Abstract
Background
Clinical outcomes in high-grade glioma (HGG) have remained relatively unchanged over the last 3 decades with only modest increases in overall survival. Despite the validation of biomarkers to classify treatment response, most newly diagnosed (ND) patients receive the same treatment regimen. This study aimed to determine whether a prospective functional assay that provides a direct, live tumor cell-based drug response prediction specific for each patient could accurately predict clinical drug response prior to treatment.
Methods
A modified 3D cell culture assay was validated to establish baseline parameters including drug concentrations, timing, and reproducibility. Live tumor tissue from HGG patients were tested in the assay to establish response parameters. Clinical correlation was determined between prospective ex vivo response and clinical response in ND HGG patients enrolled in 3D-PREDICT (ClinicalTrials.gov Identifier: NCT03561207). Clinical case studies were examined for relapsed HGG patients enrolled on 3D-PREDICT, prospectively assayed for ex vivo drug response, and monitored for follow-up.
Results
Absent biomarker stratification, the test accurately predicted clinical response/nonresponse to temozolomide in 17/20 (85%, P = .007) ND patients within 7 days of their surgery, prior to treatment initiation. Test-predicted responders had a median overall survival post-surgery of 11.6 months compared to 5.9 months for test-predicted nonresponders (P = .0376). Case studies provided examples of the clinical utility of the assay predictions and their impact upon treatment decisions resulting in positive clinical outcomes.
Conclusion
This study both validates the developed assay analytically and clinically and provides case studies of its implementation in clinical practice.
Keywords: drug response prediction, 3D culture, ex vivo, glioblastoma, glioma
Key Points.
An ex vivo assay prospectively predicted drug response for patients with HGG.
Ex vivo assay provided evidence to use dabrafenib when NGS results did not.
Importance of the Study.
HGG, including GBM, is highly aggressive with limited treatment options and short survival times. Here, we demonstrate the analytical and clinical validation of an assay to predict patient-specific response to chemotherapy within an actionable timeframe prior to treatment initiation. The clinical benefit of this assay for patients is further supported through clinical case studies demonstrating successful assay-directed use of salvage agents not traditionally used in recurrent HGG. In the future, the clinical application of assay results may direct patients and clinicians to more efficacious treatments or provide them with the information necessary to guide clinical trial enrollment.
Treatment for newly diagnosed (ND), high-grade glioma (HGG) has remained virtually the same for decades with minimally improved survival outcomes.1 For ND patients, NCCN guidelines direct clinicians to maximal safe surgical resection and clinical trial enrollment or concurrent radiation with temozolomide (TMZ).2,3 Following first-line therapy, tumor progression is nearly universal within 6–9 months.4 Time to second recurrence is generally shorter in duration. Treatment of recurrent HGG is problematic due to these time constraints and the increasingly aggressive nature of the disease. Choosing a second-line therapy is complex due to patient-specific and antineoplastic agent-specific factors combined with a lack of clear, data-driven algorithms to guide selection. Patient-specific variability in therapeutic efficacy and resultant clinical outcomes suggests significant inter-tumor heterogeneity which has been increasingly elucidated by molecular testing5 and highlights the importance of personalized treatment strategies.
Previous attempts to predict patient-specific response to chemotherapy ex vivo6–9 have not been recommended by the American Society of Clinical Oncology except for patient treatment selection in a clinical trial10 partially because they often fail to provide a clinically meaningful positive predictive value (PPV), with most only advising which drugs to avoid. The lack of success of these assays may be attributed to their failure to prospectively correlate with sufficient accuracy to clinical outcomes or their inability to present a result within an actionable timeframe.11,12
Utilizing the increased biological fidelity of 3D cell culture, we previously validated an assay that prospectively and accurately predicts (89%, P = .0004) response to first-line chemotherapy in ND ovarian cancer with results returned within 7 business days of each patient’s surgery and a PPV of 100%.13 We have modified our platform technology to facilitate testing in HGG and analytically and clinically validated the new assay for the individualized selection of chemotherapy specific for HGG. We present prospective clinical prediction in ND HGG and evidence of the clinical application of this assay in recurrent HGG as part of the observational clinical study 3D-PREDICT (ClinicalTrials.gov Identifier: NCT03561207). Together, these data provide evidence of the validation of the assay for regulatory purposes and the clinical applicability of the data generated.
Materials and Methods
Cell Lines and Reagents
SF-268, SF-539, SNB-19, and U-251 were procured from the NCI DTP, DCTD Tumor Repository. No further authentication was performed following receipt. All cell lines were used within 10 passages from the stock. Cell lines were cultured in RPMI 1640 supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Corning). Cell lines were kept at 37°C and 5% CO2 with media changes every other day and passaged at 80%–90% confluency. Drugs were sourced from Selleckchem or MedChemExpress and diluted as recommended by the manufacturers.
Specimen Collection for Assay Development
After providing written informed consent, 55 patients, ≥18 years of age with suspected or known HGG, ND and recurrent, were enrolled onto a prospective, nonintervention, Institutional Review Board (IRB) approved biology protocol at Prisma Health Cancer Institute. Tissue acquisition was carried out in accordance with the guidelines and regulations specified by the IRB. Patients received clinical care according to standard practice and results of the assay were not provided to the patient or clinician.
Clinical Study Participants and Study Design
ND and recurrent HGG patients were prospectively enrolled in the multisite, observational clinical study “3D-PREDICT REGISTRY: 3D Prediction of Patient-specific Response Using Ex Vivo Interrogation of Live Cells from Tumors,” also entitled 3D-PREDICT (ClinicalTrials.gov ID NCT03561207). IRB approval for the 3D-PREDICT protocol was obtained from a central IRB and/or each site’s local IRB. All patients provided written informed consent. Individuals underwent maximal safe resection and a portion of the tissue collected was submitted for ex vivo drug response profiling utilizing the 3D Predict Glioma assay. Clinical data were collected for each patient during follow-up visits at approximately 3-month intervals. Clinicians and patients were informed of the assay predictions according to IRB preferences. In all cases chemotherapeutic agent selection was guided by the neuro-oncologist’s clinical judgement. For recurrent patients, the clinician considered a combination of the following factors: patient’s age, performance status, comorbidities, toxicities/side effect profile of potential chemotherapy agents, and results of the 3D Predict Glioma assay. For the purposes of this study, progression-free survival (PFS) and overall survival (OS) were both defined from the time of surgical resection to the time of measured progression or death. Clinical progression was defined by radiographic progression as interpreted by the treating clinicians.
3D Predict Glioma Assay Performance
The 3D Predict Glioma assay was performed as previously described.13 Drugs were dosed at 1 of 4 dose curves from 0.005 to 100 µM, 3.9 to 2000 µM, 39 to 20 mM, or 0.0005 to 10 µM depending upon optimized response ranges. For TMZ response determination, sufficient clinical data were available to establish binary response/nonresponse predictions through ROC curve analysis (AUC = 0.8469, 95% CI 0.6727 to 1.0, P = .0112). Without sufficient clinical data to establish binary predictions, IC50 thresholds separating test-predicted response/nonresponse were determined experimentally for each drug by quartile analysis of test data from multiple patient samples. IC50 values were collated for each individual drug and cutoffs informed by the approximate 25th and 75th quartiles were used to determine predicted responders (below the lower threshold), moderate responders (between the lower and upper thresholds), and nonresponders (above the upper threshold). IC50 values above the range of concentrations tested were considered nonresponders. No responses fell below the tested drug curves.
Sequencing
DNA was isolated from tissue fragments of 3D Predict Glioma samples using QIAGEN’s DNA mini kit. Next-generation sequencing (NGS) to identify the presence of BRAF activating mutations was performed by OHSU Knight Diagnostic Laboratories using their GeneTrails Comprehensive Solid Tumor Panel.
Statistical Analysis
IC50 values were calculated using nonlinear regression normalized to vehicle treated controls. Primary analysis of assay data and clinical outcomes utilized a Fisher’s Exact test to measure statistical significance of the data distribution. Kaplan–Meier curves and log-rank tests were used to examine the statistical associations between assay results and survival.
Results
Optimization and Analytical Validation of the Assay
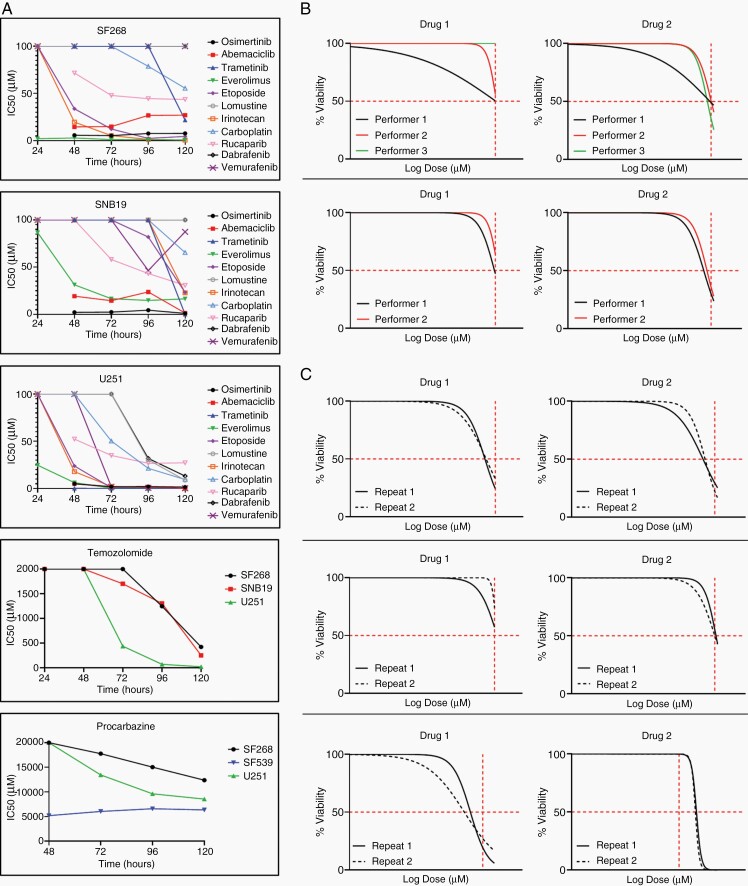
To adapt our previously validated and published assay13 for HGG, 12 drugs were validated for the drug panel based upon clinician input and potential efficacy in HGG, including agents listed in current NCCN guidelines3 and those tested in clinical trials.14–17 Optimal drug exposure duration for each drug was established against a panel of glioma cell lines (Figure 1A) to ensure assay readouts were not an artifact of extended or shortened exposure times. Drug response was measured every 24 h and optimal exposure times were defined as producing an IC50 value midway of the drug concentration range tested. Both TMZ and procarbazine required modified dose ranges. To analytically validate the assay, reproducibility was performed on primary samples (Figure 1B and C). Inter-assay reproducibility was performed by at least 2 technicians with a high level of reproducibility (Figure 1B). For intra-assay reproducibility, the assay was performed twice by the same technician (Figure 1C). The similarity of results was judged by the ability of the IC50s to fit into previously classified response/nonresponse categories.
Figure 1.
(A) Timing of optimal drug response prediction in 3 HGG cell lines. (B,C) Representative data of inter- and intra-assay reproducibility testing. Reproducibility was defined as having similar drug response readouts indicated by the vertical red hash lines in the graphs. (B) Overlapping nonlinear regression curves of assay generated drug response performed by multiple operators on 2 primary tissue samples and 2 drugs. (C) Repetition of assay generated drug response by a single operator for 3 different primary tissue samples and 2 drugs each. n = 7.
Newly Diagnosed HGG Study Design and Patient Characteristics
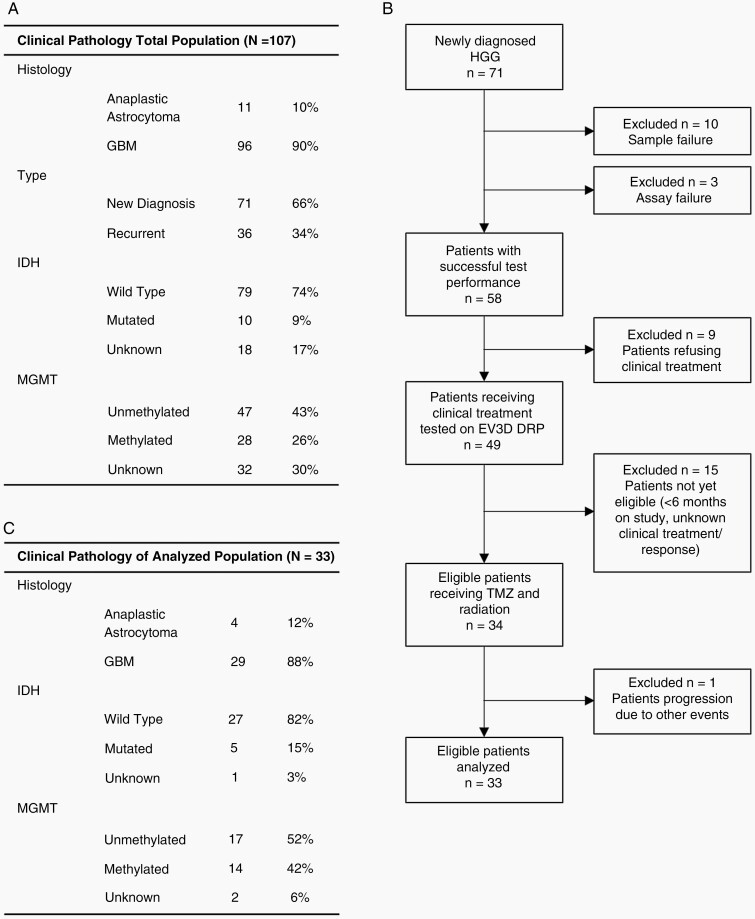
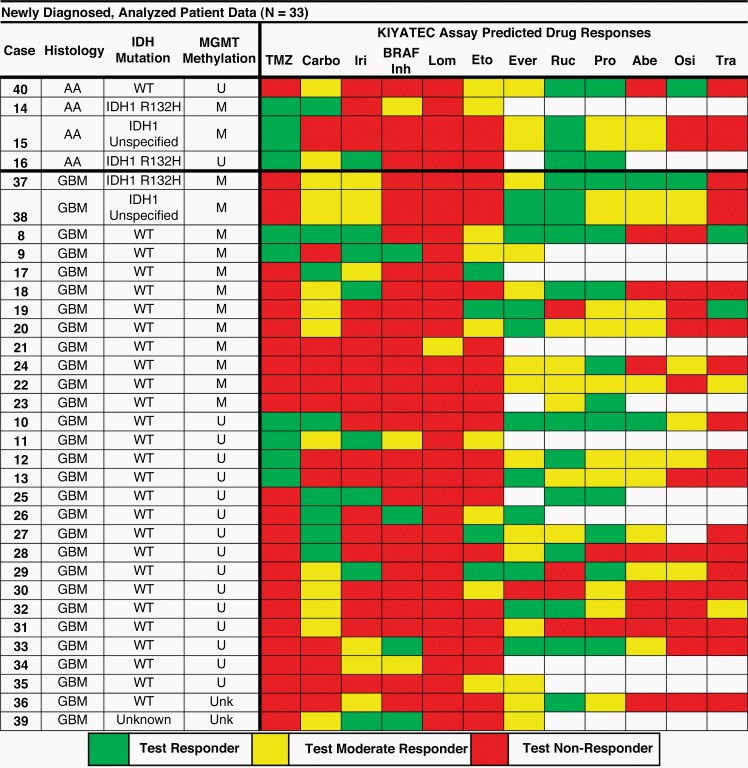
Fifty-five patients with ND or recurrent HGG were enrolled from November 2014 to July 2018. Samples from 44 patients were utilized in assay development, analytical validation, and the determination of response range categories. The remaining 11 patients were combined with an additional 96 patients enrolled from January 2019 to December 2020 as part of the clinical study 3D-PREDICT (ClinicalTrials.gov ID NCT03561207) to examine the ability of the 3D Predict Glioma assay to prospectively predict response to TMZ following maximal safe surgical resection (Figure 2A). Of the 107 samples assayed, 36 were recurrent and removed from this analysis. Ten samples failed quality control based upon cell numbers or viability, and 3 samples failed due to poor assay performance (3/61) indicating a 95% success rate for assay performance. Of the 58 remaining samples, 25 were not analyzed: 9 due to lack of clinical treatment, 15 due to insufficient follow-up at the time of this analysis, and 1 due to progression due to other events (Figure 2B and C). The 33 remaining patient tissue samples were successfully assayed for categorical response to TMZ and up to 11 other compounds (Figure 3). Every sample had a measurable level of response or moderate response to at least one agent, supporting the potential of the assay to benefit a large proportion of HGG patients during drug candidate selection. All 33 patients received clinical treatment consisting of radiation and TMZ following surgical debulking enabling a comparison between assay predicted response and clinical response.
Figure 2.
(A) Clinical pathology of all patients enrolled in the study. (B) The patient sample inclusion process. (C) Clinical pathology of the patients tested in the established assay.
Figure 3.
Clinical pathology of the 33 patients whose assay response was correlated to clinical response. Blank squares indicate drug not tested. Abbreviations: R/T = radiation + TMZ, mT = maintenance TMZ, WT = wild type, U = unmethylated, M = methylated, Carbo = carboplatin, Iri = irinotecan, Lom = lomustine, Eto = etoposide, Ever = everolimus, Ruc = rucaparib, Pro = procarbazine, Abe = abemaciclib, Osi = osimertinib, Tra = trametinib.
Prospective Correlation of Clinical Response and 3D Predict Glioma for Temozolomide in Newly Diagnosed HGG
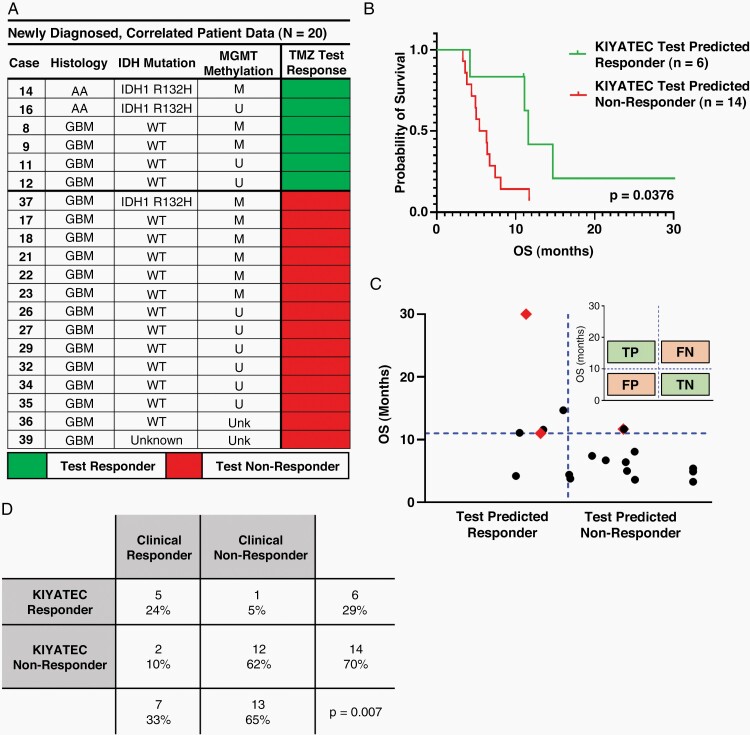
To determine the accuracy of 3D Predict Glioma prospective predictions of patient drug response, assay determined TMZ response was compared to clinical OS. Assay prediction was generated within 7 days of the patient’s debulking surgery, prior to the initiation of treatment. Of the 33 patients, 20 progressed at the time of this analysis to compare their clinical response to the test response. Six were prospectively predicted responders to TMZ and 14 were predicted nonresponders (Figure 4). TMZ test-predicted responders had a median OS of 11.6 months (4.2–30.4) compared to 5.9 months (3.3–11.7) for TMZ test-predicted nonresponders (P = .0376, HR 0.3455, 95% CI 0.1333 to 0.8954) (Figure 4B). To determine overall assay predictivity, clinical response to TMZ was defined as an OS greater than 11 months post-surgery based upon published studies for surgical debulking with radiation only.18–21 This facilitated the categorization of patients as true positives, true negatives, false positives, and false negatives (Figure 4C inset). When patients were distributed along these rules, test prediction accuracy was 85% (P = .007) (Figure 4C and D). This test accuracy rate is comparable to our previously published accuracy of 89% in high-grade ovarian cancer,13 indicating the potential of the assay to remain predictive across multiple solid tumor types.
Figure 4.
(A) Summary of the 20 patients whose TMZ test prediction data were correlated to clinical response. (B) Kaplan–Meier plot of the 20 patients who progressed enough to make a call on assay prediction following initial surgery separated by assay defined TMZ response. (C) Scatter plot of patients analyzed within this cohort. Red diamonds indicate the 4 patients who have progressed enough to assess assay prediction of OS but are still alive. The inset defines the parameters for categorizing patients as true positive (TP), true negative (TN), false positive (FP), and false negative (FN) according to both assay response and clinical response. (D) Contingency table of response cohorts based upon assay defined TMZ response.
Clinical Application of 3D Predict Glioma in Recurrent HGG
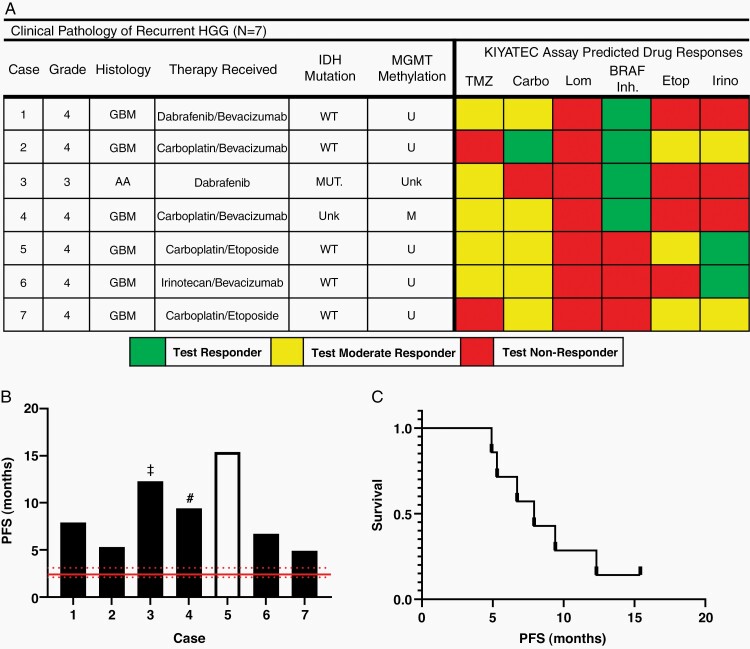
Commonly used cytotoxic chemotherapies for recurrent HGG have relatively similar response rates (<20%)22,23 and the determination of clinical use may be based upon a number of factors including patient performance status, drug toxicities, and ease of administration, none of which are associated with patient-specific biomarkers predicting efficacy. Seven patients with recurrent HGG enrolled in 3D-PREDICT for which treatment selection was influenced by the 3D Predict Glioma results and with at least 6 months of follow-up from surgical resection, were identified through physician survey and chart review at one site (Figure 5). Information obtained from 3D Predict Glioma was used along with consideration of patient-specific and agent-specific factors to facilitate selection of therapy. When considering salvage therapies, the 3D Predict Glioma assay helped guide therapy in all presented recurrent cases including agents to consider and those to avoid where no response was indicated. If 2 agents were being considered, the agent with a stronger 3D Predict Glioma response was chosen.
Figure 5.
(A) Clinical pathology of the 7 recurrent patients whose assay response was utilized as part of their clinical treatment. Abbreviations: WT = wild type, U = unmethylated, M = methylated. (B) PFS of the patients measured from time of surgery for which tumor tissue was used in the 3D Predict Glioma assay to the time of radiographic progression. Clear bar indicates the patient has not progressed at the time this work was submitted, ‡ indicates the patient tested at their third relapse, fourth surgery, and # indicates the patient tested at their second relapse, third surgery. Red lines indicate published median PFS and 95% CI (C) Kaplan–Meier plot of the patients within this cohort.
All patients underwent tissue collection at their first relapse or beyond, with one patient on second relapse/third resection, and one on third relapse/fourth resection (Figure 5A and B). Drug response was measured across the same panel of drugs as the ND patients (Figure 5A). The 7 patients had a median PFS of 7.9 months post-surgery (Figure 5C). Below we present brief case vignettes of their courses, the role the assay played, and outcomes to date.
Carboplatin
Carboplatin can cross the blood–brain barrier (BBB) and has been studied both as monotherapy and in combination with multiple agents, including bevacizumab17 and etoposide.16,24 3D Predict Glioma results were used to direct clinical use of carboplatin over other salvage agents for 4 patients with relative success. Two patients received carboplatin in combination with bevacizumab previously shown to have a median PFS of 3.5 months (2.2–3.7) with no additional clinical benefit over bevacizumab alone.17 In our clinical case studies, the PFS of each patient was 5.3 and 9.4 months, exceeding the published median PFS (Figure 6A). The other 2 patients received carboplatin in combination with etoposide, also indicated as a responder by 3D Predict Glioma. Carboplatin/etoposide combination therapy has been shown to have a median PFS of 4 months (3–5).24 In our clinical case studies, the PFS of each patient was 15.4 and 4.9 months, meeting or exceeding the published PFS (Figure 6B). Thus, outcomes of treatment decisions that incorporated the 3D Predict Glioma assay indicated drug responses met or exceeded previously published median PFS for the drug combinations with which they were treated.
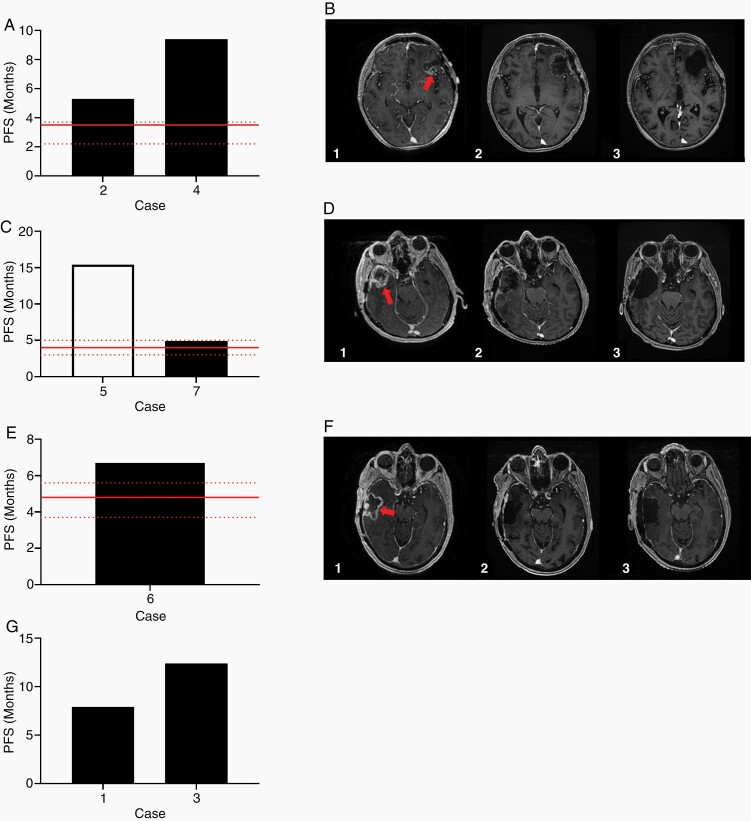
Figure 6.
(A) PFS of the patients treated with combination carboplatin/bevacizumab. Red line indicates published median PFS and 95% CI for patients treated with carboplatin/bevacizumab combination therapy. (B) MRI of the brain of one patient treated with carboplatin/bevacizumab combination therapy. T1 weighted contrasted axial images immediately preceding the patient’s third resection (1) showing enhancing nodularity (arrow) within the prior resection cavity, immediately following the third resection (2), and after 6 cycles of carboplatin/bevacizumab (3) with no evidence of enhancing disease in the cavity. (C) PFS of the patients treated with combination carboplatin/etoposide. Red line indicates published median PFS and 95% CI for patients treated with carboplatin/etoposide combination therapy. The white bar indicates that at the time of this publication, the patient has not progressed. (D) MRI of the brain of one patient treated with carboplatin/etoposide combination therapy. T1 weighted contrasted axial images immediately preceding the patient’s second resection (1) with bulky recurrent enhancing disease (arrow), immediately following the second resection (2), and after 6 cycles of carboplatin/etoposide (3) with no evidence of enhancing disease. (E) PFS of the patient treated with combination irinotecan/bevacizumab. Red line indicates published median PFS and 95% CI for patients treated with irinotecan/bevacizumab combination therapy. (F) MRI of the brain of the patient treated with irinotecan/bevacizumab combination therapy. T1 weighted contrasted axial images immediately preceding the patient’s second resection (1) showing enhancing area of recurrence (arrow), immediately following the second resection (2), and after 5 cycles of irinotecan/bevacizumab (3) without evidence of progressive disease. (G) PFS of the patients treated with dabrafenib.
Case 2, a 59-year-old male, underwent surgical resection for recurrent IDH wild-type glioblastoma (GBM) with unmethylated MGMT promoter. He was treated with concurrent chemoradiation with TMZ and 6 cycles of adjuvant TMZ. Imaging demonstrated radiographic progression at 7 months. The patient underwent re-resection with placement of carmustine wafers in the surgical cavity. Tissue sent for 3D Predict Glioma revealed a response to carboplatin, and he was treated with combination carboplatin/bevacizumab. He remained clinically and radiographically stable for a period of 5 months before radiographic progression. Case 4, a 63-year-old female, underwent resection of a GBM, IDH wild-type, with methylated MGMT promoter. She underwent concurrent chemoradiation with TMZ and 2 cycles of adjuvant TMZ. Imaging demonstrated radiographic progression 5 months later. She underwent craniotomy for re-resection, followed by 17 months of lomustine/bevacizumab. After completion of this therapy, radiographic progression was noted. Due to excellent performance status, a third craniotomy was performed for re-resection, and the tissue provided for 3D Predict Glioma revealed a moderate response to carboplatin. The patient was subsequently treated with carboplatin/bevacizumab for 9 months. She remained radiographically and clinically stable for 11 months before recurring with marked drop off in performance status (Figure 6A and B).
Case 5, a 63-year-old male, presented with a right temporal mass with significant surrounding vasogenic edema and midline shift. He underwent craniotomy for maximal safe resection. Pathology was consistent with IDH-wild type GBM, unmethylated MGMT promoter. He underwent standard concurrent chemoradiation with TMZ. Imaging at completion demonstrated bulky recurrence within the surgical cavity at 4 months. He underwent craniotomy for re-resection and placement of carmustine wafers, with 3D Predict Glioma indicating a moderate response to carboplatin and etoposide. He completed 6 cycles of combination therapy over approximately 4 months, with no recurrent disease noted over the subsequent 15 months to date off treatment, maintaining an excellent performance status. Case 7, a 60-year-old female, presented with pathology consistent with IDH-wild type GBM, unmethylated MGMT promoter. She underwent standard concurrent chemoradiation, followed by gamma knife radiosurgery to an enhancing nodule noted post-intensity-modulated radiation therapy. She was treated with 10 cycles of adjuvant TMZ before progressing at 13 months. Re-resection was performed, and tissue was sent for 3D Predict Glioma revealing a moderate response to carboplatin and etoposide with which she was treated for 4 cycles. Radiographic progression was noted at 5 months, followed by a third craniotomy. Tissue was again sent for 3D Predict Glioma revealing the tissue was no longer responsive to carboplatin and etoposide matching the clinical resistance and observed recurrence (Figure 6C and D).
Irinotecan
Irinotecan has been studied clinically as monotherapy and in combination with bevacizumab for recurrent HGG. As monotherapy it has been shown to have a median PFS of approximately 2.7 months which increases to approximately 4.8 months (3.7–5.6) when used in combination with bevacizumab.14,25–27 Case 6, a 70-year-old male, underwent craniotomy for resection of IDH-wild type GBM, unmethylated MGMT promoter. He subsequently underwent standard concurrent chemoradiation with TMZ, followed by an abbreviated course of adjuvant TMZ with disease progression after 5 months. He underwent re-resection with tissue submitted for 3D Predict Glioma which revealed a response to irinotecan. He was placed on irinotecan/bevacizumab, which he tolerated well for 8 cycles. He did experience radiographic progression with punctate areas of enhancement noted in the surgical cavity at 6 months. These relatively small areas of progression were treated with gamma knife radiosurgery. He continued combination irinotecan/bevacizumab with excellent tolerance for an additional 3 months, at which point treatment was discontinued due to further progression and declining performance status. Importantly, his PFS of 6.7 months exceeded that of those published for irinotecan with bevacizumab (Figure 6E and F).
BRAF Inhibitors
For some patients, targeted therapies for specific genetic mutations have proven successful in solid tumors. However, many of these therapies are not efficacious in HGG. Importantly, some patients benefit from these therapies without harboring the targeted mutation, demonstrating the limitations of NGS for personalized medicine and suggesting alternative signaling pathways not yet identified or described. Assays that measure functional response may identify those patients not recognized by NGS. During this observational study, 2 patients were identified by 3D Predict Glioma as responding to the BRAF inhibitor vemurafenib (Figure 6G). Since dabrafenib has an improved ability to cross the BBB, both patients received dabrafenib as their treatment.28,29
Case 1, a 54-year-old male underwent surgical resection of right temporal GBM, IDH-wild type, unmethylated MGMT promoter. Standard concurrent chemoradiation was followed by adjuvant TMZ for 6 cycles in conjunction with tumor treating fields (TTF) therapy.30 Imaging at 9 months demonstrated local progression, ultimately confirmed by surgical resection with subsequent tissue submission for 3D Predict Glioma. The patient was placed on bevacizumab/dabrafenib based upon assay determined BRAF inhibitor response. He experienced a 4-month period of stability and maintained reasonable quality of life with minimal drug toxicity. Radiographic progression was noted at 4 months followed by a clinical decline. Case 3, a 24-year-old male, underwent resection of a left temporal lobe WHO grade III anaplastic astrocytoma (AA). He completed concurrent chemoradiation with TMZ followed by 24 cycles of adjuvant TMZ then 2 additional years of disease stability off treatment. Re-resection was performed at first recurrence, and he was subsequently re-challenged with TMZ. Disease stability was achieved for 2.5 years. Additional re-resection was performed after subsequent recurrence, and he was treated with combination lomustine/bevacizumab. He was again noted to have radiographic progression at 18 months, undergoing a fourth craniotomy, during which tissue was sent for 3D Predict Glioma revealing response to vemurafenib. He was treated with dabrafenib for 12 months with disease stability and limited toxicity before again progressing, undergoing a fifth craniotomy during which a second tissue sample was assayed for drug response. This tissue sample was noted to be less responsive to BRAF inhibitor than the previous tissue specimen. Importantly, the tissues received for both patients did not harbor any known BRAF activating mutations. In one of these cases, the 3D Predict Glioma data aided the facilitation of insurance coverage for dabrafenib in the absence of molecular diagnostic indications.
Discussion
HGG treatment remains challenging and the lack of durable response in ND patients has led to guidelines recommending clinical trial enrollment.3 Guidance for treatment post-recurrence is even more limited and lacks standardization. This ambiguity combined with extremely short survival times and short recurrence cycles makes it imperative that oncologists pick the most efficacious therapy first. A testing platform with clinically actionable results providing insight into patient response to potential therapies would be invaluable to the neuro-oncologist. In this study we analytically and clinically validated an assay for individualized response predictions across a panel of 12 relevant agents and demonstrated institutional experience utilizing this personalized information in 7 patients with recurrent HGG. In ND HGG, the assay was 85% accurate (P = .007) in prediction of TMZ treatment response in combination with radiation. Assay-predicted responders had a 5.7-month extended survival time compared to assay-predicted nonresponders. In recurrent HGG, the 7 patients treated with 3D Predict Glioma defined treatments had a median PFS of 7.9 months (4.9–15.4) compared to a historical median PFS of 2.4 months (2.1–3.1).31,32
The need for accurate characterization of patient-specific outcomes before treatment initiation is generally undisputed and evident in the broad use of NGS. However, NGS only indicates the presence of an actionable mutation linked to a targeted agent; it does not assess individual patient response to that agent. Two cases in this study received the targeted agent dabrafenib due to 3D Predict Glioma, but associated NGS results failed to show an associated BRAF mutation, demonstrating the potential clinical limitations of NGS panels. The lack of widespread clinical adoption of personalized, phenotypic assays is primarily due to a paucity of demonstrated clinical accuracy. Tumor response prediction assays must include ex vivo recapitulation of in vivo biology, a high rate of assay performance success from clinically relevant patient samples, and clinically actionable turnaround times. The high correlation and positive clinical impact of this assay can be, in part, attributed to these achievements. This study has shown the feasibility of using ex vivo drug response testing in clinical care to support personalized approaches in a disease where evidence-based medicine is minimal, and the accuracy has been demonstrated in a clinically relevant patient population. The median age of GBM is 64 years with typical IDH mutation rates of 5%.33,34 Patients in this study had a median age of 66 years and a IDH mutation rate of 3% supporting the clinical relevancy of the patient population.
In this study, multiple confounding factors unrelated to assay performance may have impacted outcomes, including time between resections and chemotherapy initiation, bevacizumab/chemotherapy combinations, radiographic versus clinical assessment to define progression, other treatments including carmustine wafers, TTF, and gamma knife radiosurgery, inclusion of AA which in general has a higher survival rate than GBM, and selection bias toward patients with good performance status for resection. In the recurrent population, time between resection and treatment initiation averaged 6.7 weeks (3–14) with the longest being for the patients who received dabrafenib as time was spent waiting for insurance approval. The 3D Predict Glioma assay does not currently assess bevacizumab response. So assay response to single agent was examined in relation to published PFS for patients clinically treated by a bevacizumab combination. Future interventional studies will provide better control over other treatments, but the inherent selection bias will always be present as fresh, live tissue is required for assay performance. Future development to reduce required tissue amounts for assay performance may increase the number of patients able utilize the assay. Finally, the performance of a large, randomized controlled trial will provide better evidence of the assay’s usefulness.
This study presented results from the validation of a new assay to individually direct the treatment of patients with HGG. Rather than correlating a biomarker with the probability of treatment response, this assay assesses evidence of response by measuring the interaction of a patient’s primary tumor cells with therapy options. This translates to patient-specific, biologically driven data, eliminating probabilistic uncertainty from a population study. The prospective predictive ability of the assay was demonstrated in ND HGG treated with radiation and TMZ, and in a small number of recurrent HGG cases in which the clinician chose to use assay results to direct therapy, the selected therapy met or exceeded published median PFS. These data indicate the potential of this new assay to provide clinicians and patients with a method of determining which treatment option to use when time is not available for experimentation with therapy regimens. 3D Predict Glioma provided an additional tool to guide the neuro-oncologist’s choice of therapy for recurrent patients. Decision-making regarding salvage therapies in GBM can be challenging. In a disease with such variable responses and survival, tumor-specific information has the potential to be clinically beneficial. The ability to have individualized data to guide decisions, based on patient-specific evidence of response, realizes a shift toward individualized medicine that other approaches to HGG treatment have been unable to accomplish.
Acknowledgments
The authors would like to thank the patients for their participation and the nurses and staff of the respective institutions for assistance with patient enrollment, sample procurement, and the collection of clinical follow-up data.
Funding
This study was supported in part by the National Institutes of Health, National Cancer Institute Small Business Innovation Research Contract (HHSN261201400019C); Roswell Park Comprehensive Cancer Center and National Institutes of Health, National Cancer Institute (P30CA016056).
Conflict of interest statement. N.A.B. and J.E. have received compensation as a member of the clinical advisory board of KIYATEC, Inc. A.J.F. receives research support from Arbor Pharmaceuticals. J.E. is a consultant for Chimerix. L.O., H.E.C., and M.G. are shareholders in KIYATEC, Inc. S.S., A.M.S., M.R., L.O., J.S., H.E.C., C.R.T.V., L.H., M.G., and T.M.D. are current or former employees of KIYATEC, Inc.
Authorship Statement. Study design: H.E.C., L.H., M.G., T.M.D. In vitro, ex vivo, and in vivo experiments and downstream, analysis: S.S., L.L., A.A., A.M.S., M.R., L.O., J.S., L.L.M., A.J.F., J.E., C.K., S.G., P.H., M.L., N.A.B., S.J.H., N.R., L.H., R.A.F., T.M.D. Analysis and interpretation of data (eg, statistical analysis, clinical outcomes): L.L., A.A., M.R., T.M.D. Administrative, technical, or material support (ie, reporting or organizing data): A.M.S., L.H. Study supervision: C.R.T.V., N.A.B., S.J.H., N.R., R.A.F., T.M.D.
References
- 1. Koshy M, Villano JL, Dolecek TA, et al. Improved survival time trends for glioblastoma using the SEER 17 population-based registries. J Neurooncol. 2012;107(1):207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stupp R, Mason WP, van den Bent MJ, et al. European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group . Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 3. Network NCC. Central Nervous System Cancers (Version 3.2020). 2020. https://www.nccn.org/professionals/physician_gls/pdf/cns.pdf.
- 4. Franceschi E, Ermani M, Bartolini S, et al. Post progression survival in glioblastoma: where are we? J Neurooncol. 2015;121(2):399–404. [DOI] [PubMed] [Google Scholar]
- 5. Wlodarczyk A, Grot D, Stoczynska-Fidelus E, Rieske P. Gaps and doubts in search to recognize glioblastoma cellular origin and tumor initiating cells. J Oncol. 2020;2020:6783627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ono A, Kanno H, Hayashi A, et al. Collagen gel matrix assay as an in vitro chemosensitivity test for malignant astrocytic tumors. Int J Clin Oncol. 2007;12(2):125–130. [DOI] [PubMed] [Google Scholar]
- 7. Ranjan T, Howard CM, Yu A, et al. Cancer stem cell chemotherapeutics assay for prospective treatment of recurrent glioblastoma and progressive anaplastic glioma: a single-institution case series. Transl Oncol. 2020;13(4):100755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Linz U, Ulus B, Neuloh G, et al. Can in-vitro chemoresponse assays help find new treatment regimens for malignant gliomas? Anticancer Drugs. 2014;25(4):375–384. [DOI] [PubMed] [Google Scholar]
- 9. Howard CM, Valluri J, Alberico A, et al. Analysis of chemopredictive assay for targeting cancer stem cells in glioblastoma patients. Transl Oncol. 2017;10(2):241–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burstein HJ, Mangu PB, Somerfield MR, et al. American Society of Clinical Oncology . American Society of Clinical Oncology clinical practice guideline update on the use of chemotherapy sensitivity and resistance assays. J Clin Oncol. 2011;29(24):3328–3330. [DOI] [PubMed] [Google Scholar]
- 11. Kapałczyńska M, Kolenda T, Przybyła W, et al. 2D and 3D cell cultures - a comparison of different types of cancer cell cultures. Arch Med Sci. 2018;14(4):910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Imamura Y, Mukohara T, Shimono Y, et al. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol Rep. 2015;33(4):1837–1843. [DOI] [PubMed] [Google Scholar]
- 13. Shuford S, Wilhelm C, Rayner M, et al. Prospective validation of an ex vivo, patient-derived 3D spheroid model for response predictions in newly diagnosed ovarian cancer. Sci Rep. 2019;9(1):11153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Møller S, Grunnet K, Hansen S, et al. A phase II trial with bevacizumab and irinotecan for patients with primary brain tumors and progression after standard therapy. Acta Oncol. 2012;51(6):797–804. [DOI] [PubMed] [Google Scholar]
- 15. Vredenburgh JJ, Desjardins A, Reardon DA, Friedman HS. Experience with irinotecan for the treatment of malignant glioma. Neuro Oncol. 2009;11(1):80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Francesconi AB, Dupre S, Matos M, et al. Carboplatin and etoposide combined with bevacizumab for the treatment of recurrent glioblastoma multiforme. J Clin Neurosci. 2010;17(8):970–974. [DOI] [PubMed] [Google Scholar]
- 17. Field KM, Simes J, Nowak AK, et al. CABARET/COGNO investigators . Randomized phase 2 study of carboplatin and bevacizumab in recurrent glioblastoma. Neuro Oncol. 2015;17(11):1504–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cabrera AR, Kirkpatrick JP, Fiveash JB, et al. Radiation therapy for glioblastoma: Executive summary of an American Society for Radiation Oncology Evidence-Based Clinical Practice Guideline. Pract Radiat Oncol. 2016;6(4):217–225. [DOI] [PubMed] [Google Scholar]
- 19. Andersen AP. Postoperative irradiation of glioblastomas. Results in a randomized series. Acta Radiol Oncol Radiat Phys Biol. 1978;17(6):475–484. [DOI] [PubMed] [Google Scholar]
- 20. Kristiansen K, Hagen S, Kollevold T, et al. Combined modality therapy of operated astrocytomas grade III and IV. Confirmation of the value of postoperative irradiation and lack of potentiation of bleomycin on survival time: a prospective multicenter trial of the Scandinavian Glioblastoma Study Group. Cancer. 1981;47(4):649–652. [DOI] [PubMed] [Google Scholar]
- 21. Walker MD, Strike TA, Sheline GE. An analysis of dose-effect relationship in the radiotherapy of malignant gliomas. Int J Radiat Oncol Biol Phys. 1979;5(10):1725–1731. [DOI] [PubMed] [Google Scholar]
- 22. Seystahl K, Wick W, Weller M. Therapeutic options in recurrent glioblastoma–An update. Crit Rev Oncol Hematol. 2016;99:389–408. [DOI] [PubMed] [Google Scholar]
- 23. Nieder C, Grosu AL, Molls M. A comparison of treatment results for recurrent malignant gliomas. Cancer Treat Rev. 2000;26(6):397–409. [DOI] [PubMed] [Google Scholar]
- 24. Franceschi E, Cavallo G, Scopece L, et al. Phase II trial of carboplatin and etoposide for patients with recurrent high-grade glioma. Br J Cancer. 2004;91(6):1038–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cloughesy TF, Filka E, Kuhn J, et al. Two studies evaluating irinotecan treatment for recurrent malignant glioma using an every-3-week regimen. Cancer. 2003;97(9 Suppl):2381–2386. [DOI] [PubMed] [Google Scholar]
- 26. Friedman HS, Prados MD, Wen PY, et al. Bevacizumab alone and in combination with irinotecan in recurrent glioblastoma. J Clin Oncol. 2009;27(28):4733–4740. [DOI] [PubMed] [Google Scholar]
- 27. Vredenburgh JJ, Desjardins A, Herndon JE 2nd, et al. Phase II trial of bevacizumab and irinotecan in recurrent malignant glioma. Clin Cancer Res. 2007;13(4):1253–1259. [DOI] [PubMed] [Google Scholar]
- 28. Arozarena I, Wellbrock C. Overcoming resistance to BRAF inhibitors. Ann Transl Med. 2017;5(19):387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maraka S, Janku F. BRAF alterations in primary brain tumors. Discov Med. 2018;26(141):51–60. [PubMed] [Google Scholar]
- 30. Stupp R, Taillibert S, Kanner AA, et al. Maintenance therapy with tumor-treating fields plus temozolomide vs temozolomide alone for glioblastoma: a randomized clinical trial. JAMA. 2015;314(23):2535–2543. [DOI] [PubMed] [Google Scholar]
- 31. Wong ET, Hess KR, Gleason MJ, et al. Outcomes and prognostic factors in recurrent glioma patients enrolled onto phase II clinical trials. J Clin Oncol. 1999;17(8):2572–2578. [DOI] [PubMed] [Google Scholar]
- 32. Kamiya-Matsuoka C, Gilbert MR. Treating recurrent glioblastoma: an update. CNS Oncol. 2015;4(2):91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ostrom QT, Gittleman H, Farah P, et al. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 2013;15(Suppl 2):ii1–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]