Abstract
Background
Oral cavity is the most prevalent site of head and neck squamous cell carcinomas (HNSCCs). Most often diagnosed at a locally advanced stage, treatment is multimodal with surgery as the cornerstone. The aim of this study was to explore the molecular landscape of a homogenous cohort of oral cavity squamous cell carcinomas (OCSCCs), and to assess the prognostic value of tumor mutational burden (TMB), along with classical molecular and clinical parameters.
Patients and methods
One hundred and fifty-one consecutive patients with OCSCC treated with upfront surgery at the Institut Curie were analyzed. Sequencing of tumor DNA from frozen specimens was carried out using an in-house targeted next-generation sequencing panel (571 genes). The impact of molecular alterations and TMB on disease-free survival (DFS) and overall survival (OS) was evaluated in univariate and multivariate analyses.
Results
Pathological tumor stage, extranodal spread, vascular emboli, and perineural invasion were associated with both DFS and OS. TP53 was the most mutated gene (71%). Other frequent molecular alterations included the TERT promoter (50%), CDKN2A (25%), FAT1 (17%), PIK3CA (14%), and NOTCH1 (15%) genes. Transforming growth factor-β pathway alterations (4%) were associated with poor OS (P = 0.01) and DFS (P = 0.02) in univariate and multivariate analyses. High TMB was associated with prolonged OS (P = 0.01 and P = 0.02, in the highest 10% and 20% TMB values, respectively), but not with DFS. Correlation of TMB with OS remained significant in multivariate analysis (P = 0.01 and P = 0.005 in the highest 10% and 20% TMB values, respectively). Pathological tumor stage combined with high TMB was associated with good prognosis.
Conclusion
Our results suggest that a high TMB is associated with a favorable prognosis in patients with OCSCC treated with upfront surgery.
Key words: oral cavity squamous cell carcinoma, tumor mutational burden, next-generation sequencing, prognostic marker
Highlights
-
•
High TMB is associated with a favorable prognosis in patients with OCSCC treated with upfront surgery
-
•
Pathological tumor stage combined with high TMB is associated with good prognosis
-
•
TP53 was the most mutated gene (71%). Other frequent molecular alterations included the TERT promoter (50%)
-
•
TGFβ pathway alterations were associated with poor outcomes, although it was only observed in 4% of the patients
Introduction
Head and neck cancer is the seventh most common type of cancer.1 More than 90% of head and neck malignancies are squamous cell carcinomas (HNSCCs). Oral cavity squamous cell cancer (OCSCC) is the most common site of HNSCC with an estimated 354 900 new cases and 177 400 deaths worldwide in 2018.1 Classical risk factors of OCSCC include smoking and excessive alcohol consumption with a synergistic effect, implicated in 75% of all HNSCCs.2 While 30%-35% of oropharyngeal cancers are attributable to human papillomavirus (HPV), and associated with a better prognosis,3 HPV-positive OCSCCs are rare (<6%).4
Most HNSCC patients are diagnosed at a locally advanced stage and are treated with a multimodal therapy that usually includes surgery followed by adjuvant radiotherapy with or without chemotherapy for OCSCC patients. Despite this intense treatment, ∼25% of OCSCC patients recurred. While the 5-year overall survival (OS) for early-stage disease is 90%, survival remains poor in patients with locally advanced disease with only about half of the patients alive at 5 years.5 Despite advances in research and therapy, survival has not improved significantly in the last few decades.2
The well-established prognostic factors in OCSCC are mainly clinical and histological with disease stage, nodal involvement, extracapsular spread, invasion of resection margins, and perineural and lymphovascular invasion.6 Margin invasion and extracapsular spread are used for recurrence risk stratification and to guide post-operative treatment decision.7,8
Apart from the HPV status, and mainly in oropharyngeal cancer, there is to date a paucity of robust clinically useful prognostic markers in HNSCC.9 In the recurrent and/or metastatic setting, cytotoxic chemotherapy, with epidermal growth factor receptor-targeting antibody and, more recently, immunotherapy are used without any molecular selection except for pembrolizumab that is only approved for patients with programmed death-ligand 1 (PD-L1) combined positive score >1.10, 11, 12, 13
The development of next-generation sequencing (NGS) has helped to decipher tumor genomic landscapes revealing molecular alterations with possible predictive or prognostic significance. Several studies have shown that tumor mutational burden (TMB) has a predictive value for response to immunotherapy.14, 15, 16, 17, 18 Nevertheless, others suggested that TMB might also have a prognostic value.19, 20, 21
In this study, we aimed to explore the prognostic value of TMB in OCSCC patients treated with upfront surgery.
Patients and methods
Samples and clinical data
The cohort exclusively consisted of patients with OCSCC treated with upfront surgery at the Institut Curie between February 1991 and November 2016. Samples came from resected specimens. According to the French regulations, patients were informed about the research carried out on tissue specimens and did not express opposition. The study was authorized by the French CPP (Comité de Protection des Personnes n°2019-A01234-26).
Clinical parameters were collected and categorized as follows: age (<64/≥64 years), sex (male/female), alcohol consumption [yes (defined as >2 drinks/day everyday)/no], smoking history (yes/no), HPV status (PCR positive/negative), American Joint Committee on Cancer (AJCC) stage (I/II/III/IV), margin invasion (negative/positive), extracapsular spread (absent/present), vascular emboli (absent/present), perineural invasion (absent/present), differentiation (poorly = 1, moderately = 2, well = 3), and mitotic index (high if ≥10 mitoses/field; mid if 5-10 mitoses/field; low if <5 mitoses/field) as previously reported.22 Positive margins were defined as margins invaded with infiltrating carcinoma. Pathological tumor stage (I/II/III/IV) has been classified according to the eighth TNM (tumor–node–metastasis) staging edition.
Genomic DNA extraction
Tumor samples were frozen in liquid nitrogen in a cryotube as soon as the sample was made. All the frozen samples were stored at −80°C, under temperature control. Tumoral cellularity was verified systematically by performing cryosections and by macrodissecting tumoral zones. Samples with <50% of tumoral cells were not extracted. We used a classical phenol–chloroform protocol for DNA extractions. The quality of DNA was verified by migration on agarose gel. DNA concentration was determined by nanodrop measurement.
Next-generation sequencing
Sequencing was carried out using an in-house NGS panel of 571 genes, called DRAGON Dx (Detection of Relevant Alterations in Genes involved in Oncogenetics). Indexed paired-end libraries of tumor DNA were generated using the Agilent SureSelect XT2 library prep kit (Agilent Technologies, Santa Clara, CA). The kit supports sequencing targeted regions of the genome spanning 2.7 Mb. About 50 ng of input DNA was used to build the libraries according to manufacturer’s protocol. The pool was finally sequenced on a NovaSeq 6000 (Illumina) S2x150 bp flow cell (Illumina Inc., San Diego, CA).
Bioinformatics analysis
Read mapping
In the first analysis part, reads were mapped using ‘BWA’ mem software (v0.7.15)23 on the human reference genome (hg19 assembly) using default parameters. As a second quality control, statistics regarding the mapping (percentage of aligned reads total and falling into the capture, percentage of PCR duplicates) and the capture coverage were produced using a combination of ‘SAMtools flagstat’, ‘BEDtools coverage’, and ‘PicardTools MarkDuplicates’.
Variant calling
Variant calling of both single-nucleotide variants (SNVs) and small insertion/deletions (indels) was then carried out on the processed alignment files using a combination of SAMtools mpileup24 and ∗VarScan2∗ ∗mpileup2cns∗ (v2.4.3).25
Annotations
Annotations from several databases [RefSeq, dbsnp v150, COSMIC v86, 1000g project 08/2015 version, ESP6500, gnomAD (all and ethnicities), ICGC v21, and dbnsfp v35 predictions] were provided by ANNOVAR26 to annotate small variants. Only the RefSeq database was used for intermediate indel. During this step, all variants present in ±10 bp of each exon junction were defined as splicing.
Coverage quality control
A more detailed notion of each gene percentage, per barcode, covered by at least 100×, in the processed alignments, was also provided using a combination of ‘awk’, ‘SAMtools mpileup’, ‘BEDtools intersect’, ‘multiinter’, and ‘merge’. Bases covered by <100× were reported per barcode using the same strategy. Genes belonging to patient pathology were tagged in these two files to facilitate the search for genes of interest that might be badly covered.
Gene classification
Genes from the DRAGON Dx panel were classified according to the literature and databases (cBioPortal,27 OncoKB24) into three categories: tumor suppressor genes, oncogenes, and genes considered as both an oncogene and a tumor suppressor gene. We also categorized these genes according to the cellular pathway in which they were involved (Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2021.100178).
Variant selection algorithm
Stringent selection algorithm was applied to remove a maximum of irrelevant variants. We considered a minimal allelic ratio of 5% and a maximal frequency in the population of 0.1%. Only truncating mutations (frameshift deletion and insertion, stopgain, splicing alteration, and hotspot mutations from the Cancer Hotspot database) with a minimal coverage of 200 reads were retained for tumor suppressor gene variants. All missense variants known to be hotspot mutations from the Cancer Hotspot database28 and with no minimal coverage were retained for oncogene variants. For genes classified as both oncogenes and tumor suppressor genes (such as NOTCH1) or with known missense hotspots (like TP53), truncating mutations with a minimal coverage of 200 and known hotspot mutations with no minimal coverage were selected.
Tumor mutational burden
TMB was defined as the number of non-synonymous somatic mutations (SNVs and small indels) per megabase in coding regions (mut/Mb). Coding variants (except for intronic splicing ones, therefore exons-only which represent 1.59 Mb), without synonymous variants or polymorphisms (>0.1% minor allele frequency) and recurrent variants covered enough (not tagged as Low_Depth) were considered in all those calculations. Because the median and range of mutational load have been shown to vary across tumor types, we subsequently identified cases in the top 10th and 20th percentiles of TMB in our own cohort, and determined the log-rank P value for difference in survival as well as the direction of the effect with a hazard ratio determined from a Cox model.
Oncoprints
Oncoprints were drawn using the ComplexHeatmap package and were carried out with the Maftools package for 4.00 version of R.
Statistical analysis
The start date was set at the day of initial surgery. Survivals were measured from the surgery date to the time of death or most recent follow-up for OS, and to the first recurrence not amenable to salvage surgery or re-irradiation or death for disease-free survival (DFS).
Univariate statistical analyses were made on clinical, pathological, and molecular features considering the TMB value and genes mutated at a frequency ≥4% in the cohort (TP53, CDKN2A, FAT1, PIK3CA, NOTCH1, KMT2D, NFE2L2, FAT2, KMT2B, NSD1, HRAS, CASP8, TERT), using the log-rank (Mantel–Cox) test on GraphPad Prism 8.
All variables for which the P value was <0.05 in univariate analysis were included into a Cox model, using the coxph function on R (version 3.6.3), to determine their independent effect on survival, except for margin invasion and HPV status that we forced into the model despite a P value exceeding 0.05 given their established prognostic impact that is considered for adjuvant treatment decision. Some variables such as vascular emboli and perineural invasion were removed from multivariate analysis due to missing data.
Results
Patient characteristics
One hundred and fifty-one consecutive OCSCC patients were included. Eighty-six patients (57%) received post-operative radiotherapy with or without chemotherapy. Median follow-up was 42 months (range: 1-258 months). Mean age at diagnosis was 64 years (range: 23-91 years). Patient characteristics are described in Table 1. Most patients were male (n = 92, 61%), 48 consumed alcohol (37%), and 73 had a smoking history (53%). Seven patients (5%) had HPV-positive tumors. Most patients had a stage IV disease (n = 65, 43%), whereas 30 patients had a stage III disease (20%). A total of 110 patients (76%) presented well-differentiated tumors (grade 1). Forty-eight patients had a high mitotic index (40%). While 49 patients (38%) had vascular emboli, 60 patients (47%) had perineural invasion and 24 patients (16%) had invaded margins. High pathological AJCC stage, presence of vascular emboli, extracapsular spread, and perineural invasion were associated with a poor DFS (P < 0.0001, P = 0.001, P = 0.002, and P = 0.0008, respectively) and OS (P = 0.003, P = 0.0003, P = 0.006, and P = 0.0003, respectively). A higher age correlated with a poor OS (P = 0.04) (Table 1).
Table 1.
Clinical, biological, and pathological characteristics of the 151 patients and correlation with survival
| Disease-free survival |
Overall survival |
||||
|---|---|---|---|---|---|
| Patients n (%) | Recurrence n (%) | P valueg | Death n (%) | P valueg | |
| Total | 151 (100) | 50 (33) | 77 (51) | ||
| Age (years) | 0.90 (NS) | 0.04 | |||
| <64 | 76 (50) | 26 (52) | 34 (44) | ||
| ≥64 | 75 (50) | 24 (48) | 43 (56) | ||
| Sex | 0.90 (NS) | 0.59 (NS) | |||
| Male | 92 (61) | 27 (54) | 29 (38) | ||
| Female | 59 (39) | 23 (46) | 48 (62) | ||
| Alcohol consumptiona | 0.89 (NS) | 0.75 (NS) | |||
| No | 81 (63) | 30 (67) | 40 (63) | ||
| Yes | 48 (37) | 15 (33) | 23 (37) | ||
| Smoking historyb | 0.32 (NS) | 0.29 (NS) | |||
| No | 65 (47) | 27 (56) | 29 (43) | ||
| Yes | 73 (53) | 21 (44) | 39 (57) | ||
| HPV | 0.30 (NS) | 0.25 (NS) | |||
| Positive | 7 (5) | 1 (2) | 2 (3) | ||
| Negative | 144 (95) | 49 (98) | 75 (97) | ||
| AJCC stage | <0.0001 | 0.003 | |||
| Stage I | 20 (13) | 1 (2) | 5 (6) | ||
| Stage II | 36 (24) | 8 (16) | 17 (22) | ||
| Stage III | 30 (20) | 9 (18) | 16 (21) | ||
| Stage IV | 65 (43) | 32 (64) | 39 (51) | ||
| Invaded margins | 0.23 (NS) | 0.17 (NS) | |||
| Negative | 127 (84) | 40 (80) | 63 (82) | ||
| Positive | 24 (16) | 10 (20) | 14 (18) | ||
| Extranodal spreadc | 0.002 | 0.006 | |||
| Absent | 111 (74) | 31 (62) | 51 (67) | ||
| Present | 38 (26) | 19 (38) | 25 (33) | ||
| Vascular embolia | 0.001 | 0.0003 | |||
| Absent | 80 (62) | 21 (47) | 31 (49) | ||
| Present | 49 (38) | 24 (53) | 32 (51) | ||
| Perineural invasiond | 0.0008 | 0.0003 | |||
| Absent | 68 (53) | 16 (36) | 24 (39) | ||
| Present | 60 (47) | 28 (64) | 38 (61) | ||
| Differentiatione | 0.25 (NS) | 0.25 (NS) | |||
| Well | 110 (76) | 35 (57) | 56 (74) | ||
| Moderately | 27 (19) | 22 (36) | 15 (20) | ||
| Poorly | 8 (6) | 4 (7) | 5 (6) | ||
| Mitotic indexf | 0.84 (NS) | 0.99 (NS) | |||
| Low | 42 (34) | 15 (38) | 22 (37) | ||
| Mid | 32 (26) | 10 (26) | 14 (24) | ||
| High | 48 (40) | 14 (36) | 23 (39) | ||
AJCC, American Joint Committee on Cancer; HPV, human papillomavirus; NS, not significant.
Significant values are indicated in bold.
Information available for 129 patients.
Information available for 138 patients.
Information available for 149 patients.
Information available for 128 patients.
Information available for 145 patients.
Information available for 122 patients.
Log-rank test.
Genomic alterations
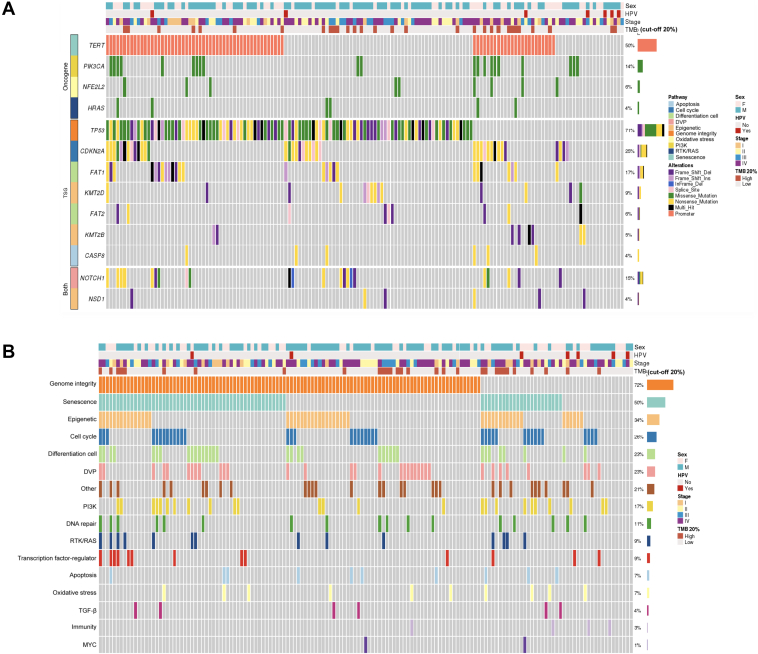
All patients had at least one molecular alteration (Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2021.100178). One hundred and seven patients (71%) of the whole cohort had an alteration in TP53, 38 (25%) in CDKN2A, 26 (17%) in FAT1, 21 (14%) in PIK3CA, 22 (15%) in NOTCH1, 14 (9%) in KMT2D, 9 (6%) in NFE2L2, 9 (6%) in FAT2, 8 (5%) in KMT2B, and 6 (4%) in NSD1, HRAS, and CASP8. Pathogenic promoter mutations in TERT were found in 76 patients (50%) (Supplementary Table S3, available at https://doi.org/10.1016/j.esmoop.2021.100178; Figure 1A). All variants detected in our population are reported in Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2021.100178.
Figure 1.
Oncoprints showing most frequently altered genes and pathways. (A) Oncoprint showing an integrated analysis of genomic alterations in the 151 oral cavity squamous cell carcinoma (OCSCC) patients. This figure provides an overview of the most frequent genomic alterations (left column) with their respective frequencies (right column) combined with clinical information and tumor mutational burden (TMB) data (heading). Each column represents a patient. Each type of genomic alteration is represented by a color code. (B) Oncoprint showing an integrated analysis of the most altered molecular pathways in the 151 OCSCC patients. Each column represents a patient. Altered pathways are listed in the left column and are ranked according to frequency in the population (right column). DVP, development pathway; HPV, human papillomavirus; PI3K, Phosphatidylinositol 3 kinase; RTK, receptor tyrosine kinase; TGF-β, transforming growth factor-β; TSG, tumor suppressor gene.
In univariate analysis, patients with TP53 mutation had a poorer OS while those with FAT2 mutation had a prolonged OS although statistical significance was not reached (P = 0.06) but with no impact on DFS. The other genes were not associated with DFS or OS (Supplementary Table S3 and Figure S1, available at https://doi.org/10.1016/j.esmoop.2021.100178).
Signaling pathways
Alterations were observed in the genome integrity pathway in 108 patients (72%). Seventy-six patients (50%) had an alteration in the senescence pathway, 51 (34%) in the epigenetic pathway, 39 (26%) in the cell cycle, 34 (23%) in the cell differentiation and development pathways, and 17 (11%) in the DNA repair pathway (Supplementary Table S4, available at https://doi.org/10.1016/j.esmoop.2021.100178; Figure 1B).
In univariate analysis, alterations in the transforming growth factor-β (TGF-β) signaling pathway were associated with a poorer DFS (P = 0.02) and OS (P = 0.01). No other pathway alterations were found to correlate with DFS or OS (Supplementary Table S4 and Figure S2, available at https://doi.org/10.1016/j.esmoop.2021.100178).
Tumor mutational burden
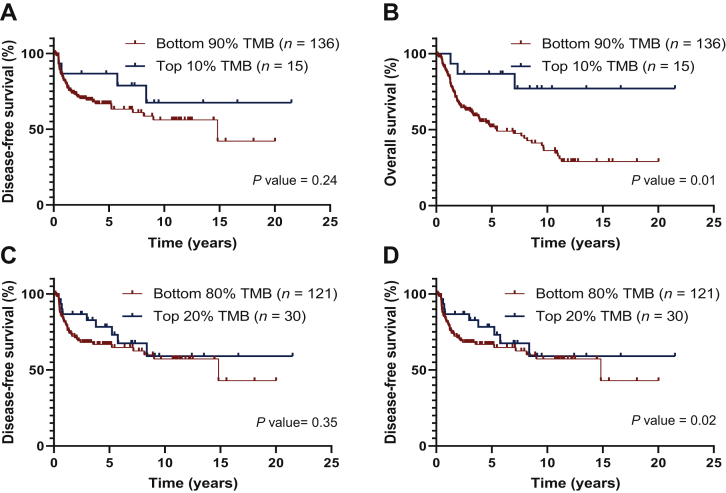
When using the 10% top TMB and 90% bottom TMB subgroups of patients, TMB values ranged between 15.1 and 126.1 mut/Mb and between 2.5 and 14.5 mut/Mb, respectively, corresponding to a threshold value of TMB of ∼15 mut/Mb. For the 20% top TMB and 80% bottom TMB subgroups, TMB values varied between 12 and 126.1 mut/Mb and between 2.5 and 11.3 mut/Mb, respectively, with a threshold value of 11.5 mut/Mb (Supplementary Table S5, available at https://doi.org/10.1016/j.esmoop.2021.100178).
Higher somatic TMB values (i.e. higher than the respective threshold) were associated with a prolonged OS (P = 0.01 and P = 0.02, in the highest 10% and 20% TMB, respectively), but not with DFS (Figure 2).
Figure 2.
Correlation of tumor mutational burden (TMB) with disease-free survival (A and C) and overall survival (B and D) in univariate analysis, using a cut-off of 10% (A and B) or 20% (C and D) for the high TMB group.
Multivariate analysis
Pathological AJCC stage, HPV status, extracapsular spread, margin invasion, TMB, and TGF-β pathway alterations were integrated into a Cox model to determine their independent effect on survival (Tables 2 and 3; Supplementary Figure S2, available at https://doi.org/10.1016/j.esmoop.2021.100178).
Table 2.
Multivariate analysis of survival according to pathological characteristics, TGF-β pathway, and TMB (cut-off 10%)
| DFSa |
OSa |
||||||
|---|---|---|---|---|---|---|---|
| Patients n (%) | HR | 95% CI (HR) | P value | HR | 95% CI (HR) | P value | |
| Total | 151 (100) | ||||||
| TMB (cut-off 10%) | 0.49 | 0.17-1.4 | 0.19 | 0.21 | 0.06-0.7 | 0.01 | |
| Low | 136 (90) | ||||||
| High | 15 (10) | ||||||
| AJCC stage | 1.90 | 1.33-2.7 | <0.001 | 1.37 | 1.07-1.8 | 0.01 | |
| Stage I | 20 (13) | ||||||
| Stage II | 36 (24) | ||||||
| Stage III | 30 (20) | ||||||
| Stage IV | 65 (43) | ||||||
| HPV | 0.42 | 0.06-3.1 | 0.40 | 0.56 | 0.14-2.3 | 0.43 | |
| Positive | 7 (5) | ||||||
| Negative | 144 (95) | ||||||
| Extranodal spreadb | 1.53 | 0.82-2.9 | 0.19 | 1.58 | 0.92-2.7 | 0.10 | |
| Negative | 111 (74) | ||||||
| Positive | 38 (26) | ||||||
| Invaded margins | 0.96 | 0.46-2.0 | 0.92 | 1.08 | 0.659-2.0 | 0.81 | |
| Negative | 127 (86) | ||||||
| Positive | 24 (16) | ||||||
| TGF-β pathway | 4.52 | 1.53-13.3 | 0.006 | 4.13 | 1.57-10.9 | 0.004 | |
| Unaltered | 6 (4) | ||||||
| Altered | 145 (96) | ||||||
95% CI, 95% confidence interval; AJCC, American Joint Committee on Cancer; DFS, disease-free survival; HPV, human papillomavirus; HR, hazard ratio; OS, overall survival; TGF-β, transforming growth factor-β; TMB, tumor mutational burden.
Significant values are indicated in bold.
Cox model.
Information available for 149 patients.
Table 3.
Multivariate analysis of survival according to pathological characteristics, TGF-β pathway, and TMB (cut-off 20%)
| DFSa |
OSa |
||||||
|---|---|---|---|---|---|---|---|
| Patients n (%) | HR | 95% CI (HR) | P value | HR | 95% CI (HR) | P value | |
| Total | 151 (100) | ||||||
| TMB (cut-off 20%) | 0.57 | 0.27-1.2 | 0.13 | 0.36 | 0.18-0.74 | 0.005 | |
| Low | 121 (80) | ||||||
| High | 30 (20) | ||||||
| AJCC stage | 1.91 | 1.34-2.7 | <0.001 | 1.40 | 1.09-1.80 | 0.008 | |
| Stage I | 20 (13) | ||||||
| Stage II | 36 (24) | ||||||
| Stage III | 30 (20) | ||||||
| Stage IV | 65 (43) | ||||||
| HPV | 0.40 | 0.06-2.9 | 0.37 | 0.52 | 0.13-2.14 | 0.37 | |
| Positive | 7 (5) | ||||||
| Negative | 144 (95) | ||||||
| Extranodal spreadb | 1.61 | 0.86-3.0 | 0.14 | 1.63 | 0.96-2.76 | 0.07 | |
| Negative | 111 (74) | ||||||
| Positive | 38 (26) | ||||||
| Invaded margins | 1.07 | 0.50-2.3 | 0.87 | 1.25 | 0.68-2.30 | 0.47 | |
| Negative | 127 (86) | ||||||
| Positive | 24 (16) | ||||||
| TGF-β pathway | 4.03 | 1.39-11.7 | 0.01 | 3.26 | 1.26-8.40 | 0.01 | |
| Unaltered | 6 (4) | ||||||
| Altered | 145 (96) | ||||||
95% CI, 95% confidence interval; AJCC, American Joint Committee on Cancer; DFS, disease-free survival; HPV, human papillomavirus; HR, hazard ratio; OS, overall survival; TGF-β, transforming growth factor-β; TMB, tumor mutational burden.
Significant values are indicated in bold.
Cox model.
Information available for 149 patients.
Regardless of the TMB threshold used (10% or 20%), a high TMB correlated with a prolonged OS (P = 0.01 and P = 0.005 in the highest 10% and 20% TMB, respectively) (Tables 2 and 3; Supplementary Figures S3 and S4, available at https://doi.org/10.1016/j.esmoop.2021.100178).
Among the other parameters, pathological AJCC stage had a worse impact on DFS and OS at both the 10% threshold (P < 0.001 and P = 0.01, respectively) (Table 2; Supplementary Figure S3A and B, available at https://doi.org/10.1016/j.esmoop.2021.100178) and the 20% threshold (P < 0.001 and P = 0.008, respectively) (Table 3; Supplementary Figure S4A and B, available at https://doi.org/10.1016/j.esmoop.2021.100178).
TGF-β pathway alteration was associated with both shorter DFS and OS at both the TMB 10% threshold (P = 0.006 and P = 0.004, respectively) and the TMB 20% threshold (P = 0.01).
By combining pathological AJCC stages with TMB status, we identified four separate prognostic groups with significantly different DFS and OS at both the 10% threshold (P = 0.002 and P = 0.003, respectively) and the 20% threshold (P = 0.002 and P = 0.002, respectively) (Supplementary Figure S5, available at https://doi.org/10.1016/j.esmoop.2021.100178). Patients with the poorest prognosis had low TMB values and AJCC TNM III/IV.
Discussion
Despite numerous studies that investigated the predictive value of TMB in response to checkpoint inhibitor, insufficient data are available regarding its prognostic value.18 We report that high TMB is associated with a favorable prognosis in patients with OCSCC treated with upfront surgery.
The median follow-up in our cohort was 3.5 years, allowing to accurately evaluate prognostic factors, since recurrences mostly occur within the first 2 years following primary treatment in HNSCC patients. Patient characteristics and related prognostic factors were in line with those reported in the literature in OCSCC.29,30 Around two-thirds of patients in our series had locally advanced OCSCC with pathological stage III or IV disease. Association with both DFS and OS was consistent with the literature.31 Extracapsular spread, vascular emboli, and perineural invasion are also associated with survival. The presence of vascular emboli precedes the passage of tumor cells into the vascular compartment, which is one of the first steps for the potential development of metastases. Lymphovascular invasion has been shown to correlate with the risk of local, regional, and distant recurrence.32 Finally, extracapsular spread is also one of the most well-established prognostic factors in HNSCC. In the study by Wreesmann et al. reviewing the survival of 266 patients treated primarily with surgery for a tongue cancer, the 5-year OS was 75% in patients without invaded lymph nodes, 50% in patients with invaded lymph nodes without extracapsular spread, and 30% in patients with extracapsular spread.33 In our study, extracapsular spread was not an independent prognostic factor in multivariate analysis, probably due to an overlap with the disease stage, since the vast majority of extranodal spread was observed in stage IV tumors.
The mutational profile observed in our cohort was similar to what was previously reported by The Cancer Genome Atlas (TCGA).34 The frequencies of the main genes involved in HNSCC like TP53, CDKN2A, FAT1, and PIK3CA were like those reported in the literature with slight differences in tumor suppressor genes like NSD1, which was less frequent in our series. The chosen minimal coverage of 200 reads might have underestimated the number of relevant variants found in poorly covered genes like NSD1. Another difference observed was the number of TERT mutations (50% in our OCSCC compared to 24% reported in TCGA35). Our cohort only included OCSCC, whereas TCGA included samples from different head and neck locations. Among all molecular alterations identified, TP53 and FAT2 gene mutations were associated with survival, although statistical significance was not reached. Mutations in TP53 are known to be associated with poor prognosis in HNSCC, whereas the prognostic value of FAT2 mutations has not been reported so far in the literature.36
The main altered signaling pathways in our cohort (genome integrity, senescence, epigenetic, cell cycle, and DNA repair) were consistent with those found in HNSCC in previous studies.37 TGF-β pathway alterations were associated with poor outcomes, although it was only observed in 4% of the patients. TGF-β is a multi-functional cytokine that regulates cell growth and differentiation, apoptosis, cell motility, extracellular matrix (ECM) production, angiogenesis, and cellular immune response.38 TGF-β has been shown to exert dual and opposed roles in oncogenesis explained by the pleiotropic nature of TGF-β, ranging from cytostatic and apoptotic tumor-suppressive effects in early-stage tumors to proliferative, invasive, angiogenic, and oncogenic effects in advanced cancer.38 A pan-cancer analysis involving 9125 tumor samples found at least one genomic alteration in mediators or TGF-β regulator signaling in 39% of samples, the highest frequencies being reported in gastrointestinal cancers. Alterations in the TGF-β superfamily correlated positively with the expression of metastasis-associated genes and with decreased survival.39
Similar conclusions were obtained in clinical studies.40 A genomic and transcriptomic analysis revealed enrichment in markers known to be regulated by TGF-β like cell adhesion and ECM remodeling in patients with melanoma non-responding to programmed cell death protein 1 (PD-1) therapy compared with responding patients.41 Another transcriptomic analysis of human tumors from TCGA suggested that up-regulation of ECM gene expression was linked to the activation of TGF-β target genes and this pan-cancer signature predicted unresponsiveness to PD-1 blockade.42 Additionally, gene-set enrichment analysis identified TGFB1 and TGFBR2 to be associated with non-response to anti-PD-L1 therapy and reduced OS in patients with urothelial cancer.43 Altogether, these studies support the use of TGF-β signaling pathway inhibitors to induce responses in otherwise unresponsive tumor. New strategies using bispecific drugs targeting PD-L1 and TGF-β are currently being investigated in HNSCC (NCT04428047, NCT04428047, NCT04220775).
In our series, we found that high TMB had a positive prognostic value confirmed in multivariate analysis. Several studies have suggested that TMB might be a predictor of response to anti-PD-1/PD-L1 agents,14, 15, 16, 17,44, 45, 46, 47 and TMB is now a Food and Drug Administration (FDA)-approved biomarker for pembrolizumab treatment in noncolorectal high microsatellite instability/mismatch repair-deficient cancers (KEYNOTE-15818). Other studies suggested that TMB might also have a prognostic value.19, 20, 21 In our series, we found that high TMB had a positive and independent prognostic value confirmed in multivariate analysis. Contrasting results have been reported in the literature on the prognostic value of TMB and varied across cancer types. A study in a population of patients with locally advanced HNSCC treated with exclusive radiochemotherapy reported a negative prognostic value of high TMB.48 In the patient population of the KEYNOTE-158 clinical trial not treated with immunotherapy, TMB had no obvious prognostic value.18 In another study, the association between TMB and survival seemed to vary according to cancer type and had no impact in HNSCC.49 Other studies are consistent with our findings. Low TMB in patients with metastatic colorectal cancer was a negative prognostic factor.50 Similar results were reported in early colorectal cancer.19 In human epidermal growth factor receptor 2-positive refractory metastatic breast cancer, high TMB was associated with a prolonged survival.51
The variability in the prognostic value of TMB may be explained by several factors. Although the threshold of 10 mut/Mb has been used to grant FDA approval of pembrolizumab across cancer types, there is no clear consensus on what threshold for high TMB is the most relevant. We defined the high TMB subgroups by using the cut-offs of 10% and 20% highest TMB levels of the whole population, corresponding in our study to TMBs higher than 15 and 11.5 mut/Mb, respectively. In both cases, a high TMB correlated with a prolonged OS in both univariate and multivariate analyses. Data gathered from several studies conducted in non-small-cell lung cancer (NSCLC) and urothelial carcinomas estimated that the TMB threshold required to benefit from the checkpoint inhibitors stands around 10 mut/Mb according to the FoundationOne panel and 7 mut/Mb according to the MSK-IMPACT panel. Higher thresholds were used, including 16.2 mut/Mb in studies with atezolizumab and 15 mut/Mb in studies with ipilimumab and nivolumab in NSCLC, but reached similar results in terms of the predictive value of TMB.52
In total, the discordant results across studies support the need for a qualitative evaluation of the tumor mutational profile, which should be complementary to the quantitative evaluation provided by the TMB. The TMB calculation depends on the sequencing panel used and on the mutations considered relevant for its calculation. Most of the targeted sequencing panels focus on driver mutations of theranostic interest. TMB is not an easy tool to handle both in its definition and its interpretation. Indeed, the variables on which the calculation of the TMB depends (i.e. threshold, type of mutations taken into account, targeted genes, sequencing depth and coverage, selection of variants, filters chosen in bioinformatics analysis) are multiple and require standardization. Our study offered optimal conditions for the evaluation of the TMB in a homogeneous cohort of patients, with no possible impact of treatments since it was evaluated from the outset at diagnosis with the use of a stringent algorithm for quantitative and qualitative evaluation of the tumor mutation profile.
Conclusion
In our series of OCSCC patients treated with upfront surgery, a high TMB was an independent favorable prognostic factor associated with a prolonged OS.
Acknowledgments
Funding
This work was supported by Institut Curie funding. High-throughput sequencing was carried out by the ICGex NGS platform of the Institut Curie supported by the grants from the Agence Nationale de la Recherche (‘Investissements d’Avenir’ program) [grant numbers ANR-10-EQPX-03 (Equipex), ANR10-INBS-09-08 (France Génomique Consortium)]; by the ITMO-Cancer Aviesan (Plan Cancer III); and by the SIRIC-Curie program (SIRIC Grant INCa-DGOS-4654).
Disclosure
CLT participated in advisory boards from MSD, BMS, Merck Serono, AstraZeneca, Nanobiotix, Celgene, GSK, Roche, Rakuten, and Seattle Genetics. All other authors have declared no conflict of interest.
Data sharing
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supplementary data
References
- 1.Ferlay J., Colombet M., Soerjomataram I. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 2.Johnson D.E., Burtness B., Leemans C.R. Head and neck squamous cell carcinoma. Nat Rev Dis Primers. 2020;6:92. doi: 10.1038/s41572-020-00224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ang K.K., Harris J., Wheeler R. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taberna M., Mena M., Pavón M.A., Alemany L., Gillison M.L., Mesía R. Human papillomavirus-related oropharyngeal cancer. Ann Oncol. 2017;28(10):2386–2398. doi: 10.1093/annonc/mdx304. [DOI] [PubMed] [Google Scholar]
- 5.Koo B.S., Lim Y.C., Lee J.S., Choi E.C. Recurrence and salvage treatment of squamous cell carcinoma of the oral cavity. Oral Oncol. 2006;42(8):789–794. doi: 10.1016/j.oraloncology.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 6.Jones A.S. Prognosis in mouth cancer: tumour factors. Eur J Cancer B Oral Oncol. 1994;30B(1):8–15. doi: 10.1016/0964-1955(94)90044-2. [DOI] [PubMed] [Google Scholar]
- 7.Bernier J., Domenge C., Ozsahin M. Postoperative irradiation with or without concomitant chemotherapy for locally advanced head and neck cancer. N Engl J Med. 2004;350(19):1945–1952. doi: 10.1056/NEJMoa032641. [DOI] [PubMed] [Google Scholar]
- 8.Cooper J.S., Pajak T.F., Forastiere A.A. Postoperative concurrent radiotherapy and chemotherapy for high-risk squamous-cell carcinoma of the head and neck. N Engl J Med. 2004;350(19):1937–1944. doi: 10.1056/NEJMoa032646. [DOI] [PubMed] [Google Scholar]
- 9.Licitra L., Perrone F., Bossi P. High-risk human papillomavirus affects prognosis in patients with surgically treated oropharyngeal squamous cell carcinoma. J Clin Oncol. 2006;24(36):5630–5636. doi: 10.1200/JCO.2005.04.6136. [DOI] [PubMed] [Google Scholar]
- 10.Vermorken J.B., Mesia R., Rivera F. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med. 2008;359(11):1116–1127. doi: 10.1056/NEJMoa0802656. [DOI] [PubMed] [Google Scholar]
- 11.Ferris R.L., Blumenschein G., Fayette J. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med. 2016;375:1856–1867. doi: 10.1056/NEJMoa1602252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burtness B., Harrington K.J., Greil R. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet. 2019;394:1915–1928. doi: 10.1016/S0140-6736(19)32591-7. [DOI] [PubMed] [Google Scholar]
- 13.Cohen E.E.W., Soulières D., Le Tourneau C. Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head-and-neck squamous cell carcinoma (KEYNOTE-040): a randomised, open-label, phase 3 study. Lancet. 2019;393:156–167. doi: 10.1016/S0140-6736(18)31999-8. [DOI] [PubMed] [Google Scholar]
- 14.Yarchoan M., Hopkins A., Jaffee E.M. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377:2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodman A.M., Kato S., Bazhenova L. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16:2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Allen E.M., Miao D., Schilling B. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350(6257):207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hellmann M.D., Ciuleanu T.-E., Pluzanski A. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med. 2018;378(22):2093–2104. doi: 10.1056/NEJMoa1801946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marabelle A., Fakih M., Lopez J. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020;21:1353–1365. doi: 10.1016/S1470-2045(20)30445-9. [DOI] [PubMed] [Google Scholar]
- 19.Domingo E., Camps C., Kaisaki P.J. Mutation burden and other molecular markers of prognosis in colorectal cancer treated with curative intent: results from the QUASAR 2 clinical trial and an Australian community-based series. Lancet Gastroenterol Hepatol. 2018;3:635–643. doi: 10.1016/S2468-1253(18)30117-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Owada-Ozaki Y., Muto S., Takagi H. Prognostic impact of tumor mutation burden in patients with completely resected non-small cell lung cancer: brief report. J Thorac Oncol. 2018;13:1217–1221. doi: 10.1016/j.jtho.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 21.Lee D.-W., Han S.-W., Bae J.M. Tumor mutation burden and prognosis in patients with colorectal cancer treated with adjuvant fluoropyrimidine and oxaliplatin. Clin Cancer Res. 2019;25:6141–6147. doi: 10.1158/1078-0432.CCR-19-1105. [DOI] [PubMed] [Google Scholar]
- 22.Hoffmann C., Vacher S., Sirven P. MMP2 as an independent prognostic stratifier in oral cavity cancers. Oncoimmunology. 2020;9:1754094. doi: 10.1080/2162402X.2020.1754094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013 [Google Scholar]
- 24.Chakravarty D., Gao J., Phillips S.M. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017;2017 doi: 10.1200/PO.17.00011. PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koboldt D.C., Chen K., Wylie T. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25(17):2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao J., Aksoy B.A., Dogrusoz U. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang M.T., Asthana S., Gao S.P. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34(2):155–163. doi: 10.1038/nbt.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woolgar J.A., Scott J., Vaughan E.D., Brown J.S., West C.R., Rogers S. Survival, metastasis and recurrence of oral cancer in relation to pathological features. Ann R Coll Surg Engl. 1995;77(5):325–331. [PMC free article] [PubMed] [Google Scholar]
- 30.Vincent N., Dassonville O., Chamorey E. Clinical and histological prognostic factors in locally advanced oral cavity cancers treated with primary surgery. Eur Ann Otorhinolaryngol Head Neck Dis. 2012;129(6):291–296. doi: 10.1016/j.anorl.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Woolgar J.A., Rogers S., West C.R., Errington R.D., Brown J.S., Vaughan E.D. Survival and patterns of recurrence in 200 oral cancer patients treated by radical surgery and neck dissection. Oral Oncol. 1999;35(3):257–265. doi: 10.1016/s1368-8375(98)00113-4. [DOI] [PubMed] [Google Scholar]
- 32.Close L.G., Brown P.M., Vuitch M.F., Reisch J., Schaefer S.D. Microvascular invasion and survival in cancer of the oral cavity and oropharynx. Arch Otolaryngol Head Neck Surg. 1989;115(11):1304–1309. doi: 10.1001/archotol.1989.01860350038011. [DOI] [PubMed] [Google Scholar]
- 33.Wreesmann V.B., Katabi N., Palmer F.L. Influence of extracapsular nodal spread extent on prognosis of oral squamous cell carcinoma. Head Neck. 2016;38(suppl 1):E1192–E1199. doi: 10.1002/hed.24190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cancer Genome Atlas Network Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barthel F.P., Wei W., Tang M. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat Genet. 2017;49(3):349–357. doi: 10.1038/ng.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou G., Liu Z., Myers J.N. TP53 mutations in head and neck squamous cell carcinoma and their impact on disease progression and treatment response. J Cell Biochem. 2016;117(12):2682–2692. doi: 10.1002/jcb.25592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanchez-Vega F., Mina M., Armenia J. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell. 2018;173(2):321–337.e10. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Derynck R., Akhurst R.J., Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29(2):117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 39.Korkut A., Zaidi S., Kanchi R.S. A pan-cancer analysis reveals high-frequency genetic alterations in mediators of signaling by the TGF-β Superfamily. Cell Syst. 2018;7:422–437.e7. doi: 10.1016/j.cels.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Groeneveldt C., van Hall T., van der Burg S.H. Immunotherapeutic potential of TGF-β inhibition and oncolytic viruses. Trends Immunol. 2020;41:406–420. doi: 10.1016/j.it.2020.03.003. [DOI] [PubMed] [Google Scholar]
- 41.Hugo W., Zaretsky J.M., Sun L. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165(1):35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chakravarthy A., Khan L., Bensler N.P. TGF-β-associated extracellular matrix genes link cancer-associated fibroblasts to immune evasion and immunotherapy failure. Nat Commun. 2018;9:4692. doi: 10.1038/s41467-018-06654-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mariathasan S., Turley S.J., Nickles D. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554:544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ott P.A., Bang Y.-J., Piha-Paul S.A. T-cell-inflamed gene-expression profile, programmed death ligand 1 expression, and tumor mutational burden predict efficacy in patients treated with pembrolizumab across 20 cancers: KEYNOTE-028. J Clin Oncol. 2019;37(4):318–327. doi: 10.1200/JCO.2018.78.2276. [DOI] [PubMed] [Google Scholar]
- 45.Ready N., Hellmann M.D., Awad M.M. First-line nivolumab plus ipilimumab in advanced non-small-cell lung cancer (CheckMate 568): outcomes by programmed death ligand 1 and tumor mutational burden as biomarkers. J Clin Oncol. 2019;37:992–1000. doi: 10.1200/JCO.18.01042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rizvi N.A., Hellmann M.D., Snyder A. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rizvi N.A., Cho B.C., Reinmuth N. Durvalumab with or without tremelimumab vs standard chemotherapy in first-line treatment of metastatic non-small cell lung cancer: the MYSTIC phase 3 randomized clinical trial. JAMA Oncol. 2020;6(5):661–674. doi: 10.1001/jamaoncol.2020.0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eder T., Hess A.K., Konschak R. Interference of tumour mutational burden with outcome of patients with head and neck cancer treated with definitive chemoradiation: a multicentre retrospective study of the German Cancer Consortium Radiation Oncology Group. Eur J Cancer. 2019;116:67–76. doi: 10.1016/j.ejca.2019.04.015. [DOI] [PubMed] [Google Scholar]
- 49.Wu H.-X., Wang Z.-X., Zhao Q. Tumor mutational and indel burden: a systematic pan-cancer evaluation as prognostic biomarkers. Ann Transl Med. 2019;7(22):640. doi: 10.21037/atm.2019.10.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Innocenti F., Ou F.-S., Qu X. Mutational analysis of patients with colorectal cancer in CALGB/SWOG 80405 identifies new roles of microsatellite instability and tumor mutational burden for patient outcome. J Clin Oncol. 2019;37:1217–1227. doi: 10.1200/JCO.18.01798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park S.E., Park K., Lee E. Clinical implication of tumor mutational burden in patients with HER2-positive refractory metastatic breast cancer. Oncoimmunology. 2018;7:e1466768. doi: 10.1080/2162402X.2018.1466768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chan T.A., Yarchoan M., Jaffee E. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30(1):44–56. doi: 10.1093/annonc/mdy495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.