and growing area of research. These studies are providing fundamental knowledge about the importance of transkingdom interactions in health and disease, and they may identify new therapeutic targets for viral infections. The enhancement of the infectivity of viruses by direct interactions with bacteria, fungi, or macrobiota can have significant consequences beyond those highlighted here, including increasing the effective multiplicity of infection and enabling complementation of defective viral particles. Therefore, understanding the impact of coinfecting pathogens and of the symbiotic microbial communities on viral pathogenesis and on immune responses will inform antibiotic choices and vaccine design to take advantage of host-beneficial interactions and minimize viral-beneficial interactions. Moreover, the implementation of bacteriophage therapy and the use of precision antibiotics that are designed to spare commensal bacteria will be key advances but might also have unforeseen consequences for infections with pathogenic viruses. These observations underscore the importance of understanding the complex microbial community encountered by pathogenic viruses and the synergistic and antagonistic interactions that occur therein.
Notes
Acknowledgments. We thank Rebekah Honce for graphic design and Dr. Keith A. Laycock for scientific editing of the manuscript.
Financial support. J. R. is partially funded through American Lebanese Syrian Associated Charities (ALSAC), National Institutes of Health National Institute of Allergy and Infectious Diseases (Grants 1U01-AI124302 and 1R01-AI110618). S. S.-C. is partially funded through ALSAC, National Institute of Allergy and Infectious Diseases (Grants R21 AI125254-01, R03 AI133527-02, R01 AI140766-01, HHSN272201400006C, and 75N93019C00052). V. C. is partially funded through National Institute of Allergy and Infectious Diseases (Grant T32 AI106700).
Supplement sponsorship. This work is part of a supplement sponsored by the National Institute of Allergy and Infectious Diseases (NIAID), part of the National Institutes of Health (NIH) and the Centers for Disease Control and Prevention (CDC).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Filyk HA, Osborne LC. The multibiome: the intestinal ecosystem’s influence on immune homeostasis, health, and disease. EBioMedicine 2016; 13:46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell 2014; 157:121–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pullen LC, Park SH, Miller SD, Dal Canto MC, Kim BS. Treatment with bacterial LPS renders genetically resistant C57BL/6 mice susceptible to Theiler’s virus-induced demyelinating disease. J Immunol 1995; 155:4497–503. [PubMed] [Google Scholar]
- 4. Kuss SK, Best GT, Etheredge CA, et al. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science 2011; 334:249–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jones MK, Watanabe M, Zhu S, et al. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 2014; 346:755–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Robinson CM, Jesudhasan PR, Pfeiffer JK. Bacterial lipopolysaccharide binding enhances virion stability and promotes environmental fitness of an enteric virus. Cell Host Microbe 2014; 15:36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Basic M, Keubler LM, Buettner M, et al. Norovirus triggered microbiota-driven mucosal inflammation in interleukin 10-deficient mice. Inflamm Bowel Dis 2014; 20:431–43. [DOI] [PubMed] [Google Scholar]
- 8. Kane M, Case LK, Kopaskie K, et al. Successful transmission of a retrovirus depends on the commensal microbiota. Science 2011; 334:245–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baldridge MT, Nice TJ, McCune BT, et al. Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection. Science 2015; 347:266–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nice TJ, Baldridge MT, McCune BT, et al. Interferon-λ cures persistent murine norovirus infection in the absence of adaptive immunity. Science 2015; 347:269–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grau KR, Zhu S, Peterson ST, et al. The intestinal regionalization of acute norovirus infection is regulated by the microbiota via bile acid-mediated priming of type III interferon. Nat Microbiol 2020; 5:84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cortez V, Boyd DF, Crawford JC, et al. Astrovirus infects actively secreting goblet cells and alters the gut mucus barrier. Nat Commun 2020; 11:2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Robinson CM, Woods Acevedo MA, McCune BT, Pfeiffer JK. Related enteric viruses have different requirements for host microbiota in mice. J Virol 2019; 93:e01339–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Looft T, Allen HK. Collateral effects of antibiotics on mammalian gut microbiomes. Gut Microbes 2012; 3:463–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Woods Acevedo MA, Erickson AK, Pfeiffer JK. The antibiotic neomycin enhances coxsackievirus plaque formation. mSphere 2019; 4:e00632–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guandalini S. Probiotics for prevention and treatment of diarrhea. J Clin Gastroenterol 2011; 45(Suppl):S149–53. [DOI] [PubMed] [Google Scholar]
- 17. Kumar R, Seo BJ, Mun MR, et al. Putative probiotic Lactobacillus spp. from porcine gastrointestinal tract inhibit transmissible gastroenteritis coronavirus and enteric bacterial pathogens. Trop Anim Health Prod 2010; 42:1855–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Varyukhina S, Freitas M, Bardin S, et al. Glycan-modifying bacteria-derived soluble factors from Bacteroides thetaiotaomicron and Lactobacillus casei inhibit rotavirus infection in human intestinal cells. Microbes Infect 2012; 14:273–8. [DOI] [PubMed] [Google Scholar]
- 19. Zhang B, Chassaing B, Shi Z, et al. Viral infection. Prevention and cure of rotavirus infection via TLR5/NLRC4-mediated production of IL-22 and IL-18. Science 2014; 346:861–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shi Z, Zou J, Zhang Z, et al. Segmented filamentous bacteria prevent and cure rotavirus infection. Cell 2019; 179:644–58.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pezoldt J, Yang JH, Zou MG, Huehn J. Microbiome and gut immunity: T cells. Gut Microbiome Health Dis 2018; 119–40. [Google Scholar]
- 22. Molberg O, Nilsen EM, Sollid LM, et al. CD4+ T cells with specific reactivity against astrovirus isolated from normal human small intestine. Gastroenterology 1998; 114:115–22. [DOI] [PubMed] [Google Scholar]
- 23. Yokoyama CC, Loh J, Zhao G, et al. Adaptive immunity restricts replication of novel murine astroviruses. J Virol 2012; 86:12262–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Malm M, Hyöty H, Knip M, Vesikari T, Blazevic V. Development of T cell immunity to norovirus and rotavirus in children under five years of age. Sci Rep 2019; 9:3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tomov VT, Osborne LC, Dolfi DV, et al. Persistent enteric murine norovirus infection is associated with functionally suboptimal virus-specific CD8 T cell responses. J Virol 2013; 87:7015–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hoffmann C, Dollive S, Grunberg S, et al. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One 2013; 8:e66019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jha AK, Uppal B, Chadha S, et al. Clinical and microbiological profile of HIV/AIDS cases with diarrhea in North India. J Pathog 2012; 2012:971958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen Y, Chen Z, Guo R, et al. Correlation between gastrointestinal fungi and varying degrees of chronic hepatitis B virus infection. Diagn Microbiol Infect Dis 2011; 70:492–8. [DOI] [PubMed] [Google Scholar]
- 29. Das S, Gupta PK, Das RR. Efficacy and safety of Saccharomyces boulardii in acute rotavirus diarrhea: double blind randomized controlled trial from a developing country. J Trop Pediatr 2016; 62:464–70. [DOI] [PubMed] [Google Scholar]
- 30. Savioli L, Albonico M. Soil-transmitted helminthiasis. Nat Rev Microbiol 2004; 2:618–9. [DOI] [PubMed] [Google Scholar]
- 31. Leung JM, Loke P. A role for IL-22 in the relationship between intestinal helminths, gut microbiota and mucosal immunity. Int J Parasitol 2013; 43:253–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nutman TB. Looking beyond the induction of Th2 responses to explain immunomodulation by helminths. Parasite Immunol 2015; 37:304–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Varyani F, Fleming JO, Maizels RM. Helminths in the gastrointestinal tract as modulators of immunity and pathology. Am J Physiol Gastrointest Liver Physiol 2017; 312:G537–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Actor JK, Shirai M, Kullberg MC, Buller RM, Sher A, Berzofsky JA. Helminth infection results in decreased virus-specific CD8+ cytotoxic T-cell and Th1 cytokine responses as well as delayed virus clearance. Proc Natl Acad Sci U S A 1993; 90:948–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Osborne LC, Monticelli LA, Nice TJ, et al. Coinfection. Virus-helminth coinfection reveals a microbiota-independent mechanism of immunomodulation. Science 2014; 345:578–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reese TA, Wakeman BS, Choi HS, et al. Helminth infection reactivates latent γ-herpesvirus via cytokine competition at a viral promoter. Science 2014; 345:573–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Loffredo-Verde E, Bhattacharjee S, Malo A, et al. Dynamic, helminth-induced immune modulation influences the outcome of acute and chronic hepatitis B virus infection. J Infect Dis 2020; 221:1448–61. [DOI] [PubMed] [Google Scholar]
- 38. Rolot M, Dougall AM, Chetty A, et al. Helminth-induced IL-4 expands bystander memory CD8+ T cells for early control of viral infection. Nat Commun 2018; 9:4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bradbury RS, Piedrafita D, Greenhill A, Mahanty S. Will helminth co-infection modulate COVID-19 severity in endemic regions? Nat Rev Immunol 2020; 20:342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hays R, Pierce D, Giacomin P, Loukas A, Bourke P, McDermott R. Helminth coinfection and COVID-19: an alternate hypothesis. PLoS Negl Trop Dis 2020; 14:e0008628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. David SC, Norton T, Tyllis T, et al. Direct interaction of whole-inactivated influenza A and pneumococcal vaccines enhances influenza-specific immunity. Nat Microbiol 2019; 4:1316–27. [DOI] [PubMed] [Google Scholar]
- 42. Rowe HM, Meliopoulos VA, Iverson A, Bomme P, Schultz-Cherry S, Rosch JW. Direct interactions with influenza promote bacterial adherence during respiratory infections. Nat Microbiol 2019; 4:1328–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rowe HM, Livingston B, Margolis E, et al. Respiratory bacteria stabilize and promote airborne transmission of influenza A virus. mSystems 2020; 5:e00762–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McCullers JA, Rehg JE. Lethal synergism between influenza virus and Streptococcus pneumoniae: characterization of a mouse model and the role of platelet-activating factor receptor. J Infect Dis 2002; 186:341–50. [DOI] [PubMed] [Google Scholar]
- 45. Ghoneim HE, Thomas PG, McCullers JA. Depletion of alveolar macrophages during influenza infection facilitates bacterial superinfections. J Immunol 2013; 191:1250–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Duvigneau S, Sharma-Chawla N, Boianelli A, et al. Hierarchical effects of pro-inflammatory cytokines on the post-influenza susceptibility to pneumococcal coinfection. Sci Rep 2016; 6:37045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Weinberger DM, Klugman KP, Steiner CA, Simonsen L, Viboud C. Association between respiratory syncytial virus activity and pneumococcal disease in infants: a time series analysis of US hospitalization data. PLoS Med 2015; 12:e1001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hament JM, Aerts PC, Fleer A, et al. Direct binding of respiratory syncytial virus to pneumococci: a phenomenon that enhances both pneumococcal adherence to human epithelial cells and pneumococcal invasiveness in a murine model. Pediatr Res 2005; 58:1198–203. [DOI] [PubMed] [Google Scholar]
- 49. Tashiro M, Ciborowski P, Klenk HD, Pulverer G, Rott R. Role of Staphylococcus protease in the development of influenza pneumonia. Nature 1987; 325:536–7. [DOI] [PubMed] [Google Scholar]
- 50. Sajjan US, Jia Y, Newcomb DC, et al. H. influenzae potentiates airway epithelial cell responses to rhinovirus by increasing ICAM-1 and TLR3 expression. FASEB J 2006; 20:2121–3. [DOI] [PubMed] [Google Scholar]
- 51. Bellinghausen C, Gulraiz F, Heinzmann AC, et al. Exposure to common respiratory bacteria alters the airway epithelial response to subsequent viral infection. Respir Res 2016; 17:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sencio V, Barthelemy A, Tavares LP, et al. Gut dysbiosis during influenza contributes to pulmonary pneumococcal superinfection through altered short-chain fatty acid production. Cell Rep 2020; 30:2934–47.e6. [DOI] [PubMed] [Google Scholar]
- 53. Groves HT, Higham SL, Moffatt MF, Cox MJ, Tregoning JS. Respiratory viral infection alters the gut microbiota by inducing inappetence. mBio 2020; 11:e03236–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Antunes KH, Fachi JL, de Paula R, et al. Microbiota-derived acetate protects against respiratory syncytial virus infection through a GPR43-type 1 interferon response. Nat Commun 2019; 10:3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Haak BW, Littmann ER, Chaubard JL, et al. Impact of gut colonization with butyrate-producing microbiota on respiratory viral infection following allo-HCT. Blood 2018; 131:2978–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee JR, Huang J, Magruder M, et al. Butyrate-producing gut bacteria and viral infections in kidney transplant recipients: a pilot study. Transpl Infect Dis 2019; 21:e13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ichinohe T, Pang IK, Kumamoto Y, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A 2011; 108:5354–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bandoro C, Runstadler JA. Bacterial lipopolysaccharide destabilizes influenza viruses. mSphere 2017; 2:e00267–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Johnson BA, Hage A, Kalveram B, et al. Peptidoglycan-associated cyclic lipopeptide disrupts viral infectivity. J Virol 2019; 93:e01282–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Furze RC, Hussell T, Selkirk ME. Amelioration of influenza-induced pathology in mice by coinfection with Trichinella spiralis. Infect Immun 2006; 74:1924–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chowaniec W, Wescott RB, Congdon LL. Interaction of Nematospiroides dubius and influenza virus in mice. Exp Parasitol 1972; 32:33–44. [DOI] [PubMed] [Google Scholar]
- 62. McFarlane AJ, McSorley HJ, Davidson DJ, et al. Enteric helminth-induced type I interferon signaling protects against pulmonary virus infection through interaction with the microbiota. J Allergy Clin Immunol 2017; 140:1068–78.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Scheer S, Krempl C, Kallfass C, et al. S. mansoni bolsters anti-viral immunity in the murine respiratory tract. PLoS One 2014; 9:e112469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shah MM, Hsiao EI, Kirsch CM, Gohil A, Narasimhan S, Stevens DA. Invasive pulmonary aspergillosis and influenza co-infection in immunocompetent hosts: case reports and review of the literature. Diagn Microbiol Infect Dis 2018; 91:147–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Van De Veerdonk F Jr, Dewi I, Cunha C, et al. Inhibition of host neuraminidase increases susceptibility to invasive pulmonary aspergillosis. Open Forum Infect Dis 2018; 5:S36. [Google Scholar]
- 66. Li Z, Lu G, Meng G. Pathogenic fungal infection in the lung. Front Immunol 2019; 10:1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Seoane PI, Taylor-Smith LM, Stirling D, et al. Viral infection triggers interferon-induced expulsion of live Cryptococcus neoformans by macrophages. PLoS Pathog 2020; 16:e1008240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vitetta L, Vitetta G, Hall S. Immunological tolerance and function: associations between intestinal bacteria, probiotics, prebiotics, and phages. Front Immunol 2018; 9:2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Elbehery AHA, Feichtmayer J, Singh D, Griebler C, Deng L. The human virome protein cluster database (HVPC): a human viral metagenomic database for diversity and function annotation. Front Microbiol 2018; 9:1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Willner D, Haynes MR, Furlan M, et al. Case studies of the spatial heterogeneity of DNA viruses in the cystic fibrosis lung. Am J Respir Cell Mol Biol 2012; 46:127–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rolain JM, Fancello L, Desnues C, Raoult D. Bacteriophages as vehicles of the resistome in cystic fibrosis. J Antimicrob Chemother 2011; 66:2444–7. [DOI] [PubMed] [Google Scholar]
- 72. Miedzybrodzki R, Fortuna W, Weber-Dabrowska B, Gorski A. Bacterial viruses against viruses pathogenic for man? Virus Res 2005; 110:1–8. [DOI] [PubMed] [Google Scholar]
- 73. Górski A, Międzybrodzki R, Jończyk-Matysiak E, Weber-Dąbrowska B, Bagińska N, Borysowski J. Perspectives of phage-eukaryotic cell interactions to control Epstein-Barr virus infections. Front Microbiol 2018; 9:630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lin DM, Koskella B, Lin HC. Phage therapy: an alternative to antibiotics in the age of multi-drug resistance. World J Gastrointest Pharmacol Ther 2017; 8:162–73. [DOI] [PMC free article] [PubMed] [Google Scholar]