Summary
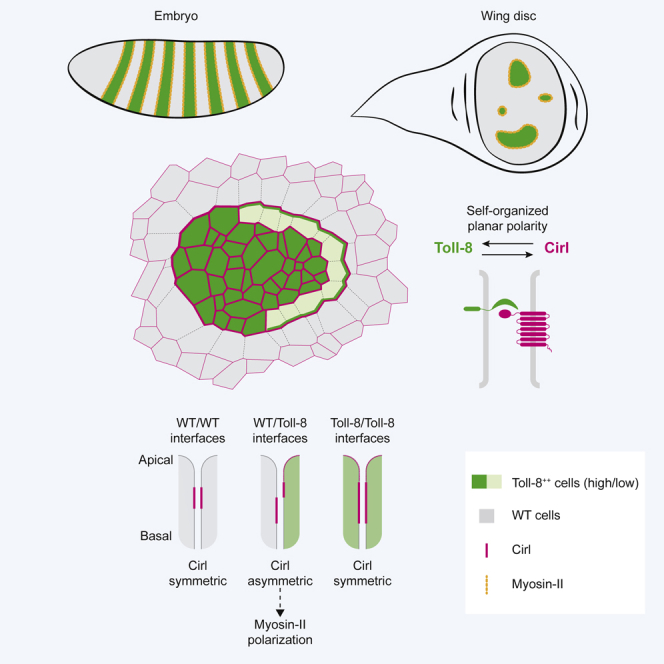
Interfaces between cells with distinct genetic identities elicit signals to organize local cell behaviors driving tissue morphogenesis. The Drosophila embryonic axis extension requires planar polarized enrichment of myosin-II powering oriented cell intercalations. Myosin-II levels are quantitatively controlled by GPCR signaling, whereas myosin-II polarity requires patterned expression of several Toll receptors. How Toll receptors polarize myosin-II and how this involves GPCRs remain unknown. Here, we report that differential expression of a single Toll receptor, Toll-8, polarizes myosin-II through binding to the adhesion GPCR Cirl/latrophilin. Asymmetric expression of Cirl is sufficient to enrich myosin-II, and Cirl localization is asymmetric at Toll-8 expression boundaries. Exploring the process dynamically, we reveal that Toll-8 and Cirl exhibit mutually dependent planar polarity in response to quantitative differences in Toll-8 expression between neighboring cells. Collectively, we propose that the cell surface protein complex Toll-8/Cirl self-organizes to generate local asymmetric interfaces essential for planar polarization of contractility.
Keywords: morphogenesis, mechanics, polarity, PCP, GPCR, Toll receptors, latrophilin
Graphical abstract
Highlights
-
•
Asymmetric expression of a single Toll receptor leads to Myo-II polarization
-
•
The adhesion GPCR Cirl binds to Toll-8 mediating Toll-8-induced Myo-II polarization
-
•
Toll-8 boundaries generate a Cirl interfacial asymmetry that can polarize Myo-II
-
•
Differences in Toll-8 levels lead to interdependent Toll-8 and Cirl planar polarity
Lavalou, Mao et al. report that Toll-8 controls myosin-II planar polarity in Drosophila embryos and wing discs via a physical interaction with the GPCR Cirl/latrophilin. They show that Toll-8 expression boundaries generate a Cirl interfacial asymmetry and propose that it is a potential signal leading to myosin-II polarization.
Introduction
Dynamic cell behaviors that drive tissue morphogenesis are often organized at local cell interfaces. On one end, surface signaling between cells with distinct genetic identities, via compartmentalized expression of ligands and receptors, can generate mechanical barriers in developmental (Dahmann et al., 2011) or pathological contexts (Bielmeier et al., 2016). For instance, expression boundaries of Eph-Ephrin or Leucine-rich repeat (LRR) proteins prevent cell mixing in vertebrate and invertebrate models (Dahmann et al., 2011; Fagotto et al., 2013; Karaulanov et al., 2006; Milán et al., 2001; Paré et al., 2019; Smith and Tickle, 2006; Tomás et al., 2011). On the other end, surface signaling can also orient fluid mechanical interfaces in the context of tissue planar polarity. Surface proteins in the core planar cell polarity (PCP) pathway, such as Flamingo/CELSR and Frizzled, translate tissue-scale cues into vectorial cell polarity known to control planar polarized actomyosin contractility (Aw and Devenport, 2017; Nishimura et al., 2012). Tissue-level gradients of the Fat/Dachsous adhesion molecules in the Fat-PCP pathway induce planar polarized accumulation of the myosin Dachs at cell interfaces to drive dynamic cell rearrangements (Bosveld et al., 2012).
The early Drosophila embryo is an excellent system to investigate how cell groups with distinct genetic identities generate planar polarized mechanical interfaces. In the ventrolateral ectoderm, myosin-II (Myo-II) is enriched at vertical interfaces between anteroposterior (AP) neighbors, which produces polarized actomyosin contractility powering AP axis extension (Bertet et al., 2004; Blankenship et al., 2006; Irvine and Wieschaus, 1994; Zallen and Wieschaus, 2004). The amplitude and polarity of actomyosin contractility appear to be controlled by diverse cell surface proteins. Levels of Myo-II activation at cell interfaces are quantitatively controlled by G protein-coupled receptor (GPCR) signaling (Garcia De Las Bayonas et al., 2019; Kerridge et al., 2016), whereas the polarized enrichment of Myo-II between AP neighbors appears to be governed by several Toll receptors, Toll-2,6,8 (Paré et al., 2014). Interactions between pair-rule genes define periodic and partially overlapping stripes of Toll receptors perpendicular to the AP axis (Paré et al., 2014). Thus, each column of cells expresses a different combination of Toll-2,6,8, which are thought to be collectively required for planar polarized Myo-II activity (Paré et al., 2014; Tetley et al., 2016). Toll receptors belong to the LRR super family and are well known for their functions in developmental patterning and innate immunity (Anthoney et al., 2018). They recently emerged as conserved molecules involved in embryonic axis elongation in different arthropod species (Benton et al., 2016; Paré et al., 2014). How Toll receptors define polarized mechanical interfaces, and whether they interact with other cell surface receptors such as GPCRs, remain largely unexplored.
Here, we investigated how Toll receptors control Myo-II planar polarity in conjugation with GPCR signaling in the Drosophila embryonic ectoderm and larval wing disc epithelium. In contrast to the combinatorial Toll code model, we show that a single Toll receptor, Toll-8, enriches Myo-II at its expression boundary. This enrichment does not require the cytoplasmic tail of Toll-8. Further, we identified a Toll-8-binding partner, the adhesion GPCR Cirl/latrophilin, which is required for Toll-8-mediated Myo-II enrichment in both embryos and wing discs. We show in wing discs that Cirl asymmetry is sufficient to enrich Myo-II, and we observed an interfacial asymmetry in Cirl apicobasal localization at Toll-8 expression boundaries. Moreover, when neighboring cells express different levels of Toll-8 in wing discs, both Toll-8 and Cirl exhibit robust and mutually dependent planar polarity. Our study thus reveals that Toll-8 and Cirl form a cell surface protein complex essential for planar polarized actomyosin activity, generate local asymmetric interfaces, and co-polarize in a self-organized manner.
Results
Asymmetric expression of a single Toll receptor leads to Myo-II polarization in embryos
In the Drosophila embryonic ectoderm, it has been hypothesized that trans-interactions between different Toll receptors across cell-cell interfaces signal to polarize junctional Myo-II (Paré et al., 2014). In light of the observation that ectopic expression of Toll-2 or Toll-8 alone induces Myo-II enrichment late in embryogenesis (Paré et al., 2014), we first tested the simplest hypothesis that a single Toll is sufficient to polarize junctional Myo-II in the embryonic ectoderm. To this end, we injected embryos with double-stranded RNAs (dsRNAs) targeting Toll-2,6,7 (toll-2,6,7 RNAi), leaving only endogenous Toll-8 expressed in vertical stripes (Figure S1A), and observed Myo-II with mCherry-tagged Myo-II regulatory light chain (MRLC-Ch). We found Myo-II specifically enriched at the interfaces between Toll-8 expressing and non-expressing cells (Figures S1A–S1D), suggesting that asymmetric expression of a single Toll, Toll-8, leads to Myo-II enrichment independent of other Toll receptors.
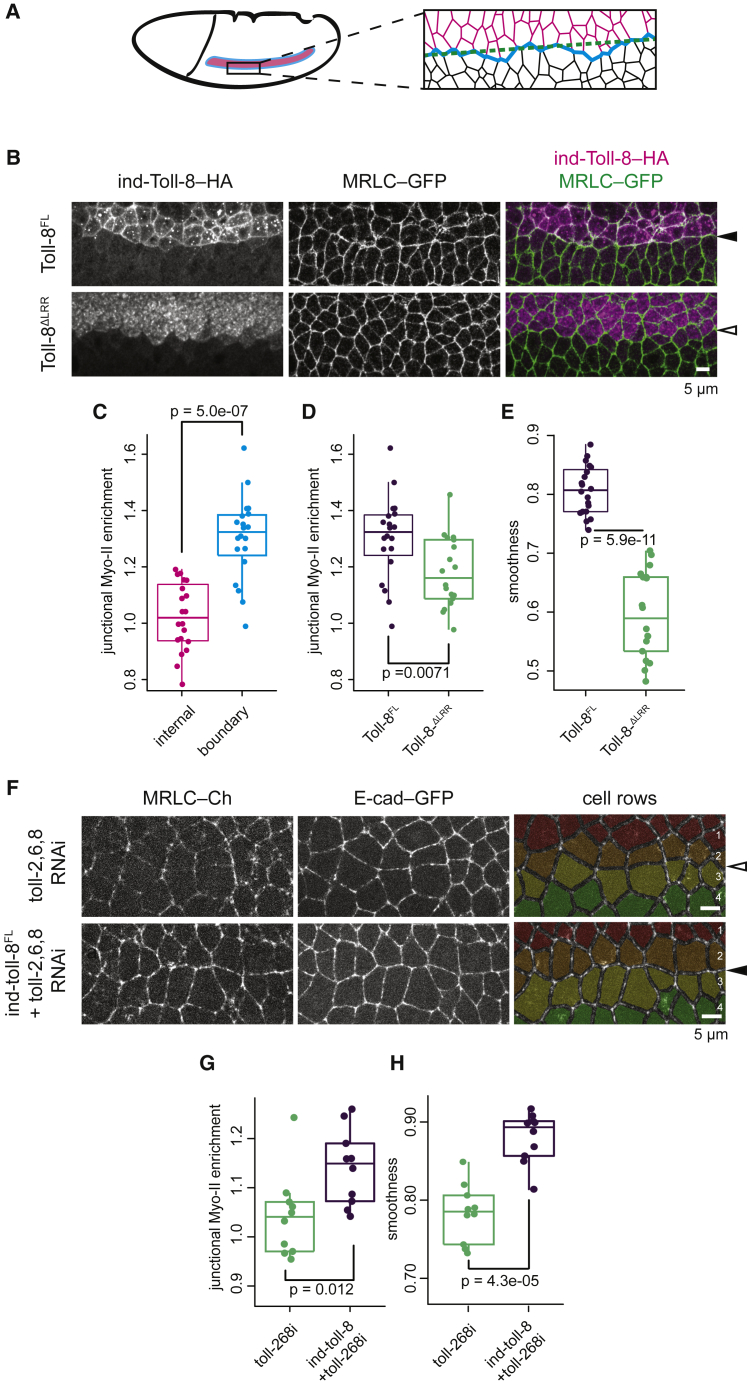
To assess the sufficiency of Toll-8 asymmetry in Myo-II polarization independent of the anteroposterior (AP) patterning system, we engineered embryos expressing a single stripe of Toll-8 along the AP axis that runs orthogonal to the endogenous Toll-2,6,8 stripes, using an intermediate neuroblasts defective (ind) enhancer (Stathopoulos and Levine, 2005) (ind-Toll-8-HA, Figure 1A). We monitored Myo-II with GFP-tagged Myo-II regulatory light chain (MRLC-GFP) and detected a Myo-II-enriched cable at the ventral boundary between ectopic Toll-8 expressing and wild-type cells (Figures 1A and 1B, Toll-8FL). Note that this ectopic Myo-II cable runs along the AP axis and perpendicular to the endogenous Myo-II planar polarity. By contrast, no Myo-II enrichment is detected at homotypic Toll-8 interfaces within the ind-Toll-8 domain (Figure 1C). Myo-II enrichment at the ventral border of ind-Toll-8 is comparable with that at the endogenous boundaries of the Toll-8 expression domain (Figure S1E). When we deleted the LRRs from the Toll-8 extracellular domain, Toll-8 was no longer localized to the plasma membrane and failed to accumulate Myo-II (Figures 1A, 1B, and 1D, Toll-8ΔLRR). Consistent with an upregulation of cortical tension, the boundary of Toll-8-expressing cells was smoother when full-length Toll-8 (Toll-8FL) was expressed compared with Toll-8ΔLRR (Figures 1B and 1E; smoothness defined by the ratio of distance between terminal vertices over total junctional length).
Figure 1.
Toll-8 asymmetry leads to Myo-II enrichment in embryos, independent of other Toll receptors
(A) Schema of an embryo expressing ind-Toll-8-HA and cell interfaces around the ventral boundary of ind-Toll-8–HA. Pink, black and blue denote interfaces within the ind-Toll-8-HA stripe, the wild-type tissue, and at the ventral boundary of the ind-Toll-8-HA, respectively. Dashed line in green: distance between the first and last vertices of the ind-Toll-8-HA ventral boundary.
(B) Anti-Toll-8-HA and anti-MRLC-GFP signals in Drosophila stage-7 embryos expressing full-length Toll-8 (Toll-8FL, top, n = 20) or Toll-8 with the extracellular LRRs removed (Toll-8ΔLRR, bottom, n = 18) driven by the ind promoter. Myo-II is enriched at the ventral boundary of the ind-Toll-8FL-HA stripe (filled arrowhead) but not for ind-Toll-8ΔLRR-HA (empty arrowhead).
(C) Junctional Myo-II enrichment relative to wild-type tissue within the ind-Toll-8FL-HA stripe (pink) or at the ventral boundary of the ind-Toll-8FL-HA stripe (blue) for the condition shown in (B, top).
(D and E) Junctional Myo-II enrichment (D) and boundary smoothness (E) at the ventral ind boundary for the conditions shown in (B).
(F) Stills from time-lapse movies in wt embryos (top, n = 10) or embryos expressing ind-Toll-8FL-HA injected with dsRNAs against Toll-2,6,8 (bottom, n = 10). Pseudo colors mark 4 cell rows. In the absence of endogenous Toll-2,6,8, Myo-II is still enriched at the ind ventral boundary (filled arrowhead).
(G and H) Junctional Myo-II enrichment (G) and boundary smoothness (H) at the ventral ind boundary for the conditions shown in (F).
When we injected ind-Toll-8-HA embryos with dsRNAs targeting only endogenous Toll-2,6,8 (toll-2,6,8 RNAi), Myo-II was still enriched at the boundary of ectopic Toll-8-expressing cells (Figures 1F, 1G, and S1F). This boundary was also smooth (Figures 1F, 1H, and S1F). Altogether, we conclude that an interface defined by asymmetric expression of a single Toll, Toll-8, is sufficient to polarize Myo-II.
Asymmetric expression of a single Toll receptor polarizes Myo-II in wing discs
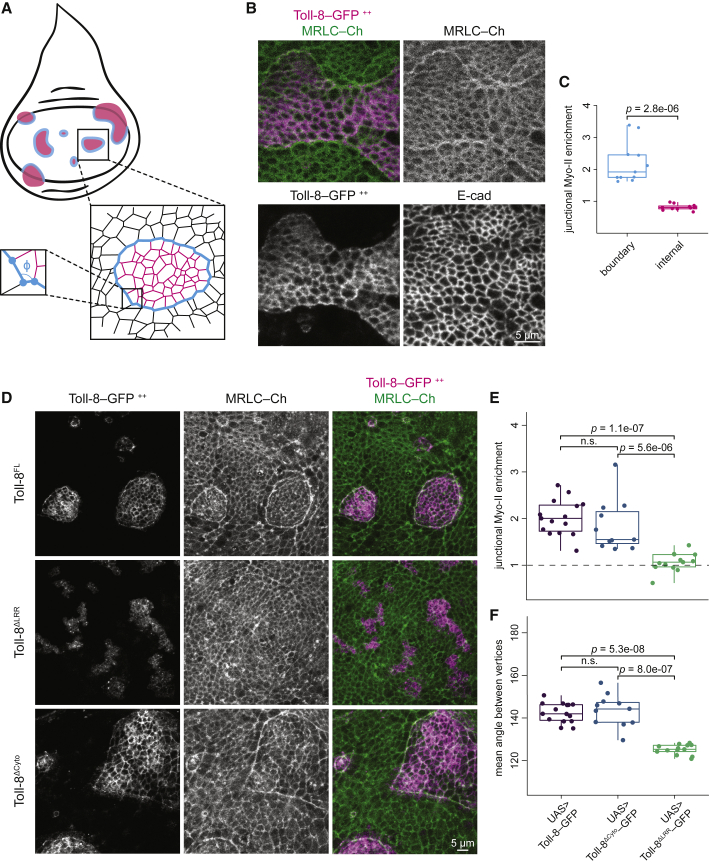
To further dissect the capacity of Toll-8 to elicit junctional Myo-II enrichment, we used clonal analysis to generate random interfaces between wild-type and Toll-8-overexpressing cells in larval wing imaginal discs since clonal analysis is not possible in early embryos. Wing imaginal discs exhibit polarized supracellular cables of Myo-II at the periphery of the pouch region near the hinge (LeGoff et al., 2013). Toll receptor overexpression causes wing developmental defects (Yagi et al., 2010), and Toll-8 is expressed in the wing hinge region but absent in most of the pouch region (Alpar et al., 2018; Yagi et al., 2010). We induced clones overexpressing Toll-8-GFP in the larval wing disc epithelium and monitored Myo-II localization with MRLC-Ch 24 h after clone induction in the wing pouch (Figure 2A). This resulted in striking junctional Myo-II enrichment specifically at the boundary of Toll-8 overexpressing clones, in contrast to homotypic interfaces within the Toll-8 clone (Figures 2A–2C). As in embryos, we found that in the absence of the LRRs, Toll-8 was no longer localized to the plasma membrane and failed to polarize Myo-II (Figures 2D and 2E, Toll-8ΔLRR). Toll-8FL clones were more compact with a smoother boundary compared with Toll-8ΔLRR clones (Figures 2D and 2F; smoothness defined by the mean angle between neighboring vertices, Figure 2A).
Figure 2.
Toll-8 asymmetry leads to Myo-II enrichment in wing discs, independent of the Toll-8 cytoplasmic tail
(A) Schema showing wing disc clones and cell interfaces around a clone. Pink, black, and blue denote interfaces within the clone, within the wild-type tissue, and at the clonal boundary, respectively. φ is the angle between neighboring vertices at the clonal boundary.
(B) Fixed Toll-8-GFP, MRLC-Ch, and E-cadherin signals from wing disc clones overexpressing full-length Toll-8 (n = 11). Myo-II is enriched at the clone boundary but not inside the clone.
(C) Junctional Myo-II enrichment relative to wild-type tissue within (pink) or at the boundary of (blue) Toll-8 overexpressing clones for the condition shown in (B).
(D) Fixed Toll-8-GFP and MRLC-Ch signals from wing disc clones overexpressing full-length Toll-8 (Toll-8FL, top, n = 15), Toll-8 with the extracellular LRRs removed (Toll-8ΔLRR, middle, n = 11), or Toll-8 with the intracellular cytoplasmic tail removed (Toll-8ΔCyto, bottom, n = 13). Myo-II enrichment does not require the cytoplasmic tail of Toll-8.
(E and F) Junctional Myo-II enrichment (E) and boundary smoothness (F) at clone boundaries for the conditions shown in (D).
The cytoplasmic domain of Toll proteins is necessary for canonical Toll signal transduction (Anthoney et al., 2018). To test whether the cytoplasmic domain of Toll-8 is required for Myo-II enrichment, we generated wing disc clones overexpressing a truncated version of Toll-8, removing its cytoplasmic domain (Toll-8ΔCyto). Surprisingly, Myo-II was still enriched at the boundary of Toll-8ΔCyto-overexpressing clones (Figures 2D and 2E), and the clonal boundary was still smooth compared with Toll-8ΔLRR clones (Figures 2D and 2F). We conclude that the cytoplasmic domain of Toll-8 is dispensable for Myo-II polarization. We observed similar results for Toll-6 and Toll-2 (Figures S2A and S2B). This indicates that the signaling events leading to Myo-II enrichment at the boundary between Toll-8-expressing and non-expressing cells may require another protein, presumably interacting with Toll-8 at the cell surface.
Cirl/latrophilin binds to Toll-8 and mediates Toll-8-induced Myo-II enrichment
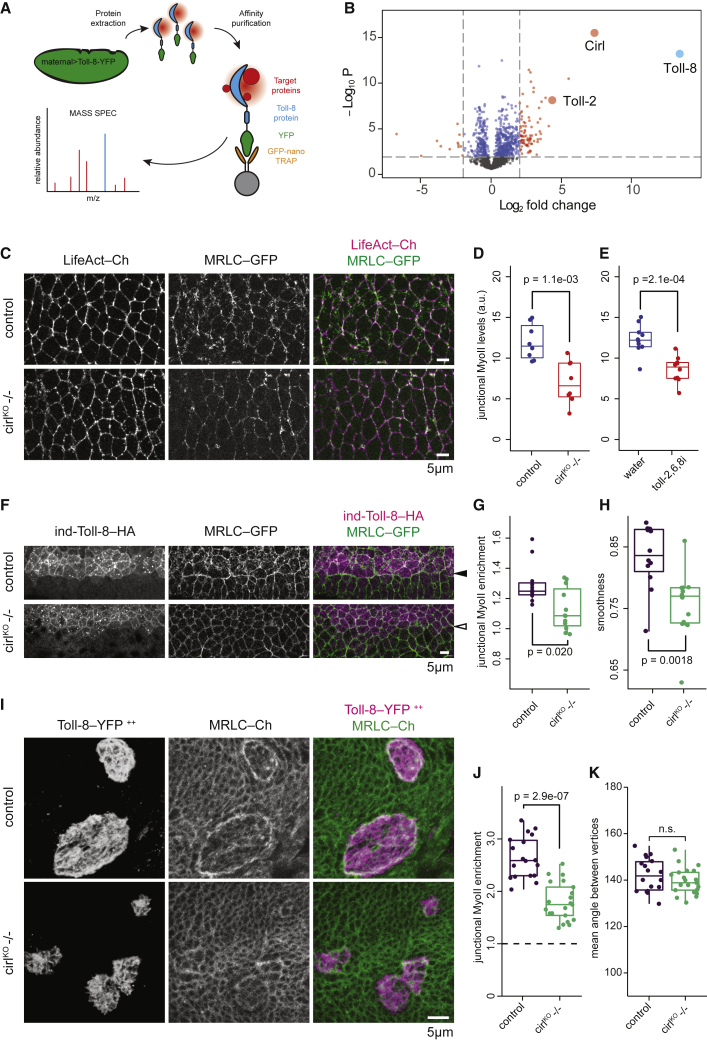
To look for binding partners of Toll-8, we performed affinity purification-mass spectrometry experiments using lysates isolated from embryos overexpressing Toll-8-YFP as a bait (Figure 3A and STAR methods). Of the specifically bound proteins that interact directly or indirectly in a complex with Toll-8, Cirl was the most abundant target (Figure 3B; Table S1) and of particular interest since it is a GPCR. Cirl is the Drosophila homolog of vertebrate latrophilin, a member of the adhesion GPCR (aGPCR) subfamily (Scholz et al., 2015; Schöneberg and Prömel, 2019). Interestingly, the Toll-8 extracellular domain shares sequence similarities with that of the human LRR protein FLRTs (Dolan et al., 2007), which are known to form protein complexes with human latrophilins (Boucard et al., 2014; Li et al., 2020; O’Sullivan et al., 2012; Sando et al., 2019; del Toro et al., 2020). We also recovered Toll-2 as a significant Toll-8-binding partner in vivo (Figure 3B; Table S1), in agreement with previous findings in vitro (Paré et al., 2014).
Figure 3.
The aGPCR Cirl interacts physically with Toll-8 and is necessary for Myo-II enrichment induced by Toll-8.
(A) Affinity purification-mass spectrometry using embryos overexpressing the Toll-8-YFP fusion protein (Toll-8 in blue, YFP in green, potential binding targets in red) and the GFP-Trap (nanobody against GFP/YFP in orange, agarose bead in gray).
(B) Volcano plot showing differential levels (x axis) and p values (y axis) of proteins identified by mass spectrometry in embryos overexpressing Toll-8-YFP versus wt embryos. Vertical dashed lines denote the cutoff for log2|Fold Change| as 2. Horizontal dashed line denotes the cutoff for p value as 0.01. Toll-8 (bait), Cirl, and Toll-2 are highlighted.
(C) Stills from time-lapse movies in wt (top, n = 8) or cirlKO−/− (bottom, n = 8) embryos. LifeAct-Ch marks cell outlines. Junctional Myo-II is reduced in cirlKO−/− null mutant embryos.
(D) Mean junctional Myo-II levels for the conditions shown in (C).
(E) Mean junctional Myo-II levels in embryos injected with water (n = 10) or dsRNAs against Toll-2,6,8 (n = 10) (related to Figures S1A and S1B).
(F) Anti-Toll-8-HA and anti-MRLC-GFP signals in wt (top, n = 12) or cirlKO−/− (bottom, n = 12) embryos expressing full-length Toll-8 under the ind promoter. Myo-II is reduced at the ind ventral boundary in cirlKO−/− embryos (empty arrowhead).
(G and H) Junctional Myo-II enrichment (G) and boundary smoothness (H) at the ventral ind boundary for the conditions shown in (F).
(I) Fixed Toll-8-YFP and MRLC-Ch signals from wing disc clones overexpressing full length Toll-8 in wt (top, n = 18) or cirlKO−/− (bottom, n = 21) wing discs. Myo-II is reduced at the boundary of Toll-8 overexpressing clones in cirlKO−/− wing discs.
(J and K) Junctional Myo-II enrichment (J) and boundary smoothness (K) at clone boundaries for the conditions shown in (I).
Using a Cirl-RFP knockin line (Scholz et al., 2017) to visualize endogenous Cirl, we found that Cirl localized to the membrane and was enriched at cell-cell interfaces around adherens junctions in both the embryo and the wing disc (Figures S3A and S3B). We next asked whether Cirl is required for Myo-II enrichment in the embryo. cirl maternal and zygotic mutant embryos showed delayed extension of the embryonic axis (Figures S3C and S3D), reduced cell intercalation (Figures S3E and S3F; Video S1), and a strong reduction of junctional Myo-II in the ectoderm (Figures 3C and 3D), resembling Myo-II reduction observed in toll-2,6,8 RNAi embryos (Figures 3E and S1A). Thus, cirl is required for junctional Myo-II enrichment in the embryo. We observed that Myo-II is reduced at vertical junctions both in toll-2,6,8 RNAi embryos and in cirl mutant embryos (Figures S3G and S3H). However, all junctions show lower Myo-II levels in cirl mutant embryos (Figure S3H). Myo-II planar polarity tends to be reduced in both conditions but is significantly reduced only in toll-2,6,8 RNAi embryos (Figure S3I). We suggest that in the absence of Cirl, Toll receptors can still polarize Myo-II via other GPCRs than Cirl.
Frames are re-aligned to keep the center of tracked cell group constant. Framer rate: 30 s/frame. Duration: 20 min. Scale bar: 5 μm, related to Figures 3 and S3.
We further tested whether Cirl is required for Toll-8-induced polarization of Myo-II by comparing the ventral boundary of ind-Toll-8-HA stripe in wild-type and cirl-null mutant embryos. In the absence of Cirl, we found a strong reduction in Myo-II enrichment at the boundary of Toll-8 stripe, which was also less smooth (Figures 3F–3H). Moreover, in cirl-null mutant wing discs, we observed a similar reduction in Myo-II enrichment at the boundary of Toll-8 clones (Figures 3I and 3J). Note that Myo-II enrichment was not completely abolished, and the clonal boundary was still smooth (Figure 3K), suggesting that other molecules in addition to Cirl may interact with Toll-8 to confer Myo-II enrichment at clone boundaries in the wing disc. Taken together, we conclude that Toll-8 and Cirl physically and functionally interact to polarize Myo-II at interfaces between Toll-8-expressing and non-expressing cells.
Cirl asymmetric interfaces lead to Myo-II enrichment
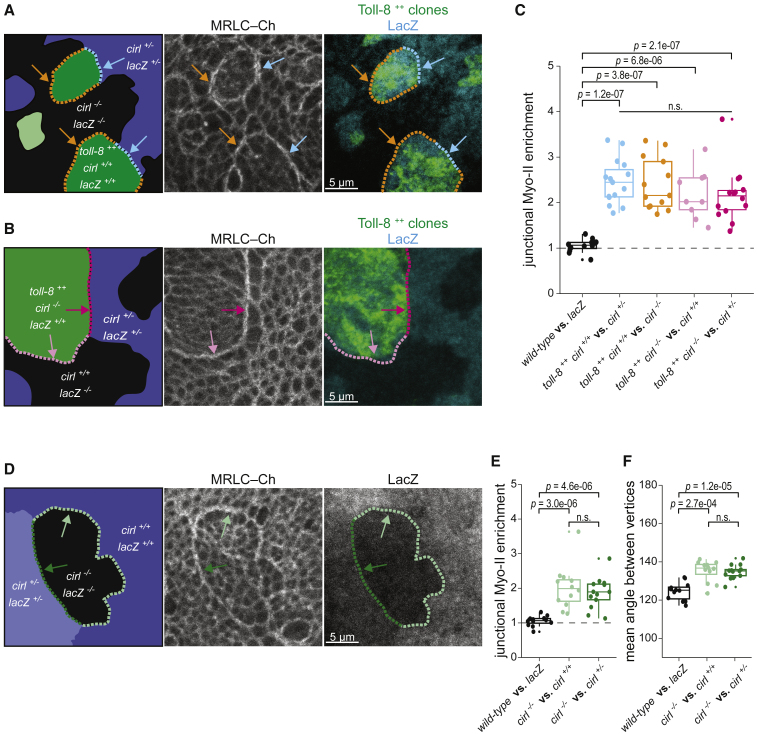
Interestingly, we observed that Myo-II was enriched on both sides of the interface between Toll-8 expressing and non-expressing cells (Figures S4A and S4A′), indicating that Myo-II is also enriched in cells that do not express Toll-8. We thus tested whether Cirl mediates Myo-II enrichment on both sides of the interface. To this end, we performed mosaic analysis with a repressible cell marker (MARCM) in wing discs and induced cirl-null mutant clones either adjacent to (Figure 4A) or coincident with (Figure 4B) Toll-8-overexpressing clones. We expected that removing Cirl from one side should lead to a reduction in Myo-II enrichment at the clone boundary due to a loss of Myo-II enrichment in cells where Cirl is absent. Surprisingly, when Cirl was only absent from neighboring cells, Myo-II was enriched at the Toll-8 boundary at similar levels to control interfaces (Figure 4A, compare orange and cyan arrows, quantified in Figure 4C) and on both sides of the clone boundary (Figure S4B). Similarly, when Cirl was only absent from cells overexpressing Toll-8, Myo-II was still enriched at similar levels at the clone boundary (Figure 4B, quantified in Figure 4C) and on both sides of the clone boundary (Figure S4C). Thus, Cirl is dispensable in either Toll-8-expressing or -responding/contacting cells. This is remarkable since Cirl must be present on at least one side of Toll-8-overexpressing clones, as the complete removal of Cirl on both sides significantly reduces Myo-II enrichment (Figure 3J). We thus hypothesized that (1) the role of Toll-8 is to induce an asymmetry of Cirl activity at the clonal boundary and that (2) Cirl asymmetric interfaces enrich Myo-II. To test the latter, we generated cirl mutant clones in the wing disc without overexpressing Toll-8. We reasoned that at the cirl mutant clonal interface Cirl localization and activity is de facto asymmetric. This is supported by the observation that Cirl is still localized in wild-type cell interfaces in contact with cirl mutant cells (Figure S4D, yellow arrowhead). We found that Myo-II was indeed enriched at the boundary of cirl mutant clones and that the boundary was smooth compared with control clones (Figures 4D–4F). This is similar to Toll-8-overexpressing clones, albeit to a lesser extent. We thus conclude that asymmetric Cirl interfaces at cirl mutant clonal boundary enrich Myo-II. Hence, we next investigated whether Toll-8 does induce a Cirl asymmetry at its expression boundary.
Figure 4.
Cirl asymmetric expression leads to Myo-II enrichment
(A) MARCM clones in a wing disc where clones overexpressing Toll-8-YFP (toll-8++ in green) are juxtaposed to control cells heterozygous (cirl+/− in blue, cyan arrows, n = 15) or null mutant (cirl−/− in black, orange arrows, n = 13) for cirl. Myo-II enrichment at the boundary of Toll-8-overexpressing cells is similar in both cases.
(B) MARCM clones in the wing disc. Myo-II is enriched at boundaries (magenta and pink arrows) of clones overexpressing Toll-8 and null mutant for cirl (toll-8++, cirl−/− in green) juxtaposed to cells heterozygous (cirl+/− in blue, n = 14) or wild type (cirl+/+ in black, n = 9) for cirl.
(C) Quantifications of junctional Myo-II enrichment at clone boundaries for the conditions shown in (A) and (B). With the exception of control clones (black boxplot, images not shown, n = 12), colors of the boxplots correspond to the arrows in (A) and (B).
(D) Myo-II is enriched at the boundary (green arrows) of cirl−/− null mutant clones (cirl−/− in black) juxtaposed to cells heterozygous (cirl+/− in light blue, n = 13) or wild type (cirl+/+ in dark blue, n = 12) for cirl.
(E and F) Junctional Myo-II enrichment (E) and boundary smoothness (F) quantified at clone boundaries for the conditions shown in (D). The black plot in (E) is the same as in (C).
Toll-8 generates a Cirl interfacial asymmetry at its expression boundary
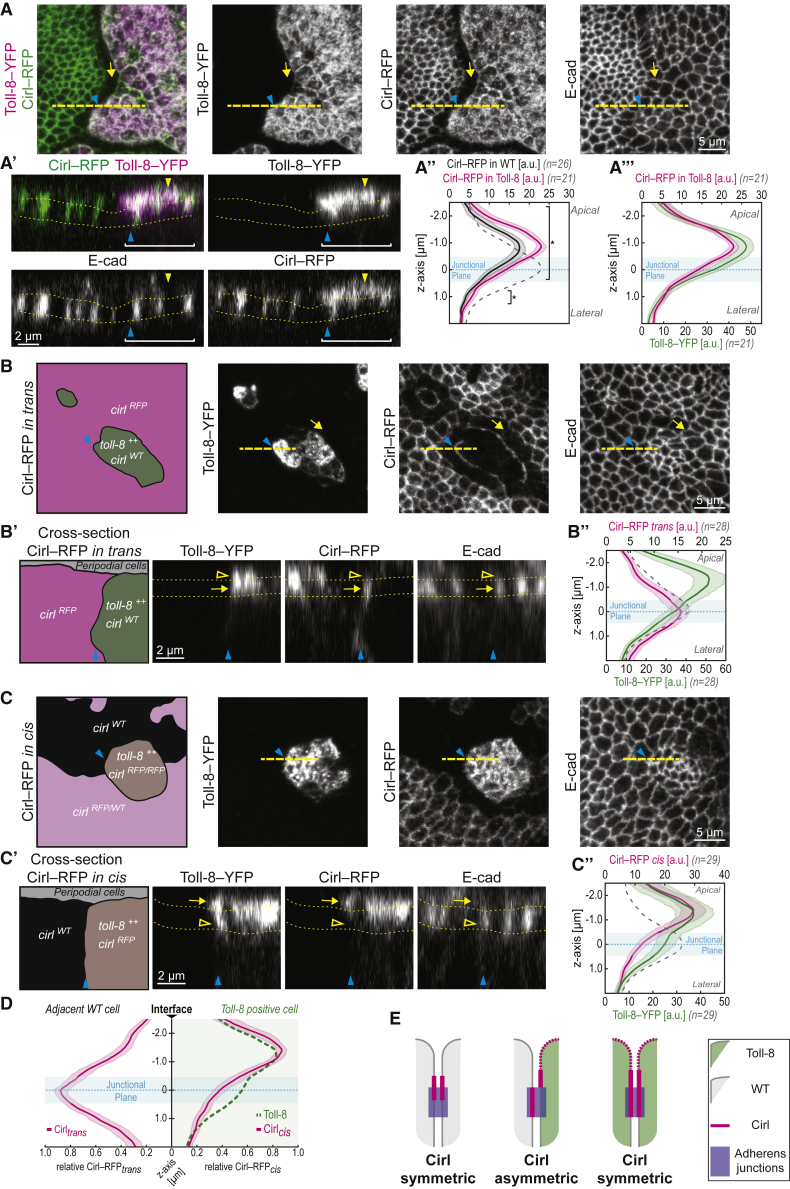
To test if Toll-8 is able to induce a Cirl asymmetry at its expression boundary, we first assessed the effect of Toll-8-overexpressing clones on Cirl localization with wing discs expressing Cirl-RFP from the endogenous locus. In control cells that do not overexpress Toll-8, Cirl-RFP was localized at adherens junctions (marked by E-cadherin localization) and in the subapical domain (i.e., the domain above adherens junctions where neighboring cells are in direct contact) (Figures 5A–5A″, black curve). In cells overexpressing Toll-8-YFP, Cirl-RFP surface levels increased both at adherens junctions and in the subapical domain (Figures 5A–5A″, magenta curve shifted compared with black curve). Cirl-RFP distribution follows that of Toll-8-YFP (Figures 5A and 5A′, see overlays, and 5A″′), suggesting that Toll-8 re-localizes and stabilizes Cirl. At high levels of expression, Toll-8-YFP was present at the free apical membrane (i.e., the domain where cells are in contact with the extracellular space), which coincided with ectopic localization of Cirl-RFP in this domain (Figure 5A′, yellow arrowheads). This suggests that Toll-8 and Cirl physically interact in cis. Strikingly, we observed that Cirl-RFP was depleted from the orthogonal junctions in wild-type cells in direct contact with the boundary of Toll-8-YFP overexpressing cells (Figure 5A, yellow arrow). Thus, Toll-8 affects Cirl localization in trans. We observed similar effects on Cirl localization with Toll-6 clonal overexpression (Figure S5A) but not with Toll-2 (Figure S5B), suggesting that Toll-6 also interacts with Cirl while Toll-2 might interact with another protein.
Figure 5.
Toll-8 generates Cirl interfacial asymmetry at the boundary of its expression domain
(A) Toll-8-YFP overexpressing clone in a Cirl-RFP wing disc. Toll-8-YFP and anti-Cirl-RFP signals colocalize inside the clone. Cirl is depleted from junctions orthogonal to the clone boundary (yellow arrow). (A′) Optical cross-section of the dashed line in (A) (apical to the top). Dotted lines mark the junctional plane based on E-cadherin signals. Both Toll-8 and Cirl are present at the free apical membrane (yellow arrowhead). (A″ and A″′) Intensity of Toll-8 and Cirl (x axis) along the apicobasal axis (y axis). The junctional plane (blue area) is defined by E-cadherin peak levels (dashed gray curve). Cirl levels increase in Toll-8 overexpressing clones both at the junctional plane and in the subapical domain (A″, square brackets, ∗p < 0.05.) following Toll-8 distribution (A″′).
(B) Toll-8-YFP overexpressing clones (toll-8++, cirlWT in green) in a wing disc where Cirl-RFP is present only outside of Toll-8 overexpressing clones (cirlRFP in magenta). Cirl is depleted from interfaces orthogonal to the clone boundary (yellow arrow) and accumulates at the clone boundary. (B′) Optical cross-section (dashed line in B) shows Cirl enrichment at the junctional plane (yellow arrow), quantified in (B″).
(C) Toll-8-YFP overexpressing clone with Cirl-RFP present inside (toll-8++, cirlRFP in brown) facing cells homozygous for untagged Cirl (cirlWT in black) or heterozygous for cirlRFP (cirlRFP/WT in light magenta). (C′) Optical cross-section (dashed line in C) shows Cirl enrichment above the junctional plane (yellow arrow), quantified in (C″).
(D) Comparison of Cirl apicobasal localization at both sides of Toll-8 clonal boundaries. Cirl is observed at the junctional plane in trans and above the junctional plane in cis creating a Cirl interfacial asymmetry at Toll-8 clonal boundaries.
(E) Model representing the interfacial asymmetry in Cirl apicobasal localization at Toll-8 expression boundary.
The depletion of Cirl from orthogonal junctions in contact with Toll-8-overexpressing cells led us to ask if Cirl is polarized at the boundary of Toll-8 overexpressing cells in trans. To test this, we used MARCM to observe endogenously tagged Cirl-RFP only adjacent to (i.e., in trans) Toll-8-YFP-overexpressing cells. Note that in Cirl-RFP-negative cells, untagged endogenous Cirl is present. When Cirl-RFP was present only in trans, Cirl-RFP was indeed localized at the clone boundary (Figure 5B, blue arrowheads) and depleted from the junctions orthogonal to the clone boundary (Figure 5B, yellow arrows). This effect did not require the presence of Cirl inside Toll-8-overexpressing cells (Figure S5C). Thus, Cirl is planar polarized in wild-type cells in direct contact with Toll-8 overexpressing cells, suggesting that Toll-8 and Cirl physically interact in trans. Surprisingly, at the clone boundary, Cirl-RFP in trans strictly colocalized with E-cadherin at adherens junctions and was absent from the subapical domain (Figure 5B′, arrow, and 5B″). We then examined how Cirl-RFP was localized at clonal boundaries in cells overexpressing Toll-8-YFP using MARCM to observe endogenously tagged Cirl-RFP only inside the clone (i.e., in cis). When Cirl-RFP was present only in cis (Figure 5C), Cirl-RFP was enriched in the subapical domain and tended to be reduced at adherens junctions (Figures 5C′, arrow, and 5C″). Therefore, at cell-cell interfaces between wild-type and Toll-8-overexpressing cells, where cells are in direct contact, Cirl is specifically localized to adherens junctions in trans (Figures 5B′ and 5B″) while it is enriched above adherens junctions in cis (Figures 5C′ and 5C″), creating an interfacial asymmetry in Cirl localization (Figure 5D). The apicobasal localization of Cirl at either side of Toll-8 clonal boundaries remained unchanged when Cirl was removed from the opposite side of the clone (Figures S5C′ and S5D″), suggesting that Cirl asymmetric localization at the boundary of Toll-8-overexpressing cells is not due to the competition for Toll-8 binding between Cirl in cis and in trans.
Toll-8 can thus induce an interfacial asymmetry in Cirl apicobasal localization at Toll-8 expression boundary. Our genetic setup does not allow us to assess Cirl localization only at one side of cell interfaces inside Toll-8 overexpressing clones. Given that inside the clone Toll-8 is symmetrically present both in cis and in trans, we assume that Cirl should be symmetrically localized at these internal interfaces, resulting in Cirl interfacial asymmetry specifically at the Toll-8 expression boundary, and not at internal interfaces (Figure 5E). Since we showed that asymmetric Cirl interfaces between wild-type and cirl mutant cells lead to Myo-II enrichment (Figures 4D and 4E), we propose that the interfacial asymmetry of Cirl induced by Toll-8 might be a signal leading to Myo-II enrichment at Toll-8 expression boundaries.
Quantitative differences in Toll-8 expression lead to mutually dependent Toll-8 and Cirl polarity
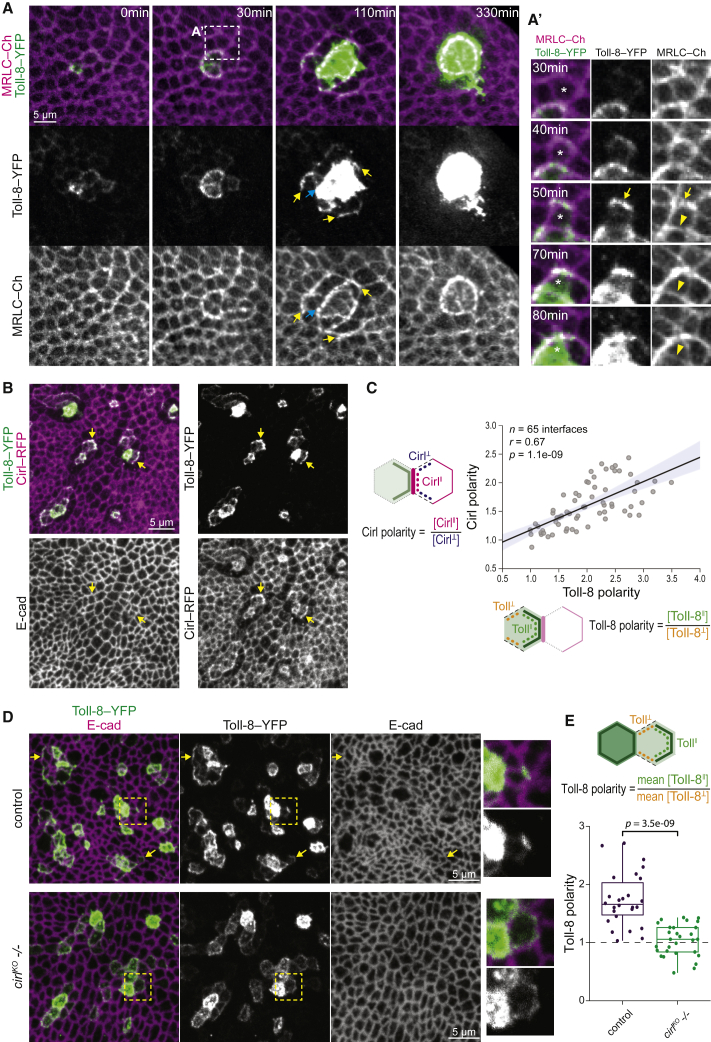
To further study the dynamic process of Myo-II planar polarization, we performed ex vivo live imaging (Dye et al., 2017) of nascent Toll-8-overexpressing clones in cultured wing discs, using a temperature-sensitive GAL80 (GAL80ts) to precisely time the onset of Toll-8 expression. Myo-II enrichment was already observed when Toll-8-YFP was initially restricted to cell-cell interfaces, prior to its subsequent accumulation in the free apical membrane (Figure S6A). This supports the idea that interfacial Toll-8-YFP, instead of Toll-8 in the free apical membrane, is responsible for the polarization of Myo-II.
Likely due to stochasticity in GAL80ts inactivation/GAL4 de-repression (Figure 6A; Videos S2, S3, and S4), the asynchronous onset of Toll-8 expression between cells in a given clone led to the generation of dynamic changes in Toll-8 expression, creating quantitative differences in Toll-8 levels between neighboring cells. We found that Myo-II was not only enriched at the boundary of Toll-8-YFP expressing cells facing Toll-8-YFP-negative cells but also enriched at interfaces between cells with different Toll-8-YFP levels (Figure 6A, 100 min, cyan arrow, and S6B; Videos S2 and S3). In this assay, the kinetics of Myo-II polarization at the boundary of cells expressing different levels of Toll-8 is in the range of 10 min (Figure 6A′, between 40 and 50 min, Myo-II became polarized at a Toll-8 interface) and the amplitude of Myo-II polarity is around 2-fold (Figure S6B), which is commensurate with the dynamics and amplitude of Myo-II polarization in the embryonic ectoderm (Bertet et al., 2004; Blankenship et al., 2006). Moreover, as the levels of Toll-8-YFP further increased, once Toll-8-YFP expression reached the same level between these contacting cells, Myo-II enrichment was no longer present at these interfaces (Figure 6A′, arrowheads), and was stabilized only at the boundary of the clone (Figure 6A, 330 min, and S6B). This argues that quantitative differences in Toll-8 expression between neighboring cells polarize Myo-II.
Figure 6.
Quantitative differences in Toll-8 expression lead to mutually dependent Toll-8 and Cirl polarity
(A) Stills from a time-lapse movie showing live dynamics of Toll-8-YFP and MRLC-Ch in a nascent Toll-8-YFP-overexpressing clone in a wing disc. Toll-8-YFP becomes detectable at 0 min and increases its levels during the time-lapse. Yellow arrows mark junctions displaying Toll-8 planar polarized enrichment associated with Myo-II enrichment. Cyan arrow indicates an interface between high and low Toll-8 cells. (A′) Zoomed view of a cell (asterisk) that dynamically upregulates expression of Toll-8-YFP during ~50 min. Myo-II is being enriched at junctions that show Toll-8 planar polarity (arrow) and this enrichment is lost once Toll-8 expression reaches similar levels between contacting cells (arrowhead).
(B) Nascent Toll-8-YFP clones in a Cirl-RFP wing disc. Cirl levels are increased at Toll-8-enriched junctions (arrows).
(C) Toll-8 planar polarity versus Cirl planar polarity in trans for the condition shown in (B) show a significant positive correlation. Error band indicates the 95% confidence interval.
(D) Toll-8 planar polarity in nascent Toll-8-YFP clones in control (top, n = 26) or cirl-null mutant (bottom, n = 29) wing discs. Zoomed views on the right. Toll-8 planar polarity is reduced in the absence of Cirl.
(E) Quantifications of Toll-8 planar polarity in cells with low Toll-8-YFP levels in direct contact with cells with high Toll-8-YFP levels for the conditions shown in (D).
Asterisks indicate a cell that dynamically upregulates expression of Toll-8–YFP during ∼50min. Myo-II is being enriched at junctions that show Toll-8–YFP planar polarity (arrow) and this enrichment is lost once Toll-8–YFP expression reaches similar levels between contacting cells (arrowheads). Scale bar: 5 μm, related to Figures 6 and S6.
Scale bar: 5 μm, related to Figures 6 and S6.
Scale bar: 5 μm, related to Figures 6 and S6.
Strikingly, between neighboring cells with quantitative differences in Toll-8 expression levels, Toll-8-YFP was initially planar polarized in cells expressing lower levels and tended to accumulate at interfaces facing away from the cells expressing higher levels (Figures 6A and 6A′, yellow arrows; Videos S2, S3, and S4). Moreover, in cells where Toll-8-YFP was planar polarized, Myo-II was specifically enriched at Toll-8-enriched interfaces, leading to Myo-II planar polarity across several rows of cells (Figure 6A, 110 min, yellow arrows; Videos S2 and S3). Therefore, Toll-8 polarization emerged when neighboring cells express different levels of Toll-8 and this is correlated with polarization of Myo-II.
We then asked how Cirl localization was affected by these nascent polarized patterns of Toll-8. Since live Cirl-RFP signals were too weak, we analyzed anti-Cirl-RFP signals in fixed wing discs. Cirl-RFP was both enriched at interfaces where Toll-8 was planar polarized (Figure 6B, arrows) and reduced in orthogonal interfaces facing cells exhibiting Toll-8 planar polarity. We found a significant correlation between Toll-8 planar polarity and Cirl planar polarity in the neighboring cells in trans (Figures 6B and 6C). We conclude that Cirl forms planar polarized patterns in response to Toll-8 polarization. Note that in this experiment, we could not assess Cirl interfacial asymmetry due to the complexity of the genetics required.
Finally, we asked if this transient Toll-8 planar polarity requires Cirl. To test this, we induced nascent Toll-8 overexpression clones in wild-type or cirl mutant wing discs. We found that Toll-8 planar polarity was strongly reduced between cells expressing different levels of Toll-8 in the absence of Cirl (Figures 6D and 6E). Thus, the planar polarities of Toll-8 and Cirl mutually depend upon one another. Since Toll-8 and Cirl form a molecular complex, we propose that Toll-8 and Cirl mutually attract each other in a positive feedback, which results in their self-organized polarity at cell interfaces.
Discussion
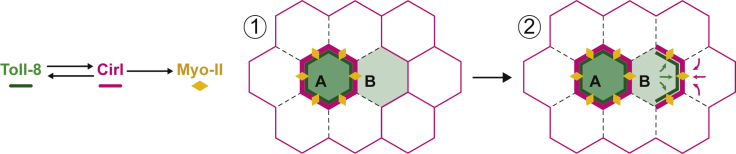
Our work sheds light on how Myo-II planar polarity emerges from the juxtaposition of cells with distinct genetic identities. We report that Toll-8 asymmetric expression leads to Myo-II polarization independent of other Toll receptors. We identified a cell surface protein complex between Toll-8 and the aGPCR Cirl/latrophilin that is required for Myo-II polarization. Though complete removal of Cirl diminishes Toll-8-induced polarization of Myo-II, removing Cirl from either side of a Toll-8 asymmetric interface does not affect Myo-II polarization. This suggests that Toll-8 expression boundary induces Cirl asymmetry and that this asymmetry itself leads to Myo-II polarization. Consistent with this, Cirl asymmetric expression alone polarizes Myo-II. Moreover, we observed that at the Toll-8 boundary, Cirl is differentially localized in trans and in cis thus creating a Cirl interfacial asymmetry. This suggests that Cirl asymmetry could be a potential signal for Myo-II enrichment. We have shown that Toll-8/Cirl co-polarity emerges from quantitative differences in Toll-8 expression between neighboring cells via mutual positive feedback. Thus, we propose a conceptual model wherein Toll-8 and Cirl generate self-organized polarity leading to Myo-II enrichment (Figure 7): the first cell expressing Toll-8 (cell A) recruits Cirl in trans from neighboring cells by depleting it from their orthogonal interfaces (Figure 7, dashed lines), resulting in Cirl planar polarity in these neighboring cells (Figure 7, panel 1). When one of these cells (cell B) initiates expression of Toll-8, Toll-8 levels are lower than in cell A. Due to this quantitative difference in Toll-8 expression and a preexisting Cirl planar polarity in cell B (where Cirl is depleted from orthogonal interfaces in contact with cell A), Toll-8 in cell B is attracted to the remaining interfaces containing Cirl in trans, which are facing away from cell A (Figure 7, panel 2, green arrows and green line in cell B). These new Toll-8-enriched interfaces will stabilize even more Cirl in trans at the expense of orthogonal interfaces in the cells adjacent to cell B that do not express Toll-8, thus propagating Cirl planar polarity one row of cells further away (Figure 7, panel 2, magenta arrows). In summary, we propose that quantitative differences in Toll-8 expression between neighboring cells translate into self-organized Toll-8/Cirl/Myo-II planar polarity due to local interactions between Toll-8 and Cirl.
Figure 7.
Toll-8 and Cirl form dynamic self-organized patterns
Model proposing mutual attraction between Toll-8 and Cirl leading to the self-organization of Toll-8/Cirl/Myo-II planar polarity. Dashed lines represent interfaces depleted of Cirl.
We propose that Toll-8 enriches Myo-II through the asymmetric localization of Cirl in the wing disc. The different localization of Cirl in cis and in trans of Toll-8 overexpressing cells is not due to cis and trans Cirl competing for binding with Toll-8. Instead, Toll-8 and Cirl may bind through different protein domains or require different unknown partners in cis and in trans that are spatially segregated along the apicobasal axis. How asymmetric Cirl enriches Myo-II on both sides of the Toll-8 expression boundary remains to be elucidated. The fact that Myo-II enrichment happens on both sides of the Toll-8 clonal boundaries, even in the absence of Cirl on one side of the boundary, is intriguing. Since Toll-8 is not expressed endogenously in the wing pouch (or expressed at undetectable levels) and given that Toll-8 without its cytoplasmic domain is able to enrich Myo-II, Myo-II polarization on the Toll-8 side is unlikely due to signaling by the Toll-8 cytoplasmic domain. The asymmetry of Cirl extracellular domain may activate other transmembrane proteins present on both sides of the clone boundary, in particular other GPCRs, which then signal bidirectionally to enrich Myo-II. Alternatively, Cirl asymmetry may elicit Myo-II polarity by differential recruitment of Rho1 regulators, akin to Cadherin2 in Ciona (Hashimoto and Munro, 2019), enriching Myo-II through its intracellular domain on one side of the interface and propagating Myo-II enrichment on the other side through mechanical feedback.
What may prevent symmetric Cirl from signaling to enrich Myo-II? Contrary to the aGPCR Flamingo (Usui et al., 1999), Cirl does not require homophilic trans interaction to localize at the membrane (Figure S4D). Interestingly in C. elegans latrophilin extracellular domains form stable dimers (Prömel et al., 2012). Hence, extracellular dimerization of Cirl in trans could be a mechanism inhibiting symmetric Cirl signaling between neighboring cells. Toll-8 expression boundary could dissociate the Cirl dimers by inducing Cirl asymmetric localization, releasing auto-inhibition and thereby eliciting signaling.
We observed in the wing disc that various quantitative and spatial patterns of Toll-8 have different outputs. Groups of cells expressing homogeneously high levels of Toll-8 form strong Myo-II cables around them similar to those present at tissue compartment boundaries. However, low quantitative differences of Toll-8 expression between neighboring cells can lead to planar polarity of Myo-II across several rows of cells. Thus, mechanical boundaries and planar polarized mechanical interfaces might be considered as a continuum, depending on quantitative inputs of cell surface protein asymmetries, such as Toll-8, interpreted by aGPCRs, such as Cirl. The role of GPCRs and G protein signaling in conveying planar polarized input quantitatively is substantiated by the observation that overexpression of heterotrimeric G protein subunits Gβ/Gγ increases Myo-II planar polarization in early Drosophila embryos and converts intercalating cell columns into boundary-like interfaces (Garcia De Las Bayonas et al., 2019).
How do Toll receptors control Cirl activity in the embryo? We were unable to detect Cirl planar polarity in the embryonic ectoderm. Likewise, Toll-8 lacks a detectable planar polarity in this system (Paré et al., 2014). This suggests that in the embryo, Toll-8/Cirl control Myo-II planar polarity without displaying detectable planar polarity. According to evidence in wing discs and our model, the signal for Myo-II polarization is asymmetrical Cirl localization across an interface. One possibility is that in the embryo, though Cirl does not show an axial asymmetry, Cirl could be nonetheless asymmetrically localized along the apicobasal axis between the two sides of vertical interfaces expressing different combinations of Toll receptors (since horizontal interfaces have a similar combination of Toll receptors expressed on each side of the interfaces).
In addition to its instructive role to polarize Myo-II in cooperation with Toll receptors, Cirl may also play a permissive role to activate Myo-II at all junctions in the embryo. Indeed, GPCR signaling is required for both global junctional Rho1/Myo-II activation (independent of planar polarization) (Kerridge et al., 2016), and for their planar polarization per se (Garcia De Las Bayonas et al., 2019). The latter requires inputs from Toll receptors, but not the former. Hence, the effect of perturbing GPCR signaling (Cirl mutant) is expected to be more global than just removing the polarity cue (Toll-2,6,8 RNAi). Moreover, Cirl may function in synergy with other GPCRs, such as Smog (Jha et al., 2018; Kerridge et al., 2016).
It was proposed that heterophilic interactions between Toll receptors were required to recruit Myo-II (Paré et al., 2014). Here, we show that asymmetric expression of a single Toll receptor, Toll-8, enriches Myo-II independent of other Toll receptors via its interaction with the aGPCR Cirl. Though we have not looked for biochemical evidence, our results indicate that Toll-6 polarizes Myo-II and Cirl similar to Toll-8 (Figures S2A and S5A). What could be the function of heterophilic interactions between Toll receptors? They could act to reinforce Myo-II planar polarity, as it was proposed in Paré et al. (2014), or alternatively they might reduce Toll receptors-induced Myo-II polarity by competing with Toll-8/Cirl binding. A detailed analysis of Myo-II levels at each interface in the tissue in every single Toll-receptor-null mutant and in each combination of double mutants would be required to answer this question. However, as Toll-2, 6, and 8 all have distinct expression patterns and boundaries in the embryo (Paré et al., 2014), each boundary of expression of a Toll receptor might polarize Myo-II independently of other Toll receptor, as suggested in Tetley et al. (2016). In this case, the multiplicity of Toll receptors expressed in spatially shifted expression patterns would allow polarization of all cells in the ectoderm.
Recently, Toll receptors were shown to be involved in cell competition (Alpar et al., 2018; Meyer et al., 2014) and growth (Germani et al., 2018). Since there is a known interplay between cell competition and mechanics (Levayer et al., 2015, 2016; Moreno et al., 2019; Vishwakarma and Piddini, 2020; Wagstaff et al., 2016), it is tantalizing to suggest a role for Cirl and Toll-8 in this context.
The vertebrate Cirl homolog latrophilins regulate synaptogenesis and repulsive neuronal migration via coincidental trans-heterophilic binding with FLRTs and teneurins (Boucard et al., 2014; O’Sullivan et al., 2012; Sando et al., 2019; del Toro et al., 2020). These processes both engage the RhoA pathway in vertebrates (Luo, 2002). Given that this pathway is critical for Myo-II activation in Drosophila embryos, it likely mediates Toll-8/Cirl signaling. FLRTs share sequence similarities with Toll-8 (Dolan et al., 2007) and FLRT3 plays an important role in tissue morphogenesis (Karaulanov et al., 2006; Smith and Tickle, 2006; Tomás et al., 2011), which might depend on its interaction with latrophilins. Moreover, in Drosophila the teneurin Ten-m interacts with the LRR protein Tartan to enrich Myo-II at compartment boundaries (Paré et al., 2019), and Cirl might play a role in this system.
The depletion of Cirl around Toll-8 overexpressing cells resembles the effect of Flamingo overexpression on Frizzled localization (Chen et al., 2008; Strutt and Strutt, 2008), both central components of the conserved core PCP pathway (Devenport, 2014). Intriguingly, Flamingo is also an aGPCR and shares sequence homology with Cirl (Langenhan et al., 2013). It is also required for junctional Myo-II activation in the chick neural plate (Nishimura et al., 2012) and during axis extension in Xenopus (Shindo and Wallingford, 2014). The self-organized planar polarization of Toll-8/Cirl identified in our study suggests the possibility that aGPCRs, such as Cirl and Flamingo, are conserved cell surface proteins involved in PCP that evolved different modalities of symmetry breaking. In the core PCP pathway, Flamingo symmetry breaking is thought to be biased by long range mechanical (Aigouy et al., 2010; Aw et al., 2016; Chien et al., 2015) or chemical gradients of adhesion molecules (Fat, Dachsous) or ligands (Wnt) (Aw and Devenport, 2017), which align Flamingo polarity across the tissue. In the Toll-8/Cirl system, Cirl symmetry breaking is potentially triggered by quantitative differences in Toll-8 transcriptional levels between neighboring cells. Directionality of spatial differences in Toll-8 transcriptional levels could define the orientation of Toll-8 and Cirl polarity, resembling the Fat-Ds PCP pathway (Bosveld et al., 2012). Thus, our work illustrates a previously unknown protein complex that generates planar polarized mechanical interfaces instructed by tissue-level cues.
Limitations of the study
One limitation of our study is that it is technically challenging to distinguish Cirl-RFP signals from both sides of an interface in the embryo, since we do not have a way to remove Cirl-RFP signal only from the cis or trans side as performed via mosaic analysis in the wing disc (which cannot be done in embryos). Analyzing the localization of Cirl with such precision in the embryo will require super-resolution microscopy and the generation of photoactivatable tools.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat Monoclonal anti-E-Cad | DHSB | RRID: AB_528120 |
| Mouse Monoclonal anti-β-catenin | DHSB | RRID: AB_528089 |
| Mouse Monoclonal anti-LacZ | DHSB | RRID: AB_2314509 |
| Rat Monoclonal anti-HA | Roche | RRID: AB_390918 |
| Rabbit Polyclonal anti-GFP | Thermo Fisher Scientific | RRID: AB_221569 |
| Rabbit Polyclonal anti-GFP | Abcam | RRID: AB_305564 |
| Rabbit Polyclonal anti-RFP | Rockland | RRID: AB_2209751 |
| Donkey anti-Rabbit Alexa Fluor 488 IgG | Thermo Fisher Scientific | RRID: AB_2535792 |
| Donkey anti-Rabbit Alexa Fluor 568 IgG | Thermo Fisher Scientific | RRID: AB_2534017 |
| Donkey anti-Mouse Alexa Fluor 568 IgG | Thermo Fisher Scientific | RRID: AB_2534013 |
| Donkey anti-Mouse Alexa Fluor 647 IgG | Jackson ImmunoResearch Labs | RRID: AB_2340863 |
| Donkey anti-Rat Alexa Fluor 647 IgG | Jackson ImmunoResearch Labs | RRID: AB_2340694 |
| Critical commercial assays | ||
| Direct-zol RNA MiniPrep | Zymo Research | Cat #R2050 |
| iScript™ Reverse Transcription Supermix | Bio-rad | Cat #1708841 |
| TaqMan Fast Advanced Master Mix | Thermo Fisher Scientific | Cat #4444557 |
| RiboMAX™ Large Scale RNA Production System | Promega | Cat #P1300 |
| GFP-Trap® Agarose | ChromoTek | Cat #gta-20; RRID: AB_2631357 |
| Deposited data | ||
| Proteomics Identifications (PRIDE) | EMBL-EBI | RRID: SCR_003411 |
| Experimental models: Organisms/strains | ||
| D.melanogaster: ind-Toll-8FL::HA | This paper | N/A |
| D.melanogaster: ind-Toll-8ΔLRR::HA | This paper | N/A |
| D.melanogaster: UASp-Toll-8FL::eGFP | This paper | N/A |
| D.melanogaster: UASp-Toll-8ΔLRR::eGFP | This paper | N/A |
| D.melanogaster: UASp-Toll-8ΔCyto::eGFP | This paper | N/A |
| D.melanogaster: UASt-Toll-8::sYFP2 | This paper | N/A |
| D.melanogaster: UASp-Toll-6FL::eGFP | This paper | N/A |
| D.melanogaster: UASp-Toll-6ΔLRR::eGFP | This paper | N/A |
| D.melanogaster: UASp-Toll-6ΔCyto::eGFP | This paper | N/A |
| D.melanogaster: UASt-Toll-6::sYFP2 | This paper | N/A |
| D.melanogaster: UASp-Toll-2FL::eGFP | This paper | N/A |
| D.melanogaster: UASp-Toll-2ΔLRR::eGFP | This paper | N/A |
| D.melanogaster: UASp-Toll-2ΔCyto::eGFP | This paper | N/A |
| D.melanogaster: UASt-Toll-2::sYFP2 | This paper | N/A |
| D.melanogaster: Toll-8::sYFP2BAC | Paré et al., 2014 | N/A |
| D.melanogaster: CirlKO[w-]/CyO | Scholz et al., 2015 | N/A |
| D.melanogaster: CirlKO[w+]/CyO | Scholz et al., 2015 | N/A |
| D.melanogaster: endo-Cirl::RFP | Scholz et al., 2017 | N/A |
| D.melanogaster: sqh-Lifeact::mCherry | Gift from P.F. Lenne | N/A |
| D.melanogaster: endo-E-cad::eGFP | Bloomington Drosophila Stock Center | BL_60584 |
| D.melanogaster: eve::sYFP2BAC | Ludwig et al., 2011 | Flybase: FBal0279504 |
| D.melanogaster: Act5C>STOP>GAL4 | Bloomington Drosophila Stock Center | BL_4780 |
| D.melanogaster: FRT42D | Bloomington Drosophila Stock Center | BL_1802 |
| D.melanogaster: FRT42D arm-LacZ | Bloomington Drosophila Stock Center | BL_7372 |
| D.melanogaster: FRT42D tub-GAL80 | Bloomington Drosophila Stock Center | BL_9917 |
| D.melanogaster: tub-GAL80ts | Bloomington Drosophila Stock Center | BL_7108 and BL_7017 |
| D.melanogaster: P{mata4-GAL-VP16}67 | Gift from E. Wieschaus | Flybase: FBti0016178 |
| D.melanogaster: sqh-Sqh::mCherry | This paper | N/A |
| D.melanogaster: sqh-Sqh::mCherry | Gift from A. Martin | N/A |
| D.melanogaster: sqh-Sqh::eGFP | Gift from R Karess | N/A |
| Oligonucleotides | ||
| Primers for dsRNA against Toll-2: T7 5’-Tag plus Left: 5’-AGTTTGAATCGAAAC GCGAGGC-3' Right: 5’-TGGAAGA TGTGGCTGGCAT-3' |
This paper | N/A |
| Primers for dsRNA against Toll-6: T7 5’-Tag plus Left: 5’-TCGAAAATCAGC CAACGTGC-3' Right: 5’-CGATTCACG GTTTAGCTGCG-3' |
This paper | N/A |
| Primers for dsRNA against Toll-7: T7 5’-Tag plus Left: 5’-TGGCAACCGTCTG GTTACTC-3' Right: 5’- CACGCA GAGCATCATGAACG-3’ |
This paper | N/A |
| Primers for dsRNA against Toll-8: T7 5’-Tag plus Left: 5’-CGTTTGTCGT TCAGCGGATG-3' Right: 5’-ATCGGGGAGGTTATGAGGGG-3' |
This paper | N/A |
| Recombinant DNA | ||
| Plasmid DNA ind-Toll-8FL::HA | This paper | N/A |
| Plasmid DNA ind-Toll-8ΔLRR::HA | This paper | N/A |
| Plasmid DNA UASp-Toll-8FL::eGFP | This paper | N/A |
| Plasmid DNA UASp-Toll-8ΔLRR::eGFP | This paper | N/A |
| Plasmid DNA UASp-Toll-8ΔCyto::eGFP | This paper | N/A |
| Plasmid DNA UASp-Toll-6FL::eGFP | This paper | N/A |
| Plasmid DNA UASp-Toll-6ΔLRR::eGFP | This paper | N/A |
| Plasmid DNA UASp-Toll-6ΔCyto::eGFP | This paper | N/A |
| Plasmid DNA UASp-Toll-2FL::eGFP | This paper | N/A |
| Plasmid DNA UASp-Toll-2ΔLRR::eGFP | This paper | N/A |
| Plasmid DNA UASp-Toll-2ΔCyto::eGFP | This paper | N/A |
| Plasmid DNA UASt-Toll-8::sYFP2 | This paper | N/A |
| Plasmid DNA UASt-Toll-6::sYFP2 | This paper | N/A |
| Plasmid DNA UASt-Toll-2::sYFP2 | This paper | N/A |
| Software and algorithms | ||
| Fiji | Schindelin et al., 2012 | RRID: SCR_002285 |
| Tissue Analyzer | Aigouy et al., 2010 | https://grr.gred-clermont.fr/labmirouse/software/WebPA/ |
| R Project for Statistical Computing | The R Foundation | RRID: SCR_001905 |
| Python Programming Language | Python Software Foundation | RRID:SCR_008394 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Thomas Lecuit (thomas.lecuit@univ-amu.fr).
Materials availability
Plasmids, FASTA sequences and transgenic fly lines generated in this study are all available on request.
Data and code availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PRIDE: PXD017895. Codes used in this study are available upon request.
Experimental model and subject details
The experiments were performed on Drosophila melanogaster embryos and larvae. The adult flies were maintained under the standard lab conditions in plastic vials at 18°C or 25°C with yeast food. Embryo collection was done in fly cages with agar plates made with apple juice, supplemented with yeast paste. Larvae were grown in plastic vials at 18°C or 25°C and collected at the third instar larvae wandering stage. Please refer to the Key resources table for the details of the fly lines being used.
Method details
Fly strains and genetics
All fly constructs are listed in the Key resources table. The following mutant alleles and insertions were used: ind-Toll-8FL::HA (attP40 on 2L), ind-Toll-8ΔLRR::HA (attP40 on 2L), UASp-Toll-8FL::eGFP (attP40 on 2L), UASp-Toll-8ΔLRR::eGFP (attP40 on 2L), UASp-Toll-8ΔCyto::eGFP (attP40 on 2L), UASt-Toll-8::sYFP2 (VK27 (attP9744) on 3R), UASp-Toll-6FL::eGFP (attP40 on 2L), UASp-Toll-6ΔLRR::eGFP (attP40 on 2L), UASp-Toll-6ΔCyto::eGFP (attP40 on 2L), UASt-Toll-6::sYFP2 (VK27 on 3R), UASp-Toll-2FL::eGFP (attP40 on 2L), UASp-Toll-2ΔLRR::eGFP (attP40 on 2L), UASp-Toll-2ΔCyto::eGFP (attP40 on 2L), UASt-Toll-2::sYFP2 (attP40 on 2L), Toll-8::sYFP2BAC (gift from J. Zallen) (Paré et al., 2014), CirlKO[w+], CirlKO[w-] (gifts from T. Langenhan) (Scholz et al., 2015), Cirl::RFP knock-in (Scholz et al., 2017) (Cirl::RFPKIN, gift from T. Langenhan), sqh-Lifeact::mCherry (VK27 on 3R, gift from P.F. Lenne), E-cad::eGFP knock-in (Huang et al., 2009) (E-cad::eGFPKIN), eve::sYFP2BAC (BAC construct, S2E.MSE.eve.YFP, FBal0279504, gift from M. Ludwig) (Ludwig et al., 2011), hs-FLP (Chan et al., 2017), Ubx-FLP (Bloomington BL 42718), Act5C>STOP>GAL4 (Bloomington BL 4780), FRT42D (Bloomington BL 1802), FRT42D arm-LacZ (Bloomington BL 7372), FRT42D tub-GAL80 (Bloomington BL 9917), tub-GAL80ts (Bloomington BL 7108) and tub-GAL80ts (Bloomington BL 7017). 67-Gal4 ({matαtub-GAL4-VP16}67) is a ubiquitous, maternally supplied GAL4 driver (gift from E. Wieschaus). MRLC is encoded by the gene spaghetti squash (sqh, Genebank ID: AY122159). Sqh was visualized using sqh-Sqh::mCherry (VK18 (attP9736) on 2R or VK27 (attP9744) on 3R for experiments in the wing disc or a construct on chromosome 2 from A. Martin (Martin et al., 2009) for live-imaging experiments in the embryo) and sqh-Sqh::eGFP transgenics (gift from R. Karess).
A list of all the experimental genotypes is given in Table S2.
Constructs and transgenesis
Expression vectors drivers
UASp expression vector driver was generated by inserting a PhiC31 attB sequence downstream from the K10 3’-UTR of pUASp. UASt expression vector driver corresponds to pUASTattB which contains a PhiC31 attB sequence inserted downstream from the SV40 3’-UTR (Bischof et al., 2007).
ind is an early horizontal stripe expression vector driver generated by modifying the pbphi-evePr-MS2-yellow (Bothma et al., 2014) (gift from T. Gregor) as follows. First, EVEstr2 enhancer sequence was replaced by the ind1.4 enhancer (Lim et al., 2013) (gift from Gerardo Jiménez). Second, a part of Hsp70Bb 5'-UTR was added after the eve basal promoter. Third, MS2-yellow sequence was replaced by a small polylinker for further cloning.
Expression vectors constructs and transgenics
Toll-8 (Tollo, CG6890), Toll-6 (CG7250) and Toll-2 (18 wheeler, CG8896) whole ORFs were amplified using specific pACMAN genomic plasmids and cloned inside each expression vectors. UASp driven Tolls ORFs were all tagged Cterminally by mEGFP with a GSAGSAAGSGEF flexible aa linker in between.UASp-Toll-8FL::eGFP is the full length Toll-8 (1346aa). UASp-Toll-8ΔCyto::eGFP is a cytoplasmic truncated version of this vector (deletion from aa H1052 to M1346 last aa). In UASp-Toll-8ΔLRR::eGFP, all LRR repeats were removed (deletion from aa E99 to L917). UASp-Toll-6FL::eGFP is the full length Toll-6 (1514aa). UASp-Toll-6ΔCyto::eGFP is a cytoplasmic truncated version of this vector (deletion from aa H1088 to A1514 last aa). In UASp-Toll-6ΔLRR::eGFP, all LRR repeats were removed (deletion from aa A139 to G964). UASp-Toll-2FL::eGFP is the full length Toll-2 (1385aa). UASp-Toll-2ΔCyto::eGFP is a cytoplasmic truncated version of this vector (deletion from aa F1026 to V1385 last aa). In UASp-Toll-2ΔLRR::eGFP, all LRR repeats were removed (deletion from aa F110 to L900). UASt-Toll-2::sYFP2, UASt-Toll-6::sYFP2 and UASt-Toll-8::sYFP2 are Cter sYFP2 tag construct of full length Toll-2,6 and 8 ORFs cloned into UASt using the same GSAGSAAGSGEF flexible aa linker in between. ind-Toll-8FL::HA is a Cter HA tag construct of full length Toll-8 with no linker in between. In ind-Toll-8ΔLRR::HA, all LRR repeats were removed (deletion from aa E99 to L917).
All recombinant expression vectors were built using “In-Fusion cloning” (Takara Bio), verified by sequencing (Genewiz) and sent to BestGene Incorporate for PhiC31 site specific mediated transgenesis into attP40 (2L, 25C7) or VK27 (attP9744, 3R, 89E11).
Antibodies
All antibodies that are used in this study are listed in the Key resources table. The following primary antibodies were used: rat-anti-E-Cad (1:200, DHSB DCAD2 concentrate), mouse-anti-β-catenin (1:400, DHSB N2 7A1 Armadillo concentrate), mouse-anti-LacZ (1:100, DHSB 40-1a concentrate), rat-anti-HA (1:100, Anti-HA High Affinity rat IgG1, Roche ROAHAHA). Sqh::eGFP was detected with rabbit-anti-GFP (1:500, Life Technologies A11122 or 1:1000 Abcam ab6556). Cirl::RFP was detected with rabbit-anti-RFP (1:1000, Rockland 600-401-379). The following secondary antibodies were used: donkey-anti-rabbit Alexa Fluor 488 IgG (Life Technologies A 21206), donkey-anti-rabbit Alexa Fluor 568 IgG (Life Technologies A10042), donkey anti-mouse Alexa Fluor 568 IgG (Life Technologies A10037), donkey-anti-mouse Alexa Fluor 647 IgG (Jackson ImmunoResearch 715 605 151) and donkey-anti-rat Alexa Fluor 647 IgG (Jackson ImmunoResearch 712 605 153). All secondary antibodies were used at 1:500.
Affinity purification mass spectrometry
Protein purification and mass spectrometry
Roughly 600 embryos for each sample were collected from overnight cages kept at 25°C for the following crosses: yw (control) or females ; 67-GAL4/+; UASt-Toll-8::sYFP2/+; x males ; 67-GAL4/+; UASt-Toll-8::sYFP2/+; (Toll-8::YFP maternal and zygotic overexpression), dechorionated with bleach, transferred directly to lysate buffer (10 mM Tris/Cl pH 7.5; 150 mM NaCl; 0.5 mM EDTA; 0.5% NP-40, supplemented with protease and phosphatase inhibitors) and crushed manually on ice over 30 minutes. Lysates were centrifuged to clear debris and protein concentrations of post-centrifugation supernatants were determined. The crude protein yield per lysate sample is usually 1000∼3000 μg. In each experiment, lysates of comparable protein concentration were incubated with pre-rinsed GFP nano-trap agarose resin (Chromotek, gta-20) at 4°C for 90 min, rinsed 3 x and resuspended in 2x Laemmli buffer with DTT. Protein extraction and purification was performed 3 times each for each cross and verified with silver staining. Protein samples were further purified on NuPAGE 4-12% Bis-Tris acrylamide gels (Life Technologies) and treated with in-gel trypsin digestion (Shevchenko et al., 1996) with minor modifications. Peptides were harvested with two extractions, first in 5% formic acid and then in 5% formic acid in 60% acetonitrile. Samples were reconstituted with 0.1% trifluoroacetic acid in 4% acetonitrile and analyzed by liquid chromatography (LC)-tandem mass spectrometry (MS/MS) with an Orbitrap Fusion Lumos Tribrid Mass Spectrometer (Thermo Electron, Bremen, Germany) online with an Ultimate 3000RSLCnano chromatography system (Thermo Fisher Scientific, Sunnyvale, CA). A detailed mass spectrometry protocol is available upon request.
Protein identification and quantification
Relative intensity-based label-free quantification (LFQ) was processed using the MaxLFQ algorithm (Cox et al., 2014) from the freely available MaxQuant computational proteomics platform (Cox and Mann, 2008). Spectra were searched against a Drosophila melanogaster database (UniProt Proteome reference, date 2017.08; 21982 entries). The false discovery rate (FDR) at the peptide and protein levels were set to 1% and determined by searching a reverse database. For protein grouping, all proteins that could not be distinguished based on their identified peptides were assembled into a single entry according to the MaxQuant rules. The statistical analysis was done with Perseus program (version 1.5.1.6) from the MaxQuant environment (www.maxquant.org). Quantifiable proteins were defined as those detected in at least 67% of samples in at least one condition. Protein LFQ normalized intensities were base 2 logarithmized to obtain a normal distribution. Missing values were replaced using data imputation by randomly selecting from a normal distribution centered on the lower edge of the intensity values that simulates signals of low abundant proteins using default parameters (a downshift of 1.8 standard deviation and a width of 0.3 of the original distribution). To determine whether a given detected protein was specifically differential, a two-sample t-test was done using permutation-based false discovery rate (pFDR) with a threshold at 0.1% (5000 permutations). The p-value was adjusted using a scaling factor s0=1 (Table S1). In Figure 3, differential proteins are highlighted by a cut-off for log2|Fold change|>2 and a p-value<0.01. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaíno et al., 2014) partner repository with the dataset identifier PXD017895.
Bright-field live imaging in embryos
Images of wild-type or mutant embryos were collected on an inverted microscope (Zeiss, AxioVision software) equipped with a programmable motorized stage to record different positions over time (Mark&Find module from Zeiss). Images were acquired every 2 min for 60 minutes from post dorsal movement of the posterior midgut primordium (0 min). The extent of elongation was measured by dividing the travel distance of the posterior midgut primordium at 40 min and normalized to the maximum travel distance.
RNA interference in embryos
dsRNA probes
dsRNA probes were made using PCR product containing the sequence of the T7 promoter (TAATACGACTCACTATAGGG) followed by 18-21 nucleotides specific to the gene. The dsRNA probe against Toll-2 (18w, CG8896) is 393-bp long and located in the 5’UTR region (Forward primer: AGTTTGAATCGAAACGCGAGGC; Reverse primer: ATGCCAGCCACATCTTCCA). The dsRNA probe against Toll-6 (CG7250) is 518-bp long and located in the 5’UTR region (Forward primer: TCGAAAATCAGCCAACGTGC; Reverse primer: CGATTCACGGTTTAGCTGCG). The dsRNA probe against Toll-7 (CG8595) is 749-bp long and located in the coding region (Forward primer: TGGCAACCGTCTGGTTACTC; Reverse primer: CGTTCATGATGCTCTGCGTG). The dsRNA probe against Toll-8 (Tollo, CG6890) is 423-bp long and located in the 5’UTR region (Forward primer: CGTTTGTCGTTCAGCGGATG; Reverse primer: CCCCTCATAACCTCCCCGAT) and does not target the ind-Toll-8::HA transgenes. Gel purified PCR products were subsequently used as a template for the in vitro RNA synthesis with T7 polymerase using Ribomax (Promega, P1300). The dsRNA probes were purified using Sure-Clean (Bioline, BIO-37047). Triple dsRNA probes against Toll-2,6,8 and Toll-2,6,7 were prepared and injected at a final concentration of 5 μM each in RNAse-free water.
Embryo injections
Embryos were collected from fresh agar plates in cages kept at 25°C allowed for 30-min egg laying. Embryos were then dechorionated in 50% bleach, rinsed and aligned on cover slips (#1.5) covered with heptane-glue. After a few minutes of desiccation, embryos were covered with Halocarbon 200 oil and injected with dsRNA or RNase-free water. Post-injection embryos were stored at 25°C until live imaging.
RT-qPCR experiment
Toll-2 and Toll-6 knock-down efficiency was estimated by measuring endogenous mRNA level using RT-qPCR by comparing embryos injected or not with triple dsRNA MIX T268. Total RNA extraction from 5 different samples of ∼100 gastrulating yw embryos each injected or not was performed using the Direct-zol RNA miniprep kit (Zymo Research, R2050) plus a on-column DNAse I treatment to remove genomic DNA contamination as recommended. Reverse transcription was performed with the iScript kit (Biorad, 1708841) according to the manufacturer’s protocol. Real-time PCR was done on a CFX96 QPCR detection system (Bio-RAD) using TaqMan Fast Advanced Master Mix with the following TaqMan probes (Applied Biosystems, Life Technologies), following classical TaqMan probes protocol:
Toll-2=18W= Dm01841837_s1 (position 5228 / GenBank NM_057466, amplicon = 152bp)
Toll-6= Dm01822826_s1 (position 5259 / GenBank NM_001259836, amplicon = 109bp)
RPl32: House-keeping gene reference: Dm02151827_g1: (exon 2-3 boundary, 377 / GenBank NM_001144655, amplicon = 72bp)
RPII140: House-keeping gene: Dm02134593_g1 (exon 2-3 boundary, 2347 / GenBank NM_001300394, amplicon = 78bp)
Act42A: House-keeping gene: Dm02362162_g1 (one exon, 1439 / GenBank NM_078901, amplicon = 108bp)
RT-qPCR conditions were as follows: 50° for 2min; 95° for 10min; 40 cycles [95°C for 15 s and 60°C for 1min]. Analyses were performed in duplicate using five independent injected embryos and controls samples. Beforehand, the three housekeeping genes (Rpl32, RPlII40 and Act42A) were compared to verify absence of any variation between samples (not shown). Transcript levels were first normalized to the chosen housekeeping gene RPI32; and then to the control group. ΔΔCq method was used to estimate relative amounts using the Bio-RAD CFX Maestro software. Further statistical analyses were performed by unpaired t-tests by using the qbasePLUS software version 2 (Biogazelle).
Fluorescence live imaging and image processing in embryos
Live embryos analyzed ranged from Stage 7 to early Stage 8 according to egg laying time (3h∼3h30 post egg laying at 25°C) and further morphological criteria (posterior mid-gut primordium displacement <1/4 of egg length). Embryos were aligned on cover slips (#1.5) with heptane-glue and were covered with Halocarbon 200 oil. Dual channel time-lapse imaging was performed on a Nikon Eclipse Ti inverted spinning disc microscope (Roper) with a 100x/1.4 oil-immersion objective at 22°C, controlled by the Metamorph software. Z stacks (step size: 0.5 μm) of 6∼10 slices were acquired every 30 seconds, for 15∼45 minutes starting from stage 6. Laser power was measured and kept constant across all experiments.
To generate 2D projections in experiments with E-Cad::GFP (Figures 1F and S1A), a custom FIJI macro (Bailles et al., 2019) integrating the ‘stack focuser’ plugin from M. Umorin was used to perform maximum intensity projection for all channels with 3 Z planes around the junctional plane (labeled by E-cad::GFP). For Figure 3C, a single plane at the junction level is manually selected based on maximum junctional sqh::GFP signals. The resulting 2D images were subjected to a background subtraction procedure using the rolling ball tool (radius 50 pixels). The 2D images were segmented on E-cad::GFP or LifeAct::Ch channels semi-automatically with manual corrections in the FIJI plug-in Tissue Analyzer (Aigouy et al., 2010). The resulting segmentation masks were then dilated by 5 pixels on either side of the junction and used as masks for subsequent quantifications. Cell tracking and quantifications of T1 transitions are performed semi-automatically with Tissue Analyzer with manual correction.
Immunofluorescence and image processing in embryos
Embryos were fixed with 8% formaldehyde for 20 min at room temperature. Embryos were processed and stained according to standard procedures (Müller, 2008). Embryos were mounted in Aqua-Polymount (Polysciences). Images were acquired on a Leica SP8 inverted confocal microscope with a 63x/1.4 NA oil-immersion objective (with exception of Figure S3A acquired on a Zeiss LSM780 with a 63x/1.4 NA oil-immersion objective). Z stacks with step size of 0.25-0.4 μm were collected.
2D images were generated by maximum intensity projections followed by the same procedure as for live imaging experiments in embryos (except for 3-pixel dilatation in segmentation masks generated from β-catenin stainings).
Clonal analysis in wing discs
Flies were allowed to lay eggs in vials for ∼8h at 25°C and vials were kept at 25°C until heat-shock.
For clonal overexpression of Tolls (Figures 2, 3I, 5A, S2, S4A, S5A, and S5B) 72h AEL (after egg laying) old larvae were heat-shocked at 37° for 10-14 minutes and dissected after 24h.
For GAL80ts experiments (Figures 6 and S6), 72hr AEL larvae were heat-shocked at 37° for 12 minutes, kept at 18° for 48 hours and subsequently incubated at 30°C for 2h15min in order to inactivate GAL80ts and allow expression of Toll-8::YFP.
For MARCM experiments (Mosaic Analysis with a Repressible Cell Marker, Figures 4A, 4B, 5B, 5C, S4B, S4C, S5C, and S5D), 72h AEL larvae were heat-shocked at 37° for 1h, kept at 18° and heat-shocked again 7 hours later at 37° for 1h. Larvae were kept at 18° for 20 hours, shifted to 25° and dissected 24h later. Keeping the larvae at 18° allowed growth of the clones in the presence of no/low levels of Toll-8 expression. Larvae to observe cirl mutant clones (Figure 4D) were treated the same way.
Immunofluorescence and image processing in wing discs
Staged larvae were dissected in PBS, transferred to 4% PFA in PBS and fixed under agitation for 18 min at room temperature. After fixation, wing discs were first rinsed with PBS, then extensively washed with PBT (PBS plus 0.2% Triton-X100) and blocked in PBT with 5% normal donkey serum (NDS, Jackson Immuno Research Laboratories, 017-000-001) for at least 30 min at room temperature, followed by incubation with primary antibody in 2% NDS overnight at 4°C. The next day wing discs were washed in PBT and incubated in secondary antibody with 2% NDS for 1h30min at room temperature. After six rounds of washes with PBT, samples were mounted in Mowiol (Sigma-Aldrich, 324590). Larval mouth hooks were used as spacers in the experiments where Myo-II was observed. Images were acquired on a Leica SP8 inverted confocal microscope with a 63x/1.4 NA oil-immersion objective. Toll-8::YFP and Sqh::Ch were visualized with their endogenous fluorescence. Image stacks with step size of 0.25-0.5 μm were collected.
Peripodial signal was masked from the image stacks in ImageJ to avoid interference with signals from the wing disc proper. 2D projections were generated using the aforementioned custom stack focuser macro in ImageJ, projecting two z planes around the junctional plane of each cell (detected by E-cad staining, except Figures 4A and 4B, projected on Sqh::Ch signals). This allows to project the entire wing pouch independently of the shape of the wing disc. The 2D-projected stacks were then segmented on E-cad stainings (except Figures 4A and 4B segmented on Sqh::Ch signals) using Tissue Analyzer (Aigouy et al., 2010).
Ex vivo live imaging and image processing in wing discs
The culture medium used for long-term time lapse imaging of wing imaginal disc explants is described in Dye et al. (Dye et al., 2017). In short, Grace’s insect medium (Sigma G9771, without sodium bicarbonate) was buffered with 5mM BisTris and the pH adjusted to 6.6-6.7. Subsequently the medium was sterile filtered (0.2μm pore size) and kept at 4°C for up to 4 weeks. At the day of the experiment the medium was supplemented with 5% fetal bovine serum (FBS), Penicillin-Streptomycin (final 1x from a 100x stock solution, Sigma P4333) and warmed to 30°C in a water bath. Just before dissection of the larvae, 20-Hydroxyecdysone (20E, Sigma, H5142) was added to yield a total concentration of 20nM. 20E was kept as a 1000x stock solution in ethanol at -20°C. For the experiment, 72h AEL larvae were heat-shocked at 37° for 12 minutes, kept at 18° for 48 hours and subsequently incubated at 30°C for 2h15min in a water bath. Subsequently, larvae were floated out of the food using 30% glycerol and washed in sterile water twice. Surface sterilization in 70% Ethanol was followed by another wash in sterile water and then in medium. Larvae were dissected in culture medium, wing discs isolated and mounted on a round cover slip. In order to restrict disc movement during imaging, discs were covered by a porous membrane (Whatman cyclopore polycarbonate membranes; Sigma, WHA70602513) using two stripes of double-sided tape as spacers. Finally, this sandwich was mounted in an Attofluor cell chamber (A7816, Invitrogen) and filled with 1ml of medium and covered with Parafilm M (P7793, Sigma-Aldrich) to avoid evaporation. Discs were imaged on a Nikon Eclipse Ti inverted spinning disc microscope (Roper) equipped with an incubation chamber heated to 30°C. Imaging was done using a 60x/1.2 NA water-immersion objective (Figures 6A and S6; Videos S2 and S3) or a 100x/1.4 NA oil-immersion objective (Video S4). Dual imaging of Toll-8::YFP and Sqh::Ch was performed by simultaneous excitation of fluorophores with 515nm and 561nm laser lines using a dichroic mirror to image on two cameras (Evolve 512, Photometrics). Stacks of 40 slices with 0.7μm spacing were acquired every 10min (60x movies) or every 5min (100x movies).
A maximum projection of the disc proper junctional plane was obtained by masking the peripodial epithelium and the lateral portion of the disc proper manually in ImageJ based on sqh::Ch signals. Background subtraction was done using a rolling ball (50px radius) in ImageJ.
Data analysis
Definition of expression interfaces
When references channels (Toll-8::HA, Toll-8::YFP, Toll-8::GFP, or LacZ staining) were available, expression interfaces were defined from reference channels in Tissue Analyzer.
To define horizontal cell rows in Figure 1F, cell rows were counted from the ventral midline, with the 4th cell row (most ventral) being 2 rows away from the ventral midline. The boundary between the 2nd and the 3rd cell rows is consistent with the position of ind ventral expression boundary.
To define vertical cell columns in Figures S1A and S1B, parasegment boundaries were visualized with Eve::YFP, with the anterior boundary of Eve::YFP signal defined as the parasegment boundary between even- and odd-numbered parasegments. Thus, cell columns 1-4 belong to odd-numbered parasegments (Eve::YFP+), while 5-8 belong to even-numbered parasegments (Eve::YFP-).
Quantification of junctional Myo-II intensities
Raw pixel intensities from segmented junctions were measured in Tissue Analyzer. To extract data tables containing raw pixel intensities from Tissue Analyzer, a customized R procedure was developed using the RSQLite package. Adjusted junctional pixel intensities were obtained by subtracting mean cytoplasmic intensity value measured on each image. Enrichment was calculated as ratios of adjusted junctional intensity values between junctions of interest and those in nearby wild-type tissues (Figures 1A and 2A).
Quantification of boundary smoothness
Boundary smoothness for ventral ind expression boundary in the embryo was calculated as the ratio between distance between two end vertices over total junctional length (Figure 1A). Boundary smoothness value approaches 1 as the boundary gets smoother.
For clone smoothness in the wing disc, an original method developed by P. Villoutreix (Centuri, France) was implemented in Tissue Analyzer by B. Aigouy (IBDM, France) under the plugin ‘Clone wiggliness’. In brief, the boundary of the clone was extracted, the vertices present at the clone boundary were ordered, and an angle was calculated for each vertex with its two neighboring vertices present at the clone boundary (Figure 2A). A mean value per clone was then calculated and this value is getting closer to 180° if the clone is smooth.
Quantification of apical-basal protein localization (Figure 5)
Protein localization and concentration along the apical-basal axis was quantified as described in Harmansa et al. (Harmansa et al., 2017) from high-z-resolution image stacks (0.35μm slice spacing, acquired on a Leica SP8 confocal microscope using a 63x/1.4 NA oil-immersion objective) of wing discs expressing Toll-8::YFP, Cirl::RFP and stained for the junctional marker E-Cadherin (E-cad). Optical cross-sections were obtained by using the ‘reslice’ option in Fiji software (ImageJ; National Institute of Health) and subsequently background was subtracted (‘rolling ball’ radius of 50px). From these cross sections, junctional fluorescence intensity profiles of E-cad, Toll-8::YFP and Cirl::RFP were extracted along a line of 0.6μm width (corresponding to approximately the width of a junction) using the ‘plot profile’ function in Fiji. Average profiles from different junctions/discs were computed by using the peak of the E-cad profile to align individual profiles. Average profiles were calculated in Excel software (Microsoft) and plotted in Python software using the Seaborn library (line plot function, error bands show the 95% confidence interval computed by bootstrapping). In the plots the junctional plane is visualized by a blue band that is defined as the 0.5μm above and below the average E-cad peak.
Quantification of Toll-8 and Cirl planar polarity (Figures 6B–6E)
We restricted our analysis to single cells expressing low Toll-8::YFP levels in the vicinity of a high Toll-8::YFP expressing cell. This ensured that each junction included in our computation only contained Toll-8::YFP originating from the single cell that was quantified. For each single cell, junctional Toll-8::YFP levels were extracted (using the segmented line tool in ImageJ, line width = 6px) along the parallel junctions (Toll-8‖, the junctions not being in contact with the high Toll-8 expressing cell) and along the two junctions being in direct contact with high Toll-8 expressing cell (Toll-8⊥). Toll-8::YFP enrichment at parallel junctions was computed by calculating the ratio between mean Toll-8‖ and mean Toll-8⊥ levels (Figure 6E, scheme). Cirl planar polarity has been computed in an analogous way (Figure 6C, scheme). To test for a correlation between Cirl and Toll-8 planar polarities, polarity values for each doublet of cell was plotted and linear fitting was performed using Python (Seaborn library for plotting and the Stats library to compute the least square regression and p-value).
Data visualization
With the exception of Figures 5, 6C, S5, and S6, data visualization was performed in R with customized scripts. The following custom packages were used: “fields” and “ggplot2”. Plots shown in Figures 5, 6C, S5, and S6 were plotted in Python using the Seaborn library.
Quantification and statistical analysis
All measurements were performed in 8-100 independent experiments. Each embryo and clone in the wing disc is considered as an independent experiment. Box-plot elements are defined as follows: center line represents the median; box limits represent the first and third quartiles (the 25th and 75th percentiles); upper whisker extends from the box limit to the largest value no further than 1.5x interquartile range from the box limit and lower whisker extends from the box limit to the smallest value at most 1.5x interquartile range from the box limit; all data points are plotted on the graph. The exact values of n for each graph are reported in the figure legends or in the figure. p-values were calculated using the Mann-Whitney U test in R, except from Figure 5A″(two-sided t-test with unequal variance) and Figure 6C (two-sided t-test performed using the Python Stats library). In all figures, n.s.: p > 0.05. No statistical method was used to predetermine sample size. The experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment.
Acknowledgments
We thank all members of the Lecuit team, B. Aigouy (IBDM, France) for stimulating discussions during the course of this project, the IBDM imaging facility for microscopy assistance, FlyBase for maintaining curated databases and the Bloomington Stock Center for providing fly stocks. We thank T. Langenhan and N. Scholz (Leipzig, Germany) for sharing information about Cirl and fly reagents, and for stimulating discussions. We thank B. Aigouy (IBDM, France) and P. Villoutreix (Centuri, France) for developing the method for quantifying clone smoothness in wing discs. We thank B. Habermann (IBDM, France) for her valuable guidance on performing pairwise alignment between Toll-8 and FLRTs. We are grateful to M. Ludwig (Birmingham, UK), P.F. Lenne (IBDM, France), E. Wieschaus (Princeton, USA), A. Martin (MIT, USA), R. Karess (IJM, France), G. Jiménez (IBMB, Spain) and T. Gregor (Pasteur Institute, France) for the gift of flies and vectors. This work was supported by grants from the European Research Council, European Union (Biomecamorph no. 323027) and the Ligue Nationale Contre le Cancer, France (Equipe Labellisée, 2018). J.L. was supported by the Fondation Bettencourt Schueller, France and the Collège de France. S.H. was supported by an EMBO Long-Term Fellowship (EMBO ALTF 217-2017) and by a Centuri Postdoctoral Fellowship. The Marseille proteomics (IBiSA) is supported by Institut Paoli-Calmettes (IPC) and Canceropôle PACA. Proteomic analyses were done using the mass spectrometry facility of Marseille proteomics (marseille-proteomique.univ-amu.fr) supported by IBISA (Infrastructures Biologie Santé et Agronomie), Plateforme Technologique Aix-Marseille, the Cancéropôle PACA, the Provence-Alpes-Côte d'Azur Région, the Institut Paoli-Calmettes and the Centre de Recherche en Cancérologie de Marseille. We acknowledge the France-BioImaging infrastructure supported by the French National Research Agency (ANR–10–INBS-04-01, Investments for the future).
Author contributions
J.L., Q.M., and T.L. conceived the project. Q.M., J.L., and S.K. performed experiments in embryos, Q.M. analyzed them. J.L. performed all the experiments and data analysis in fixed wing discs. S.H. performed and analyzed live experiments in wing discs and quantified data in Figures 5 and 6. Q.M. and A.L. prepared the samples for the mass spectrometry, which was done and analyzed by S.A. and L.C. J.-M.P. designed and generated the molecular constructs. J.L., Q.M., and T.L. wrote the manuscript, and all authors made comments.
Declaration of interests
The authors declare no competing interests.
Published: April 30, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.devcel.2021.03.030.
Contributor Information
Qiyan Mao, Email: qiyan.mao@univ-amu.fr.
Thomas Lecuit, Email: thomas.lecuit@univ-amu.fr.
Supplemental information
The proteomes of embryos overexpressing Toll-8–YFP maternally and zygotically (N = 3 protein isolations: T3, T4, T5, each with 3 replicas: T3a, T3b, T3c, …) were compared with yw embryos (N = 3 protein isolations: C3, C4, C5, each with 3 replicas: C3a, C3b, C3c, …) with affinity purification mass spectrometry (PRIDE dataset identifier PXD017895). Protein list is displayed by order of Difference log2(T/C). Rows highlighted in green denote significant hits from a two-sample t-test using permutation-based false discovery rate (pFDR) controlled at 0.1% (5000 permutations), related to Figure 3.
References
- Aigouy B., Farhadifar R., Staple D.B., Sagner A., Röper J.C., Jülicher F., Eaton S. Cell flow reorients the axis of planar polarity in the wing epithelium of drosophila. Cell. 2010;142:773–786. doi: 10.1016/j.cell.2010.07.042. [DOI] [PubMed] [Google Scholar]
- Alpar L., Bergantiños C., Johnston L.A. Spatially restricted regulation of Spätzle/toll signaling during cell competition. Dev. Cell. 2018;46:706–719.e5. doi: 10.1016/j.devcel.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthoney N., Foldi I., Hidalgo A. Toll and toll-like receptor signalling in development. Development. 2018;145:1–6. doi: 10.1242/dev.156018. [DOI] [PubMed] [Google Scholar]
- Aw W.Y., Devenport D. Planar cell polarity: global inputs establishing cellular asymmetry. Curr. Opin. Cell Biol. 2017;44:110–116. doi: 10.1016/j.ceb.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aw W.Y., Heck B.W., Joyce B., Devenport D. Transient tissue-scale deformation coordinates alignment of planar cell polarity junctions in the mammalian skin. Curr. Biol. 2016;26:2090–2100. doi: 10.1016/j.cub.2016.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailles A., Collinet C., Philippe J.M., Lenne P.F., Munro E., Lecuit T. Genetic induction and mechanochemical propagation of a morphogenetic wave. Nature. 2019;572:467–473. doi: 10.1038/s41586-019-1492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton M.A., Pechmann M., Frey N., Stappert D., Conrads K.H., Chen Y.T., Stamataki E., Pavlopoulos A., Roth S. Toll genes have an ancestral role in axis elongation. Curr. Biol. 2016;26:1609–1615. doi: 10.1016/j.cub.2016.04.055. [DOI] [PubMed] [Google Scholar]
- Bertet C., Sulak L., Lecuit T. Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature. 2004;429:667–671. doi: 10.1038/nature02590. [DOI] [PubMed] [Google Scholar]
- Bielmeier C., Alt S., Weichselberger V., La Fortezza M., Harz H., Jülicher F., Salbreux G., Classen A.K. Interface contractility between differently fated cells drives cell elimination and cyst formation. Curr. Biol. 2016;26:563–574. doi: 10.1016/j.cub.2015.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R.K., Hediger M., Karch F., Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship J.T., Backovic S.T., Sanny J.S.S.P., Weitz O., Zallen J.A. Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev. Cell. 2006;11:459–470. doi: 10.1016/j.devcel.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Bosveld F., Bonnet I., Guirao B., Tlili S., Wang Z., Petitalot A., Marchand R., Bardet P.L., Marcq P., Graner F., Bellaïche Y. Mechanical control of morphogenesis by fat/dachsous/four-jointed planar cell polarity pathway. Science. 2012;336:724–727. doi: 10.1126/science.1221071. [DOI] [PubMed] [Google Scholar]
- Bothma J.P., Garcia H.G., Esposito E., Schlissel G., Gregor T., Levine M. Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos. Proc. Natl. Acad. Sci. USA. 2014;111:10598–10603. doi: 10.1073/pnas.1410022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucard A.A., Maxeiner S., Südhof T.C. Latrophilins function as heterophilic cell-adhesion molecules by binding to teneurins: regulation by alternative splicing. J. Biol. Chem. 2014;289:387–402. doi: 10.1074/jbc.M113.504779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E.H.Y., Chavadimane Shivakumar P.C., Clément R., Laugier E., Lenne P.F. Patterned cortical tension mediated by N-cadherin controls cell geometric order in the drosophila eye. eLife. 2017;6:1–27. doi: 10.7554/eLife.22796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.S., Antic D., Matis M., Logan C.Y., Povelones M., Anderson G.A., Nusse R., Axelrod J.D. Asymmetric homotypic interactions of the atypical cadherin flamingo mediate intercellular polarity signaling. Cell. 2008;133:1093–1105. doi: 10.1016/j.cell.2008.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien Y.H., Keller R., Kintner C., Shook D.R. Mechanical strain determines the axis of planar polarity in ciliated epithelia. Curr. Biol. 2015;25:2774–2784. doi: 10.1016/j.cub.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Dahmann C., Oates A.C., Brand M. Boundary formation and maintenance in tissue development. Nat. Rev. Genet. 2011;12:43–55. doi: 10.1038/nrg2902. [DOI] [PubMed] [Google Scholar]
- del Toro D., Carrasquero-Ordaz M.A., Chu A., Ruff T., Shahin M., Jackson V.A., Chavent M., Berbeira-Santana M., Seyit-Bremer G., Brignani S. Structural basis of teneurin-latrophilin interaction in repulsive guidance of migrating neurons. Cell. 2020;180:323–339.e19. doi: 10.1016/j.cell.2019.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devenport D. The cell biology of planar cell polarity. J. Cell Biol. 2014;207:171–179. doi: 10.1083/jcb.201408039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan J., Walshe K., Alsbury S., Hokamp K., O’Keeffe S., Okafuji T., Miller S.F., Tear G., Mitchell K.J. The extracellular leucine-Rich Repeat superfamily; a comparative survey and analysis of evolutionary relationships and expression patterns. BMC Genomics. 2007;8:320. doi: 10.1186/1471-2164-8-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye N.A., Popović M., Spannl S., Etournay R., Kainmüller D., Ghosh S., Myers E.W., Jülicher F., Eaton S. Cell dynamics underlying oriented growth of the drosophila wing imaginal disc. Development. 2017;144:4406–4421. doi: 10.1242/dev.155069. [DOI] [PubMed] [Google Scholar]
- Fagotto F., Rohani N., Touret A.S., Li R. A molecular base for cell sorting at embryonic boundaries: contact inhibition of cadherin adhesion by ephrin/Eph-dependent contractility. Dev. Cell. 2013;27:72–87. doi: 10.1016/j.devcel.2013.09.004. [DOI] [PubMed] [Google Scholar]
- Garcia De Las Bayonas A., Philippe J.M., Lellouch A.C., Lecuit T. Distinct RhoGEFs activate apical and junctional contractility under control of G proteins during epithelial morphogenesis. Curr. Biol. 2019;29:3370–3385.e7. doi: 10.1016/j.cub.2019.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germani F., Hain D., Sternlicht D., Moreno E., Basler K. The toll pathway inhibits tissue growth and regulates cell fitness in an infection-dependent manner. eLife. 2018;7:1–10. doi: 10.7554/eLife.39939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmansa S., Alborelli I., Bieli D., Caussinus E., Affolter M. A nanobody-based toolset to investigate the role of protein localization and dispersal in Drosophila. eLife. 2017;6:1–22. doi: 10.7554/eLife.22549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H., Munro E. Differential expression of a classic cadherin directs tissue-level contractile asymmetry during neural tube closure. Dev. Cell. 2019;51:158–172.e4. doi: 10.1016/j.devcel.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Zhou W., Dong W., Watson A.M., Hong Y. From the cover: directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc. Natl. Acad. Sci. USA. 2009;106:8284–8289. doi: 10.1073/pnas.0900641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine K.D., Wieschaus E. Cell intercalation during Drosophila germband extension and its regulation by pair-rule segmentation genes. Development. 1994;120:827–841. doi: 10.1242/dev.120.4.827. [DOI] [PubMed] [Google Scholar]
- Jha A., van Zanten T.S., Philippe J.M., Mayor S., Lecuit T. Quantitative control of GPCR organization and signaling by endocytosis in epithelial morphogenesis. Curr. Biol. 2018;28:1570–1584.e6. doi: 10.1016/j.cub.2018.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaulanov E.E., Böttcher R.T., Niehrs C. A role for fibronectin-leucine-rich transmembrane cell-surface proteins in homotypic cell adhesion. EMBO Rep. 2006;7:283–290. doi: 10.1038/sj.embor.7400614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerridge S., Munjal A., Philippe J.M., Jha A., De Las Bayonas A.G., Saurin A.J., Lecuit T. Modular activation of Rho1 by GPCR signalling imparts polarized myosin II activation during morphogenesis. Nat. Cell Biol. 2016;18:261–270. doi: 10.1038/ncb3302. [DOI] [PubMed] [Google Scholar]
- Langenhan T., Aust G., Hamann J. Sticky signaling--adhesion class G protein-coupled receptors take the stage. Sci. Signal. 2013;6:re3. doi: 10.1126/scisignal.2003825. [DOI] [PubMed] [Google Scholar]
- LeGoff L., Rouault H., Lecuit T. A global pattern of mechanical stress polarizes cell divisions and cell shape in the growing Drosophila wing disc. Development. 2013;140:4051–4059. doi: 10.1242/dev.090878. [DOI] [PubMed] [Google Scholar]
- Levayer R., Dupont C., Moreno E. Tissue crowding induces Caspase-dependent competition for space. Curr. Biol. 2016;26:670–677. doi: 10.1016/j.cub.2015.12.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levayer R., Hauert B., Moreno E. Cell mixing induced by myc is required for competitive tissue invasion and destruction. Nature. 2015;524:476–480. doi: 10.1038/nature14684. [DOI] [PubMed] [Google Scholar]
- Li J., Xie Y., Cornelius S., Jiang X., Sando R., Kordon S.P., Pan M., Leon K., Südhof T.C., Zhao M., Araç D. Alternative splicing controls teneurin-latrophilin interaction and synapse specificity by a shape-shifting mechanism. Nat. Commun. 2020;11:2140. doi: 10.1038/s41467-020-16029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B., Samper N., Lu H., Rushlow C., Jiménez G., Shvartsman S.Y. Kinetics of gene derepression by ERK signaling. Proc. Natl. Acad. Sci. USA. 2013;110:10330–10335. doi: 10.1073/pnas.1303635110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig M.Z., Manu, Kittler R., White K.P., Kreitman M. Consequences of eukaryotic enhancer architecture for gene expression dynamics, development, and fitness. PLOS Genet. 2011;7 doi: 10.1371/journal.pgen.1002364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L. Actin cytoskeleton regulation in neuronal morphogenesis and structural plasticity. Annu. Rev. Cell Dev. Biol. 2002;18:601–635. doi: 10.1146/annurev.cellbio.18.031802.150501. [DOI] [PubMed] [Google Scholar]
- Martin A.C., Kaschube M., Wieschaus E.F. Pulsed contractions of an actin-myosin network drive apical constriction. Nature. 2009;457:495–499. doi: 10.1038/nature07522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S.N., Amoyel M., Bergantiños C., de la Cova C., Schertel C., Basler K., Johnston L.A. An ancient defense system eliminates unfit cells from developing tissues during cell competition. Science. 2014;346:1258236. doi: 10.1126/science.1258236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milán M., Weihe U., Pérez L., Cohen S.M. The LRR proteins Capricious and Tartan mediate cell interactions during DV boundary formation in the Drosophila wing. Cell. 2001;106:785–794. doi: 10.1016/s0092-8674(01)00489-5. [DOI] [PubMed] [Google Scholar]
- Moreno E., Valon L., Levillayer F., Levayer R. Competition for space induces cell elimination through compaction-driven ERK downregulation. Curr. Biol. 2019;29:23–34.e8. doi: 10.1016/j.cub.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H.-A.J. Immunolabeling of embryos. Methods Mol Biol. 2008;420:207–218. doi: 10.1007/978-1-59745-583-1_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T., Honda H., Takeichi M. Planar cell polarity links axes of spatial dynamics in neural-tube closure. Cell. 2012;149:1084–1097. doi: 10.1016/j.cell.2012.04.021. [DOI] [PubMed] [Google Scholar]
- O’Sullivan M.L., de Wit J., Savas J.N., Comoletti D., Otto-Hitt S., Yates J.R., Ghosh A. FLRT proteins are endogenous latrophilin ligands and regulate excitatory synapse development. Neuron. 2012;73:903–910. doi: 10.1016/j.neuron.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paré A.C., Naik P., Shi J., Mirman Z., Palmquist K.H., Zallen J.A. An LRR receptor-teneurin system directs planar polarity at compartment boundaries. Dev. Cell. 2019;51:208–221.e6. doi: 10.1016/j.devcel.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paré A.C., Vichas A., Fincher C.T., Mirman Z., Farrell D.L., Mainieri A., Zallen J.A. A positional Toll receptor code directs convergent extension in Drosophila. Nature. 2014;515:523–527. doi: 10.1038/nature13953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prömel S., Frickenhaus M., Hughes S., Mestek L., Staunton D., Woollard A., Vakonakis I., Schöneberg T., Schnabel R., Russ A.P., Langenhan T. The GPS motif is a molecular switch for bimodal activities of adhesion class G protein-coupled receptors. Cell Rep. 2012;2:321–331. doi: 10.1016/j.celrep.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sando R., Jiang X., Südhof T.C. Latrophilin GPCRs direct synapse specificity by coincident binding of FLRTs and teneurins. Science. 2019;363 doi: 10.1126/science.aav7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz N., Gehring J., Guan C., Ljaschenko D., Fischer R., Lakshmanan V., Kittel R.J., Langenhan T. The adhesion GPCR latrophilin/CIRL shapes mechanosensation. Cell Rep. 2015;11:866–874. doi: 10.1016/j.celrep.2015.04.008. [DOI] [PubMed] [Google Scholar]
- Scholz N., Guan C., Nieberler M., Grotemeyer A., Maiellaro I., Gao S., Beck S., Pawlak M., Sauer M., Asan E. Mechano-dependent signaling by latrophilin/CIRL quenches cAMP in proprioceptive neurons. eLife. 2017;6 doi: 10.7554/eLife.28360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöneberg T., Prömel S. Latrophilins and teneurins in invertebrates: no love for each other? Front. Neurosci. 2019;13:1–11. doi: 10.3389/fnins.2019.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Shindo A., Wallingford J.B. PCP and septins compartmentalize cortical actomyosin to direct collective cell movement. Science. 2014;343:649–652. doi: 10.1126/science.1243126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.G., Tickle C. The expression of Flrt3 during chick limb development. Int. J. Dev. Biol. 2006;50:701–704. doi: 10.1387/ijdb.062192ts. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A., Levine M. Localized repressors delineate the neurogenic ectoderm in the early Drosophila embryo. Dev. Biol. 2005;280:482–493. doi: 10.1016/j.ydbio.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Strutt H., Strutt D. Differential stability of flamingo protein complexes underlies the establishment of planar polarity. Curr. Biol. 2008;18:1555–1564. doi: 10.1016/j.cub.2008.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetley R.J., Blanchard G.B., Fletcher A.G., Adams R.J., Sanson B. Unipolar distributions of junctional myosin II identify cell stripe boundaries that drive cell intercalation throughout Drosophila axis extension. eLife. 2016;5:956–963. doi: 10.7554/eLife.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomás A.R., Certal A.C., Rodríguez-León J. FLRT3 as a key player on chick limb development. Dev. Biol. 2011;355:324–333. doi: 10.1016/j.ydbio.2011.04.031. [DOI] [PubMed] [Google Scholar]
- Usui T., Shima Y., Shimada Y., Hirano S., Burgess R.W., Schwarz T.L., Takeichi M., Uemura T. Flamingo, a seven-pass transmembrane cadherin, regulates planar cell polarity under the control of Frizzled. Cell. 1999;98:585–595. doi: 10.1016/s0092-8674(00)80046-x. [DOI] [PubMed] [Google Scholar]
- Vishwakarma M., Piddini E. Outcompeting cancer. Nat. Rev. Cancer. 2020;20:187–198. doi: 10.1038/s41568-019-0231-8. [DOI] [PubMed] [Google Scholar]
- Vizcaíno J.A., Deutsch E.W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J.A., Sun Z., Farrah T., Bandeira N. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff L., Goschorska M., Kozyrska K., Duclos G., Kucinski I., Chessel A., Hampton-O’Neil L., Bradshaw C.R., Allen G.E., Rawlins E.L. Mechanical cell competition kills cells via induction of lethal p53 levels. Nat. Commun. 2016;7:11373. doi: 10.1038/ncomms11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y., Nishida Y., Ip Y.T. Functional analysis of Toll-related genes in Drosophila. Dev. Growth Differ. 2010;52:771–783. doi: 10.1111/j.1440-169X.2010.01213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen J.A., Wieschaus E. Patterned gene expression directs bipolar planar polarity in Drosophila. Dev. Cell. 2004;6:343–355. doi: 10.1016/s1534-5807(04)00060-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Frames are re-aligned to keep the center of tracked cell group constant. Framer rate: 30 s/frame. Duration: 20 min. Scale bar: 5 μm, related to Figures 3 and S3.
Asterisks indicate a cell that dynamically upregulates expression of Toll-8–YFP during ∼50min. Myo-II is being enriched at junctions that show Toll-8–YFP planar polarity (arrow) and this enrichment is lost once Toll-8–YFP expression reaches similar levels between contacting cells (arrowheads). Scale bar: 5 μm, related to Figures 6 and S6.
Scale bar: 5 μm, related to Figures 6 and S6.
Scale bar: 5 μm, related to Figures 6 and S6.
The proteomes of embryos overexpressing Toll-8–YFP maternally and zygotically (N = 3 protein isolations: T3, T4, T5, each with 3 replicas: T3a, T3b, T3c, …) were compared with yw embryos (N = 3 protein isolations: C3, C4, C5, each with 3 replicas: C3a, C3b, C3c, …) with affinity purification mass spectrometry (PRIDE dataset identifier PXD017895). Protein list is displayed by order of Difference log2(T/C). Rows highlighted in green denote significant hits from a two-sample t-test using permutation-based false discovery rate (pFDR) controlled at 0.1% (5000 permutations), related to Figure 3.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PRIDE: PXD017895. Codes used in this study are available upon request.