Abstract
A newly emerged strain of SARS‐CoV‐2 of B.1.1.7 lineage has caused a significant surge in the SARS‐CoV‐2 infections in the UK. In this study, changes in the epitopes of spike and orf8 proteins in SARS‐CoV‐2 of B.1.1.7 lineage were investigated. Genomic alignment of the SARS‐CoV‐2/B.1.1.7 with SARS‐CoV‐2/Wuhan showed the presence of several mutations in orf1a/b, spike, orf8, and N proteins of SARS‐CoV‐2/B.1.1.7. Molecular models of spike and orf8 proteins were constructed by homology modeling. Superimposition between the spike proteins of SARS‐CoV‐2/Wuhan and SARS‐CoV‐2/B.1.1.7 showed noticeable variations in the spatial orientation in Val70‐Asn74 and Thr250‐Ser255 regions. This may have also resulted in the extension of the epitopic region at Ser244‐Gly249 in the SARS‐CoV‐2/B.1.1.7 spike protein. Superimposition of the SARS‐CoV‐2/B.1.1.7 spike protein over Fab‐spike protein complexes of SARS‐CoV‐2/Wuhan also showed subtle variations in the antibody binding affinity targeting the N‐terminal domain of the spike protein. Epitopic variations were also observed between the corresponding orf8 regions of SARS‐CoV‐2/Wuhan and SARS‐CoV‐2/B.1.1.7. Moreover, the presence of a stop codon at position 27 in orf8 connotes the emergence of two frames (orf8a and orf8b) in SARS‐CoV‐2, which further hampers its extracellular secretion, and in turn, immunogenicity. The findings of the present study could further be used to develop targeted immunotherapeutics.
Keywords: B.1.1.7, COVID‐19, Orf8, SARS‐CoV‐2, spike protein
1. INTRODUCTION
Evasion from the host immune response is one of the most common yet profoundly effective strategies that pathogens including viruses employ to win the evolutionary arms race against their hosts. The emergence of SARS‐CoV‐2 is the result of this grand battle, and after a little over a year of the COVID‐19 pandemic, nearly 100 million cases have been reported with 2.3 million lives having been lost due to the viral infection. 1 During the yearlong course of the pandemic, several strains of the virus have emerged and many have a selective advantage over the antecedent strains. This, in turn, has resulted in the emergence of multiple lineages of SARS‐CoV‐2 with variable demographic distribution. 2 Of note, the newly reported SARS‐CoV‐2 variant that has caused a recent surge in viral infections in the UK 3 is yet another example of the dynamic interplay of evolution by means of natural selection between humans and SARS‐CoV‐2.
Phylogenetic analysis of SARS‐CoV‐2 variant from the UK, dubbed as the Variant of Concern 202012/01 (VOC‐202012/01) has placed it in a separate lineage referred to as B.1.1.7. 2 For the sake of simplicity, we have referred to this variant as SARS‐CoV‐2/B.1.1.7. The variant was first reported in the UK on September 20, 2020, but since then its presence has also been observed in other parts of the world. 4 , 5 The transmissibility and virulence of the new SARS‐CoV‐2/B.1.1.7 have not been thoroughly compared with the other variants of SARS‐CoV‐2. However, its emergence parallels the significant rise in the SARS‐CoV‐2 infections in the UK with reported cases of infection and related deaths spiked 1.4‐ and 1.3‐folds, respectively, from October 2020 to December 2020. 6
Preliminary genomic analysis showed the presence and/or accumulation of significant numbers of non‐synonymous mutations in the SARS‐CoV‐2/B.1.1.7 compared to SARS‐CoV‐2/Wuhan. 4 Mutations in SARS‐CoV‐2/B.1.1.7 are dispersed in orf1a/b (T1001I, A1708D, I2230T, S2625X, and ΔSGF3675‐3677), spike (ΔHV69‐70, ΔY144, N501Y, A570D, D614G, P681H, T716I, S982A, and D1118H), orf8 (Q27X, R52I, ΔK68, Y73C, L118V, F120L, and I121F) and N (D3L, R203K, G204R, and S235F) proteins of the virus. 4 Out of these proteins, the immunogenic potential of SARS‐CoV‐2 spike and orf8 has been widely reported and seropositivity against both these proteins has been consistently shown in the SARS‐CoV‐2‐infected individuals. 7 , 8 Moreover, complex structures of antibody Fab fragment binding with N‐terminal domain (NTD) and receptor‐binding domain (RBD) of SARS‐CoV‐2 spike have also been reported. 9 , 10 As SARS‐CoV‐2/B.1.1.7 harbors mutations in both NTD and RBD of the spike protein and in orf8, this raises some interesting possibilities. First, do the variations in the spike protein and orf8 result in epitopic changes in SARS‐CoV‐2/B.1.1.7 compared to SARS‐CoV‐2/Wuhan? Second, has potential truncation of orf8 (due to the stop codon at position 27) in SARS‐CoV‐2/B.1.1.7 resulted in the emergence of two proteins (orf8a and orf8b) as previously witnessed in an earlier phase of the SARS‐CoV epidemic in 2003? 11 Third, does the orf8 of SARS‐CoV‐2/B.1.1.7 still hold the potential of making a structurally stable dimer? Based on bioinformatic analyses in this study, we have explored all these possibilities. In summary, the findings provide important insights into the structurally driven immunogenic changes in SARS‐CoV‐2/B.1.1.7.
2. METHODOLOGY
2.1. Sequence retrieval and alignment
A representative genomic sequence of SARS‐CoV‐2/B.1.1.7 (hCoV‐19/England/205090260/2020|EPI_ISL_728343|2020‐12‐12) was retrieved from GISAID and subjected to sequence alignment with the genome sequence of SARS‐CoV‐2/Wuhan (NC_045512.2) using CLUSTALW under default parameters. 12 Based on the sequence alignment, different open reading frames (where sequence variations were observed) were separated from the SARS‐CoV‐2/B.1.1.7 genome and subjected to in silico translation using the Expasy Server Translate tool. 13 The existence of the reported mutations in SARS‐CoV‐2/B.1.1.7 4 was verified by the sequence alignment of orf1a/b, spike, orf8, and N protein sequences of SARS‐CoV‐2/Wuhan and SARS‐CoV‐2/B.1.1.7.
2.2. Protein molecular modeling
The molecular model of SARS‐CoV‐2/B.1.1.7 spike was developed using the Swiss Model 14 taking atomic coordinates of spike protein (PDBid: 6XR8) as a template. 15 Orf8 proteins of SARS‐CoV‐2/B.1.1.7 and SARS‐CoV were also modeled via the Swiss Model on the basis of a template (PDBid: 7JX6). All models were assessed for structural and thermodynamic plausibility by MolProbity 16 and Gibb's free energy using Swiss‐PdbViewer v 4.1.0. 17 All models were superimposed over their respective templates and deviations in the Cα backbone were estimated in Å.
2.3. Antibody binding site prediction
Epitopes were predicted by DiscoTope v2.0 18 on the basis of three‐dimensional structures using original templates (in the case of SARS‐CoV‐2/Wuhan) and structural models (in the case of SARS‐CoV‐2/B.1.1.7) of spike proteins and orf8 under the threshold scores −3.7 and −10.0, respectively. DiscoTope gathers amino acid statistics, spatial information, and surface accessibility of the target protein from the three‐dimensional structures to develop structure‐based epitope prediction.
2.4. Molecular complex superimposition
The spike protein model of SARS‐CoV‐2/B.1.1.7 was superimposed over Spike–ACE2 complex (PDBid: 6M0J) 19 to monitor its interaction with the human ACE2. Similarly, superimposition of SARS‐CoV‐2/B.1.1.7 spike protein was also carried out over Fab–SARS‐CoV‐2 complexes, PDBid: 7C2L and PDBid: 7K8S, corresponding to the interactions of antibodies with NTD and RBD of the spike protein, respectively. 9 , 10 DS visualizer 2016 was used to visualize all protein models, superimpositions, molecular complexes, intermolecular interactions, spatial variations in the residues, and for three‐dimensional mapping of the epitopic sites.
3. RESULTS
Sequence alignment verified the existence of mutations in orf1a/b (T1001I, A1708D, I2230T, S2625X, and ΔSGF3675‐3677), spike (ΔHV69‐70, ΔY144, N501Y, A570D, D614G, P681H, T716I, S982A, and D1118H), orf8 (Q27X, R52I, ΔK68, Y73C, L118V, F120L, and I121F) and N (D3L, R203K, G204R, and S235F) proteins in SARS‐CoV‐2/B.1.1.7 compared to the SARS‐CoV‐2/Wuhan (Figures 1 and S1–S5). Among the mutations found in orf1a/b, T1001I, A1708D, I2230T, and S2625X correspond to the region that encodes NSP3 protein of the virus, whereas ΔSGF3675‐3677 (deletion) was found in NSP6 encoding region (Figures 1 and [Link], [Link]). Importantly, a mutation in spike protein, D614G, was also detected in SARS‐CoV‐2/B.1.1.7 which has been found in most lineages of SARS‐CoV‐2. 2
Figure 1.
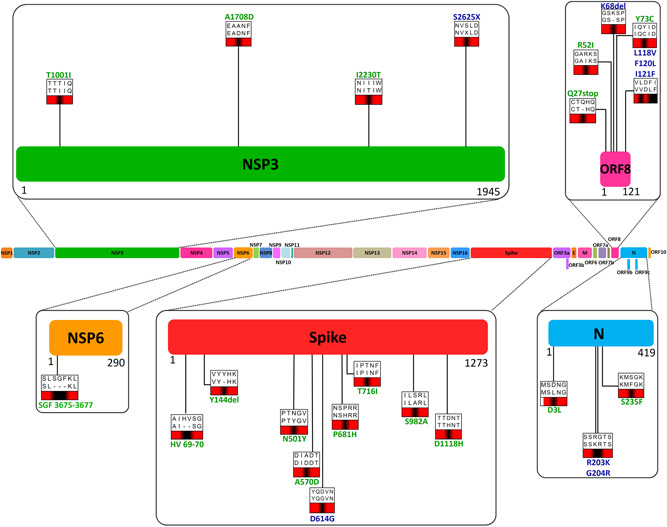
Mutations in SARS‐CoV‐2/B.1.1.7. Schematic and scaled representation of SARS‐CoV‐2 genome with mutations found in different protein‐encoding genes are shown at corresponding positions
Structural superimposition of spike proteins of SARS‐CoV‐2/Wuhan and SARS‐CoV‐2/B.1.1.7 showed considerable conservation in the Cα backbone (RMSD: 0.16 Å) in the overall structure and RBD (Arg319–Phe541) region of the proteins (Figure 2A) with subtle changes in the spatial orientation of substituting amino acids (Figure 2B). To date, several cocrystal complex structures of ACE2–Spike protein have been resolved. 19 , 20 , 21 To monitor the potential change in the intermolecular interactions due to the spike protein mutations in SARS‐CoV‐2/B.1.1.7, docking poses were developed by the superimposition of the variant spike protein over ACE2‐SARS‐CoV‐2 spike protein complex (PDBid: 6M0J). No significant differences were found in the intermolecular interactions between the ACE2 receptor and spike proteins of both the virus. However, a marginal increase in the free energy was predicted in the case of the ACE2‐SARS‐CoV‐2/B.1.1.7 spike complex (−11.8Kcal/mol) compared to the ACE2‐SARS‐CoV‐2/Wuhan spike complex (−11.9Kcal/mol) (Figure 2C). With the exception of a decrease in the numbers of charged‐polar interaction, the numbers of all other forms of intermolecular interactions (charged‐apolar, polar‐polar, polar‐apolar, and apolar‐apolar) were found increased in the SARS‐CoV‐2/B.1.1.7 spike–ACE2 complex compared to SARS‐CoV‐2/Wuhan spike–ACE2, whereas the charged‐charged interaction remained unchanged (Supporting Information Table 1). SARS‐CoV‐2 spike protein has been shown to interact with human neutralizing antibodies at NTD and RBD regions. 9 , 10 Therefore, it is conceivable that the presence of deletions in the NTD (ΔHV69‐70 and ΔY144) and amino acid substitutions in RBD (N501Y) could possibly change the epitopic sites of the SARS‐CoV‐2 spike protein. To examine this possibility, we carried out the structure‐based prediction of epitopic regions of the spike proteins of SARS‐CoV‐2/Wuhan and SARS‐CoV‐2/B.1.1.7. The location and span of the residues predicted to be epitopic were found nearly identical in both the proteins (Figure 2D and Table S2). However, in SARS‐CoV‐2/Wuhan, the sites predicted to be epitopic at the region, Thr250‐Gly252, were found extended (Ser244‐Gly249) in SARS/CoV‐2/B.1.1.7 (Figure 2D). The regions were also found to vary in their electrostatic properties where the former was more polar compared to the latter (Figure 2E). This could potentially be due to the change in the spatial conformation of the loop present between β14 and β15. Moreover, in three‐dimensional conformations, the region was also found shadowing the N‐terminal region of the protein, where ΔHV69‐70 is present. This deletion has changed the spatial orientation of turn present between β3 and β4 (Figure 2F). Of note, the region comes in the immediate proximity to the antibody (plasma isolated) binding site targeting NTD of the spike protein. 9 To explore this further, spike protein structure of SARS‐CoV‐2/B.1.1.7 was individually superimposed over co‐crystal complex structures of neutralizing antibodies against NTD and RBD of SARS‐CoV‐2 spike protein (Figure 3A). 9 , 10 The docking poses thus generated showed that the interaction between SARS‐CoV‐2/B.1.1.7 spike protein and NTD‐antibody may be less stable (ΔG = −11.9 Kcal/mole) compared to the interaction between SARS‐CoV‐2/Wuhan spike protein (ΔG = −12.2 Kcal/mole) and antibody targeting the NTD (Figure 3B). In comparison, the RBD targeting antibody showed relatively stronger interactions with the spike proteins of SARS‐CoV‐2/B.1.1.7 (ΔG = −12.8 Kcal/mole) compared to the SARS‐CoV‐2/Wuhan spike (ΔG = −12.1 Kcal/mole) (Figure 3C).
Figure 2.
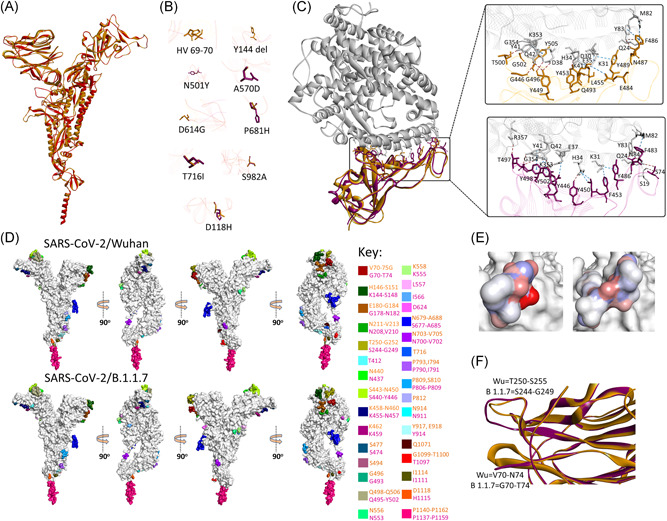
Structure and epitopes of SARS‐CoV‐2/B.1.1.7 spike protein. (A) Structural comparison in ribbon conformation of spike proteins of SARS‐CoV‐2/Wuhan (gold) and SARS‐CoV‐2/B.1.1.7 (magenta). (B) Stick representation of the spatial orientation of amino acids of SARS‐CoV‐2/Wuhan spike protein (gold) compared with substituting amino acids in SARS‐CoV‐2/B.1.1.7 spike protein (magenta). (C) Comparison of the intermolecular interactions between ACE2 receptor (gray ribbon) and RBD domain of SARS‐CoV‐2/Wuhan spike (gold ribbon) and SARS‐CoV‐2/B.1.1.7 spike (magenta ribbon). Intermolecular hydrogen bonds and non‐hydrogen bond intermolecular interactions (electrostatic and hydrophobic) are shown with brown and blue‐dotted lines, respectively, in the enlarged views. (D) Surface topology view with 360° rotation of epitope distribution in the spike proteins of SARS‐CoV‐2/Wuhan and SARS‐CoV‐2/B.1.1.7, as labeled. Corresponding epitopes in terms of position within spike protein are colored differently as mentioned in key. (E) Electrostatic surface of the corresponding epitope found variable between SARS‐CoV‐2/Wuhan (left) and SARS‐CoV‐2/B.1.1.7 spike proteins (right). (F) Structural variation in the loops between β3 and β4 (bottom) and β14 and β15 (top) are shown in ribbon conformation, where spike protein of SARS‐CoV‐2/Wuhan and SARS‐CoV‐2/B.1.1.7 are shown with gold and magenta colors, respectively
Figure 3.
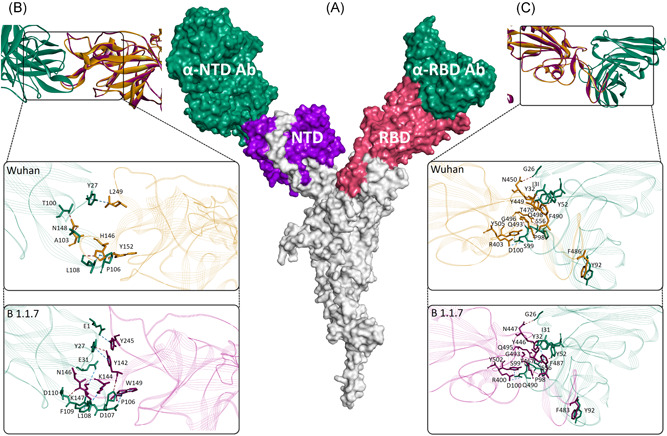
Interaction between SARS‐CoV‐2 spike protein and antibodies. (A) Cartoon showing surface topology of antibodies (sea green) interaction with the NTD (purple) and RBD (pink) of the SARS‐CoV‐2 spike protein. Intermolecular interactions between, (B) NTD and, (C) RBD of SARS‐CoV‐2/Wuhan (gold) and SARS‐CoV‐2/B.1.1.7 (magenta) spike proteins in ribbon conformation. The enlarged views in each showing stick models of intermolecular interactions between the antibody residues (sea green) and amino acids of spike proteins of SARS‐CoV‐2/Wuhan (gold) and SARS‐CoV‐2/B.1.1.7 (magenta). Intermolecular hydrogen bonds and non‐hydrogen bond intermolecular interactions (electrostatic and hydrophobic) are shown with brown and blue‐dotted lines, respectively
A stop codon mutation (Q27X) was observed in orf8 of SARS‐CoV‐2/B.1.1.7, which may result in the premature truncation of the orf8 protein. However, this premature truncation could possibly result in encoding two different chains, orf8a and orf8b, in SARS‐CoV‐2/B.1.1.7. Orf8 of SARS‐CoV‐2/Wuhan and SARS‐CoV‐2/B.1.1.7 shared 93% sequence identity, with all cysteines (Cys20, 37, 61, 83, and 102) and the C‐terminal region (Ile74‐Val117) found strongly conserved (Figure 4A). 8 For further comparison, structural models of orf8b of SARS‐CoV and SARS‐CoV‐2/B.1.1.7 were superimposed over SARS‐CoV‐2/Wuhan orf8 (PDBid: 7JX6). As expected the molecular structure of orf8 shared relatively more similarities between SARS‐CoV‐2 variants (RMSD = 0.07 Å) than with SARS‐CoV (RMSD = 0.25‐0.29 Å) (Figure 4B). However, the intramolecular disulfide bonds were found conserved at the corresponding positions in both orf8 of SARS‐CoV2 variants (Figure 4A). Like the spike protein, orf8 of SARS‐CoV‐2 has been reported for its antigenic properties. 8 Therefore, we studied whether sequence variations between orf8 of SARS‐CoV‐2/Wuhan and SARS‐CoV‐2/B.1.1.7 may have resulted in variations in the antigenicity of the protein. Structure‐based prediction of epitopic sites showed that orf8 of SARS‐CoV‐2/Wuhan is considerably more antigenic compared to the respective proteins encoded by SARS‐CoV and SARS‐CoV‐2/B.1.1.7. Despite strong structural similarities, some of the major antigenic sites of SARS‐CoV‐2/Wuhan orf8 were predicted to be lost in SARS‐CoV‐2/B.1.1.7 (Figure 4C and Table S3). The crystal structure of orf8, of SARS‐CoV‐2 (PDB id: 7JX6), heterologously expressed in E. coli, has shown homodimerization of the protein where both monomers are connected with disulfide bonds between Cys20 on each side. Stop codon mutation (Q27X) as located slightly downstream to this position (Cys20) in SARS‐CoV‐2/B.1.1.7 orf8 could inhibit its dimerization. Moreover, this may also reduce the size of the molecule, and thereby may further drop the immunogenic potential of the orf8 in SARS‐CoV‐2/B.1.1.7 in comparison to SARS‐CoV‐2/Wuhan.
Figure 4.
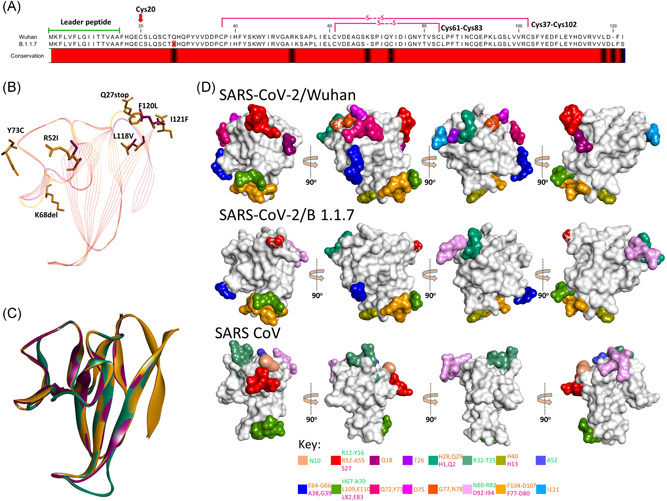
Comparison of SARS‐CoV‐2/Wuhan and SARS‐CoV‐2/B.1.1.7 orf8. (A) Sequence alignment of orf8 of SARS‐CoV‐2/Wuhan and SARS‐CoV‐2/B.1.1.7 showing leader peptide region and cysteine involved in intramolecular disulfide bonds. Note the presence of the stop codon at position 27 in SARS‐CoV‐2/B.1.1.7. The variable amino acids are indicated by the black color in the conservation bar. (B) Stick representation of the spatial orientation of amino acids of SARS‐CoV‐2/Wuhan orf8 (gold) compared with substituting amino acids in SARS‐CoV‐2/B.1.1.7 orf8 (magenta). (C) Structural comparison of orf8 in ribbon conformation of SARS‐CoV (green), SARS‐CoV‐2/Wuhan (gold), and SARS‐CoV‐2/B.1.1.7 (magenta). (D) Surface topology view with 360° rotation of epitope distribution in the orf8 of SARS‐CoV‐2/Wuhan and orf8b of SARS‐CoV and SARS‐CoV‐2/B.1.1.7, as labeled. Corresponding epitopes in terms of position within orf8/8b are colored differently (key given at the bottom)
4. DISCUSSION
Since the start of the recent pandemic of SARS‐CoV‐2 infection, the virus has evolved in several strains with variable demographic distributions. 2 The recently emerged SARS‐CoV‐2 variant, SARS‐CoV‐2/B.1.1.7 has caused a significant rise in viral infection in different parts of the UK and its presence has regularly been reported in many different countries. 2 , 3 , 4 Alarmingly, compared to the estimated accumulation of 2.5 nucleotide substitutions per month in SARS‐CoV‐2 lineages, 22 , 23 the SARS‐CoV‐2/B.1.1.7 has 29 nucleotide substitutions in comparision to SARS‐CoV‐2/Wuhan. Moreover, most of these mutations are nonsynonymous and stationed among the most immunogenic proteins, spike, and orf8 of the virus. 7 , 8 Therefore, it is tempting to speculate that these changes in SARS‐CoV‐2/B.1.1.7 may have resulted in antigenic changes in the virus. In the present investigation, changes in the antigenic architecture of the spike and orf8 proteins of SARS‐CoV‐2/B.1.11.7 have been explored and compared with that of SARS‐CoV‐2/Wuhan.
RBD of the SARS‐CoV‐2 spike protein establishes the first physical interaction between the virus and host via binding with ACE2 receptor. 19 The mutation (N501Y) in SARS‐CoV‐2/B.1.1.7 is stationed within the RBD domain, however, no major conformational change in the spike protein variant and in its interaction with the ACE2 has been observed. This implies that the affinity of the variant virus with the host primary receptor may have remained unchanged. Although it has been observed that the transmissibility rate of SARS‐CoV‐2/B.1.1.7 is considerably higher compared to SARS‐CoV‐2/Wuhan, 24 to date there is no report suggesting that the variant strain is more virulent in terms of clinical manifestation and mortality compared to SARS‐CoV‐2/Wuhan. 4 SARS‐CoV‐2 spike protein is immunogenic and host antibodies raised against SARS‐CoV‐2 have been shown to bind with the NTD and RBD of the viral protein. 9 , 10 Taking the number and location of changes in the spike protein into account, it is possible that these variations may, in turn, alter the antigenic frame of the protein, which could adversely affect the outcome of various immunotherapeutic agents. However, interactions between SARS‐CoV‐2 spike protein and antibody against its RBD have not been found adversely affected. This could possibly be due to the potential strong negative selection at the RBD domain of the viral spike protein. This notion of evolutionary restrain could further be strengthened by the observation that nonsynonymous mutations residing at the RBD (K417, S477, V484, and N501) of different SARS‐CoV‐2 lineages have divergence scores, 0.132, 0.216, 0.174, and 0.180, respectively. In comparison, spike protein mutation, D614G, present downstream to the RBD and has a divergence score of 0.307 (Figure S6). 2 Nevertheless, we have found that the antigenic span of at least one region (Ser244‐Gly249) that neighbors the NTD antibody binding site 9 may have been altered due to the conformational changes in SARS‐CoV‐2/B.1.1.7 spike protein. Additionally, NTD antibody interaction was also predicted to be slightly less stable with the spike of SARS‐CoV‐2/B.1.1.7 compared to the SARS‐CoV‐2/Wuhan spike. Reduction in the efficacy of convalescent sera and monoclonal antibodies has been demonstrated against several emerging variant viral strains along with the variations in clinical outcome. 25 , 26 , 27 Moreover, it has also been pointed that convalescent plasma actually increases the genetic variations (via selection pressure) in the virus within the host. 25 In summary, this may indicate that the immunogenic potential of SARS‐CoV‐2/B.1.1.7 spike protein may in part be different than SARS‐CoV‐2/Wuhan.
Orf8 is an accessory protein of SARS‐CoV‐2 and has not been found in many other coronaviruses that infect humans. However, the protein has been found in both SARS‐CoV and SARS‐CoV‐2 and showed sequence similarity with some of the bat coronaviruses (RaTG13, ZXC21, and ZC45). The functionality of SARS‐CoV‐2 orf8 is not known to its full extent, however, it has been suggested that the protein may be involved in the immune evasion and cytokine response mimicking. The presence of N‐terminal signal peptide (Met1‐Ala15) allows orf8 of SARS‐CoV‐2 to be released from the infected cells and also leads to the immune response. Seropositivity and circulating peptides of orf8 have been observed along with anti orf8 IgG, IgM, and IgA within the infected individuals. 8 However, orf8 of SARS‐CoV has not been reported for any seropositivity, this is possibly because of the deletion of 29 nucleotides. This in turn has resulted in the formation of two chains of orf8 during the earlier phase of the SARS‐CoV epidemic, potentially impairing its extracellular secretion. 11 The presence of the stop codon mutation in orf8 in SARS‐CoV‐2/B.1.1.7 could then also lead to the formation of two chains, orf8a and orf8b, where the latter (larger chain) does not harbor the leader peptide required for the extracellular secretion, thus reducing its immunogenicity and/or seropositivity. Therefore, it is possible to conceive that orf8 of SARS‐CoV‐2/B.1.1.7 may not be as immunogenic as compared to SARS‐CoV‐2/Wuhan orf8. Moreover, sequence variations may have resulted in the loss of many epitopic sites in the orf8 of SARS‐CoV‐2/B.1.1.7 compared to SARS‐CoV‐2/Wuhan. For SARS‐CoV, it has been shown that viruses containing orf8b replicate more efficiently in the presence of interferon compared to the virus with intact orf8. 28 Therefore, it is possible that this potential emergence of orf8a and orf8b in SARS‐CoV‐2/B.1.1.7, may also confer a similar evolutionary advantage. Furthermore, Val77 of SARS‐CoV orf8b has been shown critical to induce orf8b intracellular aggregation, lysosomal stress, autophagy and interleukin‐mediated inflammatory response via triggering NLRP3. 29 This Val77 is conserved in both the compared SARS‐CoV2/Wuhan and SARS‐CoV‐2/B.1.1.7. Therefore, it is possible that as in SARS‐CoV, its (Val77) exclusive presence in a separate and nonsecretory chain in SARS‐CoV‐2/B.1.1.7 may also contribute to the pathological manifestation of the infection. 29 Taken together, it is likely that SARS‐CoV‐2/B.1.1.7 may bear an evolutionary advantage over SARS‐CoV‐2/Wuhan, mediated not only by the antigenic changes in its spike and orf8 proteins but could also by an elevated cytokine‐mediated inflammatory response in the host due to the intracellular aggregation.
New variant lineages of SARS‐CoV‐2, like P.1 and B.1.351, have been recently reported, 2 additional studies in relation to their genetic and antigenic variations are warranted. Similarly, looking into the evolutionary road map of the accumulation of these mutations could also provide important insights. Like this study, many other investigations have successfully employed bioinformatic tools to predict the epitopic regions of different proteins in SARS‐CoV‐2/Wuhan. 30 , 31 However, experimental validations of these predictions are essentially required. Nevertheless, these findings direct further empirical investigations to explore the neutralizing capabilities of the existing repertoire of immunotherapies against SARS‐CoV‐2. In addition, cell culture‐based assay and/or serum mass spectrometry profiles could confirm the existence of the two chains of orf8 in the variant virus. These findings are expected to not only further our understanding of the disease biology of the SARS‐CoV‐2 but could also lead to the development of more refined and efficient immunotherapeutic interventions.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Mushtaq Hussain conceived and supervised the project. Mushtaq Hussain, Sanya Shabbir, Anusha Amanullah, Fozia Raza, Muhammad J. Imdad, and Sahar Zahid executed the study and were involved in the analysis and writing of the manuscript.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1002/jmv.26931
Supporting information
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
ACKNOWLEDGEMENT
The study in part is supported by the Higher Education Commission, Government of Pakistan (NRPU‐3857).
Hussain M, Shabbir S, Amanullah A, Raza F, Imdad MJ, Zahid S. Immunoinformatic analysis of structural and epitope variations in the spike and Orf8 proteins of SARS‐CoV‐2/B.1.1.7. J Med Virol. 2021;93:4461‐4468. 10.1002/jmv.26931
DATA AVAILABILITY STATEMENT
Data available in article supplementary material.
REFERENCES
- 1. WHO . Weekly operational update on COVID‐19 ‐ 13 February 2021. https://www.who.int/publications/m/item/weekly-operational-update-on-covid-19-13-february-2021
- 2. GISAID . https://www.gisaid.org/epiflu-applications/phylodynamics/
- 3. Chand M, Hopkins S, Dabrera G, et al. Investigation of novel SARS‐COV‐2 variant: Variant of Concern 202012/01. Public Health England. 2020. https://www.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variant-variant-of-concern-20201201 [Google Scholar]
- 4. Rambaut A, Loman N, Pybus O, et al. Preliminary genomic characterisation of an emergent SARS‐CoV‐2 lineage in the UK defined by a novel set of spike mutations. Written on behalf of COVID‐19 Genomics Consortium UK. 2020. https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563
- 5. Umair M, Ikram A, Salman M, et al. Importation of SARS‐CoV‐2 variant B.1.1.7 in Pakistan. J Med Virol. 2021:jmv.26869. 10.1002/jmv.26869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Worldometer . https://www.worldometers.info/coronavirus/country/uk/
- 7. Liu W, Liu L, Kou G, et al. Evaluation of nucleocapsid and spike protein‐based enzyme‐linked immunosorbent assays for detecting antibodies against SARS‐CoV‐2. J Clin Microbiol. 2020;58(6):e00461‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang X, Lam JY, Wong WM, et al. Accurate Diagnosis of COVID‐19 by a Novel Immunogenic Secreted SARS‐CoV‐2 orf8 Protein. mBio. 2020;11(5):e02431‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chi X, Yan R, Zhang J, et al. A neutralizing human antibody binds to the N‐terminal domain of the Spike protein of SARS‐CoV‐2. Science. 2020;369(6504):650‐655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barnes CO, Jette CA, Abernathy ME, et al. SARS‐CoV‐2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682‐687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oostra M, De Haan CA, Rottier PJ. The 29‐nucleotide deletion present in human but not in animal severe acute respiratory syndrome coronaviruses disrupts the functional expression of open reading frame 8. J Virol. 2007;81(24):13876‐13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Larkin MA, Blackshields G, Brown NP, et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23(21):2947‐2948. [DOI] [PubMed] [Google Scholar]
- 13. Artimo P, Jonnalagedda M, Arnold K, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40(W1):W597‐W603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Biasini M, Bienert S, Waterhouse A, et al. SWISS‐MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42(W1):W252‐W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cai Y, Zhang J, Xiao T, et al. Distinct conformational states of SARS‐CoV‐2 spike protein. Science. 2020;369(6511):1586‐1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Davis IW, Leaver‐Fay A, Chen VB, et al. MolProbity: all‐atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35(suppl_2):W375‐W383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johansson MU, Zoete V, Michielin O, Guex N. Defining and searching for structural motifs using DeepView/Swiss‐PdbViewer. BMC Bioinformatics. 2012;13(1):173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kringelum JV, Lundegaard C, Lund O, Nielsen M. Reliable B cell epitope predictions: impacts of method development and improved benchmarking. PLoS Comput Biol. 2012;8(12):e1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lan J, Ge J, Yu J, et al. Structure of the SARS‐CoV‐2 spike receptor‐binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215‐220. [DOI] [PubMed] [Google Scholar]
- 20. Xu C, Wang Y, Liu C, et al. Conformational dynamics of SARS‐CoV‐2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo‐EM. Sci Adv. 2020;7:eabe5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Benton DJ, Wrobel AG, Xu P, et al. Receptor binding and priming of the spike protein of SARS‐CoV‐2 for membrane fusion. Nature. 2020;588(7837):327‐330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang C, Liu Z, Chen Z, et al. The establishment of reference sequence for SARS‐CoV‐2 and variation analysis. J Med Virol. 2020;92(6):667‐674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shen Z, Xiao Y, Kang L, et al. Genomic diversity of severe acute respiratory syndrome‐coronavirus 2 in patients with coronavirus disease 2019. Clin Infect Dis. 2020;71(15):713‐720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Volz E, Mishra S, Chand M, et al. Report 42 ‐ Transmission of SARS‐CoV‐2 Lineage B.1.1.7 in England: insights from linking epidemiological and genetic data. 2020. https://www.imperial.ac.uk/mrc-global-infectious-disease-analysis/covid-19/report-42-sars-cov-2-variant/
- 25. Kemp SA, Collier DA, Datir R, et al. Neutralising antibodies drive Spike mediated SARS‐CoV‐2 evasion. medRxiv. 2020. [Google Scholar]
- 26. Weisblum Y, Schmidt F, Zhang F, et al. Escape from neutralizing antibodies by SARS‐CoV‐2 spike protein variants. eLife. 2020;9:e61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Baum A, Fulton BO, Wloga E, et al. Antibody cocktail to SARS‐CoV‐2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369(6506):1014‐1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wong HH, Fung TS, Fang S, Huang M, Le MT, Liu DX. Accessory proteins 8b and 8ab of severe acute respiratory syndrome coronavirus suppress the interferon signaling pathway by mediating ubiquitin‐dependent rapid degradation of interferon regulatory factor 3. Virology. 2018;515:165‐175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shi CS, Nabar NR, Huang NN, Kehrl JH. SARS‐coronavirus open reading frame‐8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov. 2019;5:1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Romero‐López JP, Carnalla‐Cortés M, Pacheco‐Olvera DL, et al. A bioinformatic prediction of antigen presentation from SARS‐CoV‐2 spike protein revealed a theoretical correlation of HLA‐DRB1* 01 with COVID‐19 fatality in Mexican population: an ecological approach. J Med Virol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. He J, Huang F, Zhang J, et al. Vaccine design based on 16 epitopes of SARS‐CoV‐2 spike protein. J Med Virol. 2020;93:2115‐2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Data Availability Statement
Data available in article supplementary material.


