Abstract
Cell-free DNA (cfDNA) from cerebrospinal fluid (CSF) offers unique opportunities for genomic profiling of tumors involving the central nervous system but remains uncommonly used in clinical practice. We describe our clinical experience using cfDNA from CSF for routine molecular testing using Memorial Sloan Kettering Integrated Mutation Profiling of Actionable Cancer Targets (targeting 468 cancer-related genes). In all, 148 cfDNA samples were assessed, comparing results of cfDNA versus genomic DNA (gDNA; gDNA from cell pellets) derived from the same CSF sample and the primary tumor. Of these, 71.6% (106/148) were successfully sequenced. Somatic alterations (mutations and fusions) were observed in 70.8% (75/106) of the samples; 97.3% (73/75) comprised variants confirming central nervous system involvement by a previously diagnosed tumor, 14.7% (11/75) had additional variants consistent with a therapy-related resistance mechanism, and 2.7% (2/75) had variants that independently diagnosed a new primary. Among samples with paired cfDNA and gDNA sequencing results, cfDNA was more frequently positive for at least one mutation [43.6% (55/126) versus 19.8% (25/126)] and harbored 1.6× more mutations (6.94 versus 4.65; P = 0.005), with higher mean variant allele fractions (41.1% versus 13.0%; P < 0.0001). Among mutation-positive cfDNAs, the corresponding gDNA was frequently negative (44.6%; 25/55) or failed sequencing (17.8%; 9/55). Routine molecular profiling of cfDNA is superior to gDNA from CSF, facilitating the capture of mutations at high variant allele frequency, even in the context of a negative cytology.
Genomic profiling of tumor tissue is an increasingly important component in the diagnosis and management of patients with cancer. For those patients with central nervous system (CNS) malignancies, whether primary or metastatic, obtaining tissue for comprehensive profiling carries numerous challenges, including high risk associated with the invasive procedure and limitations in the amount of tissue that can be obtained. In this context, liquid biopsies have emerged as a useful diagnostic approach, with the potential to circumvent many of the limitations of clinical tumor tissue–based testing.
Although plasma cell-free DNA (cfDNA) is often considered the prototype and most extensively studied sample type among liquid biopsies, the role of this DNA source may be limited in malignancies involving the CNS. In contrast to other tumors, CNS tumors generally deposit a lower proportion of circulating tumor DNA into the plasma, likely related to the presence of the blood-brain barrier.1,2 By contrast, cerebrospinal fluid (CSF), which also constitutes a readily accessible fluid through routine diagnostic lumbar puncture, is an appealing alternative with numerous advantages over plasma for genomic sequencing.
Because of its close contact with the brain, the CSF has been extensively studied for diagnostic purposes for >100 years. In neuro-oncology, morphologic detection of tumor cells (CSF cytology) is a routine test that is used for diagnosis, staging, and therapeutic decisions. Depending on the degree of involvement and the number of malignant cells recovered from the sample, tumor cells are also used for a wide array of diagnostic studies, including flow cytometry, immunohistochemistry, mass spectrometry, and molecular testing. The sensitivity of these studies, unfortunately, is often low because of the absence or scarcity of tumor cells. More recently, however, several studies have demonstrated CSF samples to be an important source of tumor-derived cfDNA.2, 3, 4, 5, 6, 7, 8, 9, 10 Compared with blood, the relatively acellular nature of CSF results in lower background level of nonneoplastic cfDNA, enabling mutation detection at high variant allele frequencies (VAFs), which could be detected with routine profiling assays and standard analysis pipelines, even in the context of relatively low sequence coverage because of low cfDNA levels.
At present, although several studies demonstrate the feasibility and analytical validity of CSF-derived cfDNA for genomic profiling using both targeted non-NGS and NGS approaches, most studies remain small and retrospective in nature.10, 11, 12, 13, 14, 15, 16 Reports of CSF cfDNA testing in clinical practice, although limited, have shown great promise, particularly among metastatic tumors.15 In a previous study, as a proof of principle, our group demonstrated the ability of our targeted next-generation sequencing assay [Memorial Sloan Kettering Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT)] to detect somatic mutations with high confidence in a large percentage of samples from patients with primary and metastatic CNS tumors.10 In this report, we describe our clinical experience using CSF liquid biopsies following validation and implementation in routine molecular profiling. We discuss the clinical utility of sequencing cfDNA from the fluid compartment of CSF versus genomic DNA (gDNA) derived from the cellular component and highlight important benefits and challenges associated with each approach (Figure 1).
Figure 1.
Overview of Memorial Sloan Kettering Integrated Mutation Profiling of Actionable Cancer Targets cerebrospinal fluid (CSF) cell-free DNA (cfDNA) sequencing results and utility. CNS, central nervous system.
Materials and Methods
Sample Acquisition
Our study examined CSF samples obtained as standard of care on patients with suspected CNS involvement by primary or metastatic malignancies and who were treated at Memorial Sloan Kettering Cancer Center from February 2018 to March 2019. All patients signed informed consent under protocols approved by the Memorial Sloan Kettering Cancer Center Institutional Review Board. Samples were submitted for cytologic evaluation as clinically indicated. Aliquots (3 to 10 mL) from the same procedure were collected in sterile body fluid containers or STRECK tubes and submitted for molecular studies. Specimens received in sterile containers were processed immediately on arrival, whereas those in STRECK tubes were stored at room temperature and processed within 24 hours of receipt. A blood sample was also obtained from all patients as matched normal control for sequencing.
DNA Extraction
Each CSF sample was separated from its cellular components using double centrifugation (10 minutes at 1600 × g and 10 minutes at 3000 × g at 22°C). cfDNA was manually extracted from the corresponding fluid compartment using MagMax cfDNA Kits (Thermo Fisher Scientific, Waltham, MA), according to manufacturer's protocol. Briefly, cfDNA fragments were bound to silica paramagnetic beads in the presence of proteinase K, guanidine thiocyanate (chaotropic salt), and nonionic surfactants. The supernatant was discarded, while the magnetic bead pellets underwent several washes with solutions of ethanol and guanidine thiocyanate. cfDNA was eluted from the beads in a low ionic solution or water. Isolation of gDNA from cell pellet material, formalin-fixed, paraffin-embedded tumor samples, and patient-matched normal blood was performed using published protocols.17 gDNA was quantified using the Quant-iT dsDNA Broad Range Assay Kit (Thermo Fisher Scientific, Waltham, MA) with a SpectraMax M2 Fluorescence Microplate Reader (Molecular Devices, San Jose, CA); cfDNA was quantified using High Sensitivity D1000 Screen Tape and corresponding reagents, which are loaded onto a 2200 Agilent Tape Station (Agilent, Santa Clara, CA).
Testing and Data Analysis
Molecular analysis was performed using MSK-IMPACT, a Food and Drug Administration–authorized and New York State Department on Health–approved targeted next-generation sequencing–based assay, which captures all protein-coding exons of 468 cancer-associated genes and select introns. Sequencing was performed as previously described; each sample paired with a genetically matched normal sample.16 Input cfDNA samples were not sheared before library preparation. Samples where somatic variants were detected were called positive. Those with at least 50× sequencing coverage and without detected variants were considered negative. Finally, samples were annotated as failed when no variants were detected, and coverage was <50×. Records were retrospectively reviewed for any molecular profiling previously performed on corresponding tumor tissue, and results were correlated with those of the CSF. Data source for reference sequence transcripts specifications from RefSeq (National Center for Biotechnology Information) is available (https://www.ncbi.nlm.nih.gov/refseq, last accessed December 11, 2020).18, 19, 20
Results
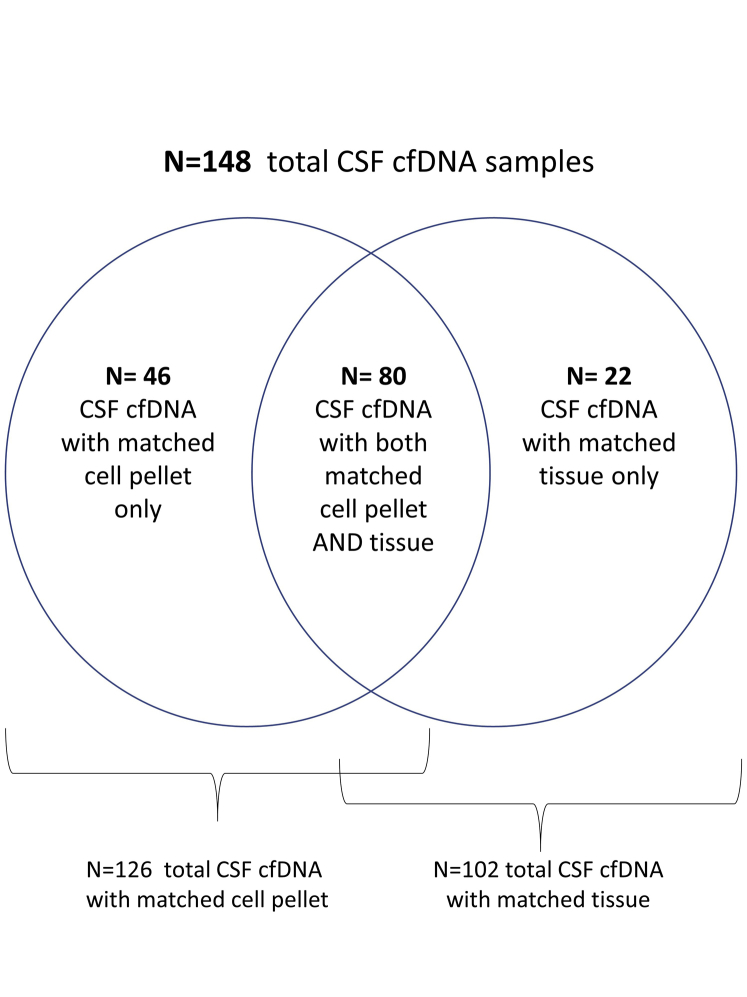
Overall, sequencing results from 148 cfDNA samples (137 patients) were compared with those of gDNA from matched CSF cell pellet (n = 126) and/or previously characterized formalin-fixed, paraffin-embedded tumor (n = 102). The entire cohort assembly is summarized in Supplemental Figure S1. A total of 75 of 148 cfDNA samples were positive for at least one tumor variant. On average, 6 variants were detected per positive cfDNA sample (range, 1 to 79 variants); 97.3% of cases (73/75) included cfDNA variants that confirmed the expected disease diagnosis, whereas 2.7% (2/75) harbored variants that independently diagnosed a new (unexpected) primary. Additional variants consistent with a therapy-related resistance mechanism were also demonstrated in 11 of 75 cases (14.7%) (Figure 1).
Overall Assay Performance
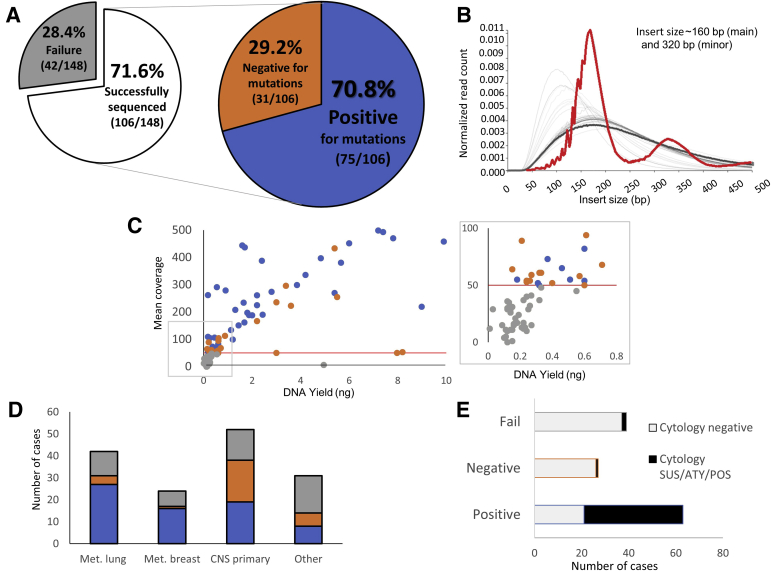
In all, the median total nucleic acid yields for cfDNA and cell pellet gDNA were 0.55 ng (range, 0.0 to 253.55 ng) and 49.3 ng (range, 0.0 to 2920.0 ng), respectively. Of the 148 cfDNA samples, 71.6% (106/148) were successfully sequenced and 28.4% failed sequencing (42/148) (Figure 1 and Figure 2A). Mean sequencing depth was 283× (range, 50× to 1192×), with mean insert sizes at approximately 160 bp (main peak) and 320 bp (minor peak), consistent with previously reported results for other cfDNA specimens (Figure 2B).21,22 A total of 70.8% (75/106) samples were positive, spanning a variety of tumor diagnoses (Figure 2, A and D). In general, positive samples had higher DNA input, compared with negative and failed cases (mean, 13.06, 2.72, and 0.31 ng, respectively), but the difference in DNA input was not statistically significant (P = 0.05, one-way analysis of variance) (Figure 2C). Of the 42 cases that failed sequencing, 39 had a DNA input of <0.3 ng.
Figure 2.
Memorial Sloan Kettering Integrated Mutation Profiling of Actionable Cancer Targets assay performance. A: Sequencing success rate was 71.6% (106/148 cases with quantifiable input DNA), whereas failure rate was 28.4% (42/148, no variants detected, coverage <50×). Somatic alterations were observed in cell-free DNA (cfDNA) in 70.8% (positive, 75/106) of successful cases. B: Insert size distribution of simultaneously sequenced samples shows that the single cerebrospinal fluid (CSF) cfDNA sample (highlighted in red) demonstrates insert sizes at approximately 160 bp (main) and 320 bp (minor), consistent with previously reported results for other cfDNA specimens, distinguishing them from formalin-fixed, paraffin-embedded tumor samples (light gray) and paired normal blood (dark gray). C: Mean coverage versus sample DNA yield (positive, blue; negative, orange; fail, gray); although failure rates were higher among low-concentration cases (less than approximately 0.3 ng), adequate coverage (>50×; red line) and positive results could still be obtained for some samples, even at low cfDNA levels (inset). D: CSF cfDNA results by tumor type (positive, blue; negative, orange; fail, gray). E: CSF cytology correlations with cfDNA results. ATY, atypical; CNS, central nervous system; Met., metastatic; POS, positive for malignant cells; SUS, suspicious.
CSF cfDNA Results by Tumor Type
Most positive cfDNA cases were from metastatic tumors [66%, 48/75: predominantly of lung (37.3%, 28/75) and breast (21.3%, 16/75) origin]. A sizable remainder (30.7%, 23/75) were from primary neuroepithelial tumors [including high-grade glioma (n = 19), atypical teratoid rhabdoid tumor (n = 1), low-grade glioma (n = 1), and pineoblastoma (n = 2)]. Among the negative cases (n = 31), those with a history of primary neuroepithelial tumors accounted for 64.5% (20/31) of the cases, followed by lung (n = 5), breast (n = 2), sarcoma (n = 2), melanoma (n = 1), and squamous cell carcinoma of unknown origin (n = 1). Failed cases included primary neuroepithelial tumors (35.7%, 15/42), followed by metastatic carcinoma [lung (n = 8), breast (n = 6), and pancreatic (n = 1)], atypical lymphoid proliferations (n = 3), melanoma (n = 1), and sarcoma (n = 1). Five cases with a history of abnormal magnetic resonance imaging without documented evidence of disease elsewhere in the body also resulted in sequencing failure (Figure 2D).
Among all patients with low-grade primary CNS tumors, 88% (15/17) were negative [low-grade glioma (n = 6) and choroid plexus papilloma (n = 1)] or failed sequencing [low-grade glioma (n = 7) and choroid plexus papilloma (n = 1)]. In contrast, among the high-grade tumors, 58.3% were positive, whereas only 25% were negative [9/36; high-grade glioma (n = 5) and medulloblastoma (n = 4)] and 16.7% failed sequencing [6/36; high-grade glioma (n = 4), choroid plexus carcinoma (n = 1), and medulloblastoma (n = 1)].
CSF Cytology Correlations with cfDNA Results
In all, 84% (63/75) of the positive cfDNA samples had a corresponding CSF sample submitted for cytologic assessment. Of these, 66.7% (42/63) demonstrated abnormal cytology (32 positive for malignant cells and 10 suspicious/atypical), whereas 33.3% (21/63) were cytologically benign. Among positive cfDNA samples with abnormal CSF cytology, 88% (37/42) were from metastatic tumors; primary neuroepithelial tumors encompassed merely 7.9% [4 positive for malignant cells (1 glioblastoma, 1 atypical teratoid rhabdoid tumor, 1 pineoblastoma, and 1 low-grade glioma); 1 atypical (glioblastoma)]. Most positive CSF cfDNA cases with benign CSF cytology were from primary neuroepithelial tumors (57%).
Among the 31 negative cfDNA samples, 27 had concurrent cytologic assessment. Of these, only one had abnormal CSF cytology (atypical, low-grade glioma). Similarly, among cfDNA samples that failed sequencing (n = 39), only two cases demonstrated abnormal CSF cytology [positive for malignant cells (lung adenocarcinoma, n = 1) and atypical (atypical lymphoid proliferation, n = 1)].
In short, sequencing of CSF cfDNA from samples with abnormal cytology (n = 45) was consistently positive for tumor mutations, with only three exceptions. However, benign CSF cytology was not a reliable indicator of the presence of tumor-derived cfDNA, as approximately a third of positive CSF cfDNA sequencing resulted from apparently cytologically benign CSF samples (Figure 2E).
Comparison of cfDNA and Baseline Tumor Sequencing Results
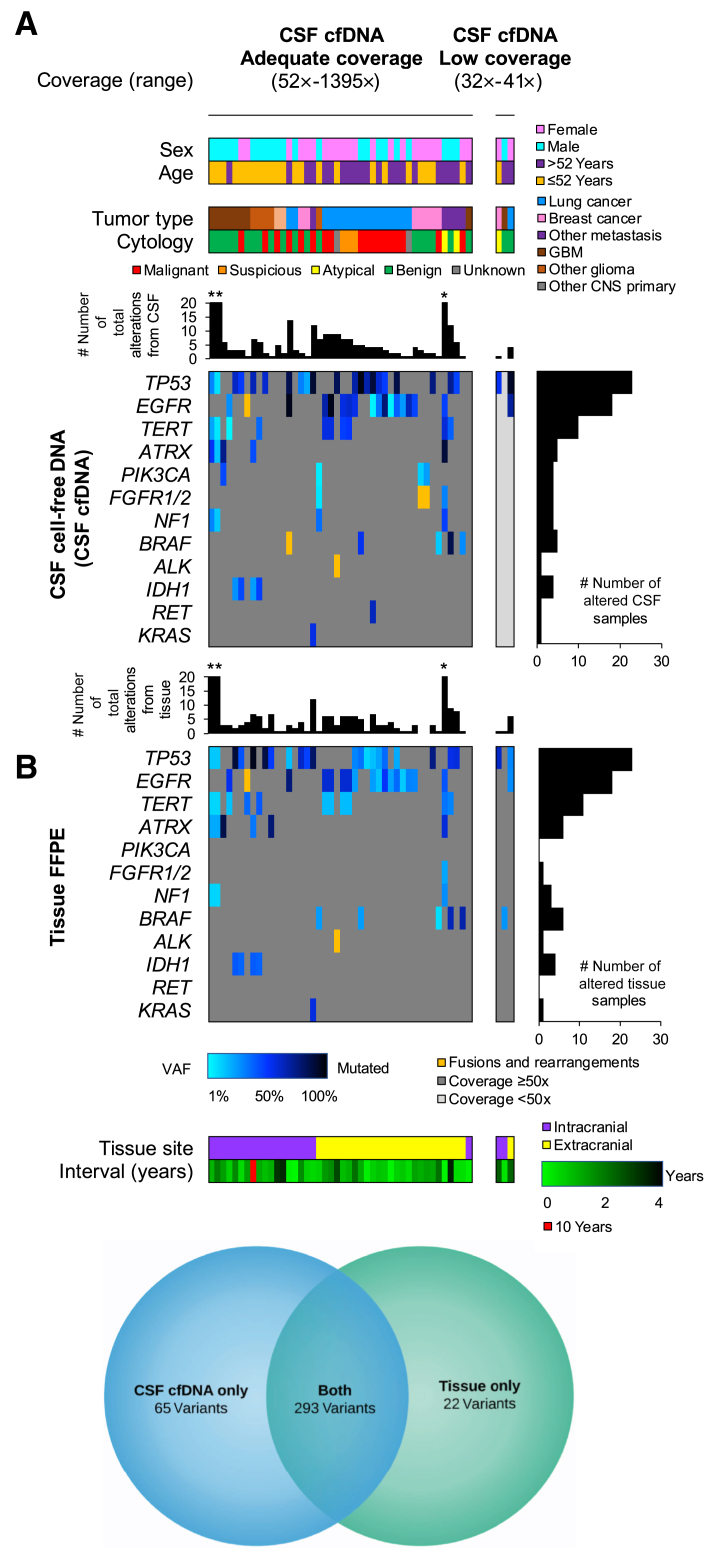
Among positive CSF cfDNA samples, 47 had a prior tumor profiled by MSK-IMPACT that could be used as baseline for comparison. This included 21 and 26 tumors from the CNS and other locations, respectively. The median interval time from biopsy to CSF sampling was 14.1 months (range, 0.30 to 121.9 months). CSF cfDNA variants were also identified in the tissue sequencing results in 93.6% cases (n = 44/47) (Figure 3A); driver mutations were also included among the identified variants. In two of three discrepant samples, CSF sampling was performed after extensive treatment history, with uncertain effect on the tumor status (Patients 1 and 2). A third sample from a patient (Patient 3) with history of tall cell variant papillary thyroid carcinoma and a brainstem mass (suspected to be metastatic) demonstrated a mutation profile that was distinct and consistent with a new primary, best classified as midline glioma, including an H3 mutation (H3FB p.K28I). Details are summarized in Supplemental Table S1. In all, of the 358 variants detected in CSF cfDNA samples with baseline tumor sequencing, 293 were also identified in the tissue. Twenty-two variants detected in tissue samples were not detected in the CSF cfDNA (Figure 3B).
Figure 3.
Comparison of cerebrospinal fluid (CSF) cell-free DNA (cfDNA) results with prior tumor sequencing. A: Integrated pathologic and molecular data for 47 positive CSF cases with paired cfDNA and genomic DNA (gDNA) from formalin-fixed, paraffin-embedded (FFPE) tissue. Cases are grouped according to the cfDNA results: cases with ≥50× (top left panel) and <50× (top right panel) median sample coverage. Each column represents a single CSF and tissue pair; the paired CSF cfDNA (middle panels) and FFPE tissue gDNA (bottom panels) data are provided. The total number of alterations includes sequence mutations and structural variants (ie, fusions), whereas the gene panel shows only the recurrently altered genes. The asterisk indicates three individual cases with >20 alterations (left to right): 31, 22, and 79 alterations for CSF cfDNA samples (middle panel) and 23, 23, and 81 alterations for FFPE tissue samples (bottom panel). B: Venn diagram demonstrating that of the 358 variants detected in CSF cfDNA samples with baseline tumor sequencing, 293 were also identified in the tissue. Twenty-two variants detected in tissue samples were not detected in the CSF cfDNA. CNS, central nervous system; GBM, glioblastoma; VAF, variant allele frequency.
Comparison of Paired CSF cfDNA and gDNA from CSF Cell Pellet
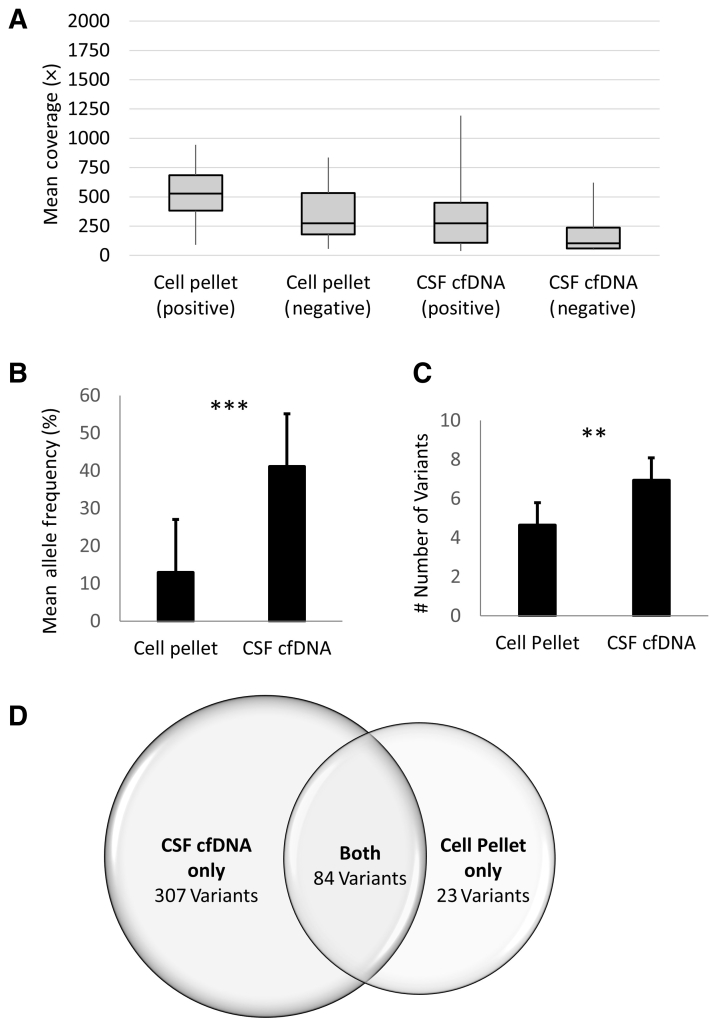
Mean coverage for the CSF-cfDNA samples (317× positive, 183× negative) was slightly lower than that achieved for matched cell pellets (345× positive, 359× negative), but the difference was not statistically significant (P = 0.02, one-way analysis of variance) (Figure 4A). In total, 391 somatic variants were detected in all cfDNA samples (range, 1 to 79) compared with 107 (range, 1 to 16) in the pellets. The cfDNA was positive in 44.4% (56/126), compared with 19.8% (25/126), of the cell pellets. In cases where both cell pellet and cfDNA samples were positive, shared mutations were identified in 100% of the cases. Among positive cfDNA samples, 55.4% (31/56) of the corresponding pellets yielded a negative result or failed sequencing; only one cell pellet harbored additional mutations not identified in the corresponding cfDNA (variants of uncertain significance in FGF4 and KMT2B) (Patient 4) (Figure 5 and Supplemental Table S1). Outside of this single exception, in samples with both positive cfDNA and positive cell pellet, all variants detected in cell pellets were present, and at a significantly higher variant allele fraction in the corresponding cfDNA (mean VAF ± SEM, 42.5% ± 0.54% CSF cfDNA versus 12.0% ± 0.25% cell pellet; P < 0.0001, paired t-test) (Figures 4B and5). Moreover, among positive samples, an average of 1.6× more variants were detected from cfDNA (means ± SEM, 6.98 ± 1.31 versus 4.28 ± 0.9; P = 0.005, paired t-test) (Figure 4C). In all, of the 391 variants detected in CSF cfDNA, only 84 were also detected in the corresponding cell pellet; only 23 variants detected in the cell pellet were not detected in cfDNA (Figure 4D).
Figure 4.
Comparison of the 126 cell-free DNA (cfDNA) samples paired with the corresponding cell pellet. A: Mean coverage for the cerebrospinal fluid (CSF)–cfDNA samples (317× positive, 183× negative) was slightly lower than that achieved for matched cell pellets (345× positive, 359× negative), but the difference was not statistically significant (P = 0.02, one-way analysis of variance). B: Despite similar coverage, variants in cfDNA samples were detected at significantly higher variant allele fraction compared with matched cell pellet (42.5% ± 0.54% CSF cfDNA versus 12.0% ± 0.25% cell pellet). C: On average, 1.6× more variants were detected from CSF cfDNA (6.98 ± 1.31 versus 4.28 ± 0.9). In total, 391 somatic variants were detected in all cfDNA samples (range, 1 to 79) compared with 107 (range, 1 to 16) in the pellets. D: Venn diagram demonstrating that only 84 of the 391 variants detected in cfDNA were also identified in the cell pellet, whereas 23 variants detected in cell pellets were not detected in CSF cfDNA. Data are given as means ± SEM (B and C). ∗∗P = 0.005, ∗∗∗P < 0.0001 (paired t-test).
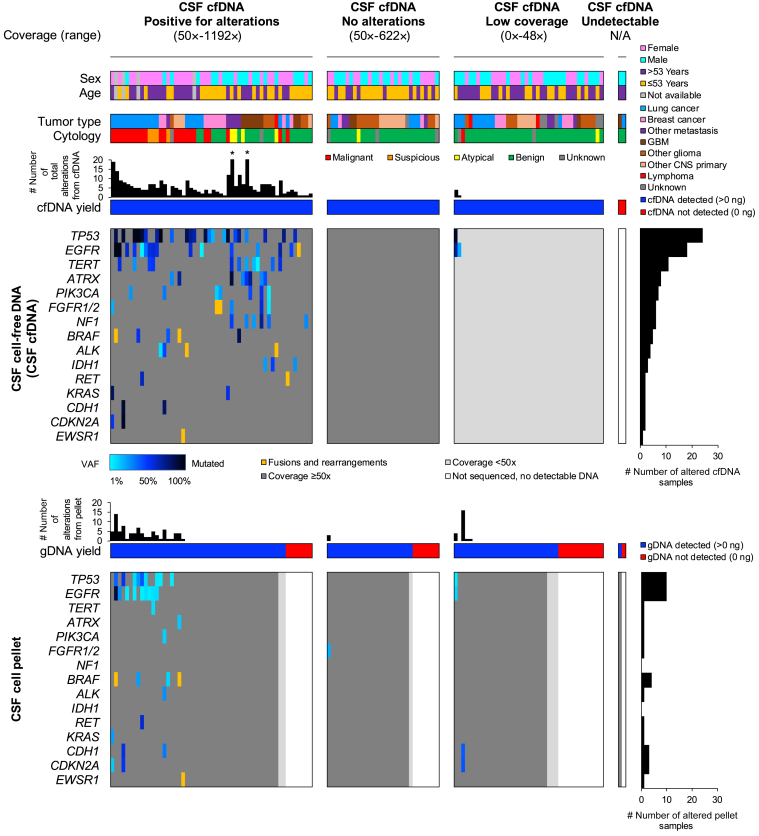
Figure 5.
Integrated pathologic and molecular data for 126 cerebrospinal fluid (CSF) cases with paired cell-free DNA (cfDNA) and cell pellet genomic DNA (gDNA). Top panels: Cases are grouped according to the cfDNA results (left to right): i) alterations detected with ≥50× median sample coverage, ii) no alterations detected with ≥50× median sample coverage, iii) all cases with <50× median sample coverage, and iv) cases not sequenced because of undetectable cfDNA (0 ng). Each column represents a single CSF sample; the paired cfDNA (middle panels) and cell pellet gDNA (bottom panels) data are provided. The total number of alterations includes sequence mutations and structural variants (ie, fusions), whereas the gene panel shows only the most recurrently altered genes. The asterisk indicates cases with >20 alterations: 79 and 31 alterations (left and right, respectively). CNS, central nervous system; GBM, glioblastoma; N/A, not available; VAF, variant allele frequency.
Comparison of the 126 cfDNA samples paired with the corresponding cell pellet is presented in Figure 5. Overall, 23.8% (30/126) and 53.2% (67/126) of the cfDNA and cell pellet samples, respectively, were negative; 31.7% (40/126) of cfDNA and 26.2% (33//126) cell pellets failed sequencing or had insufficient input DNA. Despite low coverage (<50×), two of the cfDNA samples demonstrated variants that could still be called with confidence because of their high VAF. Only three cell pellets harbored tumor variants when the corresponding cfDNA sample was negative or failed; two were low-grade gliomas (pilomyxoid astrocytoma and oligodendroglioma), and one was from a lung adenocarcinoma with leptomeningeal metastases (Patients 5, 6, and 7) (Supplemental Table S1). In all three instances, cfDNA input (0.21, 0.18, and 0.12 ng) and coverage were low (median, 89×, 17×, and 30×).
Last, two cases were observed where cfDNA sequencing was positive, which highlighted the clinical utility of cfDNA testing even in the absence of available tissue or cell pellet sequencing results for comparison. First, in a sample from a patient with a known history of metastatic colon adenocarcinoma (without molecular characterization) and negative cell pellet sequencing, cfDNA sequencing demonstrated variants consistent with a new unexpected primary high-grade glioma (Patient 8) (Supplemental Table S1). Similarly, in a patient with no past medical history, magnetic resonance imaging evidence of diffuse leptomeningeal enhancing disease, and an atypical polymorphous lymphoid population on CSF cytology (flow cytometry negative), CSF cfDNA demonstrated an ALK rearrangement, consistent with anaplastic large-cell lymphoma; diagnosis was later confirmed on nerve fascicle biopsy (Patient 9) (Supplemental Table S1).
Discussion
Although CSF assessment has been a key component of the routine evaluation of patients with suspected CNS involvement by primary or metastatic disease for over a century, the overall clinical utility has been limited by the reliance on microscopic tumor cell detection only. Even in the context of significant disease involvement, tumor cells in CSF are commonly scant or absent and, even when present, the morphologic features may be insufficient to establish a definitive diagnosis. Although the fluid itself has been traditionally neglected in the context of malignancy workup, recent studies demonstrate that this is a rich source of tumor-derived DNA.12,13,23,24 Herein, almost a third of the positive cfDNA cases resulted from cases with a corresponding normal CSF cytology. Even for those samples in which the cell pellet was positive for tumor cells on corresponding cytology, sequencing of cell pellet gDNA frequently failed because of low yield, was mutation negative, or harbored mutations in significantly lower number and at lower VAF, compared with the corresponding cfDNA. Inasmuch as VAF is a surrogate for tumor purity, this dilutional effect could be, at least partially, related to the presence of T cells and/or contaminating cells from relatively bloody taps. The rare exceptions where the cell pellet sample was more informative than the cfDNA could be attributed to low input DNA from the cfDNA specimen. Two of the three instances where this was the case were from low-grade glial tumors, where cell turnover, and consequently cfDNA shedding, is expected to be low and may be more amenable to alternative testing modalities.16 Across the vast majority of samples in our cohort, our findings support that the sequencing of cell pellets, even when morphologically positive for tumor and with higher nucleic acid yield, has minimal clinical utility compared with the cfDNA and, as a result, paired sequencing of the cell pellet has been discontinued in our laboratory.
As cancer diagnosis and therapy is increasingly guided by genomic sequencing results, CSF liquid biopsies can offer unique opportunities to recapitulate mutation profile of tumors as they exist in the central nervous system space. CSF can be more readily and safely accessed compared with tissue biopsy, both at diagnosis and at multiple time points throughout the course of therapy and disease monitoring. Several small, retrospective, and proof-of-concept studies have been published, demonstrating the utility of this approach.9,12,15,24 Reports of incorporation of CSF cfDNA analysis into routine molecular diagnostics are limited but encouraging, particularly among patients with progressive, metastatic disease.15
This current study demonstrates that despite the common challenges of low nucleic acid yield, implementation of comprehensive sequencing of CSF cfDNA for routine molecular testing is feasible and successful in a high proportion of cases. Given the relative acellular nature of CSF, the nonneoplastic background cfDNA component is low in these samples, allowing the detection of mutations at high VAFs with routine profiling assays, depending on the design. Unlike plasma-based liquid biopsy approaches, our existing clinical hybridization capture-based assay designed for tumor tissue profiling, MSK-IMPACT, was utilized for CSF cfDNA sequencing, without the need for additional technical modifications or disruptions to the laboratory workflow and with the standard analysis pipeline already employed for formalin-fixed, paraffin-embedded tissue and matched normal specimens.10,16
Although an extensive validation of MSK-IMPACT had been previously performed for tumor tissue profiling assay, as previously described, based on New York State Department on Health requirements, additional validation procedures had to be performed to incorporate the use of cfDNA as a sample type for our assay.17,25 Given the known limitations of cfDNA yield extracted from clinical CSF samples, standard accuracy, reproducibility, and sensitivity studies that confirm results using an alternate platform, or multiple replicate testing on the same sample, were precluded. Instead, a modified validation study was conducted, encompassing the initial 33 samples in this cohort that demonstrated similar accuracy and performance characteristics of the larger set described herein (Supplemental Tables S2 and S3). On the basis of these data and, as of March 2019, clinical sequencing of CSF cfDNA using the MSK-IMPACT platform was approved by New York State Department on Health.
The clinical implementation of CSF cfDNA sequencing offers unique opportunities for increased diagnostic precision. In patients with clinical suspicion of metastatic disease, sequencing confirmed CNS involvement by their primary tumor in 66% of cases, 96% of which confirmed the original mutational profile when the primary tumor was available for comparison. In several cases, additional alterations that were absent in baseline tumor were identified in the CSF, reflecting the development of a resistance mechanism, clonal evolution, or tumor heterogeneity.26
In several cases, sequencing uncovered new and unexpected primaries that changed the type and course of treatment. For some patients, results helped establish a diagnosis based on the unique alterations identified, including both mutations and fusions. With clinical implementation of this assay, we have continued to expand the sequencing efforts as part of routine clinical care and for clinical trials with sampling increasingly being incorporated as an adjunct and, in some cases, as a suitable alternative to traditional diagnostics when a biopsy is not feasible, or sequencing of the tumor yielded a failure.
One important consideration for CSF cfDNA testing is the balancing of the established sensitivity of a given assay with the clinical need for higher sensitivity assays. Although sequencing by MSK-IMPACT provides the capability to study samples at the time of high clinical suspicion of CNS involvement, for monitoring of diseases with low shedding and for tracking low disease level and alterations with low allele frequencies, higher sensitivity methods may be required.27 Further studies that systematically address pre-analytic factors (including specimen handling and processing times) with the goal of further optimizing sample yield, quality, and sequencing results are underway.
Finally, although the validity and utility of positive CSF cfDNA results in this study have been emphasized, the clinical utility of negative results should not be mitigated. Prior work has suggested that the presence of cfDNA alone could be a negative prognostic indicator.13,16 Meanwhile, sustained responses to therapy have been associated with clearance of tumor cfDNA from the CSF.28 This is similar to findings seen in cfDNA applications in other liquid biopsy specimens.1,29, 30, 31, 32 In addition, during the course of therapy, CNS lesions are frequently observed to increase in size and change in magnetic resonance imaging characteristics; prospective studies addressing CSF cfDNA findings in distinguishing true disease progression from treatment-related changes, independent of tissue biopsy, are now possible and needed.33, 34, 35 The standards for incorporation of CSF cfDNA sequencing results are rapidly evolving as this testing modality is increasingly routinely employed in clinical care.
Conclusions
Cell-free DNA from CSF is emerging as a powerful tool in the assessment of patients with tumors of the central nervous system, with both primary and metastatic disease. Because of the enrichment of tumor-derived cfDNA in CSF, despite low overall nucleic acid yield, mutations can be captured at high VAF even in the context of a negative cytology.
Acknowledgments
We thank the members of the Molecular Diagnostics Service in the Department of Pathology; the Memorial Sloan Kettering (MSK) Kids Pediatric Translational Medicine Program for support with cerebrospinal fluid collections and matched MSK Integrated Mutation Profiling of Actionable Cancer Targets tumor analyses, with support from the Scarlett Fund; and Shadia Carlo and Maureen Flaherty for administrative assistance in preparation of this article.
Footnotes
Supported in part by the Marie-Josée and Henry R. Kravis Center for Molecular Oncology and the National Cancer Institute Cancer Center Core grant P30-CA008748.
R.B. and M.E.A. contributed equally to this work.
Disclosures: S.C. has received personal consulting fees from Lilly, Novartis, and Paige.ai. and research funds to the institution from Daiichi-Sankyo. H.Y. has consulted for AstraZeneca, Daiichi, Blueprint Medicine, and Janssen. She has research funding to the institution from AstraZeneca, Daiichi, Pfizer, Lilly, Novartis, and Cullinan.
Supplemental material for this article can be found at https://doi.org/10.1016/j.jmoldx.2021.03.001.
Supplemental Data
Supplemental Figure S1.
Cohort assembly. Overall, sequencing results from 148 cell-free DNA (cfDNA) samples from 137 patients were compared with those of genomic DNA (gDNA) from matched cerebrospinal fluid (CSF) cell pellet and/or previously characterized formalin-fixed, paraffin-embedded (FFPE) tumor. n = 126 results of gDNA from matched CSF cell pellet; n = 100 results of previously characterized FFPE tumor.
References
- 1.Bettegowda C., Sausen M., Leary R.J., Kinde I., Wang Y., Agrawal N. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra224. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y., Springer S., Zhang M., McMahon K.W., Kinde I., Dobbyn L., Ptak J., Brem H., Chaichana K., Gallia G.L., Gokaslan Z.L., Groves M.L., Jallo G.I., Lim M., Olivi A., Quinones-Hinojosa A., Rigamonti D., Riggins G.J., Sciubba D.M., Weingart J.D., Wolinsky J.P., Ye X., Oba-Shinjo S.M., Marie S.K., Holdhoff M., Agrawal N., Diaz L.A., Jr., Papadopoulos N., Kinzler K.W., Vogelstein B., Bettegowda C. Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc Natl Acad Sci U S A. 2015;112:9704–9709. doi: 10.1073/pnas.1511694112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhodes C.H., Honsinger C., Sorenson G.D. PCR-detection of tumor-derived p53 DNA in cerebrospinal fluid. Am J Clin Pathol. 1995;103:404–408. doi: 10.1093/ajcp/103.4.404. [DOI] [PubMed] [Google Scholar]
- 4.Swinkels D.W., de Kok J.B., Hanselaar A., Lamers K., Boerman R.H. Early detection of leptomeningeal metastasis by PCR examination of tumor-derived K-ras DNA in cerebrospinal fluid. Clin Chem. 2000;46:132–133. [PubMed] [Google Scholar]
- 5.Shingyoji M., Kageyama H., Sakaida T., Nakajima T., Matsui Y., Itakura M., Iuchi T., Yokoi S., Kimura H., Iizasa T. Detection of epithelial growth factor receptor mutations in cerebrospinal fluid from patients with lung adenocarcinoma suspected of neoplastic meningitis. J Thorac Oncol. 2011;6:1215–1220. doi: 10.1097/JTO.0b013e318219aaae. [DOI] [PubMed] [Google Scholar]
- 6.Chen W.W., Balaj L., Liau L.M., Samuels M.L., Kotsopoulos S.K., Maguire C.A., Loguidice L., Soto H., Garrett M., Zhu L.D., Sivaraman S., Chen C., Wong E.T., Carter B.S., Hochberg F.H., Breakefield X.O., Skog J. BEAMing and droplet digital PCR analysis of mutant IDH1 mRNA in glioma patient serum and cerebrospinal fluid extracellular vesicles. Mol Ther Nucleic Acids. 2013;2:e109. doi: 10.1038/mtna.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y., Pan W., Connolly I.D., Reddy S., Nagpal S., Quake S., Gephart M.H. Tumor DNA in cerebral spinal fluid reflects clinical course in a patient with melanoma leptomeningeal brain metastases. J Neurooncol. 2016;128:93–100. doi: 10.1007/s11060-016-2081-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan W., Gu W., Nagpal S., Gephart M.H., Quake S.R. Brain tumor mutations detected in cerebral spinal fluid. Clin Chem. 2015;61:514–522. doi: 10.1373/clinchem.2014.235457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Mattos-Arruda L., Mayor R., Ng C.K.Y., Weigelt B., Martinez-Ricarte F., Torrejon D., Oliveira M., Arias A., Raventos C., Tang J., Guerini-Rocco E., Martinez-Saez E., Lois S., Marin O., de la Cruz X., Piscuoglio S., Towers R., Vivancos A., Peg V., Ramon y., Cajal S., Carles J., Rodon J., Gonzalez-Cao M., Tabernero J., Felip E., Sahuquillo J., Berger M.F., Cortes J., Reis-Filho J.S., Seoane J. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun. 2015;6:8839. doi: 10.1038/ncomms9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pentsova E.I., Shah R.H., Tang J., Boire A., You D., Briggs S., Omuro A., Lin X., Fleisher M., Grommes C., Panageas K.S., Meng F., Selcuklu S.D., Ogilvie S., Distefano N., Shagabayeva L., Rosenblum M., DeAngelis L.M., Viale A., Mellinghoff I.K., Berger M.F. Evaluating cancer of the central nervous system through next-generation sequencing of cerebrospinal fluid. J Clin Oncol. 2016;34:2404–2415. doi: 10.1200/JCO.2016.66.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Y., He J.Y., Zou Y.L., Guo X.S., Cui J.Z., Guo L., Bu H. Evaluating the cerebrospinal fluid ctDNA detection by next-generation sequencing in the diagnosis of meningeal carcinomatosis. BMC Neurol. 2019;19:331. doi: 10.1186/s12883-019-1554-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan C., Diplas B.H., Chen X., Wu Y., Xiao X., Jiang L., Geng Y., Xu C., Sun Y., Zhang P., Wu W., Wang Y., Wu Z., Zhang J., Jiao Y., Yan H., Zhang L. Molecular profiling of tumors of the brainstem by sequencing of CSF-derived circulating tumor DNA. Acta Neuropathol. 2019;137:297–306. doi: 10.1007/s00401-018-1936-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nevel K.S., DiStefano N., Lin X., Skakodub A., Ogilvie S.Q., Reiner A.S., Pentsova E., Boire A. A retrospective, quantitative assessment of disease burden in patients with leptomeningeal metastases from non-small-cell lung cancer. Neuro Oncol. 2020;22:675–683. doi: 10.1093/neuonc/noz208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Baumgarten L., Kumbrink J., Jung A., Reischer A., Flach M., Liebmann S., Metzeler K.H., Holch J.W., Niyazi M., Thon N., Straube A., von Bergwelt-Baildon M., Heinemann V., Kirchner T., Westphalen C.B. Therapeutic management of neuro-oncologic patients - potential relevance of CSF liquid biopsy. Theranostics. 2020;10:856–866. doi: 10.7150/thno.36884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villatoro S., Mayo-de-Las-Casas C., Jordana-Ariza N., Viteri-Ramirez S., Garzon-Ibanez M., Moya-Horno I., Garcia-Pelaez B., Gonzalez-Cao M., Malapelle U., Balada-Bel A., Martinez-Bueno A., Campos R., Reguart N., Majem M., Blanco R., Blasco A., Catalan M.J., Gonzalez X., Troncone G., Karachaliou N., Rosell R., Molina-Vila M.A. Prospective detection of mutations in cerebrospinal fluid, pleural effusion, and ascites of advanced cancer patients to guide treatment decisions. Mol Oncol. 2019;13:2633–2645. doi: 10.1002/1878-0261.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller A.M., Shah R.H., Pentsova E.I., Pourmaleki M., Briggs S., Distefano N., Zheng Y., Skakodub A., Mehta S.A., Campos C., Hsieh W.Y., Selcuklu S.D., Ling L., Meng F., Jing X., Samoila A., Bale T.A., Tsui D.W.Y., Grommes C., Viale A., Souweidane M.M., Tabar V., Brennan C.W., Reiner A.S., Rosenblum M., Panageas K.S., DeAngelis L.M., Young R.J., Berger M.F., Mellinghoff I.K. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature. 2019;565:654–658. doi: 10.1038/s41586-019-0882-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zehir A., Benayed R., Shah R.H., Syed A., Middha S., Kim H.R. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brister J.R., Ako-Adjei D., Bao Y., Blinkova O. NCBI viral genomes resource. Nucleic Acids Res. 2015;43:D571–577. doi: 10.1093/nar/gku1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tatusova T., DiCuccio M., Badretdin A., Chetvernin V., Nawrocki E.P., Zaslavsky L., Lomsadze A., Pruitt K.D., Borodovsky M., Ostell J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lo Y.M., Chan K.C., Sun H., Chen E.Z., Jiang P., Lun F.M., Zheng Y.W., Leung T.Y., Lau T.K., Cantor C.R., Chiu R.W. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med. 2010;2:61ra91. doi: 10.1126/scitranslmed.3001720. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez C., Snyder M.W., Tanos R., Shendure J., Thierry A.R. New insights into structural features and optimal detection of circulating tumor DNA determined by single-strand DNA analysis. NPJ Genom Med. 2018;3:31. doi: 10.1038/s41525-018-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McEwen A.E., Leary S.E.S., Lockwood C.M. Beyond the blood: CSF-derived cfDNA for diagnosis and characterization of CNS tumors. Front Cell Dev Biol. 2020;8:45. doi: 10.3389/fcell.2020.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mouliere F., Mair R., Chandrananda D., Marass F., Smith C.G., Su J., Morris J., Watts C., Brindle K.M., Rosenfeld N. Detection of cell-free DNA fragmentation and copy number alterations in cerebrospinal fluid from glioma patients. EMBO Mol Med. 2018;10:e9323. doi: 10.15252/emmm.201809323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng D.T., Mitchell T.N., Zehir A., Shah R.H., Benayed R., Syed A., Chandramohan R., Liu Z.Y., Won H.H., Scott S.N., Brannon A.R., O'Reilly C., Sadowska J., Casanova J., Yannes A., Hechtman J.F., Yao J., Song W., Ross D.S., Oultache A., Dogan S., Borsu L., Hameed M., Nafa K., Arcila M.E., Ladanyi M., Berger M.F. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Priestley P., Baber J., Lolkema M.P., Steeghs N., de Bruijn E., Shale C., Duyvesteyn K., Haidari S., van Hoeck A., Onstenk W., Roepman P., Voda M., Bloemendal H.J., Tjan-Heijnen V.C.G., van Herpen C.M.L., Labots M., Witteveen P.O., Smit E.F., Sleijfer S., Voest E.E., Cuppen E. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature. 2019;575:210–216. doi: 10.1038/s41586-019-1689-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Escudero L., Llort A., Arias A., Diaz-Navarro A., Martinez-Ricarte F., Rubio-Perez C., Mayor R., Caratu G., Martinez-Saez E., Vazquez-Mendez E., Lesende-Rodriguez I., Hladun R., Gros L., Ramon Y.C.S., Poca M.A., Puente X.S., Sahuquillo J., Gallego S., Seoane J. Circulating tumour DNA from the cerebrospinal fluid allows the characterisation and monitoring of medulloblastoma. Nat Commun. 2020;11:5376. doi: 10.1038/s41467-020-19175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grommes C., Tang S.S., Wolfe J., Kaley T.J., Daras M., Pentsova E.I., Piotrowski A.F., Stone J., Lin A., Nolan C.P., Manne M., Codega P., Campos C., Viale A., Thomas A.A., Berger M.F., Hatzoglou V., Reiner A.S., Panageas K.S., DeAngelis L.M., Mellinghoff I.K. Phase 1b trial of an ibrutinib-based combination therapy in recurrent/refractory CNS lymphoma. Blood. 2019;133:436–445. doi: 10.1182/blood-2018-09-875732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rice C.A., Tokowicz N., Fraundorf S.H., Liburd T.L. The complex interactions of context availability, polysemy, word frequency, and orthographic variables during lexical processing. Mem Cognit. 2019;47:1297–1313. doi: 10.3758/s13421-019-00934-4. [DOI] [PubMed] [Google Scholar]
- 30.Chen Z., Miao H., Zeng Q., Xu S., Chen Z., Liu K. Circulating cell-free DNA as a diagnostic and prognostic biomarker for non-small-cell lung cancer: a systematic review and meta-analysis. Biomark Med. 2020;14:587–597. doi: 10.2217/bmm-2018-0093. [DOI] [PubMed] [Google Scholar]
- 31.Thierry A.R., Mouliere F., Gongora C., Ollier J., Robert B., Ychou M., Del Rio M., Molina F. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res. 2010;38:6159–6175. doi: 10.1093/nar/gkq421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkinson C.A., Gale D., Piskorz A.M., Biggs H., Hodgkin C., Addley H., Freeman S., Moyle P., Sala E., Sayal K., Hosking K., Gounaris I., Jimenez-Linan M., Earl H.M., Qian W., Rosenfeld N., Brenton J.D. Exploratory analysis of TP53 mutations in circulating tumour DNA as biomarkers of treatment response for patients with relapsed high-grade serous ovarian carcinoma: a retrospective study. Plos Med. 2016;13:e1002198. doi: 10.1371/journal.pmed.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Wit M.C., de Bruin H.G., Eijkenboom W., Sillevis Smitt P.A., van den Bent M.J. Immediate post-radiotherapy changes in malignant glioma can mimic tumor progression. Neurology. 2004;63:535–537. doi: 10.1212/01.wnl.0000133398.11870.9a. [DOI] [PubMed] [Google Scholar]
- 34.Fujimoto D., von Eyben R., Gibbs I.C., Chang S.D., Li G., Harsh G.R., Hancock S., Fischbein N., Soltys S.G. Imaging changes over 18 months following stereotactic radiosurgery for brain metastases: both late radiation necrosis and tumor progression can occur. J Neurooncol. 2018;136:207–212. doi: 10.1007/s11060-017-2647-x. [DOI] [PubMed] [Google Scholar]
- 35.Narloch J.L., Farber S.H., Sammons S., McSherry F., Herndon J.E., Hoang J.K., Yin F.F., Sampson J.H., Fecci P.E., Blackwell K.L., Kirkpatrick J.P., Kim G.J. Biopsy of enlarging lesions after stereotactic radiosurgery for brain metastases frequently reveals radiation necrosis. Neuro Oncol. 2017;19:1391–1397. doi: 10.1093/neuonc/nox090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.