Abstract
Background
The present study was aimed at establishing the prevalence, epidemiology and molecular characterization of major haemoprotozoons (Babesia and Theileria) and rickettsia (Anaplasma) of cattle in Jammu region (North India) using microscopy and Polymerase Chain Reaction (PCR). Hematology, microscopy and PCR based prevalence studies were undertaken with 278 whole blood samples from cattle. Molecular prevalence studies were followed by genetic characterization of the isolates of Babesia, Anaplasma and Theileria spp. based on 18S rRNA, 16S rRNA and Tams1 gene, respectively. The data related to metrology and epidemiological variables like temperature, rainfall, season, age and type of livestock rearing was analyzed and correlated with occurrence of disease by statistical methods.
Results
The prevalence based on microscopy was 12.9% (36/278) whereas PCR recorded 30.22% (84/278) animals positive for haemoparasitic infections. All the samples found positive by microscopy were also recorded positive by PCR. Thus the study revealed prevalence of Babesia bigemina, Anaplasma marginale and Theileria annulata to be 9.7, 16.5 and 0.7% respectively. The metrological and epidemiological variables made inroads for the propagation of vector ticks and occurrence of infection. Haematological alterations predominantly related to decrease in haemoglobin, red blood cell count and packed cell volume were evident in diseased animals and collaterally affected the productivity. Further the genetic characterization of Babesia bigemina. (MN566925.1, MN567603, MN566924.1), Anaplasma marginale. (MH733242.1, MN567602.1) and Theileria annulata (MT113479) provided a representative data of the isolates circulating in the region and their proximity with available sequences across the world.
Conclusions
Despite holding much significance to the animal sector, comprehensive disease mapping has yet not been undertaken in several parts of India. The present study provides a blue print of disease mapping, epidemiological correlations and genomic diversity of Babesia bigemina, Anaplasma marginale and Theileria annulata circulating in the region.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12917-021-02915-9.
Keywords: Anaplasma, Babesia, Cattle, Haematology, Molecular characterisation, Theileria
Background
India is primarily an agricultural country and the losses incurred as a result of tick borne diseases (TBDs) to livestock in India are huge, estimated to be 8.7 million USD, predominantly affecting the small and marginal farmers. Among the various tick transmitted diseases babesiosis, theileriosis, anaplasmosis and ehrlichiosis are the significant ones that drive attention. Babesiosis is considered as economically important tick-borne haemoprotozoan disease in tropical and subtropical countries, the most prevalent species being Babesia bovis and Babesia bigemina [1]. Babesia bigemina, transmitted by brevirostrate tick, Rhipicephalus (Boophilus microplus) is usually associated with high parasitemia. Anaplasmosis is a rickettsial disease transmitted biologically through ticks mainly by Rhipicephalus (Boophilus) microplus [2]. It is endemic in some parts of the world [3, 4] while in India incidence of A. marginale infection has been recorded in livestock [5–7]. The Theileria species that infect bovines in India are T. annulata and T. orientalis, and both species are transmitted by Hyalomma anatolicum. The most common clinical signs observed in TBDs are fever, inappetence and mortality, with peculiar hemoglobinuria and anemia for bovine babesiosis [4], and enlarged lymph nodes for theileriosis.
Conventional microscopy is often used for the diagnosis of these diseases [5] but, cryptic cases and carrier states are often misdiagnosed or not diagnosed at all, besides low sensitivity and lack of species level differentiation [8]. Similarly, sero-diagnosis suffers from the problem of cross-reactivity and sometimes low antigen output. Microscopy along with sensitive and specific molecular tools like PCR may help in mapping the diseases and various risk factors for the transmission of major TBDs of livestock viz. babesiosis, theileriosis and anaplasmosis. In the light of the fact that 39 million crossbred cattle of India, meant to boost productivity, are at potential risk of contracting TBDs; it becomes imperative to model the prevalence, identify the risk factors and devise control strategies for the control of tick and TBDs of livestock.
The present study was aimed to generate baseline data about the prevalence of tick borne haemoprotozoan and rickettsial infections in cattle in Jammu province of North India, based on conventional and molecular techniques, assess the risk factors and haematological alterations associated with the disease. Moreover, phylogenetic relatedness and genetic diversity of the isolates infecting cattle was also investigated to aid in epidemiological surveillance.
Results
Prevalence studies based on microscopy and PCR
All the samples found positive by microscopy were also recorded positive by PCR. Microscopy revealed infection in 12.94% animals (36/278) (Fig. 1a, b, c) whereas 30.21% samples (84/278) were diagnosed as positive through PCR assay. The PCR assay could effectively diagnose cryptic and sub clinical cases and was found significantly sensitive (χ2 23.475, odds ratio, OR = 2.911, 95% CI = 1.886–4.491) as compared to microscopy. The prevalence of A. marginale was (46/278) 16.5% followed by Babesia bigemina (27/278, 9.7%) and T. annulata (2/278, 0.7%). Concurrent haemoprotozoan and rickettsial infections were detected in 9 (3.2%) animals (Table 1).
Fig. 1.
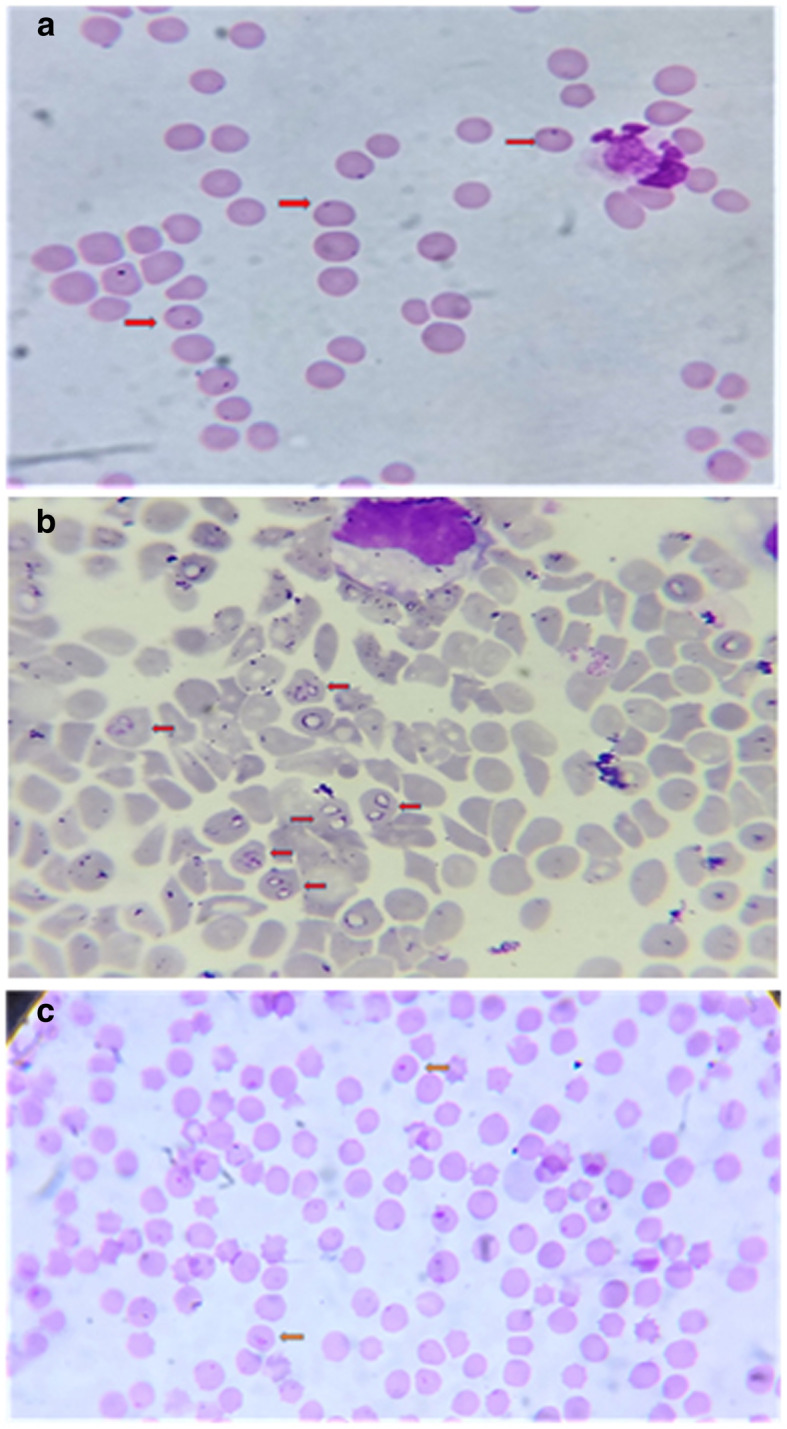
a Blood smear positive for Anaplasma spp (1000x). b Blood smear positive for Babesia piroplasms (1000x). 1c Blood smear positive for Theileria piroplasms (1000x)
Table 1.
Prevalence of haemoparasites by microscopy and PCR
| Positive by Giemsa staining of thin blood smear | Positive by PCR | |||||
|---|---|---|---|---|---|---|
| Sample Size | Positive (%) | 95% CI | Sample Size | Positive (%) | 95% CI | |
| Babesia | 278 | 12 (4.3) | 2.32–6.32 | 278 | 27 (9.7) | 1.78–2.91 |
| Anaplasma | 278 | 20 (7.1) | 4.65–9.74 | 278 | 46 (16.5) | 2.23–3.66 |
| Theileria | 278 | 1 (0.3) | −0.23 - 0.95 | 278 | 2 (0.7) | 0.51–0.83 |
| Mixed infection | 278 | 3 (1.0) | 0.06–2.10 | 278 | 9 (3.2) | 1.06–1.74 |
| Total | 278 | 36 (12.9) | 9.65–16.25 | 278 | 84 (30.2) | 2.75–4.52 |
Attribution of clinical signs to the disease
The main clinical signs exhibited were pyrexia, decreased milk production, pale mucous membrane, tick infestation and anorexia. Haemoglobinuria was observed in babesiosis and lymph node enlargement manifested in theileriosis. Tick infestation was recorded in 64/84 (76.19%) of animals positive for haemoparasites. Decreased milk production was found in 72/84 (85.71%) animals. Among 31 animals showing symptoms of pica, 30 were found positive for anaplasmosis.
Assessment of risk and impact of environmental variables
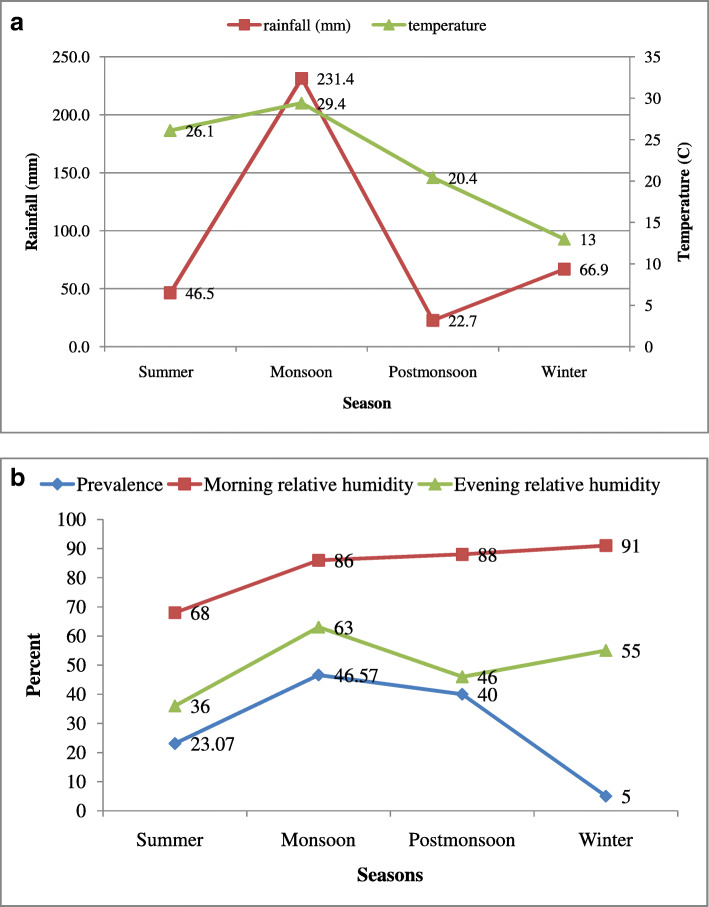
Evaluation of risk factors for occurrence of disease in the present study was undertaken by modelling effect of age, season, breed, sex and type of farm. A significant effect of season and host age on occurrence of diseases was indicated. The highest prevalence of haemoprotozoan and rickettsial infection was found in animals having > 3 years of age (45/120, 37.5%) followed by 1–3 years (30/105, 28.5%) and < 1 year of age (9/53, 16.9%). The overall prevalence did not vary significantly (p = 0.629) between male and female animals. Unorganised farms without proper flooring, cracked walls and poor sanitation showed significantly (p = 0.007) higher prevalence of infection (60/160, 37.5%,) as compared to organised farms (24/118, 20.3%, Cross bred animals revealed higher infection (33.6%) compared to indigenous animals (15.3%), reiterating the fact that cross bred animals are at higher risk than native breeds. The probability of occurrence of infections in monsoon was 16.56 times higher than winter. The correlation of environmental variables with the prevalence presented a clear picture of epidemiological interventions in the disease occurrence (Fig. 2 a and b, supplementary data 1). The detailed table of risk factors associated with prevalence of haemoprotozoon and rickettsial infections among cattle is presented in Table 2.
Fig. 2.
a: Graph depicting rainfall (in mm) and temperature (°C) during different seasons of the year in Jammu region. b: Graphical representation of morning and evening relative humidity and prevalence during different seasons of the year in Jammu region
Table 2.
Risk factors associated with prevalence of haemoparasites among cattle in Jammu, North India according to different variables
| Variable | Samples | No of animals positive (% positive for pathogen) | Total animals Positive (%) | 95% CI | Chi Square | df | P value | Odds ratio | 95% CI | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Anaplasma marginale | Babesia bigemina | Theileria annulata | Mixed | ||||||||||
| Farm Management | Organized | 118 | 11 (9.3) | 10 (8.4) | 0 | 3(2.5) | 24 (20.3) | 14.2–26.4 | 9.49 | 1 | (P < 0.01) | constant | – |
| Unorganized | 160 | 35 (21.8) | 17(10.6) | 02 (1.2) | 6(3.7) | 60(37.5) | 31.2–43.7 | 2.35 | 1.3–4.0 | ||||
| Age | < 1 year | 53 | 6(11.3) | 2(3.7) | 0 | 1(1.8) | 9(16.9) | 8.5–25.4 | 7.56 | 2 | (P < 0.05) | constant | – |
| 1–3 year | 105 | 15(14.2) | 11(10.4) | 1(0.9) | 3(2.8) | 30(28.5) | 21.3–35.8 | 0.84 | 0.3–2.1 | ||||
| > 3 year | 120 | 25(20.8) | 14(20.8) | 1(0.8) | 5(4.1) | 45(37.5) | 30.2–44.7 | 2.93 | 1.3–6.5 | ||||
| Season | Winter | 60 | 2(3.3) | 1(1.6) | 0 | 0 | 3(5.0) | 0.3–9.6 | 32.56 | 3 | (P < 0.01) | constant | – |
| Summer | 65 | 8(12.3) | 6(9.2) | 0 | 1(1.5) | 15(23.0) | 14.5–31.6 | 5.7 | 1.5–20.8 | ||||
| Monsoon | 73 | 20(27.3) | 9(12.3) | 1(1.3) | 4(5.4) | 34(46.5) | 37.0–56.1 | 16.56 | 4.7–57.7 | ||||
| Post monsoon | 80 | 16(20.0) | 11(13.7) | 1(1.2) | 4(5.0) | 32(40.0) | 31.0–48.9 | 12.6 | 3.6–43.9 | ||||
| Breed | Indigenous | 52 | 5(9.6) | 3(5.7) | 0 | 0 | 8(15.3) | 7.1–23.5 | 6.67 | 1 | (P < 0.01) | constant | – |
| Cross bred | 226 | 41(18.1) | 24(10.6) | 2(0.8) | 9(3.9) | 76(33.6) | 28.4–38.7 | 2.79 | 1.2–6.2 | ||||
| Sex | Male | 12 | 1(8.3) | 1(8.3) | 0 | 0 | 2(16.6) | −0.9 – 34.3 | 1.09 | 1 | (P > 0.05) | constant | – |
| Female | 266 | 45(16.9) | 26(9.7) | 2(0.7) | 9(3.3) | 82(30.8) | 26.1–35.4 | 2.29 | 0.4–10.4 | ||||
Haematological alterations
Haematological findings of infected animals (n = 84) were compared with healthy control (n = 10) animals. A significant decrease in the Hb (p = 0.013), PCV (p = 0.001) and RBC (p = 0.027) count of animals with babesiosis was recorded (Table 3). The RBC count was also found significantly decreased in animals harbouring mixed infection.
Table 3.
Haematological alterations in animals with haemoprotozoon and rickettsial infection
| Haematological Parameter | Non infected control (n = 10) | Babesia spp.(n = 27) | Anaplasma spp.(n = 46) | Theileria spp.(n = 2) | Mixed infection (n = 9) |
|---|---|---|---|---|---|
| Hb (g/dl) | 10.65 ± 1.02b | 6.16 ± 0.48a | 7.41 ± 0.55ab | 7.08 ± 2.07ab | 7.22 ± 1.09ab |
| PCV (%) | 28.79 ± 1.32c | 17.67 ± 1.39a | 22.91 ± 1.26bc | 19.50 ± 4.5ab | 18.22 ± 2.22ab |
| RBC (106/mm3) | 5.82 ± 0.56b | 3.07 ± 0.22a | 4.27 ± 0.43ab | 3.40 ± 0.70ab | 3.28 ± 0.84a |
| WBC (×103/ μl) | 10.52 ± 0.96ab | 6.39 ± 0.31a | 11.98 ± 1.07b | 9.00 ± 2.0ab | 9.83 ± 1.41ab |
| LYM (%) | 62.96 ± 3.13 | 59.93 ± 2.76 | 60.24 ± 2.26 | 73.50 ± 7.5 | 59.89 ± 4.40 |
| MON (%) | 1.54 ± 0.1 | 1.55 ± 0.21 | 1.56 ± 0.14 | 1.00 ± 0.5 | 1.60 ± 0.36 |
| GRA (%) | 36.77 ± 1.54 | 40.62 ± 2.12 | 40.19 ± 1.3 | 27.50 ± 6.5 | 39.67 ± 3.46 |
| MCH (pg) | 15.82 ± 1.30 | 14.54 ± 1.25 | 14.63 ± 1.01 | 14.50 ± 3.5 | 13.22 ± 2.22 |
| MCHC (g/dl) | 34.62 ± 1.23a | 45.69 ± 2.54b | 32.02 ± 1.0a | 36.50 ± 7.5ab | 34.67 ± 2.62a |
| MCV (fl) | 46.42 ± 1.97 | 47.54 ± 2.47 | 48.21 ± 1.21 | 51.50 ± 5.50 | 45.11 ± 3.35 |
Values of Hb (g/dl), PCV (%), RBC (106/mm3), WBC (×103/ μl), and MCHC (g/dl) with different superscripts a,b,c differ significantly in a row. Hb Haemoglobin; PCV Packed cell volume; RBC Red blood cells; WBC White blood cells; LYM Lymphocytes; MON Monocytes; GRA Granulocytes; MCH Mean corpuscular haemoglobin; MCHC Mean corpuscular haemoglobin concentration; MCV Mean corpuscular volume
Polymerase chain reaction and phylogenetic analysis
In the PCR assays, specific amplifications of 504 bp, 270 bp and 751 bp products were obtained for 18S rRNA gene of Babesia, 16S rRNA gene of A. marginale and Tams1 gene of T.annulata (Fig.3 a, b and c). Six PCR products comprising of three B. bigemina, two A. marginale and one T. annulata, representing different isolates of Jammu region were sequenced. The sequence similarity searches in BLAST revealed that Jammu isolate (MN566925.1) of Babesia spp. was 99.8% identical to Kathua (MN567603), Udhampur (MN566924.1) and Meghalaya (KF606864.1) isolates of Babesia bigemina and had marked similarity with B. bigemina from Argentina (HQ688688.1) and USA (MH050387.1). At position 363 C was replaced with G in Udhampur and Kathua isolates (Babesia bigemina alignment, supplementary data 2). Two sequences of A. marginale from our study, submitted to GenBank (MH733242.1, MN567602.1), revealed 100% nucleotide identity with published sequences of A. marginale of French (MN317256.1), Iranian (MK310488.1) and Vietnamese (MH686047.1) origin. The only sequence of T.annulata from Jammu region (MT113479) was found to be 99% similar to Indian isolates from Izatnagar (MF346013.1) and Hissar (AF214840.1). In Jammu isolate at position 137 C was replaced by A, A by G (607) and C by A (611) in comparison to Izatnagar isolate (T.annulata alignment, supplementary data 3). Moreover there were 6 nucleotide substitutions in Jammu isolate vis-à-vis Hisar isolate at position 365 (C to A), 368(C to A), 389 (A to G), 399 (A to C), 414 (G to C) and 425 (T to C). Furthermore, the Jammu isolate showed 99.07% sequence homology to United Kingdom W1_2 isolate (KX981026.1).
Fig. 3.
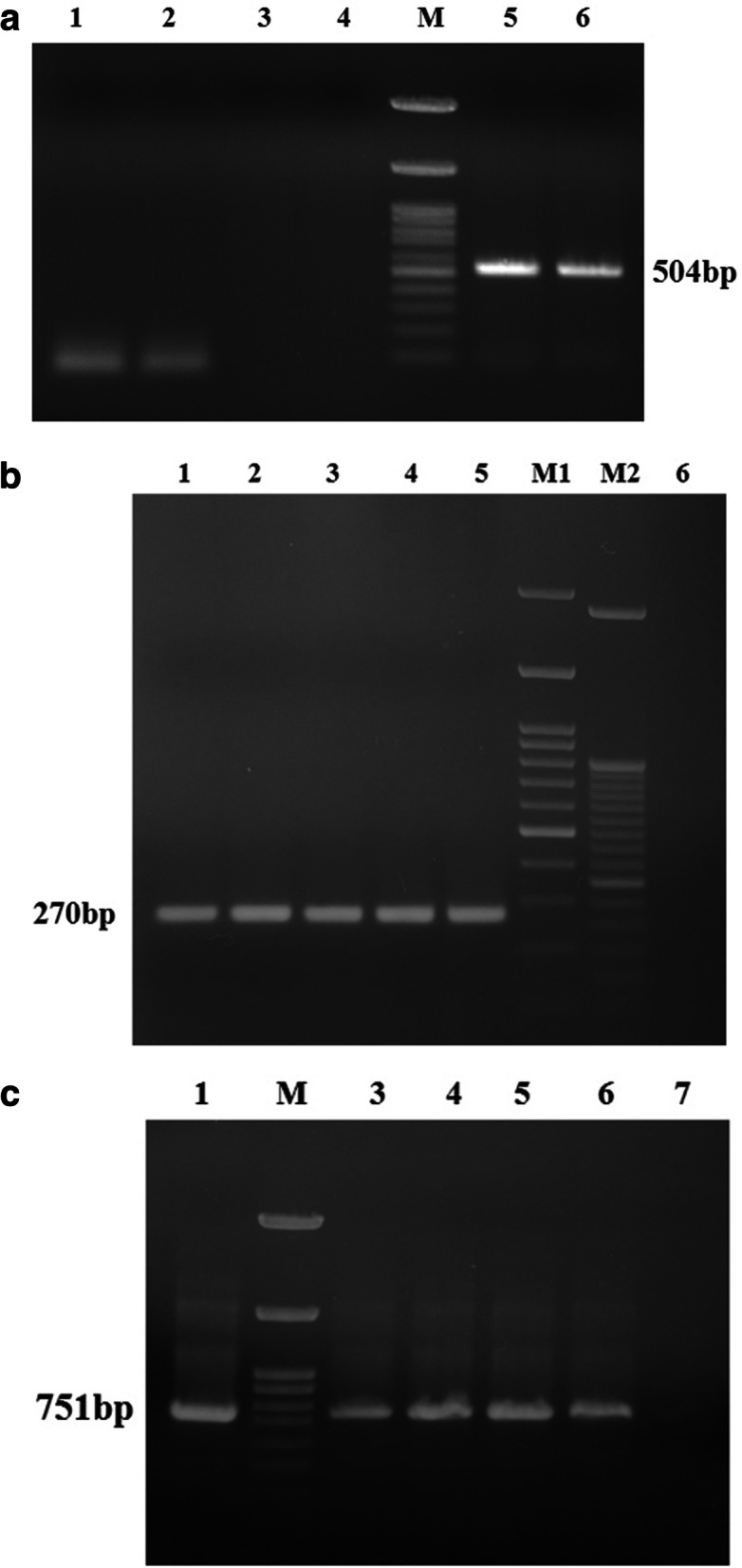
a: PCR amplification of 504 bp Babesia spp. 18S rRNA, Lane 1, 2 negative control, Lane 3, 4, 5: test samples M: 100 bp ladder, Lane 6: positive control. b PCR amplification of 270 bp Anaplasma marginale 16 s rRNA, Lane 1–4 test samples, M1: 100 bp ladder, M2: 50 bp ladder, Lane 5: positive control, Lane 6 negative control. c PCR amplification of 751 bp Theileria annulata Tams1 gene, Lane 1: positive control, M: 100 bp ladder, Lane 3,4,5,6,: test samples at gradient, Lane 7: negative control
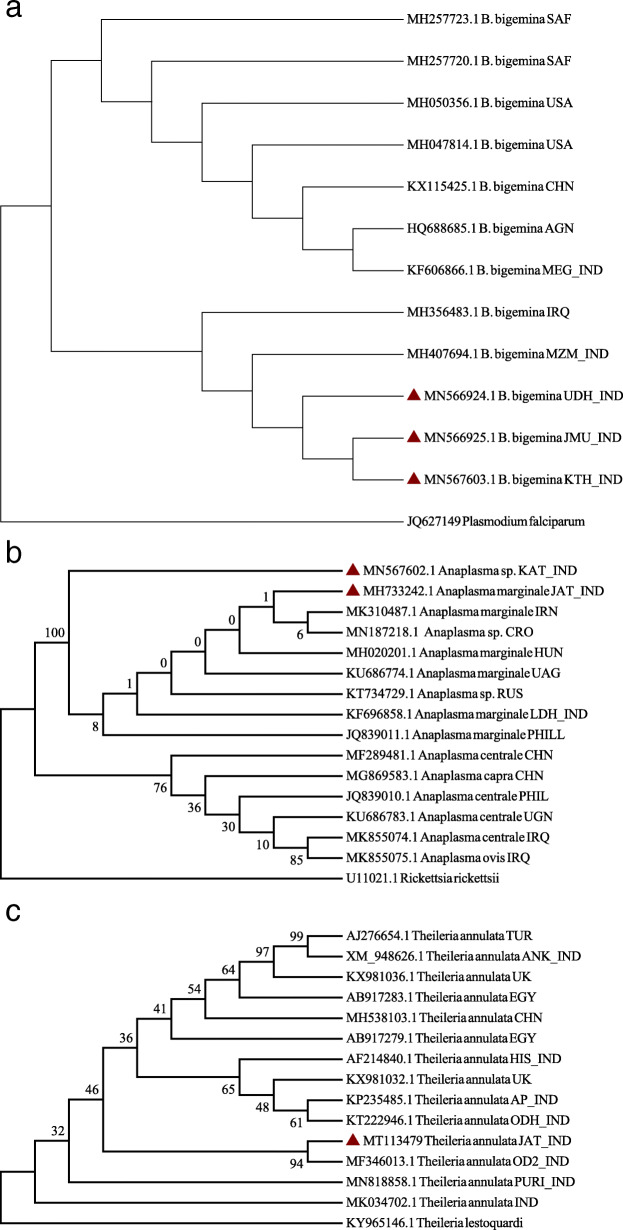
In the phylogenetic tree, our sequences clubbed together with Mizoram isolate (MH407694.1) forming separate Indian subclade (Fig. 4a). The phylogenetic tree of A. marginale (Fig. 4b) showed species level diversity in which Kathua isolate branched away from Ludhiana isolate (India, KF696858.1). The only T. annulata sequence from the present study formed a separate clade with Indian isolate of T. annulata (MF346013.1) on the phylogenetic tree (Fig. 4c).
Fig. 4.
a Phylogenetic relationship of B. bigemina isolates based on 18S rRNA gene. All accession numbers correspond to different B. bigemina isolates followed by their country of origin. The sequences generated in the present study are marked as red triangle. b Phylogenetic relationship of A. marginale isolates based on 16S rRNA gene. All accession numbers corresponds to different Anaplasma isolates followed by their country of origin. The sequences generated in the present study are marked as red triangle. c. Phylogenetic relationship of T. annulata isolates based on Tams1gene. All accession numbers corresponds to different T. annulata isolates followed by their country of origin. The sequence generated in the present study is marked as red triangle
Discussion
In developing countries like India considerable economic losses occurs in large and small holding livestock productivity farming system due to TBDs [6]. Prevalence studies are of immense importance for disease mapping and investigating the epidemiological triad. The agro-climatic conditions of the Jammu region are highly favourable for growth and multiplication of ticks which act as natural vectors of theileriosis, babesiosis and anaplasmosis. In the present study, high prevalence of Babesia bigemina (14.02%, 39/278), A. marginale (23.74%, 66/278) and low prevalence of Theileria annulata (1.079%, 3/278) can be attributed to Rhipicephalus (Boophilus) microplus being the only tick which infests the bovines of Jammu region [12]. A thorough review of literature reveals varying incidence of babesiosis ranging from 0.76 to 18.50% in India [13–15]. Anaplasma is also associated with a long term/life long carrier state [16] and so the probability of detecting positive animals, particularly in areas of endemic instability, is increased, primarily with use of molecular tools [6, 17]. The animals found positive for theileriosis had a history of importation from the neighbouring state, Punjab, having high prevalence of H. anatolicum [18] and T. annulata [5]. Owing to meagre presence of vector tick (H. anatolicum), considerably low prevalence of theileriosis was found in union territory of Jammu and Kashmir [19]. Livestock owners import high yielding animals from bordering states of Punjab and Haryana without proper quarantine and thus carrier cattle get transported to non endemic areas, increasing the likelihood of transboundary outbreaks. PCR assay has been employed for sensitive detection of haemoparasites in many diagnostic laboratories and is considered as an alternative to microscopy [20] particularly in latent infections. In our study as well, PCR was able to identify more than twice the number of cases as detected by microscopy.
There were no uncharacteristic clinical signs in the animals found positive for haemoprotozoan and rickettsial infection. High temperature, pale mucous membranes and decrease in milk production were however prominent signs. The endogenous pyrogens released in the blood due to cellular lysis lead to fever and consequently, inappetence [17]. Anemia occurs due to erythrophagocytosis, lysis of RBCs due to parasite multiplication and subsequent removal by reticuloendothelial system [21, 22]. Consequently, the vital blood parameters which include level of haemoglobin, PCV, MCHC, MCV, TLC and TEC get deranged and further aggravated by the continuous loss of blood sucked out by the ticks [23]. The impact of haemoprotozoan and rickettsial infection on the milk production is a major economical set back. Our results reflect the deviations in the blood parameters following the same cascade and are in agreement with [15, 17, 24].
The study suggests that outbreaks are likely to occur in the rainy and post rainy season due to interplay of epidemiological factors. During rainy season the epizootiological determinants such as ambient temperature and atmospheric humidity and microclimate of grazing lands are favourable for growth and development of ticks. This is unlikely to differ in other parts of country as well, based on earlier reports viz. 29.31% [25] and 58.55% [26]. The vector population was observed as a major differential factor in the prevalence of TBDs in organised and unorganized farms (p = 0.007), latter showing increased prevalence. These farms are Kuchha houses (non cemented) or made of bricks only, having cracks and crevices, with improper drainage and poor ventilation, favouring survival and breeding of vector ticks. Moreover, the owners of unorganized farms are comparatively uneducated, so lack of awareness regarding use of acaricidals and sustainable managemental practices against ticks was observed at the time of sample collection. The other determinants recorded for disease occurrence are age and breed of animal. Inverse age resistance shields the young population from clinical outbreak of diseases [27]. The highest prevalence (37.50%) was observed in animals > 3 years of age which is supported by studies from [26, 28]. Also, the adult cattle are predisposed to various stresses due to cycling heat, production, vaccination and reproduction which may augment the pathogenesis of diseases. Cross bred animals revealed higher prevalence of infection (33.62%) than indigenous breeds (15.38%), in agreement with earlier reports [26, 29]. Some workers have attributed susceptibility to the difference in the immune response to produce pro-inflammatory cytokine, which is higher in exotic animals or native breeds harbouring genetic loci for greater tolerance [30, 31].
Intracellular haemoprotozoons are under constant pressure from the immune system of their hosts, leading to emergence of genetic variants [32]. The genetic polymorphism of Babesia spp. has been reported earlier using sequence information of 18S rRNA gene [1, 33, 34]. In the phylogenetic tree, the isolates of the present study formed a separate sub clade but clustered with an isolate from Iraq. However, these distinguished itself from South African and American isolates, simulating reports from Kerala [1], wherein the isolates appeared in two different clades. In another study from India [34], close genetic relatedness was observed between B. bigemina isolates from North Eastern India with Argentina and Kenya rather than with China. Sequence information of A. marginale isolates from the present study revealed a marked divergence from A. capra, A. central and A. ovis, although, small length of nucleotide sequence couldn’t reveal any marked genetic heterogeneity from isolates of A. marginale across the world. In a study from south India, minimal heterogeneity was revealed within 16S rRNA and msp4 genes among the field isolates from Kerala [1].
The key mechanism responsible for genetic diversity among Theileria spp., is recombination during sexual reproduction [35]. The Tams1 gene has been shown to be a promising candidate for carrying antigenic diversity studies in T. annulata parasites [36, 37], however some studies have suggested no geographic specificity and other showing region specificity based on the gene polymorphism [36–38]. In the present study, T. annulata showed 99% nucleotide homology with Indian isolates and clustered together with an Indian isolate in the phylogenetic clade. Parasite diversity reports from India suggests that the Indian isolates were distributed into two groups along with other countries like Spain, Italy, Tunisia, Iran, Bahrain, Turkey and Iraq [39, 40]. The genetic diversity among the parasite strains can be one of the reasons for vaccination failures and inability to constraint the disease.
Conclusions
In conclusion the study provided a holistic picture about the prevalence of haemoprotozoon and rickettsial infection in cattle and its correlation with the environmental variables and epidemiological determinants. In addition, the hematological data may be presumed as a marker of health status of animal and its implications on the productivity of animal. The present study is probably the first report of molecular characterization of the haemoprotozoon and rickettsial infection from Jammu region of North India. It provides pioneering information about the circulating genotypes and possible diversities which are quintessential for developing vaccine and diagnostic strategies in future.
Methods
Study area and sample collection strategy
The study was conducted in Jammu province of Union Territory of Jammu and Kashmir, India, located at 32.73 oN 74.87 °E. To collect maximum number of samples, the expected prevalence of 20% with confidence limits of 95% and a desired absolute precision of 5% was considered. The number of samples thus calculated was adjusted for finite population [9] and correlated with 278 samples between October, 2017 and September 2019. At the time of blood sample collection, data related to clinical signs, age, sex, breed and type of farm were also recorded. The dairy animals were examined thoroughly and observed for pyrexia, pale mucous membrane, respiratory distress, diarrhea, tick infestation, haemoglobinurea, lymph node enlargement and nasal discharges. Also, the history of anorexia and decrease in milk production was confirmed from the animal owners. The housing system (organized or unorganized) was also taken into consideration as a variable for disease prevalence taking into account the type of flooring, drainage and ventilation of the shed. Blood was collected from the jugular vein into EDTA-coated vacutainer tubes, transported to the laboratory on ice. Thin blood smears were made, fixed in methanol, air dried and stained with Giemsa stain (1:20) as per standard protocol. Stained smears were examined microscopically under oil immersion and approximately 20–35 fields were examined. Blood was stored at − 20 °C until further analysis.
Disease prevalence and correlation with environmental variables
According to Indian meteorological department Pune, Ministry of earth sciences, Government of India, seasonal variability in the region for the year can be classified into summer (March to June), monsoon/rainy (July to September), postmonsoon/post rains (October to November) and winter (December to February). Hence study period was divided as per climatic variables and correlations were drawn with environmental variables like temperature, relative humidity and rainfall. Metrological data was obtained from the metrology observatory installed at Division of Agrometeorology, SK University of agricultural sciences and technology of Jammu.
Haematological parameters
The various blood parameters of clinically suspected blood samples were measured with automatic blood analyser (Mythic™ 18 Vet, Orphee, Switzerland). The various attributes included white blood cell count (WBC) (× 103/μl), lymphocytes (%), monocytes (%), granulocytes (%), red blood cell count (RBC) (106/mm3), haemoglobin (Hb) (g/dl), platelet count (× 103/μl), mean corpuscular haemoglobin (MCH) (picogram), mean corpuscular haemoglobin concentration (MCHC) (g/dl), mean corpuscular volume (MCV) (femtolitre) and packed cell volume (PCV) (%).
Genomic DNA extraction and polymerase chain reaction
Genomic DNA was extracted from 200 μl of the whole blood using DNA extraction kit (DNeasy blood kit, Qiagen) following the manufacturer’s protocols. Concentration of isolated DNA was measured using spectrophotometer (Eppendorf, India) and purity was checked in agarose gel electrophoresis (1.5% gel). Aliquots of extracted DNA were stored at − 20 °C until further use.
Each blood sample was subjected to three PCR reactions for detection of Babesia spp., A. marginale and T. annulata targeting 18S rRNA, 16S rRNA and Tams1 gene, respectively [10] with some minor modifications. The oligonucleotide primers used are as follows:
| Organism | Primers |
| Babesia spp. | Bb 18S F 5 ′ -TCCATTCAAGTTTCTGCCCCCATCA- 3′ |
| Bb 18S R 5 ′-CCATTACCAAGGCTCAAAAGCAACAA- 3′ | |
| A. marginale | Amar 16SF 5′ - GGCGGTGATCTGTAGCTGGTCTGA- 3′ |
| Amar 16SR 5 ′- GCCCAATAATTCCGAACAACGCTT- 3′ | |
| T. annulata | Tamulti-F 5′ -CCGTTAATGCTGCAAATGAGGAGG- 3′ |
| Tamulti-R 5′ -GAGGCGAAGACTGCAAGGGGAG- 3′ |
The PCR reaction was carried out in 25 μl total volume containing 4 μl of template DNA, 2.5 μl of 10× PCR Green buffer (Thermo Scientific, USA), 0.5 μl of 10 mM dNTP, 0.5 μl of each forward and reverse primer (20 pmol/μl), 0.2 μl Dream Taq DNA polymerase and nuclease free water to make the volume 25 μl. Amplification was performed using a S1000 thermal cycler (Bio-Rad, USA) under following conditions: For Babesia spp. initial denaturation at 94 °C for 5 min, followed by 35 amplification cycles (94 °C for 1 min, 57 °C for 1 min and 72 °C for 1 min). For A. marginale, initial denaturation at 94 °C for 5 min, followed by 35 amplification cycles (94 °C for 45 s, 58 °C for 45 s and 72 °C for 45 s). For T. annulata, initial denaturation at 95 °C for 5 min, followed by 37 amplification cycles (95 °C for 30s, 55 °C for 30s and 72 °C for 30s). A final extension step at 72 °C for 15 min was followed after amplification cycles in all three reactions. Known positive genomic DNA of Babesia spp., A. marginale and T. annulata was used as positive control, while nuclease free water was used as negative control. The PCR products were electrophoresed in 1.5% agarose gel (Tris-borate-EDTA), incorporated with ethidium bromide (0.5 μg/ml) and visualized under transilluminator (Eppendorf, India).
Sequencing and phylogenetic analysis
PCR amplified product of each selected isolate was gel purified using PCR clean up system (Promega, USA). The eluted product was commercially sequenced in an automated DNA sequencer at Agrigenome Pvt. Ltd., Kochi, Kerala. Nucleotide sequences (n = 6) generated in the study were primarily analysed using BioEdit software and submitted to GenBank. The sequences were compared with the available sequences in GenBank using BLAST program of NCBI. The sequences were aligned with clustal W programme of MEGA 6 software using gap opening penalty of 10 and gap extension penalty of 0.1 and 0.2 for the pair wise and multiple alignments, respectively. Phylogenetic trees were constructed using maximum parsimony (MP) with the tree–bisection–regrafting (TBR) algorithm and tested at 1000 bootstrap replications. The sequences were initially truncated at both ends, so as to obtain sequences that started and ended at the homologous nucleotide positions. Plasmodium falciparum (JQ627149), Rickettsia rickettsii (U11021.1) and Theileria lestoquardi (KY965146.1) sequences were used as outgroup members to root the respective trees.
Statistical analysis
Chi-square test and univariate logistic regression models were used to draw inferences from risk factors associated with prevalence of haemoparasitic infections. Haematological alterations were analyzed by one-way analysis of variance (ANOVA) using SPSS 16 software [11].
Supplementary Information
Acknowledgements
The authors are grateful to Sher-e-Kashmir University of Agricultural Sciences and Technology of Jammu, Jammu and Kashmir for the encouragement and facilities provided. The authors also thank all the farmers and field veterinarians of Jammu region for assisting in sample collection.
Availability of data and material
The datasets used and/or analyzed during the current study are included in the published article as supplementary material. The nucleotide sequence data has been deposited in the NCBI GenBank and accession numbers are cited in the manuscript. Further, data regarding animal studies can be made available on reasonable request to corresponding author.
Abbreviations
- TTBD
Tick and tick borne diseases
- RNA
Ribonucleic acid DNA: deoxyribonucleic acid
- PCR
Polymerase chain reaction
- Hb
Haemoglobin
- PCV
Packed cell volume
- RBC
Red blood cells
- WBC
White blood cells
- LYM
Lymphocytes
- MON
Monocytes
- GRA
Granulocytes
- MCH
Mean corpuscular haemoglobin
- MCHC
Mean corpuscular haemoglobin concentration
- MCV
Mean corpuscular volume.
Authors’ contributions
All authors have read and approved the manuscript. RK undertook literature search, clinical studies, experimental studies and data acquisition. AY conceptualised and designed the study, aided in manuscript preparation, editing and review. SIR performed the experiments, interpreted the results, prepared and edited the manuscript. RG did the data analysis, statistical analysis and manuscript editing. VS performed data analysis and manuscript editing. DC did statistical analysis of data. RK conceptualised the study and reviewed the final manuscript.
Funding
The funds were provided by SKUAST Jammu for PhD research and some funds were utilized from the ICAR-National Fellow Project (F.N.Agri.Edn./27/06/NP/2017-HRD). SKUAST university funds were utilized for procuring commercial kits used in research whereas nucletide sequencing and other chemicals were procured using ICAR National Fellow Project funds.
Declarations
Ethics approval and consent to participate
The cattle owners graciously permitted animal examination and blood collection through verbal consent. The permission for blood collection was granted as per order no. AU/FVSJ/AGB/17–18/415 by institutional animal ethics committee (Institutional animal ethics committee of Faculty of Veterinary Sciences and Animal Husbandry, SKUAST-Jammu, registration ID: 862/CPC/SEA).
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pradeep RK, Nimisha M, Sruthi MK, Vidya P, Amrutha BR, Kurbet PS, et al. Molecular characterization of South Indian field isolates of bovine Babesia spp. and Anaplasma spp. Parasitol Res 2018; 10.1007/s00436-018-6172-4. . [DOI] [PubMed]
- 2.Aubry P, Geale DW. A review of bovine anaplasmosis. Transbound Emerg Dis. 2011;58(1):1–30. doi: 10.1111/j.1865-1682.2010.01173.x. [DOI] [PubMed] [Google Scholar]
- 3.Marufu MC, Chimonyo M, Dzama K, Mapiye C. Seroprevalence of tick-borne diseases in communal cattle reared on sweet and sour rangelands in a semi-arid area of South Africa. Vet J. 2010;184(1):71–76. doi: 10.1016/j.tvjl.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 4.Mtshali MS, Mtshali PS. Molecular diagnosis and phylogenetic analysis of Babesia bigemina and Babesia bovis hemoparasites from cattle in South Africa. BMC Vet Res. 2013;9(1):154. doi: 10.1186/1746-6148-9-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tuli A, Singla LD, Sharma A, Bal MS, Filia G, Kaur P. Molecular epidemiology, risk factors and hematochemical alterations induced by Theileria annulata in bovines of Punjab (India) Acta Parasitol. 2015;60(3):378–390. doi: 10.1515/ap-2015-0053. [DOI] [PubMed] [Google Scholar]
- 6.Kolte SW, Larcombe SD, Jadhao SG, Magar SP, Warthi G, Kurkure NV. PCR diagnosis of tick-borne pathogens in Maharashtra state, India indicates fitness cost associated with carrier infections is greater for crossbreed than native cattle breeds. PLoS One. 2017;12:e0174595. doi: 10.1371/journal.pone.0174595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jayalakshmi K, Sasikala M, Veeraselvam M, Venkatesan M, Yogeshpriya S, Ramkumar PK, Selvaraj P, Vijayasarathi MK. Prevalence of haemoprotozoan diseases in cattle of Cauvery delta region of Tamil Nadu. J Parasit Dis. 2019;43(2):308–312. doi: 10.1007/s12639-019-01094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bock R, Jackson L, de Vos A, Jorgensen W. Babesiosis of cattle. Parasitology. 2004;129:247–269. doi: 10.1017/S0031182004005190. [DOI] [PubMed] [Google Scholar]
- 9.Thrusfield M. Veterinary epidemiology. 2. United Kingdom: Blackwell Science Ltd; 2005. [Google Scholar]
- 10.Kundave VR, Ram H, Banerjee PS, Garg R, Mahendran K, Ravikumar GVPPS, Tiwari AK. Development of multiplex PCR assay for concurrent detection of tick borne haemoparasitic infections in bovines. Acta Parasitol. 2018;63(4):759–765. doi: 10.1515/ap-2018-0090. [DOI] [PubMed] [Google Scholar]
- 11.Snedecor GW, Cochran WG. Statistical methods. 8. Ames: Iowa State University Press; 1994. [Google Scholar]
- 12.Khajuria V, Godara R, Yadav A, Katoch R. Prevalence of ixodid ticks in dairy animals of Jammu region. J Parasit Dis. 2015;39(3):418–421. doi: 10.1007/s12639-013-0354-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muraleedharan K, Syed ZK, Mallikarjun GB, Seshadri SJ. Incidence of Anaplasma spp., Babesia spp. and Trypanasoma spp. in cattle of Karnataka. J Vet Parasitol. 2005;19:135–137. [Google Scholar]
- 14.Singh NK, Singh H, Jyoti HM, Rath SS. Prevalence of parasitic infections in cattle of Ludhiana district, Punjab. J Parasit Dis. 2012;36(2):256–259. doi: 10.1007/s12639-012-0119-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma A, Singla LD, Tuli A, Kaur P, Batth BK, Javed M, et al. Molecular prevalence of Babesia bigemina and Trypanosoma evansi in dairy animals from Punjab, India, by duplex PCR: a step forward to the detection and management of concurrent latent infections. Biomed Res Int. 2013:1–8. [DOI] [PMC free article] [PubMed]
- 16.Eriks IS, Stiller D, Palmer G. Impact of persistent Anaplasma marginale rickettsemia on tick infection and transmission. J Clin Microbiol. 1993;31(8):2091–2096. doi: 10.1128/JCM.31.8.2091-2096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashuma SA, Singla LD, Kaur P, Bal MS, Batth BK, Juyal PD. Prevalence and haemato-biochemical profile of Anaplasma marginale infection in dairy animals of Punjab (India) Asian Pac J Trop Med. 2013;6(2):139–144. doi: 10.1016/S1995-7645(13)60010-3. [DOI] [PubMed] [Google Scholar]
- 18.Singh NK, Rath SS. Prevalence of ixodid ticks in buffaloes (Bubalus bubalis) of various agro-climatic zones of Punjab state, India. J Vet Parasitol. 2013;27(2):105–109. [Google Scholar]
- 19.Farooq U, Tufani NA, Malik HU, Mir MS. Clinical and morpho molecular epidemiology of bovine theileriosis in Kashmir, India. Indian J Ani Res. 2019;53:375–381. [Google Scholar]
- 20.Almeria S, Castella J, Ferrer D, Ortuno A, Estrada-Pena A, Gutierrez JF. Bovine piroplasms in Minorca (Balearic Islands, Spain): a comparison of PCR-based and light microscopy detection. Vet Parasitol. 2001;99(3):249–259. doi: 10.1016/S0304-4017(01)00464-2. [DOI] [PubMed] [Google Scholar]
- 21.Singh A, Singh J, Grewal AS, Brar RS. Study on some blood parameters of crossbred calves with experimental Theileria annulata infections. Vet Res Commun. 2001;25(4):289–300. doi: 10.1023/A:1010678625336. [DOI] [PubMed] [Google Scholar]
- 22.Modi DV, Bhadesiya CM, Mandali GC. Hematobiochemical changes in crossbred cattle infected with Theileria annulata in Banakantha district of Gujarat. Int J Sci. 2015;5:1–4. [Google Scholar]
- 23.Debbarma A, Soumitra P, Ruma J, Surajit B, Mandal SC, Ralte L, Jana PS, Shit N. Haematological impact of naturally occurring tick borne haemoparasitic infections in cattle of West Bengal, India. Explor Anim Medical Res. 2017;7:175–178. [Google Scholar]
- 24.Aulaich GS, Singla LD, Kaur P, Alka Bovine babesiosis due to Babesia bigemina: haematobiochemical and therapeutic studies. Indian J Animal Sci. 2005;75:617–611. [Google Scholar]
- 25.Maharana BR, Kumar B, Prasad A, Patbandha TK, Sudhakar NR, Joseph JP, Patel BR. Prevalence and assessment of risk factors for haemoprotozoan infections in cattle and buffaloes of south-West Gujarat, India. Indian J Anim Res. 2016;50:733–739. [Google Scholar]
- 26.Kumar B, Mondal DB, Jithin MV. Prevalence of babesiosis in cattle in Patna region, India. Int J Curr Microbiol Appl Sci. 2018;7:5167–5174. [Google Scholar]
- 27.Soulsby EJL. Helminths, Arthopods and Protozoa of Domestic Animal.; 7th. Elsevier. 2005.
- 28.Muhanguzi D, Ikwap K, Picozzi K, Waiswa C. Molecular characterization of Anaplasma and Ehrlichia species in different cattle breeds and age groups in Mbarara district (Western Uganda) Int J Anim Vet Adv. 2010;2:76–88. [Google Scholar]
- 29.Alam TH, Nasr SM. Hematological and biochemical investigation in bovine babesiosis and theileriosis. Benha Veterinary Med J. 2011;22:118–126. [Google Scholar]
- 30.Jensen K, Paxton E, Waddington D, Talbot R, Darghouth MA, Glass EJ. Differences in the transcriptional responses induced by Theileria annulata infection in bovine monocytes derived from resistant and susceptible cattle breeds. Int J Parasitol. 2008;38(3-4):313–325. doi: 10.1016/j.ijpara.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 31.Glass EJ, Crutchley S, Jensen K. Living with the enemy or uninvited guests: functional genomics approaches to investigating host resistance or tolerance traits to a protozoan parasite, Theileria annulata, in cattle. Vet Immunol Immunopathol. 2012;148(1-2):178–189. doi: 10.1016/j.vetimm.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pedroni MJ, Sondgeroth KS, Gallego-Lopez GM, Echaide I, Lau AO. Comparative transcriptome analysis of geographically distinct virulent and attenuated Babesia bovis strains reveals similar gene expression changes through attenuation. BMC Genomics. 2013;14(1):763. doi: 10.1186/1471-2164-14-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lack JB, Reichard MV, van den Bussche RA. Phylogeny and evolution of the piroplasmida as inferred from 18S rRNA sequences. Int J Parasitol. 2012;42(4):353–363. doi: 10.1016/j.ijpara.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 34.Laha R, Mondal B, Biswas SK, Chand K, Das M, Sarma D, Goswami A, Sen A. Detection of Babesia bigemina infection in cattle from North-Eastern India by polymerase chain reaction and its genetic relatedness with other isolates. Trop Anim Health and Prod. 2015;47(3):633–636. doi: 10.1007/s11250-015-0769-8. [DOI] [PubMed] [Google Scholar]
- 35.Henson S, Bishop RP, Morzaria S, Spooner PR, Pelle R, Poveda L, Ebeling M, Küng E, Certa U, Daubenberger CA, Qi W. High-resolution genotyping and mapping of recombination and gene conversion in the protozoan Theileria parva using whole genome sequencing. BMC Genomics. 2012;13(1):503. doi: 10.1186/1471-2164-13-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gubbels MJ, Katzer F, Hide G, Jongejan F, Shiels BR. Generation of a mosaic pattern of diversity in the major merozoite-piroplasm surface antigen of Theileria annulata. Mol Biochem Parasitol. 2000;110(1):23–32. doi: 10.1016/S0166-6851(00)00253-X. [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Yang X, Wang Y, Jing Z, Meng K, Liu J, Guo H, Xu R, Cheng Z. Experimental immunology genetic diversity and phylogenetic analysis of Tams1 of Theileria annulata isolates from three continents between 2000 and 2012. Cent Eur J Immunol. 2014;4(4):476–484. doi: 10.5114/ceji.2014.47732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paliwal S, Sudan V, Shanker D, Srivastava M. Coexistence of multiple Theileria annulata genotypes circulating in neonatal calves in semi-arid India. Acta Parasitol. 2020;65(3):679–685. doi: 10.2478/s11686-020-00196-0. [DOI] [PubMed] [Google Scholar]
- 39.Roy S, Bhandari V, Dandasena D, Murthy S, Sharma P. Genetic profiling reveals high allelic diversity, heterozygosity and antigenic diversity in the clinical isolates of the Theileria annulata from India. Front Physiol. 2019;10:673. doi: 10.3389/fphys.2019.00673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kundave VR, Ram H, Shahzad M, Garg R, Banerjee PS, Nehra AK, et al. Genetic characterization of Theileria species infecting bovines in India. Infect Gen Evol. 2019;75:103962. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are included in the published article as supplementary material. The nucleotide sequence data has been deposited in the NCBI GenBank and accession numbers are cited in the manuscript. Further, data regarding animal studies can be made available on reasonable request to corresponding author.