Abstract
Background
COVID-19 has resulted in significant morbidity and mortality worldwide. Lateral flow assays can detect anti-Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) antibodies to monitor transmission. However, standardized evaluation of their accuracy and tools to aid in interpreting results are needed.
Methods
We evaluated 20 IgG and IgM assays selected from available tests in April 2020. We evaluated the assays’ performance using 56 pre-pandemic negative and 56 SARS-CoV-2-positive plasma samples, collected 10–40 days after symptom onset, confirmed by a molecular test and analyzed by an ultra-sensitive immunoassay. Finally, we developed a user-friendly web app to extrapolate the positive predictive values based on their accuracy and local prevalence.
Results
Combined IgG + IgM sensitivities ranged from 33.9 to 94.6%, while combined specificities ranged from 92.6 to 100%. The highest sensitivities were detected in Lumiquick for IgG (98.2%), BioHit for both IgM (96.4%), and combined IgG + IgM sensitivity (94.6%). Furthermore, 11 LFAs and 8 LFAs showed perfect specificity for IgG and IgM, respectively, with 15 LFAs showing perfect combined IgG + IgM specificity. Lumiquick had the lowest estimated limit-of-detection (LOD) (0.1 μg/mL), followed by a similar LOD of 1.5 μg/mL for CareHealth, Cellex, KHB, and Vivachek.
Conclusion
We provide a public resource of the accuracy of select lateral flow assays with potential for home testing. The cost-effectiveness, scalable manufacturing process, and suitability for self-testing makes LFAs an attractive option for monitoring disease prevalence and assessing vaccine responsiveness. Our web tool provides an easy-to-use interface to demonstrate the impact of prevalence and test accuracy on the positive predictive values.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12879-021-06257-7.
Keywords: SARS-CoV-2, Coronavirus, COVID-19, Antibodies, Lateral flow assays
Background
Coronavirus disease 2019 (COVID-19), caused by infection with the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was declared a global pandemic by on March 11th, 2020 [1], with a second wave of the pandemic well underway [1]. However, accurate estimates of transmission rely on accurate and widely distributed population immunosurveillance tools to measure SARS-CoV-2 infection in diverse community settings. Among SARS-CoV-2-infected individuals, 40–45% are estimated to remain asymptomatic [2], suggesting that prevalence is likely underestimated [3]. Therefore, detecting prior exposure to SARS-CoV-2 as opposed to other viral infections or other coronavirus strains is crucial [4].
There are different types of clinical SARS-CoV-2 tests. Diagnostic testing relies on reverse-transcriptase quantitative polymerase chain reaction (RT-qPCR) and antigen-based immunodiagnostics to detect active infection [5]. Conversely, serological tests are useful for monitoring population prevalence and prior exposure by measuring antibodies against SARS-CoV-2 [6–8]. These include enzyme-linked immunosorbent assays (ELISAs), chemiluminescence assays, and lateral flow assays (LFAs) [5, 9, 10]. LFAs are attractive for home testing and population surveillance, since they are affordable, scalable, rely on easily accessible specimens such as fingerstick whole blood and give a result readout within minutes [7]. Since multiple vaccines received emergency use authorization [11], serological assays could be used to determine whether vaccines elicit a detectable and durable immune response [12–16]. Hence, easy-to-use LFAs will have important applications in the upcoming phases of the pandemic. Since the onset of the COVID-19 epidemic, multiple studies evaluated the accuracy of serological tests [9, 17–21]. Many of these tests received Emergency Use Authorization (EUA) through the Food and Drug Administration (FDA) [22].
Despite the utility of SARS-CoV-2 antibody tests, misinterpretation of results is very likely [23]. A negative serological test result does not preclude prior infection since seroconversion occurs 9–11 and 18–20 days after symptoms onset for IgM and IgG antibodies, respectively [10, 24]. Conversely, positive results do not indicate active infection [23]. Furthermore, the prevalence of SARS-CoV-2 is highly variable [1, 3], and known to directly impact the predictive value of a test result. A higher prevalence increases the likelihood that a positive test result indicates a real infection (i.e. higher positive predictive value) [25], but will also decrease the negative predictive value, resulting in more false negative results [25]. Therefore, accessible tools to assist the public with interpreting results based on test accuracy and different prevalence scenarios are critical [23, 26].
In April 2020, the Mass General and Brigham Center for COVID Innovation direct-to-consumer working group scanned available serological assays and selected 20 lateral flow assays, based on reported assay characteristics and supply chain availability [27]. The LFAs were evaluated by blinded operators using the same samples to standardize the evaluation of their accuracy. Additionally, we developed a user-friendly web-tool to assist the end user to interpret their results. This study provides both the evaluation data to serve as a public resource to guide implementation of LFAs, and the tool to aid the interpretation of home testing results.
Methods
Sample procurement
We procured 56 SARS-CoV-2-positive, 46 pre-pandemic SARS-CoV-2-negative, and 10 SARS-CoV-2-negative HIV-positive EDTA plasma samples. SARS-CoV-2 positive EDTA plasma samples were obtained from clinical discards banked within 24–72 h of collection at the Crimson Core of the MGB Biobank, which was composed of hospitalized symptomatic patients. All samples had positive SARS-CoV-2 PCR results using an EUA approved test at Brigham and Women’s Hospital (Panther Fusion SARS-CoV-2 assay, Hologic, Inc., San Diego, CA or Xpert Xpress SARS-CoV-2, Cepheid, Inc., Sunnyvale, CA) or the Clinical Research Sequencing Platform at the Broad Institute of MIT and Harvard (in house Laboratory Developed Test) 10–40 days prior to sample collection. The participants’ charts were reviewed by study staff to identify samples collected ≥10 days after onset of symptoms and to exclude immunosuppressed participants, after which samples were anonymized and stripped of protected health information. Pre-pandemic negative control samples were randomly selected from healthy participants with a Charlson Age-Comorbidity Index [28] score ≤ 2 and EDTA plasma banked in the MGB Biobank between Jan 1-Dec 1, 2019 from inpatients. HIV-positive control samples were obtained from EDTA plasma samples banked prior to January 2020 in a study on neuropathic pain in HIV. All HIV-positive participants were on antiretroviral therapy. For 8 out of the 10 HIV-positive samples, viral load quantification was available and showed 256 copies/ml or less, and 5 showed loads either undetectable or under 20 copies/ml. All samples were collected from consented individuals. Sample and data collection conformed to Good Clinical Practice guidelines and Declaration of Helsinki. The study was approved under the Massachusetts General Brigham (MGB) Institutional Review Board (protocol no. 2020P001204).
Lateral flow assays (LFAs)
Twenty commercial IgG/IgM lateral flow assays (LFAs) from 18 manufacturers were evaluated (Supplementary Table 1). LFAs were analyzed by blinded operators according to manufacturer instructions for use (IFU), with the exception of using micropipettes instead of manufacturer-provided droppers to minimize technical variability. Samples were thawed on ice, randomized, and brought to room temperature. Kit components were also brought to room temperature. The IFU-specified volumes of sample and buffer were added to the cassette. Specified sample volumes varied for different LFAs but were typically in the 5-20 μL range. The cassettes were run at room temperature on a flat surface and results read immediately after the time interval defined in the IFU (typically ranging from 10 to 15 min). Each cassette was independently scored by two blinded raters as either “positive,” “negative,” or “invalid”. Ratings were designated according to the interpretation guidelines outlined in each individual IFU. Each cassette was photographed under four standardized illumination conditions and viewpoints for future analysis.
Reproducibility testing
For inter-operator reproducibility analysis, separate pools of EDTA plasma were obtained from > 30 pre-pandemic healthy individuals (negative pool) and > 30 convalescent participants collected after symptom resolution at the Massachusetts General Hospital (MGH) respiratory illness clinic (positive pool). Convalescent samples for the positive pool were confirmed to be positive using the COBAS SARS-CoV-2 PCR test (Roche Diagnostics, Indianapolis, IN) at MGH. A total of 20 replicates per pool were run by two independent pairs of blinded operators, alternating between positive and negative pools (10 replicates per pair). Reproducibility was calculated according to agreement between operator ratings as well as concordance of readout with sample pool COVID status.
Sensitivity and specificity testing
Our cohort of 112 EDTA plasma samples was used across all 20 LFAs to evaluate performance: sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV). Samples were sub-aliquoted throughout the analysis to minimize freeze-thaw cycles. Binary presence/absence classifications were used, and discordant calls were resolved by a third operator inspecting photographs taken of the relevant LFA.
LFA usability
In addition to initial screening [27], each LFA kit was assessed for consumer usability based on complexity of kit materials, sample requirements, and IFU clarity. Supplied kit components were documented for completeness and examined for ease-of-use. IFU protocols were rated on a scale from 0 to 14 according to a predefined rubric (Supplementary Table 2) by three independent raters. Usability evaluations are shown in Supplementary Table 3. Sample input requirements for each LFA are in Supplementary Table 1.
Ultrasensitive Simoa serology assays
Plasma samples were diluted 4000-fold, and the total IgG and IgM levels against the SARS-CoV-2 spike protein were measured using a custom Single Molecule Array (Simoa) assay as described [29] on an automated HD-X Analyzer (Quanterix, Billerica, MA, USA), to provide a quantitative reference for anti-spike antibody titers in the plasma samples. Normalized mean Average Enzymes per Bead (AEB) levels were calculated using a standard set of calibrators produced by serially diluting a large volume of plasma from seroconverted individuals. Antibody concentrations were estimated using a calibration curve of recombinant anti-SARS-CoV-2 antibodies [30].
Analysis and Webapp
Detailed methods are in the supplementary.
Results
Study population
We obtained plasma samples from 56 pre-pandemic patients, including 10 HIV+, and 56 symptomatic inpatients in March and April 2020. HIV-negative samples were matched for sex and age. The overall study population included 25.9% Blacks, 4.5% Latinx, 9.8% Asians, and 43.8% Non-Hispanic Whites (Table 1). COVID+ samples were from individuals between 10 and 40 days post symptom onset, with 60.7% samples taken between 2 and 4 weeks. Of all the COVID+ participants, 7 (12.5%) were symptomatic outpatients and 49 (87.5%) were hospitalized. Among those hospitalized, 25 (51%) required intensive care unit (ICU) treatment and 4 (8.2%) were deceased at the time of chart review (Supplementary Table 4). Additional mortalities were possible after chart review since some participants were in critical condition in the ICU.
Table 1.
Demographic information of all individuals whose plasma samples were used for this study. Individuals are broken out by COVID+/− and HIV+/− status. Black includes one mixed African American in the COVID-HIV+ group
| COVID- HIV- | COVID- HIV+ | COVID+ HIV- | Overall | |
|---|---|---|---|---|
| n = 46 | n = 10 | n = 56 | n = 112 | |
| Sex | ||||
| Female | 28 (60.9%) | 1 (10%) | 31 (55.4%) | 60 |
| Male | 18 (39.1%) | 9 (90%) | 25 (44.6%) | 52 |
| Age | ||||
| Mean (SD) | 46.7 (13.4) | 58.6 (5.3) | 58.7 (20.4) | 53.8 (17.8) |
| Median [Min, Max] | 48.0 [19.0, 68.0] | 58.0 [48.0,65.0] | 57.5 [24.0, 98.0] | 54.5 [19.0,98.0] |
| Race | ||||
| Asian | 8 (17.4%) | 0 (0%) | 3 (5.4%) | 11 (9.8%) |
| Black | 10 (21.7%) | 4 (40%) | 16 (28.6%) | 30 (26.8%) |
| LatinX | 0 (0%) | 0 (0%) | 5 (8.9%) | 5 (4.5%) |
| Unknown/Other | 8 (17.4%) | 1 (10%) | 8 (14.3%) | 17 (15.2%) |
| White | 20 (43.5%) | 5 (50%) | 24 (42.9%) | 49 (43.8%) |
LFA performance: reproducibility
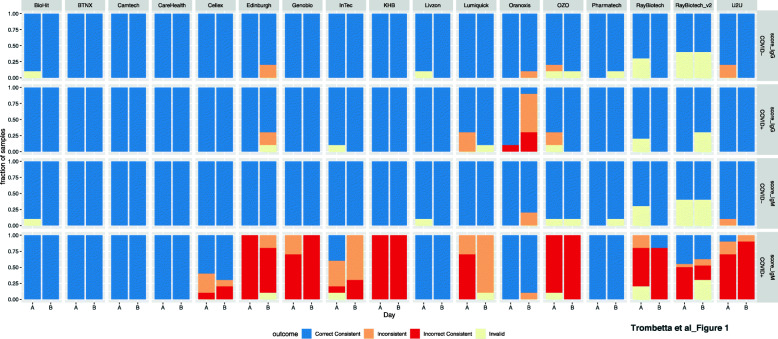
Four independent operators working in teams of two on separate days applied each pool 10 times to each LFA, with processing as dictated by the instructions for use (IFU). API version1 LFAs were not assessed for reproducibility due to limited cassette availability and the high volume required. Of the remaining 19 LFAs, three (BTNX, Camtech, and Carehealth) had 100% consistent correct outcomes across both isotypes, with an additional three (BioHit, Zhuhai Livzon, and Phamatech) having no incorrect or inconsistent outcomes with one or two invalid tests (Fig. 1). IgG was the more reproducible isotype. The majority of incorrect consistent calls came from operators calling a COVID+ sample IgM negative (Fig. 1).
Fig. 1.
LFA reproducibility. Colors represent LFA outcome performed across two days (A & B) by two independent pairs of operators on a COVID positive (COVID+) pooled sample and a COVID negative (COVID-) pooled sample. On each day ten technical replicates of each pool were performed. Blue represents replicates where both operators that day agreed on the outcome, and the outcome was correctly called positive or negative. Red represents occasions where both operators agreed, but the outcome was incorrectly called. Orange represents replicates where the operators did not agree on the LFA outcome. Light yellow represents an invalid test where control bands did not meet criteria for a valid test. IgG was the most reproducible isotype for all LFAs except Oranoxis, and consistent false negatives were common occurrences for IgM for the majority of LFAs
LFA performance: practicalities of use
We assessed the LFAs according to this rubric (Supplementary Table 2) by three independent raters and assigned a composite score on a scale of 0–14 (Supplementary Table 3). Five LFAs (BioHit, InTec, Lumiquick, Phamatech, and U2U) received full scores for IFU clarity. LFAs frequently lost points for imprecise instructions regarding time between adding sample and having sufficient sample flow to read the results. Important kit usability criteria, such as whether the included pipette droppers show clear volume markings, were also recorded (Supplementary Table 3). The IFU ambiguity led to administration of too much or little sample for a valid test. While these IFUs may not yet be intended for the general public, it will be important to clarify the instructions moving forward and include droppers to minimize potential for sample volume errors.
LFA performance: sensitivity and specificity
To focus on LFA specificity, given the likely use of these tests in low-prevalence settings, disagreement between operators was interpreted as a negative call. Across all but three of the LFAs (Biohit, BTNX, and Vivacheck), sensitivity was higher for IgG than IgM. Sensitivity for IgG ranged from 98.2% (Lumiquick) down to 72.7% (Oranoxis), and for IgM from 96.4% (BioHit) to 23.2% (Oxo and U2U) (Table 2). LFA specificity was much higher for both isotypes, with 11 LFAs having a specificity of 100% for IgG (API, API v2, BTNX, Camtech, Genobio, Oranoxis, Phamatech, Ray Biotech, Ray Biotech v2, U2U, and Zhuhai Livzon), and 8 LFAs having a specificity of 100% for IgM (API, CareHealth, Cellex, Lumiquick, Oranoxis, Ray Biotech v2, U2U, and Zhuhai Livzon) (Table 2). Under the assumption that the likelihood of two randomly occurring false positives for any one individual is low, an IgG/IgM composite score (averaged operator scores, see Supplementary Methods) was produced to maximize test specificity. Using this composite score, all LFAs except BioHit, Cellex, Edinburgh, InTec, and Vivachek achieved a specificity of 100%. This result underscores the potential of considering the outcome in both isotypes to minimize false positives, although it is more likely that a single isotype will be used in clinical testing.
Table 2.
LFA performance in terms of sensitivity and specificity
| IgG lateral flow assay performance | Manufacturer | N | TP | TN | FP | FN | sensitivity | specificity | PPV | NPV | sensitivity_CI95 | specificity_CI95 | PPV_CI95 | NPV_CI95 |
| API | 112 | 52 | 56 | 0 | 4 | 0.929 | 1 | 1 | 0.933 | (0.853–1) | (0.991–1) | (0.99–1) | (0.861–1) | |
| API (v2) | 112 | 52 | 56 | 0 | 4 | 0.929 | 1 | 1 | 0.933 | (0.853–1) | (0.991–1) | (0.99–1) | (0.861–1) | |
| BioHit | 110 | 52 | 50 | 4 | 4 | 0.929 | 0.926 | 0.929 | 0.926 | (0.853–1) | (0.847–1) | (0.853–1) | (0.847–1) | |
| BTNX | 110 | 48 | 56 | 0 | 6 | 0.889 | 1 | 1 | 0.903 | (0.796–0.982) | (0.991–1) | (0.99–1) | (0.821–0.985) | |
| Camtech | 111 | 48 | 55 | 0 | 8 | 0.857 | 1 | 1 | 0.873 | (0.756–0.958) | (0.991–1) | (0.99–1) | (0.783–0.963) | |
| CareHealth | 112 | 53 | 53 | 3 | 3 | 0.946 | 0.946 | 0.946 | 0.946 | (0.878–1) | (0.878–1) | (0.878–1) | (0.878–1) | |
| Cellex | 112 | 53 | 52 | 4 | 3 | 0.946 | 0.929 | 0.93 | 0.945 | (0.878–1) | (0.853–1) | (0.855–1) | (0.876–1) | |
| Edinburgh | 108 | 47 | 51 | 2 | 8 | 0.855 | 0.962 | 0.959 | 0.864 | (0.753–0.957) | (0.901–1) | (0.893–1) | (0.768–0.96) | |
| Genobio | 111 | 42 | 56 | 0 | 13 | 0.764 | 1 | 1 | 0.812 | (0.643–0.885) | (0.991–1) | (0.988–1) | (0.713–0.911) | |
| InTec | 111 | 49 | 54 | 1 | 7 | 0.875 | 0.982 | 0.98 | 0.885 | (0.779–0.971) | (0.938–1) | (0.931–1) | (0.797–0.973) | |
| KHB | 112 | 53 | 54 | 2 | 3 | 0.946 | 0.964 | 0.964 | 0.947 | (0.878–1) | (0.906–1) | (0.906–1) | (0.88–1) | |
| Lumiquick | 111 | 54 | 53 | 3 | 1 | 0.982 | 0.946 | 0.947 | 0.981 | (0.938–1) | (0.878–1) | (0.88–1) | (0.935–1) | |
| Oranoxis | 110 | 40 | 55 | 0 | 15 | 0.727 | 1 | 1 | 0.786 | (0.6–0.854) | (0.991–1) | (0.988–1) | (0.683–0.889) | |
| OZO | 112 | 43 | 54 | 2 | 13 | 0.768 | 0.964 | 0.956 | 0.806 | (0.649–0.887) | (0.906–1) | (0.885–1) | (0.704–0.908) | |
| Phamatech | 95 | 38 | 47 | 0 | 10 | 0.792 | 1 | 1 | 0.825 | (0.667–0.917) | (0.989–1) | (0.987–1) | (0.718–0.932) | |
| Ray Biotech | 112 | 43 | 56 | 0 | 13 | 0.768 | 1 | 1 | 0.812 | (0.649–0.887) | (0.991–1) | (0.988–1) | (0.713–0.911) | |
| Ray Biotech (v2) | 111 | 51 | 55 | 0 | 5 | 0.911 | 1 | 1 | 0.917 | (0.827–0.995) | (0.991–1) | (0.99–1) | (0.839–0.995) | |
| U2U | 112 | 43 | 56 | 0 | 13 | 0.768 | 1 | 1 | 0.812 | (0.649–0.887) | (0.991–1) | (0.988–1) | (0.713–0.911) | |
| Vivachek | 111 | 52 | 55 | 1 | 3 | 0.945 | 0.982 | 0.981 | 0.948 | (0.876–1) | (0.938–1) | (0.935–1) | (0.882–1) | |
| Zhuhai Livzon | 109 | 47 | 54 | 0 | 8 | 0.855 | 1 | 1 | 0.871 | (0.753–0.957) | (0.991–1) | (0.989–1) | (0.779–0.963) | |
| IgM lateral flow assay performance | Manufacturer | N | TP | TN | FP | FN | sensitivity | specificity | PPV | NPV | sensitivity_CI95 | specificity_CI95 | PPV_CI95 | NPV_CI95 |
| API | 112 | 27 | 56 | 0 | 29 | 0.482 | 1 | 1 | 0.659 | (0.342–0.622) | (0.991–1) | (0.981–1) | (0.552–0.766) | |
| API (v2) | 112 | 51 | 55 | 1 | 5 | 0.911 | 0.982 | 0.981 | 0.917 | (0.827–0.995) | (0.938–1) | (0.934–1) | (0.839–0.995) | |
| BioHit | 110 | 54 | 49 | 5 | 2 | 0.964 | 0.907 | 0.915 | 0.961 | (0.906–1) | (0.82–0.994) | (0.835–0.995) | (0.898–1) | |
| BTNX | 110 | 48 | 53 | 3 | 6 | 0.889 | 0.946 | 0.941 | 0.898 | (0.796–0.982) | (0.878–1) | (0.867–1) | (0.812–0.984) | |
| Camtech | 111 | 42 | 52 | 3 | 14 | 0.75 | 0.945 | 0.933 | 0.788 | (0.628–0.872) | (0.876–1) | (0.849–1) | (0.682–0.894) | |
| CareHealth | 112 | 50 | 56 | 0 | 6 | 0.893 | 1 | 1 | 0.903 | (0.803–0.983) | (0.991–1) | (0.99–1) | (0.821–0.985) | |
| Cellex | 112 | 31 | 56 | 0 | 25 | 0.554 | 1 | 1 | 0.691 | (0.415–0.693) | (0.991–1) | (0.984–1) | (0.584–0.798) | |
| Edinburgh | 108 | 29 | 49 | 4 | 26 | 0.527 | 0.925 | 0.879 | 0.653 | (0.386–0.668) | (0.845–1) | (0.753–1) | (0.539–0.767) | |
| Genobio | 111 | 31 | 52 | 4 | 24 | 0.564 | 0.929 | 0.886 | 0.684 | (0.424–0.704) | (0.853–1) | (0.766–1) | (0.573–0.795) | |
| InTec | 111 | 43 | 54 | 1 | 13 | 0.768 | 0.982 | 0.977 | 0.806 | (0.649–0.887) | (0.938–1) | (0.921–1) | (0.704–0.908) | |
| KHB | 112 | 31 | 55 | 1 | 25 | 0.554 | 0.982 | 0.969 | 0.688 | (0.415–0.693) | (0.938–1) | (0.893–1) | (0.58–0.796) | |
| Lumiquick | 111 | 37 | 56 | 0 | 18 | 0.673 | 1 | 1 | 0.757 | (0.54–0.806) | (0.991–1) | (0.986–1) | (0.653–0.861) | |
| Oranoxis | 110 | 48 | 55 | 0 | 7 | 0.873 | 1 | 1 | 0.887 | (0.776–0.97) | (0.991–1) | (0.99–1) | (0.8–0.974) | |
| OZO | 112 | 13 | 54 | 2 | 43 | 0.232 | 0.964 | 0.867 | 0.557 | (0.113–0.351) | (0.906–1) | (0.662–1) | (0.453–0.661) | |
| Phamatech | 95 | 38 | 46 | 1 | 10 | 0.792 | 0.979 | 0.974 | 0.821 | (0.667–0.917) | (0.927–1) | (0.911–1) | (0.712–0.93) | |
| Ray Biotech | 112 | 40 | 55 | 1 | 16 | 0.714 | 0.982 | 0.976 | 0.775 | (0.587–0.841) | (0.938–1) | (0.917–1) | (0.671–0.879) | |
| Ray Biotech (v2) | 111 | 47 | 55 | 0 | 9 | 0.839 | 1 | 1 | 0.859 | (0.734–0.944) | (0.991–1) | (0.989–1) | (0.766–0.952) | |
| U2U | 112 | 13 | 56 | 0 | 43 | 0.232 | 1 | 1 | 0.566 | (0.113–0.351) | (0.991–1) | (0.962–1) | (0.463–0.669) | |
| Vivachek | 111 | 51 | 55 | 1 | 4 | 0.927 | 0.982 | 0.981 | 0.932 | (0.849–1) | (0.938–1) | (0.934–1) | (0.859–1) | |
| Zhuhai Livzon | 109 | 46 | 54 | 0 | 9 | 0.836 | 1 | 1 | 0.857 | (0.729–0.943) | (0.991–1) | (0.989–1) | (0.763–0.951) | |
| Combined IgG + IgM performance | Manufacturer | N | TP | TN | FP | FN | sensitivity | specificity | PPV | NPV | sensitivity_CI95 | specificity_CI95 | PPV_CI95 | NPV_CI95 |
| API | 112 | 36 | 56 | 0 | 20 | 0.643 | 1 | 1 | 0.737 | (0.509–0.777) | (0.991–1) | (0.986–1) | (0.631–0.843) | |
| API (v2) | 112 | 51 | 56 | 0 | 5 | 0.911 | 1 | 1 | 0.918 | (0.827–0.995) | (0.991–1) | (0.99–1) | (0.841–0.995) | |
| BioHit | 110 | 53 | 50 | 4 | 3 | 0.946 | 0.926 | 0.93 | 0.943 | (0.878–1) | (0.847–1) | (0.855–1) | (0.871–1) | |
| BTNX | 110 | 47 | 56 | 0 | 7 | 0.87 | 1 | 1 | 0.889 | (0.771–0.969) | (0.991–1) | (0.989–1) | (0.803–0.975) | |
| Camtech | 111 | 44 | 55 | 0 | 12 | 0.786 | 1 | 1 | 0.821 | (0.67–0.902) | (0.991–1) | (0.989–1) | (0.722–0.92) | |
| CareHealth | 112 | 51 | 56 | 0 | 5 | 0.911 | 1 | 1 | 0.918 | (0.827–0.995) | (0.991–1) | (0.99–1) | (0.841–0.995) | |
| Cellex | 112 | 46 | 55 | 1 | 10 | 0.821 | 0.982 | 0.979 | 0.846 | (0.712–0.93) | (0.938–1) | (0.927–1) | (0.751–0.941) | |
| Edinburgh | 108 | 35 | 51 | 2 | 20 | 0.636 | 0.962 | 0.946 | 0.718 | (0.5–0.772) | (0.901–1) | (0.86–1) | (0.606–0.83) | |
| Genobio | 111 | 39 | 56 | 0 | 16 | 0.709 | 1 | 1 | 0.778 | (0.58–0.838) | (0.991–1) | (0.987–1) | (0.675–0.881) | |
| InTec | 111 | 46 | 54 | 1 | 10 | 0.821 | 0.982 | 0.979 | 0.844 | (0.712–0.93) | (0.938–1) | (0.927–1) | (0.747–0.941) | |
| KHB | 112 | 39 | 56 | 0 | 17 | 0.696 | 1 | 1 | 0.767 | (0.567–0.825) | (0.991–1) | (0.987–1) | (0.663–0.871) | |
| Lumiquick | 111 | 40 | 56 | 0 | 15 | 0.727 | 1 | 1 | 0.789 | (0.6–0.854) | (0.991–1) | (0.988–1) | (0.687–0.891) | |
| Oranoxis | 110 | 43 | 55 | 0 | 12 | 0.782 | 1 | 1 | 0.821 | (0.664–0.9) | (0.991–1) | (0.988–1) | (0.722–0.92) | |
| OZO | 112 | 19 | 56 | 0 | 37 | 0.339 | 1 | 1 | 0.602 | (0.206–0.472) | (0.991–1) | (0.974–1) | (0.497–0.707) | |
| Phamatech | 95 | 36 | 47 | 0 | 12 | 0.75 | 1 | 1 | 0.797 | (0.617–0.883) | (0.989–1) | (0.986–1) | (0.686–0.908) | |
| Ray Biotech | 112 | 41 | 56 | 0 | 15 | 0.732 | 1 | 1 | 0.789 | (0.607–0.857) | (0.991–1) | (0.988–1) | (0.687–0.891) | |
| Ray Biotech (v2) | 111 | 48 | 55 | 0 | 8 | 0.857 | 1 | 1 | 0.873 | (0.756–0.958) | (0.991–1) | (0.99–1) | (0.783–0.963) | |
| U2U | 112 | 22 | 56 | 0 | 34 | 0.393 | 1 | 1 | 0.622 | (0.256–0.53) | (0.991–1) | (0.977–1) | (0.516–0.728) | |
| Vivachek | 111 | 52 | 55 | 1 | 3 | 0.945 | 0.982 | 0.981 | 0.948 | (0.876–1) | (0.938–1) | (0.935–1) | (0.882–1) | |
| Zhuhai Livzon | 109 | 45 | 54 | 0 | 10 | 0.818 | 1 | 1 | 0.844 | (0.707–0.929) | (0.991–1) | (0.989–1) | (0.747–0.941) |
N Total number of valid assays; TP True Positive; TN True Negative; FP False Positive; FN False Negative; PPV Positive Predictive Value; NPV Negative Predictive Value; CI95: 95% confidence interval
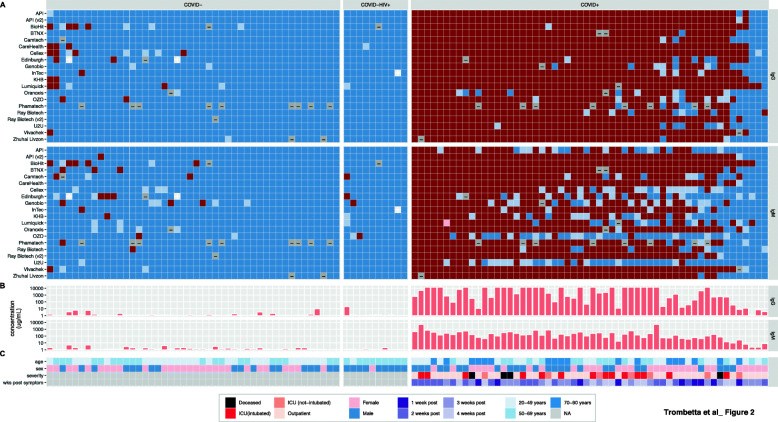
We created heatmaps to visualize individual sample outcomes across all LFAs to assess whether we systematically detected the same miscalls across multiple LFAs (Fig. 2). False negatives (blue squares in the COVID+ panel) amongst COVID+ patients were somewhat reproducible, with three COVID+ samples called negative in both isotypes by all, or all but one LFA. These miscalls were not clearly explained by known demographics (age, sex) or clinical variables (disease severity, weeks post symptom onset) (Fig. 2, bottom panel). To investigate whether these miscalls were related to low titers of anti-SARS-CoV-2 antibodies from participants with a suppressed immune response, all 112 samples were analyzed for anti-spike IgG and IgM antibodies using a custom quantitative Simoa assay [29]. The three samples that were called negative across almost all LFAs, which were also 2–3 weeks post-symptoms, had the lowest levels of anti-spike antibodies in COVID+ samples for both IgG and IgM, suggesting these participants had a slower or suppressed immune response to SARS-CoV-2 infection.
Fig. 2.
Per-individual LFA performance. Colors represent scores computed for IgG (top panel) and IgM (lower panel), where dark red = + 1 (operators all agree a band is present) and dark blue = −1 (operators all agree there is no band) with intermediate colors (pink and light blue) representing varying degrees of operator disagreement. Grey represents invalid runs. Samples are ordered within the COVID-, COVID-HIV+, and COVID+ groups in order of decreasing average score across all LFAs for both antibodies. Clinical variables include age (< 50 yrs.: light blue, 50-69 yrs.: intermediate blue, ≥70 yrs.: dark blue), sex (blue: male, pink: female), disease severity (hospitalized: light green, ICU: orange, ICU + respirator: red, deceased: black), and weeks post symptom-onset (dark green: 1–2 weeks, lightest green: ≥5 weeks)
Unlike false negatives, most false positives (Fig. 2, red squares in COVID- panel) amongst COVID- individuals appear largely uncorrelated between LFAs. However, two samples showed IgG false positives across multiple LFAs, which may suggest long lasting antibodies from exposure to coronaviruses other than SARS-CoV-2. Nonetheless, this observation is not reflected in the antibody levels from the Simoa analysis, which showed barely detectable anti-spike antibodies in these two samples.
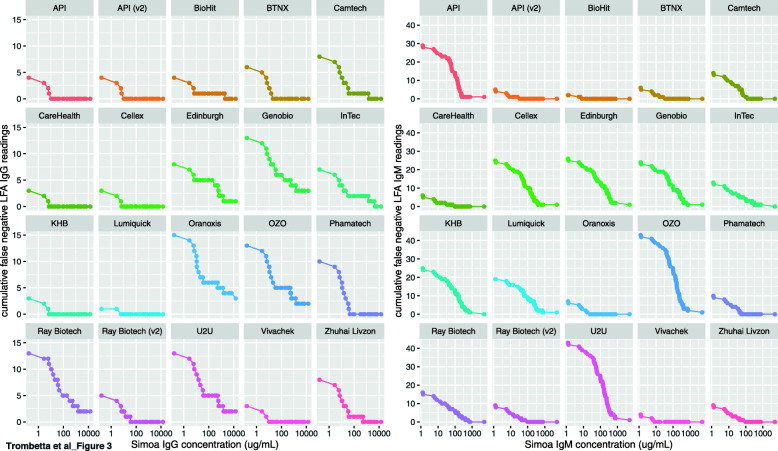
Defining the limit of detection for qualitative LFAs
The custom Simoa anti-spike IgG and IgM antibody assays use a standard curve to determine standardized antibody concentration in each sample [29]. The results obtained from this assay can therefore be used to estimate a limit of detection (LOD) for each of the qualitative LFAs. The cumulative number of false negative LFA calls in COVID+ samples (Fig. 3, y-axis) were computed as a function of decreasing antibody concentrations (Fig. 3, x-axis) separately for IgG and IgM. We define the LOD for each LFA/antibody as the concentration at which ≥95% of the COVID+ samples are called unambiguously positive (Supplementary Table 5). Using this definition, for IgG all LFAs (except Genobio, Oranoxis, OZO, Ray Biotech, and U2U) have an LOD within the linear range of the SIMOA assay (1–10,000 μg/mL). Lumiquick has the lowest LOD at 0.1 μg/mL, which was extrapolated by dilution to be within the linear range of the SIMOA standards, and CareHealth, Cellex, KHB, and Vivachek all have an LOD of 1.5 μg/mL (Supplementary Table 5). Considering the generally lower sensitivity observed with the IgM assays (Fig. 2), IgM assays consistently have higher LODs, with 9 exceeding 1000μg/mL. BioHit has the lowest IgM LOD at 0.6 μg/mL, and API v2, BTNX, CareHealth, and Vivachek all have LODs under 10 μg/mL.
Fig. 3.
Determining the limit of detection of qualitative LFAs. Samples were ranked from highest concentration of anti-spike antibody (determined by Simoa) on the right, to lowest concentration (x-axis). As the sample concentration decreases to the left, a cumulative count of false negatives is shown on the y-axis. IgG is shown on the left, IgM on the right. Note the difference in magnitude of the y-axes between IgG and IgM; these LFAs are generally more sensitive to IgG than IgM
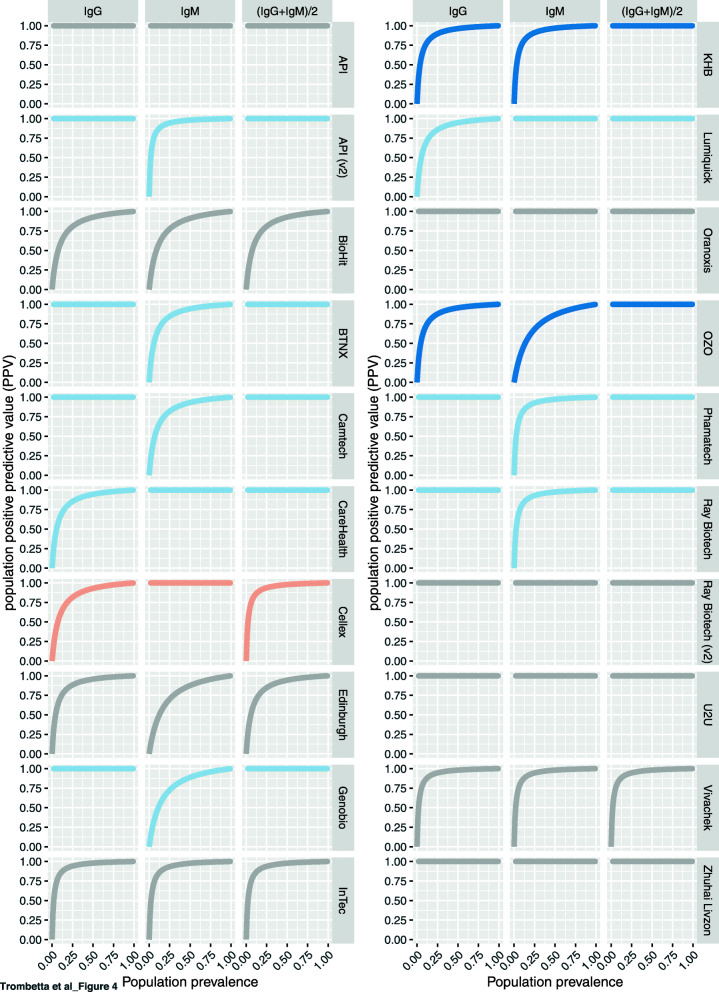
Interpreting test positivity with low prevalence in the general population
Low prevalence places a high burden on specificity [25]. Given the high proportion of true negative individuals in the population being studied, prevalence increases the ratio of false positive to true positive test outcomes [31]. Positive predictive values (PPV) correspond to the likelihood that a positive test result reflect true positivity as measured by a gold standard PCR result. We computed PPVs as a function of the fraction of the population infected with (and assumed to have produced antibodies to) SARS-CoV-2 (Fig. 4). Here we see that even with the conservative interpretation of LFA outcome (requiring the majority of operators to see a band to call a sample positive for either IgG or IgM), when the population seroprevalence is ~ 5% the PPV for these assays spans a large range (from ~ 30 to 100%). As the population prevalence increases, the burden on specificity is decreased, and at 50% prevalence the PPV of all LFAs is above 87.5%. Posterior PPV can be improved for most LFAs by requiring both IgG and IgM to be read as positive in order to count an individual as positive (Fig. 4, right panels).
Fig. 4.
Effect of a changing disease prevalence (x axis) on the population-level PPV of each test for scores derived from IgG (left), IgM (middle), and the average score of both antibodies (right). As disease prevalence increases, the high burden on specificity of LFAs is reduced. Color coding: Grey: no improvement; Light blue: rescues one of the two antibodies; Dark blue: rescues both of the antibodies; Orange: performs worse than one of the antibodies
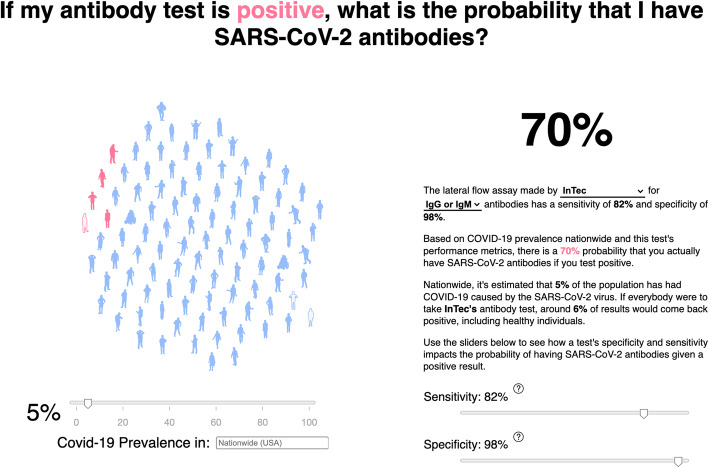
To visualize the effect of changing population prevalence on the PPV, we created an interactive webapp (https://covid.omics.kitchen; Fig. 5), which allows the user to extrapolate the likelihood that they do in fact have SARS-CoV-2 antibodies if they have a positive LFA result, given the infection prevalence and test accuracy. The app includes benchmark performance of all 20 LFAs evaluated here, as well as those reported in Whitman et al. [10] (filtered to remove samples taken under 10 days post symptom-onset). In an effort to further generalize the utility of this tool, we allow the user to explore the effects of assay performance under difference prevalence scenarios, and to input the reported prevalence data from specific geographic locations within the US [1], based on national, state, and county records.
Fig. 5.
Example image of the interactive webapp hosted at https://covid.omics.kitchen. Using the sliders, one is able to visualize the effect of changing the disease prevalence (and the test performance) on the resulting probability that, given a positive LFA test, the individual does in fact have SARS-CoV-2 antibodies. The figure shows an example of positive predictive value using the US national prevalence of 5% (on November 27th, 2020) and the performance characteristics of the InTec LFA
Discussion
In this study, we report a standardized cross-evaluation of LFAs on the same pre-pandemic SARS-CoV-2-negative and PCR-confirmed SARS-CoV-2-positive samples, and rate their reproducibility, usability, and performance characteristics. Overall, the LFAs showed a higher propensity for false negative than false positive readings. Results are public: https://covidinnovation.partners.org/evaluation/. We use the Simoa technology [29] to measure the concentrations of anti-spike protein IgG and IgM antibodies and extrapolate the assay limit of detection. We also established a web tool to aid users in understanding the likelihood they have anti-SARS-CoV-2 antibodies given a positive test result. This resource of performance characteristics of several LFAs and a tool for result interpretation, can both be used for immunosurveillance and future home testing applications.
LFAs are tractable tools to estimate community seroprevalence, especially with anticipated seasonal fluctuations in the transmission dynamics of SARS-CoV-2 and other viruses that cause the common cold, which confound the symptomatologic diagnosis of COVID-19 [32]. As new waves of the COVID-19 pandemic resurge around the globe [1], and with commencing vaccinations against SARS-CoV-2 infections [12–14, 16], there is a renewed interest in serological tests to detect anti-SARS-CoV-2 antibodies [20, 33]. Affordable LFAs offer an attractive option for monitoring the presence and longevity of anti-SARS-CoV-2 antibodies, and determining population-level herd immunity [34]. LFAs also obviate the need for complex laboratories to process the samples [35]. As the pandemic expands to previously unexposed communities, it is critical to use simple tools to monitor exposure dynamics and seroconversion in SARS-CoV-2-exposed individuals, as well as vaccine-induced immunity [16]. We tested a mixture of LFAs targeting SARS-CoV-2 nucleocapsid, spike proteins, or both. Moderna’s mRNA-1273 and Pfizer-BioNTech COVID-19 vaccines encode SARS-CoV-2 spike proteins to induce anti-spike antibodies [14, 36]. Therefore, LFAs targeting the spike and nucleocapsid proteins of SARS-CoV-2 could be used to differentiate vaccine- and infection-induced antibody responses, respectively.
Rigorous evaluation of these LFAs by manufacturer-independent parties is important. The US FDA independently reviews medical products before commercialization. The FDA used Emergency Use Authorization (EUA) authority to accelerate the implementation of diagnostic products during the pandemic. Commercial manufacturers were required to submit a completed EUA request [22]. Unfortunately, the rush to market introduced many tests that did not meet typical US or international standards [37]. Therefore, the FDA and international regulatory agencies continue to update guidelines for authorization of new serological tests. Our evaluation plan mirrored the FDA guidelines for evaluating serological tests [22]. We included 10 HIV-positive samples to test whether they have higher false positive results in SARS-CoV-2-negative samples [38], and did not detect higher false positive results.
The mere detection of IgG or IgM responses does not guarantee that neutralizing antibodies are present at protective titers [7, 39]. The study demographics suggest a slight over-representation of African Americans among cases, as reported [40]. However, the sample size was underpowered to formally determine the effect of race on test performance. In our analysis, IgM detectability was less sensitive and reproducible than IgGs across multiple LFAs, possibly due to both lower IgM titres, and lower limits of detection for the IgM LFAs. Waning antibody responses have been reported in some SARS-CoV-2-infected individuals [41–44]. Furthermore, reported cases of re-infection with SARS-CoV-2 suggest that prior exposure, and even seroconversion, do not universally protect against SARS-CoV-2 infection [44, 45]. This could result from low antibody titers as shown in an immunocompromised patient [46], low durability of infection-induced antibodies [42–44], or low neutralizing potential of SARS-CoV-2 infection-induced antibodies in some individuals [34]. IgG and IgM antibodies may also target irrelevant epitopes outside the spike and receptor binding domains, and consequently be less efficient at intercepting infection by the virus [47]. The WHO cautions against interpreting presence of antibodies, even neutralizing ones, as lower risk of re-infection and transmission [48]. The presence of antibodies could be however used for rapid immunosurveillance to monitor extent of population transmission, particularly in asymptomatic but SARS-CoV-2-exposed individuals [3, 33].
One major concern about the deployment of these tests is the misinterpretation of positive results [7, 39]. As more tests move towards FDA approval of home use, clear scientific communication about the result interpretation becomes more crucial [23, 39]. A positive serological result does not necessarily mean active infection [23, 31, 49]. Although combined use of molecular and seroconversion results can be used to confirm the diagnosis of symptomatic and hospitalized individuals [35], a positive serological test in the absence of symptoms dissociates the presence of the antibodies from the time of infection [44]. Additionally, it is important to understand the implication of false positive and false negative results, particularly in the context of a low to mid-prevalence disease such as COVID-19 [25]. Low prevalence decreases the negative predictive value of a test, but increases the false positive rates [25]. A false positive serological test result may prematurely instill confidence that one has immunity against SARS-CoV-2 infection, thus resulting in behavioral changes that increase risk of transmission [50]. Hence, the probability that a person without antibodies will test negative on a serological test is more important than test sensitivity [48–50].
Our study presents a few limitations. Although we successfully benchmarked the performance of the LFAs to a quantitative assay [29], we did not determine the neutralizing potential of these antibodies. Secondly, samples were acquired when PCR testing was restricted to severely ill patients. For epidemiological studies and population surveillance, it will be important to evaluate assay performance on asymptomatic individuals.
Conclusions
Our study provides a public resource to aid researchers, healthcare providers, public health professionals, and industries impacted by the pandemic such as airlines, in choosing the appropriate serological LFAs for their intended use cases.
Supplementary Information
Additional file 1: Supplementary Methods. Supplementary Table 1. LFA commercial kit information, sample requirements, and protocol details. Supplementary Table 2. IFU clarity rubric. Supplementary Table 3. Usability ratings and description of kit components. Supplementary Table 4. Clinical information for the COVID-positive individuals whose plasma was used in this study. Supplementary Table 5. LFA Limits of detection estimated from anti-spike antibodies concentrations measured by Simoa.
Acknowledgements
This study was possible through the Diagnostics Pillar of the Mass General Brigham COVID Center for Innovation (MGBCCI) (https://covidinnovation.partners.org/), predominantly staffed by volunteers. The LFA horizon scan and selection was performed by the Direct-to-Consumer working group, and the Diagnostics Accelerator and Validation working group developed the protocols and performed the evaluations. The study is supported by funding from the Massachusetts Consortium on Pathogen Readiness (MassCPR) (https://masscpr.hms.harvard.edu/).
Abbreviations
- AEB
Average Enzymes per Beads
- COVID-19
Coronavirus Disease of 2019
- EDTA
Ethylenediaminetetraacetic acid
- ELISA
Enzyme-linked immunosorbent assay
- EUA
Emergency Use Authorization
- FDA
Food and Drug Administration
- HIV
Human Immunodeficiency Virus
- ICU
Intensive Care Units
- IFU
Instructions-For-Use
- IgG
Immunoglobulin G
- IgM
Immunoglobulin M
- LFA
Lateral Flow Assay
- LOD
Limit of Detection
- MGB
Mass General Brigham
- MGH
Mass General Hospital
- NPV
Negative Predictive Value
- PPV
Positive Predictive Value
- RT-qPCR
Reverse Transcriptase Quantitative Polymerase Chain Reaction
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus-2
- Simoa
Single Molecule Array
Authors’ contributions
BAT, SEK, RRK, BCC, and SS wrote the manuscript. BAT, SEK, and KG led the project management, volunteer coordination, and lab operations. BAT, SEK, BCC, PK, and LLR established the Diagnostics Accelerator lab, acquired samples, and established biosafety and IRB protocols. SS and LLR managed the COVID Diagnostics Accelerator lab. BAT and RRK led sample and data management. RRK and BCC led the analysis and data interpretation. RA, JT, and GM corresponded with LFA providers and down-selected assays for evaluation. DRW led the Diagnostics Pillar of the Mass General Brigham COVID Center for Innovation. All authors planned and/or performed experiments, edited, and approved the final version of the manuscript.
Funding
This study was funded by a grant from the Massachusetts Consortium for Pathogen Readiness (MassCPR).
Availability of data and materials
Evaluation data are available at: https://covidinnovation.partners.org/evaluation/.
Web application is available at: https://covid.omics.kitchen/
Declarations
Ethics approval and consent to participate
All samples were collected from participants who provided informed consent. Sample and data collection conformed to Good Clinical Practice guidelines and Declaration of Helsinki. The study was approved under the Massachusetts General Brigham (MGB) Institutional Review Board (protocol no. 2020P001204).
Consent for publication
Not applicable.
Competing interests
DRW has a financial interest in Quanterix Corporation, a company that develops an ultra-sensitive digital immunoassay platform. He is an inventor of the Simoa technology and a founder of the company and also serves on its Board of Directors. DRW's interests were reviewed and are managed by Brigham and Womens Hospital and Partners HealthCare in accordance with their policies on competing interests. The assays in this publication have been licensed by Brigham and Womens Hospital to Quanterix Corporation. MN, TG and DRW have filed intellectual property on the technology described in this manuscript.
All other authors did not declare a conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Bianca A. Trombetta, Savannah E. Kandigian, Robert R. Kitchen, Korneel Grauwet, Lauren L. Ritterhouse and Sara Suliman contributed equally to this work.
References
- 1.Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oran DP, Topol EJ. Prevalence of asymptomatic SARS-CoV-2 infection : a narrative review. Ann Intern Med. 2020;173(5):362–367. doi: 10.7326/M20-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Havers FP, Reed C, Lim T, et al. Seroprevalence of Antibodies to SARS-CoV-2 in 10 Sites in the United States, March 23-May 12, 2020. [published online ahead of print, 2020 Jul 21]. JAMA Intern Med. 2020. 10.1001/jamainternmed.2020.4130. [DOI] [PubMed]
- 4.Ehrlich H, Boneva D, Elkbuli A. The intersection of viral illnesses: A seasonal influenza epidemic amidst the COVID-19 pandemic. Ann Med Surg (Lond) 2020;60:41–43. doi: 10.1016/j.amsu.2020.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D'Cruz RJ, Currier AW, Sampson VB. Laboratory testing methods for novel severe acute respiratory syndrome-Coronavirus-2 (SARS-CoV-2) Front Cell Dev Biol. 2020;8:468. doi: 10.3389/fcell.2020.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pecora ND, Zand MS. Measuring the serologic response to severe acute respiratory syndrome coronavirus 2: methods and meaning. Clin Lab Med. 2020;40(4):603–614. doi: 10.1016/j.cll.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peeling RW, Wedderburn CJ, Garcia PJ, Boeras D, Fongwen N, Nkengasong J, Sall A, Tanuri A, Heymann DL. Serology testing in the COVID-19 pandemic response. Lancet Infect Dis. 2020;20(9):e245–e2e9. doi: 10.1016/S1473-3099(20)30517-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.La Marca A, Capuzzo M, Paglia T, Roli L, Trenti T, Nelson SM. Testing for SARS-CoV-2 (COVID-19): a systematic review and clinical guide to molecular and serological in-vitro diagnostic assays. Reprod BioMed Online. 2020;41(3):483–499. doi: 10.1016/j.rbmo.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pickering S, Betancor G, Galao RP, Merrick B, Signell AW, Wilson HD, et al. Comparative assessment of multiple COVID-19 serological technologies supports continued evaluation of point-of-care lateral flow assays in hospital and community healthcare settings. PLoS Pathog. 2020;16(9):e1008817. doi: 10.1371/journal.ppat.1008817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitman JD, Hiatt J, Mowery CT, Shy BR, Yu R, Yamamoto TN, Rathore U, Goldgof GM, Whitty C, Woo JM, Gallman AE, Miller TE, Levine AG, Nguyen DN, Bapat SP, Balcerek J, Bylsma SA, Lyons AM, Li S, Wong AWY, Gillis-Buck EM, Steinhart ZB, Lee Y, Apathy R, Lipke MJ, Smith JA, Zheng T, Boothby IC, Isaza E, Chan J, Acenas DD, II, Lee J, Macrae TA, Kyaw TS, Wu D, Ng DL, Gu W, York VA, Eskandarian HA, Callaway PC, Warrier L, Moreno ME, Levan J, Torres L, Farrington LA, Loudermilk RP, Koshal K, Zorn KC, Garcia-Beltran WF, Yang D, Astudillo MG, Bernstein BE, Gelfand JA, Ryan ET, Charles RC, Iafrate AJ, Lennerz JK, Miller S, Chiu CY, Stramer SL, Wilson MR, Manglik A, Ye CJ, Krogan NJ, Anderson MS, Cyster JG, Ernst JD, Wu AHB, Lynch KL, Bern C, Hsu PD, Marson A. Evaluation of SARS-CoV-2 serology assays reveals a range of test performance. Nat Biotechnol. 2020;38(10):1174–1183. doi: 10.1038/s41587-020-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oliver SE, Gargano JW, Marin M, Wallace M, Curran KG, Chamberland M, et al. The advisory committee on immunization Practices' interim recommendation for use of Pfizer-BioNTech COVID-19 vaccine - United States, December 2020. MMWR Morb Mortal Wkly Rep. 2020;69(50):1922–1924. doi: 10.15585/mmwr.mm6950e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, McCullough M, Chappell JD, Denison MR, Stevens LJ, Pruijssers AJ, McDermott A, Flach B, Doria-Rose NA, Corbett KS, Morabito KM, O'Dell S, Schmidt SD, Swanson PA, 2nd, Padilla M, Mascola JR, Neuzil KM, Bennett H, Sun W, Peters E, Makowski M, Albert J, Cross K, Buchanan W, Pikaart-Tautges R, Ledgerwood JE, Graham BS, Beigel JH, mRNA-1273 Study Group An mRNA vaccine against SARS-CoV-2 - preliminary report. N Engl J Med. 2020;383(20):1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson EJ, Rouphael NG, Widge AT, Jackson LA, Roberts PC, Makhene M, Chappell JD, Denison MR, Stevens LJ, Pruijssers AJ, McDermott A, Flach B, Lin BC, Doria-Rose NA, O'Dell S, Schmidt SD, Corbett KS, Swanson PA, 2nd, Padilla M, Neuzil KM, Bennett H, Leav B, Makowski M, Albert J, Cross K, Edara VV, Floyd K, Suthar MS, Martinez DR, Baric R, Buchanan W, Luke CJ, Phadke VK, Rostad CA, Ledgerwood JE, Graham BS, Beigel JH, mRNA-1273 Study Group Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N Engl J Med. 2020;383(25):2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walsh EE, Frenck RW, Jr, Falsey AR, Kitchin N, Absalon J, Gurtman A, Lockhart S, Neuzil K, Mulligan MJ, Bailey R, Swanson KA, Li P, Koury K, Kalina W, Cooper D, Fontes-Garfias C, Shi PY, Türeci Ö, Tompkins KR, Lyke KE, Raabe V, Dormitzer PR, Jansen KU, Şahin U, Gruber WC. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N Engl J Med. 2020;383(25):2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dagotto G, Yu J, Barouch DH. Approaches and challenges in SARS-CoV-2 vaccine development. Cell Host Microbe. 2020;28(3):364–370. doi: 10.1016/j.chom.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poland GA, Ovsyannikova IG, Kennedy RB. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet. 2020;396(10262):1595–1606. doi: 10.1016/S0140-6736(20)32137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, Ai J, Loeffelholz MJ, Tang YW, Zhang W. Meta-analysis of diagnostic performance of serology tests for COVID-19: impact of assay design and post-symptom-onset intervals. Emerg Microbes Infect. 2020;9(1):2200–2211. doi: 10.1080/22221751.2020.1826362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caini S, Bellerba F, Corso F, Diaz-Basabe A, Natoli G, Paget J, et al. Meta-analysis of diagnostic performance of serological tests for SARS-CoV-2 antibodies up to 25 April 2020 and public health implications. Euro Surveill. 2020;25(23):2000980. 10.2807/1560-7917.ES.2020.25.23.2000980. [DOI] [PMC free article] [PubMed]
- 19.Lisboa Bastos M, Tavaziva G, Abidi SK, Campbell JR, Haraoui LP, Johnston JC, et al. Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ. 2020;370:m2516. doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moura DTH, McCarty TR, Ribeiro IB, Funari MP, Oliveira P, Miranda Neto AA, et al. Diagnostic Characteristics of Serological-Based COVID-19 Testing: A Systematic Review and Meta-Analysis. Clinics (Sao Paulo) 2020;75:e2212. doi: 10.6061/clinics/2020/e2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutiérrez-Cobos A, Gómez de Frutos S, Domingo García D, et al. Evaluation of diagnostic accuracy of 10 serological assays for detection of SARS-CoV-2 antibodies. Eur J Clin Microbiol Infect Dis. 2021;40(5):955-61. 10.1007/s10096-020-04092-3. [DOI] [PMC free article] [PubMed]
- 22.FDA. EUA Authorized Serology Test Performance. United States Food and Drug Administration. 2020. https://www.fda.gov/medicaldevices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/eua-authorized-serology-test-performance/. Accessed 25 Dec 2020.
- 23.Bermingham WH, Wilding T, Beck S, Huissoon A. SARS-CoV-2 serology: test, test, test, but interpret with caution! Clin Med (Lond) 2020;20(4):365–368. doi: 10.7861/clinmed.2020-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long QX, Liu BZ, Deng HJ, Wu GC, Deng K, Chen YK, Liao P, Qiu JF, Lin Y, Cai XF, Wang DQ, Hu Y, Ren JH, Tang N, Xu YY, Yu LH, Mo Z, Gong F, Zhang XL, Tian WG, Hu L, Zhang XX, Xiang JL, du HX, Liu HW, Lang CH, Luo XH, Wu SB, Cui XP, Zhou Z, Zhu MM, Wang J, Xue CJ, Li XF, Wang L, Li ZJ, Wang K, Niu CC, Yang QJ, Tang XJ, Zhang Y, Liu XM, Li JJ, Zhang DC, Zhang F, Liu P, Yuan J, Li Q, Hu JL, Chen J, Huang AL. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26(6):845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 25.Tenny S, Hoffman MR. Prevalence. Treasure Island (FL): StatPearls; 2020. [Google Scholar]
- 26.Theel ES, Slev P, Wheeler S, Couturier MR, Wong SJ, Kadkhoda K. The Role of Antibody Testing for SARS-CoV-2: Is There One? J Clin Microbiol. 2020;58(8):e00797–20. Published 2020 Jul 23. 10.1128/JCM.00797-20. [DOI] [PMC free article] [PubMed]
- 27.Jorfi M, Luo NM, Hazra A, Herisson F, Miller M, Toombs JA, et al. Diagnostic technology for COVID-19: comparative evaluation of antigen and serology-based SARS-CoV-2 immunoassays, and contact tracing solutions for potential use as at-home products. MedRxiv. 2020.
- 28.Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40(5):373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 29.Norman M, Gilboa T, Ogata AF, Maley AM, Cohen L, Busch EL, Lazarovits R, Mao CP, Cai Y, Zhang J, Feldman JE, Hauser BM, Caradonna TM, Chen B, Schmidt AG, Alter G, Charles RC, Ryan ET, Walt DR. Ultrasensitive high-resolution profiling of early seroconversion in patients with COVID-19. Nat Biomed Eng. 2020;4(12):1180–1187. doi: 10.1038/s41551-020-00611-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian X, Li C, Huang A, Xia S, Lu S, Shi Z, Lu L, Jiang S, Yang Z, Wu Y, Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. 2020;9(1):382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hernaez R, Thrift AP. High negative predictive value, low prevalence, and Spectrum effect: caution in the interpretation. Clin Gastroenterol Hepatol. 2017;15(9):1355–1358. doi: 10.1016/j.cgh.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 32.Kissler SM, Tedijanto C, Goldstein E, Grad YH, Lipsitch M. Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science. 2020;368(6493):860–868. doi: 10.1126/science.abb5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alter G, Seder R. The power of antibody-based surveillance. N Engl J Med. 2020;383(18):1782–1784. doi: 10.1056/NEJMe2028079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neagu M. The bumpy road to achieve herd immunity in COVID-19. J Immunoassay Immunochem. 2020;41(6):928–45. 10.1080/15321819.2020.1833919. [DOI] [PubMed]
- 35.Nilles EJ, Karlson EW, Norman M, Gilboa T, Fischinger S, Atyeo C, et al. Evaluation of two commercial and two non-commercial immunoassays for the detection of prior infection to SARS-CoV-2. medRxiv. 2020;2020.06.24.20139006. Published 2020 June 26. 10.1101/2020.06.24.20139006.
- 36.Corbett KS, Edwards DK, Leist SR, Abiona OM, Boyoglu-Barnum S, Gillespie RA, Himansu S, Schäfer A, Ziwawo CT, DiPiazza AT, Dinnon KH, Elbashir SM, Shaw CA, Woods A, Fritch EJ, Martinez DR, Bock KW, Minai M, Nagata BM, Hutchinson GB, Wu K, Henry C, Bahl K, Garcia-Dominguez D, Ma LZ, Renzi I, Kong WP, Schmidt SD, Wang L, Zhang Y, Phung E, Chang LA, Loomis RJ, Altaras NE, Narayanan E, Metkar M, Presnyak V, Liu C, Louder MK, Shi W, Leung K, Yang ES, West A, Gully KL, Stevens LJ, Wang N, Wrapp D, Doria-Rose NA, Stewart-Jones G, Bennett H, Alvarado GS, Nason MC, Ruckwardt TJ, McLellan JS, Denison MR, Chappell JD, Moore IN, Morabito KM, Mascola JR, Baric RS, Carfi A, Graham BS. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586(7830):567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Badnjevic A, Pokvic LG, Dzemic Z, Becic F. Risks of emergency use authorizations for medical products during outbreak situations: a COVID-19 case study. Biomed Eng Online. 2020;19(1):75. doi: 10.1186/s12938-020-00820-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan SS, Chew KL, Saw S, Jureen R, Sethi S. Cross-reactivity of SARS-CoV-2 with HIV chemiluminescent assay leading to false-positive results. J Clin Pathol. 2020:jclinpath-2020-206942. 10.1136/jclinpath-2020-206942. [DOI] [PMC free article] [PubMed]
- 39.Gill D, Ponsford MJ. Testing for antibodies to SARS-CoV-2. BMJ. 2020;371:m4288. doi: 10.1136/bmj.m4288. [DOI] [PubMed] [Google Scholar]
- 40.Kullar R, Marcelin JR, Swartz TH, Piggott DA, Macias Gil R, Mathew TA, Tan T. Racial disparity of coronavirus disease 2019 in African American communities. J Infect Dis. 2020;222(6):890–893. doi: 10.1093/infdis/jiaa372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Long QX, Tang XJ, Shi QL, Li Q, Deng HJ, Yuan J, Hu JL, Xu W, Zhang Y, Lv FJ, Su K, Zhang F, Gong J, Wu B, Liu XM, Li JJ, Qiu JF, Chen J, Huang AL. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26(8):1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 42.Ibarrondo FJ, Fulcher JA, Goodman-Meza D, Elliott J, Hofmann C, Hausner MA, Ferbas KG, Tobin NH, Aldrovandi GM, Yang OO. Rapid decay of anti-SARS-CoV-2 antibodies in persons with mild Covid-19. N Engl J Med. 2020;383(11):1085–1087. doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kutsuna S, Asai Y, Matsunaga A. Loss of anti-SARS-CoV-2 antibodies in mild Covid-19. N Engl J Med. 2020;383(17):1695–1696. doi: 10.1056/NEJMc2027051. [DOI] [PubMed] [Google Scholar]
- 44.Milani GP, Dioni L, Favero C, Cantone L, Macchi C, Delbue S, et al. Serological follow-up of SARS-CoV-2 asymptomatic subjects. Sci Rep. 2020;10(1):20048. doi: 10.1038/s41598-020-77125-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.To KK, Hung IF, Ip JD, Chu AW, Chan WM, Tam AR, et al. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis: Off Publ Infect Dis Soc Am 2020. [DOI] [PMC free article] [PubMed]
- 46.Mulder M, van der Vegt D, Oude Munnink BB, GeurtsvanKessel CH, van de Bovenkamp J, Sikkema RS, et al. Reinfection of SARS-CoV-2 in an immunocompromised patient: a case report. Clin Infect Dis: Off Publ Infect Dis Soc Am. 2020. 10.1093/cid/ciaa1538. [DOI] [PMC free article] [PubMed]
- 47.Hussain A, Hasan A, Nejadi Babadaei MM, Bloukh SH, Chowdhury MEH, Sharifi M, Haghighat S, Falahati M. Targeting SARS-CoV2 spike protein receptor binding domain by therapeutic antibodies. Biomed Pharmacother. 2020;130:110559. doi: 10.1016/j.biopha.2020.110559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.WHO. Immunity passports in the context of COVID-19. World Health Organization commentaries. 2020;https://www.who.int/news-room/commentaries/detail/immunity-passports-in-the-context-of-covid-19. (Accessed: Friday, December 25th, 2020).
- 49.Waller J, Rubin GJ, Potts HWW, Mottershaw AL, Marteau TM. Immunity Passports' for SARS-CoV-2: an online experimental study of the impact of antibody test terminology on perceived risk and behaviour. BMJ Open. 2020;10(8):e040448. doi: 10.1136/bmjopen-2020-040448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown RCH, Kelly D, Wilkinson D, Savulescu J. The scientific and ethical feasibility of immunity passports. Lancet Infect Dis. 2021;21(3):e58-e63. 10.1016/S1473-3099(20)30766-0. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Methods. Supplementary Table 1. LFA commercial kit information, sample requirements, and protocol details. Supplementary Table 2. IFU clarity rubric. Supplementary Table 3. Usability ratings and description of kit components. Supplementary Table 4. Clinical information for the COVID-positive individuals whose plasma was used in this study. Supplementary Table 5. LFA Limits of detection estimated from anti-spike antibodies concentrations measured by Simoa.
Data Availability Statement
Evaluation data are available at: https://covidinnovation.partners.org/evaluation/.
Web application is available at: https://covid.omics.kitchen/