Abstract
An obligately anaerobic, non-motile and Gram-positive rod bacterium, strain SW219 was isolated from ceacum of feral chickens. Based on 16S rRNA sequence analysis, the strain SW219 exhibited 97.88% similarity to Collinsella massiliensis strain GD3 strain, the closest valid species. The genome size of SW219 was 2.58 Mbp with 64.5 mol% of G+C content. The phenotypic and genotypic analysis suggested that the strain SW219 is a new species belonging to the family Coriobacteriaceae within the Actinobacteria phylum, which the name Collinsella avium sp. nov. is proposed. The type strain of Collinsella avium is SW219 (= DSM 109235T and = CCOS 1884T).
Keywords: Chicken, Collinsella, gut health, microbiome, new species, taxonogenomics, whole genome
Introduction
Animal gut harbours a high diversity of microbial community which plays a significant role in host metabolism and immune system, including pathogen protection [1,2]. Although gut microbiota composition and diversity has been accessed using metagenomics analysis, huge number of bacterial species remains uncultured [[3], [4], [5]]. To study interaction and function of gut bacteria, isolation and cultivation of uncultured bacteria is an important step. Taxonomical characterisation of new bacterial species allows the study of their phenotype and genotype, which may potentially impact host health. Here we report the isolation of new strain SW219 from the pooled ceacal contents of feral chicken. We further characterised the strain using taxonogenomics approach and identified it as a new species.
Methods
Isolation and growth conditions
The strain SW219 was isolated from ceacum of feral chicken by culturing in strict anaerobic conditions using a Coy Lab anaerobic chamber containing 85% nitrogen, 10% hydrogen and 5 % carbon dioxide. Modified brain heart infusion (BHI-M) medium, which contained 37 g/L of BHI, 5 g/L of yeast extract, 1 mL of 1 mg/mL menadione, 0.3 g of L-cysteine, 1 mL of 0.25 mg/L of resazurin, 1 mL of 0.5 mg/mL hemin, 10 mL of vitamin and mineral mixture, 1.7 mL of 30 mM acetic acid, 2 mL of 8 mM propionic acid, 2 mL of 4 mM butyric acid, 100 μl of 1 mM isovaleric acid, and 1% of pectin and inulin was used for strain culturing and maintenance.
Strain identification and phylogenetic analysis
The strain SW219 initially was identified using MALDI-TOF (Bruker, Germany). Later, genomic DNA of the strain SW219 was extracted using DNeasy Blood & Tissue kit (Qiagen), in accordance with the manufacturer's instructions. 16S rRNA gene sequences was amplified using universal primer set 27F (5′- AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′- ACCTTGTTACGACTT- 3′) [6,7]. The PCR amplicon was sequenced using Sanger dideoxy method (ABI 3730XL; Applied Biosystems). The full-length of 16S rRNA gene of strain SW219 was assembled using Geneious software [8]. The 16S rRNA gene sequence of strain SW219 was compared with closely related strains in the GenBank (www.ncbi.nlm.nih.gov/genbank/) and EZBioCloud databases (www.ezbiocloud.net/eztaxon) [9]. The 16S rRNA sequences of valid related species were obtained from EZBioCloud databases for phylogenetic analysis using MEGA7 software [10]. Multiple alignments were generated using the CLUSTAL-W algorithm [11]. Reconstruction of phylogenetic tree was built using the maximum-likelihood [12], maximum-parsimony [13] and neighbour-joining [14] methods. The distance matrices were generated using Kimura's two-parameter model. Bootstrap resampling analysis of 1000 replicates was performed to estimate the confidence of tree topologies.
Phenotypic tests
Colony morphology of strain SW219 was determined after 2-3 days of incubation on BHI-M agar plates. Gram-staining was performed using a Gram-Staining kit set (Difco), in accordance with the manufacturer's instructions. Cell morphologies of cultures during exponential growth were examined by scanning electron microscopy. Aerotolerance was examined by incubating cultures for 2 days under aerobic and anaerobic conditions. Motility of this microorganism was verified using motility medium with triphenyltetrazolium chloride [15]. The growth was indicated by the presence of red colour, reduced form of triphenyltetrazolium chloride after it is absorbed into bacterial cell wall. Other biochemical tests including utilisation of various substrates and enzyme activities were investigated using the AN MicroPlate (Biolog) and API ZYM (bioMérieux) in accordance with the manufacturer's instructions. Cellular fatty acids were determined following Microbial Identification System (MIDI) protocols from cell biomass using gas chromatography (Agilent 7890A) [16].
Genome sequencing and analysis
The library for genome sequencing was prepared using Nextera XT kit (Illumina Inc). The library was sequenced on a MiSeq platform using Illumina 2 × 250 paired end chemistry. The reads were assembled using Unicycler that builds an initial assembly graph from short reads using the de novo assembler SPAdes 3.11.1 [17]. The quality assessment for the assembly was performed using QUAST [18]. The circular map of SW219 genome was generated using DNAPlotter software [19]. Genome annotation was performed using Rapid Annotation using Subsystem Technology server [20]. The analysis of functional categories of strain SW219 and other members of the genus Collinsella was analysed by Rapid Annotation using Subsystem Technology and was presented as a heatmap generated using Explicet software [21]. Furthermore, the average nucleotide identity (ANI) was calculated between SW219 and the related genomes using OrthoANI software [22]. The digital DNA-DNA hybridisation (dDDH) was performed between strain SW219 and the closet phylogenetic neighbour using Genome-to Genome Distance Calculator (GGDC 2.1) web server (http://ggdc.dsmz.de) [23].
Results
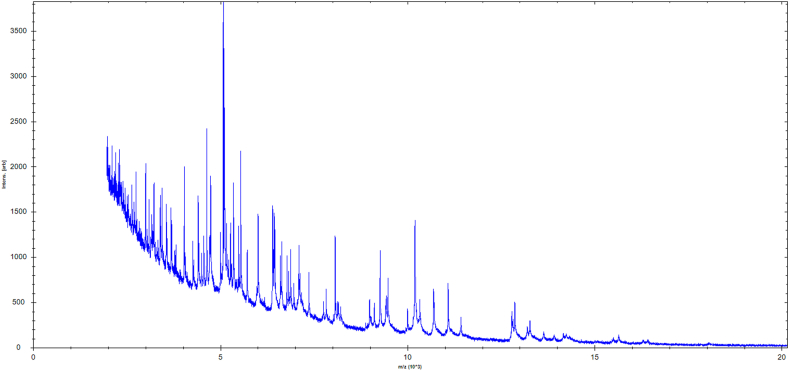
Strain SW219 was isolated from cecal content of feral chicken. Colonies of strain SW219 were white convex, translucent and entire circular margin with a mean diameter of 0.1-0.3 mm after 2-3 days of incubation on BHI-M agar (Fig. 1). For an initial identification, the score of MALDI-TOF MS was less than 1.7 suggesting no match organism in the database (Fig. 2). Based on 16S rRNA phylogenic analysis, strain SW219 was clustered within the Collinsella genus. It exhibited 97.88% similarity to Collinsella massiliensis strain GD3 (GenBank accession number JX424766.1), the closest species with standing in nomenclature (Fig. 3). The similarity less than 98% of 16S rRNA sequences suggested that it is a new species. The characteristics of this strain were further investigated using multiple tests and analyses.
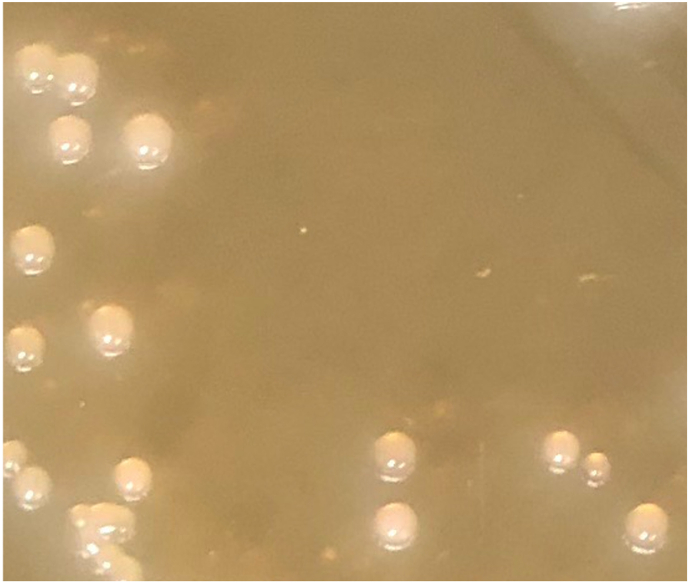
Fig. 1.
Colony morphology of Collinsella avium sp. nov. strain SW219. Cells were cultured for 48-72 hours at 37o C in BHI-M medium.
Fig. 2.
MALDI-TOF MS reference spectrum of Collinsella avium sp. nov. strain SW219. The reference spectrum was generated by comparison of spectra from 12 individual colonies.
Fig. 3.
Phylogenetic tree showing position of Collinsella avium sp. nov. strain SW219 within closely related taxa in the genus Collinsella of Coriobacteriaceae family. GenBank accession numbers of the 16S rRNA gene sequences are given in parentheses. Sequences were aligned using CLUSTALW. The phylogenetic tree was obtained using the maximum-likelihood method and generated using MEGA 7 software. Bootstrap values obtained by repeating the analysis 1000 times to generate a majority consensus tree are indicated at the nodes. The scale bar indicates a 2% nucleotide sequence divergence.
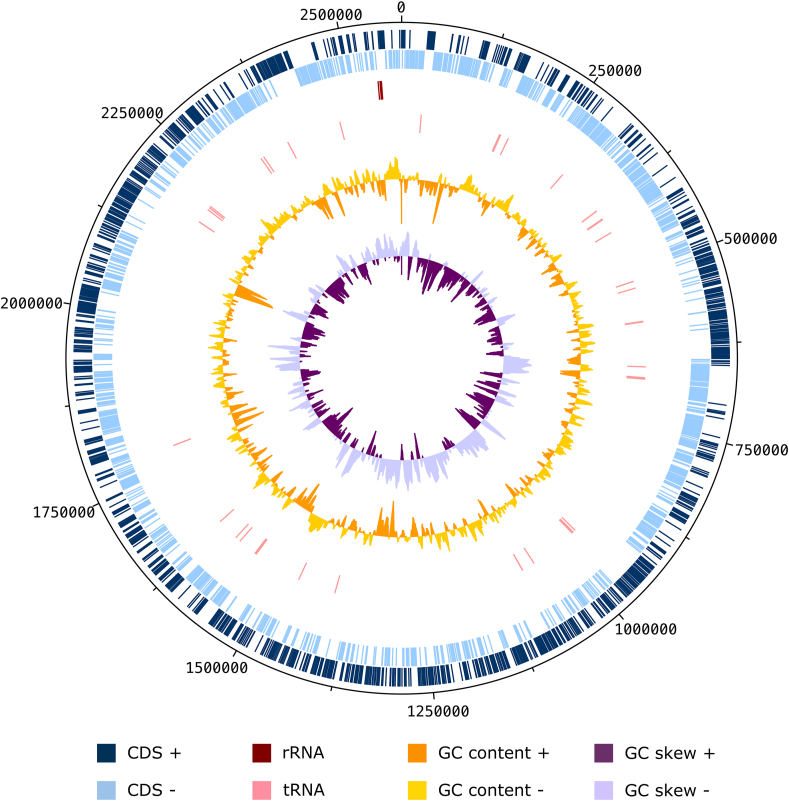
The strain SW219 grew in BHI-M medium at 37°C for 24 h under anaerobic condition. Cells were Gram-positive bacilli and non-motile with a diameter of 0.5-2.0 μm (Fig. 4). General features and biochemical characteristics of the strain SW219 were distinct from other members of the genus Collinsella (Table 1). The major cellular fatty acids included C16: 0 DMA, C16: 0 aldehyde, C16: 0 and C12: 0 (Table 2). The genome of strain SW219 is 2.58 Mbp with 64.5 mol% of G+C content (Fig. 5). A total of 2162 coding sequences (CDS) and 56 RNAs were identified in the genome. Based on the analysis of functional categories (COGs), the dominant categories were amino acids and derivatives, carbohydrate and protein mechanism (Fig. 6). Overall, the COGs pattern of the strain SW219 was very similar to other species of Collinsella. The stain SW219 showed the similar level of multiple categories such as amino acids and derivatives, phosphorus metabolism, and stress response to the closest relative, C. massiliensis while other categories were similar to other members of the genus Collinsella. For example, DNA metabolism and nucleosides and nucleotides categories were similar to those of C. tanakaei and C. stercoris, respectively. In addition, stain SW219 contained the same percentage of protein metabolism genes as C. aerofaciens, C. stercoris, C. tanakaei, and C. vaginalis. The ANI values of genome sequences between the stain SW219 and other members of the genus Collinsella ranged from 75-77% (Fig. 6). These values were significantly less than the proposed ANI cutoff of 95–96 % for the identical species [24]. Species cutoff value for a new species is 70% based dDDH analysis. When the stain SW219 is compared with its closely related species, all values were less than 70%, which clearly shows the need for the designation of this strain as a new species (Table 3) (Fig. 7).
Fig. 4.
Scanning electron microscopy (SEM) of Collinsella avium strain SW219. Cell were cultured for 24 hours at 37o C in BHI-M medium. Bar, 2.0 μm.
Table 1.
Characteristics of strain SW219 and other members of the genus Collinsella
| Characteristic | SW219 | C. massiliensis | C. intestinalis | C. aerofaciens | C. tanakei | C. stercoris |
|---|---|---|---|---|---|---|
| Habitat | chicken gut | human gut | human gut | human gut | human gut | NA |
| Cell morphology | rods | rods | rods | cocci to rods | rods | rods |
| Cell diameter (μm) | 0.5-2.0 | 0.57 | 0.3-0.5 | 0.3-0.7 | 0.5-1.0 | 0.3-0.5 |
| Gram stain | + | + | + | + | + | + |
| Motility | — | — | — | — | — | — |
| DNA G+C content (mol%) | 64.5 | 65.8 | 64.4 | 61 | 60.8 | 61.2 |
| API test | ||||||
| Alkaline phosphatase | + | + | + | — | + | + |
| Indirect ELISA Leucine arylamidase |
+ | — | NA | NA | NA | NA |
| Valine arylamidase | + | + | NA | NA | NA | NA |
| Cystine arylamidase | + | — | NA | NA | NA | NA |
| Trypsin | — | — | NA | NA | NA | NA |
| Acid phosphatase | + | + | + | — | + | + |
| α−galactosidase | + | NA | NA | NA | NA | NA |
| β−galactosidase | — | — | — | + | — | + |
| β−glucuronidase | — | — | — | — | + | — |
| α−glucuronidase | + | — | — | + | — | — |
| β−glucosidase | — | + | — | — | + | + |
| N-acetyl-β-glucosaminidase | + | — | + | — | — | + |
| Utilisation of | ||||||
| Glucose | + | — | + | + | + | + |
| Mannose | + | — | + | + | + | + |
| Galactose | + | — | + | + | NA | + |
| Fructose | + | — | + | + | NA | + |
| Maltose | — | — | — | + | + | + |
| Sucrose | + | NA | NA | NA | NA | NA |
| Maltotriose | + | NA | NA | NA | NA | NA |
| Cellubiose | — | — | — | — | + | + |
| Lactose | — | — | — | + | + | + |
| Rhamnose | — | — | — | NA | — | — |
| Ribose | NA | — | — | + | + | + |
| Raffinose | — | — | — | NA | NA | — |
| Sorbitol | + | + | — | — | — | — |
NA, data not available; +/-, depending on tests used.
Table 2.
Cellular fatty acid compositions of Collinsella avium strain SW219
| Fatty acid | Mean relative % |
|---|---|
| Straight chain | |
| 12:00 | 10.98 |
| 14:00 | 14.62 |
| 16:00 | 12.26 |
| 18:0 | 2.12 |
| 16:0 aldehyde | 15.36 |
| 18:0 aldehyde | 2.21 |
| Demethylacetal (DMA) | |
| 12:0 DMA | 7.01 |
| 14:0 DMA | 2.58 |
| 16:0 DMA | 22.05 |
| 18:0 DMA | 5.13 |
| Unsaturated | |
| 13:1 at 12-13 | 2.11 |
| 18:1 w9c | 2.45 |
| 18:2 w6,9c | 1.13 |
| Summed feature 1 | 2.11 |
∗Summed features are fatty acids that could not be separated using the MIDI System. Summed feature 1 contains C13: 1 and/or C14: 0 aldehyde.
Fig. 5.
Graphical circular map of Collinsella avium strain SW219 genome. From outside to the centre: coding sequences on the forward strand (CDS +), coding sequences on the reverse strand (CDS -), tRNAs, rRNAs, GC content and GD skew. This map was generated using DNAPlotter software.
Fig. 6.
Distribution of functional features of predicted coding sequences of Collinsella avium strain SW219 and closely related genomes. The functional features were predicted based on the clusters of orthologous groups. Heatmap was generated from genome annotation of individual species by RAST using Explicet software.
Table 3.
Digital DNA-DNA hybridisation comparing the genomic similarity between members of the genus Collinsella
| % DDH estimate | C. massiliensis | C. tanakaei | C. vaginalis | C. intestinalis | C. stercoris | C. aerofaciens |
|---|---|---|---|---|---|---|
| HSP length/total length | 18.30 | 15.8 | 15.6 | 16.60 | 16.5 | 16 |
| Identities/HSP length | 2.32 | 2.27 | 22.3 | 22.90 | 2.36 | 22.3 |
| Identities/total length | 2.32 | 2.27 | 15.7 | 22.90 | 16.5 | 16.1 |
| Difference in % G+C | 1.35 | 4.23 | 0.1 | 2 | 1.27 | 3.92 |
DDH,
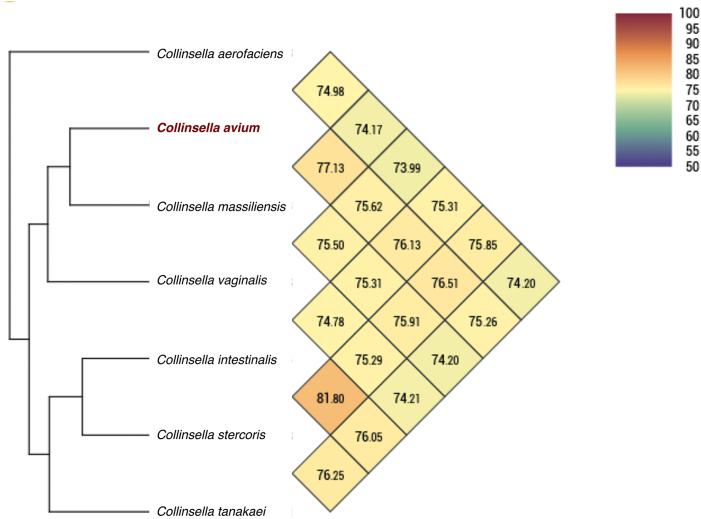
Fig. 7.
Average nucleotide identity comparison of Collinsella avium strain SW219 and related species with a valid taxonomy. Heatmap generated with OrthoANI values calculated using the OAT software between Collinsella avium sp. nov. and the closely related species.
Discussion
Recently, the high throughput culturing of gut microbiota known as “culturomics” have been developed to characterise and identify unculturable bacteria, allowing isolation of several new species from the gut microbiota [25,26]. Although metagenomics is the primary strategy for the study of gut microbiota [27], the large portion of gut microbiota are uncultured organisms [5]. Culturing of bacteria is important to understand their functions, and contributions in health and diseases [28,29]. In this study, culturomics based isolation recovered a previously uncultured bacterium SW219 from the ceacum of feral chickens.
Generally, 16S rRNA gene sequence similarity is primarily used to determine the taxonomic threshold for determining new species [24]. The 16S rRNA-based phylogenetic analysis showed that C. massiliensis was the closest related species of the strain SW219 with 97.88% similarity. A 16S rRNA identity cutoff value of up to 98.6% can be used to designate new isolate as a distinct species [30]. Furthermore, the ANI and dDDH values of the strain SW219 is markedly different when compared to its closest genomes (Fig. 5, Fig. 6). In addition, strain SW219 exhibited distinct phenotypic properties to its closest valid strain, including carbon source utilisation, enzymatic activity and cellular fatty acid components (Table 1, Table 2).
Therefore, strain SW219 could be classified as a new species within the genus Collinsella.
Conclusion
Based on the genotypic and phenotypic features of the strain SW219, this organism identified herein should be described as a new species of the genus Collinsella. This organism is proposed as the type strain of the new species, for which the name is Collinsella avium is proposed.
Description of Collinsella avium sp. nov
Collinsella avium sp. nov. (a'vi.um. L. gen. pl. n. avium, of birds). Collinsella avium is an obligately anaerobic, non-motile and Gram-positive rod bacterium. The optimal growth of this strain is at 37o C and pH 7 in BHI-M medium. The average size of each cell is 0.5-2.0 μm. Colonies are visible on BHI-M agar after 2-3 days and are approximately 0.1-0.3 cm in diameter, with white translucent, round and convex with entire circular margin. Positive enzyme activity of this organism includes alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, valine arylamidase, cystine arylamidase, acid phosphatase, naphthol-As-Bi-phosphopydrolase, α-galactosidase, α-glucosidase and N-acetyl-β-glucosaminidase. The major fatty acids are C16: 0 DMA, C16: 0 aldehyde, C16: 0 and C12: 0. The genome of strain SW219 is 2.58 Mbp with 64.5 mol% of G+C content. The type strain is SW219 (= DSM 109235T) isolated from ceacum of feral chickens.
Nucleotide sequence accession number
The 16S rRNA gene was deposited in GenBank under accession number MN099104. Whole genome sequence was deposited in NCBI Sequence Read Archive under the accession number SRR9967483.
Deposit in culture collection
Strain SW219 was deposited in The Leibniz Institute DSMZ—German Collection of Microorganisms and Cell Cultures GmbH and The national Culture Collection of Switzerland (CCOS) under numbers DSM 109235 and CCOS 1884, respectively.
Transparency declaration
None to declare.
This work was supported in part by the USDA National Institute of Food and Agriculture, Hatch projects SD00H532-14 and SD00R540-15, and a grant from the South Dakota Governor’s Office of Economic Development awarded to JS.
Acknowledgements
SW would like to acknowledge the Science Achievement Scholarship of Thailand. The authors acknowledge Electron Microscopy Core Facility at the Bowling Green State University, Ohio, USA, for assistance with scanning electron microscopy. The authors would like to thank Linto Antony and Roshan Kumar, Veterinary and Biomedical Science Department, South Dakota State University, USA, for their assistance in genome sequencing and analysis.
References
- 1.Brestoff J.R., Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14(7):676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khosravi A., Yanez A., Price J.G., Chow A., Merad M., Goodridge H.S. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe. 2014;15(3):374–381. doi: 10.1016/j.chom.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morgan X.C., Huttenhower C. Meta'omic analytic techniques for studying the intestinal microbiome. Gastroenterology. 2014;146(6):1437–14348 e1. doi: 10.1053/j.gastro.2014.01.049. [DOI] [PubMed] [Google Scholar]
- 4.Greub G. Culturomics: a new approach to study the human microbiome. Clin Microbiol Infect. 2012;18(12):1157–1159. doi: 10.1111/1469-0691.12032. [DOI] [PubMed] [Google Scholar]
- 5.Lagier J.C., Khelaifia S., Alou M.T., Ndongo S., Dione N., Hugon P. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Lane D.J. 16S/23S rRNA sequencing. In: Stackebrandt E., Goodfellow M., editors. Nucleic acid techniques in bacterial systematic. John Wiley and Sons; New York: 1991. [Google Scholar]
- 7.Stackebrandt E., Liesack W. Nucleic acids and classification. In: Goodfellow M., O'Donnell A.G., editors. Handbook of new bacterial systematics. Academic Press; London, England: 1993. [Google Scholar]
- 8.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim O.S., Cho Y.J., Lee K., Yoon S.H., Kim M., Na H. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012;62(Pt 3):716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- 10.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson J.D., Higgins D.G., Gibson T.J., Clustal W. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17(6):368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 13.Fitch W.M. Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool. 1971;20(10):406–416. [Google Scholar]
- 14.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 15.Tittsler R.P., Sandholzer L.A. The use of semi-solid agar for the detection of bacterial motility. J Bacteriol. 1936;31:575–580. doi: 10.1128/jb.31.6.575-580.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sasser M. MIDI technical note 101 Newark. DE: MIDI Inc; 1990. Identification of bacteria by gas chromatography of cellular fatty acids. [Google Scholar]
- 17.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13(6) doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carver T., Thomson N., Bleasby A., Berriman M., Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25(1):119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Overbeek R., Olson R., Pusch G.D., Olsen G.J., Davis J.J., Disz T. The SEED and the Rapid annotation of microbial genomes using subsystems Technology (RAST) Nucleic Acids Res. 2014;42(Database issue):D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robertson C.E., Harris J.K., Wagner B.D., Granger D., Browne K., Tatem B. Explicet: graphical user interface software for metadata-driven management, analysis and visualization of microbiome data. Bioinformatics. 2013;29(23):3100–3101. doi: 10.1093/bioinformatics/btt526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee I., Ouk Kim Y., Park S.C., Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66(2):1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- 23.Meier-Kolthoff J.P., Auch A.F., Klenk H.P., Goker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim M., Oh H.S., Park S.C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64(Pt 2):346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 25.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28(1):237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaeberlein T., Lewis K., Epstein S.S. Isolating "uncultivable" microorganisms in pure culture in a simulated natural environment. Science. 2002;296(5570):1127–1129. doi: 10.1126/science.1070633. [DOI] [PubMed] [Google Scholar]
- 27.Dhakan D.B., Maji A., Sharma A.K., Saxena R., Pulikkan J., Grace T. The unique composition of Indian gut microbiome, gene catalogue, and associated fecal metabolome deciphered using multi-omics approaches. Gigascience. 2019;8(3) doi: 10.1093/gigascience/giz004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goodman A.L., Kallstrom G., Faith J.J., Reyes A., Moore A., Dantas G. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci U S A. 2011;108(15):6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rettedal E.A., Gumpert H., Sommer M.O. Cultivation-based multiplex phenotyping of human gut microbiota allows targeted recovery of previously uncultured bacteria. Nat Commun. 2014;5:4714. doi: 10.1038/ncomms5714. [DOI] [PubMed] [Google Scholar]
- 30.Yarza P., Yilmaz P., Pruesse E., Glockner F.O., Ludwig W., Schleifer K.H. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol. 2014;12(9):635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]