Abstract
When sequencing small RNA libraries derived from whole blood, the most abundant microRNAs (miRs) detected are often miR-486-5p, miR-451a, and miR-92a-3p. These highly expressed erythropoietic miRs are released into the sample from red blood cell hemolysis. Next-generation sequencing of these unwanted miRs leads to a waste in sequencing cost and diminished detection of lowly expressed miRNAs, including many potential miRNA biomarkers. Previous work has developed a method to reduce targeted miRNAs using oligonucleotides that bind their target miRNA and prevent its ligation during library construction, although the extent to which oligonucleotides can be multiplexed and their effect on larger cohorts has not been thoroughly explored. We present a method for suppressing detection of three highly abundant heme miRs in a single multiplexed blocking oligonucleotide reaction. In a small paired-sample pilot (n = 8) and a large cohort of samples (n = 901), multiplexed oligos reduced detection of their target miRNAs by approximately 70%, allowing for an approximately 10-fold increase in reads mapping to nonheme miRs and increased detection of very lowly expressed miRs, with minimal off-target effects. By removing all three highly expressed erythropoietic miRNAs from next-generational sequencing libraries, this commercially available multiplexed blocking oligonucleotide method allows for greater detection of lowly expressed biomarkers, improving the efficacy, cost-efficiency, and sensitivity of biomarker studies and diagnostic tests.
miRNAs regulate cellular processes by modulating gene expression at the posttranscriptional level, degrading their target mRNAs or suppressing protein translation.1,2 Recently, miRNAs have also been studied as potentially powerful biomarkers of disease. Because these modulatory molecules are secreted into the extracellular space and circulating blood, they can be easily detected in whole blood and serum/plasma samples, making them the perfect candidates for biomarker studies. Recent miRNA biomarker studies have demonstrated their potential utility across a wide variety of human diseases, including Alzheimer disease, obesity, attention-deficit disorder, and cancers.3, 4, 5, 6, 7, 8 The clinical application of these biomarker studies is an area of active research and may contribute to earlier diagnosis and treatment.9
Several technologies have been used to detect miRNAs in blood-derived samples and other tissues, such as real-time quantitative PCR, microarrays, and next-generation sequencing (NGS) among others.10 Because of its sensitivity, efficiency, and high-throughput nature, NGS is often the technique of choice to study miRNAs. However, NGS has several limitations that restrict its use in measuring miRNA levels. During library construction of small RNA libraries, the T4 RNA Ligase 2 ligates highly expressed miRNAs in a biased way, leading to their increased detection during sequencing.11,12 This issue is further exacerbated by the possibility of hemolysis of red blood cells (RBCs) within the sample during extraction. RBCs highly express several miRNA species (eg, hsa-miR-486-5p, hsa-miR-451a, and hsa-miR-92a-3p) that become highly abundant within the pool of miRNA after hemolysis3,13, 14, 15 and consume a greater fraction of the per sample sequencing output. These miRNA species are generally not of interest in miRNA studies, creating a large waste of sequencing costs and limiting detection of disease-relevant biomarkers, which are often expressed at lower levels. In addition, several studies reported that the concentration of the three erythropoietic miRNAs increases with increasing levels of hemolysis, which would further contribute to sequencing bias and dilution of disease-relevant biomarkers in sequencing data.16,17
Two previous studies have suggested a solution to this issue by preventing library construction of highly abundant miRs using blocking oligonucleotides complementary to miRNA species of interest. These short oligonucleotides hybridize to the target miR, preventing 5′ ligation during library construction and greatly reducing detection of the targeted abundant miRNA species.14,18However, these studies have several limitations that remain to be addressed. The potential of multiplexing multiple blocking oligonucleotides into one reaction has not been thoroughly explored. The effect of blocking oligonucleotides on detection of their isomiRs has also not been closely examined. miRNA isomiRs are still not fully understood; some seem to function in the same mechanism as their mature counterpart, whereas others may have alternative functions.19,20 Detection of isomiRs during sequencing may also be attributable to sequencing errors.21 In addition to this, the effect of blocking oligonucleotides on a large cohort of samples has not been thoroughly explored. This is an important limitation because larger cohorts for small RNA sequencing can have a range of additional considerations, such as batch to batch variability, broader range of sample types, variable sample quality, and varying levels of hemolysis.22,23 To support the results of the previous studies, as well as address these limitations, we present a method of preventing ligation and library construction of three miRNA species highly expressed in whole blood samples.
Materials and Methods
Total RNA Isolation
Whole blood samples from adults were collected for both the pilot samples (A through D) and the larger cohort, which is part of the COPDGene Study. COPDGene was approved by the institutional review boards at Partners Healthcare and all participating centers. Pilot samples were extracted from fresh samples, whereas the large cohort used archival samples. Total RNA was isolated from whole blood samples using the PAXgene Blood miRNA Kit (Preanalytix, Hombrechtikon, Switzerland) or the miRNeasy kit (Qiagen, Germantown, MD) according to the manufacturer's instructions.
Small RNA Library Construction and Sequencing
Blocking oligonucleotides were obtained from Perkin Elmer (Waltham, MA) and are now commercially available as a pool of oligonucleotides (catalog number NOVA-513103). Oligonucleotides are complementary to the full sequence of the mature heme miRs: miR-92a-3p: 5′-ACAGGCCGGGACAAGTGCAATA-3′; miR-486-5p: 5′-CTCGGGGCAGCTCAGTACAGGA-3′; and miR-451a: 5′-AACTCAGTAATGGTAACGGTTT-3′.
Isolated total RNA was converted to cDNA libraries using the NEXTFLEX Small RNA-Seq version 3 Automation Kit (Perkin Elmer, Waltham, MA). Library construction was performed in a single batch for pilot samples and across 13 batches for the cohort samples. Automated library preparation was performed for all samples on a Sciclone G3 NGS and NGSx automated liquid handler (Perkin Elmer). For pilot samples, varying amounts of RNA (200, 400, 600, or 800 ng) were used as input into library construction; all values were within the manufacturer recommended range of 200 ng to 2 μg. Approximately 100 ng of input total RNA was used for cohort library construction. The blocking oligonucleotides are provided by the vendor as a blocker mix at an overall concentration of 5 μmol/L and are added at a volume of 1 μL of blocker mix to 9.5 μL of RNA before library construction regardless of the RNA concentration.
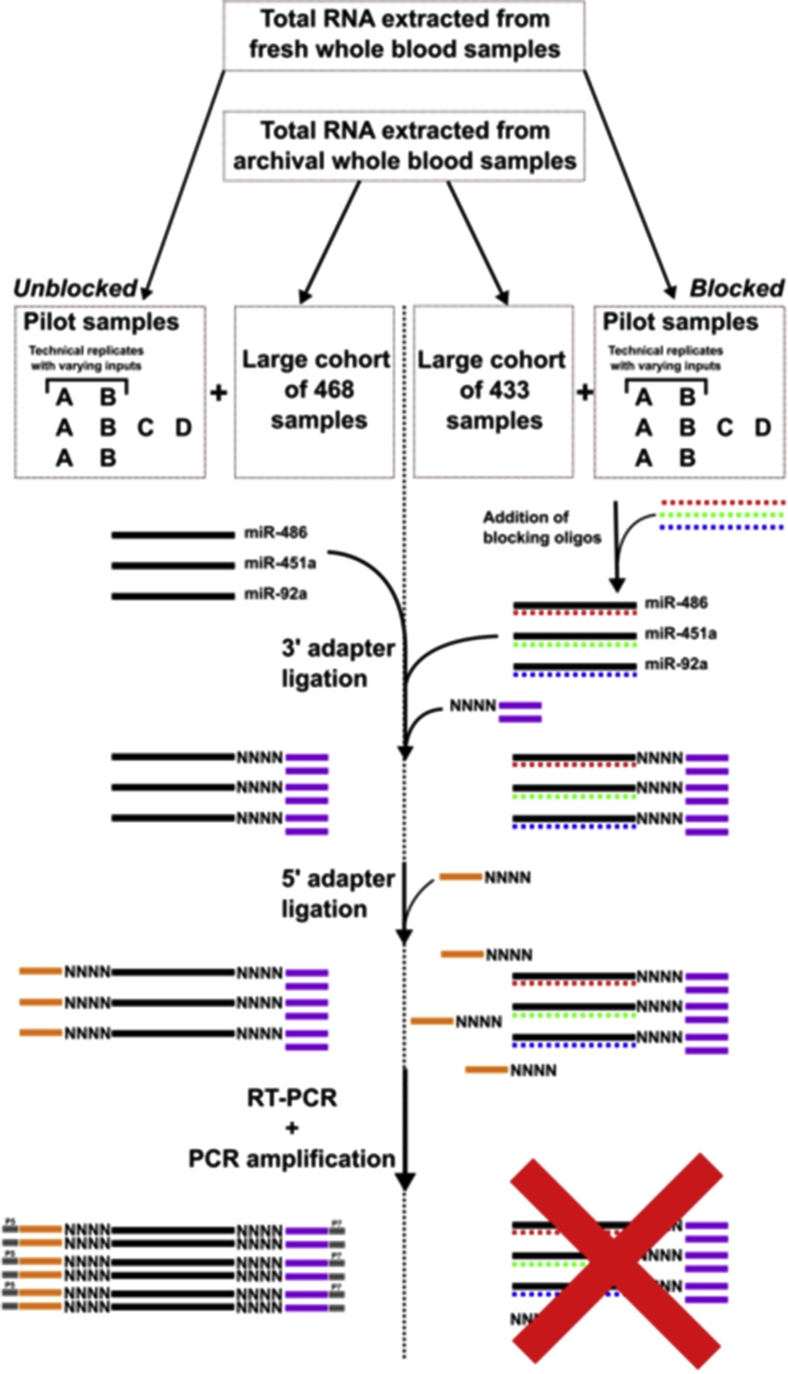
Library construction for all samples was then performed according to manufacturer's automated protocol. Briefly, 3′ 4N adenylated adapters are ligated by denaturation followed by 2 hours of incubation at 25°C.23 After excess 3′ adapter is removed by ethanol wash, adapters are inactivated by addition of NEXTFLEX Adapter Inactivation Enzyme and incubation at 12°C for 15 minutes then 50°C for 20 minutes. 5′ 4N adapters are ligated at 20°C for 1 hour. M-MulV Reverse Transcriptase is added and samples incubated at 42°C for 30 minutes followed by 10 minutes of incubation at 90°C. Cleanup is performed using NEXTFLEX cleanup beads with ethanol and isopropanol washes. Libraries were amplified using the following protocol: 2 minutes at 95°C; 19 cycles of 20 seconds at 60°C, 30 seconds at 60°C, and 15 seconds at 72°C; and final extension at 72°C for 2 minutes. Gel-Free size selection and cleanup were performed using NEXTFLEX cleanup beads and an additional ethanol wash step. Library construction process for the pilot and large cohort is summarized in Figure 1.
Figure 1.
Library construction process using multiplexed pool of blocking oligonucleotides. Schematic for NEXTFLEX small RNA library construction, with and without the use of blocking oligonucleotides. Fresh whole blood samples were used for the pilot, but the large cohort used RNA extracted from archival whole blood samples. For blocked samples, blocking oligonucleotides were added to extracted RNA samples before library preparation. Pilot samples and large cohort were processed in the same way.
Libraries were quantified using the Real-Time Quantitative PCR Library Quantification Kit–Illumina/ABI Prism (KAPA Biosystems, Wilmington, MA). Libraries were analyzed for correct size using BioAnalyzer High Sensitivity DNA chips (Agilent, Santa Clara, CA). Libraries were sequenced on a HiSeq 2500 (Illumina, San Diego, CA) according to standard Illumina protocols.
Data Preparation and Filtering
Adapters were trimmed from raw FASTQ files using the BaseSpace Sequence Hub application FASTQ Toolkit (https://basespace.illumina.com, last accessed May 7, 2021). An additional four nucleotides were trimmed at the 5′ and 3′ ends because of the addition of extranucleotides added at this location by the NEXTFLEX Small RNA-Sequencing kit. Trimmed reads were aligned in the BaseSpace Sequence Hub Small RNA version 1.0 application. With Bowtie2, reads were aligned to the hg19/miRBase 21 database.
Count matrices from BaseSpace contain both mature and isomiR hits. Isomir hits are defined as a subsequence on the same strand of a precursor sequence but not the 5′ or 3′ mature miRNA sequence. The precursor sequences are obtained from the miRNA database (miRBase 21). For Figure 2, A–C, mature and isomiR hits were collapsed together by removing the suffix from the miRNA ID (ie, removing -1 from mir-486-1) and then adding counts into a single category. For Figure 2D and Supplemental Figure S1D, mature and isomiR hits were plotted separately. Figures 3, 4, and 5 also use mature and isomiR hits collapsed together, as for Figure 2, A–C.
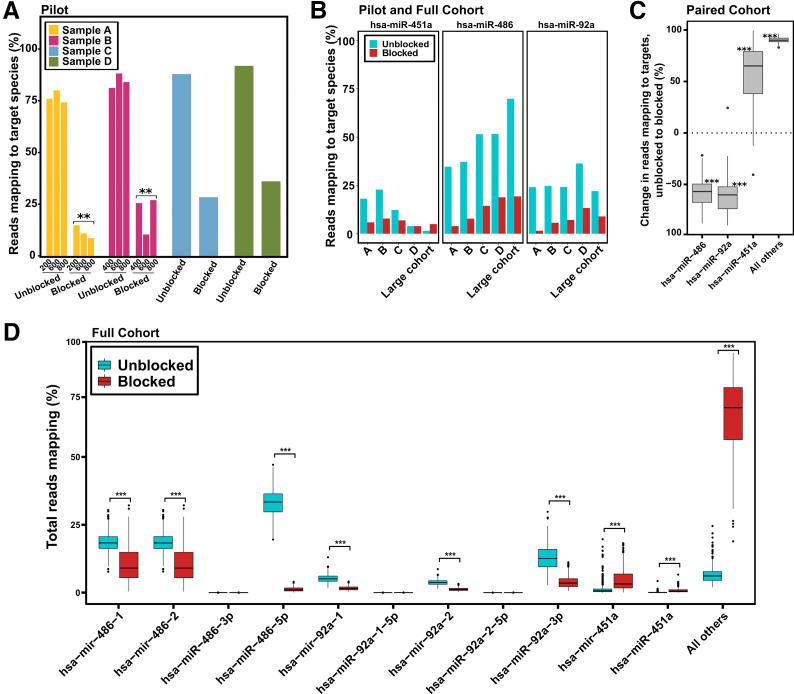
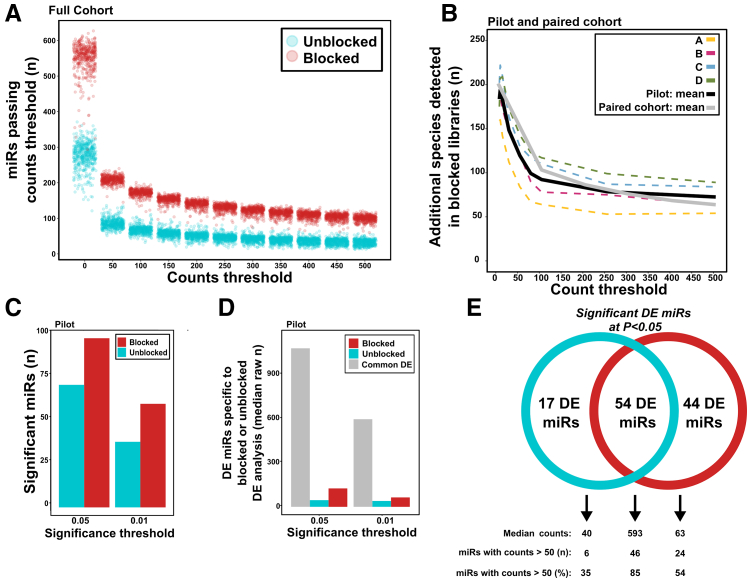
Figure 2.
Blocking oligonucleotides reduce detection of highly abundant hemolysis microRNAs (miRNAs) at multiple RNA input levels A: The percentage of total reads mapping to any of the three targeted hemolysis markers for paired samples A, B, C, and D. Three replicates for each condition were used for samples A and B with varying levels of RNA input into library construction. No significant difference was found within any of the four sets of three replicates. Blocked samples A and B had significantly lower levels of the three hemolysis markers. B: The percentage of total reads mapping to the three targets (or its isomiRs) was calculated for each pilot sample and for the large cohort of samples. Technical replicates for samples A and B are collapsed. The mean percentage mapping across all samples is used for the large cohort. C: Of the 901 large cohort samples, 150 consist of a blocked/unblocked pair from the same individuals (75 pairs). The percentage change in mapping to each target (or its isomiRs) and to all nontarget miRs was calculated for each blocked/unblocked pair. The mean percentage change across all 75 pairs is shown here (paired t-test comparing blocked/unblocked in each sample pair for miR-486, miR-451a, miR-92a, and nontarget miRs was performed). D: The effect of blocking oligonucleotides on the full large cohort of unblocked and blocked samples was determined by calculating the percentage of total reads mapping to each of the three targets or one of their isomiRs. Mature (both -3p/-5p forms) and isomiR/precursor forms of the three targets are shown separately. Large cohort, n = 901 (unblocked samples, n = 468; blocked samples, n = 433). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
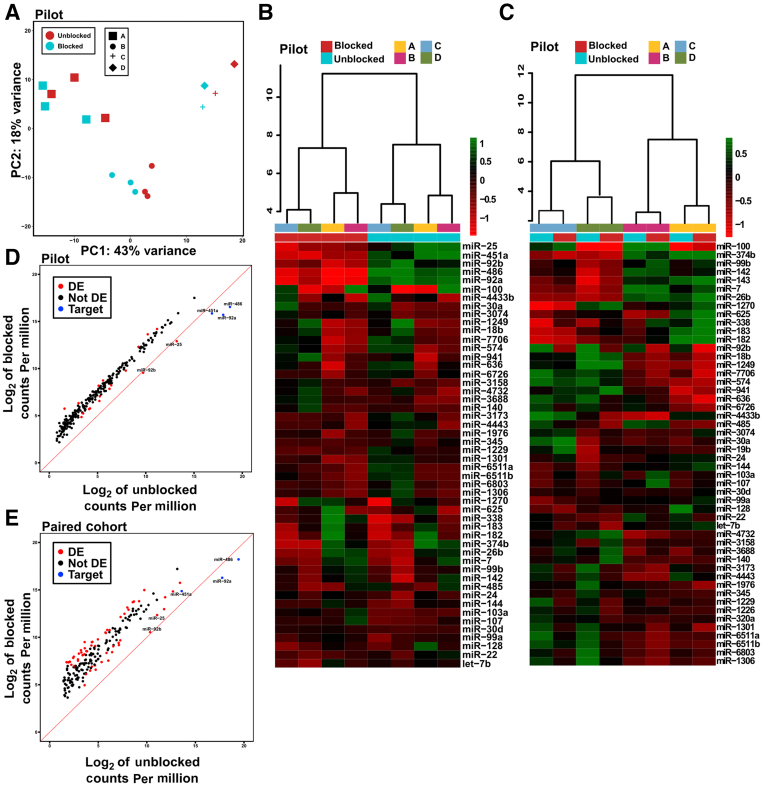
Figure 3.
Overall microRNA (miRNA) expression patterns are largely unaffected by blocking oligonucleotides. A: Principal component analysis (PCA) of all 16 pilot libraries. Points colored by library construction method; shapes depict sample used. B: Pairwise differential expression (DE) comparing blocked and unblocked pilot libraries was performed using DESeq2. Technical replicates for samples A and B were collapsed. The top 50 differentially expressed miRNAs are plotted on a heatmap. Hierarchical clustering dendrogram is shown at the top [approximately unbiased (au) value = 54 for blocked samples; au value = 55 for unblocked samples]. C: Pairwise DE comparing blocked and unblocked libraries was performed using DESeq2. Technical replicates for samples A and B were collapsed. The three target miRs as well as miR-25 were removed from the count matrix before analysis. The top 50 differentially expressed miRNAs are plotted on a heatmap. Hierarchical clustering dendrogram is shown at the top (au value = 100 for samples A and B; au value = 93 for sample C; and au value = 95 for sample D). D: For the mean across the four pilot samples (A–D), log10 counts per million (CPM) values were plotted for unblocked and blocked libraries. Any miR with <1 CPM in >50% of all libraries was considered not detected and was excluded from analysis. The red line denotes a slope of 1; miRNA species falling beneath the line have lower CPM values in blocked libraries compared with unblocked libraries. Significantly differentially expressed miRs from the DESeq2 analysis described in panel C (padj <0.01) are colored in red. miRNA species that are both differentially expressed and have a lower CPM value in blocked libraries are labeled. Target heme miRs are colored in blue. E: The same analysis as described in panel D was performed for the paired large cohort of miRNA libraries.
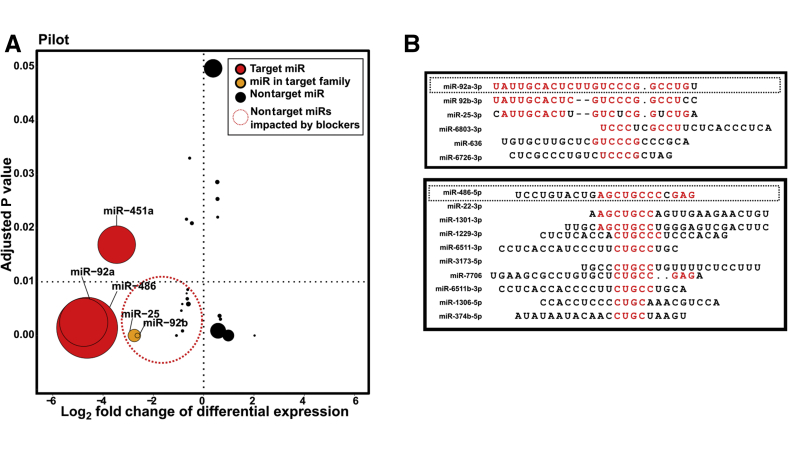
Figure 4.
Some nonheme microRNAs (miRNAs) are reduced by blocking oligonucleotides and share modest sequence similarity with target miRs A: Graph summarizing factors involved in determining true off-targets (see Table 1 for values). All miRs to the right of the vertical dotted line (positive fold change) were significantly increased in blocked libraries. All miRs below the horizontal dotted line were significantly (adjusted P < 0.01) differentially expressed. Red points are targets miRs, and orange points are miRs in the same family as a target miR. Size of point scales with the total number of mean counts per million that support the miRNA. The dotted circled portion denotes miRNA species that may be of concern as off-target effects. B: Sequence similarity of potential off-target effects with target sequences. 3p/5p designation added based on which version of the miR shared sequence similarity with a target miR. Red text denotes regions of alignment between target heme miR and off target miR. Dotted boxes denote sequences of miR92a and miR-486 used for blocking oligonucleotides.
Figure 5.
Sensitivity of microRNA (miRNA) detection is increased in blocked libraries A: For each sample in the large unpaired cohort, the number of species passing several raw count thresholds was determined. B: A similar analysis to that in panel A was performed for the pilot and paired large cohort samples. For each of these samples, the number of miRNAs passing several raw count thresholds was determined. This value for unblocked samples was subtracted from the number passing thresholds for blocked samples. The mean across all four pilot samples is shown in black; the mean across all 75 paired large cohort samples is shown in gray. At every threshold, more miRNA species pass the threshold in blocked samples than in unblocked samples, particularly at low thresholds C: For the pilot samples, differential expression (DE) using DESeq2 was performed on two groups: three unblocked A samples versus three unblocked B samples and three blocked A samples versus three blocked B samples. The number of differentially expressed miRNAs at two significance cutoffs is plotted here. D: Using the same DE analyses as described in panel C, the median counts for blocked-specific differentially expressed miRs, unblocked-specific differentially expressed miRs, or differentially expressed miRs found in both analyses are plotted. E: Differentially expressed miRs were subset into three groups: miRs differentially expressed in only the blocked DE analysis, miRs differentially expressed in only the unblocked DE analysis, and miRs differentially expressed in both analyses. For these three subsets, the median raw counts and the proportion of DE miRs with counts >50 are shown.
For all analyses, miRNA counts were downsampled to a common level (to the level of the sample with the lowest number of read counts: for large cohort, downsampled to 2.1 million total counts; for pilot samples, downsampled to 7.97 million total counts) using the R package metaseqR (R Foundation for Statistical Computing, Indianapolis, IN). For samples A and B, technical replicates were collapsed with the DESeq2 collapseReplicates function in R (Figure 3, A—C) or by averaging the counts for each miR.
Quantitative Analysis of Blocker Performance
The effect of RNA input on miRNA expression was determined using one-way ANOVA testing in R. Principal component analysis was conducted using the DESeq2 package plotPCA in blind mode.
Before differential expression (DE), the count matrix was filtered by removing any miRNA species without at least 1 count per million (CPM) in at least 50% of blocked and unblocked samples. Normalization and DE were performed using the package DESeq2 in R using default (closed-form) dispersion estimates and Wald tests.24 Pairwise DE was performed, controlling for sample differences, for both the pilot samples and the paired large cohort. Multiple testing correction was performed using the default DESeq2 Benjamini and Hochberg test. All P values listed in article are the adjusted P values. Downsampled blocked and unblocked counts for all miRNAs were converted to counts per million and plotted, with coloring based on significance (Figure 3, D and E). miRs were considered significant if adjusted P < 0.05; for the paired cohort, an additional filter of absolute [log2 fold change (log2FC)] >1 was also applied.
To prioritize off-target effects, adjusted P values were plotted against the log2FC (Figure 4A). Sequence similarity between highest-priority off-targets and target heme miRs was assessed using blastn-short (Figure 4B). When determining overlap of significantly differentially expressed miRs between the pilot and paired cohort, only a P value threshold was applied to determine significance (Table 1).
Table 1.
Differential Expression Results from DESeq2 Analysis: Pilot and Cohort Comparison
| Species | Pilot data |
Paired cohort |
Sequence similarity | ||||
|---|---|---|---|---|---|---|---|
| Log2 fold change | padj | Mean CPM | Log2 fold change | padj | Mean CPM | ||
| miR-451a | −3.46 | 1.69 × 10−02 | 101,918 | −1.96 | 1.67 × 10−36 | 21,132 | |
| miR-486 | −4.63 | 1.44 × 10−03 | 275,553 | −4.95 | 2.62 × 10−83 | 507,823 | |
| miR-92a | −4.77 | 2.46 × 10−03 | 169,866 | −5.13 | 0.00 | 150,568 | |
| miR-25 | −2.76 | 1.04 × 10−59 | 9376 | −2.18 | 2.41 × 10−186 | 3668 | miR-92a |
| miR-92b | −2.63 | 1.04 × 10−59 | 875 | −3.45 | 0.00 | 1392 | miR-92a |
| miR-1301 | −1.08 | 3.82 × 10−08 | 74 | −0.80 | 3.79 × 10−39 | 117 | miR-486 |
| miR-7706 | −0.89 | 4.63 × 10−03 | 53 | −0.82 | 1.29 × 10−24 | 55 | miR-486 |
| miR-636 | −0.87 | 2.77 × 10−03 | 27 | −0.83 | 1.95 × 10−19 | 41 | miR-92a |
| miR-1229 | −0.85 | 5.86 × 10−03 | 35 | −1.20 | 2.91 × 10−42 | 78 | miR-486 |
| miR-3173 | −0.84 | 8.61 × 10−04 | 160 | −1.62 | 1.00 × 10−135 | 359 | miR-486 |
| miR-6511a | −0.68 | 2.17 × 10−02 | 143 | −1.15 | 3.92 × 10−68 | 311 | miR-486 |
| miR-6726 | −0.68 | 7.84 × 10−03 | 27 | −1.04 | 2.39 × 10−29 | 69 | miR-92a |
| miR-6803 | −0.65 | 6.84 × 10−03 | 203 | −1.26 | 3.14 × 10−48 | 408 | miR-92a |
| miR-1306 | −0.64 | 8.55 × 10−03 | 94 | −1.16 | 2.06 × 10−114 | 191 | miR-486 |
| miR-22 | −0.61 | 5.86 × 10−03 | 603 | 0.37 | 8.35 × 10−11 | 440 | miR-486 |
| miR-6511b | −0.56 | 3.30 × 10−02 | 113 | −0.84 | 1.82 × 10−38 | 258 | miR-486 |
| miR-374b | −0.47 | 2.09 × 10−02 | 317 | 0.94 | 4.55 × 10−11 | 56 | miR-486 |
| miR-30d | 0.38 | 4.97 × 10−02 | 20,846 | 0.36 | 1.24 × 10−14 | 13,383 | Not tested for sequence similarity |
| miR-941 | 0.54 | 2.86 × 10−02 | 451 | 1.69 | 1.87 × 10−112 | 368 | |
| miR-24 | 0.55 | 2.54 × 10−02 | 350 | 1.43 | 5.09 × 10−47 | 191 | |
| miR-625 | 0.56 | 2.20 × 10−02 | 97 | −0.53 | 1.82 × 10−07 | 65 | |
| miR-182 | 0.57 | 8.89 × 10−04 | 14,595 | ||||
| miR-183 | 0.6 | 4.88 × 10−04 | 3337 | ||||
| miR-100 | 0.63 | 3.67 × 10−03 | 352 | 0.91 | 1.62 × 10−13 | 229 | |
| miR-99b | 0.65 | 5.79 × 10−04 | 183 | 0.49 | 2.09 × 10−12 | 153 | |
| miR-30a | 0.67 | 3.04 × 10−03 | 147 | ||||
| miR-107 | 0.72 | 1.14 × 10−04 | 258 | 1.93 | 6.56 × 10−63 | 173 | |
| miR-3688 | 0.95 | 1.74 × 10−05 | 49 | ||||
| miR-103a | 0.98 | 5.93 × 10−06 | 7411 | 1.73 | 2.12 × 10−175 | 4654 | |
| miR-338 | 2.02 | 2.77 × 10−11 | 32 | ||||
All miRs that were differentially expressed in the pairwise DESeq2 analysis of pilot data (Figure 3B) are shown. For each miR, data are given for the pilot data analysis (green) as well as in the large cohort, if it was detected as DE in the paired large cohort (blue). miRs are ordered based on log2 fold change, adjusted P value, and mean raw counts supporting the miRNA species. For miRs significantly decreased in blocked libraries, sequence similarity to a target heme miR is reported (see Figure 4C for sequences).
CPM, count per million; DE, differential expression; miR, microRNA.
Expression data were normalized using the DESeq2 rlog function before hierarchical clustering (euclidean method) and heatmap visualization using the R package pheatmap. Only the top 50 genes (ordered by adjusted P value) are shown on the heatmaps. Approximately unbiased P values for clusters were calculated using the R package pvclust using the euclidean method. All other plotting was performed using the R package ggplot2.
Significance testing was conducted in R using a paired or unpaired t-test where appropriate. For Figure 5D, a Dunn post hoc test was conducted in R to compare medians rather than means.
All analyses in R are available at Github (https://github.com/jenna-labelle/BlockerProject, last accessed September 1, 2020).
Results
Blocking Oligonucleotides Reduce Expression of Targeted Hemolysis Markers Regardless of Sample Input Levels
Three commercially available small oligonucleotides that bind directly to the entire mature miRNA sequence of three abundant hemolysis markers were obtained: hsa-miR-486-5p, hsa-miR-451a, and hsa-miR-92a-3p (Perkin Elmer, sequence proprietary; catalog number NOVA-513103). Binding of these oligonucleotides prevents 5′ ligation during library construction, removing these miRNA species from the library. To evaluate the effect of the three blocking oligos on expression of the targeted hemolysis markers, the NEXTFLEX Small RNA-Seq Kit version 3 we used to perform library construction and sequencing of 16 pilot samples as well as a large cohort of samples. For the pilot, 16 whole blood total RNA samples from four different samples (A–D) were used. Four varying RNA inputs in library construction were used for samples A and B, all within the optimal range recommended by the manufacturer (200 ng to 2 μg). Eight samples in total (three replicates each of A and B plus samples C and D) were extracted from fresh whole blood samples and processed through library construction according to the standard protocol. An additional eight paired samples were generated using the mixture of three blocking oligos. In addition to this pilot study, 468 previously sequenced unblocked small RNA libraries and 433 blocked small RNA libraries extracted from archival samples were also compared (Figure 1).
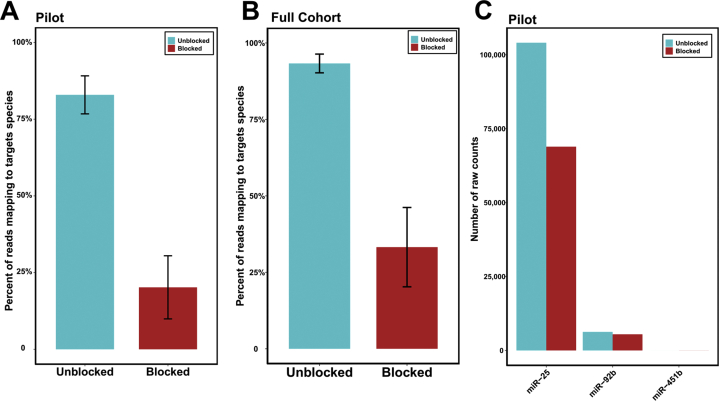
After sequencing these libraries, the proportion of total reads mapping to any of the three target miRNAs was quantified. In the libraries constructed according to the standard protocol, the three targeted hemolysis markers represent a mean of 83% and 93% of all reads after sequencing for the paired pilot samples and large cohort, respectively (Supplemental Figure S1, A and B). Libraries constructed using blocking oligonucleotides had a 76% and 63% reduction in the proportion of reads mapping to any of the three targeted hemolysis markers (a mean of 20% and 35% of total reads) for the pilot samples and large cohort, respectively.
In the paired sample pilot, four varying levels of RNA (200, 400, 600, and 800 ng) were used as input into blocked and unblocked library construction for samples A and B, resulting in 12 libraries total. The blocker mix was added to extracted RNA before library construction. Overall, the proportion of reads mapping to the three hemolysis markers remained relatively consistent, regardless of the input level (Figure 2A). The use of blocking oligos resulted in a similar reduction in target detection at every input level tested (P > 0.05). For the remainder of the analysis, the three input levels for samples A and B were treated as technical replicates and collapsed.
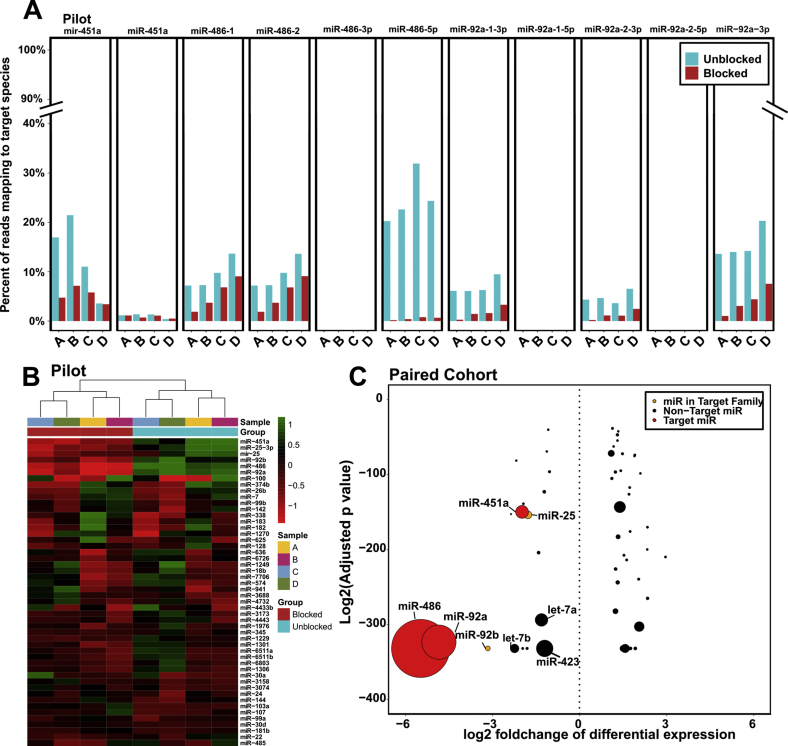
In analysis, the reads mapping to both mature miRNA sequences and isomiR sequences, defined as a subsequence on the same strand of a precursor sequence, were examine but the 5′ or 3′ mature miRNA sequence was excluded. These isomiR/precursors are denoted by the prefix mir rather than miR, which is used for mature miRNAs. All isomiRs for each miRNA species have been collapsed into a single category for each precursor (see Materials and Methods). For both the pilot and large cohorts, a large decrease in the detection of miR-486 (reduced by 76% and 68%; odds ratios = 6.24 and 5.64, respectively) and miR-92a (reduced by 76% and 69%; odds ratios = 4.8 and 3.19, respectively) was observed. However, high levels of miR-451a in unblocked libraries (>10% of total reads) were only detected in pilot samples A through C but not in the large cohort of samples or pilot sample D. Across the pilot samples A through C, miR-451a was decreased by a mean of 59% (odds ratio = 2.87) (Figure 2B).
Within the large cohort of 901 samples, 150 consist of an unblocked/blocked library pair from the same individuals (75 unique individuals/sample pairs). For each of these paired samples, the effect of blocking oligonucleotides on target detection was determined by calculating the percent change in target mapping with and without blocking oligonucleotides. This change was calculated for each pair individually; miR-486 and miR-92a were decreased by a mean of 58% and 61%, respectively (Figure 2C). As in the unpaired large cohort (Figure 2B), miR-451a was not decreased in blocked samples and instead was increased by a mean of 58% (Figure 2C). This finding may be attributable to a combination of its low levels within unblocked samples and increased detection of lowly expressed miRs with the use of blockers; this possibility is explored later. Detection of nontarget miRs was markedly increased in the blocked samples (90% increase) (Figure 2C).
Although there are only three desired heme miR targets, each target also has at least one isomiR category, for a total of 11 isomiR or mature targets. In the complete large cohort data set (including unpaired samples), several of these miRs were virtually undetected: miR-486-3p, miR-92a-1-5p, and miR-92a-2-5p. Both miR-486-5p and miR-92a-3p were detected at high levels in unblocked samples (330,000 CPM and 130,000 CPM, respectively) and significantly decreased in blocked samples (12,000 CPM and 38,000 CPM, respectively; adjusted P < 10 × 10−174). This reduction was particularly marked for miR-486-5p; blocking oligonucleotides resulted in a mean 96% decrease in detection. Surprisingly, miR-451a was virtually undetected (approximately 1100 CPM). The isomiR group of miR-451a was detected at slightly higher levels (approximately 13,000 CPM) but was not decreased in blocked samples. Although levels of mir-451a varied across samples, detection of this miR was significantly higher in blocked libraries than in unblocked libraries (P = 3.09 × 10−46). In addition, we found that the two miR-486 isomiR groups (mir-486-1 and mir-486-2) were significantly reduced by blocking oligonucleotides (adjusted P < 10 × 10−83), although with less efficiency than its mature form (approximately 42%). Similarly, the two miR-92a isomiR groups (mir-92a-1 and mir-92a-2) were significantly reduced by blocking oligos (adjusted P < 10 × 10−219, approximately 68%). The percentage of reads mapping to nonheme miRs were also compared. As expected, there was a large, significant increase in the percentage of reads mapping to nontarget miRs in blocked libraries (approximately 65,000 CPM in unblocked libraries increased to approximately 650,000 CPM in blocked libraries; P = 7.72 × 10−307). For the remainder of the analysis, the isomiR/mature forms of each of the three targets were collapsed together.
Global miRNA Expression Patterns Are Largely Unchanged by Blocking Oligonucleotides
To analyze the effects of the blocking oligonucleotides on overall miRNA expression as well as any significant off-target effects, blind principal component analysis was performed to obtain a global view of miRNA expression across the eight paired pilot samples (Figure 3A). Samples cluster by sample rather than by library construction method (standard or blocking oligonucleotide method).
To analyze the effects of blocking oligonucleotides on the expression of all nontarget miRNAs, DESeq2 was used to perform pairwise DE, comparing the four paired unblocked pilot libraries with the four blocked pilot libraries (technical replicates for samples A and B were collapsed). Before DE, any miRNA species without at least 1 CPM in 50% of blocked and unblocked library was considered not detected and was removed. Of the 195 miRs tested, 27 nontarget miRs that were differentially expressed between blocked and unblocked libraries (adjusted P < 0.05; Benjamini and Hochberg multiple testing correction) were identified. After performing hierarchical clustering and plotting the top 50 differentially expressed miRs on a heatmap, samples clearly cluster by blocker status not by sample type (Figure 3B). However, this clustering appears to be driven primarily by only a few top species (the three target heme miRs and miR-25), shown at the top of the heatmap. The strength of each cluster is denoted by an approximately unbiased P value at the top of each cluster; the low P values (54 and 55) of the two blocked/unblocked clusters indicate relatively low confidence clusters (values >95 indicate clusters of high confidence) (Figure 3B).
To focus on nontarget miRs, the three target miRs were removed from the count matrix before performing an additional DE analysis. In addition miR-25 was removed from the input count matrix: this miR is in the same family as miR-92a and shares significant sequence similarity, as discussed in the section entitled Blocking Oligonucleotides May Cause Reduced Detection of a Small Number of Nonheme miRs. Counts for this miR were clearly lower in blocked libraries compared with unblocked libraries (Supplemental Figure S1C). Overall, this finding seemed to suggest that blocking oligonucleotides are having an unintended but unavoidable effect on miR-25 detection and that excluding it from the DE analysis would allow us to focus more closely on nontarget miRs. Interestingly, this effect existed for both the mature (miR-25-3p) and isomiR/precursor (mir-25) forms; when both were included as input to DE, they were identified as significantly decreased, albeit with a slightly stronger effect for the mature form (P = 7.19 × 10−55/2.39 × 10−49 and log2FC = ˗2.99/˗2.17) (Supplemental Figure S2B). After removing miR-25, performing DE on this subset count matrix and visualizing by plotting the top 50 differentially expressed miRs on a heatmap, it was found that samples cluster tightly by sample type rather than by blocker status (Figure 3C). Supporting this idea, approximately unbiased P values for the four sample clusters were high (100, 100, 93, and 95 for samples A, B, C, and D), indicating clusters of very high confidence.
Overall, CPM values averaged across all eight blocked and unblocked pilot libraries tightly follow a similar trend (Figure 3D). Although several nontarget miRs are significantly decreased in blocked libraries, only two actually have lower counts in blocked libraries: miR-92b and miR-25. Both these species share significant sequence similarity to miR-92a, as discussed in the next section, and their reduced detection may be unavoidable because of the limits of the technology. Of note, the slope of the lines is shifted above one for most miRs in blocked libraries, reflecting the overall increase in nonheme miRs.
We performed a similar DE analysis was performed with the 75 sample pairs in the cohort (150 samples total). Pairwise DESeq2 identified 57 nontarget significantly differentially expressed miRs (log2FC >1, adjusted P < 0.05; of 195 total miRs tested). The mean CPM values in the paired large cohort follow the same trend for blocked and unblocked libraries (Figure 3E). Although the spread of the data is slightly wider than the pilot, it could be attributed to the much larger sample size of this cohort. Controlling for batch effects may also reduce some of the noise observed, but this is not possible given the limitations of this cohort because there is no overlap in batches between blocked and unblocked groups. In addition, with a larger number of samples, only target heme-miR counts are lower in blocked than unblocked samples (miR-486 and miR-92a). For both the pilot and the paired cohort, although several miRs were identified as DE, the overall effect appears to be proportional throughout the count distribution.
Blocking Oligonucleotides May Cause Reduced Detection of a Small Number of Nonheme miRs
Overall, the expression of miRNA species across all libraries follows a similar pattern in blocked and unblocked samples (Figure 3, D and E). Most miRNA species, with the exception of the three target miRNAs, have a similar expression level in blocked and unblocked samples. However, there are clearly several nontarget, differentially expressed miRNA species that are of potential concern as off-target effects; in the pilot data, 27 nontarget miRs were identified as DE (adjusted P < 0.05). Of these, 14 were decreased in blocked libraries, only one of which had a log2FC <˗1 (miR-1301).
Of the 57 nontarget miRs identified as DE in the paired cohort (log2FC > 1; adjusted P < 0.05), only 20 were decreased in blocked samples. All significantly decreased miRs identified in the pilot data were also decreased in the large cohort. However, 38 miRs were identified as DE in the paired cohort but not in the pilot, which may be because of the extraction batch effects or a broader range of miRNA content in the paired cohort. The fold change and CPM values of these 57 nontarget differentially expressed miRs are considerably lower than for the target miRs (a mean of 1 times fold change and 2800 CPM for nontarget differentially expressed miRs vs 5 times fold change and 325,000 CPM in miR-92a and miR-486) (Supplemental Figure S1E). In addition, odds ratios for even the top three nontarget DE miRs (miR-25, miR-92b, and miR-1301) were barely >1 or just <1.5, 1.1, and 0.5, respectively, compared with 6.24 for miR-486.
Because most (22/27 all nontarget differentially expressed miRs; 14/14 of decreased miRs) of the differentially expressed miRs in the pilot data were also identified as differentially expressed in the large cohort, potential off-target miRs identified using the pilot data were analyzed for the sake of simplicity. Some of these miRs are within the same family as the target miRNAs or share significant sequence similarity. Another consideration is the direction of the fold change of the differentially expressed miR; a positive value indicates that expression is increased in blocked samples, suggesting that it is not of significant concern as an off-target blocking oligo effect. The degree of significance (adjusted P value) also affects our confidence that the differentially expressed miR is a true off-target effect. Finally, another consideration is the raw read count for the differentially expressed miR. Although a CPM threshold was implemented before DE analysis, some miRNA species still have very low overall counts, decreasing our confidence and concern regarding these potential off-target effects (Table 1).
Taking into account P value (<0.01) and log2FC (<0) of all 27 of the potential off-targets, the list can be filtered down to just 12 of high confidence (Figure 4A). Of these, nine were detected at high levels (CPM >50, mean across all libraries). Similarly, for the paired large cohort, of 57 total nontarget differentially expressed miRs, only 18 are of high confidence (CPM >50, log2FC <˗1, adjusted P < 0.01) (Supplemental Figure S2C). All significantly decreased miRNAs from the pilot analysis (adjusted P < 0.05) share moderate sequence similarity with one of the target heme miRs; their reduced detection may be unavoidable with this technology (Figure 4B). Two miRs (miR-25 and miR-92b) share significant sequence similarity with miR-92a; more than half of the sequence of these two off-target miRs mirror the heme miR sequence. Three more miRs share moderate sequence similarity with miR-92a. Nine off-target miRNA species have moderate sequence similarity to miR-486. Interestingly, all these miRs share the same short motif of CUGCC (Figure 4B).
Benefits of Use of Blocking Oligonucleotides: Increased Sensitivity and DE Capability
These results have hinted at the possibility that the use of blocking oligonucleotides increases the number of reads for most miRNA species. To examine this possibility more quantitatively, the number of miRs that passed a total count threshold across blocked and unblocked libraries within the full large cohort were determined (Figure 5A). At every threshold tested, the blocked samples identified far more miRNA species passing that threshold than the unblocked. This is particularly true for lower thresholds (>1, 50, and 100 counts). In the full, unpaired large cohort, blocked libraries identified a mean of 274 additional species compared with unblocked libraries.
A similar analysis performed the pilot and the paired cohort, comparing the increase in number of species detected for each individuals. For each individual, the number of species reaching each count threshold in unblocked samples is subtracted from the number reaching each threshold in blocked samples. As for the unpaired cohort, the blocked samples had far more miRNA species reaching each count threshold than the unblocked (Figure 5B). Blocked samples had a mean of 120 more miRs with at least 50 counts than unblocked samples. Additionally, pilot libraries constructed with blockers identified a mean of 111 new miRNA species that were not detected at all in samples without blockers.
One of the most popular downstream analyses of miRNA sequencing data is DE. With the increased sensitivity that blocking oligonucleotides appear to provide, the use of blockers will likely increase the ability of DE to detect significantly DE miRNA species. To test this hypothesis DESeq2 was used to perform two DE analyses on the paired pilot libraries: three A libraries (without blocker) versus three B libraries (without blocker) and three A libraries (with blocker) versus three B libraries (with blocker). In the first analysis (unblocked libraries), 71 significantly differentially expressed miRNA species were found (at adjusted P < 0.05; Benjamini and Hochberg multiple testing correction; of 1705 miRs tested). A substantial (28%) improvement was observed in the second analysis using blocker samples, with 98 of 1705 significantly differentially expressed miRNA species being detected (Figure 4B).
At both significance cutoffs tested (0.05 and 0.01), the blocked group had a higher number of differentially expressed miRNAs. Most differentially expressed miRs identified in the unblocked analysis were also found in the blocked analysis (60/98) (Figure 5C). However, 17 miRs were identified as differentially expressed in the unblocked analysis but were not detected as differentially expressed in the blocked analysis. For miRs that were found only in the blocked or unblocked DE analysis, the median count is clearly lower than that of miRs found in both analyses (adjusted P < 0.01) (Figure 5D). Most miRs found in both analyses had a mean of at least 50 counts (85%). Blocked-specific miRs had fewer high-count miRs (54%), whereas unblocked-specific miRs had even fewer high-count miRs (35%). Figure 5E summarizes the overlaps between the two DE analyses.
Discussion
Erythropoietic miRNAs released from blood cells significantly alter the circulating miRNA content in serum and plasma samples, thereby decreasing sensitivity of biologically relevant circulating miRNAs. When the commonly used PAXgene RNA extraction system is used, virtually complete hemolysis of RBCs occurs.25 Although it is difficult to control for variables related to sample handling that lead to an overabundance of hemolytic miRNAs, blocking oligonucleotides targeting these miRNAs is effective in their removal. This study expands on this application through the use of a commercially available multiplexed pool of blocking oligonucleotides that target three of the most abundant hemolytic-associated miRNAs (hsa-miR-486, hsa-miR-451a, and hsa-miR-92a) and assesses their effectiveness on a large whole blood RNA cohort.3,13,14 In the unblocked samples, miR-486 and mir-92a represented 70% and 22% of total miRNA hits, respectively. However, miR-451 accounted for only 1.5%. Levels of hemolysis-associated miRNAs vary across samples and may reflect the level of RBC contamination in our samples.26 Indeed high levels of miR-451a (14%) were found in three unblocked pilot samples (A–C) (Figure 2B). In addition, it was determined that optimizaiton of input sample/blocking oligonucleotide ratios was not needed. Using varying amounts of input RNA, comparable blocking was achieved regardless of input levels (Figure 2A). Notably, the blocker is provided in the blocker mix at an overall concentration of 5 μmol/L, and when using 1 μL of blocker the system is expected to be saturated and is independent of the amount. As such, the absence of a dose effect (ie, the same level of reduction of target miRNAs regardless of input RNA) observed in the pilot study indicates that the blocker mix concentration is sufficient for a range of inputs. This flexibility would allow for quick implementation of this method to any target miR of interest at a broad range of RNA inputs.
A mean 90% increase was found in the number of reads supporting nonheme miRs (Figure 2D). This increase in nonheme miRs was proportionate to unblocked libraries and was found for virtually all nonheme miRs (Figure 3, D and E) and allowed for the detection of 201 miRs that were not previously detected (for large cohort; 111 additional miRs for pilot data) (Figure 5, A and B). Because many miRNA biomarkers are expressed at very low levels within the blood, the use of all three multiplexed blockers could allow for increased detection of these lowly expressed miRs, leading to improved efficacy of this new diagnostic tool. Additionally improved sensitivity was observed in DE with the use of blocking oligonucleotides, A comparison of blocked samples A and B found 28% more significantly differentially expressed miRs than a comparison of unblocked samples A and B.
Off-target effects of blocking oligonucleotide on nonheme miRs are minimal (Figure 3, D and E, and Figure 4A), and the impacted nontarget miRs identified in pilot samples share moderate sequence similarity with miR-486, miR-451a, or miR-92a (Figure 4B), which has also been reported in other blocking oligonucleotide designs targeting hemolytic miRNAs.14,18 Of the 14 nontarget miRs identified as significantly decreased in blocked pilot samples, all were also identified as significantly decreased in the paired large cohort analysis, supporting the idea that reduction of these miRs is unavoidable given the limits of the technology and driven by sequence similarity of these miRs to target miRs. Most notably, the significantly reduced miR-25, which has been associated with cell cycle regulation, shares 72% sequence homology with miR-92a.27 Interestingly, the same motif in miR-486 is found in nine off-target miRs, suggesting that this region within miR-486 may play an important role in binding and ligation prevention. Blockers appear to be more effective for miR-486 against the exact mature sequence, not allowing for any mismatches. An approximately 97% reduction in miR-486-5p detection was observed in pilot and large cohort samples, which matches exactly the full sequence of miR-486-5p, compared with an approximately 69% reduction if isomiRs are included (Figure 2D and Supplemental Figure S1D). Using miR-486 blocking oligonucleotides with a shorter sequence may result in more effective targeting because it accounts for minor differences in sequences within a pool of molecules. In contrast to miR-486, miR-451a had very few matches to the exact mature sequence in miRBase in pilot samples (approximately 1% of unblocked reads) (Supplemental Figure S1D). Instead, most reads assigned to this miRNA species had one or two insertions/deletions, especially at the 5′ and 3′ end (data not shown). The blocker for this miR was also the least effective of the three (44% reduced compared with 76% reduced for the two other targets) (Supplemental Figure S1D) and did not reduce miR-451a levels at all in the large cohort, although that is likely because of the low levels of miR-451a present within the samples (Figure 2D). Because most miR-451a molecules in the sample actually differ by several bases from the mature sequence, the blocking oligonucleotides could be less effective. It may be possible to minimize off-target effects for specific miRNAs by modifying the blocking oligonucleotide design; however, the short target regions offered by miRNAs make it difficult to completely avoid off-target hybridization.
When comparing DE in blocked and unblocked libraries, most DE miRs detected in unblocked libraries were also detected in the blocked libraries (Figure 5E). However, 17 miRs were only identified as DE in the unblocked analysis. The overall counts for these miRs found only in unblocked samples were quite low (40) compared with the counts for miRs found DE in both (593), suggesting technical variability in library preparation as a reason. However, blocking oligonucleotides may impede the detection of very lowly expressed species making less reliable for these miRs.
Overall, removal of unwanted erythropoietic miRNAs from whole blood samples before sequencing results in higher coverage of other miRNAs and detection of very lowly expressed miRs. Further work may determine the efficacy of this technique to samples derived from plasma or serum or other miRNAs from tissue types. In addition, although this study focused on three heme-related miRNAs that were highly abundant in the selected cohort, other highly abundant heme miRs (eg, miR-16, miR-21) have been reported.23,28 This approach, however, can easily be extended to these or other highly abundant miRs of interest. Thus, a key technical element to the successful design and application of these blockers is a priori knowledge of the abundant heme miRNAs present in samples from the study cohort by sequencing a few samples from that cohort without any blockers. A limitation of this application is the off-target effects that may block other mature miRNAs or nonmature miRNAs at varying levels. The off-target effects are driven mainly by sequence similarity to target miRs, and the level of inhibition of these nontarget miRNAs can be assessed by comparing to an initial unblocked miRNA sequencing run.
Despite limitations, this method offers highly multiplexed sequencing compatibilities while detecting biomarkers present at very low levels within whole blood samples. As clinical applications of miRNA biomarkers from whole blood are discovered, this technique may help reduce the cost and improve sensitivity of these assays.
Acknowledgment
Library construction and sequencing were performed at the Translational Genomics Core at Mass General Brigham Personalized Medicine.
Footnotes
Supported by NIH grants R01 HL127332 (K.T.), R01 HL129935 (K.T.), R01HL130512 (C.H.), R01HL125583 (C.H.), and R01HL139634 (M.J.M.) and COPDGene Study grants U01 HL089856 and U01 HL089897.
Disclosures: S.P. and K.A. are employees of PerkinElmer. J.L., M.B., A.B., S.T.W., and S.A. are employees of Mass General Brigham Personalized Medicine.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2021.03.006.
Supplemental Data
Supplemental Figure S1.
A: Percentage of reads mapping to any of the three target heme miRs in blocked and unblocked groups; pilot data used. B: Same analysis as in panel A performed for full large cohort (unpaired). C: Mean number of raw counts for each species in the same family as any of the targets; pilot data used. P = 7.8 × 10−9 (A); P = 2.2 × 10−16 (B).
Supplemental Figure S2.
A: Same analysis as in Figure 2D is performed for the pilot data. The percentage of reads mapping to every target miR, both mature and isomiR, is plotted for each sample, comparing blocked and unblocked percentages B: Both mature and isomiR forms of miR-25 are retained in the count matrix rather than collapsing together. Pairwise differential expression (DE) is performed, and the top 50 most significantly differentially expressed miRs are plotted on a heatmap. C: Off-target summary of DE from panel A. Log2 fold change (log2FC) and log2 adjusted P value plotted for each miR identified as DE (adjusted P < 0.05 and absolute log2FC > 1, with the size of points corresponding to mean raw counts of each species).
References
- 1.Lytle J.R., Yario T.A., Steitz J.A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5' UTR as in the 3' UTR. Proc Natl Acad Sci U S A. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 3.Pritchard C.C., Kroh E., Wood B., Arroyo J.D., Dougherty K.J., Miyaji M.M., Tait J.F., Tewari M. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 2012;5:492–497. doi: 10.1158/1940-6207.CAPR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stojanovic J., Tognetto A., Tiziano D.F., Leoncini E., Posteraro B., Pastorino R., Boccia S. MicroRNAs expression profiles as diagnostic biomarkers of gastric cancer: a systematic literature review. Biomarkers. 2019;24:110–119. doi: 10.1080/1354750X.2018.1539765. [DOI] [PubMed] [Google Scholar]
- 5.Wu H.Z., Ong K.L., Seeher K., Armstrong N.J., Thalamuthu A., Brodaty H., Sachdev P., Mather K. Circulating microRNAs as biomarkers of Alzheimer's disease: a systematic review. J Alzheimers Dis. 2016;49:755–766. doi: 10.3233/JAD-150619. [DOI] [PubMed] [Google Scholar]
- 6.Oses M., Margareto Sanchez J., Portillo M.P., Aguilera C.M., Labayen I. Circulating miRNAs as biomarkers of obesity and obesity-associated comorbidities in children and adolescents: a systematic review. Nutrients. 2019;11:2890. doi: 10.3390/nu11122890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srivastav S., Walitza S., Grunblatt E. Emerging role of miRNA in attention deficit hyperactivity disorder: a systematic review. Atten Defic Hyperact Disord. 2018;10:49–63. doi: 10.1007/s12402-017-0232-y. [DOI] [PubMed] [Google Scholar]
- 8.Adhami M., Haghdoost A.A., Sadeghi B., Malekpour Afshar R. Candidate miRNAs in human breast cancer biomarkers: a systematic review. Breast Cancer. 2018;25:198–205. doi: 10.1007/s12282-017-0814-8. [DOI] [PubMed] [Google Scholar]
- 9.Murray M.J., Watson H.L., Ward D., Bailey S., Ferraresso M., Nicholson J.C., Gnanapragasam V.J., Thomas B., Scarpini C.G., Coleman N. “Future-proofing” blood processing for measurement of circulating miRNAs in samples from biobanks and prospective clinical trials. Cancer Epidemiol Biomarkers Prev. 2018;27:208–218. doi: 10.1158/1055-9965.EPI-17-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez-Gutierrez A.D., Catalan O.M., Vázquez-Romo R., Porras Reyes F.I., Alvarado-Miranda A., Lara Medina F., Bargallo-Rocha J.E., Orozco Moreno L.T., Cantú De León D., Herrera L.A., Lopez-Camarillo C., Perez-Plasencia C., Campos-Parra A. l. miRNA profile obtained by next-generation sequencing in metastatic breast cancer patients is able to predict the response to systemic treatments. Int J Mol Med. 2019;44:1267–1280. doi: 10.3892/ijmm.2019.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vigneault F., Sismour A.M., Church G.M. Efficient microRNA capture and bar-coding via enzymatic oligonucleotide adenylation. Nat Methods. 2008;5:777–779. doi: 10.1038/nmeth.1244. [DOI] [PubMed] [Google Scholar]
- 12.Hafner M., Renwick N., Brown M., Mihailovic A., Holoch D., Lin C., Pena J.T., Nusbaum J.D., Morozov P., Ludwig J., Ojo T., Luo S., Schroth G., Tuschl T. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. Rna. 2011;17:1697–1712. doi: 10.1261/rna.2799511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin H., Shannon C.P., Fishbane N., Ruan J., Zhou M., Balshaw R., Wilson-McManus J.E., Ng R.T., McManus B.M., Tebbutt S.J. Variation in RNA-Seq transcriptome profiles of peripheral whole blood from healthy individuals with and without globin depletion. PLoS One. 2014;9:e91041. doi: 10.1371/journal.pone.0091041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juzenas S., Lindqvist C.M., Ito G., Dolshanskaya Y., Halfvarson J., Franke A., Hemmrich-Stanisak G. Depletion of erythropoietic miR-486-5p and miR-451a improves detectability of rare microRNAs in peripheral blood-derived small RNA sequencing libraries. NAR Genom Bioinformat. 2020;2:lqaa008. doi: 10.1093/nargab/lqaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shkurnikov M.Y., Knyazev E.N., Fomicheva K.A., Mikhailenko D.S., Nyushko K.M., Saribekyan E.K., Samatov T.R., Alekseev B.Y. Analysis of plasma microRNA associated with hemolysis. Bull Exp Biol Med. 2016;160:748–750. doi: 10.1007/s10517-016-3300-y. [DOI] [PubMed] [Google Scholar]
- 16.Pizzamiglio S., Zanutto S., Ciniselli C.M., Belfiore A., Bottelli S., Gariboldi M., Verderio P. A methodological procedure for evaluating the impact of hemolysis on circulating microRNAs. Oncol Letters. 2017;13:315–320. doi: 10.3892/ol.2016.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirschner M.B., Edelman J.J.B., Kao S.C.H., Vallely M.P., van Zandwijk N., Reid G. The impact of hemolysis on cell-Free microRNA biomarkers. Front Genet. 2013;4:94. doi: 10.3389/fgene.2013.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts B.S., Hardigan A.A., Kirby M.K., Fitz-Gerald M.B., Wilcox C.M., Kimberly R.P., Myers R.M. Blocking of targeted microRNAs from next-generation sequencing libraries. Nucleic Acids Res. 2015;43:e145. doi: 10.1093/nar/gkv724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu C.W., Evans J.M., Huang S., Mahoney D.W., Dukek B.A., Taylor W.R., Yab T.C., Smyrk T.C., Jen J., Kisiel J.B., Ahlquist D.A. A Comprehensive Approach to Sequence-oriented IsomiR annotation (CASMIR): demonstration with IsomiR profiling in colorectal neoplasia. BMC Genomics. 2018;19:401. doi: 10.1186/s12864-018-4794-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhanoa J.K., Verma R., Sethi R.S., Arora J.S., Mukhopadhyay C.S. Biogenesis and biological implications of isomiRs in mammals- a review. ExRNA. 2019;1:3. [Google Scholar]
- 21.Wright C., Rajpurohit A., Burke E.E., Williams C., Collado-Torres L., Kimos M., Brandon N.J., Cross A.J., Jaffe A.E., Weinberger D.R., Shin J.H. Comprehensive assessment of multiple biases in small RNA sequencing reveals significant differences in the performance of widely used methods. BMC Genomics. 2019;20:513. doi: 10.1186/s12864-019-5870-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myklebust M.P., Rosenlund B., Gjengstø P., Bercea B.S., Karlsdottir Á., Brydøy M., Dahl O. Quantitative PCR measurement of miR-371a-3p and miR-372-p is influenced by hemolysis. Front Genetics. 2019;10:463. doi: 10.3389/fgene.2019.00463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edelman M.B.K.B., Kao S.C.-H., Vallely M.P., Zandwijk Nv, Reid1 G. The impact of hemolysis on cell-Free microRNA biomarkers. Front Genet. 2013;94 doi: 10.3389/fgene.2013.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Love M.I., Huber W., anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer A., Paroni F., Günther K., Dharmadhikari G., Ahrens W., Kelm S., Maedler K. Evaluation of existing methods for human blood mRNA isolation and analysis for large studies. PloS One. 2016;11:e0161778. doi: 10.1371/journal.pone.0161778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirschner M.B., Kao S.C., Edelman J.J., Armstrong N.J., Vallely M.P., van Zandwijk N., Reid G. Haemolysis during sample preparation alters microRNA content of plasma. PLoS One. 2011;6:e24145. doi: 10.1371/journal.pone.0024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petrocca F., Visone R., Onelli M.R., Shah M.H., Nicoloso M.S., de Martino I., Iliopoulos D., Pilozzi E., Liu C.G., Negrini M., Cavazzini L., Volinia S., Alder H., Ruco L.P., Baldassarre G., Croce C.M., Vacchione A. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 28.Yamada A., Cox M.A., Gaffney K.A., Moreland A., Boland C.R., Goel A. Technical factors involved in the measurement of circulating microRNA biomarkers for the detection of colorectal neoplasia. PLoS One. 2014;9:e112481. doi: 10.1371/journal.pone.0112481. [DOI] [PMC free article] [PubMed] [Google Scholar]