Abstract
CRISPR-Cas systems are found widely in prokaryotes where they provide adaptive immunity against virus infection and plasmid transformation. We describe a minimal functional CRISPR-Cas system, comprising a single ~70 kilodalton protein, CasΦ, and a CRISPR array, encoded exclusively in the genomes of huge bacteriophages. CasΦ employs a single active site for both CRISPR RNA (crRNA) processing and crRNA-guided DNA cutting to target foreign nucleic acids. This hypercompact system is active in vitro and in human and plant cells with expanded target recognition capabilities relative to other CRISPR-Cas proteins. Useful for genome editing and DNA detection but with a molecular weight half that of Cas9 and Cas12a genome-editing enzymes, CasΦ offers advantages for cellular delivery that expand the genome editing toolbox.
One Sentence Summary:
Phage-derived CasΦ uses a single active site to process guide RNA and cut DNA for genome editing and nucleic acid detection.
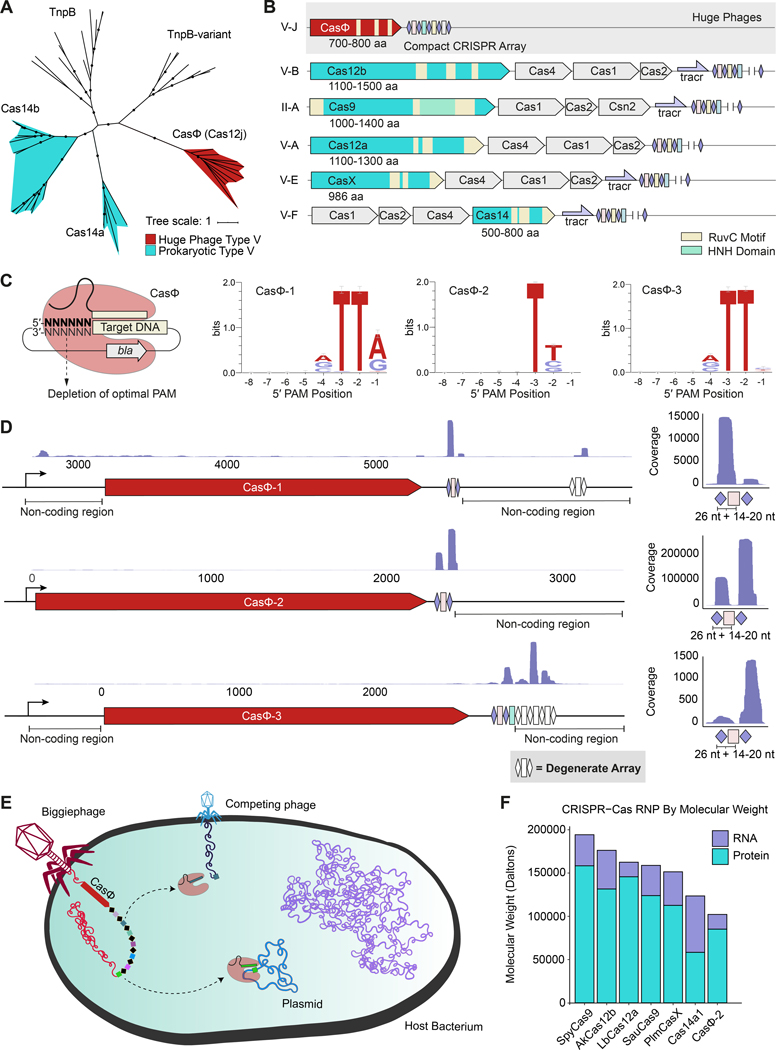
Competition between viruses and their host microbes fostered the evolution of CRISPR-Cas systems that employ nucleases and non-coding CRISPR RNAs (crRNAs) to target foreign nucleic acids by complementary base pairing (1). Processing of CRISPR array transcripts, consisting of repeats and spacer sequences acquired from viruses or other mobile genetic elements (MGEs) (2), generates mature crRNAs that guide Cas proteins (3) to detect and destroy previously encountered viruses. Although found almost exclusively in microbial genomes, the recent discovery of ubiquitous huge bacteriophages (viruses of bacteria) revealed the surprising prevalence of CRISPR-Cas systems encoded in their genomes (4). These systems notably lack CRISPR spacer acquisition machinery (Cas1, Cas2 and Cas4 proteins) and generally harbor compact CRISPR arrays (median: 5 spacers per array), some of which target the genes of competing phages or phage hosts. CasΦ (Cas12j) is a family of Cas proteins encoded in the Biggiephage clade (4). CasΦ contains a C-terminal RuvC domain with remote homology to that of the TnpB nuclease superfamily from which type V CRISPR-Cas proteins are thought to have evolved (4, 5) (fig. S1). However, CasΦ shares <7% amino acid identity with other type V CRISPR-Cas proteins and is most closely related to a TnpB group distinct from miniature type V (Cas14) proteins (Fig. 1A).
Fig. 1.
CasΦ is a bona fide CRISPR-Cas system from huge phages. (A) Maximum Likelihood phylogenetic tree of type V effector proteins and respective predicted ancestral TnpB nucleases. Bootstrap and approximate likelihood-ratio test values ≥ 90 are denoted on the branches with black circles. (B) Illustrations of genomic CRISPR-Cas loci of CasΦ, Cas14, and systems previously employed in genome editing applications. (C) Graphical representation of the PAM depletion assay and the resulting PAMs for three CasΦ orthologs. (D) RNA-sequencing results (left) mapped onto the native genomic loci of CasΦ orthologs and their upstream and downstream non-coding regions as cloned with reduced CRISPR-arrays into expression plasmids. Enlarged view of RNA mapped onto the first repeat-spacer pair (right). (E) Schematic of the hypothesized function of Biggiephage-encoded CasΦ in an instance of superinfection of its host. CasΦ may be used by the huge phage to eliminate competing mobile genetic elements. (F) Predicted molecular weights of the ribonucleoprotein (RNP) complexes of small CRISPR-Cas effectors and those functional in editing of mammalian cells.
CasΦ’s unusually small size of ~70–80 kDa, about half the size of the Cas9 and Cas12a (Fig. 1B), and its lack of co-occurring genes raised the question of whether CasΦ functions as a bona fide CRISPR-Cas system. We investigated three divergent CasΦ orthologs from metagenomic assemblies (fig. S2), hereafter referred to as CasΦ−1, CasΦ−2 and CasΦ−3. To examine CasΦ’s ability to recognize and target DNA in bacterial cells, we tested whether CasΦ could protect Escherichia coli from plasmid transformation. CRISPR–Cas systems target DNA sequences following or preceding a 2–5 base pair (bp) Protospacer Adjacent Motif (PAM) for self-versus-non-self discrimination (6). To determine whether CasΦ uses a PAM, we transformed a library of plasmids containing randomized regions adjacent to crRNA-complementary target sites, thereby depleting plasmids harboring functional PAMs. This revealed the crRNA-guided double-strand DNA (dsDNA) targeting capability of CasΦ and minimal T-rich PAM sequences, including 5′-TBN-3′ PAMs (where B is G, T, or C) depleted for CasΦ−2 (Fig. 1C).
We next used the E. coli expression system and plasmid interference assay to determine the components required for CRISPR-CasΦ system function. RNA-sequencing analysis revealed transcription of the casΦ gene and the reduced CRISPR array but no evidence of other non-coding RNA such as a trans-activating CRISPR RNA (tracrRNA) within the locus (Fig. 1D). In addition, CasΦ activity could be readily reprogrammed to target other plasmid sequences by altering the guide RNA (fig. S3). These findings suggest that in its native environment, CasΦ is a functional phage protein and bona fide CRISPR-Cas effector capable of cleaving crRNA-complementary DNA such as other phage (Fig. 1E). Furthermore, these results demonstrate that this single-RNA system is much more compact than other active CRISPR-Cas systems (Fig. 1F).
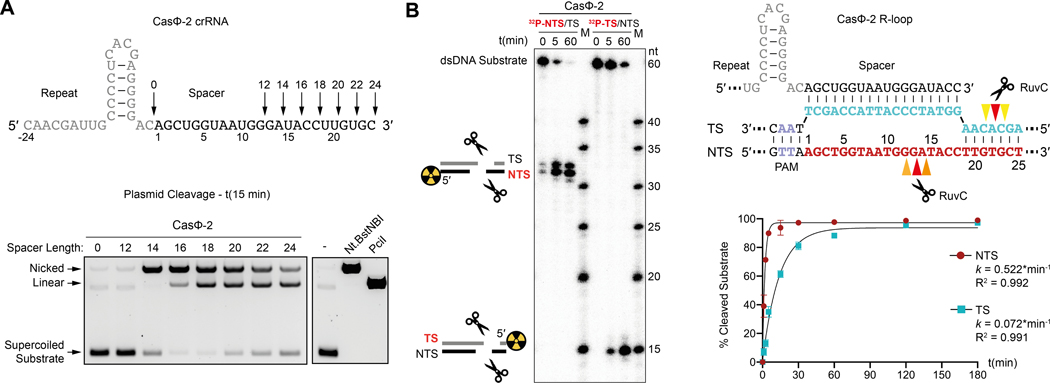
We next investigated the DNA recognition and cleavage requirements of CasΦ in vitro. RNA-seq revealed that the crRNA spacer, which is complementary to DNA targets, is 14–20 nucleotides (nt) long (Fig. 1D). Incubation of purified CasΦ (fig. S4) with crRNAs of different spacer sizes along with supercoiled plasmid or linear dsDNA revealed that DNA cleavage requires the presence of a cognate PAM and a spacer of ≥ 14 nt (Fig. 2A; fig. S5A). Analysis of the cleavage products showed that CasΦ generated staggered 5′-overhangs of 8–12 nt (Fig. 2B, C; fig. S5B, C), similar to the staggered DNA cuts observed for other type V CRISPR-Cas enzymes including Cas12a and CasX (7, 8). We also observed that CasΦ−2 and CasΦ−3 were more active in vitro than CasΦ−1, and the non-target strand (NTS) was cleaved faster than the target-strand (TS) within the RuvC active site (Fig. 2D; figs. S6A, S7; Supplementary Text). Furthermore, CasΦ was found to cleave ssDNA but not ssRNA in cis and in trans (fig. S6B, S8), suggesting that CasΦ may also target ssDNA MGEs or ssDNA intermediates. The trans-cleavage activity of CasΦ, observed only upon DNA recognition in cis (fig. S8), coupled with a minimal PAM requirement (Fig. 1C), may be useful for broader nucleic acid detection as previously demonstrated for type V and type VI Cas proteins (9–11).
Fig. 2.
CasΦ cleaves DNA. (A) Supercoiled plasmid cleavage assay testing CasΦ RNPs reconstituted with crRNAs of different spacer lengths. (B) Cleavage assay targeting dsDNA oligo-duplices for mapping of the cleavage structure. (C) Scheme illustrating the cleavage pattern. (D) NTS and TS DNA cleavage efficiency (n = 3 each, mean ± s.d.). Data is shown in fig. S7B.
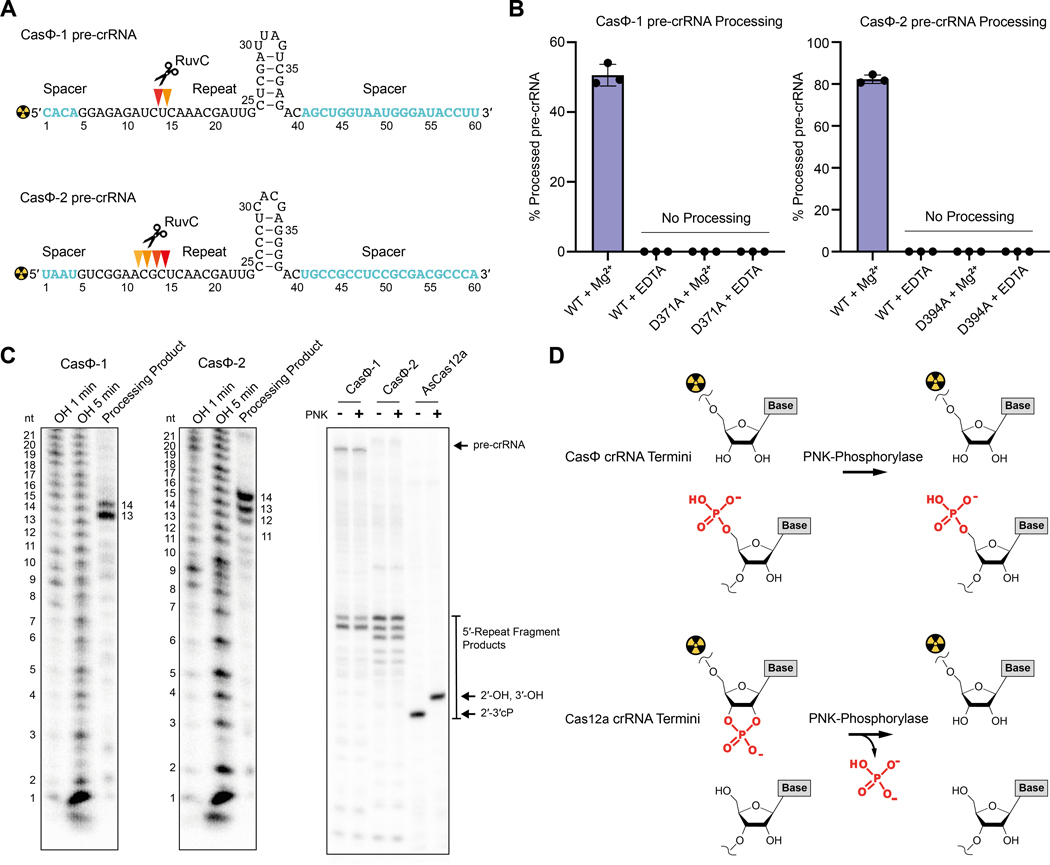
CRISPR-CasΦ systems must produce mature crRNA to guide foreign DNA cleavage. Other type V CRISPR-Cas proteins process pre-crRNAs using an internal active site distinct from the RuvC domain (12) or by recruiting Ribonuclease III to cleave a pre-crRNA:tracrRNA duplex (13–16). The absence of a detectable tracrRNA for CasΦ hinted that CasΦ may catalyze crRNA maturation on its own. To test this possibility, we incubated purified CasΦ with substrates designed to mimic the pre-crRNA structure (Fig. 3A). Reaction products corresponding to a 26–29 nt-long repeat and 20 nt spacer sequence of the crRNA were observed only in the presence of wild type CasΦ, corroborated by RNA-seq analysis of native loci (Figs. 1D; 3A, C; fig. S9). In control experiments, we found that pre-crRNA processing is strictly magnesium-dependent (Fig. 3B; fig. S9), which is different from other CRISPR-Cas RNA processing reactions and suggested a distinct cleavage mechanism. Notably, the RuvC domain requires magnesium to cleave DNA (17), and some RuvC domains have been reported to have endoribonucleolytic activity (15). Based on these observations, we tested CasΦ containing a RuvC-inactivating mutation and found it to be incapable of processing pre-crRNAs (Fig. 3B; fig. S9A, B). Both wild-type and catalytically inactivated CasΦ proteins bind crRNA, and their reconstituted complexes with pre-crRNA have similar elution profiles from a size exclusion column, suggesting no pre-crRNA binding or protein stability defect resulting from the RuvC mutation (fig. S10).
Fig. 3.
CasΦ processes pre-crRNA within the RuvC active site. (A) pre-crRNA substrates and processing sites (red triangles) as derived from the OH-ladder in panel C. (B) Pre-crRNA processing assay for CasΦ−1 and CasΦ−2 in dependence of Mg2+ and RuvC active site residue variation (D371A and D394A) (n = 3 each, mean ± s.d.; t = 60 min). Data is shown in fig. S9B. (C) Left and middle: Alkaline hydrolysis ladder (OH) of the pre-crRNA substrate. Right: PNK-phosphatase treatment of the CasΦ and Acidaminococcus sp. Cas12a cleavage products. (D) Graphical representation of the mature crRNA termini chemistry of CasΦ and Cas12a and PNK-phosphorylase treatment outcomes.
We hypothesized that if the RuvC domain is responsible for pre-crRNA processing, the products should contain 5′-phosphate and 2′- and 3′-hydroxyl moieties as observed in RNAs generated by the RuvC-related RNase HI enzymes (17). In contrast, other type V CRISPR-Cas enzymes process pre-crRNA by metal-independent acid-base catalysis in an active site distinct from the RuvC, generating 2′−3′-cyclic phosphate crRNA termini, as observed for Cas12a (18). Phosphatase treatment of CasΦ-generated crRNA followed by denaturing acrylamide gel analysis showed no change in the crRNA migration, distinct from the change in mobility detected for crRNA generated by Cas12a (Fig. 3C; fig. S9C). This result implies that no 2′−3′-cyclic phosphate was formed during the reaction catalyzed by CasΦ, in contrast to the acid-base catalyzed processing reaction by Cas12a (Fig. 3C, D). Together, these data demonstrate that CasΦ uses a single RuvC active site for both pre-crRNA processing and DNA cleavage.
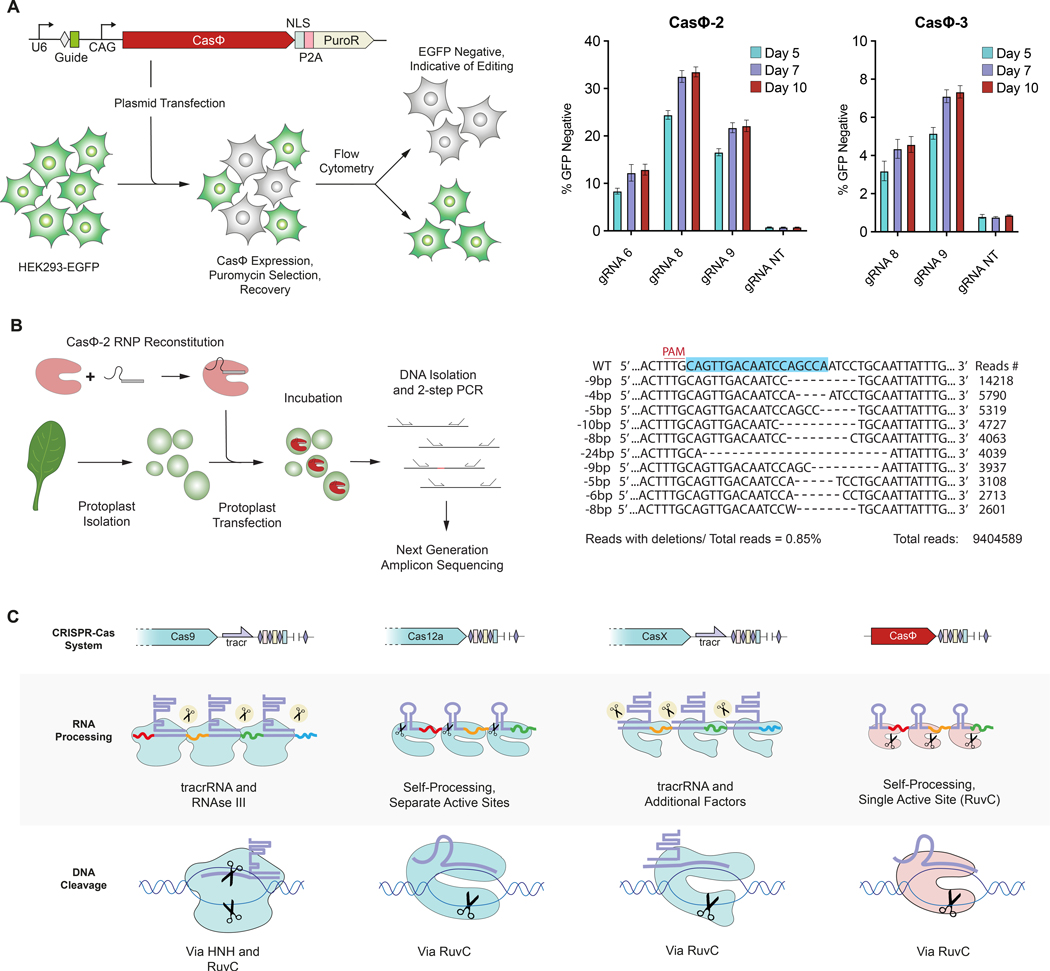
The versatility and programmability of CRISPR-Cas systems for genome editing in virtually any organism have sparked a revolution in biotechnology and fundamental research (19). To investigate whether CasΦ can be harnessed for human genome editing, we performed a gene disruption assay (8) using CasΦ co-expressed with a crRNA in HEK293 cells (Fig. 4A). We found that CasΦ−2 and CasΦ−3, can induce targeted disruption of a genomically integrated EGFP gene (Fig. 4A; fig. S11). In one case, CasΦ−2 with an individual guide RNA was able to edit up to 33% of cells (Fig. 4A), comparable to levels initially reported for CRISPR–Cas9, CRISPR–Cas12a, and CRISPR–CasX (7, 8, 20). We next tested if CasΦ−2 can be delivered as RNPs into plant protoplasts to edit the endogenous Arabidopsis thaliana PDS3 gene (Fig. 4B; fig. S12). Next generation sequencing revealed that CasΦ−2 introduces primarily 8–10 bp deletions (Fig. 4B), consistent with the cleavage pattern observed in vitro (Fig. 2C). The small size of CasΦ in combination with its minimal PAM requirement will be particularly advantageous for both vector-based delivery into cells and a wider range of targetable genomic sequences, providing a powerful addition to the CRISPR-Cas toolbox.
Fig. 4.
CasΦ is functional for genome editing. (A) Experimental workflow of the GFP disruption assay (left) and GFP disruption using CasΦ−2 and CasΦ−3 and a non-targeting (NT) guide as a negative control (n = 3 each, mean ± s.d.). (B) Experimental workflow of CasΦ−2 RNP-mediated genome-editing in A. thaliana mesophyll protoplasts (left) and amplicon sequencing data (right) showing the most frequent deletions for gRNA33 in the targeted region (blue) within the AtPDS3 gene. (C) Scheme illustrating the differences in RNA processing and DNA cutting for Cas9, Cas12a, CasX, and CasΦ.
Three other well-characterized Cas enzymes Cas9, Cas12a, and CasX, use one (Cas12a and CasX) or two active sites (Cas9) for DNA cutting and rely on a separate active site (Cas12a) or additional factors (CasX and Cas9) for crRNA processing (Fig. 4C). The finding that a single RuvC active site in CasΦ is capable of crRNA processing and DNA cutting suggests that size limitations of phage genomes, possibly in combination with large population sizes and higher mutation rates in phages compared to prokaryotes (21–23), led to a consolidation of chemistries within one catalytic center. Such compact proteins may be particularly amenable to engineering and laboratory evolution to create new functionalities for genome manipulation, and highlight huge phages as an exciting forefront for discovery and biotechnological applications for human health.
Supplementary Material
Acknowledgements:
We thank Anabel Schweitzer and Hannah Spinner for providing plasmids (pBFC545, pBFC546, pBFC547 and the PAM library plasmid) and Juan Hurtado for providing the EGFP-HEK293 cell line. We thank Doudna Lab members as well as Alexander Crits-Christoph and Jacob West-Roberts for helpful discussions, and Rohan Sachdeva for bioinformatic support.
Funding: PP is supported by the Paul G. Allen Frontiers Group and NIH Somatic Cell Genome Editing consortium (NIH U01AI142817–02). BAS is supported by an NSF Graduate Research Fellowship (No. DGE 1752814). GJK is supported by an NHMRC Investigator Grant (EL1, APP1175568) and previously an American Australian Association Fellowship. JAD receives funding from the William M. Keck Foundation, the National Multiple Sclerosis Society, the NIH (RM1HG009490, U01AI142817–02), NSF (1817593) JAD and SEJ are Investigators of the Howard Hughes Medical Institute. JAD is a Paul Allen Distinguished Investigator.
Footnotes
Competing Interests: The Regents of the University of California, Berkeley and Los Angeles have patents pending for CRISPR technologies on which the authors are inventors. JAD is a co-founder of Caribou Biosciences, Editas Medicine, Intellia Therapeutics, Scribe Therapeutics, and Mammoth Biosciences. JAD is a scientific advisory board member of Caribou Biosciences, Intellia Therapeutics, eFFECTOR Therapeutics, Scribe Therapeutics, Synthego, and Mammoth Biosciences. JAD is a director at Johnson & Johnson and has sponsored research projects by Pfizer, Roche Biopharma, and Biogen. JFB is a founder of Metagenomi. SEJ is a scientific co-founder of Inari Agriculture and SEJ and JAD are members of its scientific strategy board.
Data and materials availability: All data are available in the manuscript or the supplementary material. Reagents are available through Addgene and upon request from JAD.
References and Notes:
- 1.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P, CRISPR provides acquired resistance against viruses in prokaryotes. Science. 315, 1709–1712 (2007). [DOI] [PubMed] [Google Scholar]
- 2.McGinn J, Marraffini LA, Molecular mechanisms of CRISPR–Cas spacer acquisition. Nat. Rev. Microbiol 17, 7–12 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Hille F, Richter H, Wong SP, Bratovič M, Ressel S, Charpentier E, The Biology of CRISPR-Cas: Backward and Forward. Cell. 172, 1239–1259 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Al-Shayeb B, Sachdeva R, Chen L-X, Ward F, Munk P, Devoto A, Castelle CJ, Olm MR, Bouma-Gregson K, Amano Y, He C, Méheust R, Brooks B, Thomas A, Lavy A, Matheus-Carnevali P, Sun C, Goltsman DSA, Borton MA, Sharrar A, Jaffe AL, Nelson TC, Kantor R, Keren R, Lane KR, Farag IF, Lei S, Finstad K, Amundson R, Anantharaman K, Zhou J, Probst AJ, Power ME, Tringe SG, Li W-J, Wrighton K, Harrison S, Morowitz M, Relman DA, Doudna JA, Lehours A-C, Warren L, Cate JHD, Santini JM, Banfield JF, Clades of huge phages from across Earth’s ecosystems. Nature (2020), doi: 10.1038/s41586-020-2007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, Severinov K, Zhang F, Koonin EV, Diversity and evolution of class 2 CRISPR–Cas systems. Nat. Rev. Microbiol 15, 169–182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gleditzsch D, Pausch P, Müller-Esparza H, Özcan A, Guo X, Bange G, Randau L, PAM identification by CRISPR-Cas effector complexes: diversified mechanisms and structures. RNA Biology. 16 (2019), pp. 504–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F, Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 163, 759–771 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J-J, Orlova N, Oakes BL, Ma E, Spinner HB, Baney KLM, Chuck J, Tan D, Knott GJ, Harrington LB, Al-Shayeb B, Wagner A, Brötzmann J, Staahl BT, Taylor KL, Desmarais J, Nogales E, Doudna JA, CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature. 566, 218–223 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, Doudna JA, CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 360, 436–439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.East-Seletsky A, O’Connell MR, Knight SC, Burstein D, Cate JHD, Tjian R, Doudna JA, Two Distinct RNase Activities of CRISPR-C2c2 Enable Guide RNA Processing and RNA Detection. Nature. 538, 270–273 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, Myhrvold C, Bhattacharyya RP, Livny J, Regev A, Koonin EV, Hung DT, Sabeti PC, Collins JJ, Zhang F, Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 356, 438–442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E, The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 532, 517–521 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Burstein D, Harrington LB, Strutt SC, Probst AJ, Anantharaman K, Thomas BC, Doudna JA, Banfield JF, New CRISPR-Cas systems from uncultivated microbes. Nature. 542, 237–241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrington LB, Burstein D, Chen JS, Paez-Espino D, Ma E, Witte IP, Cofsky JC, Kyrpides NC, Banfield JF, Doudna JA, Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 362, 839–842 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan WX, Hunnewell P, Alfonse LE, Carte JM, Keston-Smith E, Sothiselvam S, Garrity AJ, Chong S, Makarova KS, Koonin EV, Cheng DR, Scott DA, Functionally diverse type V CRISPR-Cas systems. Science. 363, 88–91 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, Zhang F, Koonin EV, Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol. Cell 60, 385–397 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nowotny M, Retroviral integrase superfamily: the structural perspective. EMBO Rep. 10, 144–151 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swarts DC, van der Oost J, Jinek M, Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRISPR-Cas12a. Mol. Cell 66, 221–233.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knott GJ, Doudna JA, CRISPR-Cas guides the future of genetic engineering. Science. 361, 866–869 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM, RNA-guided human genome engineering via Cas9. Science. 339, 823–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duffy S, Shackelton LA, Holmes EC, Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet 9, 267–276 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Lee M-C, Marx CJ, Repeated, selection-driven genome reduction of accessory genes in experimental populations. PLoS Genet. 8, e1002651 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lynch M, Streamlining and simplification of microbial genome architecture. Annu. Rev. Microbiol 60, 327–349 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, Charpentier E, Cheng D, Haft DH, Horvath P, Moineau S, Mojica FJM, Scott D, Shah SA, Siksnys V, Terns MP, Venclovas Č, White MF, Yakunin AF, Yan W, Zhang F, Garrett RA, Backofen R, van der Oost J, Barrangou R, Koonin EV, Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol, 1–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knott GJ, Thornton BW, Lobba MJ, Liu J-J, Al-Shayeb B, Watters KE, Doudna JA, Broad-spectrum enzymatic inhibition of CRISPR-Cas12a. Nat. Struct. Mol. Biol, 1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE, Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol 34, 339–344 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Yoo S-D, Cho Y-H, Sheen J, Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc 2, 1565–1572 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Swarts DC, Jinek M, Mechanistic Insights into the cis- and trans-Acting DNase Activities of Cas12a. Mol. Cell 73, 589–600.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cofsky JC, Karandur D, Huang CJ, Witte IP, Kuriyan J, Doudna JA, CRISPR-Cas12a exploits R-loop asymmetry to form double-strand breaks. bioRxiv (2020), p. 2020.02.10.937540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.